Abstract
AprA and CfaD are secreted proteins that function as autocrine signals to inhibit cell proliferation in Dictyostelium discoideum. Cells lacking AprA or CfaD proliferate rapidly, and adding AprA or CfaD to cells slows proliferation. Cells lacking the ROCO kinase QkgA proliferate rapidly, with a doubling time 83% of that of the wild type, and overexpression of a QkgA-green fluorescent protein (GFP) fusion protein slows cell proliferation. We found that qkgA− cells accumulate normal levels of extracellular AprA and CfaD. Exogenous AprA or CfaD does not slow the proliferation of cells lacking qkgA, and expression of QkgA-GFP in qkgA− cells rescues this insensitivity. Like cells lacking AprA or CfaD, cells lacking QkgA tend to be multinucleate, accumulate nuclei rapidly, and show a mass and protein accumulation per nucleus like those of the wild type, suggesting that QkgA negatively regulates proliferation but not growth. Despite their rapid proliferation, cells lacking AprA, CfaD, or QkgA expand as a colony on bacteria less rapidly than the wild type. Unlike AprA and CfaD, QkgA does not affect spore viability following multicellular development. Together, these results indicate that QkgA is necessary for proliferation inhibition by AprA and CfaD, that QkgA mediates some but not all of the effects of AprA and CfaD, and that QkgA may function downstream of these proteins in a signal transduction pathway regulating proliferation.
Physiological processes that define and maintain the sizes of tissues are poorly understood. Although a number of characterized gene products negatively regulate the sizes of tissues (21, 23), the mechanism by which the activities of such gene products are controlled is unclear. One potential mechanism for tissue size regulation consists of tissue-specific autocrine signals that inhibit proliferation in a concentration-dependent manner (18). Since the extracellular concentration of such factors increases as a function of cell density and/or cell number, the proliferation-inhibiting function of these factors can limit tissue size. Considerable evidence for such factors has been reported. For instance, full hepatectomy in one of two rats with conjoined circulatory systems stimulated proliferation in the intact liver of the conjoined rat, suggesting the existence of a systemic factor produced by the liver that inhibits the proliferation of hepatocytes (16). However, only a small number of factors with analogous functional roles, such as myostatin, which regulates skeletal muscle size (30), and Gdf11, which negatively regulates neurogenesis in the olfactory epithelium (38), have been identified. The mechanisms by which such signals inhibit proliferation are not well understood. As such autocrine signals may serve to limit tumor growth (14, 20), elucidation of the identities of such factors and their associated signal transduction pathways may yield novel cancer therapies.
We have identified two such autocrine proliferation-repressing signals in the social amoeba Dictyostelium discoideum, a genetically and biochemically tractable model organism. The proteins AprA and CfaD are secreted by Dictyostelium and inhibit the proliferation of Dictyostelium cells in a concentration-dependent manner (4, 12). Cells in which the genes encoding either AprA or CfaD have been disrupted by homologous recombination proliferate rapidly, and cells overexpressing AprA or CfaD proliferate slowly (4, 11). Adding recombinant AprA (rAprA) or recombinant CfaD (rCfaD) to cells slows proliferation, demonstrating that these proteins function as extracellular signals (4, 12). In addition to exhibiting rapid proliferation, aprA− and cfaD− cells exhibit a multinucleate phenotype, strongly suggesting that AprA and CfaD are negative regulators of mitosis (4, 11). aprA− cells are insensitive to the proliferation-inhibiting effects of CfaD (12), and cfaD− cells are insensitive to AprA (4), indicating the necessity of both genes for proliferation inhibition and suggesting a common proliferation-inhibiting mechanism. The G protein complex subunits Gα8, Gα9, and Gβ are necessary for proliferation inhibition by AprA, and the addition of recombinant AprA to purified cell membranes increases binding of GTP to wild-type and gα9− cell membranes but not gα8− or gβ− membranes, indicating that AprA activates a proliferation-inhibiting signal transduction pathway of which Gα8 and Gβ are components (5). The signal transduction pathway downstream of Gα8 and the associated mechanism of proliferation inhibition are unknown.
Although the selective forces that have maintained functional autocrine proliferation inhibitors in proliferating Dictyostelium cells are unclear, AprA and CfaD may provide an advantage during the multicellular portion of the Dictyostelium life cycle. Upon starvation, Dictyostelium cells secrete pulses of the chemoattractant cyclic AMP, leading to cells streaming toward aggregation centers (15, 27). This process causes the formation of multicellular groups regulated in size by a secreted protein complex that stimulates stream breakup (9, 10). These groups develop into multicellular fruiting body structures composed of a mass of stress-resistant spores supported by an approximately 1-mm-high stalk (24). While the stalk cells inevitably die in an act of apparent altruism (31), the presence of nutrients stimulates spore germination and a continuation of proliferation (13). Following development, aprA− and cfaD− cells form fewer viable spores than the wild type (4, 11), suggesting that AprA and CfaD increase the fitness of Dictyostelium during development.
Like aprA− and cfaD− cells, Dictyostelium cells lacking the ROCO family kinase QkgA have an abnormally rapid proliferation (1). The ROCO protein family is widely conserved and is defined by the presence of a Ras of complex protein (Roc) domain followed by a C terminus of Roc (Cor) domain, which mediates homodimerization (19). In eukaryotes, these domains are commonly followed C terminally by a kinase domain with similarity to the tyrosine kinase-like (TKL) group of kinases (3, 26, 29). In Dictyostelium, other ROCO proteins function in cyclic GMP signaling (8, 35) and cytokinesis (2), and a total of 11 predicted ROCO proteins are present in the genome, 10 of which, including QkgA, encode kinase domains predicted to be catalytically active (17). The human genome encodes two ROCO kinases, which are expressed in a wide range of tissues (25, 40). Little is known regarding the physiological functions of these proteins, although the ROCO protein LRRK2 is implicated in a dominantly inherited form of Parkinson's disease (40) and negatively regulates neurite growth in rat cortical cultures (28).
In this report, we show that, like aprA− and cfaD− cells, qkgA− cells proliferate to a higher cell density than the wild type and tend to be multinucleate. Additionally, we show that qkgA− cells are insensitive to exogenous AprA and CfaD, indicating that QkgA is required for AprA and CfaD signal transduction.
