Abstract
Phosphoinositides are important regulators of diverse cellular functions, and phosphatidylinositol 3-monophosphate (PI3P) is a key element in vesicular trafficking processes. During its intraerythrocytic development, the malaria parasite Plasmodium falciparum establishes a sophisticated but poorly characterized protein and lipid trafficking system. Here we established the detailed phosphoinositide profile of P. falciparum-infected erythrocytes and found abundant amounts of PI3P, while phosphatidylinositol 3,5-bisphosphate was not detected. PI3P production was parasite dependent, sensitive to a phosphatidylinositol-3-kinase (PI3-kinase) inhibitor, and predominant in late parasite stages. The Plasmodium genome encodes a class III PI3-kinase of unusual size, containing large insertions and several repetitive sequence motifs. The gene could not be deleted in Plasmodium berghei, and in vitro growth of P. falciparum was sensitive to a PI3-kinase inhibitor, indicating that PI3-kinase is essential in Plasmodium blood stages. For intraparasitic PI3P localization, transgenic P. falciparum that expressed a PI3P-specific fluorescent probe was generated. Fluorescence was associated mainly with the membrane of the food vacuole and with the apicoplast, a four-membrane bounded plastid-like organelle derived from an ancestral secondary endosymbiosis event. Electron microscopy analysis confirmed these findings and revealed, in addition, the presence of PI3P-positive single-membrane vesicles. We hypothesize that these vesicles might be involved in transport processes, likely of proteins and lipids, toward the essential and peculiar parasite compartment, which is the apicoplast. The fact that PI3P metabolism and function in Plasmodium appear to be substantially different from those in its human host could offer new possibilities for antimalarial chemotherapy.
Phosphatidylinositol is a crucial phospholipid in eukaryotic cells. It is a structural membrane lipid, and phosphorylation of the hydroxyl groups of its inositol head group by specific lipid kinases leads to the production of seven different phosphoinositide species, which have been found to be enriched in distinct cellular compartments. They play key roles in a multitude of cellular processes, such as membrane traffic, cell motility, cytoskeletal reorganization, DNA synthesis, the cell cycle, adhesion, and signal transduction (9). Approximately 1% of total lipids in mammalian cells are phosphoinositides, mainly phosphatidylinositol 4-monophosphate (PI4P) and phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] (45). Derivatives phosphorylated at the 3 position are considerably less abundant in mammalian cells. Phosphatidylinositol 3-monophosphate (PI3P) is a ubiquitous lipid in eukaryotic cells and is present in small amounts in mammalian cells (classically <15% of PI4P), while PI3P is as abundant as PI4P in the yeast Saccharomyces cerevisiae (2). It has been suggested that one of the functions of these lipids is to establish membrane identity (46); PI4P predominates at the Golgi apparatus (33), PI(4,5)P2 at the plasma membrane (62), PI3P on early endosomes (20), and phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2] on late endocytic organelles (48). Certain phosphoinositides have been shown to play important roles in constitutive membrane traffic. PI3P is involved in endocytosis and vesicular trafficking toward the lysosome in yeast and mammalian cells (23, 39). PI3P has also been shown to be implicated in the processes of retrograde trafficking from the endosome to the Golgi apparatus and in autophagy (7, 42). PI(3,5)P2 is found in yeast, mammalian, and plant cells (11) and is essential for protein sorting in multivesicular bodies (38), which are an intermediate compartment for the degradation of cell surface receptors within lysosomes.
PI3P is the product of PI3-kinase class III (16), also termed Vps34 (vacuolar protein sorting), and PI(3,5)P2 is synthesized by the phosphorylation of PI3P by PIKfyve (phosphoinositide kinase, FYVE finger containing; Fab1 in yeast) (38). Both enzymes are conserved across eukaryotic evolution from yeast to mammals. Vps34-type enzymes are the only PI3-kinases in unicellular organisms. In contrast, metazoan cells possess three classes of PI3-kinases (I, II, and III), which differ from each other by their activation mode and their substrate specificity (56), leading to the synthesis of phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3], phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2], and PI3P, respectively. The synthesis of PI(3,4,5)P3 and PI(3,4)P2 is controlled by agonist stimulation of cells, resulting in strong fluctuations in their cellular levels. These phosphoinositides are generally not observed in unicellular organisms.
Plasmodium falciparum is the causal agent of the most severe form of human malaria. During the symptomatic phase of the disease, the parasite resides within a vacuole in mature erythrocytes, cells that are devoid of protein and lipid biosynthesis and intracellular compartments. In addition to the classically observed organelles of eukaryotic cells, Plasmodium contains certain particular compartments. These include the apicoplast, a four-membrane bounded plastid-like organelle derived from an ancestral secondary endosymbiosis event (61), and specialized secretory organelles, rhoptries and micronemes, located at the apical pole and involved in host cell invasion. Plasmodium blood stages internalize host cell hemoglobin that is degraded in a specialized compartment, the food vacuole (17). Some characteristics of the food vacuole, such as low pH and the presence of proteolytic enzymes, could qualify it as a lysosome-like organelle (21). However, this very peculiar compartment is present only during Plasmodium blood stages, is absent from mosquito and liver stages, and is not found in other apicomplexan parasites. Targeting proteins and lipids to these numerous cellular compartments requires sophisticated vesicular trafficking. Certain classical actors in vesicular trafficking in eukaryotic cells, such as Rab GTPases and SNARE-like proteins, are present in Plasmodium (3, 44).
Here we addressed the question of whether the second group of classical actors in vesicle transport, i.e., the phosphoinositides PI3P and PI(3,5)P2, are synthesized by the parasite. For this purpose, we established, for the first time, a detailed phosphoinositide profile of P. falciparum-infected red blood cells and detected important amounts of PI3P, which we found located at the food vacuole membrane and the apicoplast. In contrast, PI(3,5)P2 could not be detected, indicating possible differences in protein sorting between Plasmodium and other eukaryotes. PI3-kinase activity is essential for Plasmodium, suggesting that PI3P-dependent functions might be an interesting target for future drug development.
MATERIALS AND METHODS
Parasites.
P. falciparum strains 3D7 and 3D7attB (37) were maintained in continuous culture in human leukocyte-free erythrocytes (obtained from the local blood bank). Parasites were cultured at 5% hematocrit in RPMI 1640 medium containing 25 mM HEPES, pH 7.4 (Gibco), supplemented with 5 mg/ml AlbuMAX type I (Gibco), 88 μM hypoxanthine (Sigma), and 0.04 mg/ml gentamicin (Gibco). Cultures were incubated in modular chambers under 5% CO2, 5% O2, and 90% N2 at 37°C. Parasites were synchronized by 5% sorbitol treatment (30) and were regularly enriched for late stages using Plasmion (32). Growth inhibition assays were performed as described previously (10) with 48 h of drug contact followed by 18 h of [3H]hypoxanthine incorporation.
Plasmodium berghei strain ANKA (clone 2.34) was used and was maintained in female NMRI mice (Charles River) by manual passage. This research adhered to the Principles of Laboratory Animal Care. Animal studies were approved by the local animal use committee (comité régional d'éthique sur l'expérimentation animale).
Cloning of GFP2×FYVE expression constructs.
The green fluorescent protein (GFP)-2×FYVE and GFP-2×FYVEC215S coding sequences were amplified from the pEGFP2×FYVE and pGEX-2×FYVEC215S plasmids (kindly supplied by H. Stenmark) using primer pairs 244/245 and 272/273 (see Table S1 in the supplemental material). The amplicons were cloned as AvrII/AflII fragments into the pENR-GFP-attP vector (37), replacing the ENR-GFP gene. The resulting plasmids, pLN-GFP-2×FYVE and pLN-GFP-2×FYVEC215S, were verified by sequencing. DNA for transfection was prepared using the Qiagen Plasmid Maxi kit. For each transfection, 75 μg of DNA and 50 μg of pINT (each in 30 μl Tris-EDTA [TE] buffer) were resuspended in 240 μl Cytomix (120 mM KCl, 0.15 mM CaCl2, 2 mM EGTA, 5 mM MgCl2, 10 mM K2HPO4/KH2PO4, 25 mM HEPES [pH 7.6]).
P. falciparum transfection.
One hundred microliters of 3D7attB-infected red blood cells from a synchronized 5% parasitemia ring-stage culture was washed in Cytomix and added to 300 μl of a plasmid preparation in electroporation cuvettes (Bio-Rad). Parasites were transfected by electroporation (0.31 kV, 950 μF; Bio-Rad Gene Pulser II) and were immediately transferred to culture plates containing 5 ml complete medium and 120 μl fresh red blood cells. Five hours after transfection, selection was started using 2.5 nM WR99210 and 2.5 μg/ml blasticidin. The medium was changed daily for the first 6 days, and then every 2 days until ring-stage parasites were detected. Cultures were diluted weekly by the addition of 30% fresh red blood cells. Drug pressure was maintained until correct integration at the genomic attB locus was verified.
