Abstract
Eisosomes are subcortical organelles implicated in endocytosis and have hitherto been described only in Saccharomyces cerevisiae. They comprise two homologue proteins, Pil1 and Lsp1, which colocalize with the transmembrane protein Sur7. These proteins are universally conserved in the ascomycetes. We identify in Aspergillus nidulans (and in all members of the subphylum Pezizomycotina) two homologues of Pil1/Lsp1, PilA and PilB, originating from a duplication independent from that extant in the subphylum Saccharomycotina. In the aspergilli there are several Sur7-like proteins in each species, including one strict Sur7 orthologue (SurG in A. nidulans). In A. nidulans conidiospores, but not in hyphae, the three proteins colocalize at the cell cortex and form tightly packed punctate structures that appear different from the clearly distinct eisosome patches observed in S. cerevisiae. These structures are assembled late during the maturation of conidia. In mycelia, punctate structures are present, but they are composed only of PilA, while PilB is diffused in the cytoplasm and SurG is located in vacuoles and endosomes. Deletion of each of the genes does not lead to any obvious growth phenotype, except for moderate resistance to itraconazole. We could not find any obvious association between mycelial (PilA) eisosome-like structures and endocytosis. PilA and SurG are necessary for conidial eisosome organization in ways that differ from those for their S. cerevisiae homologues. These data illustrate that conservation of eisosomal proteins within the ascomycetes is accompanied by a striking functional divergence.
In mammalian cells and in Saccharomyces cerevisiae there is cogent evidence that membrane proteins are organized in discrete domains. In the latter organism, some transporters, such as Can1p, Tat2p, and Fur1p, are organized in discrete domains on the plasma membrane. This specific domain has been named MCC, for membrane compartment occupied by Can1p (17, 26, 27). As many as 21 proteins share the MCC localization pattern. These include MCC integral components, such as the membrane protein Sur7 and the MCC-associated cytosolic proteins Pil1 and Lsp1 (see below) (18). Sur7 is a transmembrane protein consisting of four putative transmembrane domains and remains associated with the MCC compartments even under physiological conditions in which all other MCC components are dispersed (17). The pair of homologous proteins, Pil1 and Lsp1, are components of subcortical punctate assemblies named “eisosomes” (52) which show localization identical to that of MCC proteins. Both MCC and eisosome components were shown to localize to furrow-like invaginations of the plasma membrane (43). The biological functions of MCC and eisosomes are quite elusive. They were initially characterized to be sites of lipid and protein endocytosis (53), but this function is by no means certain (18). In Candida albicans, Sur7 (CaSur7) has additional roles in cell wall synthesis, actin cytoskeleton organization, and septin localization (3, 4), while in S. cerevisiae, sur7 mutants show a diminished efficiency of sporulation (54). Eisosomes are synthesized de novo in the bud during cell division (28). The Pil1/Lsp1 cytoplasmic components of the eisosome, together with the membrane protein Sur7, are conserved throughout the ascomycetes (3) (see below).
Recent work on eisosomal proteins has mainly dealt with two issues. First, both Pil1 and Lsp1 are phosphorylated. Phosphorylation is mediated by a pair of redundant kinases, Pkh1p and Pkh2p. The evidence linking phosphorylation with eisosome assembly and disassembly is, however, contradictory (25, 53). The Pkh1/2 kinases are conserved throughout the eukaryotes and strictly conserved in the ascomycetes, with one putative orthologue of the Pkh1/2 pair being present in all Aspergillus genomes sequenced. The second avenue of research is the identification of eisosome-associated proteins, including those necessary for eisosome assembly. Two recent reports have characterized a second conserved protein with four transmembrane domains, Nce102p, to be essential for eisosome assembly (15, 43). The striking conservation in all available ascomycete genomes of proteins involved in eisosome structure or assembly posits an interesting paradox. No obvious macroscopic growth phenotype is seen in S. cerevisiae cells from which PIL1, LSP1, or SUR7 is deleted in mutants with single, double, or triple mutations (52). The only report dealing with eisosomal proteins in an organism other than S. cerevisiae concerns the Sur7 homologue of Candida albicans. CaSur7 is organized in punctate eisosome-like structures. At variance with S. cerevisiae, a deletion of the cognate gene results in a clear growth phenotype (see above) which resembles that resulting from the inhibition of β-glucan synthesis (2).
Model filamentous ascomycetes, such as Neurospora crassa, Aspergillus nidulans, Sordaria macrospora, and Podospora anserina, together with a host of plant and animal pathogens and an increasing number of opportunistic human pathogens, belong to the subphylum Pezizomycotina, which may have diverged from the subphylum Saccharomycotina (such as S. cerevisiae and C. albicans) between 650 million and more than 1,000 million years ago (34). All these organisms are characterized by a highly polarized growth pattern and developmental processes which include the alternation of asexual and sexual cycles involving two different types of spores: conidiospores and ascospores. Differently from the Saccharomycotina (see below), the sexual cycle involves a dikaryotic stage and a transient diploid, which never divides as such but which enters meiosis as soon as it is formed (see reference 55 and references therein).
A. nidulans is arguably, among the Pezizomycotina, the organism where membrane proteins and endocytosis have been better studied. In this organism, a number of transporters driven by their physiological promoters have been visualized in the cell membrane (UapC [48], PrnB [45], UapA and AzgA [35], AgtA [5], MstE [14], FcyB [51], UreA [M. Sanguinetti and A. Ramón, personal communication], and FurD [G. Diallinas and F. Borbolis, personal communication]). Load-and-chase experiments have shown that when the dye FM4-64 is endocytosed, it first appears in cortical punctate structures (36). Active research concerning membrane proteins (see reference 12 for a review), determination and maintenance of polarity during development (including the possible involvement of sphingolipids [23], which may be involved in signaling eisosomal protein phosphorylation in S. cerevisiae), and ongoing recent work on endocytosis (1, 5, 6, 16, 36, 40) make A. nidulans an obvious model within the Pezizomycotina with which to study eisosomal presence, structure, and function. A description of the presence and fate of these organelles during asexual development is presented below.
(The work of I. Vangelatos was in partial fulfillment of Ph.D. thesis requirements at NCSRD.)
MATERIALS AND METHODS
Media and growth conditions.
Minimal medium (MM) and complete medium (CM) as well as the growth conditions for A. nidulans were described by Cove (10). pH 5.5 and 8.2 MM were made using MM without salt solution and with the addition of 25 mM NaH2PO4 and 25 mM Na2HPO4, respectively. When necessary, supplements were added at the adequate concentrations (http://www.gla.ac.uk/acad/ibls/molgen/aspergillus/supplement.html). Glucose (1%) was used throughout as a carbon source. Urea (5 mM) or ammonium l-(+)-tartrate (10 mM) was used as the nitrogen source. The antifungal drugs caspofungin (MSD, Merck & Co Inc., Whitehouse Station, NJ) and itraconazole (Sigma) were used at final concentrations of 10 μg/ml and 15 μM, respectively, in CM. Caffeine, calcofluor white, SDS, Congo red, and CaCl2 were purchased from Sigma and used at final concentrations of 10 mM, 100 μg/ml, 50 μg/ml, 50 μg/ml, and 100 mM, respectively, in CM (20). Staining with the dye FM4-64 (Molecular Probes, Inc.) was as described by Peñalva (36). In particular, coverslips with germinated conidia (12 h of growth at 25°C) adhering to the glass surface were placed on top of plastic covers, covered with 0.1 ml of 10 μM FM4-64, incubated on ice for 15 min, washed in 5 ml MM, and immediately observed. Vacuole staining with 7-amino-4-chloromethyl coumarin (CMAC; Molecular Probes, Inc.) was as described by Tavoularis at al. (45). Coverslips with germinated conidia were placed on top of plastic covers, covered with 0.1 ml of 1/1,000 dilution of CMAC (5-mg/ml stock solution), incubated at 25°C for 30 min, washed twice with phosphate-buffered saline (PBS), and observed under a phase-contrast fluorescent microscope with a 4′,6-diamidino-2-phenylindole filter (excitation, 365/10 nm; UV-1A filter; model DM 400 digital microscope and model BA 400 light microscope [Leica]).
Crosses between A. nidulans strains were carried out as described by Pontecorvo et al. (37). For growth tests, conidiospores were inoculated on solid CM or MM supplemented with the appropriate substrates and incubated at 25°C, 37°C, or 42°C for 2 to 4 days. To monitor PilA, PilB, and SurG mRNA steady-state levels in different developmental stages from ungerminated conidia to young mycelia, strains were grown on liquid MM containing 1% (wt/vol) glucose and 5 mM urea as the sole carbon and nitrogen sources, respectively, for 0, 4, 8, 12, 16, and 20 h at 25°C.
Fungal and bacterial strains. (i) A. nidulans strains.
The different auxotrophic mutations of A. nidulans strains are compiled by A. J. Clutterbuck (http://www.gla.ac.uk/acad/ibls/molgen/aspergillus/index.html). In particular, pantoB100, pabaA1, pabaB22, riboB2, pyroA4, pyrG89, and argB2 indicate auxotrophies for d-pantothenic acid, p-aminobenzoic acid, riboflavin, pyridoxine hydrochloride, uracil/uridine, and l-arginine, respectively. The nkuAΔ mutation results in a dramatically decreased frequency of heterologous integration events into the A. nidulans genome. The LO1516 (Table 1) strain expresses functional chimeric histone H1 molecules fused with the monomeric red fluorescent protein (mRFP). The VS125 (agtA::sgfp) strain expresses functional chimeric AgtA molecules fused with the green fluorescent protein (sGFP). These markers do not affect the localization of eisosomal proteins. All strains used in this work are listed in Table 1. In every case, MM indicates minimal medium supplemented with the requirements relevant to the strains used in the experiment. pabaA1 was used as the wild-type (wt) strain. The VS79 to VS81 and VS83 strains (pilA::sgfp pilB::sgfp and surG::sgfp pilA::mrfp, respectively; for complete genotypes, see Table 1) were isolated after transformation of protoplasts of the nkuAΔ pyrG89 pyroA4 or nkuAΔ pyrG89 riboB2 strain with the pilA::sgfp::AfpyrG+ (pyrG gene of A. fumigatus), pilB::sgfp::AfpyrG+, surG::sgfp::AfpyrG+, or pilA::mrfp::AfpyrG+ translational fusion cassettes (see below). The VS84 to VS86 (surGΔ::Afpyro+ [pyro gene of A. fumigatus], pilBΔ::AfpyrG+, and pilAΔ::Afribo+ [ribo gene of A. fumigatus], respectively) strains were isolated after transformation of protoplasts of the nkuAΔ pyrG89 pyroA4 or nkuAΔ pyrG89 riboB2 recipient strain with the surGΔ, pilBΔ, and pilAΔ deletion cassettes (see deletion of the pilA, pilB, and surG genes). The VS87 (pilAΔ::Afribo+ pilBΔ::AfpyrG+) strain was isolated by crossing the VS85 and VS86 strains. The VS91 (pilA::mrfp pilB::sgfp) and VS94 (pilA::mrfp surG::sgfp) strains were isolated by crossing the VS83 strain with the VS80 and VS81 strains, respectively. The VS128 (pilB::sgfp pilAΔ::Afribo+) and VS129 (surG::sgfp pilAΔ::Afribo+) strains were isolated by crossing the VS84 strain with the VS80 and VS81 strains, respectively. The VS118 (pilA::mrfp surGΔ::Afpyro+) and VS132 (pilB::sgfp surGΔ::Afpyro+) strains were isolated by crossing the VS84 strain with the VS79 and VS80 strains, respectively. The VS145 (surG::sgfp hhoA::mrfp) and VS153 (pilB::sgfp hhoA::mrfp) strains were isolated by crossing the LO1516 strain with the VS80 and VS81 strains, respectively. The VS172 (agtA::sgfp pilAΔ::Afribo+) strain was isolated by crossing the VS86 and VS125 strains. The VS186 strain was isolated by crossing the VS83 strain with the TpA4 strain (45). The nkuAΔ pyrG89 riboB2 and nkuA pyrG89 pyroA4 strains were kindly provided by M. Peñalva and were used for the deletion and the in-locus fusions of the pilA, pilB, and surG genes. For plasmid details, see below and Table 2.
