Abstract
Colonization and infection of the human host by opportunistic pathogen Candida albicans derive from an ability of this fungus to colonize mucosal tissues and prosthetic devices within the polymicrobial communities present. To determine the functions of C. albicans cell wall proteins in interactions with host or bacterial molecules, Saccharomyces cerevisiae was utilized as a surrogate host to express C. albicans cell wall proteins Als3p, Eap1p, Hwp1p, and Rbt1p. Salivary pellicle and fibrinogen were identified as novel substrata for Als3p and Hwp1p, while only Als3p mediated adherence of S. cerevisiae to basement membrane collagen type IV. Parental S. cerevisiae cells failed to form biofilms on salivary pellicle, polystyrene, or silicone, but cells expressing Als3p or Hwp1p exhibited significant attachment to each surface. Virulence factor Rbt1p also conferred lower-level binding to salivary pellicle and polystyrene. S. cerevisiae cells expressing Eap1p formed robust biofilms upon polystyrene surfaces but not salivary pellicle. Proteins Als3p and Eap1p, and to a lesser degree Hwp1p, conferred upon S. cerevisiae the ability to bind cells of the oral primary colonizing bacterium Streptococcus gordonii. These interactions, which occurred independently of amyloid aggregate formation, provide the first examples of specific C. albicans surface proteins serving as receptors for bacterial adhesins. Streptococcus gordonii did not bind parental S. cerevisiae or cells expressing Rbt1p. Taken collectively, these data suggest that a network of cell wall proteins comprising Als3p, Hwp1p, and Eap1p, with complementary adhesive functions, promotes interactions of C. albicans with host and bacterial molecules, thus leading to effective colonization within polymicrobial communities.
Candida albicans is a pleiomorphic fungus found on mucosal surfaces of the gastrointestinal and genitourinary tracts, skin, and oral cavity (2). As an opportunistic pathogen, C. albicans can form potentially lethal fungal masses in the kidney, heart, and brain upon gaining access to the bloodstream (4), and invasive fungal infections are becoming increasingly problematic in the clinical setting (34). Candida species are now the third most common cause of nosocomial bloodstream infections. In the United States alone there are an estimated 70,000 cases per year of disseminated candidiasis (34), with an associated health care cost of $2 billion to $4 billion/year (44, 45). C. albicans is also responsible for >90% of oral fungal diseases derived from polymicrobial biofilms, and ≤90% of HIV-infected individuals suffer from oral candidiasis, which may progress to advanced esophageal candidiasis (10).
C. albicans can colonize a wide variety of sites within the host in addition to mucosal tissues, such as catheters, stents, surgical implants, and dentures. This ability can be attributed, at least in part, to the large number of proteins expressed on the candidal cell surface, which mediate adhesion to a range of substrata. Cell wall proteins (CWPs) in C. albicans also play a critical role in biofilm formation. Within the host, Candida species are frequently found as part of polymicrobial biofilms, in which antagonistic, synergistic, and mutualistic interactions among microbes significantly influence composition of the community microflora (17). This is particularly pertinent for colonization of the oral cavity, where up to 100 different microbial species may be isolated from a single site at any given time. To successfully colonize the host and cause disease, C. albicans must therefore not only attach directly to host tissues or medical devices but also navigate interactions with a diverse microflora to ensure the availability of suitable binding sites, nutrients, and growth conditions.
It has been shown that C. albicans coaggregates (coadheres) strongly with Streptococcus bacteria indigenous to the human oral cavity such as Streptococcus gordonii and Streptococcus sanguinis (13, 18). These bacteria are pioneer colonizers of oral cavity surfaces, and it is hypothesized that interactions with these streptococci may promote oral carriage and persistence of C. albicans, thereby supporting candidal reservoirs for opportunistic infections following disruption of the oral ecology. Previous work by Holmes et al. (13, 14) identified Streptococcus gordonii cell wall-associated polypeptides SspA, SspB, and CshA, together with linear cell wall phosphopolysaccharides, as potential targets for C. albicans binding streptococcal cells. However, the reciprocal receptors on the surface of C. albicans recognized by streptococci have yet to be identified.
This work utilizes Saccharomyces cerevisiae, which does not bind streptococci, as a heterologous host for expression and identification of candidal surface proteins targeted by Streptococcus gordonii. Four surface proteins were selected that had been previously implicated in C. albicans colonization and pathogenesis: Als3p, Eap1p, Hwp1p, and Rbt1p. Als3p (comprehensively reviewed by Hoyer et al. [15]), Hwp1p (29, 40), and Eap1p (20, 22) are associated with mediating interactions of C. albicans with host epithelial cells and with biofilm formation in catheter models. Expression of Als3p or Hwp1p has been shown to be hypha specific, while Eap1p is expressed by each morphological form (16, 20, 41). Rbt1p shares 43% sequence identity with Hwp1p and has been associated with virulence in mouse and rabbit models of C. albicans infection (6). Using a recombinase-based Gateway cloning system (Invitrogen), each of the C. albicans proteins was expressed on the surface of S. cerevisiae. Their functional properties in adherence and biofilm formation were determined, and proteins Als3p and Eap1p were identified as potential Streptococcus gordonii receptors on the surface of C. albicans.
MATERIALS AND METHODS
Microbial strains and culture conditions.
The bacterial and yeast strains used in this study are listed in Table 1. Streptococci were routinely grown in brain heart infusion broth (Lab M) supplemented with 5 g/liter yeast extract (BD) in candle jars at 37°C without shaking. Escherichia coli cells were grown with shaking at 37°C in Luria-Bertani medium (Lab M). When required, antibiotics were added to the medium at the following concentrations: gentamicin, 10 μg/ml; ampicillin, 100 μg/ml. Candida albicans cells were grown with shaking in YPD broth (1% yeast extract, 2% neopeptone [BD], 2% glucose) at 37°C. Saccharomyces cerevisiae cells were grown with shaking at 30°C in complete synthetic medium (CSM) without uracil (ForMedium) supplemented with 0.67% yeast nitrogen base (YNB) (Difco) and 2% glucose. Cultures of parent strain S. cerevisiae BY4742 were supplemented with 25 μg/ml uracil.