MATERIALS AND METHODS
Cell culture, immunoblotting, AprA/CfaD inhibition assays, and spore viability.
qkgA− cells (1) were a kind gift from Jeff Williams. Ax2 wild-type, qkgA− clone DBS0236839 (1), aprA− clone DBS0235509 (11), and cfaD− clone DBS0302444 (4) cells were grown in HL5 medium (Formedium Ltd., Norwich, England) as previously described (10). To generate cells expressing the QkgA C terminus-green fluorescent protein (GFP) fusion, cells were transformed with the extrachromosomal vector QkgA-pDM323 (36), a gift from Wouter van Egmond and Peter van Haastert, and selected in 15 μg/ml Geneticin. For each transformation, at least two clones were analyzed and had similar proliferative phenotypes. For proliferation curve experiments, all cells were grown in the absence of selective drugs. To compare levels of extracellular AprA and CfaD, cells in axenic shaking culture at 3 × 106 cells/ml or 12 × 106 cells/ml were collected by centrifugation at 3,000 × g, and a sample of the supernatant was boiled in an equivalent volume of 2× loading buffer. Twenty microliters of each sample was run on a 4 to 15% polyacrylamide gel, and Western blots were stained for AprA according to reference 11 or for CfaD according to reference 4. Proliferation inhibition assays were done according to reference 12, with an incubation time of 16 h. For spore viability assays, 107 cells were washed once in 8 ml PDF (20 mM KCl, 9.2 mM K2HPO4, 13.2 mM KH2PO4, 1 mM CaCl2, 2.5 mM MgSO4, pH 6.4), resuspended in 750 μl PDF, deposited on halved black filter pads (8-μm pore size, 47-mm diameter; Millipore) on a pad of Whatman no. 3 filter paper soaked in PDF, and allowed to develop for 48 h. Filters were placed in an Eppendorf tube containing 1 ml PDF plus 0.5% Triton X-100 and briefly vortexed to disaggregate fruiting bodies. The filter was then removed, and cells were incubated in PDF plus 0.5% Triton X-100 for 5 min. Spores were collected by centrifugation at 6,000 × g for 1 min, resuspended in 1 ml PDF, and counted with a hemocytometer. After dilution in PDF, 50 spores were plated in triplicate on SM/5 plates with Klebsiella aerogenes bacteria, and the number of resultant plaques was counted after 4 days to assess the number of viable spores. For measurement of colony diameter as a function of time, cells growing in HL5 medium at densities of 1 × 106 to 3 × 106 cells/ml were serially diluted, mixed with K. aerogenes bacteria, and spread on SM/5 plates. Plates with well-spaced colonies were imaged daily along with a ruler, and the diameters of at least 3 colonies per genotype per day were measured using ImageJ software (32).
Mass and protein determination.
To determine the mass of 107 cells, 5 × 107 cells from shaking cultures in HL5 medium at densities of 2 × 106 to 5 × 106 cells/ml were collected by centrifugation for 3 min at 1,500 × g, and the pellet was resuspended in approximately 1 ml of residual medium and transferred to a microcentrifuge tube. The cells were then collected by centrifugation for 3 min at 3,000 × g. The supernatant was removed, and the cell pellets were weighed. To determine the mass of 107 cells, the measured value was divided by five. To determine the protein content of cells, the pellets were then resuspended to a total volume of 1 ml in PBM (20 mM KH2PO4, 0.01 mM CaCl2, 1 mM MgCl2, pH 6.1, with KOH) and were frozen at −80°C. Following thawing, cells were resuspended by vortexing them, and 100 μl of the lysed cells was mixed with 100 μl of PBM containing 0.2% SDS and 0.2% Triton X-100. Lysates were rotated at room temperature for 10 min, and protein concentrations were measured by a Bio-Rad (Hercules, CA) protein assay by combining 20 μl of lysate sample with 1 ml of 1× Bio-Rad dye. Solutions of 1.0, 0.5, 0.25, and 0.125 mg/ml bovine serum albumin (BSA) in an amount of buffer equivalent to the amount of the lysate were used as standards.
Calculation of doubling times.
To calculate the doubling time (td) of cell cultures during exponential growth, the following equation was used: td = ln(2)/[ln(Pn/P0)/t], where P0 is the initial cell density, Pn is the final cell density, and t is the time interval in hours. Doubling times were calculated during growth from day 1 to day 5 from cultures inoculated at 1 × 105 cells/ml.
Fluorescence microscopy.
DAPI (4′,6-diamidino-2-phenylindole) staining and imaging of nuclei were done as described previously (11). For imaging of cells expressing QkgA-GFP, cells growing in HL5 medium were washed and resuspended in low-fluorescence axenic medium (7), grown overnight in shaking culture, collected by centrifugation at 1,500 × g, and resuspended in low-fluorescence medium at a volume equivalent to that of the starting volume. A drop of cells was placed on a glass slide and imaged with a Zeiss Axioplan II microscope. For deconvolution imaging, cells growing in shaking culture in HL5 medium at densities of 1 × 106 to 4 × 106 cells/ml were allowed to settle on a glass coverslip for 10 min, fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min, permeabilized in 0.2% Triton X-100 in PBS for 2 min, washed thrice in PBS for 5 min, and mounted in Vectashield mounting medium with DAPI (Vector, Burlingame, CA). Samples were then imaged with an Olympus FV1000 microscope and three-dimensionally deconvoluted using Autodeblur software (Media Cybernetics, Bethesda, MD).
Video microscopy and colony edge imaging.
To examine random cell motility, serial dilutions of Dictyostelium cells were mixed with K. aerogenes bacteria in SM broth (per liter, 10 g glucose, 10 g proteose peptone, 1 g yeast extract, 1 g MgSO4·7H2O, 1.9 g KH2PO4, 0.6 g K2HPO4, pH 6.4) and grown in 8-well glass slides overnight. Conditions were selected in which Dictyostelium cells were spaced approximately 40 μm apart and the ratio of bacteria to Dictyostelium cells was approximately 10:1. Cells were then imaged using an inverted microscope with a 10× objective, and cell movement was recorded using a Panasonic CCTV VW-BC200 camera. Distances between the approximate centroids of cells during 5-min intervals were measured and scaled to a stage micrometer to determine random motility values in micrometers. For colony edge imaging, 1-μl volumes of serially diluted Dictyostelium cells in HL5 medium were allowed to settle as a small spot in the middle of the well on 8-well glass slides for 15 min, and then 300 μl of K. aerogenes culture in SM medium was added to the well and cells were grown overnight and subsequently imaged with an inverted microscope using a 20× phase-contrast objective.
