Abstract
Trypanosomatid protozoans are reliant on posttranscriptional processes to control gene expression. Regulation occurs at the levels of mRNA processing, stability, and translation, events that may require the participation of the poly(A) binding protein (PABP). Here, we have undertaken a functional study of the three distinct Leishmania major PABP (LmPABP) homologues: the previously described LmPABP1; LmPABP2, orthologous to the PABP described from Trypanosoma species; and LmPABP3, unique to Leishmania. Sequence identity between the three PABPs is no greater than 40%. In assays measuring binding to A-rich sequences, LmPABP1 binding was poly(A) sensitive but heparin insensitive; LmPABP2 binding was heparin sensitive and less sensitive to poly(A), compatible with unique substitutions observed in residues implicated in poly(A) binding; and LmPABP3 displayed intermediate properties. All three homologues are simultaneously expressed as abundant cytoplasmic proteins in L. major promastigotes, but only LmPABP1 is present as multiple isoforms. Upon transcription inhibition, LmPABP2 and -3 migrated to the nucleus, while LmPABP1 remained predominantly cytoplasmic. Immunoprecipitation assays showed an association between LmPABP2 and -3. Although the three proteins bound to a Leishmania homologue of the translation initiation factor eukaryotic initiation factor 4G (eIF4G) (LmEIF4G3) in vitro, LmPABP1 was the only one to copurify with native LmEIF4G3 from cytoplasmic extracts. Functionality was tested using RNA interference (RNAi) in Trypanosoma brucei, where both orthologues to LmPABP1 and -2 are required for cellular viability. Our results indicate that these homologues have evolved divergent functions, some of which may be unique to the trypanosomatids, and reinforces a role for LmPABP1 in translation through its interaction with the eIF4G homologue.
Trypanosomatids are flagellated protozoans, which are best represented by pathogenic species belonging to the genera Leishmania and Trypanosoma, responsible for diseases with worldwide impact, such as leishmaniasis, sleeping sickness, and Chagas' disease. These organisms display unique biological and molecular features rarely found in other eukaryotes, including polycistronic transcription (33) and coupled trans-splicing and polyadenylation (41), to produce typical monocistronic mRNAs with a 5′ cap and a 3′ poly(A) tail. The lack of one promoter for one protein-coding gene implies a much reduced role for transcription in regulating gene expression. Therefore, it is assumed that such regulation occurs mainly at the posttranscriptional level, possibly through mechanisms that act upon mRNA processing, stability, and translation (14, 15).
Initiation is the most likely target for mechanisms involved in the regulation of translation. During translation initiation, ribosomes are recruited to the mRNA and positioned at the first adequate AUG codon, a process mediated by the translation initiation factors (eukaryotic initiation factors [eIFs]) (reviewed in references 27 and 52). Poly(A) binding protein (PABP), the major cytoplasmic mRNA binding protein (also known as PABPC or pab1p in Saccharomyces cerevisiae) binds to the 3′-end poly(A) tails of the mRNAs and participates in an extensive range of cellular functions (reviewed in references 8, 24, 39, and 42). In the initiation of translation, it promotes an interaction between the 3′ and 5′ ends of the mRNA by binding to the 3′ poly(A) tail and to eIF4G (32, 40, 59), a component of the cap binding complex eIF4F (reviewed in references 23 and 53). The PABP/eIF4G interaction seems to enhance translation initiation and mRNA stability and may be a checkpoint for mRNA integrity prior to translation (3, 58). PABP also interacts with the release factor eRF3, an event that may couple translation termination with ribosome recycling and mRNA decay (29, 60). Aside from translation, PABP participates in several steps of mRNA biogenesis, processing, and degradation (11, 12, 30). In the last, dissociation of PABP from poly(A) may be required for deadenylation, the first step in the major decay pathway for many mRNAs. Furthermore, this protein seems to be involved in mRNA transport from the nucleus to the cytoplasm (9, 12, 22, 63).
The PABP structure is highly conserved (reviewed in reference 39), with its N-terminal two-thirds consisting of four tandem RNA recognition motifs (RRMs). Each RRM is defined by the presence of two unique RNP motifs and, at the structural level, assumes a canonical RNA binding fold of four antiparallel β-strands backed by two α-helixes (18). The first two RRMs are sufficient for specific poly(A) binding (38) and have been implicated in the interaction between PABP and eIF4G (13, 32, 35). RRM3 and -4 lack poly(A) binding specificity and may mediate recognition of non-poly(A) sequences (36, 57). The PABP C-terminal region, although it does not bind RNA, contains a conserved domain, named PABC, which has been shown to be responsible for the binding of release factor eRF3, the PABP-interacting proteins Paip1 and Paip2, and the poly(A) nuclease PAN (37, 55). Connecting the RRMs to the PABC domain is a proline- and glutamine-rich linker region, the least conserved segment of the protein, which is important for the multimerization of PABP molecules on poly(A) (44) and has been implicated in the PABP function in mRNA deadenylation and decay in yeast (56, 64).
So far, PABP homologues have been described from several species of trypanosomatids (4, 5, 51). The first homologue identified was Trypanosoma cruzi PABP1, encoded by two tandemly repeated genes and constitutively expressed (5). The Trypanosoma brucei orthologue has also been characterized and found to be 86.4% identical (88.7% similar) to the T. cruzi protein (31). Leishmania major PABP1 (LmPABP1) is encoded by a single-copy gene constitutively expressed during the L. major life cycle and is only 35% identical to either of the Trypanosoma PABP homologues, a level of identity comparable to the 36% identity between the Leishmania and human proteins. LmPABP1 is predominantly cytoplasmic and has at least two isoforms, one of which is phosphorylated. Complementation assays have demonstrated that it does not rescue the lethality of pab1 deletion in S. cerevisiae (4).
We have previously identified, within available trypanosomatid genome sequences, multiple conserved homologues of the subunits of eIF4F, an observation that hints at a higher degree of complexity in translation initiation than might be expected for unicellular organisms (20). Three PABP homologues are also found in L. major genome sequences (LmPABP1, LmPABP2, and LmPABP3), with only the first two being conserved in Trypanosoma species. Here, we set out to characterize them functionally and to investigate potential roles in translation. The three L. major proteins are simultaneously expressed but differ in protein and RNA binding properties and in subcellular localization under conditions of transcription inhibition. Coupled with in vivo data for the two orthologues conserved in T. brucei, which are both essential for viability, these results are compatible with functional distinctions between these homologues that may have implications for the cellular translation apparatus.
MATERIALS AND METHODS
Protein sequences used in this study and sequence analysis methods.
All of the L. major and T. brucei sequences described in the text were originally derived from the respective genome sequences and confirmed later through sequencing of the cloned fragments. The original annotation of the genomic LmPABP1 sequence deposited in GenBank (accession no. XP_843606) lacks the first 17 residues, which have subsequently been included in the sequence deposited at GeneDb (ID, LmjF35.5040). The sequence that defines the full-length protein, then, is the one available at GeneDb. The genomic LmPABP1 sequence and the one described previously (AAC 64372) differ further by 9 amino acids. These might reflect intraspecies polymorphism; however, we consider it an unlikely possibility, since the differences between the Leishmania infantum (XP_001469326/LinJ35_V3.5360) and L. major genomic sequences for PABP1 are limited to only 5 positions. LmPABP2 (XP_843520/LmjF35.4130) and LmPABP3 (XP_001683760/LmjF25.0080) were the second and third Leishmania PABP homologues identified and were named accordingly. TbPABP1 is the T. brucei sequence that encodes the LmPABP1 orthologue (XP_827237/Tb09.211.0930). To avoid confusion, the previously described T. brucei PABPI (31), which is 100% identical to the second genomic PABP from T. brucei, will be referred to throughout the text and figures as TbPABP2 or PABP2 (XP_827358/Tb09.211.2150). For the same reasons, PABP2 or TcPABP2 will be used to refer, when necessary, to the original T. cruzi PABP1 (5). Sequence analysis and alignments were carried out essentially as previously reported (20). Nuclear localization signals (NLS) were investigated using the PredictNLS program (16; http://www.rostlab.org/services/predictNLS/).