Table 1.
Microbial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Streptococcus gordonii DL1 (Challis) | Wild type | 32 |
| S. gordonii OB235 | Fibril-deficient mutant; cshA::ermAM | 27 |
| Escherichia coli OmniMAX 2-T1R | F′ {proAB+lacIqlacZΔM15 Tn10(Tetr) Δ(ccdAB)} mcrA Δ(mrr-hsdRMS-mcrBC) φ80(lacZ)ΔM15 Δ(lacZYA-argF) U169 endA1 recA1 supE44 thi-1 gyrA96 relA1 tonA panD | Invitrogen |
| Candida albicans SC5314 | Wild-type | 12 |
| Saccharomyces cerevisiae BY4742 | MATα leu2Δ0 lys2Δ0 ura3Δ0 his3Δ1 | 5 |
| UB2155 | pBC542 | This study |
| UB2156 | pBC542-ALS3sm | This study |
| UB2157 | pBC542-ALS3lg | This study |
| UB2158 | pBC542-EAP1 | This study |
| UB2159 | pBC542-HWP1 | This study |
| UB2160 | pBC542-RBT1 | This study |
| UB2161 | pBC542-CWP1 | This study |
| Plasmids | ||
| pDONR207 | 5.6 kb; Genr; pUC ori; Gateway entry vector | Invitrogen |
| pBC542 | 8.3 kb; Ampr; pMB1 ori; Gateway destination vector | 50 |
Heterologous expression of C. albicans proteins.
Routine cloning procedures were performed according to the methods described by Sambrook et al. (38). Plasmids (listed in Table 1) were purified from E. coli cells using the QIAquick spin miniprep purification kit (Qiagen). Oligonucleotides (listed in Table 2) were synthesized by MWG Biotech. Genomic DNA was extracted from yeast cells using GeneReleaser (BioVentures). PCR products were purified using the QIAquick PCR purification kit (Qiagen).
Table 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| ALS3attB1 | ATGCTACAACAATATACATTGTTACTC |
| ALS3attB2 | TCATAAATAATCACAGTATCAGTTCCACC |
| EAP1attB1 | ATGAAAGTTTCTCAAATTTTACCATTAGCT |
| EAP1attB2 | GCTTCAACTTCAGGACCATTGGTAGG |
| HWP1attB1 | ATGAGATTATCAACTGCTCAACTTATT |
| HWP1attB2 | GCACCTTCAAATGTAGAAATAGGAGC |
| RBT1attB1 | ATGAGATTTGCAACTGCCCAACTCGCT |
| RBT1attB2 | GAAGCAATAGTGTATGAGGAGTTAGC |
| CWP1attB1 | ATGAAATTCTCCACTGCTTTGTCTGTC |
| CWP1attB2 | TGTTCGTAAACTGTGTTTGGAGCTTG |
All forward (attB1) primers comprise the sequence GGGGACAAGTTTGTACAAAAAAGCAGGCTAAAACC followed by the indicated sequence; all reverse (attB2) primers comprise the sequence GGGGACCACTTTGTACAAGAAAGCTGGGT followed by the indicated sequence.
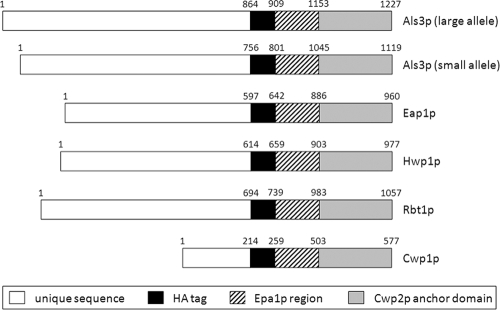
C. albicans hypha-specific proteins Als3p and Hwp1p, together with CWPs Eap1p and Rbt1p, were expressed on the surface of surrogate host S. cerevisiae using the Gateway recombinase-based cloning system (Invitrogen) according to the manufacturer's instructions. In brief, fragments corresponding to the small (ALS3sm; GenBank accession no. AY223551) and large (ALS3lg; GenBank accession no. AY223552) alleles of ALS3 that terminated after the tandem repeat domain (2,267 bp and 2,591 bp, respectively) were amplified by PCR from C. albicans SC5314 genomic DNA, using the primers listed in Table 2. The resulting PCR products were gel purified and mixed with Gateway BP enzymes for insertion into donor vector pDONR207. The resulting entry vectors were transformed into E. coli, and predicted insertions were confirmed by sequencing. This process was then repeated for C. albicans genes EAP1 (1,790 bp; GenBank GeneID 3643814), HWP1 (1,841 bp; GenBank GeneID 3645372), and RBT1 (2,081 bp; GenBank accession no. AF254142), terminated upstream of the predicted GPI anchor site. Using Gateway LR enzymes, C. albicans gene fragments from successful entry vectors were recombined with destination vector pBC542 (50) to generate appropriate expression vectors. These were transformed into E. coli, and predicted recombinations were confirmed by sequencing. Expression vectors were then purified and electroporated into S. cerevisiae BY4742. S. cerevisiae transformed with pBC542 alone was also generated as a control. Having originally been designed to investigate adhesive functions of C. glabrata adhesin family Epa, proteins are expressed from destination vector pBC542 as fusions with a hemagglutinin (HA) tag, the Ser/Thr-rich region of C. glabrata adhesin Epa1p, and the C-terminal domain of S. cerevisiae cell wall protein Cwp2p (50) (Fig. 1). An additional control was therefore constructed by cloning CWP1 (641 bp; GenBank GeneID 853766) from S. cerevisiae BY4742 into pBC542. This enabled any phenotypic effects of the Epa1p domain to be identified.
Fig. 1.
