Abstract
The anaphase-promoting complex (APC), a large evolutionarily conserved ubiquitin ligase complex, regulates cell cycle progression through mitosis and G1. Here, we present data suggesting that APC-dependent cell cycle progression relies on a specific set of posttranslational histone-modifying enzymes. Multiple APC subunit mutants were impaired in total and modified histone H3 protein content. Acetylated H3K56 (H3K56Ac) levels were as reduced as those of total H3, indicating that loading histones with H3K56Ac is unaffected in APC mutants. However, under restrictive conditions, H3K9Ac and dimethylated H3K79 (H3K79me2) levels were more greatly reduced than those of total H3. In a screen for histone acetyltransferase (HAT) and histone deacetylase (HDAC) mutants that genetically interact with the apc5CA (chromatin assembly) mutant, we found that deletion of GCN5 or ELP3 severely hampered apc5CA temperature-sensitive (ts) growth. Further analyses showed that (i) the elp3Δ gcn5Δ double mutant ts defect was epistatic to that observed in apc5CA cells; (ii) gcn5Δ and elp3Δ mutants accumulate in mitosis; and (iii) turnover of the APC substrate Clb2 is not impaired in elp3Δ gcn5Δ cells. Increased expression of ELP3 and GCN5, as well as genes encoding the HAT Rtt109 and the chromatin assembly factors Msi1 and Asf1, suppressed apc5CA defects, while increased APC5 expression partially suppressed elp3Δ gcn5Δ growth defects. Finally, we demonstrate that Gcn5 is unstable during G1 and following G1 arrest and is stabilized in APC mutants. We present our working model in which Elp3/Gcn5 and the APC work together to facilitate passage through mitosis and G1. To progress into S, we propose that at least Gcn5 must then be targeted for degradation in an APC-dependent fashion.
All cells grow and divide through a mechanism conserved in virtually all eukaryotic organisms. Arguably the most critical event in cell division is the transmission of an error-free genetic copy of parental chromosomes to all descendants. Thus, all activities that promote genomic stability are absolutely crucial to replicative fidelity. The evolutionarily conserved anaphase-promoting complex (APC), a large multisubunit ubiquitin ligase (E3), plays a critical role in maintaining genomic stability by controlling transit through mitosis and G1. This is accomplished primarily by targeting proteins that inhibit different steps in mitosis for degradation (28, 47, 69). For example, Pds1, the Saccharomyces cerevisiae securin, is targeted for destruction to allow sister chromatid separation, while Clb2, a B type cyclin, is targeted for destruction in order to exit mitosis. The yeast APC contains at least 13 subunits, but the function of individual subunits remains mostly unknown. The APC's role in promoting genomic stability is highlighted by the finding that defects in APC activity are associated with cancer development and premature aging (3, 18, 23, 24, 27, 29, 41, 57), and this may occur through APC influence on chromatin structure. We have shown that the yeast APC is required for chromatin assembly specifically during mitosis (21), via an intracellular signaling pathway involving the E3's Rsp5 and the SCF (Skp/Cdc53/F-box), the E2 Ubc7 (1), and the individual chromatin assembly factors Cac1, Cac2, Msi1, Asf1, Hir1, and Hir2 (25, 26). However, the extent to which the APC controls chromatin structure and the mechanism(s) adhered to remain utterly unknown.
A thorough understanding of how the APC influences chromatin structure may improve our understanding of disease onset and premature aging. Recent studies in mammalian systems have demonstrated physical interactions between the APC and chromatin-modifying enzymes and transcriptional activators (5, 61). However, in yeast, links between the APC and chromatin-modifying enzymes are lacking. Nonetheless, at least two histone acetyltransferases (HATs) in yeast have been associated with mitotic progression, namely, Gcn5, the HAT component of the SAGA transcriptional initiator complex, and Rtt109 (17, 20, 35, 63). Cells lacking GCN5 experience (i) increased centromere-based plasmid loss, (ii) increased G2 cells with unsegregated nuclei, (iii) increased sensitivity to microtubule-depolymerizing agents, (iv) hypersensitivity to Clb2 overexpression, and (v) delayed entrance to mitosis (35, 63). Gcn5 is recruited to centromeres, likely throughout the cell cycle (63), as well as to promoters of genes expressed in late mitosis (35). Furthermore, many genes expressed during mitosis are highly enriched for Gcn5-dependent genes. Thus, it appears that transit through mitosis requires Gcn5-dependent acetylation of centromeric histones and/or acetylation of histones within the promoters of late-mitosis-specific genes, suggesting that Gcn5 may be required for the expression of genes necessary for mitotic exit and passage through G1/S. Complete transcriptional initiation and elongation, however, appear to require both Gcn5 and the HAT component of the Elongator complex, Elp3 (36, 67, 68). Elp3 was reported to preferentially acetylate H3K14 and H4K8 (66), while Gcn5 has a more robust substrate population, including H3K9, H3K14, H3K18, and H3K23, but not H3K56 (17, 21). Elp3 and Gcn5 were shown to act in a redundant manner to activate transcription; they both target H3K9 and H3K14, and double mutant phenotypes were dramatically impaired compared to those of single mutants, being characterized by extreme slow growth and severe hypoacetylation of multiple H3K residues (36, 68). Thus, if global histone acetylation is important for APC activity and entry into G1, then Gcn5 and Elp3 may be crucial for this activity.
The second HAT demonstrated to play a role in mitotic progression is Rtt109, the yeast orthologue of human CBP (14), which acetylates histone H3K56 in concert with the chromatin assembly factor (CAF) Asf1 (10). Human APC5 physically and functionally interacts with CBP (61), and yeast apc5CA phenotypes are exacerbated by deletion of ASF1 (27). Thus, genetic interactions between apc5CA and rtt109Δ mutants would indicate that the interaction between the APC and histone-modifying enzymes may be conserved. In yeast, deletion of RTT109 delays passage through mitosis, inducing susceptibility to DNA damage, and delays activation of the DNA damage checkpoint (17). A gcn5Δ rtt109Δ double mutant was shown to be hypersensitive to DNA damage and could not acetylate H3K9, whereas the single mutants retained remnants of these activities (17). Furthermore, a recent report demonstrated that Gcn5 was involved in replication-coupled chromatin assembly in conjunction with Rtt109 (8). Genetic interactions between gcn5Δ and mutations in replication-coupled CAFs (CAF-I and Asf1) supported this observation. Acetylated H3K9 (H3K9Ac) and H3K56Ac are important marks in newly synthesized histone H3 (37, 43). An attractive model was proposed to explain Asf1/Rtt109/Gcn5 interactions; in this model, Asf1 presents H3 and H4 separately to Rtt109 and Gcn5 for acetylation of H3K56 and H3K9, respectively, prior to passage of the acetylated histones to CAF-I for deposition into chromatin (17).
In this report, we provide evidence supporting the hypothesis that the APC is required for histone synthesis and histone metabolism in mitotically active cells. We observed that total and modified histone H3 levels were reduced in different APC mutants. We provide evidence that Elp3 and Gcn5 are important for mitotic progression by functioning in a pathway that is likely vital for APC-dependent mitotic exit. Our data suggest that to exit G1, at least Gcn5 is required to be degraded in an APC-dependent manner.
MATERIALS AND METHODS
Yeast strains and plasmids.
The Saccharomyces cerevisiae strains used in this study are shown in Table 1. The HAT and histone deacetylase (HDAC) mutant strains were derived from the ResGen collection of yeast deletion strains (generous gifts from W. Xiao, University of Saskatchewan). The Research Genetics (ResGen) mutants were backcrossed repeatedly with our laboratory S288c background strain. Strains were considered congenic when multiple isolates from a cross displayed identical phenotypes. This typically required five or six backcrosses. The strain with the apc11-13 allele (YTH1085), in the S288c background, was kindly provided by T. Hunter. The cdc16-1 (YTH370) and cdc23-1 (YTH371) W303 strains and the isogenic wild type (WT; YTH361) were obtained from D. Stuart. The plasmids encoding GAL-driven APC5-HA (hemagglutinin epitope tag), APC10-HA, GCN5-HA, ELP3-HA, and RTT109-HA were obtained from Open Biosystems and kindly provided by W. Xiao. The copper-driven GST-ASF1 and GST-MSI1 plasmids were described earlier (25). The plasmid pTH55 (YCp50-APC5) was described previously (23).
Table 1.