Fluorescence microscopy.
For GFP imaging, infected red blood cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) (EM Sciences) overnight at 4°C. After 2 washing steps with PBS, cells were allowed to settle on polylysine-coated 10-well slides (Fisher Scientific) for 1 h at room temperature. Cells were permeabilized with 0.1% Triton X-100 for 5 min. After three washes with PBS, cells were incubated with a primary antibody (rabbit anti-acyl carrier protein [anti-ACP; kindly provided by G. McFadden] or rabbit anti-chloroquine resistance transporter [anti-CRT; MR4 MRA-308], both at 1:600 in PBS) for 1 h, washed again three times with PBS, and incubated with a secondary antibody (anti-rabbit Alexa Fluor 488) for 30 min. Cells were washed 3 times with PBS and were then treated for 5 min with 2 nM Hoechst 33342 (Sigma) diluted with PBS for nuclear staining. Slides were mounted with Vectashield. Images were taken using a Zeiss inverted, Apotome-equipped Axiovert 200M microscope, with a 63× apochromat objective and differential interference contrast (DIC) and Axiovision software.
Immunoelectron microscopy.
Highly enriched, late-stage-infected cells were fixed with 4% paraformaldehyde in 0.2 M sodium phosphate buffer for 90 min and were then washed in PBS-10% fetal calf serum (FCS; Sigma), and infused in 2.3 M sucrose containing 10% polyvinylpyrrolidone before being frozen in liquid nitrogen. Sections were obtained on a Leica Ultracut instrument equipped with an FCS cryoattachment operating at −100°C. Sections were floated successively on PBS-FCS, rabbit anti-GFP antibodies (Abcam) diluted 1:200 in PBS-FCS, and 10 nm protein A-gold (Utrecht) diluted in PBS to an optical density at 525 nm (OD525) of 0.05, with five 3-min washes in PBS between each step. Sections were then embedded in methylcellulose (2%)-uranyl acetate (0.4%) and observed with a Zeiss EM10 electron microscope.
Labeling, identification, and quantification of phosphoinositides.
Late stages of infected red blood cells were enriched to 80 to 90% parasitemia using Plasmion (32). For this purpose, 1 volume of packed cells was mixed with 1.4 volumes of complete culture medium and 2.4 volumes of Plasmion and was then incubated for 30 min at 37°C, and the late-stage parasites were recovered from the supernatant by centrifugation. Packed cells (volume, 100 μl) were washed once with complete medium and twice with phosphate-free PL buffer (137 mM NaCl, 4 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM glucose, 30 mM HEPES [pH 7.4]). Cells were labeled with 500 μCi H332PO4 (MP Biomedicals) in 1 ml (final volume) in PL buffer for 1 h at 37°C. Excess radioactivity was removed by three rapid washes with 0.9% NaCl. The cells were resuspended in 500 μl of 0.9% NaCl and were lysed by stepwise addition and intensive vortexing of 2.4 ml methanol (4 volumes), 1.2 ml chloroform, 200 μl HCl (12 M), 1.2 ml chloroform, and 1.2 ml KCl (2 M). After centrifugation at 3,000 rpm for 5 min, the organic phase was collected and pooled with the wash of the aqueous phase with 4 ml chloroform. The extracts were dried under a nitrogen stream at room temperature. Dried lipids were resuspended in 40 μl of chloroform-methanol (2:1, vol/vol) and resolved by thin-layer chromatography (TLC) using chloroform-acetone-methanol-acetic acid-water (80:30:26:24:14, vol/vol) as a solvent. The TLC plate (silica gel 60 with a concentrating zone [Merck]) had been pretreated with 1% potassium oxalate in 2 mM EDTA-methanol (vol/vol) for 15 min and activated at 100°C for 1 h. Spots corresponding to PI monophosphates and PI bisphosphates were visualized with a PhosphorImager scanner and identified by comparison with the migration of authentic standards. The zones encompassing PI monophosphates and PI bisphosphates/PI trisphosphates were scraped off separately. The phosphoinositides were then deacylated by the addition of 1 ml of a methylamine reagent, composed of 40% methylamine-methanol-n-butanol-H2O (26.8:45.7:11.4:16, vol/vol), to the silica powder. After incubation at 53°C for 50 min, the methylamine reagent was completely evaporated under a nitrogen stream at 37°C. The samples were resuspended in 1.2 ml H2O and were separated by high-performance liquid chromatography (HPLC) using a Whatman Partisphere 5 SAX column (4.6 mm by 125 mm) with guard cartridge anion-exchanger units (reference no. 4641 0005; Whatman) and a gradient of 1 M (NH4)2HPO4 (pH 3.8) and double-distilled water as described previously (41).
P. berghei PI3-kinase knockout.
To knock out the P. berghei PI3-kinase (PbPI3-kinase) gene in P. berghei strain ANKA, the recently described PCR-based method was used (12), by which a gene deletion is achieved by transfection with linear DNA obtained by PCR amplification without prior cloning. In the transfection construct, the resistance gene is flanked by two short regions homologous to the target locus. Two sequential PCRs were performed. Two 500-bp PbPI3K homologous sequences were amplified with primers 161/162 and 168/164 (see Table S1 in the supplemental material). The amplicons obtained were then combined with the linearized pDEFhDHPEA vector (12), which supplied the human dihydrofolate reductase (hDHFR) resistance cassette, and were PCR amplified with primers 161 and 164 to yield a final construct of 3 kbp. All PCRs were carried out with the Advantage 2 polymerase mixture (BD Biosciences) and the following amplification program: 95°C for 2 min; 35 cycles of 30 s at 95°C, 45 s at 52°C, and 4 min at 62°C; and a final extension at 62°C for 10 min (51). Ten micrograms of the PCR product was digested with 10 U DpnI for 2 h at 37°C to eliminate any contaminating pDEFhDHPEA plasmid. The DNA was ethanol precipitated and resuspended for transfection at 2 μg/μl in water.
Plasmid construction for PbPI3-kinase tagging.
The transfection constructs for C-terminal tagging of the PbPI3-kinase were based on plasmid pDEFhDHPEA (pOB116; kindly provided by Oliver Billker). This plasmid carries the hDHFR gene, conferring resistance to pyrimethamine. For our purposes, the following elements were added: 1.5 kb of the 3′ end of the PbPI3K gene with or without the stop codon, either the GFP coding sequence or a triple hemagglutinin (HA) tag fused to the four-cysteine motif (3HA-Cys), and the 3′ untranslated region (3′ UTR) sequence of the PbDHFR gene. The PbPI3K gene was amplified from genomic DNA with primers 159/160 or 159/274, the 3′ UTR with primers 155/156 from plasmid pOB116, the GFP tag with primers 181/182 from plasmid pRON4-GFP (provided by Maryse Lebrun), and the 3HA-Cys tag (34) with primers 157/158 from a 2HA tag-containing plasmid (see Table S1 in the supplemental material). Both tags were designed to contain a GGPGG spacer separating the PbPI3K coding sequence from the tag. All reactions were done with Phusion polymerase (Finnzymes), and all amplicons were subcloned into pCR-blunt TOPO (Invitrogen). First the 3′ UTR was cloned as a ClaI/PstI fragment into pOB116. Then either the GFP sequence or the 3HA-Cys tag was added as a BglII/ClaI fragment. Finally, the PbPI3-kinase sequence was cloned as a SacII/BglII fragment. All constructs were verified by sequencing.
For transfection, plasmids were prepared using the Promega Mini-wizard plus kit and were linearized in the center of the PbPI3-kinase sequence by digestion with BclI for 2 h at 50°C. Following ethanol precipitation, the DNA was resuspended in water at 2 μg/μl.
Transfection of P. berghei.
Transfection was done by following the Amaxa transfection protocol (12). Mature P. berghei schizonts were obtained by culturing blood from an infected mouse overnight in complete medium as described previously (55). Schizonts were collected by centrifugation on a Nycodenz density gradient as described previously (55), washed, and used immediately for transfection. Ten micrograms (5 μl) of the PCR amplicon or digested plasmid DNA was resuspended in 100 μl of Amaxa buffer, mixed with 1 × 107 to 3 × 107 schizonts per transfection, and electroporated using the high-efficiency Nucleofactor transfection device along with the Human T-cell Nucleofactor kit (Lonza, Germany). Immediately after electroporation, 200 μl of fresh mouse blood was added and incubated 15 min at 37°C with gentle agitation to allow cell invasion by the merozoites. The suspension was injected intraperitoneally into female NMRI mice that had been treated with 100 μl of phenylhydrazine (25 mg/ml) 3 days before infection (5). Pyrimethamine selection in drinking water (70 μg/ml) was started 24 h postinfection and was maintained for 4 days. Parasites were harvested 6 days later and were mechanically passaged to two mice that were maintained under drug pressure until final bleeding for further analysis.