Schematic representation of fusion proteins expressed on the surface of S. cerevisiae using Gateway expression vector pBC542. Specific CWP sequences from C. albicans (Als3p, Eap1p, Hwp1p, Rbt1p) or S. cerevisiae (Cwp1p) are fused to an HA-tagged construct with the Ser/Thr-rich region of C. glabrata protein Epa1p and the C-terminal domain of S. cerevisiae protein Cwp2p carrying a GPI anchor. Amino acid residues for each segment are indicated.
Immunofluorescence.
Successful expression of HA-tagged fusion proteins on the surface of S. cerevisiae was confirmed by immunofluorescence. Each fusion protein was expressed under the control of the TEF1 promoter. Cell populations in mid-exponential phase were therefore utilized to ensure optimal protein expression. Yeast cells were harvested (5,000 × g, 5 min) at an optical density at 600 nm (OD600) of 1.0 (approximately 1 × 107 cells/ml), washed in phosphate-buffered saline (PBS), and fixed in 4% paraformaldehyde for 30 min at 25°C. Fixed cells were blocked with rabbit serum (Sigma) for 30 min at 25°C, washed, and incubated for 1 h at 4°C with 10 μg/ml mouse anti-HA antibody (ZYMED Laboratories). Cells were washed again, incubated for a further 1 h with rabbit anti-mouse fluorescein isothiocyanate (FITC)-conjugated antibody (Dako; 1:200 dilution), and then visualized by fluorescence microscopy.
To quantify relative fusion protein expression levels, cells were fluorescently labeled, as described above, and aliquots (100 μl) were applied to wells of a black 96-well microtiter plate (MTP; Greiner). Fluorescence intensity (relative fluorescence units [RFU]) was then measured using a Molecular Devices SpectraMax M2 MTP reader. This was correlated with total protein levels, as determined by the Lowry assay (23) on equivalent cell aliquots (100 μl). In brief, cells were boiled for 10 min in 1 N NaOH (0.5 ml), cooled, and mixed with copper tartrate (2.5 ml) and Folin-Ciocalteu (0.5 ml) reagents. The A750 after 30 min was determined as a measure of total protein. S. cerevisiae cells expressed Als3p (small and large alleles), Eap1p, and Rbt1p at a level of 1 RFU/μg protein, while Hwp1p and Cwp1p were expressed at 2 RFU/μg protein and 4 RFU/μg protein, respectively.
Interactions of S. cerevisiae with streptococci.
Yeast cells were grown for 16 h, washed in PBS, and adjusted to an OD600 of 0.2 in CSM. Suspensions (2 ml) were transferred to glass bijou bottles and incubated with shaking for 3 h at 30°C. Streptococcus gordonii DL1 cells were grown for 16 h, washed in PBS, and incubated in the dark in 1.5 mM FITC for 1 h at 25°C. FITC-labeled cells were washed thoroughly in 0.05 M Na2CO3 containing 0.1 M NaCl (pH 9) and adjusted to an OD600 of 0.5 (approximately 5 × 108 cells/ml) in CSM, and then aliquots (1 ml) were added to Saccharomyces suspensions following the 3-h incubation period. Cells were then incubated together for a further 1 h before being harvested (3,000 × g, 2 min), resuspended in 1/10 original volume, and visualized by light and fluorescence microscopy. Where appropriate, interactions were performed in the presence of 30 μM Congo red (Sigma). To semiquantify streptococcal attachment, cellD imaging software (Olympus) was used to calculate the mean number of fluorescent pixels bound per S. cerevisiae cell (minimum cell count, 200).
Preparation of saliva-coated coverslips.
Human whole saliva was collected from a minimum of five adult volunteers (in accordance with the Human Tissue Act, University of Bristol) into a chilled tube on ice, pooled, treated for 10 min with 2.5 mM dithiothreitol (DTT), and clarified by centrifugation (10,000 × g, 10 min). Whole salivary supernatant was then passed through a 0.2-μm filter and diluted to 10% with distilled water. Aliquots (1 ml) were transferred to wells of a 12-well plate containing 19-mm-diameter glass coverslips, and plates were incubated for 16 h at 4°C. Saliva-coated coverslips were then washed in PBS and transferred to wells of a fresh plate.
Binding to salivary pellicle.
Yeast cells were grown for 16 h, washed in PBS, and adjusted to an OD600 of 1.0 in CSM. Suspensions (1 ml) were transferred to wells containing saliva-coated coverslips and incubated with gentle shaking for 4 h at 30°C. Suspensions were then aspirated and the coverslips washed before being submerged in 1% crystal violet for 2 min. Excess stain was removed by washing in distilled water, and the coverslips were then transferred to a fresh 12-well plate and submerged in 10% acetic acid (1 ml) for 15 min. Suspensions (200 μl) were serially diluted, and the A595 was then determined as a measurement of biomass.
Biofilm formation on salivary pellicle.
Yeast cell suspensions were prepared as described above, transferred to wells containing saliva-coated coverslips, and incubated with gentle shaking for 4 h at 30°C. Suspensions were then aspirated and the coverslips washed in PBS before fresh CSM (1 ml) was added to each well. Plates were incubated for a further 20 h, after which time coverslips were washed and stained as described above. The A595 was determined as a measurement of biomass. Alternatively, stained coverslips were air dried, mounted in DePeX (BDH), and visualized by light microscopy.
Attachment to polystyrene.
Binding and biofilm assays were performed as described above, with the exception that yeast suspensions were applied directly to the wells of a 12-well plate in the absence of coverslips.
Attachment to silicone.
Sterile silicone squares (1.5 cm2) were pretreated with horse serum (Sigma) in wells of a 12-well plate overnight at 37°C with shaking. The silicone was washed in PBS and transferred to wells of a fresh plate. Binding and biofilm assays were performed as described above, with the exception that yeast suspensions were applied to wells containing silicone in place of coverslips.
Microtiter plate binding assay.