PCR and cloning.
The coding sequences for the three L. major PABP homologues were amplified from total DNA extracted from the Friedlin strain. The LmPABP1 and LmPABP2 sequences were both amplified through two rounds of PCR. First, the full-length sequences were amplified using primers lacking restriction sites and that annealed just before and after the translation start and stop codons, respectively (LmPABP1, 5′ primer, CAC TCA AAT GGC TGC TGC TGT CC, and 3′ primer, CTT CTC GCT TAC GCC GTC TGA TG; LmPABP2, 5′ primer, AA AGG AGA AGC AGC CAA CCA TG, and 3′ primer, GCG CGC GTG CAT GCA CGG GTG). The resulting fragments were then used as templates in a second set of PCRs, which allowed their amplification flanked by sites for BamHI/HindIII (LmPABP1) or BamHI/NotI (LmPABP2) (LmPABP1, 5′ primer, GTG GGA TCC ATG GCT GCT GCT GTC CAG GAA G, and 3′ primer, TGA AGC TTC GCC GTC TGA TGC GCC TTG AGC; LmPABP2, 5′primer, GTG GGA TCC ATG GCC TTC ACT GGT CCG, and 3′ primer, TGG CGG CCG CAA CGC TCA TGT GAC GGT TCA G; the cited restriction sites are underlined). LmPABP3 was amplified in a single PCR flanked by BamHI/NotI (5′ primer, TCC GGA TCC ATG GTG GCC CCA GCG CAA C; 3′ primer, TCC GCG GCC GCA TTG CCA GTG TGC TGC TGG). The LmPABP1 sequence was first cloned into the BamHI/HindIII sites of the plasmid vector pET21A (Novagen) for the expression of a recombinant C-terminally tagged His fusion. Later, it was recovered by partial digestion and subcloned into the BamHI/NotI sites of pGEX4T3 (GE Healthcare), which allowed the expression of recombinant protein with glutathione S-transferase (GST) at its N terminus. LmPABP2 and -3 were cloned directly into the BamHI/NotI sites of both pET21A and pGEX4T3 for the expression of similar recombinant proteins. All amplified fragments and constructs were confirmed through automatic sequencing.
For the RNA interference (RNAi) experiments, the sequences encoding the two T. brucei PABP homologues were amplified from genomic DNA flanked by sites for HindIII and BamHI and subcloned into the same sites of the transfection vector p2T7-177 (61) using exactly the approach described previously (34).
Expression and purification of recombinant proteins.
For the expression of either His- or GST-tagged recombinant proteins, plasmids were transformed into Escherichia coli BLR or BL21 cells. The transformed bacteria were grown in LB medium and induced with IPTG (isopropyl-β-d-thiogalactopyranoside). The induced cells were sedimented, resuspended in phosphate-buffered saline (PBS), and lysed by sonication. Protein purification was performed as described previously (17) with either Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) or glutathione-4B-Sepharose (Amersham Biosciences). Protein products were analyzed in 15% SDS-PAGE stained with Coomassie blue R-250. For the quantification of the recombinant proteins, serial dilutions were compared in Coomassie-stained gels with serial dilutions of known concentrations of bovine serum albumin (BSA).
Antibody production and Western blotting.
Rabbit antisera were raised against LmPABP1, -2, and -3 by immunizing adult New Zealand White rabbits with the His-tagged recombinant forms. To reduce the background, the various antibodies were affinity purified and stored in aliquots at −80°C prior to use. Western blotting was performed with the Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore), using as the second antibody peroxidase-conjugated goat anti-rabbit IgG serum (Jackson ImmunoResearch Laboratories) or protein A-peroxidase (GE Healthcare). Reactions were detected by enhanced chemiluminescence (ECL). Densitometric scanning of the various results was performed with Kodak 1D Image Analysis Software, version 3.5 for Windows.
Parasite cultures.
Promastigotes of L. major (MHOM/IL/81/Friedlin) were generally maintained in modified LIT medium prepared as described previously (20). Total protein lysates used for the expression analysis were obtained from log-phase hemocytometer-quantified parasite cell pellets resuspended directly in SDS-PAGE sample buffer. For the immunofluorescence assays, the same cells were grown in Schneider's insect medium supplemented with antibiotics and 10% fetal calf serum. Procyclic forms of T. brucei Lister 427(pLEW29, pLEW13) (62) were maintained as previously described (19).
Fluorescence microscopy.
For the indirect immunofluorescence assay, mid-log-phase L. major promastigotes (5 × 106/ml) were harvested, washed with PBS, fixed with 3% paraformaldehyde, and allowed to adhere to poly-l-lysine-coated slides. Permeabilization was carried out with 0.1% NP-40, followed by blocking with 1% BSA. Antibody detection of LmPABP1, LmPABP2, and LmPABP3 followed standard procedures. DNA was stained using TOTO-3 (Molecular Probes), and cells were visualized using a Leica SPII-AOBS confocal microscope.
EMSAs.
To generate the RNA probes used in the assays, a pTZ18R-derived plasmid containing a 61-nucleotide-long A-rich sequence (17) was linearized with XbaI prior to being used as a template for transcription with T7 RNA polymerase in the presence of [α-32P]ATP (GE Healthcare). The resulting RNAs were purified by centrifugation through ProbeQuant G-50 Micro Columns (GE Healthcare) and quantitated after precipitation with cetyltrimethylammonium bromide (CTAB). Electrophoretic mobility shift assays (EMSAs) were performed as described previously (44), with the binding reaction performed at room temperature and heparin added only when specifically stated. Electrophoresis was performed at 18°C. For the experiments with the competitor polynucleotides [poly(A), poly(U), and poly(C)], the polynucleotides were included in the binding reactions prior to the addition of the labeled probe.
RNAi and metabolic labeling.
All procedures for the creation of transgenic T. brucei cell lines and RNAi experiments were carried out essentially as described previously (19). For metabolic labeling, cultures at selected times after RNAi induction were washed with methionine-free SDM-79 medium and then resuspended at 1 × 107 cells/ml in the same medium containing 25 μCi/ml of [35S]methionine and incubated for 1 h at 27°C prior to being harvested and analyzed by SDS-PAGE and autoradiography.
Pulldown assays.
Pulldown assays were performed essentially as described previously (20), using glutathione-Sepharose beads (GE Healthcare). The beads alone or GST or GST-tagged PABP fusions immobilized on the beads were assayed for the ability to bind to 35S-labeled LmEIF4G3, obtained after transcription and translation of the full-length gene cloned on the pET21D plasmid. Alternatively, GST-tagged full-length LmEIF4G3 (GST-LmEIF4G31-636 [21]) was incubated with the 35S-labeled Leishmania PABP homologues obtained through the transcription and translation of the pET21A-derived plasmids linearized with either NotI (LmPABP2 and -3) or HindIII (LmPABP1).
IPs.
Immunoprecipitations (IPs) were carried out with L. major cytoplasmic extracts and protein A-Sepharose beads from GE Healthcare. For the cytoplasmic extracts, exponentially grown promastigote cells were harvested, washed with PBS, and resuspended in lysis buffer (20 mM HEPES-KOH, pH 7.4, 75 mM potassium acetate, 4 mM magnesium acetate, 2 mM dithiothreitol [DTT], supplemented with protease inhibitors) at a concentration of 2 × 108 cells/ml. The cells were lysed by several cycles of freeze-thawing, followed by centrifugation at 10,000 × g for 15 min to remove cellular debris. The supernatant was used for the IPs, which essentially followed standard procedures. For each reaction, 30 μl of beads (∼3 mg) was incubated overnight at 4°C with 1 to 2 μg of selected affinity-purified antibodies, or 5 μl of the total preimmune sera as a control, in 0.5 ml of PBS. After being washed, antibody-bound beads were incubated with 100 μl of the cytoplasmic extracts (2 h at 4°C). Specifically, bound proteins were eluted in SDS-PAGE sample buffer and analyzed by SDS-PAGE and Western blotting. If necessary, during the Western blotting procedures, the IgG heavy-chain band was cut out from the membrane to prevent its reaction with the secondary antibody.