List of strains used in this study
| Strain | Genotypea | Source (reference) |
|---|---|---|
| S288c derivatives | ||
| YTH5 | MATaade2 his3 lys2 ura3 (WT) | Harkness et al. (23) |
| YTH1085 (YAP201) | MATaura3-52 lys2-801 ade2-101 trp1-D63 his3-D200 apc11::HIS3 leu2-D1::apc11-13::LEU2 | T. Hunter |
| YTH1377 | MAT? ade2 his3 leu2 lys2 ura3 apc5CA-PA::His5 apc9::kanMX6 | Harkness et al. (24) |
| YTH1387 | MAT? ade2 his3 leu2 lys2 ura3 apc5CA-PA::His5 cdc26::kanMX6 | Harkness et al. (24) |
| YTH1410 | MAT? ade2 his3 leu2 lys2 ura3 apc5CA-PA::His5 apc10::kanMX6 | Harkness et al. (24) |
| YTH1636 | MATaade2 his3 leu2 lys2 ura3 (WT) | Harkness et al. (24) |
| YTH1637 | MATα ade2 his3 leu2 lys2 ura3 apc5CA-PA::His5 | Harkness et al. (24) |
| YTH1648 | MAT? ade2 his3 leu2 lys2 ura3 apc9::kanMX6 | This study |
| YTH1669 | MAT? ade2 his3 leu2 lys2 ura3 cdc26::kanMX6 | This study |
| YTH1693 | MATα ade2 his3 leu2 lys2 ura3 apc10::kanMX6 | This study |
| YTH1992 | MAT? apc5CA-PA::His5 hpa2::kanMX6 | This study |
| YTH1994 | MAT? hpa2::kanMX6 | This study |
| YTH1997 | MAT? sas2::kanMX6 | This study |
| YTH1998 | MAT? apc5CA-PA::His5 sas2::kanMX6 | This study |
| YTH2001 | MAT? apc5CA-PA::His5 rpd3::kanMX6 | This study |
| YTH2003 | MAT? rpd3::kanMX6 | This study |
| YTH2061 | MAT? apc5CA-PA::His5 gcn5::kanMX6 hpa2::kanMX6 | This study |
| YTH2072 | MATagcn5::kanMX6 | This study |
| YTH2075 | MAT? apc5CA-PA::His5 gcn5::kanMX6 | This study |
| YTH2076 | MAT? gcn5::kanMX6 hpa2::kanMX6 | This study |
| YTH2216 | MAT? apc5CA-PA::His5 elp3::kanMX6 | This study |
| YTH2217 | MAT? elp3::kanMX6 | This study |
| YTH2219 | MAT? elp3::kanMX6 | This study |
| YTH2260 | MAT? hos1::kanMX6 | This study |
| YTH2261 | MAT? apc5CA-PA::His5 hos1::kanMX6 | This study |
| YTH2305 | MAT? hda1::kanMX6 | This study |
| YTH2306 | MAT? apc5CA-PA::His5 hda1::kanMX6 | This study |
| YTH2389 | MAT? hos2::kanMX6 | This study |
| YTH2390 | MAT? apc5CA-PA::His5 hos2::kanMX6 | This study |
| YTH2504 | MAT? hat1::kanMX6 | This study |
| YTH2507 | MAT? apc5CA-PA::His5 hat1::kanMX6 | This study |
| YTH2525 | MAT? sas3::kanMX6 | This study |
| YTH2526 | MAT? apc5CA-PA::His5 sas3::kanMX6 | This study |
| YTH2804 | MAT? hos3::kanMX6 | This study |
| YTH2805 | MAT? hos3::kanMX6 | This study |
| YTH2806 | MAT? apc5CA-PA::His5 hos3::kanMX6 | This study |
| YTH2807 | MAT? apc5CA-PA::His5 hos3::kanMX6 | This study |
| YTH3037 | MAT? gcn5::kanMX6 | This study |
| YTH3038 | MATα apc5CA-PA::His5 gcn5::kanMX6 | This study |
| YTH3606 | MAT? elp3::kanMX6 gcn5::kanMX6 | This study |
| YTH3607 | MAT? apc5CA-PA::His5 elp3::kanMX6 gcn5::kanMX6 | This study |
| BF264-15Du | ||
| YTH370 | MATaade1 bar1 his3 leu2 trp1 ura3 cdc16-1 | D. Stuart |
| YTH371 | MATaade1 his2 trp1 ura3 cdc23-1 | D. Stuart |
| YTH1007 | MAT? ade1 bar1 his2 leu2 trp1-1 ura3Δns | D. Stuart |
?, mating type undetermined.
Media and genetic methods.
Media were prepared as previously described (24). Glucose and galactose were used at a final concentration of 2%. Escherichia coli strains JM109 and DH10B were used to propagate DNA plasmids. DNA manipulations such as restriction enzyme digestions, DNA minipreps, yeast and E. coli transformations, and yeast genomic DNA preparation were carried out according to standard protocols (2). Spot dilution assays were conducted by pipetting 3 μl of cells from samples generated from a 10-fold dilution series onto the various media described and grown at the temperatures indicated. The starting spot generally contained 5 × 104 cells. Genetic analysis of the yeast strains consisted of crossing the appropriate haploid strains on YPD plates (1% yeast extract, 2% peptone, 2% glucose). The resulting diploids were patched onto sporulation plates (1% potassium acetate, 0.1% yeast extract, and 0.05% glucose plus 41 μg/ml adenine) to induce meiosis. Tetrads were dissected using a dissecting microscope (Singer). Segregation of the kanMX6 cassette was determined by patching spores onto 0.2 mg/ml Geneticin-supplemented YPD plates, and all other markers were monitored by patching spores onto SD medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose, 0.13% dropout powder lacking the appropriate amino acids, and 1 sodium hydroxide [NaOH] tablet). Strains containing multiple kanMX6 integrated markers were obtained by identifying tetrads where the kanMX6 markers segregated together in a 2:2 pattern. Northern analyses were performed as previously described (24).
Quantitative real-time PCR (Q-PCR). (i) Total RNA isolation.
Frozen cells were mechanically disrupted using a Disruptor Genie (Scientific Industries). Total RNA was extracted using an RNeasy mini kit (Qiagen, Mississauga, ON, Canada) with the optional on-column DNase I digestion to remove genomic DNA. Purified RNA was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE). The purified RNA samples were stored at −80°C until analysis.
(ii) First-strand cDNA synthesis.
First-strand cDNA synthesis was performed using an iScript cDNA synthesis kit (Bio-Rad). A volume of 1 μg of total RNA was combined with 4 μl of 5× iScript reaction mix, 1 μl of iScript reverse transcriptase, and RNase-free water to a final volume of 20 μl. Reaction mixes were incubated at 25°C for 25 min and 42°C for 30 min and, on completion, were inactivated at 85°C for 5 min. The cDNA samples were stored at −20°C until required.
(iii) PCR.
Gene-specific primers targeted against yeast TFC1 and GCN5 were designed based on the full-length sequences available from the Saccharomyces Genome Database (SGD) (Table 2). Target gene expression was measured using semiquantitative RT-PCR. Q-PCR was performed in 96-well PCR plates using an ABI 7300 real-time PCR system (Applied Biosystems, Carlsbad, CA). A separate 45-μl PCR mixture consisting of SYBR green master mix (Applied Biosystems), cDNA, gene-specific primers, and nuclease-free water was prepared for each cDNA sample of interest and for each primer pair. A final reaction volume of 20 μl was transferred to each well, and reactions were performed in duplicate. The PCR mix was denatured at 95°C for 10 min before the first PCR cycle. The thermal cycle profile was as follows: 10 s at 95°C and extension for 1 min at either 56°C (TFC1) or 60°C (GCN5). A total of 40 PCR cycles were used. The efficiency of each Q-PCR assay was assessed by construction of standard curves using serial dilutions of a cDNA generated by random pooling of cDNA from each sample. To quantify Q-PCR results, the cycle at which the fluorescence signal was first significantly different from background (CT) was determined for each reaction. The expression level of GCN5 was normalized with reference to the TFC1 endogenous control gene. Changes in target gene expression were calculated using the ΔΔCT (2−ΔΔCT) method (40).
Table 2.
Q-PCR primers
| Gene name | Primer sequence |
|---|---|
| TFC1 | 5′ GCTGGCACTCATATCTTATCGTTTCACAATG |
| 3′ GAACCTGCTGTCAATACCGCCTGGAG | |
| GCN5 | 5′ TGAAGAGGATCACTTGGATGG |
| 3′ CTTTATCGGTGCCCTCTTG |
Protein stability.
Cultures were grown overnight to log phase and set back to an optical density (OD) of 0.5. The incubation was continued for three more hours to ensure that the cells were mitotically active. Cells were arrested in G1 by adding 1 μM alpha factor (ZymoResearch) to mitotically active cells for 3.5 h. Arrest was confirmed by visualizing cell shmooing and by flow cytometry (Saskatoon Cancer Center). Cells were centrifuged and washed twice with H2O prior to resuspension into fresh medium containing 10 μg/ml cycloheximide (Calbiochem) to prevent further protein synthesis. Samples were collected before and after arrest, as well as every 20 min for 3 h, for flow cytometry and Western analyses. Flow cytometry and Western analyses were performed as previously described (2, 26, 28). Antibodies against histones H2B (Upstate), H3 (Abcam), H4 (Abcam), H3K9Ac (Upstate), H3K14Ac (Abcam), H3K56Ac (Abcam), phosphorylated H3S10 (H3S10phos; Santa Cruz), dimethylated H3K79 (H3K79me2; Abcam), GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Sigma), tandem affinity purification (TAP) (Genescript), and Clb2 (Santa Cruz) were purchased from the indicated suppliers. The GST J90 antibody was generated by T. A. A. Harkness and M. C. Schultz (unpublished data). When using antibodies against H3K14Ac, 5% bovine serum albumin (BSA) was used, rather than 5% milk, in phosphate-buffered saline (PBS) containing 0.1% (vol/vol) Tween 20.
RESULTS
Total and modified histone levels are altered in APC mutants.