Substrata were diluted in coating buffer (20 mM Na2CO3, 20 mM NaHCO3, pH 9.3) and adsorbed (0.5 μg/well) onto wells of an Immulon 2HB 96-well MTP (Nunc) at 4°C for 16 h. Wells were washed with Tris-buffered saline (TBS) (100 mM Tris, 1.5 M NaCl, pH 8) containing 5 mM CaCl2 (TBSC), blocked with 1% (wt/vol) gelatin at 30°C for 1 h, and washed with TBSC. Yeast cells were grown for 16 h, harvested (5,000 × g, 5 min), washed once in TBSC, and adjusted to an OD600 of 1.0 in CSM. Cells (100-μl aliquots) were added to MTP wells and incubated for 4 h at 30°C with gentle shaking. Wells were washed twice with TBS, and adherent yeast cells were fixed with 25% (vol/vol) formaldehyde for 30 min at 25°C. Wells were washed twice again with TBS, and fixed cells were stained by the addition of 0.5% (wt/vol) crystal violet for 2 min at 25°C. Excess stain was removed by washing three times with TBS, and remaining stain was then dissolved with 10% (vol/vol) acetic acid. The A595 was determined as a measurement of biomass.
Microtiter plate biofilm assay.
MTPs and yeast cell suspensions were prepared as described above. Yeast cells (100-μl aliquots) were added to MTP wells and incubated with gentle shaking for 4 h at 30°C. Wells were washed twice with TBS before fresh CSM (100 μl) was added to each well. Plates were incubated for a further 20 h, after which nonadherent cells were removed by washing twice with TBS. Adherent cells were fixed and stained as described above. The A595 was determined as a measurement of biomass.
RESULTS
Expression of C. albicans adhesins in surrogate host S. cerevisiae.
Streptococcus species coadhere (coaggregate) strongly with C. albicans using specific protein adhesins (14). However, the receptors on the candidal cell surface that are recognized by these streptococcal adhesins are unknown. To investigate the role of CWPs from C. albicans in mediating streptococcal interactions, four candidal CWP adhesins associated with colonization and biofilm formation were expressed on the surface of surrogate host S. cerevisiae BY4742. Previous work has shown that S. cerevisiae does not form biofilms or interact with streptococci (8) (unpublished data). This heterologous host therefore provided an ideal system for determining the role of the selected adhesins in these processes in the absence of other candidal proteins.
Genes encoding each of the C. albicans adhesins were cloned into destination vector pBC542 (50) using the Gateway recombinase-based cloning system (Invitrogen). Proteins were expressed on the surface of S. cerevisiae as fusions with a HA tag toward the C terminus, followed by the Ser/Thr-rich region of Epa1p from Candida glabrata and the C-terminal domain of S. cerevisiae protein Cwp2p for efficient anchoring to the cell wall (see Fig. 1). Amplified gene fragments terminated immediately downstream of the tandem repeat domain (15) for ALS3 (small and large alleles), while all other gene fragments terminated close to the GPI anchor site (see Fig. 1). Successful recombination events were confirmed at each stage by sequencing. Surface expression by S. cerevisiae was ultimately confirmed by immunofluorescence, using antibody targeted against the HA tag within each construct.
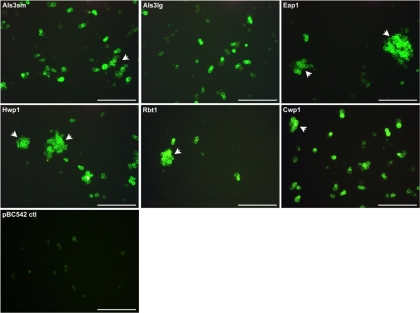
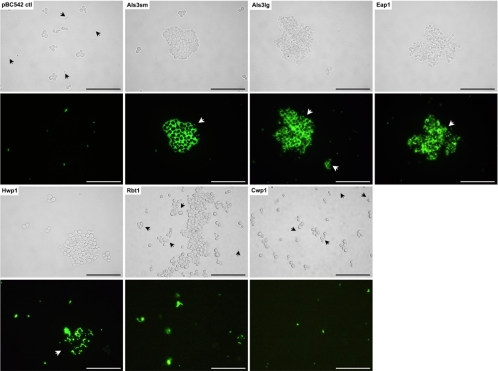
Heterologous expression of each protein was under the control of the TEF1 promoter and was demonstrated for S. cerevisiae clones expressing each of the C. albicans adhesins (Fig. 2). By correlating fluorescence intensities (RFU) with total protein levels, relative expression levels were found to be 1 RFU/μg protein for S. cerevisiae cells expressing Als3p (small and large alleles), Eap1p, and Rbt1p and 2 RFU/μg protein for cells expressing Hwp1p. The highest expression level (4 RFU/μg protein) was calculated for cells expressing Cwp1p, likely reflecting the fact that Cwp1p is derived from the S. cerevisiae host strain itself rather than C. albicans. Generally, expression of the C. albicans proteins on the S. cerevisiae cell surface led to various degrees of clumping (Fig. 2), which was most obvious with the Eap1p- and Hwp1p-expressing constructs. In contrast, no clumping or fluorescence was seen for S. cerevisiae transformed with pBC542 vector alone (Fig. 2). Because of the C. glabrata Epa1p domain engineered toward the C terminus of each protein cloned using pBC542, an additional control clone was generated by transforming S. cerevisiae with pBC542 carrying CWP1 amplified from the S. cerevisiae BY4742 genome (Fig. 1 and 2). S. cerevisiae provides an adhesion-negative background for the adhesive properties being investigated in these studies. An adhesion profile similar to that of the S. cerevisiae vector-alone control was therefore predicted for this Cwp1p-expressing strain, which would confirm that no attachment/biofilm formation could be attributed to the Epa1p fragment.
Fig. 2.
Confirmation of surface expression of C. albicans adhesins by surrogate host S. cerevisiae. C. albicans SC5314 adhesins Als3p (small [sm] and large [lg] alleles), Eap1p, Hwp1p, and Rbt1p were cloned into S. cerevisiae BY4742 together with S. cerevisiae control protein Cwp1p and pBC542 vector alone. Expression clones were then fixed in paraformaldehyde, probed with antibody directed against the HA tag incorporated within each fusion protein followed by a FITC-conjugated secondary antibody, and visualized by fluorescence microscopy (scale bar, 50 μm). Arrows indicate examples of cell clumping.