RESULTS
Identification of three distinct PABP homologues from L. major.
The low level of identity observed between the first PABP homologues described from Leishmania and Trypanosoma species implied that they were not true orthologues and that more than one PABP homologue could exist within the trypanosomatids. Indeed, the sequencing of the L. major genome confirmed the presence, not only of the previously described Leishmania PABP (LmPABP1), but also of a second homologue, here called LmPABP2, which is the orthologue of the proteins identified previously from T. cruzi and T. brucei (5, 31). These orthologues are clearly identifiable, with 70% and 64% identities, respectively, between the PABP1 and PABP2 orthologues from L. major and T. brucei. Later, with the completion of the sequencing of the first Leishmania genome (33), a third PABP homologue was identified (LmPABP3); however, it has no trypanosome counterpart.
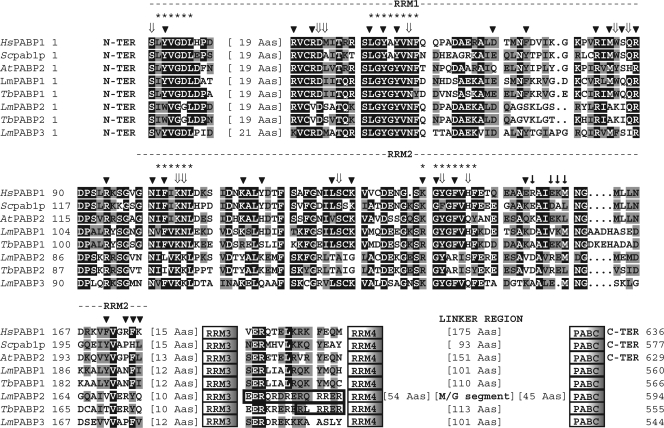
All trypanosomatid PABP homologues conform to the classical division into the N terminus, containing the four conserved RRMs; a central linker region; and the C terminus, with the PABC domain. The overall identities between the three proteins are quite low (32% for LmPABP1/LmPABP2, 33% for LmPABP1/LmPABP3, and 33% for LmPABP2/ LmPABP3) and are concentrated in the N terminus and the PABC domain, with no significant similarity within the linker region. The L. major PABP RRMs are more similar to the corresponding RRMs from other homologues than to each other, implying an early duplication process of the full-length gene. Compared with the human protein, RRM2 from LmPABP1 and -3 is the most conserved domain, followed by RRM1 and -4, with RRM3 being clearly more divergent. RRM2 is also the most conserved, and RRM3 the most divergent, when plant and yeast PABPs are compared with their human counterparts. In contrast, for LmPABP2, the most conserved domain by far is RRM1, followed by RRM3 and -4, with RRM2 being the most divergent.
When the sequences from RRM1 and -2 from the L. major and T. brucei homologues are compared with the prototype yeast and human PABP sequences (1, 26, 54), as well as Arabidopsis thaliana PABP2, representing the various plant PABP homologues (7), unique distinctions can be observed for the trypanosomatid PABP2 orthologues (Fig. 1). Several amino acid residues that perform direct contact with the RNA substrate and/or are responsible for the poly(A) binding specificity (based on reference 18) and are generally conserved are specifically replaced in these proteins. These residues are mainly positioned within RRM2, where both RNP motifs are altered, but substitutions are also found within RRM1 and the segment connecting the two RRMs. In contrast, residues that localize to the first two RRMs and have been previously defined as important for the PABP/eIF4G interaction (13, 32, 35, 48) are overall less strictly conserved, and no specific feature was observed for any of the proteins investigated. Outside of RRM1 and -2, no further distinctions were observed for any of the proteins, with the exception of the LmPABP2 linker region. This possesses an extended segment (∼50 amino acids long) very rich in methionine and glycine, sometimes arranged as multiple repeats of the triplet MGG (or VGG), which is conserved in orthologues from L. infantum and Leishmania braziliensis (not shown) but is mostly absent from Trypanosoma orthologues.
Fig. 1.
Sequence analysis of the L. major and T. brucei PABP homologues. Shown is a ClustalW alignment comparing the sequences from RRM1 and -2 of the human (Hs), yeast (Sc; S. cerevisiae), and plant (At; A. thaliana) PABP homologues with the various L. major/T. brucei proteins. The two RRMs are indicated, as well as the positions of RRM3 and -4, the linker region, and the PABC domain. Sequences between RRM3 and -4 where putative NLS were identified in the PABP2 orthologues are also shown. Within the aligned regions, amino acids identical in more than 60% of the sequences are highlighted in dark gray, while amino acids defined as similar, based on the BLOSUM 62 Matrix, in more than 60% of the sequences are shown in pale gray. When necessary, spaces were inserted within the various sequences (dots) to allow better alignment. The asterisks define the two RNP motifs. The arrowheads indicate amino acids that have been shown to interact with the RNA, while ⇓ highlights amino acids specifically involved in poly(A) recognition (18). ↓ symbols indicate the residues in yeast PABP RRM2 relevant to the interaction with eIF4G (based on reference 48). The location of the methionine/glycine-rich segment within the linker region from LmPABP2 is highlighted. The A. thaliana PABP2 homologue was included in the alignment because it is the nearest homologue to LmPABP1 within the various A. thaliana PABPs (not shown). For the GenBank/GeneDB accession numbers for the trypanosomatid PABP homologues, see Materials and Methods. The other GenBank accession numbers are as follows: HsPABP, P11940; Scpab1p, NP_011092; AtPABP2, P42731.
Quantitation of intracellular levels in L. major promastigotes.
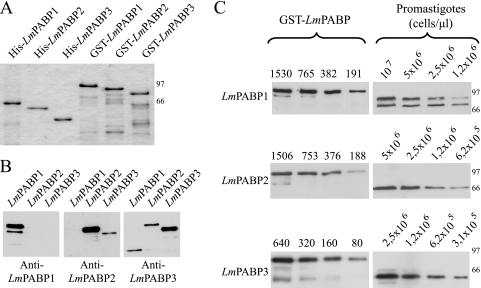
To start their functional characterization, the genes encoding the three L. major PABP homologues were cloned, and the corresponding proteins were expressed, His tagged or as GST fusions, in E. coli, and antisera were generated in rabbits against these recombinant proteins. Figure 2A shows a representative gel with aliquots of the recombinant proteins used throughout this work. LmPABP1 migrated as the largest protein, with roughly the same size as LmPABP2; LmPABP3 was smaller. The three sera were tested in Western blots against the three distinct recombinant proteins. They were found to be specific to the protein for which they were made (Fig. 2B), although the sera against LmPABP2 weakly cross-reacted with recombinant LmPABP3, producing a much weaker signal, and only when 12.5-fold more of the LmPABP3 protein was blotted. Likewise, the anti-LmPABP3 sera also produced a very weak reaction with LmPABP2. Considering the clear differences in the molecular weights of the two native proteins and the lack of any significant cross-reaction in samples from total parasite extracts, these weak cross-reaction events were not deemed to affect the results described below. Next, total protein extracts from L. major promastigotes grown exponentially were assayed in Western blots to investigate the expression of each PABP homologue, as well as to produce a rough estimate of their intracellular concentrations (Fig. 2C). LmPABP1 was represented by at least two distinctly migrating bands with apparent molecular masses of approximately 69 and 75 kDa, with the larger band representing the phosphorylated form of the protein, as previously described (4). LmPABP2 and -3 were detected as single bands with no indication of phosphorylation. Quantitation of the intracellular levels of the three proteins produced estimates of 7 × 104, 2 × 105, and 1.5 × 105 molecules/cell for LmPABP1, -2, and -3, respectively (for LmPABP1, the values obtained were derived from the sum of the quantitations of both phosphorylated and nonphosphorylated forms). Considering the number of mRNA molecules per T. brucei procyclic cell to be around 5 × 104 (19) and assuming similar levels for L. major promastigotes, the quantitation values indicate that all three PABP homologues are present in excess of the number of individual mRNAs.