Table 1.
Aspergillus nidulans strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| wt | pabaA1 | CS2498 (Fungal Genetics Stock Center) |
| nkuAΔ pyrG89 riboB2 | pyrG89 argB2 nkuAΔ::argB+ pabaB22 riboB2 | M. Peñalva |
| nkuAΔ pyrG89 pyroA4 | pyrG89 argB2 nkuAΔ::argB+ pyroA4 | M. Peñalva |
| LO1516 | pyrG89 argB2 nkuAΔ::argB+ pyroA4 hhoA::mrfp::Afribo riboB2 | C. Scazzocchio |
| VS79 | pyrG89 argB2 nkuAΔ::argB+ pabaB22 pilA::sgfp::AfpyrG+ riboB2 | This study |
| VS80 | pyrG89 pilB::sgfp::AfpyrG+ argB2 nkuAΔ::argB+ pabaB22 riboB2 | This study |
| VS81 | pyrG89 surG::sgfp::AfpyrG+argB2 nkuAΔ::argB+ pabaB22 riboB2 | This study |
| VS83 | pyrG89 argB2 nkuAΔ::argB+ pyroA4 pilA::mrfp::AfpyrG+ | This study |
| VS84 | pyrG89 argB2 surGΔ::Afpyro+ nkuAΔ::argB+ pyroA4 | This study |
| VS85 | pyrG89 pilBΔ::AfpyrG+ argB2 nkuAΔ::argB+ pabaB22 riboB2 | This study |
| VS86 | pyrG89 argB2 nkuAΔ::argB+ pabaB22 pilAΔ::Afribo+ riboB2 | This study |
| VS87 | pyrG89 pilBΔ::AfpyrG+ argB2 nkuAΔ::argB+ pabaB22 pilAΔ::Afribo+ riboB2 | This study |
| VS91 | pyrG89 argB2 surG::sgfp::AfpyrG+ nkuAΔ::argB+ pilA::mrfp::AfpyrG+ riboB2 | This study |
| VS94 | pyrG89 pilB::sgfp::AfpyrG+ argB2 nkuAΔ::argB+ pilA::mrfp::AfpyrG+ riboB2 | This study |
| VS118 | pyrG89 argB2 surGΔ::Afpyro+ nkuAΔ::argB+ pyroA4 pilA::mrfp::AfpyrG+ | This study |
| VS125 | agtA::sgfp::AfpyrG+ argB2 riboB2 | This study |
| VS128 | pyrG89 pilB::sgfp::AfpyrG+ argB2 nkuAΔ::argB+ pabaB22 pilAΔ::Afribo+ riboB2 | This study |
| VS129 | pyrG89 surG::sgfp::AfpyrG+argB2 nkuAΔ::argB+ pabaB22 pilAΔ::Afribo+ riboB2 | This study |
| VS132 | pyrG89 pilB::sgfp::AfpyrG+ argB2 surGΔ::Afpyro+ nkuAΔ::argB+ pyroA4 | This study |
| VS145 | pyrG89 surG::sgfp::AfpyrG+argB2 nkuAΔ::argB+ pabaB22 hhoA::mrfp::Afribo+ riboB2 | This study |
| VS153 | pyrG89 pilB::sgfp::AfpyrG+ argB2 nkuAΔ::argB+ pyroA4 hhoA::mrfp::Afribo+ riboB2 | This study |
| VS172 | pyrG89 agtA::sgfp::AfpyrG+ argB2 nkuAΔ::argB+ pilAΔ::Afribo+ riboB2 | This study |
| VS186 | yA2 pantoB100, prnB::sgfp::trpC 3′ terminator sequence pyrG89 argB2 nkuAΔ:: pilA::mrfp::AfpyrG+ | This study |
Table 2.
Plasmids used in this worka
| Cloning vector | Description | Source, yr (reference) |
|---|---|---|
| pRG3 | pGEM with 18S rRNA gene | Delcasso-Tremousaygue et al., 1988 (11) |
| p1548 | AfriboB | Szewczyk et al., 2006 (provided by M. Peñalva) (44) |
| p1547 | AfpyroA | Szewczyk et al., 2006 (provided by M. Peñalva) (44) |
| p1439 | 5GA-sgfp-AfpyrG | Szewczyk et al., 2006 (provided by M. Peñalva) (44) |
| p1491 | 5GA-mrfp-AfpyrG | Szewczyk et al., 2006 (provided by M. Peñalva) (44) |
See also Materials and Methods.
(ii) Escherichia coli strains.
The Escherichia coli strain used was DH5α.
Transformation methods.
Transformation of E. coli was carried out as described by Sambrook and Russell (39). Transformation of A. nidulans is described by Tilburn et al. (47).
Plasmids.
The pRG3 plasmid carries the radish 18S rRNA gene (11).The p1548 plasmid contains the riboB gene of Aspergillus fumigatus (44), which complements the riboB2 mutation of A. nidulans (kindly provided by M. Peñalva). The p1547 plasmid contains the pyroA gene of A. fumigatus (44), which complements the pyroA4 mutation of A. nidulans (kindly provided by M. Peñalva). The p1439 and p1491 plasmids contain a 5 Gly-Ala (5GA) linker fused in frame with the sGFP and with mRFP, respectively, followed by the A. fumigatus pyrG gene (kindly provided by M. Peñalva) (44).
DNA manipulations.
Plasmid preparation from E. coli strains was carried out as described by Sambrook and Russell (39). DNA digestion was carried out as described by Sambrook and Russell (39). Total DNA extraction from A. nidulans is described by Lockington et al. (24). Southern blot analysis was carried out as described by Sambrook and Russell (39). The restriction enzymes used to monitor the deletion of surG, pilB, and pilA from the genomes of the VS84, VS85, and VS86 strains, respectively, were HindIII (pilAΔ::Afribo+surGΔ::Afpyro+) and PstI (pilBΔ::AfpyrG+). AfriboB, AfpyrG, and AfpyroA probes corresponding to the purified ∼1-kb PCR fragments were obtained using as template DNA the p1548, p1439, and p1547 plasmids, respectively, and the PilAΔ-Ribo F and PilAΔ-Ribo R, PilBΔ-PyrG F and PilBΔ-PyrG R, and SurGΔ-Pyro F and SurGΔ-Pyro R primer pairs, respectively (Table 3).
Table 3.
Oligonucleotides used in this study
| Name | Sequence (5′–3′) |
|---|---|
| PilAΔ-Ribo F | GCA GAA TAT CGG CTG GTC TC CGC TCT AGA ACT AGT GGA TCC |
| PilAΔ-Ribo R | GCT CAT TCA GTG AGT GCT CG CCT CGA GGT CGA CGG TAT CG |
| PilBΔ-PyrG F | CAC CTG TCT AGC CTC AGC AA GTC GCC TCA AAC AAT GCT CTT C |
| PilBΔ-PyrG R | CAC AAA GCA CTA ATC ACC CCT T CTG AGA GGA GGC ACT GAT GC |
| SurGΔ-Pyro F | GTC TAC TCG TCT CTC ATC AGA CGC TCT AGA ACT AGT GGA TCC |
| SurGΔ-Pyro R | GTG TAT AGC CGA CAG CAG CA CCT CGA GGT CGA CGG TAT CG |
| P1 PilAΔ | CGA TCC TAG CTC TCA GGA TC |
| P3 PilAΔ | GGA TCC ACT AGT TCT AGA GCG GAG ACC AGC CGA TAT TCT GC |
| P4 PilAΔ | CGA TAC CGT CGA CCT CGA GG CGA GCA CTC ACT GAA TGA GC |
| P6 PilAΔ | ACC AAC CTA GTC GAC GTG AC |
| P1 PilBΔ | CTC AGC TGA GAG ACT GTC AG |
| P3 PilBΔ | GAA GAG CAT TGT TTG AGG CGA C TTG CTG AGG CTA GAC AGG TG |
| P4 PilBΔ | GCA TCA GTG CCT CCT CTC AG AAG GGG TGA TTA GTG CTT TGT G |
| P6 PilBΔ | TCG GAG TCA ATG TAG TAC AGC |
| P1 SurGΔ | GCT CAC ATC CAC AAT GTC TAG |
| P3 SurGΔ | GGA TCC ACT AGT TCT AGA GCG TCT GAT GAG AGA CGA GTA GAC |
| P4 SurGΔ | CGA TAC CGT CGA CCT CGA GG TGC TGC TGT CGG CTA TAC AC |
| P6 SurGΔ | GAC TGC CAC ACC TCA CCT C |
| P2 PilAΔ | GCT GAA CCA GAA GAG GCT GC |
| P5 PilAΔ | GCA TCC ATG ATG TCA GCA TAC |
| P2 PilBΔ | TCA TCA CCA GGC AAG ATC ATC |
| P5 PilBΔ | CAT CGT TCC CAT GCT CAG AC |
| P2 SurGΔ | TCT TGT GCT GAG GGA ACT AAG |
| P5 SurGΔ | CGT CCT CAT CCG TGT CTG C |
| PilA F | CGG GAG CCT GTC CCT GTC GGA GCT GGT GCA GGC GCT G |
| PilA R | GCT CAT TCA GTG AGT GCT CG CTG AGA GGA GGC ACT GAT GC |
| PilA P1 | GCA TCG TAC ATA CTC TAT GCG |
| PilA P2 | CAC CAC TCT CCT CGA CCA AG |
| PilA P3 | CAG CGC CTG CAC CAG CTC C GAC AGG GAC AGG CTC CCG |
| PilA P4 | GCA TCA GTG CCT CCT CTC AG CGA GCA CTC ACT GAA TGA GC |
| PilA P5 | GCA TCC ATG ATG TCA GCA TAC |
| PilA P6 | ACC AAC CTA GTC GAC GTG AC |
| PilB F | AGC GAG TTG CAG TGC CCA TT GGA GCT GGT GCA GGC GCT G |
| PilB R | CAC AAA GCA CTA ATC ACC CCT T CTG AGA GGA GGC ACT GAT GC |
| PilB P1 | GCA GAA GAA GGA GCT CTG TC |
| PilB P2 | CGT GCT GAT GGC AGA AAT GG |
| PilB P3 | CAG CGC CTG CAC CAG CTC C AAT GGG CAC TGC AAC TCG CT |
| PilB P4 | GCA TCA GTG CCT CCT CTC AG AAG GGG TGA TTA GTG CTT TGT G |
| PilB P5 | CAT CGT TCC CAT GCT CAG AC |
| PilB P6 | TCG GAG TCA ATG TAG TAC AGC |
| SurG F | GCA ACA AGG AAA TCG CTC CCG GAG CTG GTGCAG GCG CTG GAG |
| SurG R | GTG TAT AGC CGA CAG CAG CAC TGA GAG GAG GCA CTG ATG CGT G |
| SurG P1 | CTT CAT CGT TCA AGC TTC AGG |
| SurG P2 | TCT ACT CGT CTC TCA TCA GAG |
| SurG P3 | C TCC AGC GCC TGC ACC AGC TCC GGG AGC GAT TTC CTT GTT GC |
| SurG P4 | CAC GCA TCA GTG CCT CCT CTC AG TGC TGC TGT CGG CTA TAC AC |
| SurG P5 | TGT CGA GTT TCT GCC TCT CC |
| SurG P6 | GAC TGC CAC ACC TCA CCT C |
High-fidelity and long-fragment PCRs were carried out using an LA Taq kit (Takara). High-fidelity and small-fragment PCRs were carried out using platinum Pfx polymerase (Invitrogen), while conventional PCRs were carried out using Taq polymerase (NEB). DNA bands were purified from agarose gels using a Wizard PCR Preps DNA purification system (Promega). The [32P]dCTP-labeled DNA molecules, which were used as gene-specific probes, were prepared using a Megaprime DNA labeling system kit (Amersham Life Sciences).