Statistics.
All statistics were done with Prism (GraphPad Software, San Diego, CA). Unless otherwise noted, one-way analysis of variance (ANOVA) using Tukey's test was used to compare differences between multiple groups, and t tests were used to compare differences between two groups. Significance was defined as a P value of <0.05.
RESULTS
qkgA− cells show proliferative phenotypes like aprA− and cfaD− cells.
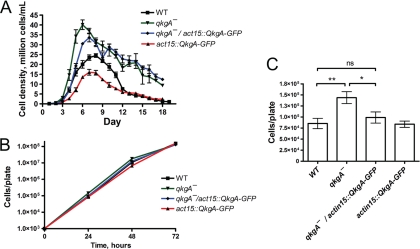
As the loss of functional components of the AprA/CfaD signal transduction pathways may result in an increased rate of proliferation, we have been examining mutants reported to have a fast-proliferation phenotype to determine if the mutants may have defects in AprA and/or CfaD signaling. Abe et al. disrupted the gene encoding the ROCO protein QkgA to validate a novel disruption strategy (1), and the resulting qkgA− cells were observed to proliferate rapidly. We also observed that qkgA− cells had a significantly faster doubling time than wild-type cells and saw that qkgA− cells, like aprA− and cfaD− cells (4, 11), reached a significantly higher stationary density than wild-type cells in axenic culture (Fig. 1A and Table 1). The observed doubling time and stationary density of qkgA− cells were not significantly different from those of aprA− cells examined simultaneously (P > 0.05, one-way ANOVA) (Table 1), further suggesting that AprA and QkgA may function in a common proliferation-inhibiting mechanism.
Fig. 1.
QkgA slows proliferation and lowers the stationary density of cells. (A) Log-phase cells were inoculated into HL5 medium at 1 × 105 cells/ml, and cell densities were measured daily. Values are means ± SEMs (n ≥ 3) for all conditions. WT, wild type. (B) One thousand cells were plated on SM/5 plates with K. aerogenes bacteria, and the total number of cells was determined daily. By 72 h, cells had begun to overgrow the bacteria. Values are mean ± SEMs (n = 4 for all conditions). (C) Cell numbers per plate at 24 h. The absence of error bars indicates that the error was smaller than the plot symbol. *, P < 0.05; **, P < 0.01 (repeated-measures one-way ANOVA, Tukey's test). ns, not significant.
Table 1.
Effect of QkgA on the doubling time and stationary density of cellsa
| Cell type | Doubling time (h) | Maximum observed cell density (106 cells/ml) |
|---|---|---|
| Wild type | 13.2 ± 0.3 | 24.5 ± 0.9 |
| aprA− | 10.9 ± 0.1** | 44.1 ± 1.7** |
| qkgA− | 11.5 ± 0.3* | 40.1 ± 2.4** |
| qkgA−/actin15::qkgA-GFP | 12.7 ± 0.2 | 33.6 ± 2.2* |
| actin15::qkgA-GFP | 16.2 ± 0.9** | 15.8 ± 1.2* |
Doubling times and stationary densities were calculated as described in Materials and Methods. Values are means ± SEMs from five or more independent experiments. *, P < 0.05 (the difference between the value and the wild-type value is significant); **, P < 0.01 (one-way ANOVA, Tukey's test).
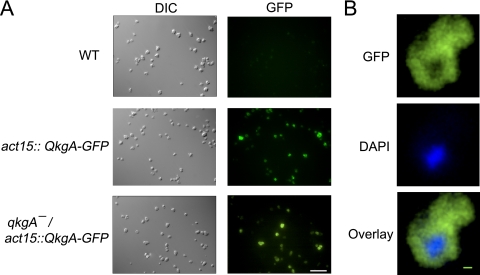
To attempt to rescue the phenotypes of qkgA− cells and to determine the phenotype of overexpression of QkgA, we transformed cells with a QkgA-GFP fusion construct (with GFP at the C terminus of QkgA) under the control of the actin15 promoter (36) and confirmed expression by fluorescence microscopy (Fig. 2). The fusion protein showed what appeared to be a cytosolic or vesicular localization, and no change in localization was observed at high density or in response to rAprA and rCfaD (Fig. 2B and data not shown). Expression of QkgA-GFP in wild-type cells resulted in slow proliferation and a low stationary density compared to those of the wild type (Fig. 1A and Table 1). Expression of QkgA-GFP in qkgA− cells resulted in a reduced rate of proliferation, with cells exhibiting a log-phase doubling time significantly less than that of qkgA− cells (P < 0.05, one-way ANOVA), consistent with recently published data (36). These results support the hypothesis that QkgA is a negative regulator of proliferation.
Fig. 2.
Expression of QkgA-GFP in wild-type and qkgA− cells. (A) Cells of the indicated genotype were grown in low-fluorescence axenic medium for 24 h and then washed once in medium and imaged by fluorescence microscopy using a 40× objective. Scale bar, 50 μm. DIC, differential inference contrast. (B) qkgA−/act15::qgkA-GFP cells grown in HL5 medium were fixed in 4% paraformaldehyde and imaged with a 60× objective. The image was subsequently three-dimensionally deconvoluted. Scale bar, 1 μm.
qkgA− cells exhibit fast proliferation on lawns of bacteria.
In addition to exhibiting fast proliferation in axenic shaking culture, aprA− cells proliferate significantly faster than wild-type cells when grown on a lawn of bacteria (11), although cfaD− cells show no significant difference in proliferation from the wild type under these conditions (4). To determine whether QkgA affects the proliferation of cells grown on bacterial lawns, we spread 1,000 cells on agar with bacteria and measured the total number of cells per plate daily. Unlike with previous results, we saw no lag phase during proliferation on bacteria, possibly due to a larger quantity of bacteria spread with the cells. After 24 h, qkgA− cells had proliferated more than the wild type, and expression of QkgA-GFP in qkgA− cells rescued this increase in proliferation (Fig. 1B and C). Expression of QkgA-GFP in wild-type cells did not significantly slow proliferation on bacteria. The levels of proliferation between genotypes during subsequent 24-hour periods were not significantly different. These results indicate that the absence of QkgA results in fast proliferation on bacteria when cell numbers are low.
qkgA− cells secrete AprA and CfaD.