Fig. 2.
Quantitation of intracellular levels of LmPABP1, -2, and -3. (A) His- and GST-tagged recombinant versions of the three L. major PABP homologues used in this work. The sizes of molecular mass markers (kDa) run on the same gel are shown on the right. (B) Specificities of the antibodies produced against the PABP homologues. The three GST fusions were blotted with three different antisera (indicated below each blot). (Left) Eight nanograms of LmPABP1 and 100 ng of LmPABP2 and -3. (Middle) Eight nanograms of LmPABP2 and 100 ng of LmPABP1 and -3. (Right) Eight nanograms of LmPABP3 and 100 ng of LmPABP1 and -2. (C) Quantitation of the expression levels of LmPABP1, -2, and -3 in extracts of exponentially grown L. major promastigotes. The different recombinant GST fusions, diluted to appropriate concentrations (in fentomoles), and different dilutions of whole parasite extracts were blotted with the corresponding antisera. The results were quantitated by densitometry, and the values were used to estimate the number of molecules per cell, as described previously (20). The results shown are representative of at least four different experiments using a minimum of three independently grown cell cultures. The larger band observed in the LmPABP3 blot represents a minor cross-reaction to one of the other two PABP homologues, probably LmPABP2.
Poly(A) binding assays.
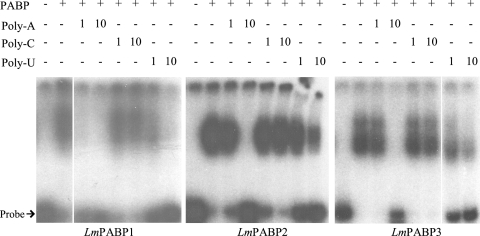
To investigate the binding of the Leishmania PABP homologues to RNA, EMSAs were carried out with the three recombinant GST fusions, using as a probe a labeled A-rich RNA derived from the 5′ untranslated region (UTR) of the human PABP mRNA. This probe consisted of sets of 5 to 8 A residues interspersed among 3 to 6 pyrimidines within a 61-nucleotide-long A-rich tract; it has been shown to be bound by human PABP in both EMSA and UV cross-linking experiments (17, 44). The three Leishmania proteins differ in the way they bind to the probe. Binding by GST-LmPABP1 produces a distinct shift pattern that is resistant to the addition of 5 mg/ml heparin. In contrast, GST-LmPABP2 produces a strong single shift that is prevented by the addition of 1 mg/ml heparin, while GST-LmPABP3 produces two shifts, with the higher one being inhibited by heparin at 1 mg/ml (Fig. 3 and data not shown). The specificity of binding to the probe was investigated by performing the assays, again using the A-rich probe, in the presence of the unlabeled competitors poly(A), poly(C), and poly(U). Binding by GST-LmPABP1 was impaired by the addition of even low levels of poly(A) (1 μg/ml), while binding by GST-LmPABP2 and -3 was affected only when 10-fold more of the polynucleotide was added. Poly(C) did not interfere with the binding of any of the three proteins even at the highest concentration added (10 μg/ml), while poly(U) produced an effect in all three proteins at 10 μg/ml, probably as a consequence of its base pairing to the probe and producing an RNA duplex (Fig. 3).
Fig. 3.
Electrophoretic mobility shift assay to investigate binding of the Leishmania PABPs to an A-rich RNA probe. The assay was performed by incubating the 32P-labeled A-rich probe described in the text with a fixed amount (100 ng) of the three recombinant GST fusions and analysis on a 5% nondenaturing polyacrylamide gel. Shifts obtained in the presence of the proteins were compared with the probe on its own. Where stated, the binding reactions were supplemented with 1 or 10 μg/ml of the polynucleotide poly(A), poly(C), or poly(U).
Subcellular localization of LmPABP1, -2, and -3.
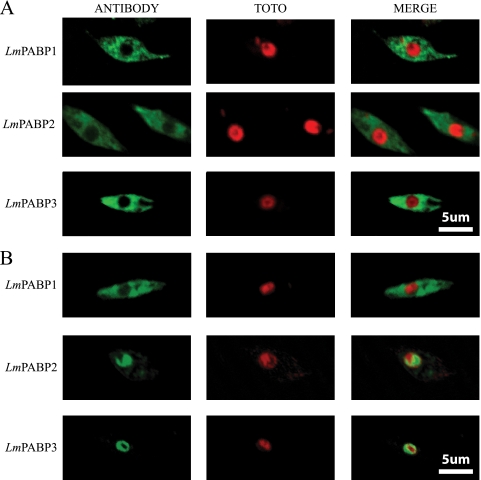
Mammalian PABP has been shown to be distributed throughout the cytoplasm, with a certain degree of localization to the perinuclear region (25). Upon transcription inhibition with actinomycin D, this distribution changes, and the protein can also be detected in the nucleus (2), a behavior that also has been shown to occur with LmPABP1 (4). To compare the subcellular localizations of the three L. major PABP homologues, the corresponding antisera were used in indirect immunofluorescence assays with exponentially grown cells and visualized under a confocal microscope. As shown in Fig. 4A, under normal growth conditions, all three proteins were localized mainly in the cytoplasm, with little or no nuclear localization and no further compartmentalization within the cytoplasm. Preincubation of the cells with actinomycin D, performed as reported previously (4), led to a minor migration of LmPABP1 to the nucleus (observed only through the fluorescence microscope) (not shown), but the protein remained predominantly within the cytoplasm (Fig. 4B). Under the same circumstances, both LmPABP2 and -3 migrated in their entirety to the nucleus with very little of either protein remaining in the cytoplasm. This differential localization of LmPABP2 and -3 is not induced upon incubation of the cells with the translational inhibitor cycloheximide (not shown). Within the nucleus, the two proteins do not colocalize with the DNA marker, suggesting a differential localization within the nucleoplasm distinct from the chromatin. A search for NLS, which are at least partially conserved in trypanosomatids (43), was then carried out using the different Leishmania PABP sequences. Putative motifs were found within both L. major and T. brucei PABP2 (Fig. 1), localized at the end of RRM3. These motifs are within a segment which, compared to other PABPs, has diverged in the PABP2 and -3 homologues. Although no predicted NLS was observed within LmPABP3, the equivalent segment was rich in positively charged residues, generally required for the nuclear localization elements (16).
Fig. 4.
Subcellular localization of LmPABP1, -2, and -3. Subcellular localization of the three L. major PABP homologues was carried out by indirect immunofluorescence using the distinct antiserum sets specific for each protein, followed by incubation with Alexa 488-conjugated secondary antibodies and analysis with a confocal microscope. Where indicated, the cells were counterstained with TOTO-3 to locate the nuclear and kinetoplast DNAs. (A) Exponentially grown L. major cells cultured under standard conditions in Schneider's insect medium. (B) Same as panel A, but with the addition of actinomycin D (10 μg/ml) to the growth medium for the last 10 h prior to harvesting and use in the assay.
Defining interactions between the different Leishmania PABP homologues.