Deletion of pilA, pilB, and surG genes.
The entire pilA, pilB, and surG open reading frames (ORFs) (1,044 bp [ANID_05217.1], 1,200 bp [ANID_3931.1], and 735 bp [ANID_4615.1], respectively) were replaced in an nkuAΔ pyrG89 riboB2 (pilA and pilB) or nkuAΔ pyroA4 (surG) strain by the riboB, pyrG, or pyroA (pilA, pilB, and surG, respectively) gene after a double crossing-over event, using the fusion PCR gene replacement method (44). The primers used are listed in Table 3. In the pilA deletion cassette, the fragment corresponding to the central part contains the riboB gene of A. fumigatus amplified from the p1548 plasmid using primers PilAΔ-Ribo F and PilAΔ-Ribo R. In the pilB deletion cassette, the fragment corresponding to the central part contains the pyrG gene of A. fumigatus amplified from the p1439 plasmid using primers PilBΔ-PyrG F and PilBΔ-PyrG R. For the surG deletion cassette, the fragment corresponding to the central part contains the pyroA gene of A. fumigatus amplified from the p1547 plasmid using primers SurGΔ-Pyro F and SurGΔ-Pyro R. The sequences 1,115 bp upstream and 1,014 bp downstream of the pilA ORF were amplified from genomic DNA of a wild-type strain (pabaA1) using the P1 PilAΔ-P3 PilAΔ and P4 PilAΔ-P6 PilAΔ primer pairs, respectively. The sequences 1,126 bp upstream and 1,112 bp downstream of the pilB ORF were amplified from genomic DNA of a wild-type strain (pabaA1) using the P1 PilBΔ-P3 PilBΔ and P4 PilBΔ-P6 PilBΔ primer pairs, respectively. The sequences 973 bp upstream and 1,073 bp downstream of the surG ORF were amplified from genomic DNA of a wild-type strain (pabaA1) using the P1 SurGΔ-P3 SurGΔ and P4 SurGΔ-P6 SurGΔ primer pairs, respectively. The whole pilA, pilB, and surG deletion cassettes used to transform the nkuAΔ pyrG89 riboB2 and nkuAΔ pyroA4 strains were amplified using the P2 PilAΔ-P5 PilAΔ, P2 PilBΔ-P5 PilBΔ, and P2 SurGΔ-P5 SurGΔ primer pairs, respectively (Table 3). Selection of transformants was carried out on urea-containing minimal medium lacking riboflavin, uracil/uridine, or pyridoxine, as required for each replacement. The in-locus replacement of pilA, pilB, and surG with riboB, pyrG, and pyroA, respectively, was confirmed by Southern blot analysis (data not shown). The deleted outcrossed strains were checked by PCR using specific primer pairs P2 PilAΔ-P5 PilA for the pilAΔ strain, P2 PilBΔ-P6 PilB for the pilBΔ strain, and P2 SurGΔ-P5 SurG for the surGΔ strain, respectively (Table 3).
Construction of pilA, pilB, and surG in-locus fusions.
Cassettes containing the pilA::sgfp, pilB::sgfp, surG::sgfp, pilA::mrfp, and pilB::mrfp sequences were constructed by joining three different PCR fragments, as described by Szewczyk et al. (44), using the PilA, PilB, and SurG pairs of primers (Table 3). To construct the pilA::sgfp, pilB::sgfp, surG::sgfp, pilA::mrfp, and pilB::mrfp translational fusions, DNA fragments corresponding to the central part of the construction were amplified from the p1439 and p1491 plasmids. These fragments contain a 5GA linker fused in frame with the sGFP and the mRFP proteins, followed by the pyrG gene of A. fumigatus. The upstream flanking sequence of the sgfp and mrfp ORFs is a 1,128-bp fragment containing a part of the pilA ORF followed by the 5GA linker, while the downstream flanking sequence is a fragment containing a 1,014-bp fragment corresponding to the 3′ end of the pilA gene, just after the chain termination codon. The upstream flanking sequence of the sgfp ORF is a fragment containing 1,105 bp of the pilB ORF, followed by the 5GA linker, while the downstream flanking sequence is a fragment containing a 1,112-bp fragment corresponding to the 3′ downstream region of the pilB gene, just after the chain termination codon. The upstream flanking sequence of the sgfp ORF is a 1,157-bp fragment of the surG ORF, followed by the 5GA linker, while the downstream flanking sequence is a fragment containing a 1,073-bp fragment corresponding to the 3′ end of the surG gene, just following the chain termination codon. All fragments were amplified from genomic DNA of a wild-type strain (pabaA1) using the P1-P3 (PilA, PilB, or SurG) and P1-P4 (PilA, PilB, or SurG) primer pairs. The whole fusion cassettes were amplified using the P2-P5 (PilA, PilB, or SurG) pair of primers and were used to transform nkuAΔ pyrG89 riboB2 (pilA::sgfp, pilB::sgfp, surG::sgfp, and pilB::mrfp) or nkuAΔ pyroA4 (pilA::mrfp) strains. Transformants were selected on MM with urea as the sole nitrogen source and without uracil/uridine. The in-locus replacement of pilA, pilB, and surG with pilA::sgfp, pilB::sgfp, surG::sgfp, or pilA::mrfp was confirmed by Southern blot analysis. Genomic DNA was restricted with EcoRV (pilA::sgfp, pilA::mrfp), NcoI (pilB::sgfp), or PstI (surG::sgfp) and hybridized with the AfriboB, AfpyrG, and AfpyroA sequence fragments, respectively. These fragments were amplified from plasmids p1439, p1548, and p1547 using the primer pairs Ribo F-Ribo R with “PilAΔ-Ribo F with PilAΔ-Ribo R,” PyrG F-PyrG R with “PilBΔ-PyrG F with PilBΔ-PyrG R,” and Pyro F-Pyro R with “SurGΔ-Pyro F with SurGΔ-Pyro R,” respectively (Tables 2 and 3). All transformants checked by Southern blot analysis contained single-copy sequence integrations at the pilA, pilB, and surG loci; and their sequences did not show any differences from those of the wild-type controls under any of the conditions tested (see Results).
RNA manipulations.
Total RNA extraction from A. nidulans was carried out using the TRIzol reagent (Invitrogen), according to the manufacturer's instructions. RNA was separated on glyoxal agarose gels, as described by Sambrook and Russell (39). The hybridization technique is described by Church and Gilbert (9). To monitor RNA loading, the radish 18S rRNA gene was used as the probe (11). This corresponds to the ∼1.5-kb EcoRI-EcoRI fragment purified from plasmid pRG3. pilA, pilB, and surG mRNA steady-state levels were monitored by hybridization with probes corresponding to the purified ∼1-kb PCR fragments obtained using as the template DNA from a wild-type (pabaA1) strain and the P1-P3 (PilA, PilB, and SurG, respectively) primer pairs.
Membrane protein extraction and Western blot analysis.
Protein extracts were prepared as described by Kafasla et al. (21). In particular, mycelia grown for 16 h in MM containing 5 mM urea as the sole nitrogen source and supplemented with the appropriate auxotrophs and conidiospores (0 h of growth) were harvested, frozen, and ground in liquid nitrogen. All subsequent steps were carried out at 4°C. Conidia and mycelial powder was resuspended in 1.5 ml ice-cold extraction buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, pH 8.0) supplemented with a protease inhibitor cocktail (Sigma) and 0.2 mM phenylmethylsulfonyl fluoride (PMSF). A volume of ∼200 μl sterile glass beads (diameter, 0.1 mm) in conidia extraction buffer was added, and the suspension was vortexed for about 2 min. After 10 min incubation on ice, unbroken cells and larger cell debris were removed by a short low-speed centrifugation (3,000 × g for 3 min). Trichloroacetic acid was added to a final volume of 5%, followed by 10 min incubation on ice. Total proteins were precipitated by a 5-min centrifugation at 13,000 × g, washed with 500 μl Tris base, and resuspended in extraction buffer. The sample protein concentration was measured by the method of Bradford (7a). Protein samples (20 μg) were fractionated on a 10% SDS-polyacrylamide gel and electroblotted (Mini Protean Tetra cell; Bio-Rad) onto a polyvinylidene difluoride (PVDF) membrane (Macherey-Nagel) for immunodetection. The membrane was treated with 2% nonfat dry milk, and immunodetection was performed using a primary mouse anti-GFP monoclonal antibody (Roche) and a secondary goat anti-mouse IgG horseradish peroxidase (HRP)-linked antibody (Cell Signaling). Blots were developed by the chemiluminescent method using an enhanced chemiluminescence reagent (Amersham Bioscience).
FM and LSCM.