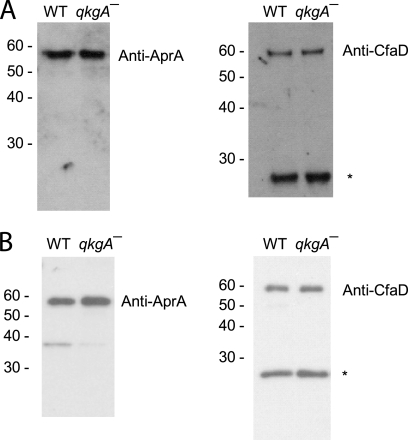
One potential explanation for a fast-proliferation phenotype is a decrease in the extracellular accumulation of AprA or CfaD relative to levels in the wild type. To determine if low extracellular levels of AprA or CfaD might be responsible for the fast proliferation of qkgA− cells, we examined extracellular levels of AprA and CfaD in qkgA− cells. Both AprA and CfaD accumulate in the medium of qkgA− cells at levels comparable to the wild-type levels (Fig. 3), strongly suggesting that the rapid proliferation of qkgA− cells is not due to altered levels of extracellular AprA or CfaD.
Fig. 3.
qkgA− cells secrete AprA and CfaD. Conditioned medium from wild-type and qkgA− cells at log phase (3 × 106 cells/ml) (A) or after log phase (12 × 106 cells/ml) (B) were assayed by Western blotting with anti-AprA antibodies (left) or anti-CfaD antibodies (right). Data are representative of three independent experiments. The asterisk indicates a breakdown product of CfaD. Numbers at the left indicate molecular masses in kDa.
qkgA− cells are insensitive to proliferation inhibition by AprA and CfaD.
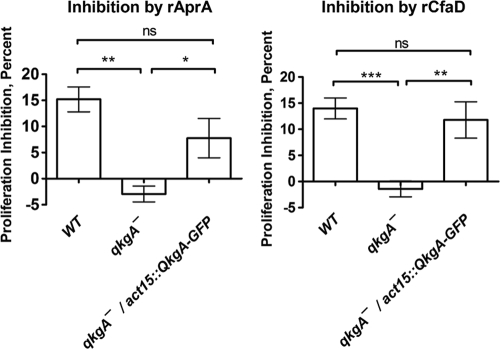
If AprA and/or CfaD signals through QkgA to repress proliferation, we expected that qkgA− cells would show some degree of insensitivity to AprA and/or CfaD. To test this prediction, we incubated proliferating cells with recombinant AprA (rAprA) or rCfaD and determined the percent decrease in cell density compared to that of a buffer control after a 16-hour incubation. Wild-type cells exhibited an approximately 15% decrease in cell density in response to rAprA or rCfaD (Fig. 4), which is on the order of what we have previously observed (4, 5, 12). However, no inhibition of qkgA− cell proliferation by either rAprA or rCfaD was detected. Although rAprA appeared to slightly increase the proliferation of qkgA− cells, paired t tests of cell densities after either rAprA/rCfaD or buffer was added showed no significant differences in cell density between the two conditions for qkgA− cells, indicating that rAprA and rCfaD have no effect on the proliferation of qkgA− cells. Expression of QkgA-GFP in qkgA− cells restored the ability of rAprA or rCfaD to inhibit proliferation, strongly suggesting that the insensitivity of qkgA− cells to rAprA and rCfaD is due specifically to a nonfunctional qkgA gene. These results support the hypothesis that QkgA is required for AprA- and CfaD-mediated inhibition of proliferation.
Fig. 4.
The expression of QkgA-GFP rescues the insensitivity of qkgA− cells to rAprA (left) and rCfaD (right). Proliferating cells were incubated for 16 h with either 1,000 ng/ml rAprA, 600 ng/ml rCfaD, or an equivalent volume of buffer, and cell densities were then determined. The percent inhibition of cell density compared to that of a buffer control is shown. Values are means ± SEMs (n ≥ 4 for all conditions). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (one-way ANOVA, Tukey's test).
qkgA− cells are multinucleate.
aprA− and cfaD− cells exhibit a multinucleate phenotype (4, 11). To determine whether qkgA− cells show this phenotype, we counted the number of nuclei per cell of DAPI-stained log-phase cells. qkgA− cells were significantly less likely to have one nucleus per cell than the wild type and significantly more likely to have either two nuclei or three or more nuclei per cell than the wild type (Table 2). Expression of QkgA-GFP in qkgA− cells rescued this multinucleate phenotype. Overexpression of QkgA-GFP in wild-type cells resulted in significantly fewer binucleate cells than wild-type cells. These results indicate that QkgA negatively regulates the number of nuclei per cell, perhaps by inhibiting mitosis. Additionally, these results show further phenotypic similarities between qkgA− cells and aprA− and cfaD− cells, supporting the hypothesis that QkgA mediates at least some aspects of AprA- and CfaD-induced signaling.
Table 2.
Effect of QkgA on the number of nuclei per cella
| Cell type | % of cells with n nuclei |
||
|---|---|---|---|
| 1 | 2 | 3+ | |
| Wild type | 78 ± 2 | 20 ± 1 | 2 ± 1 |
| qkgA− | 60 ± 2*** | 30 ± 1** | 9 ± 1** |
| qkgA−/actin15::qkgA-GFP | 81 ± 2 | 18 ± 2 | 1 ± 1 |
| actin15::qkgA-GFP | 85 ± 1 | 14 ± 1* | 1 ± 1 |
Log-phase cells were fixed and stained with DAPI, and the number of nuclei per cell for at least 200 cells per genotype was determined by fluorescence microscopy. Values are means ± SEMs from three independent experiments. *, P < 0.05 (the difference between the value and the wild-type value is significant); **, P < 0.01; ***, P < 0.001 (one-way ANOVA, Tukey's test).