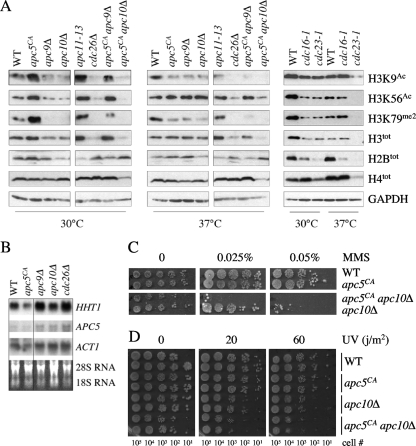
We initiated our studies into the role of the APC in histone metabolism by characterizing total histone protein and H3K9Ac and K56Ac profiles in a panel of single and double APC subunit mutants. Antibodies against H3K79me2 were also used, as H3K79me2 is linked with DNA repair and activation of genes required in late mitosis (7, 54). Whole-cell lysates were prepared from APC mutants harvested after growth at either the permissive temperature or following a shift to the restrictive temperature of 37°C. All the APC mutants used, except the apc9Δ mutant, display reduced growth at elevated temperatures and arrest at the metaphase/anaphase junction. Although APC5 is an essential gene, the apc5CA allele contains a 2-bp deletion in the 5′ end of the gene and mutants containing this allele continue to progress through mitosis at 37°C, albeit at a much lower rate, accumulating replicated and unsegregated chromosomes (1, 23, 25).
At 30°C, total H2B and H4 levels in most APC mutants were essentially unchanged in comparison to those in the wild type (Fig. 1A). Exceptions were observed with the apc10Δ, apc11-13, cdc16-1, and cdc23-1 mutants, where H2B levels were reduced. At 37°C, total H4 levels remained relatively unimpaired, except for reduced levels in cdc23-1 and apc5CA apc10Δ cells. Reduced H2B levels were also observed at 37°C in the cdc16-1 and cdc23-1 mutants. At 30°C, histone H3 total levels were reduced in cdc26Δ, cdc16-1, cdc23-1, and apc5CA apc10Δ mutants. The pattern was exaggerated at 37°C. In summary, for total H2B, H3, and H4 levels, discrete differences were observed, with H3 displaying the widest range in levels.
Fig. 1.
The APC is required for maintenance of total and modified histone H3 levels. (A) Extracts prepared from the APC mutants shown at the top of the lanes, grown at 30°C or after a 3-h shift to 37°C, were analyzed by Western analysis with the antibodies indicated on the right. Antibodies against GAPDH were used to control for protein load. (B) A Northern analysis was performed on the strains shown grown overnight at room temperature to early log phase. PCR fragments corresponding to the HHT1, APC5, and ACT1 ORFs were prepared, randomly labeled using [α-32P]CTP, and hybridized to purified and separated total RNA. (C and D) The mutants shown were grown overnight to early log phase in rich glucose media at 30°C. The next day, a 10-fold dilution series was prepared and spotted onto plates containing increasing concentrations of methyl methanesulfonate (MMS) (C) or plated onto YPD plates and exposed to an increasing UV dose (D). The plates exposed to UV were wrapped in aluminum foil and grown in the dark for 3 days. The approximate number of cells in each diluted spot is indicated below the plates.
Next, we asked whether the APC mutants influenced the modification state of histone H3. Western analyses were performed with antibodies that recognized H3K9Ac and H3K56Ac. At 30°C, the levels of acetylated K9 and K56 for the most part mirrored total H3 levels. At 37°C, the K56Ac profile followed total H3 levels, whereas K9Ac levels were further reduced in most mutants. These observations indicate that at elevated temperatures, the APC mutants have a greater impact on H3K9Ac than H3K56Ac.
We also measured H3K79me2 levels in the APC mutant panel, as K79me2 is considered necessary for expression of genes associated with late mitotic events (54). We observed that H3K79me2 levels were impaired beyond that observed for total H3, particularly at 37°C. Consistent with the association of both H3K9Ac and H3K79me2 with mitotic gene activation, the H3K79me2 profile ran parallel to the H3K9Ac profile. These observations suggest that mitotic gene activation mediated through H3K9Ac and H3K79me2 is involved with APC activity.
We next tested whether reduced histone protein levels reflected impaired transcription. We prepared total RNA from APC subunit mutants (apc5CA, apc9Δ, apc10Δ, and cdc26Δ mutants) grown overnight to early log phase at room temperature in 2% glucose. DNA fragments corresponding to HHT1 (encoding histone H3), APC5, and ACT1 open reading frames (ORFs) were randomly labeled using [α-32P]CTP and hybridized against the separated RNA from each strain. The RNA gel is shown as a load control. The results show that in mutants with decreased H3 protein levels (apc9Δ, apc10Δ, and cdc26Δ mutants), HHT1 mRNA levels are elevated (Fig. 1B). These observations indicate that the lower H3 protein levels are not due to reduced transcription.
Apc5 and Apc10 play redundant roles in histone metabolism.
We previously reported that Apc5 and Apc10, found in separate arms of the APC (60), appear to harbor redundant functions in regards to life span and growth at elevated temperatures (24). We observed that the levels of modified and unmodified H3 in apc5CA apc10Δ mutants were lower than those in the single mutants, consistent with our earlier results. To monitor the redundant nature of the apc5CA apc10Δ mutant further, we investigated whether reduced total and modified histone levels observed in this double mutant rendered cells sensitive to DNA damage. We observed that the double mutant was more sensitive to methyl methanesulfonate (MMS) and UV exposure than either of the single mutants (Fig. 1C and D). Thus, Apc5 and Apc10 may define complementary activities that respond to stress.
Deletion of genes encoding histone modifiers alter apc5CA ts defects.
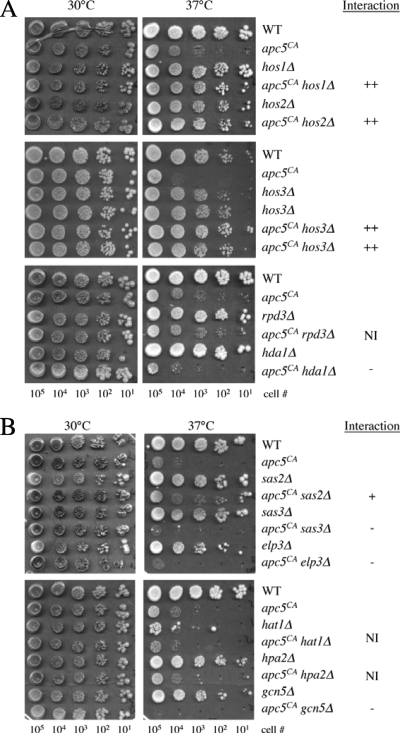
To delve further into how the APC impacts histone metabolism, we performed a screen of nonessential HAT and HDAC mutants for those that influence the apc5CA temperature-sensitive (ts) growth defect. First, we combined the apc5CA allele with the HDAC gene deletion hda1Δ, hos1Δ, hos2Δ, hos3Δ, or rpd3Δ. The apc5CA allele was used for this analysis as it has been found to be an effective bait in screens for genetic interactions (1, 24, 25). Yeast HDACs have various functions within cells, resulting in different effects on histone acetylation (50). For example, Hda1 and Rpd3 are believed to be recruited to many, yet distinct, yeast promoters, and the absence of these genes causes increased global histone acetylation (38, 49, 50, 53). Consistent with Hda1 and Rdp3 recruitment to distinct promoters, deletion of HDA1 negatively impacted the apc5CA ts defect, while deletion of RPD3 had no effect (Fig. 2A). On the other hand, Hos1, Hos2, and Hos3 work at a subset of promoters, which are mainly represented by rDNA and ribosomal protein-encoding genes (50), and do not seem to alter global acetylation levels. Deletion of HOS1, HOS2, or HOS3 in the apc5CA background completely suppressed the apc5CA ts defect. These preliminary observations suggest that it is not simply the altered histone acetylation that impacts the apc5CA phenotype; perhaps it is the specific function of the enzyme deleted that matters.
Fig. 2.
Genetic interactions of HDAC and HAT mutants with the apc5CA allele. (A) apc5CA cells were repeatedly crossed with the mutants shown to generate double mutants. Tenfold serial dilutions of the different mutants were spot diluted onto rich media, and growth levels were compared at the temperatures shown. Plates were grown from 3 to 5 days and scanned each day. (B) apc5CA cells were repeatedly crossed with the mutants shown to generate double mutants. The mutants were prepared and analyzed as described above. The effect of the interaction is indicated on the right. ++, strong suppressive interaction in double mutant that grows like wild type; +, double mutant grows better than single mutants but not at wild-type level. −, double mutant grows more poorly than the single mutants. NI, no interaction.