Ten milliliters of a suspension of 5 × 105 or 5 × 107 conidia/ml was inoculated onto sterile coverslips embedded into appropriate liquid culture media, incubated for 4 h or 16 h at 25°C, and observed by fluorescence microscopy (FM) and laser scanning confocal microscopy (CSLM), as described previously (45, 46). Two hundred microliters of a suspension of 5 × 107 conidia/ml was inoculated into sterile Eppendorf tubes and observed by FM and CSLM, as described previously (45, 46). To elicit AgtA-GFP endocytosis, ammonium l-(+)-tartrate was added to a final concentration of 10 mM at 30, 60, and 120 min before observation. Filipin was used at a concentration of 25 μg/ml at 37°C for 15 min before the observation. Samples were observed on an Axioplan Zeiss phase-contrast epifluorescent microscope with appropriate filters, and the resulting images were acquired with a Zeiss MRC5 digital camera using AxioVs40 (version 4.40.0) software. Images were then processed with Adobe Photoshop CS2 (version 9.0.2) software. Confocal scanning laser microscopy to examine PilA-sGFP, PilB-sGFP, SurG-sGFP, PilA-mRFP, PilB-mRFP, and HhoA-mRFP localization was carried out on an MRC 1024 confocal system (Laser Sharp, version 3.2 software [Bio-Rad]; zoom, ×2 to ×5; excitation, 488 nm/blue and 568 nm/yellow for sGFP and mRFP, respectively; samples at laser power 3 [PilA, SurG, and HhoA] and 30% [PilB]; Kalman filter; n = 5 to 6; 0.3-μm cut; iris, 7 to 8; crypton/argon laser; Nikon Diaphot 300 microscope with a ×60 [oil immersion] lens; emission filter, 522/DF35; lens reference, Plan Apo 60/1.40 oil immersion DM 160175, 60 DM/Ph4, 160/0.17 [Nikon, Japan]).
Bioinformatic tools and databases.
The databases consulted were http://www.fgsc.net/, through which most fungal genomes are accessible. Specifically, Aspergillus sequences were obtained through http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html. The yeast databases consulted were http://www.genolevures.org/yeastgenomes.html, http://www.yeastgenome.org/, http://www.genedb.org/genedb/pombe/, and http://www.candidagenome.org/. The Pneumocystis carinii database is http://pgp.cchmc.org/. Phylogenetic trees were constructed online using the different programs contained in http://www.phylogeny.fr/. Muscle alignments were carried out with http://www.ebi.ac.uk/Tools/muscle/index.html. T-Coffee alignments were carried out at http://www.phylogeny.fr/. Alignments were carried out with the Boxshade program (http://www.ch.embnet.org/software/BOX_form.html). Transmembrane protein topologies were predicted with the HMMTOP (http://www.enzim.hu/hmmtop/html/submit.html), PRED-TMR (http://athina.biol.uoa.gr/PRED-TMR2/input.html), TopPred (http://www.ch.embnet.org/software/TMPRED_form.html), and SPLIT4 (http://split.pmfst.hr/split/4/) programs. Coiled coils were predicted at http://toolkit.tuebingen.mpg.de/pcoils and http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_lupas.html.
RESULTS
Eisosomal core components in Aspergillus.
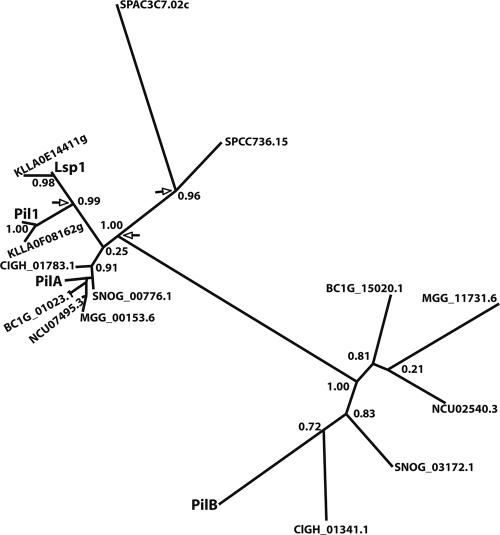
A search of the available fungal databases reveals that homologues of Pil1 and Lsp1 are present in all the ascomycetes (properly, phylum Ascomycota). In the available genomes of all the Pezizomycotina, including the aspergilli, we found proteins belonging to two clades, which we call the PilA and the PilB clades. Very stringent conservation is seen within the PilA clade, while the proteins of the PilB clade show a higher rate of divergence. Proteins of the PilA clade show a central coiled-coil domain (residues 166 to 198 for PilA), similarly to Pil1 (52) and Lsp1, while all proteins of the PilB clade show the presence of an additional carboxy-terminal coiled-coil domain (residues 147 to 178 and 225 to 244 for PilB; data not shown). The PilA and PilB clades are represented in A. nidulans by the sequences with the designations ANID_05217.1 and ANID_03931.1, respectively. The duplication which generated PilA and PilB is possibly ancestral to the Pezizomycotina, as orthologues of ANID_05217.1 (73% identity) and ANID_03931.1 (56% identity) are present in Tuber melanosporum (R. Percudani, personal communication), a member of the Pezizales, which is a basal branch of the Pezizomycotina (42). The PilA clade clusters with the Pil1-like and Lsp1-like proteins of the subphylum Saccharomycotina, with the latter arising from an independent duplication event at the root of the Saccharomycotina. For clarity, the results for proteins from only two genomes (S. cerevisiae and Kluveromyces lactis) of the latter subphylum, together with the results for proteins from some representative genomes of the Pezizomycotina, are shown in Fig. 1. A third duplication independent of those of the Saccharomycotina and Pezizomycotina is present in all the available genomes of the genus Schizosaccharomyces, represented in Fig. 1 by S. pombe. Two close homologues showing 64% to 42% identities with the two S. pombe proteins, respectively, are also present in Pneumocystis carinii, another member of the Taphrinomycotina (data not shown). The homologues present in the Taphrinomycotina are quite divergent from Pil1/PilA, but their position in the tree is consistent with the proposal that the ancestral Pil protein could have been Pil1/PilA-like. This topology, indicating the occurrence of three independent duplications, one for each subphylum, is maintained when all available genomes are included in trees constructed with a variety of algorithms (32; our unpublished data; A. Olivera-Couto and P. Aguilar, personal communication). The Meu14 protein of S. pombe, necessary for the second division of meiosis and the accurate formation of the forespore membrane (30), is clearly related to Pil1/Pil1A/Lsp1 (22% identity with both Pil1 and PilA, 24% identity with PilB) (31). When it is included in trees similar to that shown in Fig. 1, it appears as an outgroup to both the Pil1/Lsp1/PilA clade (including the S. pombe putative eisosomal proteins) and the PilB clade (data not shown) (32).
Fig. 1.
Eisosomal proteins in the subphylum Pezizomycotina. Phylogenetic unrooted tree, including the Pil1 (YGR086C) and Lsp1 (YPL004C) proteins of S. cerevisiae; the two putative eisosomal proteins of the second member of the Saccharomycotina, Kluvermyces lactis (KLLA); a member of the Taphrinomycotina, S. pombe (SPAC3C7.02c, SPCC736.15); and the PilA and PilB homologues (see text) of a number of representative fungi of the Pezizomycotina. The proteins of A. nidulans (Euriotiomycetes) referred to are PilA (ANID_05217.1) and PilB (ANID_03931.1) Other species referred to are Botritis cynerea (BC) (Leotiomycetes), Coccidioides immitis (CIGH; Euriotiomycetes), Magnaporthe grisae (MGG), Neurospora crassa (NCU; Sordariomycetes), and Stegonospora nodorum (SNOG; Dothideomycetes), followed by their systematic numbers in the relevant databases. Alignments were carried out with T-Coffee, curated with the Phylogeny site internal curation program (see Materials and Methods), and the tree was obtained with the maximum-likelihood program alLRT-PhyML (4a; http://www.phylogeny.fr/version2_cgi/alacarte.cgi) and redrawn after a Drawtree image. The digits at the nodes represent the results of the approximate likelihood ratio test and are nonparametric branch support values (http://atgc.lirmm.fr/alrt/). The PilA clade is clustered with all the homologues of the Saccharomycotina (approximate likelihood ratio test value = 1), but note the weak support (approximate likelihood ratio test value = 0.25) of the node where the PilA clade branches out from the homologues in the Saccharomycotina; in trees (data not shown) where many more species of Saccharomycotina and Pezizomycotina are included, the topology is maintained and the approximate likelihood ratio test value increases to >0. 80. For reasons of space, we do not show the branch support approximate likelihood ratio test values within the PilA clade; all nodes with the exception of the node where PilA branches out have strong support (approximate likelihood ratio test value > 0.80); the node where PilA branches out has an approximate likelihood ratio test value of 0.43. While we show only representative species in this tree, the topology of a tight cluster of PilA homologues and a looser cluster of PilB homologues is maintained in a tree that was constructed with the same or other algorithms and that includes at least one species of every genus of the Pezizomycotina for which there is an available sequence. Arrows show the nodes corresponding to the three independent duplications that gave origin to the PilA and PilB clades of the Pezizomycotina, the Pil1 and Lsp1 clades of the Saccharomycotina, and the two clades of the Taphrinomycotina.
The Sur7 transmembrane protein has been characterized as a component of eisosomes in both S. cerevisiae and C. albicans (see Introduction). Three putative members of the Sur7 family of proteins are present in A. nidulans, ANID_04615.1, ANID_01331.1, and ANID_05213.1. These proteins represent each of the three clades of Sur7-like proteins present in the aspergilli (see Fig. S1 in the supplemental material). ANID_04615.1 (to be called SurG, whose cognate gene is surG) is clearly the orthologue Sur7 protein of S. cerevisiae and C. albicans (showing 21% and 27% identities, respectively). The positions of the four transmembrane domains, typical of Sur7, are conserved in the three homologues, together with the sequence showing similarities with the Claudin-like domain (3).
The subcellular localization of eisosomal proteins is developmentally regulated.
To determine the subcellular localization of PilA, PilB, and SurG, we used strains carrying the pilA-gfp and pilA-mrfp, pilB-gfp, and surG-gfp alleles replacing the corresponding resident genes, respectively. The GFP fusion strains behave like the wild type in relation to growth on complete medium in the presence or absence of itraconazole (see below). It was also checked that the temporal pattern of expression of the mRNAs corresponding to each of the gfp fusion proteins is identical to that seen for the wild-type genes (data not shown; see below).
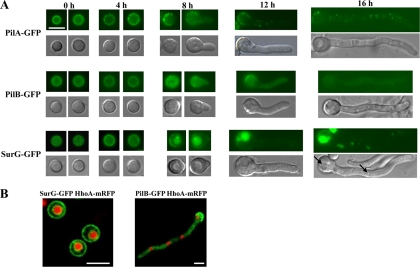
Epifluorescence microscopy of ungerminated conidia (Fig. 2) showed PilA-GFP, PilB-GFP, and SurG-GFP localized as patches in the periphery of the spores. SurG-GFP is additionally localized perinuclearly. The perinuclear location of SurG is more clearly shown by confocal microscopy in a strain carrying both SurG-GFP and the H1 histone gene fused to mRFP, suggesting localization of SurG to the endoplasmic reticulum, similar to that shown by the endoplasmic reticulum chaperone ShrA (Fig. 2B) (13). Polar and equatorial sections of confocal microscopic images of ungerminated conidia showed that the PilA-GFP patches are not as clearly separated in A. nidulans as they are in S. cerevisiae (see Fig. S2 in the supplemental material).