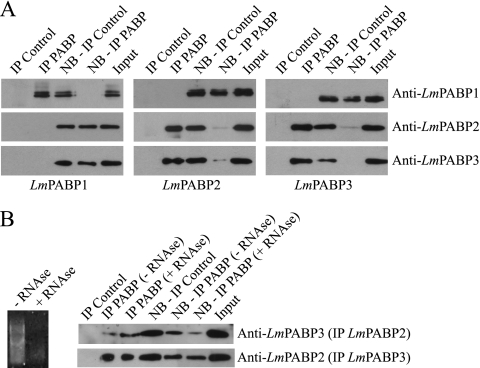
The simultaneous migration of LmPABP2 and -3 to the nucleus upon transcription inhibition suggests that the two proteins may interact directly or at least bind to the same target mRNAs. To investigate this possibility, we opted to individually immunoprecipitate the three native Leishmania PABPs from total cytoplasmic extracts and probe the precipitated immunocomplexes, or the resulting nonbound supernatants, for the presence of the remaining PABP homologues. As shown in Fig. 5A, each of the isoform-specific antibodies efficiently brought down nearly all of the corresponding proteins, with very little left in the respective nonbound supernatants, confirming the efficiency of the procedure. When the immunoprecipitated samples and supernatant of LmPABP1 were investigated for the presence of LmPABP2 and -3, no detectable amount of either protein was observed in the precipitated complexes, with both supernatant and input samples containing equivalent quantities of the two proteins. Likewise, when the samples from the immunoprecipitation of LmPABP2 and -3 were assayed for the presence of LmPABP1, no evidence for its coprecipitation was observed. In contrast, immunoprecipitation of LmPABP2 brought down LmPABP3 and depleted both LmPABP2 and -3 from the nonbound supernatant, while immunoprecipitation of LmPABP3 also brought down LmPABP2 and depleted both of them from the nonbound fraction. The LmPABP2 and -3 assays were then repeated, comparing the effects of pretreating the cytoplasmic extracts with RNase on the immunoprecipitation yields (Fig. 5B). Despite the nearly complete degradation of the endogenous RNA, both proteins continued to coprecipitate, indicating that they interact in an RNA-independent manner.
Fig. 5.
Analysis of the interactions between the three PABP homologues in vivo. (A) Distinct IP reactions were set up using total L. major cytoplasmic extract and affinity-purified antibodies directed against LmPABP1 (left), LmPABP2 (middle), and LmPABP3 (right), as well as the respective preimmune control sera (Control). Precipitated immunocomplexes, as well as the nonbound supernatant (NB) and the “Input” (equivalent to the amount of protein added to the IP reaction mixture), were then used in Western blot assays with the three distinct antisera, as indicated. (B) Effect of RNase treatment on the interaction between LmPABP2 and -3. Aliquots of the cytoplasmic extract were incubated in the presence or absence of 0.1 μg/μl of RNAse A (for 5 min at 37°C) prior to use in IP reactions carried out as for panel A with either the LmPABP2 or LmPABP3 antibody. For these assays, the IP of LmPABP2 was developed with the LmPABP3 antiserum and vice versa. On the left is shown the effect of the RNase treatment on the extract's RNAs.
Analysis of the interaction of LmPABP1, -2, and -3 with a Leishmania eIF4G homologue.
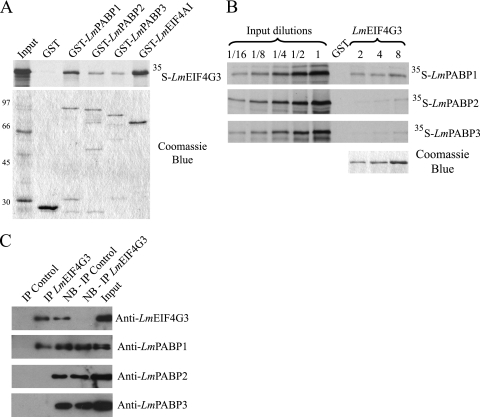
The binding between PABP and eIF4G is a critical event for the activity of PABP in translation. In Leishmania, several eIF4G homologues have been reported, one of which (LmEIF4G3) has been shown to interact with both the human and Leishmania eIF4A homologues (20) and has been shown to function within the context of an eIF4F complex (65). To investigate binding interactions between the three Leishmania PABP homologues and LmEIF4G3, pulldown assays were carried out. First, the recombinant GST fusions of LmPABP1 through -3 were immobilized on glutathione-Sepharose beads and tested for the ability to bind to the 35S-labeled full-length LmEIF4G3. In these assays GST alone and its fusion to the Leishmania eIF4A homologue, LmEIF4AI (originally named LmEIF4A1), were used as negative and positive controls, respectively. The LmEIF4G3 labeled protein bound to all three GST-PABP fusions, with the binding to GST-LmPABP1 being somewhat more efficient than those to the other two proteins (Fig. 6A). Binding to the positive-control GST-LmEIF4AI was superior to binding to any of the GST-PABP constructs, but no binding to the negative GST control was observed. A reverse assay was also carried out with full-length GST-LmEIF4G3 immobilized on the beads and incubated with the distinct 35S-labeled PABP homologues, again using GST as a negative control (Fig. 6B). Three concentrations of GST-LmEIF4G3 were used in these assays, and the input labeled proteins were diluted severalfold to produce a better picture of the different bindings. Binding by LmPABP1 was readily detected, even with the smallest amount of GST-LmEIF4G3 used. In contrast, even in the presence of 4-fold more of the recombinant GST-tagged protein, only trace amounts of labeled LmPABP2 and -3 bound to the beads. We then went on to investigate the interaction between the eIF4G homologue and LmPABP1, -2, and -3 in vivo after immunoprecipitation of native LmEIF4G3, followed by blotting with each of the three PABP antisera. As shown in Fig. 6C, only LmPABP1 was detected within the product of the immunoprecipitation reaction, with no signal produced with antibodies directed against LmPABP2 and -3.
Fig. 6.
Analysis of the interaction between an L. major eIF4G homologue (LmEIF4G3) and LmPABP1, -2, and -3. (A) Pulldown assays between GST-PABP fusions and 35S-labeled LmEIF4G3. GST-LmEIF4AI was used as a positive control. On top is shown the autoradiography with the pulldown results, while below is shown the equivalent Coomassie-blue stained gel (molecular mass markers [kDa] are shown on the left). (B) Reverse pulldown assay with GST-LmEIF4G3 immobilized on the glutathione-Sepharose beads and incubated with the labeled LmPABP1, -2, and -3. Three different concentrations of GST-LmEIF4G3 (2, 4, and 8 μg, shown on the gel stained with Coomassie blue at the bottom right) were used in equivalent pulldown reactions with the three 35S-labeled proteins. For this assay, aliquots of the “Input” (equivalent to the same amount of labeled protein used in the pulldown reactions) plus serial dilutions (up to 16-fold) were also run on the gel and autoradiography for comparison with the amount of protein bound to the beads. In both panels A and B, GST alone bound to the beads was used as a negative control. (C) Immunoprecipitation of native LmEIF4G3, performed as described for Fig. 5. The product of the reaction, the nonbound (NB) supernatant, and the “Input” were then individually blotted with the anti-LmEIF4G3 serum and each of the three PABP antisera.
The two PABP homologues conserved in Leishmania and Trypanosoma species are essential for cell viability.
The conservation of the trypanosomatid PABP1 and -2 homologues in both Leishmania and Trypanosoma species is a strong indication that they perform conserved functions within the various trypanosomatid species and that results derived from one organism can generally be extrapolated to the whole group. To investigate the requirements for these two proteins for cellular viability in T. brucei, the genes encoding their orthologues (TbPABP1 and -2) were cloned into a tetracycline-inducible RNAi vector and transfected into procyclic cell lines, and synthesis of double-stranded RNA was induced by the addition of tetracycline.