Next, we asked the following questions. (i) Will deletion of HAT genes also have differential effects on the apc5CA ts defect? (ii) Can we ascertain the role of the APC in histone modification by characterizing the genetic interactions between apc5CA and HAT mutants? The mutant HAT alleles combined with apc5CA were elp3Δ, gcn5Δ, hat1Δ, hpa2Δ, sas2Δ, and sas3Δ. As with the HDACs, a division of labor has been observed with yeast HATs (50). Gcn5 and Elp3 appear to function at many active promoters, resulting in global decreases in histone acetylation when these genes are deleted (16, 68). Interestingly, only 4% of yeast transcripts were downregulated when GCN5 was deleted (39). Sas2 appears to globally acetylate H4K16, with perhaps preferential activity at subtelomeric regions (33, 58), which has been linked to life span (13). Sas3 does not appear to affect H4 but rather acts on H3. Sas3 action on H3 overlaps with that of Gcn5, as sas3Δ gcn5Δ double mutants were lethal (31, 51). While Hat1 is required for acetylation of cytosolic H4 (52), no function has yet been ascribed to HpaI. Our results demonstrate that expression of the apc5CA ts defect does not require HAT1 or HPA2 (Fig. 2B), as no change in growth was observed when these genes were deleted in the apc5CA background. On the other hand, deletion of ELP3, GCN5, or SAS3 further compromised the growth of apc5CA cells at 37°C, whereas deletion of SAS2 partially suppressed the growth defect. Taken together, mutations in genes that promote gene silencing (hos1Δ, hos2Δ, hos3Δ, and sas2Δ) improve the apc5CA ts defect, while those that impair gene activation (elp3Δ, gcn5Δ, and sas3Δ) worsen it. The observation with hda1Δ is in contrast to this, as Hda1 is thought to silence transcription, yet the hda1Δ mutation impaired apc5CA ts growth. It is possible that some gene(s) repressed by Hda1 may be involved in APC inhibition. Based on the results presented here, targets of Rpd3, Hpa2, and Hat1 are predicted to play a limited role, if any, in APC activity.
Hpa2 and Gcn5 interact antagonistically alone but synergistically when combined with apc5CA.
To identify additional interactions between the HATs and the APC, we created triple mutants with apc5CA. Although deletion of HPA2 in apc5CA cells had no obvious effect, deletion of HPA2 in apc5CA gcn5Δ cells had a greater impact on the phenotype (Fig. 2B and 3A, 35°C). One possible scenario may be that in apc5CA cells, Gcn5 and Hpa2 have redundant functions that work in a positive manner with the APC. Deletion of HPA2 in gcn5Δ cells, however, restored gcn5Δ ts growth at 35°C and 37°C (Fig. 3A), indicating that Gcn5 and Hpa2 in a WT APC background function antagonistically. How this antagonistic interaction is altered in apc5CA cells to produce a synergistic interaction remains to be determined.
Fig. 3.
Genetic interactions between apc5CA and HAT/HDAC mutants are complex. (A) A genetic interaction between hpa2Δ and gcn5Δ mutants is revealed in apc5CA cells. The mutants shown were constructed by genetic crosses and tested for temperature sensitivity using spot dilutions. See the legend to Fig. 2 for explanation of interaction symbols. (B) The elp3Δ gcn5Δ defect is more severe than that of strains containing apc5CA, as determined by spot dilutions. (C) The mutants shown were created through multiple rounds of backcrossing and characterized using spot dilutions. (D) Increased expression of APC5 partially suppresses elp3Δ gcn5Δ ts growth. WT and elp3Δ gcn5Δ cells were transformed with an APC5-expressing construct under the control of the GAL1 promoter or the empty vector control. The transformants were spot diluted onto glucose- or galactose-supplemented plates and grown for 3 to 13 days.
Genetic interactions between ELP3/GCN5 and apc5CA.
As described above, combining the apc5CA allele with gcn5Δ or elp3Δ further exacerbated the phenotype of any of the single mutants alone. This supports the notion that Gcn5 and Elp3 share an overlapping function with at least Apc5. However, Gcn5 and Elp3, components of SAGA and Elongator, respectively, are themselves redundant transcription-associated HATs; deletion of the genes encoding the two proteins resulted in phenotypes that were more severe than those observed with either single mutant (36, 68). We found that the elp3Δ gcn5Δ mutant is more severely impaired than either the apc5CA elp3Δ or apc5CA gcn5Δ mutant (Fig. 3B). To further characterize the genetic interactions observed between these three mutants, we constructed triple mutants and assessed growth rates at various temperatures (Fig. 3C). The triple mutant phenotype was similar to that of the elp3Δ gcn5Δ mutant, indicating an epistatic interaction in which Elp3/Gcn5 may be upstream of Apc5. To test the validity of this possibility, we predicted that increased expression of a component downstream of Elp3/Gcn5 may improve elp3Δ gcn5Δ mutant phenotypes but expression of an upstream factor would not. Therefore, APC5 was expressed under the control of the GAL1 promoter in elp3Δ gcn5Δ cells on glucose- and galactose-supplemented media. Overexpression of APC5 on galactose was toxic to wild-type cells grown at 37°C and to elp3Δ gcn5Δ cells at 30°C (Fig. 3D). However, when the cells were grown on glucose, weak APC5 expression was apparent, since growth of elp3Δ gcn5Δ cells expressing APC5 was improved at both 30°C and 37°C. The above observations support the idea that the APC acts downstream of the redundant Elp3/Gcn5 function.
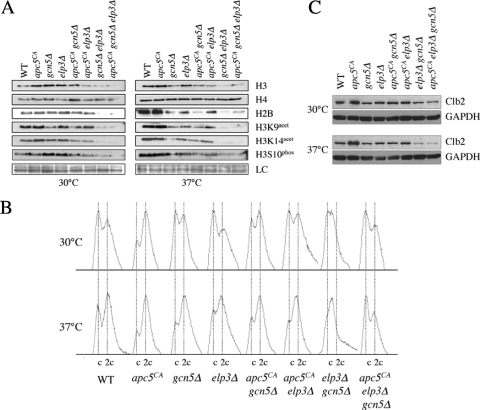
Histone H2B and H3 levels are reduced in elp3Δ gcn5Δ cells.
To determine if reduced histone levels correlate with apc5CA, gcn5Δ, and elp3Δ double and triple mutant growth limitations, we prepared whole-cell protein extracts from the different strains grown to mid-log phase at 30°C, with or without a subsequent shift to 37°C for 3 h. The extracts were analyzed for histone abundance and posttranslational modifications. Figure 4A shows representative data from this analysis. The results following a shift to 37°C are an exaggerated version of that observed at 30°C: total histone H2B and H3 levels are reduced in mutants containing both elp3Δ and gcn5Δ, whereas the total H4 level is unaffected. Acetylation of H3K9 and K14 and phosphorylation of H3S10 are also reduced but follow the total H3 profile and are therefore unlikely to represent separate defects. The H3 and H2B total protein profiles are similar, suggesting coregulation of H3 and H2B that is dependent on Elp3 and Gcn5 (64). However, the addition of the apc5CA allele modestly influenced total H3 levels only in elp3Δ cells. Thus, the total and acetylated state of histone H3 does not necessarily explain the synergistic genetic interaction observed when apc5CA is combined with elp3Δ or gcn5Δ. Alternatively, impaired histone stoichiometry observed in elp3Δ gcn5Δ cells could contribute to the growth phenotypes associated with this mutant. Previous work has shown that altered histone stoichiometry in yeast causes cell cycle and transcriptional defects (11, 44).
Fig. 4.
Histone posttranslational modifications and cell cycle profiles in apc5CA, elp3Δ, and gcn5Δ single, double, and triple mutants. (A) apc5CA, elp3Δ, and gcn5Δ mutants were used to characterize histone profiles. Extracts were prepared after growth at 30°C or following a shift to 37°C for 3 h. Proteins were separated by SDS-PAGE, and Western analysis was performed using the antibodies indicated. (B) Flow cytometry was conducted on the asynchronous cultures described for panel A. (C) Clb2 stability was assessed in the cells described for panel A. Cell cultures were prepared and proteins were extracted as described for panel A. Western analyses were performed using antibodies against endogenous Clb2 and GAPDH as a load control (LC).
Next, we asked if apc5CA cell cycle progression is altered in the presence of elp3Δ and/or gcn5Δ. We performed flow cytometry on the various mutants grown at 30°C to early log phase or followed by a 3-h shift to 37°C. The results indicate that at 37°C, with the exception of cells containing elp3Δ and gcn5Δ, all of the mutants accumulated with a large percentage of cells containing a 2c DNA content, similar to that observed with the apc5CA mutant (Fig. 4B). Our observation that strains lacking ELP3 or GCN5 accumulated replicated but undivided DNA is consistent with a defect in mitotic passage. However, mutants lacking both ELP3 and GCN5 accumulated with unreplicated or replicating DNA, suggesting a defect in G1/S progression. To determine whether elp3Δ gcn5Δ cells are in fact exiting mitosis effectively, we examined the stability of the APC substrate Clb2, which is targeted for degradation to allow mitotic exit. The various mutants were grown to early log phase at 30°C. Asynchronous cells were then harvested for protein extract preparation or were switched to 37°C for an additional 3 h before harvesting. Western analyses were performed with antibodies against endogenous Clb2p, a cyclin required for APC activation, which then becomes a target (28, 47, 69). In early-log-phase apc5CA cultures, Clb2 protein accumulated (Fig. 4C). We also observed this with apc10Δ and cdc16-1 cells (data not shown). Under similar conditions, Clb2 did not accumulate in elp3Δ or gcn5Δ single and double mutants. The elp3Δ gcn5Δ mutant suppressed the apc5CA defect, as the triple mutant turned over Clb2 similar to the way the WT did (Fig. 4C). Thus, elp3Δ gcn5Δ cells likely progress through the M/G1 boundary effectively but appear to be impaired in transiting through G1/S. The slowed progression through G1 may provide time for an impaired APC to fully turn over Clb2.