Fig. 2.
Subcellular localization of PilA, PilB, and SurG proteins at various asexual developmental stages of the A. nidulans life cycle. (A) Representative pictures from epifluorescence microscopy of strains expressing chimeric PilA-GFP, PilB-GFP, and SurG-GFP molecules in ungerminated (0 h), swollen (4 h), and germinated (8 h) conidia and young mycelia (12 h, 16 h). Strains were grown in the presence of 5 mM urea and 1% (wt/vol) glucose as the sole nitrogen and carbon sources, respectively, at 25°C. GFP fluorescence is shown in the upper panels of each row, while Nomarski pictures of the same samples are shown in the lower panels. Black arrows indicate the central vacuole and endosomes. Bar, 5 μm. (B) Representative pictures from laser scanning confocal microscopy of strains expressing both SurG-GFP and HhoA-mRFP (histone1-RFP) molecules in ungerminated wild-type conidia (left panel) and both PilB-GFP and HhoA-mRFP molecules in young mycelia (16 h) (right panel). The strains were grown as described for panel A. Bars, 5 μm.
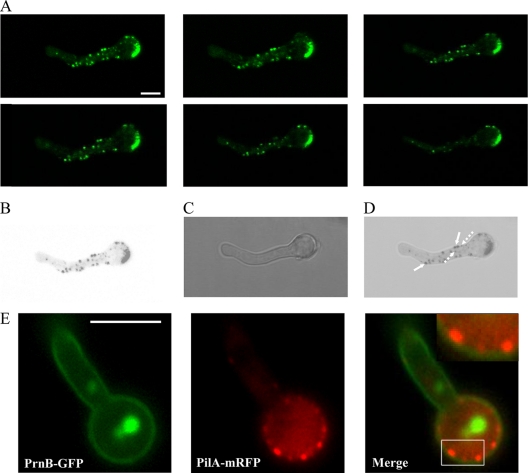
No change in the localization of PilA or PilB is seen during the period of isotropic growth. The same stands for the peripheral localization of SurG; nevertheless, the perinuclear ring disappears before the emergence of the germination tube (this is better seen in Fig. 9 [see below]). All three proteins are restricted to the periphery of the conidial head opposite the emergent germ tube, while they have different destinations in young and older mycelia. PilA is present in discrete patches in young mycelia (16 h; Fig. 2) but also older and branching mycelia (20 and 24 h; data not shown). PilA spots in hyphae are not uniform in size and are not restricted to the periphery as they are in conidiospores. This is shown in Fig. 3, where the brightest PilA spots are localized at the periphery of the hyphae, while smaller ones are present both at the interior of the hyphae and at its periphery. Furthermore, double labeling of PrnB (the major proline transporter) (41, 45) and PilA indicates that hyphal membrane-bound PilA punctate structures (which we can equate to eisosomes) are localized to the internal face of the membrane (Fig. 3E). In hyphae, PilB-GFP is seen as very low intensity, diffused fluorescence in the cytoplasm (Fig. 3A), the settings of the microscope being such that the difference in fluorescence between the ungerminated conidia and the germlings is faithfully reflected. This suggests that the protein is downregulated after germination. PilB-GFP is excluded from the nuclei, as shown in a strain carrying both PilB-GFP and the gene encoding the H1 histone fused to mRFP (Fig. 3B). Finally, in hyphae SurG-GFP is confined to the vacuole and endosomes (Fig. 3A, black arrows) (some residual signal can be seen in the membrane of the conidial head), and this is confirmed by CMAC staining (data not shown).
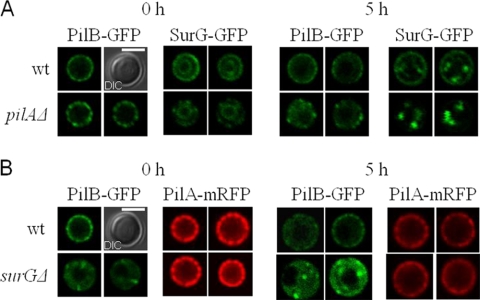
Fig. 9.
Subcellular localization of PilB-GFP and SurG-GFP proteins expressed in conidia of a pilAΔ strain and of PilA-mRFP and PilB-GFP proteins expressed in conidia of a surGΔ strain. (A) Pictures of representative equatorial sections from laser scanning confocal microscopy of strains expressing chimeric PilB-GFP and SurG-GFP molecules in ungerminated (0 h) and swollen (5 h) conidia of a wild-type and a pilAΔ strain. (B) Representative pictures from laser scanning confocal microscopy of strains expressing chimeric PilA-mGFP and PilB-GFP molecules in ungerminated (0 h) and swollen (5 h) conidia of a wild-type and a surGΔ strain. In both panels A and B, Nomarski pictures (DIC) of wild-type conidia 0 h are shown. Bars, 5 μΜ. For the PilA-GFP and PilB-GFP strains, two independent conidia are shown.
Fig. 3.
(A) Confocal z-stack sections showing PilA-GFP in a wild-type strain. The strain was grown in the presence of 5 mM urea and 1% (wt/vol) glucose as the sole nitrogen and carbon sources, respectively, for 16 h at 25°C. (B to D) Inverted black and white fluorescence of the first z-stack section merged to the corresponding DIC. Bar, 5 μm (A to D). Note that PilA spots are not uniform in size and are not restricted at the periphery of mycelia. The largest PilA eisosomes (filled arrows in panel D) are localized at the periphery, while the smaller ones are at both the interior and the periphery (dashed arrows). (E) Subcellular localization of PilA and PrnB proteins in mycelia. Representative pictures from laser scanning confocal microscopy of strains expressing both PilA-mGFP and PrnB-GFP molecules in young mycelia (12 h) are shown. The upper right inset in the Merge panel shows a magnification of the boxed region. Strains were grown in the presence of 5 mM urea and 1% (wt/vol) glucose as the sole nitrogen and carbon sources, respectively, at 25°C. To induce prnB gene expression, 20 mM l-proline was added during the last 2 h of growth (45). Bar, 5 μm.
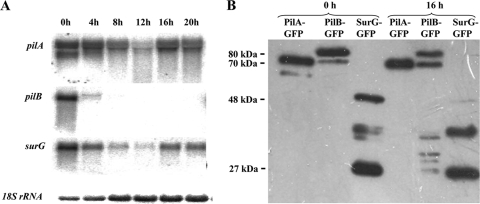
Expression of pilA, pilB, and surG genes.
The accumulation of the three eisosomal proteins in conidia led us to investigate whether this is correlated with transcription of the cognate genes, from resting conidia through young mycelia. The results presented in Fig. 4A show that both pilA and surG mRNAs accumulate more in resting conidia that at any other time point of germination tested. At variance with this, pilB mRNA is abundant in resting conidia and is not detectable within the sensitivity of the Northern blot at 4 h, 8 h, 12 h, and 16 h after the onset of germination. The transcription of the pilB homologue of A. fumigatus, Afu6g08320, is also clearly detected in dormant conidia and is barely detectable at the onset of germination (22).
Fig. 4.
(Α) Expression of the pilA, pilB, and surG genes in a wild-type strain. pilA, pilB, and surG transcript levels in ungerminated (0 h), swollen (4 h), and germinated (8 h) conidia and in young (12 h, 16 h) and older (20 h) mycelia are shown. Strains were grown in the presence of 5 mM urea and 1% (wt/vol) glucose as sole nitrogen and carbon sources, respectively, at 25°C. Steady-state 18S rRNA levels were used to monitor the amount of RNA loading in each lane. (B) Western blot analysis of the PilA, PilB, and SurG tagged proteins. Approximately 20 μg of total protein fractions of conidia (0 h) and young mycelia (16 h) derived from strains expressing PilA, PilB, or SurG tagged with GFP proteins was fractionated on a 10% SDS-polyacrylamide gel, transferred to a PVDF membrane, and probed with a primary mouse anti-GFP monoclonal antibody and a secondary goat anti-mouse IgG HRP-linked antibody. Protein markers are indicated in kDa on the left. Equal loading was checked by Coomassie blue staining.
The fluorescence signal (Fig. 2) seen in germlings and mycelia for PilB (cytosolic) and SurG (vacuolar) could be due either to the presence of the intact fusion proteins, to degradation products conserving the proteins' C termini, or simply to degradation-resistant GFP accumulating in the cytosol and vacuoles, respectively. Western blots (Fig. 4B) carried out with protein extracts of ungerminated conidia (0 h) or young mycelia (16 h) grown under the same conditions used to investigate intracellular localization show that bands corresponding to full-length PilA-GFP and PilB-GFP are present in both conidia and mycelia, while bands corresponding to full-length SurG-GFP are present in conidia and are faintly observed in mycelia. Molecular masses are indicated in kDa. The slower-migrating bands of PilA-GFP and PilB-GFP have approximate apparent molecular masses of 70 kDa and 80 kDa, respectively (calculated molecular masses, 67 kDa and 71.5 kDa, respectively). The PilA and PilB bands running just below them have apparent molecular masses of 62 and 72 kDa, respectively. We cannot conclude from these data if N-terminal proteolysis and/or other posttranslational modifications are responsible for the presence of these additional electrophoretic species, together with those seen between 48 and 27 kDa of the protein marker. The apparent molecular mass of SurG-GFP is about 48 kDa (calculated molecular mass, 54 kDa), a difference which is not surprising for a protein comprising a highly hydrophobic moiety. A band with an apparent molecular mass of 27 kDa corresponding to free GFP is seen only in strains carrying the SurG-GFP fusion. This, together with the downregulation of the full-length SurG-GFP observed after 16 h of growth, demonstrates that the vacuolar fluorescence detected in the mycelia of SurG-GFP strains (Fig. 2A) mostly derives from free GFP and degradation intermediates.
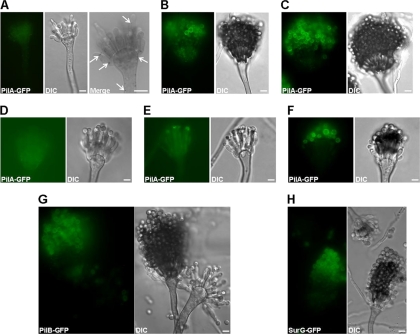
Eisosomal proteins assemble during conidiogenesis.
The presence and colocalization (see below) of the three eisosomal proteins in ungerminated conidia led us to investigate their appearance during conidial development. Figure 5 shows that during conidiogenesis the three proteins are present in late, mature conidia (Fig. 5B and 5C for PilA-GFP, Fig. 5G for PilB-GFP, and Fig. 5H for SurG-GFP). PilA spots can be detected in the stalk of the conidiophore, the vesicle, metulae, and phialides (Fig. 5A). Differently from PilB and SurG, which are seen only in mature, older conidia on the conidiophore, the appearance of PilA in conidia budding from the phialides is quite variable from one conidiophore to the other and even within the same conidiophore. Figure 5D and F shows two extremes in which none and almost all of the emerging conidia express PilA-GFP, respectively, while the micrograph in Fig. 5E shows an intermediate situation, with only some of the budding conidia expressing PilA-GFP. Control strains which did not include any GFP fusion protein do not show any conidiophore or conidiospore fluorescence under the same observation conditions (data not shown).