RESULTS
Phosphoinositide profile of P. falciparum-infected red blood cells.
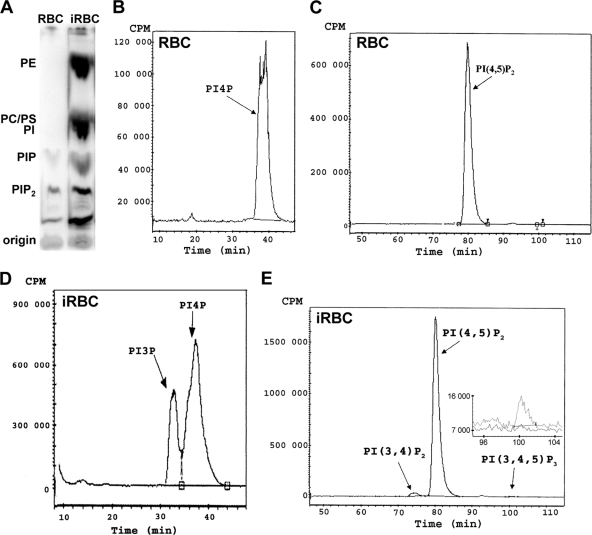
It has been shown previously that Plasmodium-infected erythrocytes produce important amounts of phosphatidylinositol mono- and bisphosphates (13); however, their molecular identity had not been determined. Here we analyzed the detailed phosphoinositide profile of P. falciparum-infected red blood cells. For this purpose, highly enriched late-blood-stage parasites were metabolically labeled with [32P]phosphate, and the lipids were then extracted and separated by thin-layer chromatography (TLC). Incorporation of [32P]phosphate into the major structural phospholipids phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol was lacking in uninfected erythrocytes but was substantial after Plasmodium infection (Fig. 1A). We found that infected erythrocytes (iRBC) produced about 13 times more total phosphoinositides than equal numbers of uninfected erythrocytes (data not shown).
Fig. 1.
Analysis of phospholipids by thin-layer chromatography (TLC) and HPLC. Equal numbers of highly enriched P. falciparum-infected red blood cells (iRBC) and uninfected red blood cells (RBC) were labeled with H332PO4 for 1 h at 37°C, and phospholipids were extracted. (A) Separation by TLC. PhosphorImager acquisition data are shown. The positions of the major radiolabeled phospholipids phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylinositol (PI), PI-monophosphates (PIP), and PI-bisphosphates (PIP2), identified with respect to the retention factor value of unlabeled authentic standards, are indicated. (B to E) The zones corresponding to the PI-monophosphates and to the PI-bis/trisphosphates were recovered from the silica plate, and the lipids were deacylated and analyzed by HPLC. Shown are the radioactivity profiles of phosphatidylinositol monophosphates (B and D) and phosphatidylinositol bis- and trisphosphates (C and E) in uninfected red blood cells (B and C) and infected red blood cells (D and E) over time. (E) (Inset) Enlargement and overlay of the zone corresponding to PI(3,4,5)P3 with the profile of infected RBC in gray and that of uninfected RBC in black.
The areas corresponding to phosphatidylinositol mono- and polyphosphates were recovered separately and analyzed by high-performance liquid chromatography (HPLC), allowing identification and quantification of phosphoinositide isomers (41). Uninfected red blood cells produced only PI4P and PI(4,5)P2. In contrast, infected red blood cells produced a complex phosphatidylinositide profile; in addition to PI4P and PI(4,5)P2, they synthesized large amounts of PI3P and small amounts of PI(3,4)P2 and PI(3,4,5)P3 (Fig. 1B to E). The quantity of PI3P produced was important, representing 29.7% ± 12.9% (mean ± standard error of the mean [SEM]) of total phosphatidylinositol monophosphates (n = 8). In contrast, in uninfected red blood cells, PI3P was undetectable even after prolonged labeling (up to 18 h), indicating that its production in infected red blood cells was indeed induced by the parasite. Concerning polyphosphoinositides in infected erythrocytes, PI(4,5)P2 was by far the most abundant, corresponding to 70% ± 11.4% (mean ± SEM) of total phosphoinositides, while PI(3,4)P2, and PI(3,4,5)P3 were present at 0.6% ± 0.3% and 0.4% ± 0.3%, respectively (n = 8). Surprisingly, we could not detect PI(3,5)P2, a lipid involved in vesicle transport toward the vacuole/lysosome and present in yeast, plant, and mammalian cells (11).
When phosphoinositides were studied during synchronized parasite maturation of a culture at 10% parasitemia, total phosphoinositide labeling increased about 3-fold between the ring and late stages (Table 1). PI3P levels were relatively low with respect to the other phosphoinositides in ring stages (about 1% of total phosphoinositides) but became more predominant in late stages (about 4%), which may indicate that the function linked to the presence of this lipid is more developed during late parasite stages. The relative abundance of the predominant phosphoinositides PI4P and PI(4,5)P2 also shifted from 19% PI4P and 80% PI(4,5)P2 in rings to 32% PI4P and 64% PI(4,5)P2 in late stages.
Table 1.
Phosphoinositide synthesis during blood-stage development of P. falciparuma
| Phosphoinositide | Radioactivity (cpm)b at the following stage: |
||
|---|---|---|---|
| Ring | Trophozoite | Schizont | |
| PI3P | 11,649 | 146,391 | 184,723 |
| PI4P | 286,958 | 827,290 | 1,336,574 |
| PI(3,4)P2 | ND | 12,823 | 16,750 |
| PI(4,5)P2 | 1,076,872 | 2,523,291 | 2,646,058 |
Highly synchronized cultures at 10% parasitemia were used.
Values correspond to counts per minute detected after HPLC analysis in one experiment of two. ND, not detected.
Given the surprisingly high levels of PI3P in P. falciparum-infected erythrocytes, we were interested in establishing a link between the phosphoinositide profile and PI3-kinase activity, and we analyzed the effect of wortmannin, a well-established inhibitor of PI3-kinases. Highly enriched, late-stage parasite cultures were pretreated with 100 nM wortmannin for 30 min, followed by metabolic labeling in the presence of wortmannin for 1 h. This treatment did not alter the synthesis of labeled structural phospholipids in comparison to mock-treated samples as analyzed by TLC (data not shown). Concerning phosphoinositides, the only significant difference observed was a 58% decrease in PI3P levels, while the levels of the major phosphoinositides, PI4P and PI(4,5)P2, remained unchanged (Fig. 2). This indicated that PI3P production in P. falciparum-infected erythrocytes was dependent on a wortmannin-sensitive PI3-kinase and that at the above indicated concentration, wortmannin inhibited PI3-kinase activity specifically, i.e., without affecting other PI-kinases.
Fig. 2.
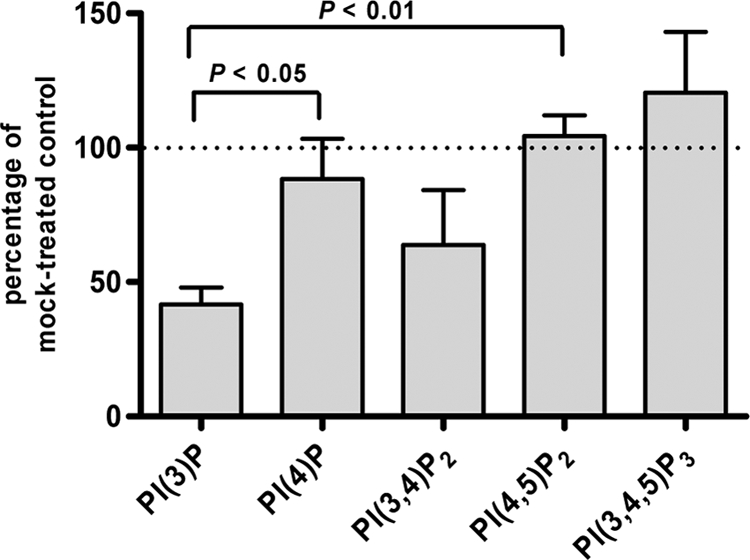
Effect of wortmannin treatment on phosphoinositide synthesis in infected erythrocytes. Highly enriched infected red blood cell cultures were pretreated for 30 min with 100 nM wortmannin and were subsequently labeled for 1 h with H332PO4 in the presence of 100 nM wortmannin. The incorporation of radioactivity into individual phosphoinositides is expressed as a percentage of that for control cells incubated without wortmannin. Error bars correspond to SEMs (n = 5). P values were determined by unpaired two-tailed t tests and are indicated. The variations in the PI(3,4)P2 and PI(3,4,5)P3 levels were not statistically significant with respect to the values for PI4P and PI(4,5)P2.
The P. falciparum genome encodes a PI3-kinase.