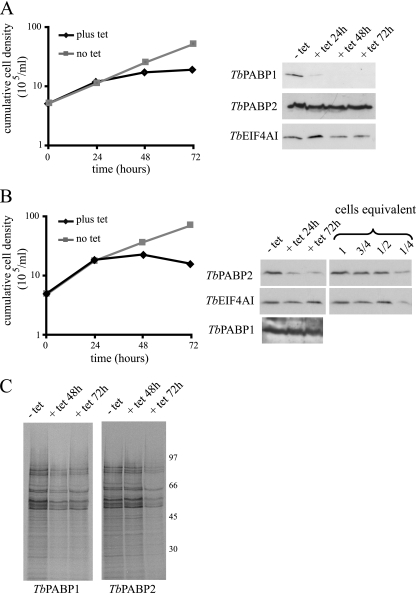
Upon induction of the RNAi, the results obtained for both TbPABP1 and -2 were in general very similar, with reduction in cellular proliferation and cessation of growth occurring between 24 and 48 h after tetracycline addition (Fig. 7A and B). Cell death ensued within a few days, although it tended to be a more immediate consequence for TbPABP2 than for TbPABP1. Western blotting using the LmPABP1 and -2 antibodies, which cross-react with their T. brucei counterparts, confirmed the efficiency of both RNAi procedures. For TbPABP1, very little of the protein was detected within 24 h of RNAi induction and at subsequent time points (Fig. 7A), while the levels of the translation initiation factor TbEIF4AI, used as a control, or TbPABP2 remained unaffected. Likewise for TbPABP2, the antibodies confirmed major decreases in levels within 24 h of RNAi induction, but in contrast to the TbPABP1 RNAi, residual levels of the protein (<25%) could be detected even 72 h after tetracycline addition. No effect was seen, however, on the levels of TbPABP1 or the control, TbEIF4AI (Fig. 7B). Metabolic labeling with [35S]methionine was also carried out to monitor changes in protein synthesis at selected time points after RNAi (Fig. 7C). For both proteins, an overall inhibition of protein synthesis was observed within 72 h of tetracycline addition, although at this stage, it is not possible to rule out for either protein a nonspecific effect on translation caused by the cell stress induced by the RNAi procedure. The lack of qualitative changes in the profiles of proteins being synthesized, nevertheless, indicates that the two proteins are not involved in the translation of different mRNA populations.
Fig. 7.
RNA interference of the T. brucei PABP homologues. (A and B) Representative growth curves after tetracycline (tet) induction of the RNAi for TbPABP1 (A) and TbPABP2 (B). Growth was monitored by counting the viable cells, and the expression of TbPABP1 and -2 or an endogenous control protein (TbEIF4AI) was assayed to monitor the efficiency of the RNAi procedure (shown on the right). Note the various dilutions of total cell extract used to estimate the efficiency of RNAi depletion for TbPABP2 (1 to 1/4 cell equivalent; 1 cell equivalent was equal to 106 cells and was used in the various RNAi lanes). The same RNAi samples were blotted on a second membrane, loaded with twice as much cellular extract, to detect TbPABP1. (C) Total protein synthesis at selected time points after tetracycline addition. Metabolic labeling was performed by incubating aliquots of the cells in the presence of [35S]methionine, followed by SDS-PAGE and autoradiography. The sizes of molecular mass markers (in kDa) run on the same gel from the autoradiography are shown on the right. The RNAi results are representative of multiple experiments performed with two distinct transfected cell lines, each submitted to at least one independent labeling experiment.
DISCUSSION
The origin of PABP, with its four-RRM arrangement plus the PABC domain, seems to predate the split of the main eukaryotic lineages. PABP homologues with the same conserved structure are present in all eukaryotes investigated so far, with the possible exception of Giardia lamblia, in which a true homologue could not be found within the sequences derived from the published genome (47). The trypanosomatid PABPs are no exception in this regard, although some features, particularly for the PABP2 homologue, are unique and might indicate novel functions that have evolved within this group of protozoa. The results presented here clearly indicate that trypanosomatid PABP1 and PABP2 bind to different targets and perform nonredundant and essential functions for cellular survival, despite being simultaneously expressed as abundant cytoplasmic proteins. It also seems very likely that the third homologue (PABP3) in Leishmania species either acts redundantly with PABP2 or is required to complement its function. At this stage, however, it is not known whether its absence from Trypanosoma species reflects an origin after the split between the Leishmania and Trypanosoma lineages or a loss from the latter.
The literature concerning protein synthesis in trypanosomatids is still scarce. The availability of selected genome sequences (33) and the analysis of the number of isotypes of subunits of eIF4F (20) have indicated a potentially complex translation initiation. The distinct PABP homologues observed may add yet a further element of complexity to the translation initiation process in trypanosomatids. So far, the results presented here, including the sequence analysis and poly(A) binding data, subcellular localization experiments, and pulldown assays, clearly implicate the PABP1 homologue as the major candidate to perform the functions already defined for PABP in translation in other eukaryotes. Other roles cannot be ruled out, and this homologue has already been shown to be able to prevent the deadenylation of RNAs in vitro (45). Lack of coprecipitation between the PABP1 and -2 homologues clearly highlights their potential to perform distinct roles; however, the abundance of PABP2, as well as the results from the RNAi experiments in T. brucei, also confirms that it plays a major role in mRNA metabolism in the trypanosomatids. The presence of yet a third homologue in Leishmania clearly deserves further attention to its function and how it relates to that of PABP2, especially considering the fact that they likely interact and/or bind to the same messages.
The sequence analysis of RRM1 and -2 from PABP2 suggested unique differences in RNA binding affinity and/or specificity, a result corroborated by the in vitro RNA binding assays. These differences may indicate novel functions that nevertheless seem to be conserved within the trypanosomatids. Considering that both RRM2 and PABC have been implicated in mediating the interaction of PABP with protein partners, such as eIF4G and eRF3 (28, 32, 35), the divergence of RRM2 in PABP2 might indicate a functional differentiation in comparison with PABP1, perhaps through interaction with novel RNA and protein partners, without losing the activities associated with the PABC domain. Indeed, in Crithidia fasciculata, the PABP2 orthologue has been found to be part of a complex that recognizes sequence elements in the untranslated regions of mRNAs that accumulate periodically during the cell cycle (46). Again, in T. cruzi, its orthologue was found to interact directly with TcUBP-1, an RNA binding protein that binds to AU-rich sequences in the 3′ UTRs of mRNAs, which are selectively destabilized in the trypomastigote stage of the parasite (21). Interestingly, TcUBP-1 can form homodimers through a glycine-rich region that is reminiscent of the central linker segment of PABP2. More recently, TcUBP-1 has been shown to migrate to the nucleus under conditions of arsenite-induced stress (10), a behavior similar to that observed for LmPABP2 and -3 under transcription inhibition. In another possible parallel with LmPABP2, this shuttling is dependent on the presence of a nuclear localization signal localized within the unique RRM of TcUBP-1. Nevertheless, the T. cruzi PABP2 protein does not shuttle to the nucleus in the presence of arsenite, and treatment with actinomycin D prevents the nuclear localization of TcUBP-1 (10), suggesting different mechanisms for the shuttling of the two proteins. Overall, it is possible that PABP2 may function as a general partner of different RNA binding proteins involved in the posttranscriptional control of the expression of selected mRNAs. Other roles could be associated with mRNA processing and export, considering the striking effect on subcellular localization of PABP2 (and also PABP3) induced upon inhibition of transcription. This may explain why the T. brucei PABP2 orthologue has been found associated with the U1 snRNP complex, required for the cis splicing of the intron-containing poly(A) polymerase message (49, 50).