QkgA does not negatively regulate cell growth on a per-nucleus basis.
Cell growth, defined as the accumulation of mass or protein, and cell proliferation, the increase in cell number, are often interdependent properties but can be regulated independently (22). Although aprA− and cfaD− cells are more massive than wild-type cells, mass and protein are not accumulated in cells of these genotypes at a rate higher than that of the wild type on a per-nucleus basis, suggesting that AprA and CfaD do not regulate growth (4, 11). To determine the effect of QkgA on the mass and protein content of cells, we examined these properties in log-phase cells. The protein content of wild-type cells was identical to that previously observed (4, 5), although we observed lower cell mass values than were previously seen, which may have been due to our measuring mass in our study using cells in HL5 medium as opposed to cells washed in phosphate buffer or water. Like aprA− and cfaD− cells, qkgA− cells were more massive than wild-type cells, though differences in protein content between genotypes were not significant (Table 3). Expression of QkgA-GFP in qkgA− cells resulted in cells with roughly wild-type levels of cell mass, and overexpression of QkgA-GFP in the wild-type background had no significant effect on cell mass or protein content. On a per-nucleus basis, no significant differences were observed between wild-type and qkgA− cells in mass or protein content.
Table 3.
Effect of QkgA on the mass and protein content of cellsa
| Cell type | Mass (mg)/107 cells | Amt of protein (mg)/107 cells | No. of nuclei/100 cells | Mass (mg)/107 nuclei | Amt of protein (mg)/107 nuclei |
|---|---|---|---|---|---|
| Wild type | 5.18 ± 0.24 | 0.32 ± 0.02 | 125 ± 2 | 4.20 ± 0.21 | 0.25 ± 0.02 |
| qkgA− | 7.15 ± 0.16** | 0.36 ± 0.02 | 152 ± 5* | 4.69 ± 0.19 | 0.23 ± 0.02 |
| qkgA−/act15::qkgA-GFP | 5.54 ± 0.64 | 0.32 ± 0.04 | 120 ± 2 | 4.61 ± 0.53 | 0.27 ± 0.03 |
| actin15::qkgA-GFP | 4.83 ± 0.62 | 0.27 ± 0.02 | 117 ± 1 | 4.15 ± 0.53 | 0.24 ± 0.02 |
Mass and protein content were determined as described in Materials and Methods. The number of nuclei per 100 cells was calculated from the data presented in Table 2. Values are means ± SEMs from three or more independent experiments. *, P < 0.05 (indicates that the difference between the value and the wild-type value is significant); **, P < 0.01 (one-way ANOVA, Tukey's test).
To examine the effect of QkgA on cell growth, we assumed a steady-state average cell size (i.e., that a doubling in cell number results in a doubling of mass, protein, or nuclei) and divided the mass, protein, or nuclear content of cells of different genotypes by the calculated log-phase doubling times to estimate the mass, protein, or nuclear increase per hour. On a per-cell basis, qkgA− cells showed a significantly higher mass accumulation than the wild type, while expression of QkgA-GFP in qkgA− cells caused a mass accumulation like that in the wild type (Table 4). qkgA− cells accumulated nuclei more rapidly than wild-type cells, and overexpression of QkgA-GFP in the wild-type background resulted in a rate of nuclear accumulation lower than that of the wild type. When examined on a per-nucleus basis, the masses and protein accumulations per hour of wild-type and qkgA− cells were not significantly different (P > 0.05, one-way ANOVA). These results suggest that, like AprA and CfaD, QkgA negatively regulates proliferation but does not significantly affect cell growth on a per-nucleus basis.
Table 4.
Effect of QkgA on the mass and protein accumulation of cellsa
| Cell type | Mass (mg)/107 cells/h | Amt of protein (μg)/107 cells/h | No. of nuclei (10−5)/107 cells/h | Mass (mg)/107 nuclei/h | Amt of protein (μg)/107 nuclei/h |
|---|---|---|---|---|---|
| Wild type | 0.39 ± 0.02 | 24 ± 2 | 9.6 ± 0.3 | 0.31 ± 0.02 | 19 ± 1 |
| qkgA− | 0.62 ± 0.02** | 31 ± 2 | 12 ± 0.5** | 0.44 ± 0.02 | 22 ± 1 |
| qkgA−/actin15::qkgA-GFP | 0.44 ± 0.05 | 25 ± 3 | 9.5 ± 0.2 | 0.36 ± 0.04 | 21 ± 3 |
| actin15::qkgA-GFP | 0.30 ± 0.04 | 17 ± 1 | 7.2 ± 0.4* | 0.26 ± 0.03 | 14 ± 1 |
Mass, protein, and nucleus values from Table 3 were divided by the observed doubling times of the respective genotypes. Doubling times were calculated as described in Materials and Methods. Values are means ± SEMs from three or more independent experiments. *, P < 0.05 (the difference between the value and the wild-type value is significant); **, P < 0.01 (one-way ANOVA, Tukey's test).
QkgA does not affect spore viability.
Following development, aprA− and cfaD− cells yield fewer detergent-resistant spores than the wild type (4, 11). We examined the ability of qkgA− cells to generate detergent-resistant spores by allowing equivalent numbers of wild-type and qkgA− cells to develop, by collecting spores, and by plating dilutions of detergent-treated spores. The number of visible phase-dark, ovoid spores collected following development was 102% ± 4% of the input cell number for the wild type and 117% ± 1% for qkgA− cells (mean ± standard error of the mean [SEM], n = 3). Although the difference between the numbers of recovered spores from wild-type and qkgA− cells was significant (P < 0.05, t test), this significance was lost when the values were considered on a per-nucleus basis by dividing the percentage of recovered spores to input cells by the observed number of nuclei per cell for each genotype (as calculated from Table 3), suggesting that this difference was the result of the multinuclearity of qkgA− cells. As judged by plaque number after cells were plated on bacteria, the number of viable spores after detergent treatment was 67% ± 5% of the number of spores plated for wild-type cells and 68% ± 9% for qkgA− cells (mean ± SEM, n = 3). Differences in detergent-resistant spore numbers between wild-type and qkgA− cells were not significant (t test). Using an alternative spore viability assay (11), we also observed no significant difference between wild-type spore viability and qkgA− spore viability (data not shown). These results suggest that QkgA does not affect the development of spores.