Adherence and biofilm formation by S. cerevisiae expressing C. albicans adhesins.
Expression of C. albicans adhesins in a surrogate host provides the opportunity to investigate the properties of individual proteins in isolation from other candidal proteins. However, there is a possibility that differences in codon usage and host glycosylation events may affect their functions. The adherence properties of each heterologous host strain were therefore investigated to confirm, or otherwise assess, the reported phenotype conferred in C. albicans.
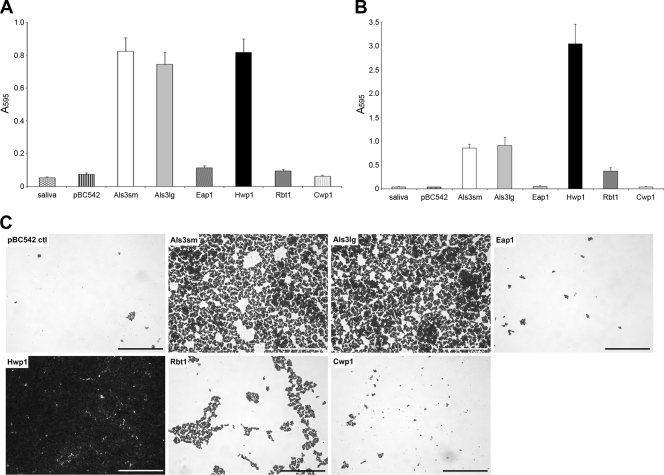
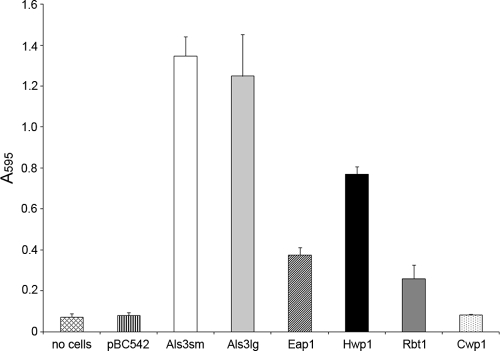
C. albicans strains with mutations in ALS3 and HWP1 have been shown to be defective in biofilm formation in both in vitro and in vivo catheter models (29, 47). Since all tissues within the oral cavity are constantly bathed in saliva, biofilm formation on a salivary pellicle by each S. cerevisiae heterologous host strain was investigated. Staining with crystal violet was used as a measurement of biomass, together with biofilm visualization by light microscopy. After 4 h of incubation at 37°C, very few cells could be seen attached to saliva-coated coverslips for S. cerevisiae transformed with pBC542 alone, and staining levels were comparable to those of wells to which no yeast cells had been added (saliva-only control) (Fig. 3A). Likewise, little adhesion was seen for S. cerevisiae cells expressing control protein Cwp1p, Rbt1p, or Eap1p. In contrast, substantial attachment was seen for S. cerevisiae cells expressing Als3p, large or small alleles, and Hwp1p, with biomass levels approximately 10-fold higher than for cells carrying pBC542 vector alone (Fig. 3A). Studies were then extended to biofilm formation over 24 h (Fig. 3B and C). A strong correlation was seen between initial attachment levels and biomass accumulation. No biofilm was formed on the salivary pellicle by S. cerevisiae cells carrying pBC542 vector alone or expressing control protein Cwp1p or Eap1p. In contrast, substantial biofilm was visualized for S. cerevisiae expressing both alleles of Als3p (Fig. 3C), although biomass levels increased only 1.2-fold from those measured at 4 h (Fig. 3B). Significant biofilm development was also seen for S. cerevisiae cells expressing Hwp1p (Fig. 3C), with an approximately 4-fold increase in biomass compared to that at 4 h (Fig. 3B). A 4-fold increase in biomass from 4 h to 24 h was also seen for S. cerevisiae cells expressing Rbt1p, which shows 43% identity to Hwp1p, although overall levels were just 12% of those for cells expressing Hwp1p (Fig. 3B).
Fig. 3.
Attachment and biofilm formation on a saliva-coated surface by S. cerevisiae cells expressing candidal adhesins. S. cerevisiae cells expressing candidal adhesins Als3p, Eap1p, Hwp1p, and Rbt1p, together with control protein Cwp1p and pBC542 vector alone, were incubated with saliva-coated coverslips for 4 h at 30°C. Nonadherent cells were removed, and total biomass was measured immediately by crystal violet staining (A) or after a further 20 h of incubation at 30°C in CSM (B). (C) Biofilm formation was visualized after 24 h by light microscopy (scale bar, 100 μm). Values given represent mean ± standard deviation (SD) of results of two independent experiments performed in triplicate.