The P. falciparum genome contains one predicted PI3-kinase gene, PFE0765w, encoding a predicted protein of 2,133 amino acids (aa) containing the characteristic domains of class III PI3-kinases: a calcium/lipid-binding C2 domain (aa 297 to 371), a PI3-kinase family accessory domain (aa 1097 to 1280), and a C-terminal PI3- and PI4-kinase catalytic domain (aa 1867 to 2130) (54) (see Fig. S1 in the supplemental material). However, the predicted P. falciparum protein is substantially longer than any PI3-kinases described so far, in part due to the presence of five different repetitive peptide motifs of six to eight amino acids in the P. falciparum sequence, repeated 5 to 12 times and accounting for about 15% of the total amino acid sequence (see Fig. S1). PI3-kinase orthologues are annotated for other Plasmodium species (PlasmoDB 6.3); however, only the sequences of the Plasmodium vivax (PvPI3K; 1,779 aa) and Plasmodium chabaudi (PcPI3K; 1,796 aa) PI3-kinases are complete. A multiple sequence alignment of the three Plasmodium PI3-kinases (see Fig. S1) highlighted the fact that sequence conservation was restricted mainly to the three predicted functional domains: amino acid identity was around 86% for the catalytic domain, 64% for the accessory domain, and 34% for the C2 domain (P. falciparum versus P. vivax/P. chabaudi). Repetitive peptide motifs were not detected in PvPI3K and PcPI3K. The catalytic domain of PfPI3-kinase contains the highly conserved motifs that participate in ATP binding and phosphate transfer, in particular the catalytic loop sequence 1987GIGDRHLDN; 1881K, which interacts with the α-phosphate of ATP; and the metal ion binding sites 1995N, 1884D, and 2008D (60). The sequence following 2008D has been shown to confer substrate specificity (43) and supports the prediction that the Plasmodium protein is a class III enzyme; these enzymes are described as having phosphatidylinositol as sole substrate producing PI3P. Furthermore, 1881K, which is essential for kinase activity and is covalently modified in mammalian PI3-kinases by the PI3-kinase inhibitor wortmannin (64), is also present in PfPI3-kinase.
The PI3-kinase gene is essential in P. berghei.
To investigate whether PI3-kinase is essential in Plasmodium species, we chose to disrupt the PI3K gene in P. berghei. Since no sequence information for the 5′ end of the PbPI3K gene was available (corresponding to the first 387 aa of the P. chabaudi sequence), we generated a PbPI3K knockout construct that, upon double homologous recombination, would lead to deletion of the accessory and catalytic domains, generating a truncated protein without a functional kinase domain (see Fig. S2A in the supplemental material). The transfection construct was generated using the recently described PCR-based method (12). In four independent transfections, pyrimethamine-resistant parasites were obtained, suggesting successful transfection. However, none of the populations showed integration of the recombinant DNA into the PbPI3K locus, as analyzed by PCR (data not shown).
In order to verify that targeting of the PbPI3K locus was possible, we generated three constructs for single homologous recombination (see Fig. S2B in the supplemental material). Two constructs aimed at tagging PbPI3-kinase at the C-terminal end with either green fluorescent protein (GFP) (plasmid pOB-PbPI3K-GFP) or a triple hemagglutinin (3HA) tag (plasmid pOB-PbPI3K-3HA). A third construct was identical to the 3HA tag plasmid except that the stop codon of the PbPI3K open reading frame was maintained (plasmid pOB-PbPI3K-stop-3HA), so that the coding sequence was not changed upon integration, while the same genetic control elements were introduced as with the authentic tagging constructs. Correct genome integration was achieved only with the control construct that maintained the stop codon (see Fig. S2C), while modification of the PI3-kinase sequence by adding a GFP or 3HA tag was unsuccessful in four independent transfections (data not shown). Taken together, these results demonstrated that the PbPI3K locus was targetable. However, gene disruption was lethal to blood-stage parasites, as was epitope tagging at the extreme C-terminal end, which likely altered PbPI3-kinase function or stability. These results strongly suggested that PI3-kinase activity is essential in P. berghei blood-stage parasites.
To analyze whether PI3-kinase activity is also essential for P. falciparum blood-stage development, we studied the effect of PI3-kinase inhibitors on P. falciparum growth in vitro. Wortmannin is known to be unstable in complex media and at a neutral pH (66), and accordingly, we could not detect any effect of a single dose as high as 25 μM wortmannin in a standard 2-day P. falciparum growth assay (10). However, the alternative PI3-kinase inhibitor LY294002 (59), which is stable under cell culture conditions, interfered with P. falciparum growth, with a 50% inhibitory concentration (IC50) of 26 ± 6 μM (mean ± SEM; n = 4), indicating that PI3-kinase might also be essential for P. falciparum development.
Generation of transgenic P. falciparum expressing a PI3P-specific fluorescent probe.
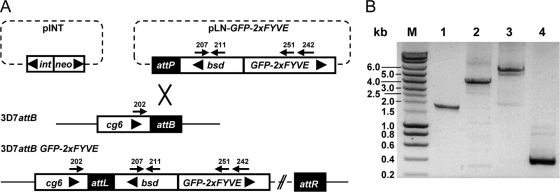
We next aimed at monitoring the intraparasitic localization of PI3P in infected red blood cells. For this purpose, we used a fusion of GFP to a tandem repeat of the FYVE (Fab1, YOTB, Vac1, EEA1) domain of the mammalian Hrs protein as a PI3P-specific probe. This construct has been reported to specifically bind PI3P (20) and has been used in various organisms to monitor the intracellular localization of PI3P (22, 58). In order to generate transgenic P. falciparum parasites expressing GFP-2×FYVE, a construct was designed in which GFP-2×FYVE was cloned under the control of the moderately strong calmodulin promoter (plasmid pLN-GFP-2×FYVE). The construct was integrated into the genome as a single-copy gene by using the recently described attB/attP site-specific recombination system (37) and strain 3D7attB (Fig. 3A). The same approach was used to generate transgenic parasites that express GFP fused to a mutated version of the FYVE domain (construct GFP-2×FYVEC215S). The cysteine-to-serine mutation at position 215 in both FYVE domains has previously been shown to completely abolish PI3P binding (20). This construct was designed as a control in order to identify and characterize the effect due to specific PI3P binding of GFP-2×FYVE. Stably transfected parasites were obtained less than 3 weeks after transfection, indicating that GFP-2×FYVE expression (wild type or mutated) did not interfere with parasite growth. Correct integration at the attB locus and the presence of GFP-2×FYVE were confirmed by PCR analysis (Fig. 3B).
Fig. 3.
Generation of GFP-2×FYVE expressing P. falciparum parasites. (A) Schematic representation of the recombination event leading to the integration of GFP-2×FYVE into P. falciparum strain 3D7attB. The two plasmids used for cotransfection are represented at the top: plasmid pINT, carrying the integrase expression unit (int) and a neomycin resistance cassette (neo), and plasmid pLN-GFP-2×FYVE, carrying the GFP-2×FYVE fusion gene under the control of the calmodulin promoter and a blasticidin resistance cassette (bsd) flanking the attP site. The attB site is present in the genomic cg6 locus in strain 3D7attB (37). Recombination mediated by the integrase between the attP and attB sites yielded strain 3D7attB-GFP-2×FYVE. The primers used in PCR analysis of transgenic parasites are indicated by arrows. Large arrowheads indicate the transcriptional directions of genes and expression cassettes. The diagram is not drawn to scale. (B) PCR analysis of genomic DNA of the transgenic parasite population was performed to confirm the correct integration of GFP-2×FYVE into the P. falciparum genome. The relative positions of the primers used are indicated in panel A. Lanes 1 to 3 demonstrate the integration of pLN-GFP-2×FYVE at the attB locus (lane 1, primers 202/211; lane 2, primers 202/251; lane 3, primers 202/242. Lane 4 shows the presence of the blasticidin resistance gene (primers 207/211). Expected fragment sizes were 1,800 bp, 4,260 bp, 4,880 bp and 346 bp for lanes 1 to 4, respectively. M, molecular weight marker, with fragment sizes indicated on the left.
Subcellular localization of PI3P.
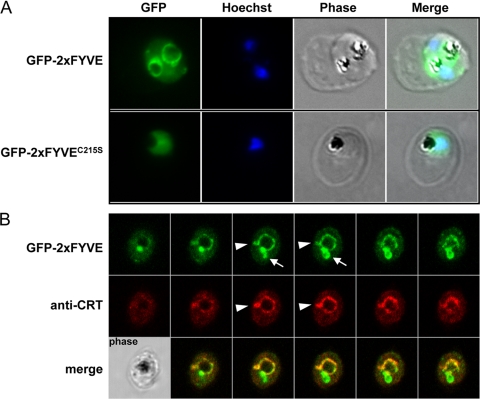
Transgenic parasites expressing either GFP-2×FYVE or its mutated version GFP-2×FYVEC215S were analyzed by fluorescence microscopy. For GFP2×FYVE the fluorescent signal was detected as a circular structure surrounding the food vacuole, which was identified by the presence of hemozoin crystals in the corresponding phase images (Fig. 4). Additionally, we observed one or a few dots of bright fluorescence, generally located on the circular structure, and some diffuse cytoplasmic staining. In contrast, fluorescence was diffuse throughout the parasite cytoplasm for GFP-2×FYVEC215S, as would be expected for a cytosolic GFP construct. These results demonstrated that the pattern observed with the nonmutated construct was due to functional FYVE domains.