Colonies of cells lacking AprA, CfaD, or QkgA expand slowly.
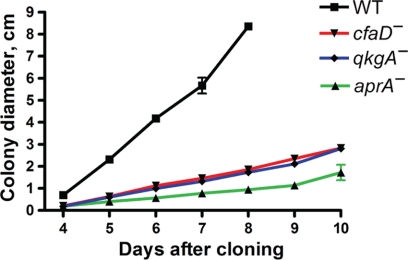
We noticed that, despite a rapid-proliferation phenotype, aprA− and cfaD− cells appeared to expand as a colony on a lawn of bacteria less rapidly than the wild type. To examine this apparent phenotype and to determine if qkgA− cells are similar, we plated serial dilutions of cells on bacterial lawns and measured the sizes of colonies. Under these conditions, the expansion of colonies of aprA− and cfaD− cells was less than that of the wild type (Fig. 5). Additionally, colonies of qkgA− cells showed a rate of expansion less than that of the wild type but similar to those of cfaD− and aprA− cells. These results suggest that, although AprA, CfaD, and QkgA inhibit proliferation, a drawback to not expressing these proteins is a decreased ability of colonies of cells to expand under at least some conditions.
Fig. 5.
AprA, CfaD, and QkgA affect the expansion of colonies on bacterial lawns. Serial dilutions of log-phase cells in shaking culture (1 × 106 to 4 × 106 cells/ml) were mixed with bacteria and spread on SM/5 plates, and the average diameters of well-spaced colonies were determined daily. Values are means ± SEMs (n ≥ 3). The absence of error bars indicates that the error was smaller than the plot symbol. For the wild type, plates were overgrown after day 8, and colony boundaries could not be determined after this point.
As a reduced-colony-expansion phenotype could be due to defects in cell motility, we examined the random motility of cells cultured at low densities with bacteria on glass slides using video microscopy. During a 5-min interval, we observed an average displacement of wild-type, aprA−, cfaD−, and qkgA− cells of 7.6 ± 0.6 μm, 10.4 ± 1.1 μm, 8.6 ± 0.6 μm, and 10.5 ± 0.7 μm, respectively (means ± SEMs from three independent experiments with at least 10 cells per condition). Differences in observed motilities were not significant between any of the genotypes tested (P > 0.05, Tukey's test). These results show that aprA−, cfaD−, and qkgA− cells exhibit random motility comparable to that of the wild type and suggest that the reduced colony expansion observed in these mutants is not due to a defect in cell motility.
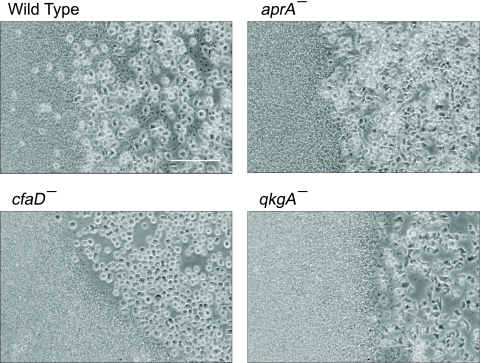
We used a similar experimental setup to examine the morphology of Dictyostelium colonies at the interface of populations of Dictyostelium and bacteria. At the edges of the wild-type colonies, some of the Dictyostelium cells were dispersed into the bacterial lawn, and the boundary between Dictyostelium cells and bacteria was poorly defined (Fig. 6). In contrast, boundaries between bacteria and aprA−, cfaD−, or qkgA− cells were well defined and invasion by Dictyostelium cells into the bacteria was not evident. Together, these results indicate that AprA, CfaD, and QkgA affect the expansion of cell colonies but not the random motility of individual cells.
Fig. 6.
AprA, CfaD, and QkgA affect cell dispersal at the borders of colonies during growth with bacteria. Small spots of wild-type, aprA−, cfaD−, or qkgA− cells were grown overnight in glass culture chambers with K. aerogenes bacteria, and interfaces between bacteria and Dictyostelium cell populations were imaged. Scale bar, 100 μm.
DISCUSSION
Like aprA− and cfaD− cells, qkgA− cells have a rapid-proliferation phenotype (1). We found that qkgA− cells, like aprA− and cfaD− cells, have a high stationary density in shaking culture, multinuclearity and rapid accumulation of nuclei, a rate of growth per nucleus like that of the wild type, and a reduced rate of colony expansion when grown with bacteria. In addition, QkgA is required for the proliferation-inhibiting activity of AprA and CfaD. Together, these results suggest that QkgA may serve as a signal-transducing component downstream of AprA and CfaD.
qkgA− cells proliferate more rapidly than wild-type cells and have a higher stationary-phase density, whereas wild-type cells overexpressing QkgA-GFP have a slower proliferation and a lower stationary-phase cell density. These results suggest that the effects of QkgA on proliferation and stationary density are sensitive to the cellular concentration of QkgA. We saw that qkgA− cells expressing QkgA-GFP proliferate like wild-type cells during log-phase growth, although qkgA−/act15::qkgA-GFP cells proliferate to a higher stationary density than the wild type. It may be the case, therefore, that native QkgA has increased activity at higher cell densities but that the ectopically expressed QkgA-GFP fusion protein does not. Alternatively, QkgA-GFP may be only partially active or not targeted correctly, or, as these experiments were done in the absence of selective drugs, QkgA-GFP expression may be reduced at high cell densities due to plasmid loss.
Like aprA− and cfaD− cells, qkgA− cells tend to be multinucleate. Although a cytokinesis defect could be responsible for this phenotype, this seems unlikely, as cells with a cytokinesis defect are expected to proliferate slowly rather than rapidly. We observed that QkgA inhibits the accumulation of nuclei in cells. If cells cannot complete cytokinesis as fast as they produce new nuclei, this could lead to an increased number of nuclei per cell, which could explain the multinucleate phenotype of qkgA− cells.