Fig. 5.
Localization of PilA, PilB, and SurG proteins during conidial development. Representative wide-field fluorescence and DIC pictures of wild-type conidiophores expressing chimeric PilA-GFP (A to F), PilB-GFP (G), and SurG-GFP (H) proteins are shown. The Merge section of panel A shows in inverted black and white an enlarged portion of the same conidial head shown in the PilA-GFP and DIC panels in order to highlight the PilA-GFP spots present in metulae and a budding phialide. Expression of PilB-GFP (G) and SurG-GFP (H) during conidial development is also presented. Young PilA-GFP-expressing conidiophores are shown in panels A and D to F, and mature PilA-GFP-expressing conidiophores are shown in panels B and C. All strains were inoculated on microscope slides covered with MM containing 2% glycerol and 0.8% agarose and grown at 37°C for 2 days. Bars, 5 μm.
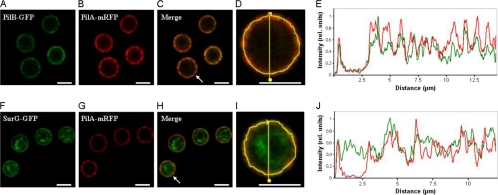
Colocalization of PilA, PilB, and SurG in quiescent conidia.
PilA, PilB, and SurG are localized at the periphery of resting conidia. We thus investigated their colocalization. To this aim we constructed strains carrying PilA-mRFP and PilB-GFP or SurG-GFP. Figure 6 shows colocalization of PilA with PilB and colocalization of PilA with SurG in the periphery of the conidia; there is obviously no colocalization of PilA with the perinuclear fraction of SurG. The PilB fluorescence intensity more consistently follows the peaks of PilA fluorescence than those of SurG. It can be seen from both Fig. 2 and Fig. 6 that the localization pattern of SurG is less discontinuous and punctate than that of PilA and PilB.
Fig. 6.
Colocalization of PilB with PilA and SurG with PilA in resting conidiospores. Representative confocal fluorescence micrographs (A to C and F to H) and fluorescence intensity profiles of PilA (red curves in panels E and J), PilB (green curve in panel E), and SurG (green curve in panel J) are shown. The fluorescence intensity, plotted along the yellow line in panel D (a magnification of panel C) and panel I (a magnification of panel H) that runs through both the inside and the periphery of the arrowed cell in panels C and H, respectively, was calculated using ImageJ software and normalized to the maximum value. Bars, 5 μm.
Phenotypic characterization of mutants with deletions. (i) Growth phenotypes.
We have constructed strains with pilA, pilB, and surG deletions (see Materials and Methods). A strain with both pilA and pilB deletions was also constructed. No growth phenotype at 25°C, 37°C, and 42°C was seen for any of the strains with deletions on either complete or minimal medium supplemented with urea, ammonium, or nitrate as the sole nitrogen source (data not shown). Moreover, conidia from strains with a pilA, pilB, or surG deletion exhibited swelling and polarity establishment (time of germination tube appearance) indistinguishable from that of a wild-type strain at 25°C, 37°C, and 42°C. This was seen after incubation of conidia in minimal medium supplemented with urea as the sole nitrogen source for 2 h, 4 h, and 6 h under the conditions described above and observation of differential interference contrast (DIC) images using a phase-contrast fluorescent microscope (data not shown). As eisosomal proteins are localized in quiescent conidia, we investigated whether they are important for conidial survival. The conidial survival for strains with a pilA, pilB, or surG deletion was not significantly different from that for the wild-type conidia after incubation of freshly harvested conidia for 4 h at 4°C (<100%), 25°C (<60 to 70%), and 45°C (<60 to 75%). Strains with deletions were checked for sensitivity to caffeine, ethanol, SDS, Congo red, CaCl2, and the antifungal drugs caspofungin and itraconazole. Figure 7 shows that pilAΔ mutants, pilAΔ pilBΔ double mutants, and surGΔ mutants but not pilBΔ strains show moderate resistance to 15 μM itraconazole. The itraconazole resistance phenotype cosegregates in crosses with the genetic markers riboB and pyroA, used to interrupt the pilA and surG genes, respectively (see Materials and Methods; data not shown). No other phenotype of resistance or hypersensitivity was observed.
Fig. 7.
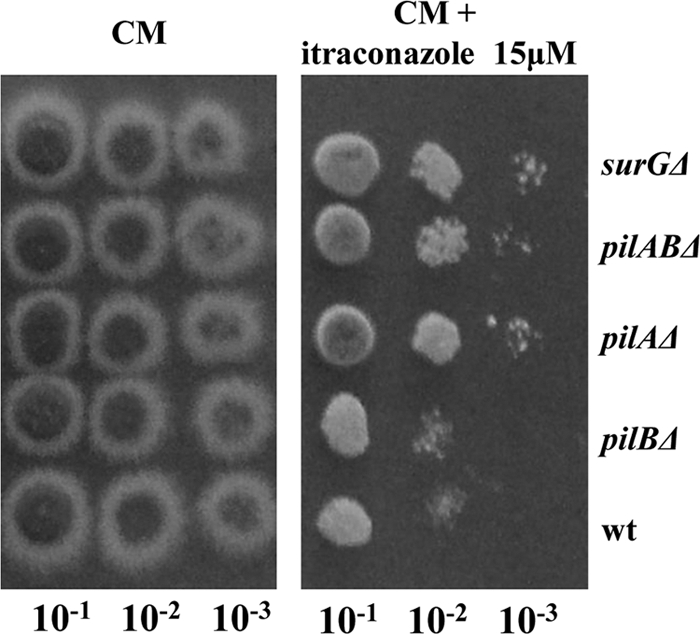
Itraconazole resistance of pilAΔ, pilABΔ, and surGΔ strains. Cotton-filtered pilAΔ, pilBΔ, pilABΔ, surGΔ, and wild-type conidiospore suspensions were prepared in PBS; counted using a Neubauer counting chamber and found to have a concentration of ∼106 conidia/ml; and then plated (5 μl) as 10−1, 10−2, and 10−3 serial dilutions on CM in the absence or the presence of 15 μM itraconazole. The plates were incubated at 37°C for 3 days.
(ii) Cell localization patterns and processes.
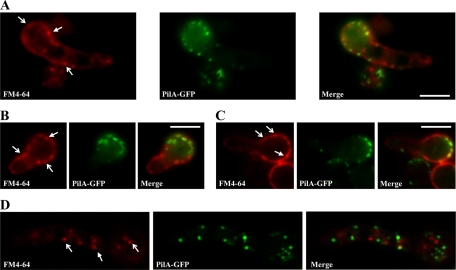
PilA (but not PilB or SurG) is present in punctate structures in mycelia. The presence of ammonium results in endocytosis in the germlings and mycelia of a number of transporters involved in the utilization of nitrogen sources (5, 35, 49). We have investigated whether the ammonium-elicited endocytosis of the dicarboxylic amino acid transporter AgtA (5) was affected in a strain with the pilA deletion. The expression of AgtA is identical in pilA+ and pilAΔ strains (data not shown). The endocytosis of AgtA-GFP, checked at time intervals of 30, 60, and 120 min after addition of ammonium, is identical in pilA+ and pilAΔ strains, within the limits of the confocal microscopic observation, being completed at 120 min in both strains (results not shown). Similarly, the rate of endocytosis of the lipophilic fluorochrome FM4-64 is no different between a pilA+ strain and a pilAΔ strain, within the limits of the epifluorescence microscopic observation (data not shown). Moreover, deletion of PilA does decrease either the uptake of FM4-64 or its early internalization pattern. As reported by Peñalva (36), the earliest FM4-64 internalization intermediates are cortical punctate structures. Figure 8 shows that these structures do not usually colocalize with PilA in germlings. Such colocalization that could be seen may well be coincidental. Thus, PilA foci are not obligatory endocytic portals for this lipid marker, as has been proposed for eisosomes in S. cerevisiae (52).
Fig. 8.
Representative epifluorescence (A to C) and confocal (D) images of a PilA-GFP strain labeled with FM4-64. Samples were taken after the dye was loaded and the samples were placed on ice for 5 min (A). Note the cortical punctate structures indicated by arrows: forked headed arrows indicate FM4-64 internalization sites free of PilA, while triangular headed arrows indicate FM4-64 internalization sites colocalizing with PilA.
We have checked if any of the strains with a deletion is affected by the distribution of filipin, a polyene antibiotic which selectively stains ergosterol and which can be detected by its blue fluorescence under UV light. No changes in filipin binding were seen in young mycelia of strains with the pilA and surG deletions (data not shown). In these strains, the strong filipin staining at the tip of the hyphae is not altered. However, preliminary results strongly suggest that the distribution of filipin staining is affected in ungerminated conidia by the deletion of either pilA or surG but not by the deletion of pilB (not shown).
(iii) Effects on eisosome assembly.
We have checked the distribution of SurG-GFP and PilB-GFP in strains with the pilA deletion and PilA-GFP and PilB-GFP in strains with the surG deletion. Figure 9A shows that in pilAΔ ungerminated conidia (0 h), PilB-GFP patches become larger and more distinctly separated from each other. After 5 h of isotropic growth, PilB is distributed in the cytoplasm as it is in the wild type, with fewer patches persisting at the conidial periphery. In a pilAΔ background, ungerminated conidia (0 h) show a significant decrease of SurG-GFP peripheral patches, while the inner perinuclear ring remains intact, indicating that PilA is required for proper localization of SurG at the conidial periphery. In the absence of SurG (Fig. 9B), the peripheral targeting and distribution of PilB-GFP are drastically affected, with the protein being localized to a few bright clusters at the periphery of ungerminated conidia, which project into the cytoplasm, and to a diffuse cytoplasmic pool, which becomes more evident after 5 h of isotropic growth. At this time point, PilB-GFP clusters not only are peripheral but also appear in the cytoplasm. On the other hand, deletion of SurG does not affect the localization of PilA-mRFP.