Fig. 4.
GFP fluorescence in parasites expressing GFP-2×FYVE or GFP-2×FYVEC215S. (A) Cells were fixed and analyzed for GFP fluorescence. Nuclei were stained with Hoechst stain (blue stain in the merged images). (B) Colocalization of GFP-2×FYVE and the food vacuole membrane marker CRT using rabbit anti-CRT. Serial images of a Z-stack acquisition (0.38 μm step) are displayed. Arrowheads indicate protrusions from the food vacuole membrane that are colabeled with CRT and GFP-2×FYVE, while arrows indicate zones of intense GFP-2×FYVE staining in the absence of the food vacuole marker. A corresponding phase-contrast image is shown in the lower left corner.
The P. falciparum chloroquine resistance transporter (PfCRT) is widely used as a marker for the food vacuole membrane (15). In colocalization studies of GFP-2×FYVE and CRT in immunofluorescence microscopy, we found a partial match of the two markers (Fig. 4), confirming the presence of PI3P on the food vacuole membrane in P. falciparum blood-stage parasites. However, the bright dot of GFP fluorescence that was present in most parasites generally did not colocalize with CRT. To gain insight into the 3-dimensional structure of the food vacuole in relation to the compartment labeled by GFP-2×FYVE expression, we acquired serial stack images that were used to reconstitute 3-dimensional profiles (Fig. 4B). The sphere-like food vacuole membrane appeared to be labeled homogenously with GFP-2×FYVE, together with zones of more intense GFP label protruding into the cytoplasm, some of which were also labeled by anti-CRT, while others were not (Fig. 4B).
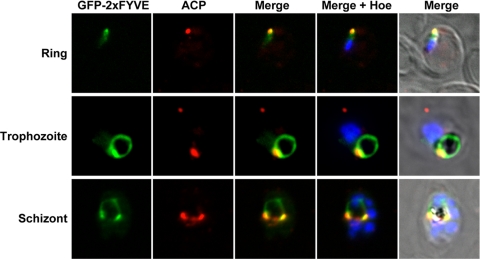
In order to determine the subcellular compartment corresponding to these bright dots, we performed colocalization studies. Using anti-ACP antibodies as a marker of the apicoplast, we found that the bright GFP signal often localized close to, or colocalized with, the apicoplast (Fig. 5). This was the case throughout the erythrocytic development of the parasite. In early-stage parasites without visible hemozoin crystal, the GFP signal was generally faint and punctate, corroborating the low PI3P levels detected in ring-stage parasites (Table 1). At the same time, GFP-2×FYVE did not colocalize with the mitochondrion or with lipid bodies (see Fig. S3 in the supplemental material); the latter have been reported to be associated with the food vacuole (25).
Fig. 5.
Colocalization of GFP-2×FYVE with the apicoplast. GFP-2×FYVE expressing parasites at the indicated developmental stages were incubated with rabbit anti-ACP as an apicoplast marker. Nuclei were stained with Hoechst stain (Hoe).
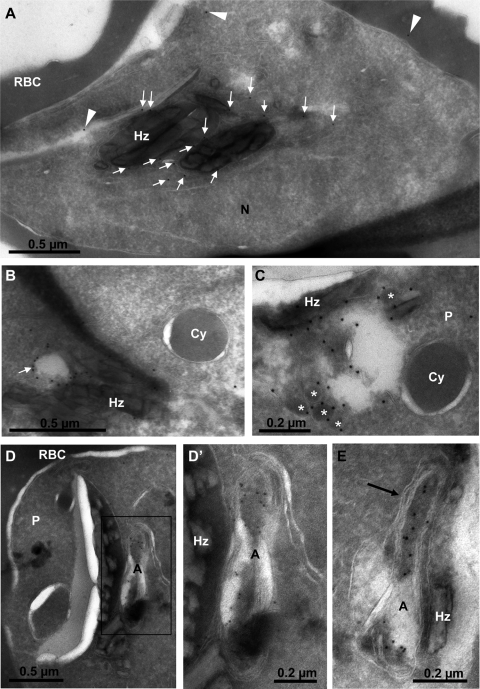
We finally analyzed the localization of GFP-2×FYVE in late-stage parasites by cryoimmunoelectron microscopy using rabbit anti-GFP antibodies. Most of the protein A-gold particles were found on the apicoplast and on clusters of vesicular structures, all of which were located in the vicinity of the food vacuole (Fig. 6). Most of the apicoplast label appeared to be in the lumen (Fig. 6D and E). GFP-2×FYVE-positive vesicles were bounded by a single membrane and were either electron dense (Fig. 6C) or electron lucent (Fig. 6B), with a diameter of about 0.1 μm or 0.2 μm, respectively. Cytostomal vesicles, identified by their double membrane and their hemoglobin content, were not labeled (Fig. 6B and C). A faint but specific label was also found on or near the food vacuole membrane (Fig. 6A). These observations confirmed that PI3P was present on the food vacuole membrane and the apicoplast, and they indicated the presence of PI3P in the membranes of small vesicles clustered near the food vacuole and/or the apicoplast.
Fig. 6.
Localization of GFP-2×FYVE by cryoimmunoelectron microscopy. (A) Section through a young schizont showing most of the gold label on or near the food vacuole membrane (arrows). Arrowheads point to what should be considered the background. Hz, hemozoin crystals; N, parasite nucleus; RBC, erythrocyte cytosol. (B and C) The membranes of electron-lucent (B) (arrow) and electron-dense (C) (asterisks) vesicles neighboring the food vacuole (Hz) are strongly labeled. Cytostomal vesicles (Cy) are not labeled. (D to E) General (D) and close-up (D′ and E) views of young schizonts showing the strong labeling of the apicoplast, mostly within the lumen. The four membranes of the apicoplast are clearly identified (E, arrow). A, apicoplast; P, parasite.
DISCUSSION
The results presented here show that infection of red blood cells by P. falciparum leads to profound changes in the phosphoinositide profile. It has long been known that in normal human erythrocytes, the only phospholipids to be labeled upon incubation with radioactive phosphate are phosphoinositides and phosphatidic acid (1). Here we show that concerning phosphoinositides, mature erythrocytes synthesize only PI(4,5)P2 and its precursor PI4P. Accordingly, the published human erythrocyte proteome revealed the presence of PI4-phosphate-5-kinase type III and diacylglycerol kinase γ, though with very low sequence coverage (26).
Upon infection with P. falciparum, the phosphoinositide profile becomes complex, and we could show that Plasmodium-infected red blood cells contain PI3P. The amounts detected in late-stage parasites were important, representing about one-third of total PI monophosphates. During the intraerythrocytic development from rings to schizonts, the proportion of PI3P with respect to total phosphoinositides increased around 4-fold, which may indicate that PI3P function is more developed in late-stage parasites. Synthesis of PI3P was sensitive to wortmannin, a well-known inhibitor of PI3-kinase, thereby establishing a link between PI3P production and PI3-kinase in infected red blood cells. We observed somewhat more than 50% inhibition of PI3P levels in the presence of 100 nM wortmannin. Inhibition at this concentration was considered specific, since production of the two main phosphoinositides, PI4P and PI(4,5)P2, was not influenced.
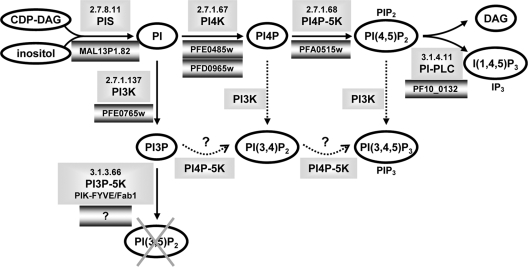
Our analysis also revealed that the parasite did not produce detectable levels of PI(3,5)P2. This lipid is readily detected in yeast and in mammalian cells, where its abundance has been shown to vary under stress conditions (11). PI(3,5)P2 is synthesized through phosphorylation of PI3P by PIKfyve/Fab1 (18). We analyzed the Plasmodium genome database PlasmoDB 6.3 for genes coding for the enzymes involved in phosphoinositide metabolism (Fig. 7). PI synthase (63) and PI4P-5-kinase (31) have been biochemically validated. We found candidates for the other enzymes except for PIKfyve/Fab1 PI3-phosphate-5-kinase. Given the lack of a Fab1 orthologue and the observed absence of PI(3,5)P2 synthesis, we conclude that this enzyme activity is indeed missing in Plasmodium. In other organisms, PI(3,5)P2 is localized mainly on multivesicular bodies (MVBs) and is implicated in the maintenance of endosome and lysosome/vacuole morphology and in endosome-to-Golgi complex and MVB-to-vacuole trafficking (18, 47). In apicomplexan parasites, MVB-like structures have so far been observed only upon disturbance of endocytic pathways by chemical or mutational manipulations (65), suggesting that endosomal trafficking in Plasmodium might be different from the classical scheme described for yeast and mammals.