Elp3 and Gcn5 inhibit passage through G1/S.
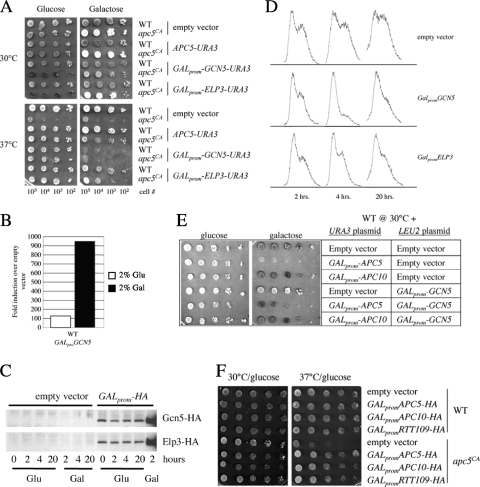
Our data suggest Elp3 and Gcn5 may be required for passage through G1. APC mutants accumulate with a G2/M DNA content, but the APC is also required for G1 progression. Thus, we questioned whether increased ELP3 or GCN5 expression is able to compensate for aberrant APC activity by expressing ELP3-HA and GCN5-HA under the control of the GAL1 promoter in wild-type and apc5CA cells. The results show that on glucose, apc5CA cells expressing ELP3 or GCN5 grow similarly to the wild-type cells and to apc5CA cells complemented with APC5 at elevated temperatures (Fig. 5A). We performed quantitative real-time PCR to confirm whether GALprom-GCN5 was expressed when grown on glucose (Fig. 5B). In WT cells grown in 2% glucose, GCN5 was expressed over 100-fold from the GAL1 promoter, compared to empty vector control cells. A Western analysis confirmed that both GCN5 and ELP3 were expressed from the GAL1 promoter in 2% glucose (Fig. 5C). When the transformants were grown on galactose to overexpress the constructs, we observed that overexpressed GCN5 was toxic to wild-type and apc5CA cells. Overexpression of ELP3 was moderately detrimental to apc5CA cells at elevated temperatures. To examine the cell cycle effects of low-level ELP3 and GCN5 expression, we grew WT cells expressing galactose-inducible GCN5 or ELP3, or an empty vector control, to early log phase in glucose. The cells were subcultured, and samples were taken after 2, 4, and 20 h of growth for flow cytometry. We observed that cells expressing the empty vector continued to cycle (Fig. 5D). However, cells expressing ELP3 or GCN5 rapidly accumulated with a G1 DNA content. Since overexpression of GCN5 caused cells to accumulate with unreplicated DNA, we asked if there was a link between Gcn5 downregulation and APC function. For this analysis, we coexpressed galactose-inducible GCN5 from a LEU2-based plasmid together with galactose-inducible APC5. The LEU2-based GALprom-GCN5 construct was not toxic (Fig. 5E), compared to the URA3-based plasmid (Fig. 5A), and partially suppressed APC5 overexpression toxicity. This observation suggests that APC overexpression toxicity may be linked to Gcn5 levels dropping below some optimum threshold point. Finally, considering the genetic interactions documented for gcn5Δ- and rtt109Δ-harboring cells (8, 17), we asked whether increased expression of the HAT gene RTT109 could also suppress the apc5CA ts defect. Consistent with our GCN5 and ELP3 studies, low-level RTT109 expression restored the apc5CA ts defect (Fig. 5F). Therefore, the apc5CA allele used in this study has allowed us to uncover previously unidentified potential interactions in yeast between the APC and a dynamic network of histone-modifying enzymes.
Fig. 5.
Low-level expression of the HAT gene ELP3, GCN5, or RTT109 suppresses the apc5CA ts defect. (A) WT and apc5CA cells were transformed with plasmids expressing GCN5 or ELP3 under the control of the GAL promoter. Transformants were spot diluted onto glucose- and galactose-supplemented media and grown for 10 days at either 30°C or 37°C. The plates were then scanned. (B) WT cells expressing an empty vector or GALprom-GCN5-HA were grown overnight in 2% glucose at 30°C, and the cultures were divided the next morning. One sample was resuspended in 2% galactose, and cultures were incubated for a further 3 h. Total RNA was then extracted and used in quantitative PCRs with primers against GCN5 and TFC1 to normalize expression. The ΔΔCT (2−ΔΔCT) method was used to determine fold changes in expression compared to that of empty vector controls. (C) Western analyses were performed on extracts obtained from WT cells expressing either an empty vector, GALprom-GCN5-HA, or GALprom-ELP3-HA. The cells were grown overnight to early log phase at 30°C. The next morning, the cultures were divided, with 2% galactose added to one and the other left in 2% glucose. Samples were taken at the times indicated and probed using antibodies against hemagglutinin (HA). (D) Low-level expression of GCN5 or ELP3 results in cells exiting the cell cycle early in G1. Cells were grown overnight to early log phase and then diluted back in glucose-supplemented media. Samples were taken at the times indicated and prepared for flow cytometry. (E) WT and apc5CA cells were cotransformed with the plasmids shown and grown at 30°C on 2% glucose- or galactose-supplemented plates. (F) RTT109 under the control of the GAL1 promoter was expressed in WT and apc5CA cells. The transformants were spot diluted onto glucose-supplemented plates and grown at 30°C or 37°C for 3 days.
Apc5 may be involved in facilitating histone H3 modification.
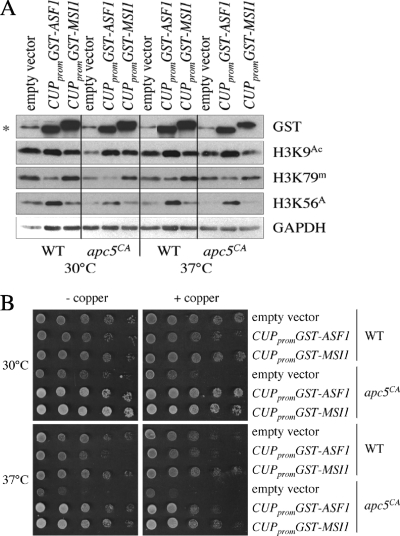
Considering that apc5CA ts defects can be suppressed by increased expression of both RTT109 (Fig. 5F) and ASF1 (22), we questioned whether reduced histone modifications in apc5CA cells were linked to the chromatin assembly deficit also observed in apc5CA cells (22). To address this, we increased expression of GST-ASF1 or GST-MSI1, via the inducible CUP1 promoter, in WT and apc5CA cells and examined modified histone H3 levels. We found that increased ASF1 expression resulted in elevated levels of H3K9Ac and H3K56Ac but reduced levels of H3K79me2 (Fig. 6A). This supports a proposed role for Asf1 in presenting histones H3 and H4 to Rtt109- and Gcn5-containing complexes for acetylation prior to passage to CAF-1 for deposition into chromatin (17). Increased expression of MSI1 had no effect on histone modifications. As a control, a spot dilution is shown to demonstrate the ability of GST-ASF1 and GST-MSI1 to suppress apc5CA defects (Fig. 6B). One possible explanation for these results is that the APC subunit Apc5 may be directly involved in facilitating histone methylation and acetylation.
Fig. 6.
Influence of increased ASF1 or MSI1 expression of histone modifications. (A) Histone modifications were assessed in WT and apc5CA cells overexpressing GST-ASF1 or GST-MSI1. Protein extracts were prepared from cells following growth at 30°C or after a shift to 37°C for 3 h. Proteins were induced by the addition of 100 μM CuSO4 for 3 h. Extracts were then analyzed using the antibodies indicated. (B) The cells described for panel A were spot diluted onto control plates, or plates containing 100 μM CuSO4, and grown for 3 days at 30°C and 37°C to confirm that apc5CA ts defects were suppressed.
Gcn5 instability depends on the APC.
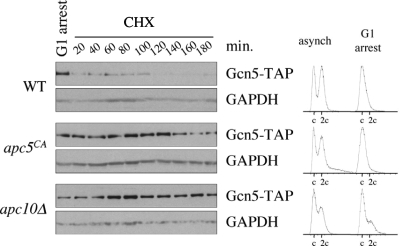
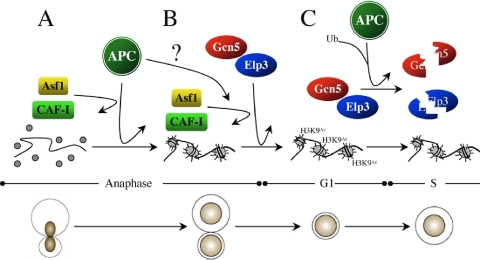
Our data indicate that gcn5Δ interacts genetically with the mutant APC subunit allele apc5CA and that Gcn5 may be targeted for degradation in order to progress through the G1/S transition. To test the hypothesis that Gcn5 is unstable during G1 in an APC-dependent manner, we arrested WT, apc5CA, and apc10Δ cells expressing an endogenous GCN5-TAP allele in G1 using α-factor. G1 arrest was confirmed using flow cytometry (Fig. 7). Following arrest, the cells were washed and resuspended into fresh media containing cycloheximide to block all further protein synthesis. Samples were removed every 20 min for Gcn5-TAP protein analysis by Western blotting. We observed that Gcn5-TAP was unstable in WT cells but stable in apc5CA and apc10Δ cells (Fig. 7). Taken together, our observations provide evidence to support our model that Gcn5 and Elp3 participate in mitotic progression in a manner that requires input from the APC (Fig. 8). We hypothesize that the APC facilitates the interaction between the HATs and the CAFs in order to correctly acetylate and assemble histones into chromatin. This mitotic acetylation pattern is likely established to allow progression through G1. Once this pattern is established, Gcn5 (and perhaps Elp3) is targeted for degradation to pass through the G1/S boundary.