DISCUSSION
Our own data (see the data in the supplemental material) and those of others (32; A. Olivera-Couto and P. Aguilar, unpublished data) establish the universal presence of homologues of Pil1 and Lsp1 in the ascomycetes. The sequence divergence between the PilA and PilB clades of the Pezizomycotina, including differences in the conservation of putative phosphorylation sites (data not shown), and the presence of a second coiled-coil domain in PilB are associated with a different fate in the course of development in A. nidulans. While Pil1 and Lsp1 are integral components of eisosomes in all stages of the S. cerevisiae cell cycle, in A. nidulans, PilB shows colocalization with PilA only in conidia. The eisosomes of the A. nidulans conidiospore can be considered equivalent to those of S. cerevisiae, as they contain both Pil paralogues and SurG, the orthologue of Sur7 (see below). However, there are some intriguing differences between the S. cerevisiae cellular eisosomes and those of the conidiospores of A. nidulans. In S. cerevisiae there is about 1 eisosome/3 μm2 of surface area, and they are well separated from each other at a distance of >0.5 μm (28). In the A. nidulans conidiospore, eisosomes are more tightly packed and seem to touch each other; thus, they cannot be easily counted (see Fig. S2 in the supplemental material). The peripheral localization of SurG does not match exactly the localization of PilA in conidia but shows a more continuous distribution (Fig. 6). The perinuclear ring of SurG has not been reported for either S. cerevisiae or C. albicans.
In S. cerevisiae, Pil1 but not Lsp1 is essential for proper eisosome assembly (28, 53). In contrast to S. cerevisiae, in A. nidulans the absence of PilA does not markedly affect the localization of its paralogue, PilB, in conidia. In the absence of PilA, PilB patches are still uniformly distributed at the conidial periphery, but they are larger and more widely spaced than in the wild type. In S. cerevisiae, Sur7 and Lsp1 are similarly affected by a pil1 deletion. In A. nidulans, the distribution of SurG in a pilAΔ background (see Fig. 9 and Results section) is drastically altered, but the scanty SurG patches are very different from the large peripheral Sur7 clusters seen in pil1 strains. A deletion of SUR7 in S. cerevisiae has no effect on the localization of Pil1 and Lsp1. This also applies to strains with additional deletions from the two paralogues, which cluster with Sur7 in the phylogenetic tree (see Fig. S1 in the supplemental material). Similar results were obtained for the deletion of Sur7 in C. albicans (3). The situation is quite different in A. nidulans. While the distribution of PilA is not affected in surGΔ strains, that of PilB is drastically affected. The distribution of PilB in surGΔ strains is partly reminiscent, in quiescent conidia, of the distribution of Lsp1 in pil1Δ strains. That SurG is necessary for PilB localization is also consistent with the localization of PilB to the cytoplasm of wild-type hyphae, where SurG is sequestered in the vacuole and endosomes and, consequently, may not be available to interact with PilB. Thus, the strong similarity of eisosomal components in S. cerevisiae cells and A. nidulans conidiospores is not correlated with the identical interactions leading to their assembly.
Eisosomes have been defined as portals of endocytosis (52). However, the data presented in this article imply that if they play a role in endocytosis in A. nidulans, this role is minor or limited to specific cargos, which remain unidentified. Neither the endocytosis of FM4-64 nor the ammonium-elicited endocytosis of the AgtA (dicarboxylic amino acid transporter [5]; data not shown) or PrnB (the major proline transporter [41]; data not shown) are visibly affected in mycelia by a deletion of pilA, encoding the only eisosomal protein present in discrete foci in hyphae. It should be noted that although the AgtA and PrnB proteins of A. nidulans and the Hxt2 protein of S. cerevisiae are transporters with a homogeneous distribution in the plasma membrane, upon endocytosis Hxt2 accumulates in discrete plasma membrane-associated foci (52), while the two A. nidulans proteins do not (5).
Endocytosis in A. nidulans has been investigated in mycelia (36), where it is involved in the cycling of transporters (5, 16, 35, 49) and in sensing the external pH (19, 38, 50), but has not been investigated in the process of conidial germination; and the possibility remains open that the yeast-like eisosomes of conidiospores have a role in early, germination-related endocytosis events. The endocytic pathway is nearly essential in A. nidulans. Deletion of components of the endosomal sorting complex required for transport III (ESCRT-III; Vps24 [38] and Vps20, Vps32, and Vsp36 [A. M. Calcagno-Pizzarelli and H. N. Arst, personal communication) results in a drastic impairment of growth. This contrasts with the absence of any obvious growth phenotype in surGΔ, pilAΔ, and pilBΔ strains. A possible explanation for this apparent paradox is that, as proposed for S. cerevisiae, endocytosis could proceed by two independent pathways, one of which would not involve eisosomes (18).
A vexing result, common to S. cerevisiae and A. nidulans, is the absence of any drastic phenotype in deletion mutants of the eisosomal soluble proteins (Pil1/Lsp1/PilA/PilB) or membrane proteins (Sur7/SurG). We did not observe any growth phenotype in any media or at any temperature tested for strains with pilA, pilB, pilA pilB, or surG deletions. Nevertheless it was reported at a recent meeting that a deletion of ANID_04615.1 (surG) results in growth impairment on minimal medium with nitrate as the nitrogen source at 37°C and 42°C, an impairment that is seen only on complete medium at 42°C (8a; D. Chung and B. Shaw, personal communication). We did not observe this phenotype on minimal medium using a variety of nitrogen sources, including nitrate, or on complete medium at any temperature. The phenotype did not appear after outcrossing our strain with the deletion, which eliminates the possibility that our strain could carry a suppressor of the growth phenotype. We do not know the reason for this difference, which merits investigation. However, a growth phenotype was observed for strains with the surG deletion and the pilA deletion. This is the mild but clear resistance to itraconazole which cosegregates in crosses with each deletion. This is surprising, as deletions of SUR7 in Candida albicans result in marked hypersensitivity to fluconazole (3), another triazole antifungal agent to which Aspergillus species are tolerant (33). This resistance probably operates at the level of conidial survival or germination, and it may be in some way related to the mislocalization of filipin staining in conidia but not in mycelia both in strains with the surG deletion and in strains with the pilA deletion. Triazole drugs act by inhibiting the cytochrome P450-dependent conversion of lanosterol to ergosterol. Filipin is a polyene macrolide antifungal agent that selectively binds and stains ergosterol at the plasma membrane.
One of the surprising results from this work is that in A. nidulans, S. cerevisiae-like eisosomes assemble during conidial formation and disassemble during germination, resulting in germlings and mycelia in eisosome-like punctate structures comprising PilA and a novel cellular distribution (cytosolic and vacuolar, respectively) for PilB and SurG. As no specific conidial survival phenotype was found in strains with eisosomal component deletions, this developmentally regulated distribution remains baffling. EglD, a putative endoglucanase comprising an expansin-like domain, also localizes at the periphery of ungerminated conidia (7). It would be of interest to study whether this localization implies a topological or functional association with eisosomes. In mycelia, PalI and PalH, involved in pH sensing, have been reported to form membrane-associated punctate structures (8). The connection between PilA and these proteins is under investigation.
The work presented above is a first study of eisosomal structure and function in a model filamentous ascomycete. The universal presence of eisosomal proteins in the ascomycetes and probably in other fungal phyla (3, 32; Olivera-Couto and Aguilar, unpublished; C. Scazzocchio, unpublished data), coupled with the paucity and diversity of the phenotypes of the deletion mutants observed in three different fungal species (S. cerevisiae, C. albicans, and A. nidulans) and the striking developmental pattern of eisosomal protein distribution seen in A. nidulans, continues to be an unsolved paradox. The role of the Meu14 protein, a relative of the Pil1/PilA proteins in the meiosis of S. pombe, suggests that eisosomal proteins may have acquired entirely new functions in different ascomycete phylogenetic groups. Eisosomes, as defined in S. cerevisiae, are present within the asexual cycle only in the conidia of A. nidulans (see also reference 29), positing the question of their functional significance in this developmental stage. The punctate structures seen in mycelia can be considered to define a new class of assemblies comprising only one Pil paralogue. Thus, it would be of interest to explore in this filamentous ascomycete the developmental fate of other conserved proteins known to share the S. cerevisiae MCC localization pattern.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dawoon Chung and Brian Shaw for open discussion relating to the phenotype found in their laboratory for an isolate with a surG deletion, Pablo Aguilar and Agustina Olivera-Couto for sharing unpublished data, and G. Diallinas for laboratory facilities. Specifically, C.S. thanks Agustina Olivera-Couto for very helpful insights concerning the phylogeny of the Pil-like proteins and Andrés Iriarte for help with interpretation of phylogenies. T.S. thanks Miguel Peñalva for laboratory facilities, helpful discussions, and encouragement at the onset of this work.
This work was supported by research grants from the NCSR Demokritos to I.V. and IKY to C.G.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 6 August 2010.