Fig. 7.
Schematic representation of the phosphoinositide metabolism in Plasmodium. Phosphoinositides and metabolites are represented by ovals. The corresponding enzymes together with their EC nomenclature are shown in unbordered shaded boxes, and the predicted or confirmed P. falciparum genes are shown in shaded boxes with dark borders. The putative synthesis of PI(3,4)P2 and PI(3,4,5)P3 through PI3-kinase or PI4P-5-kinase is indicated by dotted arrows. DAG, diacylglycerol; I(1,4,5)P3 (IP3), inositol trisphosphate; PIS, PI-synthase; PI-PLC, PI-specific phospholipase C.
Surprisingly, we also detected very low quantities of PI(3,4)P2 and PI(3,4,5)P3 in infected erythrocytes. These lipids are generally not found in unicellular organisms, where only class III PI3-kinases are present. A recent publication reports that P. falciparum PfPI3-kinase immunoprecipitated from infected red blood cell extracts was capable of phosphorylating PI, PI4P, and PI(4,5)P2 in vitro (54) and could explain the presence of PI(3,4)P2 and PI(3,4,5)P3 that we observed in P. falciparum-infected erythrocytes. Our results clearly showed that in intact P. falciparum-infected erythrocytes, PI3P is by far the most abundant phosphoinositide phosphorylated at position 3 (87.5% ± 6.1% [mean ± SEM; n = 8]). Interestingly, in our experiments, the synthesis of PI(3,4,5)P3 (and of PI(3,4)P2) was not significantly reduced by wortmannin treatment, indicating that PIP3 synthesis might be independent of PI3-kinase activity. This excludes the possibility that PIP3 synthesis might be due to a residual silent activity of the human enzyme in red blood cells that becomes activated upon parasite infection, since in this case its synthesis should be sensitive to wortmannin, a potent inhibitor of human class I PI3-kinases. To our knowledge, the only other unicellular organism for which PI(3,4,5)P3 production has been described so far is Schizosaccharomyces pombe (36). It has been shown that PI(3,4,5)P3 production in S. pombe is mediated not by the sole type III PI3-kinase (52), but by a PI4P-5-kinase, which in a sequential reaction phosphorylates PI3P first to PI(3,4)P2 and then to PI(3,4,5)P3 (36). The situation might be similar in P. falciparum (Fig. 7), a possibility additionally supported by the fact that the unique parasite PI4P-5-kinase has been shown to phosphorylate PI3P in vitro (31). It remains to be shown whether PI(3,4,5)P3 plays a physiological role in Plasmodium and what this function might be.
The design of fluorescent molecular probes that bind phosphoinositides with high specificity has allowed the imaging of phosphoinositide dynamics in living cells (reviewed in reference 57). A tandem repeat of the FYVE domain of the mammalian Hrs protein fused to GFP (20) has been successfully used in various organisms to visualize PI3P (22, 58). Expressing the same construct in P. falciparum allowed us to identify three compartments that are enriched in PI3P: the food vacuole membrane, the apicoplast, and single membrane vesicles. The food vacuole is a compartment with “lysosome-like” functions in Plasmodium, and association of PI3P with its membrane is in accordance with the PI3P-mediated function in transport toward the lysosome in other organisms. Indeed, it has very recently been shown that treatment of parasite cultures with the PI3-kinase inhibitors wortmannin and LY294002 inhibited endocytosis within the parasite, resulted in entrapment of hemoglobin vesicles in the parasite cytoplasm, and prevented their fusion with the food vacuole, suggesting that PI3-kinase is involved in endocytosis from the host and the trafficking of hemoglobin in the parasite (54). The authors also reported that PfPI3-kinase was in part exported to the red blood cell cytosol. This was demonstrated by using antibodies raised against a recombinant His-tagged PfPI3-kinase fragment showing perfect colocalization with the conserved C-terminal end of var gene products in immunofluorescence analysis. Additionally, PfPI3-kinase activity could be immunoprecipitated from the host cell fraction upon selective membrane permeabilization (54). Our construct was not aimed at exporting GFP-2×FYVE from the parasite cytosol. We therefore could not test this unexpected observation, and we did not attempt to separate parasite membranes from host cell membranes after metabolic labeling of phosphoinositides because of the prohibiting levels of radioactivity necessary for their identification. Finally, PfPI3-kinase has also been immunolocalized within the food vacuole (54). In addition, the only FYVE domain-containing protein of P. falciparum, which has been shown to bind PI3P in vitro, has also been localized to the lumen of the food vacuole; however, this localization was independent of the FYVE domain, and thus independent of PI3P-binding (35). We did not observe GFP-2×FYVE fluorescence within the food vacuole, but again, our probe was designed to be cytosolic, since this is the usual location of PI3-kinase. So far there has been no proposition as to how PfPI3-kinase exported outside the parasite or present within the food vacuole could mechanistically influence hemoglobin uptake and digestion in the parasite. In mammalian cells and other organisms, the small GTPase Rab5 is a marker for early endosomes and colocalizes with PI3P-containing membranes (8). In transgenic P. falciparum parasites that express a constitutively active mutant of Rab5a, the protein localized to small double-membrane hemoglobin-containing vesicles (14). In our analysis we did not observe any GFP-2×FYVE label on double-membrane cytostomal vesicles (Fig. 6), which have been reported to be the most commonly occurring hemoglobin-containing vacuoles in late-stage P. falciparum parasites (14). The fact that we observed only immunogold-decorated vesicles that had a single membrane indicates that these do not correspond to the described hemoglobin transport vacuoles but may constitute a distinct cellular structure.
The food vacuole is a very peculiar compartment that is present only during Plasmodium blood stages and is not present in mosquito and liver stages or in other apicomplexan parasites. Our localization studies with GFP-2×FYVE revealed that PfPI3-kinase is active in the cytosol of the parasite and that, in addition to the food vacuole membrane, PI3P was enriched at the apicoplast and in the membranes of small single-membrane vesicles. We propose that these groups of vesicles identified by electron microscopy and the apicoplast correspond to the intense dot(s) observed in late stages in immunofluorescence analysis. Late-stage Plasmodium parasites contain numerous vesicular structures, but we cannot exclude the possibility that expression of GFP-2×FYVE as a PI3P-specific probe induced (without interfering with normal parasite growth) the formation and/or accumulation of vesicles, as has been observed in other cellular systems (20). The apicoplast is a peculiar, four-membrane bounded organelle that originated from endocytosis of an alga by the ancestor of the apicomplexans (secondary endosymbiosis) (29). We could recently demonstrate an essential role for PI3P in vesicle traffic to the apicoplast in Toxoplasma gondii, a related apicomplexan parasite (L. Tawk, J. F. Dubremetz, P. Montcourrier, G. Chicanne, F. Merezegue, V. Richard, B. Payrastre, M. Meissner, H. J. Vial, C. Roy, K. Wengelnik, and M. Lebrun, submitted for publication). Strong expression of GFP-2×FYVE in Toxoplasma (and thus massive sequestration of PI3P) was not tolerated by the parasite, an effect that was found to be linked to a default in apicoplast biogenesis, leading to loss of this essential organelle, a phenotype termed “delayed death.” At low expression levels, parasites appeared undisturbed, and GFP-2×FYVE was found at the apicoplast membranes and on electron-dense and electron-lucent vesicles in the vicinity of the apicoplast. These vesicles were shown to carry PI3P and proteins destined for the outermost apicoplast membranes, such as FtsH1 and APT1 (27, 28; Tawk et al., submitted).
In Plasmodium we localized PI3P to the apicoplast; however, we did not observe a delayed death phenotype, which may be due to differences in the expression levels of GFP-2×FYVE and/or the abundance of PI3P in these two apicomplexan parasites. Immunogold particles revealing GFP-2×FYVE were detected mainly in the apicoplast lumen. This finding is intriguing, since the GFP-2×FYVE construct does not contain a signal sequence or apicoplast-targeting sequence, generally required for protein import. Its location in the lumen indicates that the protein is not in a complex with the lipid PI3P anymore. While certain aspects of protein import into the apicoplast are well understood (40, 53), little is known about lipid transport toward the organelle and lipid distribution among the four membranes. One hypothesis might be that GFP-2×FYVE reaches the apicoplast intermembrane space as a consequence of its PI3P-binding capacity and might then be trafficked further independently of lipid interaction. The GFP-2×FYVE-positive vesicles in P. falciparum are similar in size and morphology to the Toxoplasma vesicles shown to carry PI3P and apicoplast membrane proteins, and it is tempting to speculate that PI3P-containing vesicles in Plasmodium might be involved in transport processes, likely of proteins and lipids, toward the essential and peculiar parasite compartment, which is the apicoplast.