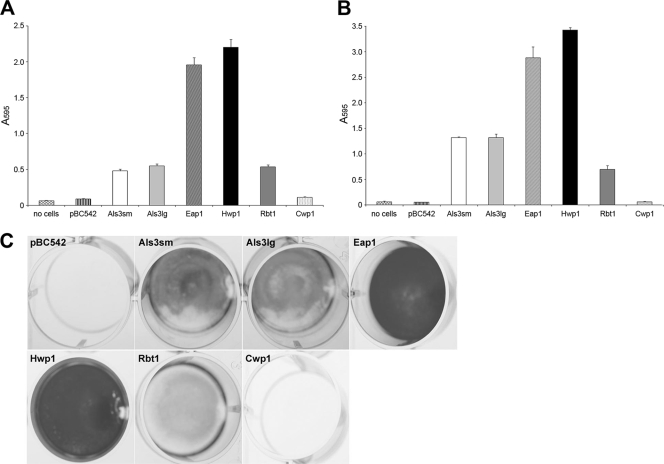
Since Eap1p did not mediate binding to the salivary pellicle, attachment of S. cerevisiae constructs to polystyrene was investigated. Adherence to plastic or polystyrene was the original phenotype by which Eap1p adhesin was identified (20). In these experiments, as for the previous ones, no initial adhesion or biofilm development was seen on polystyrene for S. cerevisiae cells transformed with pBC542 vector alone or expressing control protein Cwp1p (Fig. 4). Both allelic forms of Als3p and Rbt1p promoted significant attachment of S. cerevisiae to polystyrene at comparable levels after 4 h, and biomass measurements were approximately 5-fold higher than those for cells with vector alone (Fig. 4A). Biomass continued to accumulate over 24 h, albeit at different rates, with final biomass levels approximately 3-fold and 1.3-fold higher than those at 4 h for Als3p and Rbt1p, respectively (Fig. 4B and C). The highest levels of initial attachment were seen for S. cerevisiae cells expressing Eap1p and Hwp1p, at >20-fold higher than for cells carrying pBC542 vector alone (Fig. 4A). This correlated well with overall biofilm formation, with a further 50% increase in biomass occurring over 24 h for both heterologous host strains (Fig. 4B and C).
Fig. 4.
Attachment and biofilm formation on polystyrene by S. cerevisiae cells expressing candidal adhesins. S. cerevisiae cells expressing candidal adhesins Als3p, Eap1p, Hwp1p, and Rbt1p, together with control protein Cwp1p and pBC542 vector alone, were incubated in polystyrene wells for 4 h at 30°C. Nonadherent cells were removed and total biomass was measured immediately by crystal violet staining (A) or after a further 20 h of incubation at 30°C in CSM (B). (C) Biofilm formation was visualized after 24 h by light microscopy. Values given represent mean ± SD of results of two independent experiments performed in triplicate.
As a final confirmation of candidal protein functionality, attachment of each surrogate host strain to silicone was investigated. C. albicans proteins Als3p (47), Hwp1p (29), and Eap1p (22) have all been implicated in mediating biofilm formation on this surface, which has potentially important clinical implications for C. albicans colonization of indwelling medical devices. No adhesion to silicone above background was seen for S. cerevisiae cells carrying pBC542 vector alone or expressing control protein Cwp1p (Fig. 5). The highest levels of binding were recorded for cells expressing Als3p, with both allelic forms behaving similarly. In each case, final biomass quantities were >15-fold higher than for cells with vector alone (Fig. 5). Hwp1p and Eap1p also promoted significant attachment of S. cerevisiae to silicone, albeit at different levels. Biomass measurements after 4 h were approximately 10-fold higher for cells expressing Hwp1p than for cells with pBC542 vector alone, compared to 5-fold higher for cells expressing Eap1p (Fig. 5). S. cerevisiae cells expressing Rbt1p bound silicone in greater numbers than cells with empty vector alone, although overall biomass levels were only a third of those measured for cells expressing related protein Hwp1p (Fig. 5). Studies were then extended to investigate biofilm formation over 24 h. However, S. cerevisiae cells expressing Als3p, Eap1p, and Hwp1p readily detached from silicone over this time period, making biomass quantification inaccurate (data not shown).
Fig. 5.
Attachment to silicone by S. cerevisiae cells expressing candidal adhesins. S. cerevisiae cells expressing candidal adhesins Als3p, Eap1p, Hwp1p, and Rbt1p, together with control protein Cwp1p and pBC542 vector alone, were incubated with silicone squares for 4 h at 30°C. Nonadherent cells were removed, and total biomass was measured by crystal violet staining. Values given represent mean ± SD of results of two independent experiments performed in triplicate.
Adherence to extracellular matrix (ECM) proteins.
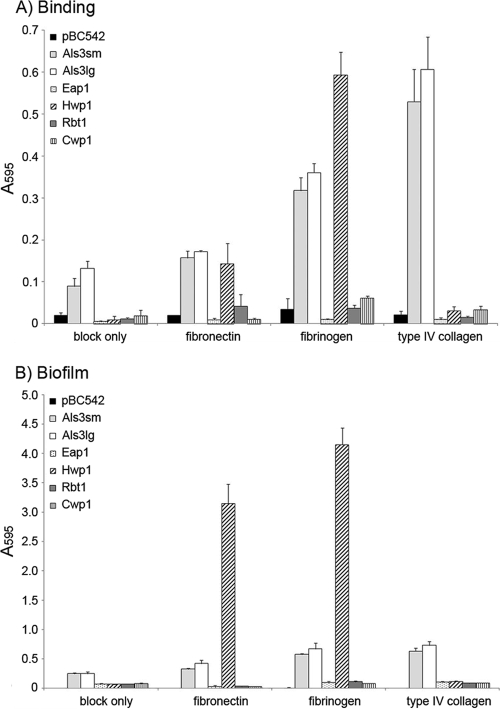
Another factor that could likely promote oral carriage of C. albicans, and may also facilitate tissue penetration and dissemination, is an ability to bind components of the host ECM. C. albicans has been shown to bind several ECM proteins, including collagen types I and IV, vitronectin, fibrinogen, laminin, and fibronectin (reviewed by Chaffin [7]). There is some evidence that Als3p may promote attachment to fibronectin (39). The abilities of each heterologous host strain to bind ECM proteins collagen type IV, fibrinogen, and fibronectin were therefore investigated, using a microtiter plate-based assay. Following 4 h of incubation with each substratum, only three candidal adhesins exhibited significant adhesion to any of the ECM proteins compared to control S. cerevisiae clones with vector alone or expressing Cwp1p (Fig. 6A). Als3p exhibited some background attachment to the blocking agent gelatin used in these assays (Fig. 6A), as has been reported previously (24). Although statistically significant, binding to fibronectin was only marginally greater (<2-fold) than adhesion to gelatin. More substantial binding was measured for fibrinogen and collagen type IV, at levels >3- and >5-fold above background, respectively. Compared to that of cells with pBC542 vector alone, adhesion to fibrinogen and collagen type IV was approximately 10- and 25-fold higher, respectively, when cells expressed Als3p (Fig. 6A). Biomass levels were consistently higher for the large allelic form of Als3p than for the small allelic form, but the differences were not statistically significant (Fig. 6A). Unlike Als3p, Hwp1p did not mediate attachment to collagen type IV. Biomass levels were, however, 8- and 18-fold higher for fibronectin and fibrinogen, respectively, for Hwp1p-expressing cells than for those carrying vector alone (Fig. 6A).