The presence of three distinct PABP homologues in Leishmania, two of which are conserved in Trypanosoma species, is remarkable when it is considered that single-cell eukaryotes normally have only one homologue (42). In metazoans, the numbers of PABP homologues vary from one in Drosophila to four in humans (reviewed in reference 24), but multiple homologues might be a consequence of the need to differentially modulate mRNA translation and gene expression in different cellular types and tissues during development. In plants, the situation is more complex, with eight distinct homologues described for Arabidopsis, which vary as to their patterns of expression, the presence or absence of the PABC domain, and requirements for viability (6, 7). Noteworthy is the fact that all of the plant PABP homologues display conserved residues within the RNP motifs of RRM1 and -2, unlike what is seen in the trypanosomatid PABP2 proteins, indicating a possible conservation of poly(A) binding specificity. Taking everything together, defining in detail the function of the trypanosomatid PABPs remains a relevant goal that can unravel novel and unique mechanisms associated with mRNA metabolism and translation in these peculiar organisms, with possible impacts on the understanding of these processes in eukaryotes in general.
ACKNOWLEDGMENTS
This work was partially funded by grants provided by the Brazilian funding agencies FACEPE (006/2003-CNPq/PPP) and CNPq (476255/2004-0). T.D.D.C.L., D.M.N.M., and C.R.S.R. received studentships from CAPES and FACEPE. J.R.C.V. received a PQ (“pesquisador visitante”) scholarship from FIOCRUZ. Work in M. Carrington's laboratory is funded by the Wellcome Trust, and L.E. received a studentship funded by the BBSRC.
Technical assistance in growing the Leishmania cells was provided by B. S. Lima. We thank C. Docena and the PDTIS-FIOCRUZ Program for Technological Development in Tools for Health for access and help with confocal microscopy and sequencing.
Footnotes
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Adam S. A., Nakagawa T., Swanson M. S., Woodruff T. K., Dreyfuss G. 1986. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol. Cell. Biol. 6:2932–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonina E., Stauber R., Pavlakis G. N. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273:13015–13021 [DOI] [PubMed] [Google Scholar]
- 3.Amrani N., Ghosh S., Mangus D. A., Jacobson A. 2008. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453:1276–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates E. J., Knuepfer E., Smith D. F. 2000. Poly(A)-binding protein I of Leishmania: functional analysis and localisation in trypanosomatid parasites. Nucleic Acids Res. 28:1211–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batista J. A., Teixeira S. M., Donelson J. E., Kirchhoff L. V., Martins de Sa C. 1994. Characterization of a Trypanosoma cruzi poly(A)-binding protein and its genes. Mol. Biochem. Parasitol. 67:301–312 [DOI] [PubMed] [Google Scholar]
- 6.Belostotsky D. A. 2003. Unexpected complexity of poly(A)-binding protein gene families in flowering plants: three conserved lineages that are at least 200 million years old and possible auto- and cross-regulation. Genetics 163:311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bravo J., Aguilar-Henonin L., Olmedo G., Guzman P. 2005. Four distinct classes of proteins as interaction partners of the PABC domain of Arabidopsis thaliana Poly(A)-binding proteins. Mol. Genet. Genomics 272:651–665 [DOI] [PubMed] [Google Scholar]
- 8.Brook M., Smith J. W., Gray N. K. 2009. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction 137:595–617 [DOI] [PubMed] [Google Scholar]
- 9.Brune C., Munchel S. E., Fischer N., Podtelejnikov A. V., Weis K. 2005. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA 11:517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassola A., Frasch A. C. 2009. An RNA recognition motif mediates the nucleocytoplasmic transport of a trypanosome RNA-binding protein. J. Biol. Chem. 284:35015–35028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang T. C., Yamashita A., Chen C. Y., Yamashita Y., Zhu W., Durdan S., Kahvejian A., Sonenberg N., Shyu A. B. 2004. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 18:2010–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chekanova J. A., Belostotsky D. A. 2003. Evidence that poly(A) binding protein has an evolutionarily conserved function in facilitating mRNA biogenesis and export. RNA 9:1476–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng S., Gallie D. R. 2007. eIF4G, eIFiso4G, and eIF4B bind the poly(A)-binding protein through overlapping sites within the RNA recognition motif domains. J. Biol. Chem. 282:25247–25258 [DOI] [PubMed] [Google Scholar]
- 14.Clayton C., Shapira M. 2007. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 156:93–101 [DOI] [PubMed] [Google Scholar]
- 15.Clayton C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cokol M., Nair R., Rost B. 2000. Finding nuclear localization signals. EMBO Rep. 1:411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Melo Neto O. P., Standart N., Martins de Sa C. 1995. Autoregulation of poly(A)-binding protein synthesis in vitro. Nucleic Acids Res. 23:2198–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deo R. C., Bonanno J. B., Sonenberg N., Burley S. K. 1999. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98:835–845 [DOI] [PubMed] [Google Scholar]
- 19.Dhalia R., Marinsek N., Reis C. R., Katz R., Muniz J. R., Standart N., Carrington M., de Melo Neto O. P. 2006. The two eIF4A helicases in Trypanosoma brucei are functionally distinct. Nucleic Acids Res. 34:2495–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhalia R., Reis C. R., Freire E. R., Rocha P. O., Katz R., Muniz J. R., Standart N., de Melo Neto O. P. 2005. Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol. Biochem. Parasitol. 140:23–41 [DOI] [PubMed] [Google Scholar]
- 21.D'Orso I., Frasch A. C. 2002. TcUBP-1, an mRNA destabilizing factor from trypanosomes, homodimerizes and interacts with novel AU-rich element- and poly(A)-binding proteins forming a ribonucleoprotein complex. J. Biol. Chem. 277:50520–50528 [DOI] [PubMed] [Google Scholar]
- 22.Dunn E. F., Hammell C. M., Hodge C. A., Cole C. N. 2005. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 19:90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gingras A. C., Raught B., Sonenberg N. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913–963 [DOI] [PubMed] [Google Scholar]
- 24.Gorgoni B., Gray N. K. 2004. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief Funct. Genomic Proteomic 3:125–141 [DOI] [PubMed] [Google Scholar]
- 25.Görlach M., Burd C. G., Dreyfuss G. 1994. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp. Cell Res. 211:400–407 [DOI] [PubMed] [Google Scholar]
- 26.Grange T., Martins de Sa C., Oddos J., Pictet R. 1987. Human mRNA polyadenylate binding protein: evolutionary conservation of a nucleic acid binding motif. Nucleic Acids Res. 15:4771–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hershey J. W. B., Merrick W. C. 2000. Pathway and mechanism of initiation of protein synthesis, p. 33–88 InSonenberg N., Hershey J. W. B., Mathews M. B. (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28.Hoshino S., Hosoda N., Araki Y., Kobayashi T., Uchida N., Funakoshi Y., Katada T. 1999. Novel function of the eukaryotic polypeptide-chain releasing factor 3 (eRF3/GSPT) in the mRNA degradation pathway. Biochemistry 64:1367–1372 [PubMed] [Google Scholar]
- 29.Hosoda N., Kobayashi T., Uchida N., Funakoshi Y., Kikuchi Y., Hoshino S., Katada T. 2003. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 278:38287–38291 [DOI] [PubMed] [Google Scholar]
- 30.Hosoda N., Lejeune F., Maquat L. E. 2006. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell. Biol. 26:3085–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hotchkiss T. L., Nerantzakis G. E., Dills S. C., Shang L., Read L. K. 1999. Trypanosoma brucei poly(A) binding protein I cDNA cloning, expression, and binding to 5 untranslated region sequence elements. Mol. Biochem. Parasitol. 98:117–129 [DOI] [PubMed] [Google Scholar]