PI3-kinase appeared to be essential in Plasmodium. P. falciparum in vitro growth was sensitive to the PI3-kinase inhibitor LY294002. In P. berghei the PbPI3K locus could be targeted successfully but could neither be tagged nor deleted, indicating that PbPI3-kinase was essential at the blood stage. Vps34 orthologues are highly conserved at their C-terminal end, containing only very few amino acids after the catalytic domain (see Fig. S1 in the supplemental material). Mutational analysis in S. cerevisiae revealed that the last amino acids of PI3-kinase are essential for kinase activity (6). On the other hand, C-terminal fusion proteins with GFP or the TAP tag have been described in the literature (24). In general, Vps34 membrane association and activation are mediated by Vps15, a serine/threonine kinase containing HEAT and WD40 repeats and acting as a regulatory subunit (4, 50). We could not identify a Vps15 orthologue in Plasmodium, which might indicate that regulation of PI3-kinase in the parasite differs from its human host. Finally, we found that very few proteins with PI3P-binding domains are actually predicted in the P. falciparum proteome. Effector proteins bind PI3P via FYVE or PX domains (19, 49). The interaction of these effector proteins with additional proteins like Rab5 finally lead to the formation of protein complexes that drive membrane fusion between the donor and corresponding acceptor membranes (4). In Plasmodium, two proteins with PX-domains (PF07_0017 and MAL7P1.108) and only one with a FYVE-domain (PF14_0574) were identified. Strikingly, this is a low number compared to the 15 PX and 5 FYVE domain proteins in S. cerevisiae, for example.
In summary, (i) the absence of a Fab1 orthologue and of PI(3,5)P2, (ii) the low number of PI3P-interacting proteins, including the absence of the important endosome marker protein EEA1 (early endosomal antigen 1), (iii) the apparent absence of a Vps15 orthologue, (iv) the structural particularities of the Plasmodium PI3-kinase itself, and (v) a possible unusual role in trafficking to the apicoplast, an essential organelle for the survival of the parasite, strongly indicate that PI3P-dependent functions might be substantially different from the well-studied systems in yeast and, most importantly, in mammalian cells. Interference with Plasmodium PI3P metabolism could thus prove to be an interesting possibility for new antimalarial chemotherapy.
Supplementary Material
ACKNOWLEDGMENTS
This work is part of the activities of the BioMalPar European Network of Excellence (LSHP-CT-2004-503578).
We thank H. Stenmark, O. Billker, and M. Lebrun for supplying plasmids, G. McFadden for providing antibodies, L. Berry-Sterkers for assistance with microscopy, and S. Besteiro and M. Lebrun for critical reading of the manuscript. We thank MR4 for providing us with antibodies contributed by R. Cooper/T. Wellems (anti-PfCRT/K, MRA-308).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 13 August 2010.
REFERENCES
- 1.Allan D. 1982. Inositol lipids and membrane function in erythrocytes. Cell Calcium 3:451–465 [DOI] [PubMed] [Google Scholar]
- 2.Auger K. R., Carpenter C. L., Cantley L. C., Varticovski L. 1989. Phosphatidylinositol 3-kinase and its novel product, phosphatidylinositol 3-phosphate, are present in Saccharomyces cerevisiae. J. Biol. Chem. 264:20181–20184 [PubMed] [Google Scholar]
- 3.Ayong L., Pagnotti G., Tobon A. B., Chakrabarti D. 2007. Identification of Plasmodium falciparum family of SNAREs. Mol. Biochem. Parasitol. 152:113–122 [DOI] [PubMed] [Google Scholar]
- 4.Backer J. M. 2008. The regulation and function of class III PI3Ks: novel roles for Vps34. Biochem. J. 410:1–17 [DOI] [PubMed] [Google Scholar]
- 5.Billker O., Dechamps S., Tewari R., Wenig G., Franke-Fayard B., Brinkmann V. 2004. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell 117:503–514 [DOI] [PubMed] [Google Scholar]
- 6.Budovskaya Y. V., Hama H., DeWald D. B., Herman P. K. 2002. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J. Biol. Chem. 277:287–294 [DOI] [PubMed] [Google Scholar]
- 7.Burda P., Padilla S. M., Sarkar S., Emr S. D. 2002. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 115:3889–3900 [DOI] [PubMed] [Google Scholar]
- 8.Christoforidis S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S. C., Waterfield M. D., Backer J. M., Zerial M. 1999. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1:249–252 [DOI] [PubMed] [Google Scholar]
- 9.Corvera S., D'Arrigo A., Stenmark H. 1999. Phosphoinositides in membrane traffic. Curr. Opin. Cell Biol. 11:460–465 [DOI] [PubMed] [Google Scholar]
- 10.Desjardins R. E., Canfield C. J., Haynes J. D., Chulay J. D. 1979. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob. Agents Chemother. 16:710–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dove S. K., Cooke F. T., Douglas M. R., Sayers L. G., Parker P. J., Michell R. H. 1997. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature 390:187–192 [DOI] [PubMed] [Google Scholar]
- 12.Ecker A., Moon R., Sinden R. E., Billker O. 2006. Generation of gene targeting constructs for Plasmodium berghei by a PCR-based method amenable to high throughput applications. Mol. Biochem. Parasitol. 145:265–268 [DOI] [PubMed] [Google Scholar]
- 13.Elabbadi N., Ancelin M. L., Vial H. J. 1994. Characterization of phosphatidylinositol synthase and evidence of a polyphosphoinositide cycle in Plasmodium-infected erythrocytes. Mol. Biochem. Parasitol. 63:179–192 [DOI] [PubMed] [Google Scholar]
- 14.Elliott D. A., McIntosh M. T., Hosgood H. D., III, Chen S., Zhang G., Baevova P., Joiner K. A. 2008. Four distinct pathways of hemoglobin uptake in the malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 105:2463–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fidock D. A., Nomura T., Talley A. K., Cooper R. A., Dzekunov S. M., Ferdig M. T., Ursos L. M., Sidhu A. B., Naude B., Deitsch K. W., Su X. Z., Wootton J. C., Roepe P. D., Wellems T. E. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6:861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster F. M., Traer C. J., Abraham S. M., Fry M. J. 2003. The phosphoinositide (PI) 3-kinase family. J. Cell Sci. 116:3037–3040 [DOI] [PubMed] [Google Scholar]
- 17.Francis S. E., Sullivan D. J., Jr., Goldberg D. E. 1997. Hemoglobin metabolism in the malaria parasite Plasmodium falciparum. Annu. Rev. Microbiol. 51:97–123 [DOI] [PubMed] [Google Scholar]
- 18.Gary J. D., Wurmser A. E., Bonangelino C. J., Weisman L. S., Emr S. D. 1998. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143:65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaullier J. M., Simonsen A., D'Arrigo A., Bremnes B., Stenmark H., Aasland R. 1998. FYVE fingers bind PtdIns(3)P. Nature 394:432–433 [DOI] [PubMed] [Google Scholar]
- 20.Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg D. E., Slater A. F., Cerami A., Henderson G. B. 1990. Hemoglobin degradation in the malaria parasite Plasmodium falciparum: an ordered process in a unique organelle. Proc. Natl. Acad. Sci. U. S. A. 87:2931–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall B. S., Gabernet-Castello C., Voak A., Goulding D., Natesan S. K., Field M. C. 2006. TbVps34, the trypanosome orthologue of Vps34, is required for Golgi complex segregation. J. Biol. Chem. 281:27600–27612 [DOI] [PubMed] [Google Scholar]
- 23.Herman P. K., Emr S. D. 1990. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:6742–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691 [DOI] [PubMed] [Google Scholar]
- 25.Jackson K. E., Klonis N., Ferguson D. J., Adisa A., Dogovski C., Tilley L. 2004. Food vacuole-associated lipid bodies and heterogeneous lipid environments in the malaria parasite, Plasmodium falciparum. Mol. Microbiol. 54:109–122 [DOI] [PubMed] [Google Scholar]
- 26.Kakhniashvili D. G., Bulla L. A., Jr., Goodman S. R. 2004. The human erythrocyte proteome: analysis by ion trap mass spectrometry. Mol. Cell. Proteomics 3:501–509 [DOI] [PubMed] [Google Scholar]