Fig. 6.
Attachment and biofilm formation on extracellular matrix proteins by S. cerevisiae cells expressing candidal adhesins. S. cerevisiae cells expressing candidal adhesins Als3p, Eap1p, Hwp1p, and Rbt1p, together with control protein Cwp1p and pBC542 vector alone, were incubated with 0.5 μg ECM protein for 4 h at 30°C. Nonadherent cells were removed, and total biomass was measured immediately by crystal violet staining (A) or after a further 20 h of incubation at 30°C in CSM (B). Values given represent mean ± SD of results of two independent experiments performed in triplicate.
Studies were then extended to examine biofilm formation upon these ECM proteins by each heterologous host strain after 24 h. A strong correlation was seen between initial adhesive capabilities and biofilm development, with only cells expressing Als3p or Hwp1p forming substantial biofilms (Fig. 6B). Levels of biomass accumulation did, however, vary considerably. Despite the extended incubation period of the biofilm assay compared to the binding assay, total biomass of Als3p-expressing cells on type IV collagen did not appear to increase substantially (Fig. 6B). An approximately 2-fold increase was seen with fibrinogen, with both allelic forms behaving similarly (Fig. 6B). In contrast, much greater biomass accumulated over the extended incubation period for cells expressing Hwp1p. Compared to results of the binding assay, biomass levels increased by approximately 7-fold in the presence of fibrinogen and 22-fold with fibronectin. As for the binding assay, no attachment or biofilm formation on type IV collagen was seen for S. cerevisiae cells expressing Hwp1p (Fig. 6B).
Role of C. albicans adhesins in mediating interactions with Streptococcus.
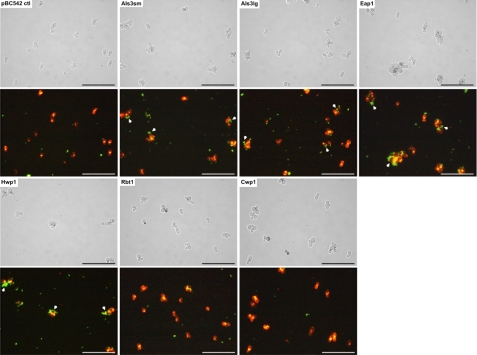
It is hypothesized that coadherence (coaggregation) between oral streptococci and C. albicans facilitates carriage and persistence of C. albicans within the oral cavity. To identify C. albicans adhesins that may mediate such coadherence events, interactions of each S. cerevisiae heterologous host strain with pioneer colonizer Streptococcus gordonii were investigated. S. cerevisiae cells were grown at 30°C for 3 h in CSM, after which time FITC-labeled Streptococcus gordonii DL1 cells were added for a further 1 h. Saccharomyces cells were then washed to remove unattached bacteria before being visualized by light and fluorescence microscopy. No interactions were seen between S. cerevisiae cells carrying pBC542 vector alone and those expressing control protein Cwp1p (Fig. 7). In contrast, Streptococcus gordonii bound strongly to cells expressing Als3p, both large and small alleles. The streptococcal cells also bound to S. cerevisiae expressing Eap1p and, to a lesser extent, Hwp1p (Fig. 7). Only weak binding of Streptococcus gordonii DL1 cells to S. cerevisiae cells expressing Rbt1p was observed (Fig. 7).
Fig. 7.
Interactions of Streptococcus gordonii DL1 with C. albicans adhesins expressed on the surface of surrogate host S. cerevisiae. S. cerevisiae cells expressing candidal adhesins Als3p, Eap1p, Hwp1p, and Rbt1p, together with control protein Cwp1p and pBC542 vector alone, were grown at 30°C in CSM for 3 h and incubated for a further 1 h with FITC-labeled streptococci. Cells were then harvested and visualized by fluorescence and light microscopy (scale bar, 50 μm). Black arrows indicate locations of streptococci in phase-contrast images. White arrows indicate examples of interactions between streptococci and aggregates of S. cerevisiae cells.
Relationship between Streptococcus gordonii adherence and amyloid formation.
It was noted that Streptococcus gordonii often appeared to interact strongly with aggregates of S. cerevisiae cells expressing Als3p, Hwp1p, or Eap1p (Fig. 7). It had been reported previously that Als family adhesins in C. albicans have the propensity to self-aggregate due to the presence of amyloid-forming sequences (31, 37). To determine if such sequences were also present in the adhesins expressed using the Gateway cloning system, each cloned gene fragment (excluding the signal sequence) was analyzed by the β-aggregation prediction program TANGO (11). Each protein derived from C. albicans was found to contain at least one sequence with high potential to form β strand-rich aggregates (Table 3). In contrast, no such peptide sequences were found within the cloned Cwp1p fragment.
Table 3.
Sequences within cloned C. albicans adhesins with high amyloid potential
| Protein | Sequence positiona | Sequenceb |
|---|---|---|
| Als3p | 261 | IFITY |
| 325 | IVIVAT | |
| Eap1p | 98 | TAYTTTVITVT |
| 119 | AVTTGVTIITVT | |
| 276 | TVITV | |
| 293 | VTTGVVTI | |
| Hwp1p | 312 | TVVTVT |
| 329 | VTTGVIVIT | |
| 377 | TVITVT | |
| 394 | VTTGVVVVT | |
| Rbt1p | 85 | FNAAFVV |
| 415 | TTVITVT | |
| 433 | VTTGVVVVT | |
| 515 | IVTI | |
| 531 | VTTGVVVVT |
Amino acid position of the first residue in the sequence.