- 32.Imataka H., Gradi A., Sonenberg N. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17:7480–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivens A. C., Peacock C. S., Worthey E. A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M. A., Adlem E., Aert R., Anupama A., Apostolou Z., Attipoe P., Bason N., Bauser C., Beck A., Beverley S. M., Bianchettin G., Borzym K., Bothe G., Bruschi C. V., Collins M., Cadag E., Ciarloni L., Clayton C., Coulson R. M., Cronin A., Cruz A. K., Davies R. M., De Gaudenzi J., Dobson D. E., Duesterhoeft A., Fazelina G., Fosker N., Frasch A. C., Fraser A., Fuchs M., Gabel C., Goble A., Goffeau A., Harris D., Hertz-Fowler C., Hilbert H., Horn D., Huang Y., Klages S., Knights A., Kube M., Larke N., Litvin L., Lord A., Louie T., Marra M., Masuy D., Matthews K., Michaeli S., Mottram J. C., Muller-Auer S., Munden H., Nelson S., Norbertczak H., Oliver K., O'Neil S., Pentony M., Pohl T. M., Price C., Purnelle B., Quail M. A., Rabbinowitsch E., Reinhardt R., Rieger M., Rinta J., Robben J., Robertson L., Ruiz J. C., Rutter S., Saunders D., Schafer M., Schein J., Schwartz D. C., Seeger K., Seyler A., Sharp S., Shin H., Sivam D., Squares R., Squares S., Tosato V., Vogt C., Volckaert G., Wambutt R., Warren T., Wedler H., Woodward J., Zhou S., Zimmermann W., Smith D. F., Blackwell J. M., Stuart K. D., Barrell B., Myler P. J. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly S., Reed J., Kramer S., Ellis L., Webb H., Sunter J., Salje J., Marinsek N., Gull K., Wickstead B., Carrington M. 2007. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol. Biochem. Parasitol. 154:103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kessler S. H., Sachs A. B. 1998. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khanam T., Muddashetty R. S., Kahvejian A., Sonenberg N., Brosius J. 2006. Poly(A)-binding protein binds to A-rich sequences via RNA-binding domains 1+2 and 3+4. RNA Biol. 3:170–177 [DOI] [PubMed] [Google Scholar]
- 37.Kozlov G., Trempe J. F., Khaleghpour K., Kahvejian A., Ekiel I., Gehring K. 2001. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. U. S. A. 98:4409–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kühn U., Pieler T. 1996. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J. Mol. Biol. 256:20–30 [DOI] [PubMed] [Google Scholar]
- 39.Kühn U., Wahle E. 2004. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 1678:67–84 [DOI] [PubMed] [Google Scholar]
- 40.Le H., Tanguay R. L., Balasta M. L., Wei C. C., Browning K. S., Metz A. M., Goss D. J., Gallie D. R. 1997. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J. Biol. Chem. 272:16247–16255 [DOI] [PubMed] [Google Scholar]
- 41.Liang X. H., Haritan A., Uliel S., Michaeli S. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2:830–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangus D. A., Evans M. C., Jacobson A. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchetti M. A., Tschudi C., Kwon H., Wolin S. L., Ullu E. 2000. Import of proteins into the trypanosome nucleus and their distribution at karyokinesis. J. Cell Sci. 113:899–906 [DOI] [PubMed] [Google Scholar]
- 44.Melo E. O., Dhalia R., Martins de Sa C., Standart N., de Melo Neto O. P. 2003. Identification of a C-terminal poly(A)-binding protein (PABP)-PABP interaction domain: role in cooperative binding to poly(A) and efficient cap distal translational repression. J. Biol. Chem. 278:46357–46368 [DOI] [PubMed] [Google Scholar]
- 45.Milone J., Wilusz J., Bellofatto V. 2004. Characterization of deadenylation in trypanosome extracts and its inhibition by poly(A)-binding protein Pab1p. RNA 10:448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mittra B., Ray D. S. 2004. Presence of a poly(A) binding protein and two proteins with cell cycle-dependent phosphorylation in Crithidia fasciculata mRNA cycling sequence binding protein II. Eukaryot. Cell 3:1185–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morrison H. G., McArthur A. G., Gillin F. D., Aley S. B., Adam R. D., Olsen G. J., Best A. A., Cande W. Z., Chen F., Cipriano M. J., Davids B. J., Dawson S. C., Elmendorf H. G., Hehl A. B., Holder M. E., Huse S. M., Kim U. U., Lasek-Nesselquist E., Manning G., Nigam A., Nixon J. E., Palm D., Passamaneck N. E., Prabhu A., Reich C. I., Reiner D. S., Samuelson J., Svard S. G., Sogin M. L. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921–1926 [DOI] [PubMed] [Google Scholar]
- 48.Otero L. J., Ashe M. P., Sachs A. B. 1999. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 18:3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palfi Z., Lane W. S., Bindereif A. 2002. Biochemical and functional characterization of the cis-spliceosomal U1 small nuclear RNP from Trypanosoma brucei. Mol. Biochem. Parasitol. 121:233–243 [DOI] [PubMed] [Google Scholar]
- 50.Palfi Z., Schimanski B., Gunzl A., Lucke S., Bindereif A. 2005. U1 small nuclear RNP from Trypanosoma brucei: a minimal U1 snRNA with unusual protein components. Nucleic Acids Res. 33:2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pitula J., Ruyechan W. T., Williams N. 1998. Trypanosoma brucei: identification and purification of a poly(A)-binding protein. Exp. Parasitol. 88:157–160 [DOI] [PubMed] [Google Scholar]
- 52.Preiss T., Hentze M. W. 2003. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays 25:1201–1211 [DOI] [PubMed] [Google Scholar]
- 53.Prévôt D., Darlix J. L., Ohlmann T. 2003. Conducting the initiation of protein synthesis: the role of eIF4G. Biol. Cell 95:141–156 [DOI] [PubMed] [Google Scholar]
- 54.Sachs A. B., Bond M. W., Kornberg R. D. 1986. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell 45:827–835 [DOI] [PubMed] [Google Scholar]
- 55.Siddiqui N., Mangus D. A., Chang T. C., Palermino J. M., Shyu A. B., Gehring K. 2007. Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J. Biol. Chem. 282:25067–25075 [DOI] [PubMed] [Google Scholar]
- 56.Simón E., Seraphin B. 2007. A specific role for the C-terminal region of the poly(A)-binding protein in mRNA decay. Nucleic Acids Res. 35:6017–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sladic R. T., Lagnado C. A., Bagley C. J., Goodall G. J. 2004. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur. J. Biochem. 271:450–457 [DOI] [PubMed] [Google Scholar]
- 58.Svitkin Y. V., Sonenberg N. 2006. Translational control by the poly(A) binding protein: a check for mRNA integrity. Mol. Biol. 40:684–693 [PubMed] [Google Scholar]
- 59.Tarun S. Z., Jr., Sachs A. B. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15:7168–7177 [PMC free article] [PubMed] [Google Scholar]
- 60.Uchida N., Hoshino S., Imataka H., Sonenberg N., Katada T. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem. 277:50286–50292 [DOI] [PubMed] [Google Scholar]
- 61.Wickstead B., Ersfeld K., Gull K. 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 125:211–216 [DOI] [PubMed] [Google Scholar]
- 62.Wirtz E., Leal S., Ochatt C., Cross G. A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 63.Woods A. J., Kantidakis T., Sabe H., Critchley D. R., Norman J. C. 2005. Interaction of paxillin with poly(A)-binding protein 1 and its role in focal adhesion turnover and cell migration. Mol. Cell. Biol. 25:3763–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao G., Chiang Y. C., Zhang C., Lee D. J., Laue T. M., Denis C. L. 2007. PAB1 self-association precludes its binding to poly(A), thereby accelerating CCR4 deadenylation in vivo. Mol. Cell. Biol. 27:6243–6253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoffe Y., Leger M., Zinoviev A., Zuberek J., Darzynkiewicz E., Wagner G., Shapira M. 2009. Evolutionary changes in the Leishmania eIF4F complex involve variations in the eIF4E-eIF4G interactions. Nucleic Acids Res. 37:3243–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]