REFERENCES
- 1.Abenza J. F., Pantazopoulou A., Rodríguez J. M., Galindo A., Peñalva M. A. 2009. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic 10:57–75 [DOI] [PubMed] [Google Scholar]
- 2.Alvarez F. J., Konopka J. B. 2007. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in. Candida albicans. Mol. Biol. Cell 18:965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez F. J., Douglas L. M., Rosebrock A., Konopka J. B. 2008. The Sur7 protein regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol. Biol. Cell 19:5214–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez F. J., Douglas L. M., Konopka J. B. 2009. The Sur7 protein resides in punctate membrane subdomains and mediates spatial regulation of cell wall synthesis in Candida albicans. Commun. Integr. Biol. 2:76–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Anisimova M., Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539–552 [DOI] [PubMed] [Google Scholar]
- 5.Apostolaki A., Erpapazoglou Z., Harispe L., Billini M., Kafasla P., Kizis D., Peñalva M. A., Scazzocchio C., Sophianopoulou V. 2009. AgtA, the dicarboxylic amino acid transporter of Aspergillus nidulans, is concertedly down-regulated by exquisite sensitivity to nitrogen metabolite repression and ammonium-elicited endocytosis. Eukaryot. Cell 8:339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araujo-Bazán L., Peñalva M. A., Espeso E. A. 2008. Preferential localization of the endocytic internalization machinery to hyphal tips underlies polarization of the actin cytoskeleton in Aspergillus nidulans. Mol. Microbiol. 67:891–905 [DOI] [PubMed] [Google Scholar]
- 7.Bouzarelou D., Billini M., Roumelioti K., Sophianopoulou V. 2008. EglD, a putative endoglucanase, with an expansin like domain is localized in the conidial cell wall of Aspergillus nidulans. Fungal Genet. Biol. 45:839–850 [DOI] [PubMed] [Google Scholar]
- 7a.Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 8.Calcagno-Pizarelli A. M., Negrete-Urtasun S., Denison S. H., Rudnicka J. D., Bussink H. J., Múnera-Huertas T., Stanton L., Hervás-Aguilar A., Espeso E. A., Tilburn J., Arst H. N., Jr., Peñalva M. A. 2007. Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot. Cell 6:2365–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Chung D., Shaw B. 2009. Abstr. 25th Fungal Genet. Meet., abstr. 322. [Google Scholar]
- 9.Church G. M., Gilbert W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. U. S. A. 81:1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cove D. J. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113:51–56 [DOI] [PubMed] [Google Scholar]
- 11.Delcasso-Tremousaygue D., Grellet F., Panabieres F., Ananiev E. D., Delseny M. 1988. Structural and transcriptional characterization of the external spacer of a ribosomal RNA nuclear gene from a higher plant. Eur. J. Biochem. 172:767–776 [DOI] [PubMed] [Google Scholar]
- 12.Diallinas G. 2007. Aspergillus transporters, p. 301–310 InGoldman G., Osmani S., (ed.), The aspergilli: genomics, medicine, biotechnology and research methods. CRC Press, Boca Raton, FL [Google Scholar]
- 13.Erpapazoglou Z., Kafasla P., Sophianopoulou V. 2006. The product of the SHR3 orthologue of Aspergillus nidulans has restricted range of amino acid transporter targets. Fungal Genet. Biol. 43:222–233 [DOI] [PubMed] [Google Scholar]
- 14.Forment J. V., Flipphi M., Ramon D., Ventura L., Maccabe A. P. 2006. Identification of the mstE gene encoding a glucose-inducible, low affinity glucose transporter in Aspergillus nidulans. J. Biol. Chem. 281:8339–8346 [DOI] [PubMed] [Google Scholar]
- 15.Fröhlich F., Moreira K., Aguilar P. S., Hubner N. C., Mann M., Walter P., Walther T. C. 2009. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J. Cell Biol. 185:1227–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gournas C., Amillis S., Vlanti A., Diallinas G. 2010. Transport-dependent endocytosis and turnover of a uric acid-xanthine permease. Mol. Microbiol. 75:246–260 [DOI] [PubMed] [Google Scholar]
- 17.Grossmann G., Opekarová M., Malinsky J., Weig-Meckl I., Tanner W. 2007. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 26:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grossmann G., Malinsky J., Stahlschmidt W., Loibl M., Weig-Meckl I., Frommer W. B., Opekarová M., Tanner W. 2008. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 183:1075–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herranz S., Rodríguez J. M., Bussink H. J., Sánchez-Ferrero J. C., Arst H. N., Jr., Peñalva M. A., Vincent O. 2005. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. U. S. A. 102:12141–12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill T. W., Loprete D. M., Momany M., Ha Y., Harsch L. M., Livesay J. A., Mirchandani A., Murdock J. J., Vaughan M. J., Watt M. B. 2006. Isolation of cell wall mutants in Aspergillus nidulans by screening for hypersensitivity to calcofluor white. Mycologia 98:399–409 [DOI] [PubMed] [Google Scholar]
- 21.Kafasla P., Frillingos S., Sophianopoulou V. 2007. The proline permease of Aspergillus nidulans: functional replacement of the native cysteine residues and properties of a cysteine-less-transporter. Fungal Genet. Biol. 44:615–626 [DOI] [PubMed] [Google Scholar]
- 22.Lamarre C., Sokol S., Debeaupuis J. P., Henry C., Lacroix C., Glaser P., Coppée J. Y., François J. M., Latgé J. P. 2008. Transcriptomic analysis of the exit from dormancy of Aspergillus fumigatus conidia. BMC Genomics 16:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X. M., Momsen M. M., Brockman H. L., Brown R. E. 2003. Sterol structure and sphingomyelin acyl chain length modulate lateral packing elasticity and detergent solubility in model membranes. Biophys. J. 85:3788–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockington R. A., Sealy-Lewis H. M., Scazzocchio C., Davies R. W. 1985. Cloning and characterization of the ethanol utilization regulon in Aspergillus nidulans. Gene 33:137–149 [DOI] [PubMed] [Google Scholar]
- 25.Luo G., Gruhler A., Liu Y., Jensen O. N., Dickson R. C. 2008. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 283:10433–10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malínská K., Malínský J., Opekarová M., Tanner W. 2003. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell 14:4427–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malínská K., Malínský J., Opekarová M., Tanner W. 2004. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 117(Pt. 25):6031–6041 [DOI] [PubMed] [Google Scholar]
- 28.Moreira K. E., Walther T. C., Aguilar P. S., Walter P. 2009. Pil1 controls eisosome biogenesis. Mol. Biol. Cell 20:809–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh Y. T., Ahn C. S., Kim J. G., Ro H. S., Lee C. W., Kim J. W. 2010. Proteomic analysis of early phase of conidia germination in Aspergillus nidulans. Fungal Genet. Biol. 47:246–253 [DOI] [PubMed] [Google Scholar]
- 30.Ohtaka A., Okuzaki D., Saito T. T., Nojima H. 2007. cp4, a meiotic coiled-coil protein, plays a role in F-actin positioning during Schizosaccharomyces pombe meiosis. Eukaryot. Cell 6:971–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okuzaki D., Satake W., Hirata A., Nojima H. 2003. Fission yeast meu14+ is required for proper nuclear division and accurate forespore membrane formation during meiosis II. J. Cell Sci. 116:2721–2735 [DOI] [PubMed] [Google Scholar]
- 32.Olivera-Couto A. 2009. Búsqueda de estructuras homólogas a los eisosomas de Saccharomyces cerevisiae en eucariotas superiores. Tesis de grado; Facultad de Ciencias, Universidad de la República, Montevideo, Uruguay [Google Scholar]
- 33.Osherov N., Kontoyiannis D. P., Romans A., May G. S. 2001. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14alpha-demethylase gene, pdmA. J. Antimicrob. Chemother. 48:75–81 [DOI] [PubMed] [Google Scholar]
- 34.Padovan A. C., Sanson G. F., Brunstein A., Briones M. R. 2005. Fungi evolution revisited: application of the penalized likelihood method to a Bayesian fungal phylogeny provides a new perspective on phylogenetic relationships and divergence dates of Ascomycota groups. J. Mol. Evol. 60:726–735 [DOI] [PubMed] [Google Scholar]
- 35.Pantazopoulou A., Lemuh N. D., Hatzinikolaou D. G., Drevet C., Cecchetto G., Scazzocchio C., Diallinas G. 2007. Differential physiological and developmental expression of the UapA and AzgA purine transporters in Aspergillus nidulans. Fungal Genet. Biol. 44:627–640 [DOI] [PubMed] [Google Scholar]
- 36.Peñalva M. A. 2005. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet. Biol. 42:963–975 [DOI] [PubMed] [Google Scholar]
- 37.Pontecorvo G., Roper J. A., Hemmons L. M., MacDonald K. D., Bufton A. W. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141–238 [DOI] [PubMed] [Google Scholar]
- 38.Rodríguez-Galán O., Galindo A., Hervás-Aguilar A., Arst H. N., Jr., Peñalva M. A. 2009. Physiological involvement in pH signaling of Vps24-mediated recruitment of Aspergillus PalB cysteine protease to ESCRT-III. J. Biol. Chem. 284:4404–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, Cold Spring Harbor, NY [Google Scholar]
- 40.Sanchez-Ferrero J. C., Peñalva M. A. 2007. Endocytosis. InOsmani S., Goldman G. (ed.), The aspergilli: genomics, medical applications, biotechnology, and research methods. CRC Press, Boca Raton, FL [Google Scholar]
- 41.Sophianopoulou V., Scazzocchio C. 1989. The proline transport protein of Aspergillus nidulans is very similar to amino-acid transporters of Saccharomyces cerevisiae. Mol. Microbiol. 3:705–714 [DOI] [PubMed] [Google Scholar]
- 42.Spatafora J. W., Sung G. H., Johnson D., Hesse C., O'Rourke M., Serdani B., Spotts R., Lutzoni F., Hofstetter V., Miadlikowska J., Reeb V., Gueidan C., Fraker E., Lumbsch T., Lücking R., Schmitt I., Hosaka K., Aptroot A., Roux C., Miller A. N., Geiser D. M., Hafellner J., Hestmark G., Arnold A. E., Büdel B., Rauhut A., Hewitt D., Untereiner W. A., Cole M. S., Scheidegger C., Schultz M., Sipman H., Schoch C. L. 2006. A five-gene phylogeny of Pezizomycotina. Mycologia 98:1018–1028 [DOI] [PubMed] [Google Scholar]
- 43.Strádalová V., Stahlschmidt W., Grossmann G., Blazíková M., Rachel R., Tanner W., Malinsky J. 2009. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J. Cell Sci. 122:2887–2894 [DOI] [PubMed] [Google Scholar]
- 44.Szewczyk E., Nayak T., Oakley C. E., Edgerton H., Xiong Y., Taheri-Talesh N., Osmani S. A., Oakley B. R. 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1:3111–3120 [DOI] [PubMed] [Google Scholar]
- 45.Tavoularis S., Scazzocchio C., Sophianopoulou V. 2001. Functional expression and cellular localization of a green fluorescent protein-tagged proline transporter in Aspergillus nidulans. Fungal Genet. Biol. 33:115–125 [DOI] [PubMed] [Google Scholar]
- 46.Tavoularis S. N., Tazebay U. H., Diallinas G., Sideridou M., Rosa A., Scazzocchio C., Sophianopoulou V. 2003. Mutational analysis of the major proline transporter (PrnB) of Aspergillus nidulans. Mol. Membr. Biol. 20:285–297 [DOI] [PubMed] [Google Scholar]
- 47.Tilburn J., Scazzocchio C., Taylor G. G., Zabicky-Zissman J. H., Lockington R. A., Davies R. W. 1983. Transformation by integration in Aspergillus nidulans. Gene 26:205–221 [DOI] [PubMed] [Google Scholar]
- 48.Valdez-Taubas J., Diallinas G., Scazzocchio C., Rosa A. L. 2000. Protein expression and subcellular localization of the general purine transporter UapC from Aspergillus nidulans. Fungal Genet. Biol. 30:105–113 [DOI] [PubMed] [Google Scholar]
- 49.Valdez-Taubas J., Harispe L., Scazzocchio C., Gorfinkiel L., Rosa A. L. 2004. Ammonium-induced internalisation of UapC, the general purine permease from Aspergillus nidulans. Fungal Genet. Biol. 41:42–51 [DOI] [PubMed] [Google Scholar]
- 50.Vincent O., Rainbow L., Tilburn J., Arst H. N., Jr., Peñalva M. A. 2003. YPXL/I is a protein interaction motif recognized by Aspergillus PalA and its human homologue, AIP1/Alix. Mol. Cell. Biol. 23:1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vlanti A., Diallinas G. 2008. The Aspergillus nidulans FcyB cytosine-purine scavenger is highly expressed during germination and in reproductive compartments and is downregulated by endocytosis. Mol. Microbiol. 68:959–977 [DOI] [PubMed] [Google Scholar]
- 52.Walther T. C., Brickner J. H., Aguilar P. S., Bernales S., Pantoja C., Walter P. 2006. Eisosomes mark static sites of endocytosis. Nature 439:998–1003 [DOI] [PubMed] [Google Scholar]
- 53.Walther T. C., Aguilar P. S., Fröhlich F., Chu F., Moreira K., Burlingame A. L., Walter P. 2007. Pkh-kinases control eisosome assembly and organization. EMBO J. 26:4946–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young M. E., Karpova T. S., Brügger B., Moschenross D. M., Wang G. K., Schneiter R., et al. 2002. The Sur7 family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol. Cell. Biol. 22:927–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zickler D. 2006. Meiosis in mycelial fungi. InKües U., Fischer R. (ed.), The mycota. I. Growth, differentiation and sexuality. Springer-Verlag, Berlin, Germany [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.