When estimating the accumulation of mass and protein per hour on a per-nucleus basis, we saw that qkgA− cells showed a higher mass and protein accumulation than the wild type, though these differences were not significant, suggesting that qkgA− cells accumulate mass and protein per nucleus at a rate like that of the wild type. The mass of wild-type cells increases as a function of cell density (33). As qkgA− cells proliferate rapidly but accumulate mass at a rate like that of the wild type, QkgA may thus affect the ability of cells to increase in mass as a function of density. It is unclear whether this increase in mass provides an advantage to wild-type cells, as our data show that qkgA− cells at stationary density do not decrease in number more rapidly than the wild type and that qkgA− cells do not show a defect in generating viable spores.
As aprA−, cfaD−, and qkgA− cells proliferate more rapidly than wild-type cells, AprA, CfaD, and QkgA almost certainly serve some fitness-increasing function, or else cells with these functional proteins would likely be selected against. Although aprA− and cfaD− cells yield fewer viable spores after development than the wild type, qkgA− cells do not show a defect in spore viability, suggesting that QkgA has not been selected for due to a role in the generation of viable spores. However, we saw that colonies of aprA−, cfaD−, and qkgA− cells showed a reduced rate of expansion when grown on bacterial lawns. The ability of a population of cells to expand rapidly almost certainly provides an advantage in an environment with low nutrient concentrations, as a population of cells must require a certain quantity of nutrients in order to continue increasing in number at an optimal rate, and the cell population depends upon expansion to attain this quantity of nutrients. Thus, although aprA−, cfaD−, and qkgA− cells may proliferate more rapidly than the wild type, these cells could potentially be at a disadvantage due to their reduced ability to expand as a colony. The mechanism by which these genes function in colony expansion is unclear. As we saw that qkgA− cells proliferate more than the wild type after a 24-hour period of growth in the presence of bacteria, it seems unlikely that qkgA− cells are deficient in utilizing bacteria as an energy source. aprA−, cfaD−, and qkgA− cells aggregate to form fruiting bodies and show random cell motility in the presence of bacteria, like wild-type cells, indicating that these genes are not essential for cell motility. It may be the case that the loss of these genes affects the ability of cells to chemotax toward bacteria or that AprA and/or CfaD functions as a chemorepellant and thus aids in the dispersal of a population of cells. Alternatively, rapid proliferation in itself may reduce colony expansion by reducing the local nutrient concentration available to cells, causing cell starvation and/or precocious development. Interestingly, cells lacking the heterotrimeric G protein complex subunit Gβ, which we have shown to be essential for AprA signaling (5), or the kinase YakA also proliferate rapidly in culture (5, 34) but show a reduced colony expansion phenotype (37, 39).
Currently, we cannot distinguish whether QkgA is actively involved in AprA/CfaD signaling (for instance, in a phospho-cascade) or whether QkgA plays a permissive role, such as the establishment or maintenance of some component of AprA/CfaD signaling. However, the fact that qkgA− cells proliferate rapidly suggests that QkgA functions specifically in negative regulation of proliferation as opposed to having a general function of cell metabolism or maintenance that affects multiple cellular processes, as one might predict that the loss of function of such a gene would have pleiotropic effects and thus lead to unfit or inviable cells. Therefore, it seems likely that QkgA functions specifically to slow proliferation and therefore may be an integral part of the AprA/CfaD-mediated mechanism of proliferation inhibition.
Together, our results indicate that the ROCO family kinase QkgA negatively regulates both proliferation and stationary density and is essential for the function of the proliferation-inhibiting autocrine signals AprA and CfaD. To our knowledge, no ROCO family kinase other than QkgA has been rigorously characterized as a regulator of proliferation. However, an RNA interference-mediated screen of the Drosophila melanogaster kinome for genes that mediate cell cycle progression identified a ROCO kinase family member as a potential regulator of cell cycle phase and centrosomal function (6). It therefore may be the case that regulation of proliferation by ROCO kinases is conserved in metazoans and that other ROCO proteins may have functions analogous to those of QkgA in Dictyostelium.
ACKNOWLEDGMENTS
We thank Jeff Williams for qkgA−cells, Peter van Haastert and Wouter van Egmond for the QkgA-GFP vector, Bill Loomis for the spore viability protocol, and Stephanie Karam for assistance with colony size measurement.
This work was supported by NIH grant GM074990.
Footnotes
Published ahead of print on 13 August 2010.
REFERENCES
- 1.Abe T., Langenick J., Williams J. G. 2003. Rapid generation of gene disruption constructs by in vitro transposition and identification of a Dictyostelium protein kinase that regulates its rate of growth and development. Nucleic Acids Res. 31:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abysalh J. C., Kuchnicki L. L., Larochelle D. A. 2003. The identification of pats1, a novel gene locus required for cytokinesis in Dictyostelium discoideum. Mol. Biol. Cell 14:14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand V. S., Braithwaite S. P. 2009. LRRK2 in Parkinson's disease: biochemical functions. FEBS J. 276:6428–6435 [DOI] [PubMed] [Google Scholar]
- 4.Bakthavatsalam D., Brock D. A., Nikravan N. N., Houston K. D., Hatton R. D., Gomer R. H. 2008. The secreted Dictyostelium protein CfaD is a chalone. J. Cell Sci. 121:2473–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakthavatsalam D., Choe J. M., Hanson N. E., Gomer R. H. 2009. A Dictyostelium chalone uses G proteins to regulate proliferation. BMC Biol. 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettencourt-Dias M., Giet R., Sinka R., Mazumdar A., Lock W. G., Balloux F., Zafiropoulos P. J., Yamaguchi S., Winter S., Carthew R. W., Cooper M., Jones D., Frenz L., Glover D. M. 2004. Genome-wide survey of protein kinases required for cell cycle progression. Nature 432:980–987 [DOI] [PubMed] [Google Scholar]