Fig. 7.
Gcn5 is unstable during G1 but stabilized in APC mutants. WT, apc5CA, and apc10Δ cells expressing endogenous GCN5-TAP were arrested in G1 using α-factor after reaching early log phase at 30°C. Flow cytometry results of the asynchronous and α-factor-arrested cells are shown on the right. Following arrest, the cells were washed and added to fresh media containing cycloheximide (CHX) to block all further protein synthesis. Samples were taken every 20 min for 3 h to assess Gcn5-TAP protein stability. GAPDH Western analyses were conducted to control for protein load.
Fig. 8.
Model of APC/HAT/CAF interactions. (A) The APC directs mitotic chromatin assembly though Asf1 and CAF-I. (B) Our data indicate that Gcn5 and Elp3 independently work redundantly with the APC to promote mitotic progression. The establishment of a transcriptional profile required for G1 progression by Gcn5/Elp3 is likely necessary for APC function. We propose that the APC facilitates the interaction between the HATs and Asf1 in order to deliver acetylated histones to the CAF-I complex for deposition into chromatin. (C) Finally, to exit G1 and enter S, we propose that the transcriptional profile established by Elp3/Gcn5 must be reset. This is likely accomplished by APC-dependent targeting of at least Gcn5 for degradation during G1.
DISCUSSION
The anaphase-promoting complex (APC) is most commonly associated with targeting proteins that inhibit sister chromatin separation (Pds1) and exit from mitosis (Clb2) for ubiquitin- and proteasome-dependent degradation (28, 47, 69). APC activity is necessary to maintain genomic stability and is inhibited by the spindle checkpoint until all chromosomes are properly aligned along the metaphase plate (18, 23, 29, 30). Our studies have also linked the APC with mitotic chromatin assembly and cellular survival (1, 23–26). Considering that aberrant APC activity is associated with defective chromosome structure, cancer development, and premature aging (3, 24, 27, 41, 57), we speculated that the involvement of APC in chromatin metabolism may be critical for maintaining genomic stability. The results presented here suggest a link between cell cycle progression and histone metabolism. We provide data supporting the hypothesis that the HATs Gcn5 and Elp3 are independently required for progression through mitosis but share an overlapping function that is necessary for APC-dependent passage through G1. This hypothesis is based on our initial findings that ELP3 or GCN5 deletions delay mitotic passage, whereas the elp3Δ gcn5Δ mutant sports a G1 phenotype that is epistatic to the apc5CA phenotype (Fig. 2B, 3B, and 3C). We propose that Gcn5 and Elp3 may be required for the reestablishment of a transcriptional profile that enables cells to proceed through G1. Early exit from the cell cycle in G1 in cells with increased GCN5 or ELP3 expression (Fig. 5D) suggests the possibility that a G1-specific transcriptional profile established by Gcn5 and Elp3 must be reset in order to efficiently exit G1 (Fig. 8). The involvement of the CAFs CAF-I, Asf1, and the Asf1 binding partner Rtt109 in this pathway suggests a possible testable mechanism. H3K56Ac is tightly linked with chromatin assembly (10), but loading of H3 with K56Ac prior to chromatin assembly is likely unaffected in apc5CA mutants (Fig. 1A), whereas the deposition of these loaded histones onto DNA is likely the compromised step (23). Our earlier results demonstrating that increased expression of ASF1 suppressed apc5CA defects, but only when the CAF-I complex was intact (25), suggest that it may possibly be the interaction between Asf1 and CAF-I that is faulty in apc5CA cells, as Asf1 is believed to pass acetylated histones onto the CAF-I complex (10).
The APC may be involved in reestablishing a transcriptional profile required for cell cycle reentry.
Previously we speculated that the APC may play a role in the initiation of transcription of genes required for cell cycle reentry (23). Our data presented here support this possibility, as deletion of genes encoding Gcn5, Elp3, or Sas3, which appear to function together at similar genes to facilitate transcription (Fig. 2A) (31, 51, 68), impairs the apc5CA ts defect (Fig. 3). On the other hand, deletion of genes involved in gene silencing (SAS2, HOS1, HOS2, or HOS3) suppressed the apc5CA ts defect (Fig. 2). A recent study using fission yeast found that treating APC mutants with HDAC inhibitors, or deleting the HDACs Clr3, Clr6, or Hos2 (all sensitive to trichostain A), restored mutant APC phenotypes (32). Phenotypic restoration was shown to coincide with increased APC complex formation, and Apc8/Cut23 was acetylated. It was suggested that Clr6/Rpd3 inhibited APC assembly, while Clr3/Hda1 and Hos2 block sister chromatid separation by loading chromatin with cohesin, both situations resulting in APC inactivity. In our study, deletion of RPD3 had no effect on the apc5CA phenotype, HDA1 deletion exacerbated the phenotype, and HOS2 deletion suppressed it (Fig. 2A). It seems that the link between chromatin and the APC in budding and fission yeasts is conserved, but the mechanisms involved may have diverged. It is not clear whether the APC influences chromatin modifications and gene expression in fission yeast, but a recent demonstration that the Atf1 transcription factor genetically and physically interacted with the fission yeast Apc5 suggests that the link between the APC and transcription is indeed conserved (45). Additional work will be required to work out the details and differences between the two yeast species and the relevance of these differences as far as human biology is concerned.
Nonetheless, evidence exists suggesting a link between budding yeast APC and chromatin dynamics. Previously, we demonstrated genetic interactions between APC mutants and mutations in genes encoding the CAFs CAF-I, Asf1, Hir1, and Hir2 (23). Asf1 appears to couple histone acetylation with histone deposition (10, 17), and here we provide evidence that the APC genetically interacts with mutations in multiple histone-modifying enzymes, such as the HATs Elp3, Gcn5, Rtt109, Sas2, and Sas3 and the HDACs Hda1, Hos1, Hos2, and Hos3 (Fig. 2 and 5F). Since Asf1 and Rtt109 are known to physically and functionally interact (10, 17), identifying both factors as genetic modifiers of apc5CA phenotypes is compelling evidence that at least Apc5 is involved in modifying and/or assembling histones into chromatin. Furthermore, suppression of apc5CA ts defects by increased Rtt109 expression provides additional evidence to suggest that the mechanism facilitating this interaction is conserved, as human APC5 and the Rtt109 orthologue CBP physically and functionally interact (14, 61).
While one target of Rtt109, H3K56, does not appear to be a primary player in APC mutant phenotypes (Fig. 1), another target, H3K9 (17), and methylation of H3K79 do appear to be associated with APC mutant phenotypes. H3K9Ac increases during mitosis and is associated with expression of late mitotic genes (17, 35). Similarly, H3K79me2, which depends upon Dot1 activity (62), accumulates during mitosis and is also associated with the expression of mitosis-specific genes (54). Both of these modifications are reduced in most APC mutants in our study. Gcn5 is a key HAT involved in the acetylation of H3K9 (17, 64). Elp3, on the other hand, acetylates primarily H3K14 and H4K8 (66) but has been shown to functionally overlap with Gcn5 at H3K9 (36). Our work shows that under restrictive conditions, H3K9 and H3K14 acetylation is decreased in elp3Δ and gcn5Δ cells but abolished in the double mutant, as is total H3 content. This indicates that the loss of H3K9 and H3K14 acetylation in the elp3Δ gcn5Δ double mutant may be due to the loss of total H3. Previously, Kristjuhan et al. (36) also showed that the majority of H3K9 and H3K14 acetylation was lost in the double mutant. However, in that study, the double mutant did not impair total H3 levels. Previous studies have also shown redundant Gcn5 and Sas3 activity at intensely transcribed genes, as the mutations are lethal when combined (31, 51). Considering that sas3Δ also exacerbates apc5CA ts defects (Fig. 2B), it appears likely that Elp3, Gcn5, and Sas3 all work together at actively transcribed genes, perhaps in an APC-dependent fashion to set the stage for cell cycle reentry following cell division. Support for this hypothesis was provided by a recent synthetic genetic array (SGA) screen using the apc5CA allele as bait (12, 34). Four genes encoding components of the nonsense mediated mRNA decay pathway, NAM7, UPF3, EBS1, and NMD2, suppressed the apc5CA ts defect when deleted. Furthermore, LSM1, which encodes a protein involved in cytoplasmic mRNA degradation, and AIR2, a gene required for nuclear RNA quality control, both suppressed the apc5CA ts defect when deleted. Together, these data suggest that increased overall RNA abundance is a benefit when at least Apc5 is compromised.
Gcn5 and Elp3 are required for G1 progression, and at least Gcn5 must be removed to exit G1.