- 27.Karnataki A., Derocher A., Coppens I., Nash C., Feagin J. E., Parsons M. 2007. Cell cycle-regulated vesicular trafficking of Toxoplasma APT1, a protein localized to multiple apicoplast membranes. Mol. Microbiol. 63:1653–1668 [DOI] [PubMed] [Google Scholar]
- 28.Karnataki A., Derocher A. E., Coppens I., Feagin J. E., Parsons M. 2007. A membrane protease is targeted to the relict plastid of Toxoplasma via an internal signal sequence. Traffic 8:1543–1553 [DOI] [PubMed] [Google Scholar]
- 29.Kohler S., Delwiche C. F., Denny P. W., Tilney L. G., Webster P., Wilson R. J., Palmer J. D., Roos D. S. 1997. A plastid of probable green algal origin in Apicomplexan parasites. Science 275:1485–1489 [DOI] [PubMed] [Google Scholar]
- 30.Lambros C., Vanderberg J. P. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65:418–420 [PubMed] [Google Scholar]
- 31.Leber W., Skippen A., Fivelman Q. L., Bowyer P. W., Cockcroft S., Baker D. A. 2009. A unique phosphatidylinositol 4-phosphate 5-kinase is activated by ADP-ribosylation factor in Plasmodium falciparum. Int. J. Parasitol. 39:645–653 [DOI] [PubMed] [Google Scholar]
- 32.Lelievre J., Berry A., Benoit-Vical F. 2005. An alternative method for Plasmodium culture synchronization. Exp. Parasitol. 109:195–197 [DOI] [PubMed] [Google Scholar]
- 33.Levine T. P., Munro S. 2002. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 12:695–704 [DOI] [PubMed] [Google Scholar]
- 34.Martin B. R., Giepmans B. N., Adams S. R., Tsien R. Y. 2005. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 23:1308–1314 [DOI] [PubMed] [Google Scholar]
- 35.McIntosh M. T., Vaid A., Hosgood H. D., Vijay J., Bhattacharya A., Sahani M. H., Baevova P., Joiner K. A., Sharma P. 2007. Traffic to the malaria parasite food vacuole: a novel pathway involving a phosphatidylinositol 3-phosphate-binding protein. J. Biol. Chem. 282:11499–11508 [DOI] [PubMed] [Google Scholar]
- 36.Mitra P., Zhang Y., Rameh L. E., Ivshina M. P., McCollum D., Nunnari J. J., Hendricks G. M., Kerr M. L., Field S. J., Cantley L. C., Ross A. H. 2004. A novel phosphatidylinositol(3,4,5)P3 pathway in fission yeast. J. Cell Biol. 166:205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nkrumah L. J., Muhle R. A., Moura P. A., Ghosh P., Hatfull G. F., Jacobs W. R., Jr., Fidock D. A. 2006. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat. Methods 3:615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Odorizzi G., Babst M., Emr S. D. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95:847–858 [DOI] [PubMed] [Google Scholar]
- 39.Odorizzi G., Babst M., Emr S. D. 2000. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem. Sci. 25:229–235 [DOI] [PubMed] [Google Scholar]
- 40.Parsons M., Karnataki A., Derocher A. E. 2009. Evolving insights into protein trafficking to the multiple compartments of the apicomplexan plastid. J. Eukaryot. Microbiol. 56:214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Payrastre B. 2004. Phosphoinositides: lipid kinases and phosphatases. Methods Mol. Biol. 273:201–212 [DOI] [PubMed] [Google Scholar]
- 42.Petiot A., Ogier-Denis E., Blommaart E. F., Meijer A. J., Codogno P. 2000. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275:992–998 [DOI] [PubMed] [Google Scholar]
- 43.Pirola L., Zvelebil M. J., Bulgarelli-Leva G., Van Obberghen E., Waterfield M. D., Wymann M. P. 2001. Activation loop sequences confer substrate specificity to phosphoinositide 3-kinase alpha (PI3Kalpha). Functions of lipid kinase-deficient PI3Kalpha in signaling. J. Biol. Chem. 276:21544–21554 [DOI] [PubMed] [Google Scholar]
- 44.Quevillon E., Spielmann T., Brahimi K., Chattopadhyay D., Yeramian E., Langsley G. 2003. The Plasmodium falciparum family of Rab GTPases. Gene 306:13–25 [DOI] [PubMed] [Google Scholar]
- 45.Rameh L. E., Cantley L. C. 1999. The role of phosphoinositide 3-kinase lipid products in cell function. J. Biol. Chem. 274:8347–8350 [DOI] [PubMed] [Google Scholar]
- 46.Roth M. G. 2004. Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 84:699–730 [DOI] [PubMed] [Google Scholar]
- 47.Rutherford A. C., Traer C., Wassmer T., Pattni K., Bujny M. V., Carlton J. G., Stenmark H., Cullen P. J. 2006. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J. Cell Sci. 119:3944–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sbrissa D., Ikonomov O. C., Shisheva A. 2002. Phosphatidylinositol 3-phosphate-interacting domains in PIKfyve. Binding specificity and role in PIKfyve. Endomembrane localization. J. Biol. Chem. 277:6073–6079 [DOI] [PubMed] [Google Scholar]
- 49.Song X., Xu W., Zhang A., Huang G., Liang X., Virbasius J. V., Czech M. P., Zhou G. W. 2001. Phox homology domains specifically bind phosphatidylinositol phosphates. Biochemistry (Mosc.) 40:8940–8944 [DOI] [PubMed] [Google Scholar]
- 50.Stack J. H., DeWald D. B., Takegawa K., Emr S. D. 1995. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129:321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su X. Z., Wu Y., Sifri C. D., Wellems T. E. 1996. Reduced extension temperatures required for PCR amplification of extremely A+T-rich DNA. Nucleic Acids Res. 24:1574–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takegawa K., DeWald D. B., Emr S. D. 1995. Schizosaccharomyces pombe Vps34p, a phosphatidylinositol-specific PI 3-kinase essential for normal cell growth and vacuole morphology. J. Cell Sci. 108(Pt 12):3745–3756 [DOI] [PubMed] [Google Scholar]
- 53.Tonkin C. J., Kalanon M., McFadden G. I. 2008. Protein targeting to the malaria parasite plastid. Traffic 9:166–175 [DOI] [PubMed] [Google Scholar]
- 54.Vaid A., Ranjan R., Smythe W. A., Hoppe H. C., Sharma P. 2010. PfPI3K, a phosphatidylinositol-3 kinase from Plasmodium falciparum, is exported to the host erythrocyte and is involved in hemoglobin trafficking. Blood 115:2500–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Dijk M. R., Waters A. P., Janse C. J. 1995. Stable transfection of malaria parasite blood stages. Science 268:1358–1362 [DOI] [PubMed] [Google Scholar]
- 56.Vanhaesebroeck B., Leevers S. J., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., Waterfield M. D. 2001. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 70:535–602 [DOI] [PubMed] [Google Scholar]
- 57.Varnai P., Balla T. 2006. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim. Biophys. Acta 1761:957–967 [DOI] [PubMed] [Google Scholar]
- 58.Vermeer J. E., van Leeuwen W., Tobena-Santamaria R., Laxalt A. M., Jones D. R., Divecha N., Gadella T. W., Jr., Munnik T. 2006. Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47:687–700 [DOI] [PubMed] [Google Scholar]
- 59.Walker E. H., Pacold M. E., Perisic O., Stephens L., Hawkins P. T., Wymann M. P., Williams R. L. 2000. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol. Cell 6:909–919 [DOI] [PubMed] [Google Scholar]
- 60.Walker E. H., Perisic O., Ried C., Stephens L., Williams R. L. 1999. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature 402:313–320 [DOI] [PubMed] [Google Scholar]
- 61.Waller R. F., McFadden G. I. 2005. The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr. Issues Mol. Biol. 7:57–79 [PubMed] [Google Scholar]
- 62.Watt S. A., Kular G., Fleming I. N., Downes C. P., Lucocq J. M. 2002. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem. J. 363:657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wengelnik K., Vial H. J. 2007. Characterisation of the phosphatidylinositol synthase gene of Plasmodium species. Res. Microbiol. 158:51–59 [DOI] [PubMed] [Google Scholar]
- 64.Wymann M. P., Bulgarelli-Leva G., Zvelebil M. J., Pirola L., Vanhaesebroeck B., Waterfield M. D., Panayotou G. 1996. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol. Cell. Biol. 16:1722–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang M., Coppens I., Wormsley S., Baevova P., Hoppe H. C., Joiner K. A. 2004. The Plasmodium falciparum Vps4 homolog mediates multivesicular body formation. J. Cell Sci. 117:3831–3838 [DOI] [PubMed] [Google Scholar]
- 66.Yuan H., Barnes K. R., Weissleder R., Cantley L., Josephson L. 2007. Covalent reactions of wortmannin under physiological conditions. Chem. Biol. 14:321–328 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.