- 7.Bokko P. B., Francione L., Bandala-Sanchez E., Ahmed A. U., Annesley S. J., Huang X., Khurana T., Kimmel A. R., Fisher P. R. 2007. Diverse cytopathologies in mitochondrial disease are caused by AMP-activated protein kinase signaling. Mol. Biol. Cell 18:1874–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosgraaf L., Waijer A., Engel R., Visser A. J., Wessels D., Soll D., van Haastert P. J. 2005. RasGEF-containing proteins GbpC and GbpD have differential effects on cell polarity and chemotaxis in Dictyostelium. J. Cell Sci. 118:1899–1910 [DOI] [PubMed] [Google Scholar]
- 9.Brock D. A., Buczynski G., Spann T. P., Wood S. A., Cardelli J., Gomer R. H. 1996. A Dictyostelium mutant with defective aggregate size determination. Development 122:2569–2578 [DOI] [PubMed] [Google Scholar]
- 10.Brock D. A., Gomer R. H. 1999. A cell-counting factor regulating structure size in Dictyostelium. Genes Dev. 13:1960–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brock D. A., Gomer R. H. 2005. A secreted factor represses cell proliferation in Dictyostelium. Development 132:4553–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe J. M., Bakthavatsalam D., Phillips J. E., Gomer R. H. 2009. Dictyostelium cells bind a secreted autocrine factor that represses cell proliferation. BMC Biochem. 10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotter D. A., Raper K. B. 1966. Spore germination in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U. S. A. 56:880–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demicheli R. 2001. Tumour dormancy: findings and hypotheses from clinical research on breast cancer. Semin. Cancer Biol. 11:297–306 [DOI] [PubMed] [Google Scholar]
- 15.Firtel R. A. 1996. Interacting signaling pathways controlling multicellular development in Dictyostelium. Curr. Opin. Genet. Dev. 6:545–554 [DOI] [PubMed] [Google Scholar]
- 16.Fisher B., Szuch P., Levine M., Fisher E. R. 1971. A portal blood factor as the humoral agent in liver regeneration. Science 171:575–577 [DOI] [PubMed] [Google Scholar]
- 17.Goldberg J. M., Manning G., Liu A., Fey P., Pilcher K. E., Xu Y., Smith J. L. 2006. The dictyostelium kinome—analysis of the protein kinases from a simple model organism. PLoS Genet. 2:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomer R. H. 2001. Not being the wrong size. Nat. Rev. Mol. Cell Biol. 2:48–54 [DOI] [PubMed] [Google Scholar]
- 19.Gotthardt K., Weyand M., Kortholt A., Van Haastert P. J., Wittinghofer A. 2008. Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO J. 27:2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guba M., Cernaianu G., Koehl G., Geissler E. K., Jauch K. W., Anthuber M., Falk W., Steinbauer M. 2001. A primary tumor promotes dormancy of solitary tumor cells before inhibiting angiogenesis. Cancer Res. 61:5575–5579 [PubMed] [Google Scholar]
- 21.Harvey K. F., Pfleger C. M., Hariharan I. K. 2003. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114:457–467 [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen P., Tyers M. 2004. How cells coordinate growth and division. Curr. Biol. 14:R1014–R1027 [DOI] [PubMed] [Google Scholar]
- 23.Justice R. W., Zilian O., Woods D. F., Noll M., Bryant P. J. 1995. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9:534–546 [DOI] [PubMed] [Google Scholar]
- 24.Kessin R. H. 2001. Dictyostelium: evolution, cell biology, and the development of multicellularity. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 25.Korr D., Toschi L., Donner P., Pohlenz H. D., Kreft B., Weiss B. 2006. LRRK1 protein kinase activity is stimulated upon binding of GTP to its Roc domain. Cell Signal. 18:910–920 [DOI] [PubMed] [Google Scholar]
- 26.Lewis P. A. 2009. The function of ROCO proteins in health and disease. Biol. Cell 101:183–191 [DOI] [PubMed] [Google Scholar]
- 27.Loomis W. F. 2008. cAMP oscillations during aggregation of Dictyostelium. Adv. Exp. Med. Biol. 641:39–48 [DOI] [PubMed] [Google Scholar]
- 28.MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. 2006. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron 52:587–593 [DOI] [PubMed] [Google Scholar]
- 29.Marin I., van Egmond W. N., van Haastert P. J. 2008. The Roco protein family: a functional perspective. FASEB J. 22:3103–3110 [DOI] [PubMed] [Google Scholar]
- 30.McPherron A. C., Lawler A. M., Lee S. J. 1997. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387:83–90 [DOI] [PubMed] [Google Scholar]
- 31.Shaulsky G., Kessin R. H. 2007. The cold war of the social amoebae. Curr. Biol. 17:R684–R692 [DOI] [PubMed] [Google Scholar]
- 32.Sheffield J. B. 2007. ImageJ, a useful tool for biological image processing and analysis. Microsc. Microanal. 13:200–201 [Google Scholar]
- 33.Soll D. R., Yarger J., Mirick M. 1976. Stationary phase and the cell cycle of Dictyostelium discoideum in liquid nutrient medium. J. Cell Sci. 20:513–523 [DOI] [PubMed] [Google Scholar]
- 34.Souza G. M., Lu S., Kuspa A. 1998. YakA, a protein kinase required for the transition from growth to development in Dictyostelium. Development 125:2291–2302 [DOI] [PubMed] [Google Scholar]
- 35.van Egmond W. N., Kortholt A., Plak K., Bosgraaf L., Bosgraaf S., Keizer-Gunnink I., van Haastert P. J. 2008. Intramolecular activation mechanism of the Dictyostelium LRRK2 homolog Roco protein GbpC. J. Biol. Chem. 283:30412–30420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Egmond W. N., van Haastert P. J. 2010. Characterization of the Roco protein family in D. discoideum. Eukaryot. Cell 9:751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Es S., Weening K. E., Devreotes P. N. 2001. The protein kinase YakA regulates g-protein-linked signaling responses during growth and development of Dictyostelium. J. Biol. Chem. 276:30761–30765 [DOI] [PubMed] [Google Scholar]
- 38.Wu H. H., Ivkovic S., Murray R. C., Jaramillo S., Lyons K. M., Johnson J. E., Calof A. L. 2003. Autoregulation of neurogenesis by GDF11. Neuron 37:197–207 [DOI] [PubMed] [Google Scholar]
- 39.Wu L., Valkema R., Van Haastert P. J., Devreotes P. N. 1995. The G protein beta subunit is essential for multiple responses to chemoattractants in Dictyostelium. J. Cell Biol. 129:1667–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R. J., Calne D. B., Stoessl A. J., Pfeiffer R. F., Patenge N., Carbajal I. C., Vieregge P., Asmus F., Muller-Myhsok B., Dickson D. W., Meitinger T., Strom T. M., Wszolek Z. K., Gasser T. 2004. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44:601–607 [DOI] [PubMed] [Google Scholar]