Increased ELP3, GCN5, and RTT109 expression was sufficient to restore the apc5CA ts growth defect to wild-type levels (Fig. 5A and F). One possible explanation for the phenotypic restoration is that the HATs could act upstream of the APC by promoting the transcription of genes required for APC activity. Alternatively, the HATs, together with the APC, may act in a redundant pathway required for mitotic progression. Our observation that Clb2, an APC substrate targeted for degradation, remains unstable in elp3Δ gcn5Δ mutants (Fig. 4C) suggests that Elp3 and Gcn5 are not upstream APC activators. Overexpression of these genes, on the other hand, particularly GCN5, was toxic to wild-type cells. Flow cytometry of cells expressing ELP3 or GCN5 at low levels, which is beneficial to apc5CA cells, showed slowed progression through G1 in comparison to that of empty vector controls (Fig. 5A and D). Delayed progression through G1 may assist a compromised APC in completing G1-dependent functions. Thus, taken together, the results suggest that Gcn5 and Elp3 have independent functions during mitosis but act redundantly to ensure G1 passage. The APC presumably remains active in elp3Δ gcn5Δ mutants but apparently has no impact on G1 progression in this background. One possible explanation for the apparent lack of effect when Apc5 is mutated in this background during G1 would be if Elp3/Gcn5 was required for the expression of an APC target or facilitator that acted during G1. If Elp3 and Gcn5 do indeed drive the expression of such a protein, then it stands that Elp3 and/or Gcn5 would need to be inhibited to exit G1. A recent report also found mitotic defects in gcn5Δ mutants, which were attributed to defective centromere/kinetochore interactions (63). Furthermore, in HeLa cells, GCN5 is required for the acetylation, and subsequent phosphorylation, of CDC6, which is necessary for the relocalization of CDC6 to the cytoplasm during S phase (46). GCN5 formed a complex with CDC6, and with cyclin A when bound to CDK2, during early S phase. Paolinelli (46) demonstrated that GCN5 levels peaked at early S phase and were decreased by mid-S phase. Thus, reports in the literature are consistent with a mitotic function for Gcn5 and that GCN5 content, at least in human cells, cycles.
The APC, chromatin, and life span.
The idea that the APC is required for histone metabolism in actively growing cells fits well with current literature on chromatin structure, cancer, and life span (6, 9, 42, 56). APC substrates continue to be identified. A recent report found that many APC substrates required for cell cycle progression are transcribed in mitosis and then targeted for degradation. Human proteins meeting these criteria were screened as APC targets, and one was found, RCS1, that interacted with the NuRD chromatin remodeling complex, which is important for transcriptional regulation (70). Among the proteins that copurified with RCS1 were HDAC1 and -2 and the histone chaperones RbAp46/48. These findings provide additional evidence in support of the idea that the evolutionarily conserved APC is involved in multiple events that require stringent chromatin-modifying activities.
Aging and cancer are tightly associated with chromatin metabolism. Cells suffering from genomic instability are more likely to prematurely age or enter an uncontrolled proliferative state than cells capable of maintaining genomic stability. Furthermore, mutations in eukaryotic model systems that enhance longevity are often involved in increasing genomic stability, either through increased oxidant scavenging or through an increased capacity to repair DNA. Our observation that histone protein levels decrease in APC mutants may be related to the reduced life span observed in APC mutants (24). Decreased histone levels in aging cells could lead to multiple problems related to aging-related diseases, most notably compromised transcription and genomic instability. We do not believe the decreased histone levels are due to reduced histone transcripts (Fig. 1B), but we cannot rule out the possibility that the capacity to store histones is compromised in APC mutants (22). Although it is also possible that histone translation is compromised in APC mutants, we have not observed global defects in protein translation.
The APC promotes genomic stability (4, 23, 44, 55) and resistance to cancer (48, 65). The effects of the APC on genomic stability are likely linked with our observations suggesting that the APC requires a specific transcriptional profile in order to promote mitotic exit and G1 maintenance. Decreased histone acetylation may impair reestablishment of this transcriptional profile as cells exit mitosis, but histone hypoacetylation is also a major player in promoting uncontrolled cell proliferation. Potent histone deacetylase inhibitors (HDACi's) have a remarkable ability to kill cancer cells in vitro. In fact, several HDACi's are currently in clinical trials (19, 59). HDACi's are known to increase global histone acetylation and induce the expression of genes involved in growth arrest, differentiation, and apoptosis (15). Thus, global hypoacetylation, a condition observed in several APC mutants, likely leads to changes in chromatin remodeling that repress expression of genes that promote genomic stability.
ACKNOWLEDGMENTS
We thank Ata Ghavidel, Tony Hunter, Dave Stuart, and Wei Xiao for the generous gifts of plasmids and strains. Stanley Moore and members of the Harkness laboratory are thanked for critical reading of the manuscript. Alison McGuire is acknowledged for her expert technical assistance with the quantitative PCR analysis. Mark Boyd assisted with the flow cytometry analysis.
E.L.T. is the recipient of a University of Saskatchewan College of Medicine Graduate Scholarship. This work was supported by grants to T.A.A.H. from the Canadian Institutes for Health Research and the Canadian Foundation for Innovation.
Footnotes
Published ahead of print on 13 August 2010.
REFERENCES
- 1.Arnason T. G., Pisclevich M. G., Dash M. D., Davies G. F., Harkness T. A. A. 2005. Novel interaction between Apc5p and Rsp5p in an intracellular signaling pathway in Saccharomyces cerevisiae. Eukaryot. Cell 4:134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. 1995. Current protocols in molecular biology. Wiley, New York, NY [Google Scholar]
- 3.Baker D. J., Jeganathan K. B., Cameron J. D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R. B., de Groen P. C., Roche P., van Deursen J. M. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744–749 [DOI] [PubMed] [Google Scholar]
- 4.Bellanger S., Blachon S., Mechali F., Bonne-Andrea C., Thierry F. 2005. High-risk but not low-risk HPV E2 proteins bind to the APC activators Cdh1 and Cdc20 and cause genomic instability. Cell Cycle 4:1608–1615 [DOI] [PubMed] [Google Scholar]
- 5.Binné U. K., Classon M. K., Dick F. A., Wei W., Rape M., Kaelin W. G., Jr., Näär A. M., Dyson N. J. 2007. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat. Cell Biol. 9:225–232 [DOI] [PubMed] [Google Scholar]
- 6.Bitterman K. J., Medvedik O., Sinclair D. A. 2003. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol. Mol. Biol. Rev. 67:376–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bostelman L. J., Keller A. M., Albrecht A. M., Arat A., Thompson J. S. 2007. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair 6:383–395 [DOI] [PubMed] [Google Scholar]
- 8.Burgess R. J., Zhou H., Han J., Zhang Z. 2010. A role for Gcn5 in replication-coupled nucleosome assembly. Mol. Cell 37:469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campisi J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120:513–522 [DOI] [PubMed] [Google Scholar]
- 10.Chen C. C., Tyler J. 2008. Chromatin reassembly signals the end of DNA repair. Cell Cycle 7:3792–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark-Adams C. D., Norris D., Osley M. A., Fassler J. S., Winston F. 1988. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 2:150–159 [DOI] [PubMed] [Google Scholar]
- 12.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St. Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. 2010. The genetic landscape of a cell. Science 327:425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang W., Steffen K. K., Perry R., Dorsey J. A., Johnson F. B., Shilatifard A., Kaeberlein M., Kennedy B. K., Berger S. L. 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459:802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das C., Lucia M. S., Hansen K. C., Tyler J. K. 2009. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459:113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummond D. C., Noble C. O., Kirpotin D. B., Guo Z., Scott G. K., Benz C. C. 2005. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu. Rev. Pharmacol. Toxicol. 45:495–528 [DOI] [PubMed] [Google Scholar]
- 16.Durant M., Pugh B. F. 2006. Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:2791–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fillingham J., Recht J., Silva A. C., Suter B., Emili A., Stagljar I., Krogan N. J., Allis C. D., Keogh M. C., Greenblatt J. F. 2008. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol. Cell. Biol. 28:4342–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Higuera I., Manchado E., Dubus P., Cañamero M., Méndez J., Moreno S., Malumbres M. 2008. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 10:802–811 [DOI] [PubMed] [Google Scholar]
- 19.Graham J. S., Kaye S. B., Brown R. 2009. The promises and pitfalls of epigenetic therapies in solid tumours. Eur. J. Cancer 45:1129–1136 [DOI] [PubMed] [Google Scholar]
- 20.Grant P. A., Duggan L., Cote J., Roberts S. M., Brownell J. E., Candau R., Ohba R., Owen-Hughes T., Allis C. D., Winston F., Berger S. L., Workman J. L. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640–1650 [DOI] [PubMed] [Google Scholar]
- 21.Grant P. A., Eberharter A., John S., Cook R. G., Turner B. M., Workman J. L. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274:5895–5900 [DOI] [PubMed] [Google Scholar]
- 22.Gunjan A., Verreault A. 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115:537–549 [DOI] [PubMed] [Google Scholar]
- 23.Harkness T. A. A., Davies G. F., Ramaswamy V., Arnason T. G. 2002. The ubiquitin-dependent targeting pathway in Saccharomyces cerevisiae plays a critical role in multiple chromatin assembly regulatory steps. Genetics 162:615–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harkness T. A. A., Shea K. A., Legrand C., Brahmania M., Davies G. F. 2004. A functional analysis reveals dependence on the anaphase-promoting complex for prolonged life span in yeast. Genetics 168:759–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harkness T. A. A., Arnason T. G., Legrand C., Pisclevich M. G., Davies G. F., Turner E. L. 2005. Contribution of CAF-I to anaphase-promoting-complex-mediated mitotic chromatin assembly in Saccharomyces cerevisiae. Eukaryot. Cell 4:673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harkness T. A. A. 2005. Chromatin assembly from yeast to man: Conserved factors and conserved molecular mechanisms. Curr. Genomics 6:227–240 [Google Scholar]
- 27.Harkness T. A. A. 2006. The anaphase promoting complex and aging: the APCs of longevity. Curr. Genomics 7:263–272 [Google Scholar]
- 28.Harper J. W., Burton J. L., Solomon M. J. 2002. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 16:2179–2206 [DOI] [PubMed] [Google Scholar]
- 29.Hartwell L. H., Smith D. 1985. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 110:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herzog F., Primorac I., Dube P., Lenart P., Sander B., Mechtler K., Stark H., Peters J. M. 2009. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science 323:1477–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howe L., Auston D., Grant P., John S., Cook R. G., Workman J. L., Pillus L. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15:3144–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimata Y., Matsuyama A., Nagao K., Furuya K., Obuse C., Yoshida M., Yanagida M. 2008. Diminishing HDACs by drugs or mutations promotes normal or abnormal sister chromatid separation by affecting APC/C and adherin. J. Cell Sci. 121:1107–1118 [DOI] [PubMed] [Google Scholar]
- 33.Kimura A., Umehara T., Horikoshi M. 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32:370–377 [DOI] [PubMed] [Google Scholar]
- 34.Koh J. L., Ding H., Costanzo M., Baryshnikova A., Toufighi K., Bader G. D., Myers C. L., Andrews B. J., Boone C. 2010. DRYGIN: a database of quantitative genetic interaction networks in yeast. Nucleic Acids Res. 38:D502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krebs J. E. 2007. Moving marks: dynamic histone modifications in yeast. Mol. Biosyst. 3:590–597 [DOI] [PubMed] [Google Scholar]
- 36.Kristjuhan A., Walker J., Suka N., Grunstein M., Roberts D., Cairns B. R., Svejstrup J. Q. 2002. Transcriptional inhibition of genes with severe histone H3 hypoacetylation in the coding region. Mol. Cell 10:925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo M. H., Brownell J. E., Sobel R. E., Ranalli T. A., Cook R. G., Edmondson D. G., Roth S. Y., Allis C. D. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269–272 [DOI] [PubMed] [Google Scholar]
- 38.Kurdistani S. K., Robyr D., Tavazoie S., Grunstein M. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31:248–254 [DOI] [PubMed] [Google Scholar]
- 39.Lee T. I., Causton H. C., Holstege F. C., Shen W. C., Hannett N., Jennings E. G., Winston F., Green M. R., Young R. A. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701–704 [DOI] [PubMed] [Google Scholar]
- 40.Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 41.Luo J., Emanuele M. J., Li D., Creighton C. J., Schlabach M. R., Westbrook T. F., Wong K. K., Elledge S. J. 2009. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137:835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maslov A. Y., Vijg J. 2009. Genome instability, cancer and aging. Biochim. Biophys. Acta 1790:963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masumoto H., Hawke D., Kobayashi R., Verreault A. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436:294–298 [DOI] [PubMed] [Google Scholar]
- 44.Meeks-Wagner D., Hartwell L. H. 1986. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44:43–52 [DOI] [PubMed] [Google Scholar]
- 45.Ors A., Grimaldi M., Kimata Y., Wilkinson C. R., Jones N., Yamano H. 2009. The transcription factor Atf1 binds and activates the APC/C ubiquitin ligase in fission yeast. J. Biol. Chem. 284:23989–23994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paolinelli R., Mendoza-Maldonado R., Cereseto A., Giacca M. 2009. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat. Struct. Mol. Biol. 16:412–420 [DOI] [PubMed] [Google Scholar]
- 47.Peters J. M. 1999. Subunits and substrates of the anaphase-promoting complex. Exp. Cell Res. 248:339–349 [DOI] [PubMed] [Google Scholar]
- 48.Pray T. R., Parlati F., Huang J., Wong B. R., Payan D. G., Bennett M. K., Issakani S. D., Molineaux S., Demo S. D. 2002. Cell cycle regulatory E3 ubiquitin ligases as anticancer targets. Drug Resist. Updat. 5:249–258 [DOI] [PubMed] [Google Scholar]
- 49.Robert F., Pokholok D. K., Hannett N. M., Rinaldi N. J., Chandy M., Rolfe A., Workman J. L., Gifford D. K., Young R. A. 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16:199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robyr D., Suka Y., Xenarios I., Kurdistani S. K., Wang A., Suka N., Grunstein M. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109:437–446 [DOI] [PubMed] [Google Scholar]
- 51.Rosaleny L. E., Ruiz-García A. B., García-Martínez J., Pérez-Ortín J. E., Tordera V. 2007. The Sas3p and Gcn5p histone acetyltransferases are recruited to similar genes. Genome Biol. 8:R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruiz-García A. B., Sendra R., Galiana M., Pamblanco M., Pérez-Ortín J. E., Tordera V. 1998. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J. Biol. Chem. 273:12599–12605 [DOI] [PubMed] [Google Scholar]
- 53.Rundlett S. E., Carmen A. A., Kobayashi R., Bavykin S., Turner B. M., Grunstein M. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. U. S. A. 93:14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulze J. M., Jackson J., Nakanishi S., Gardner J. M., Hentrich T., Haug J., Johnston M., Jaspersen S. L., Kobor M. S., Shilatifard A. 2009. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell 35:626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Searle J. S., Schollaert K. L., Wilkins B. J., Sanchez Y. 2004. The DNA damage checkpoint and PKA pathways converge on APC substrates and Cdc20 to regulate mitotic progression. Nat. Cell Biol. 6:138–145 [DOI] [PubMed] [Google Scholar]
- 56.Sedivy J. M., Banumathy G., Adams P. D. 2008. Aging by epigenetics—a consequence of chromatin damage? Exp. Cell Res. 314:1909–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song S. J., Song M. S., Kim S. J., Kim S. Y., Kwon S. H., Kim J. G., Calvisi D. F., Kang D., Lim D. S. 2009. Aurora A regulates prometaphase progression by inhibiting the ability of RASSF1A to suppress APC-Cdc20 activity. Cancer Res. 69:2314–2323 [DOI] [PubMed] [Google Scholar]
- 58.Suka N., Luo K., Grunstein M. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32:378–383 [DOI] [PubMed] [Google Scholar]
- 59.Thomas S., Munster P. N. 2009. Histone deacetylase inhibitor induced modulation of anti-estrogen therapy. Cancer Lett. 280:184–191 [DOI] [PubMed] [Google Scholar]
- 60.Thornton B. R., Ng T. M., Matyskiela M. E., Carroll C. W., Morgan D. O., Toczyski D. P. 2006. An architectural map of the anaphase-promoting complex. Genes Dev. 20:449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turnell A. S., Stewart G. S., Grand R. J., Rookes S. M., Martin A., Yamano H., Elledge S. J., Gallimore P. H. 2005. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature 438:690–695 [DOI] [PubMed] [Google Scholar]
- 62.van Leeuwen F., Gafken P. R., Gottschling D. E. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745–756 [DOI] [PubMed] [Google Scholar]
- 63.Vernarecci S., Ornaghi P., Bâgu A., Cundari E., Ballario P., Filetici P. 2008. Gcn5p plays an important role in centromere kinetochore function in budding yeast. Mol. Cell. Biol. 28:988–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verzijlbergen K. F., Faber A. W., Stulemeijer I. J., van Leeuwen F. 2009. Multiple histone modifications in euchromatin promote heterochromatin formation by redundant mechanisms in Saccharomyces cerevisiae. BMC Mol. Biol. 10:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wasch R., Engelbert D. 2005. Anaphase-promoting complex-dependent proteolysis of cell cycle regulators and genomic instability of cancer cells. Oncogene 24:1–10 [DOI] [PubMed] [Google Scholar]
- 66.Winkler G. S., Kristjuhan A., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. 2002. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. U. S. A. 99:3517–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wittschieben B. O., Otero G., de Bizemont T., Fellows J., Erdjument-Bromage H., Ohba R., Li Y., Allis C. D., Tempst P., Svejstrup J. Q. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4:123–128 [DOI] [PubMed] [Google Scholar]
- 68.Wittschieben B. O., Fellows J., Du W., Stillman D. J., Svejstrup J. Q. 2000. Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J. 19:3060–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zachariae W., Nasmyth K. 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13:2039–2058 [DOI] [PubMed] [Google Scholar]
- 70.Zhao W. M., Coppinger J. A., Seki A., Cheng X. L., Yates J. R., III, Fang G. 2008. RCS1, a substrate of APC/C, controls the metaphase to anaphase transition. Proc. Natl. Acad. Sci. U. S. A. 105:13415–13420 [DOI] [PMC free article] [PubMed] [Google Scholar]