Residues with β-aggregation potential of >20% were included, as calculated using TANGO algorithm; peptide sequences located within signal sequences were excluded.
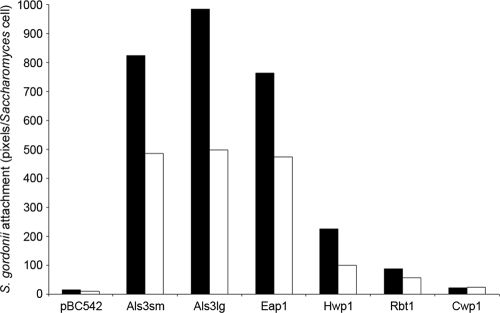
Rbt1p was found to contain multiple amyloid-forming sequences (Table 3) and to form large aggregates, yet it did not interact strongly with Streptococcus gordonii (Fig. 7). This implied that streptococcal attachment did not simply reflect the capabilities of S. cerevisiae cells to form β strand-rich aggregates but rather was dependent upon the specific surface protein profile of each surrogate host strain. To further confirm this specificity of binding, studies were performed using Congo red, a dye compound that is able to disrupt amyloid formation (37). Congo red was successfully employed to disrupt formation of large aggregates (>10 cells) by the S. cerevisiae heterologous host strains. Interactions of Streptococcus gordonii with each heterologous host strain were then visualized in the presence of Congo red (Fig. 8), and levels of streptococcal attachment were semiquantified by counting the number of fluorescent pixels associated with each surrogate host strain in the presence and absence of Congo red (Fig. 9). Disruption of amyloid-forming aggregates reduced attachment of Streptococcus gordonii DL1 cells to S. cerevisiae expressing Als3p (small and large alleles) or Eap1p by 40 to 50% compared to cells in the absence of Congo red (Fig. 9). This suggested that clumps of cells might promote streptococcal interactions. Importantly, however, levels of Streptococcus gordonii binding to Als3p- or Eap1p-expressing cells in the presence of Congo red remained approximately 50-fold higher than to cells with pBC542 vector alone or expressing control protein Cwp1p (Fig. 8 and 9). Likewise, levels of Streptococcus gordonii attachment to cells expressing Hwp1p were inhibited by approximately 50% in the presence of Congo red but remained 10-fold higher than to control cells carrying empty vector (Fig. 8 and 9). Adhesion of Streptococcus gordonii to cells expressing Rbt1p was approximately 5-fold higher than to cells with vector alone in both the presence and absence of Congo red (Fig. 8 and 9). It was clear, therefore, that high levels of Streptococcus gordonii attachment to S. cerevisiae cells were specific to expression of Als3p and Eap1p and that these interactions were independent of large aggregate formation.
Fig. 8.
Effects of Congo red on the interactions of Streptococcus gordonii DL1 with S. cerevisiae expressing C. albicans adhesins. S. cerevisiae cells expressing candidal adhesins Als3p, Eap1p, Hwp1p, and Rbt1p, together with control protein Cwp1p and pBC542 vector alone, were grown in CSM for 3 h in the presence of 30 μM Congo red and incubated for a further 1 h with FITC-labeled streptococci. Cells were then harvested and visualized by fluorescence and light microscopy (scale bar, 50 μm). Arrows indicate examples of streptococci attached to yeast cells.
Fig. 9.
Binding of Streptococcus gordonii DL1 to S. cerevisiae expressing C. albicans adhesins in the presence or absence of Congo red. S. cerevisiae cells expressing candidal adhesins Als3p, Eap1p, Hwp1p, and Rbt1p, together with control protein Cwp1p and pBC542 vector alone, were grown in CSM for 3 h in the presence (open bars) or absence (filled bars) of 30 μM Congo red and incubated for a further 1 h with FITC-labeled streptococci. Cells were then harvested and visualized by fluorescence and light microscopy. Levels of streptococcal attachment were defined as the number of fluorescent pixels bound per Saccharomyces cell.
We also tested the ability of the Streptococcus gordonii cshA mutant, which is reduced in attachment to C. albicans (14), to bind S. cerevisiae expressing C. albicans adhesins. It is apparent that the CshA surface protein, which forms fibrils and confers hydrophobic properties to Streptococcus gordonii cells (26, 27), contains seven motifs with high amyloid-forming potential. We hypothesized, therefore, that it might be an interaction between the amyloid-forming sequences of CshA and those of candidal proteins Als3p or Eap1p, which promote Streptococcus-Candida interactions. However, the Streptococcus gordonii cshA mutant was unaffected in binding S. cerevisiae expressing Als3p, Eap1p, or Hwp1p compared to wild-type Streptococcus gordonii DL1 (data not shown). These results indicate that Streptococcus gordonii CshA does not primarily recognize C. albicans proteins Als3p, Eap1p, or Hwp1p and that possession of motifs with β-aggregation potential is not sufficient to mediate interactions with other proteins bearing similar amyloid-forming sequences. Other adhesin-receptor interactions must be occurring between Streptococcus gordonii and C. albicans. These are currently under investigation.
DISCUSSION
A critical first step in successful colonization of the oral cavity by any microorganism is attachment to a surface. This enables retention and subsequent growth, which in turn may lead to biofilm formation, incorporation within a polymicrobial community, and, in the case of pathogens, disease progression. For C. albicans, the expression of multiple surface protein adhesins with differing receptor specificities is essential to the attachment processes (7). Such adhesins mediate binding of C. albicans to mucosal tissues, to saliva-coated surfaces, and to other members of the oral microflora. In many instances, however, the adhesin-receptor mechanisms have yet to be defined.
To study the interactive properties of C. albicans CWPs, selected members of this protein family were expressed in surrogate host S. cerevisiae using expression vector pBC542 (50). Expression of each protein was under the control of the TEF1 promoter, and immunofluorescence microscopy confirmed successful surface presentation of the HA-tagged C. albicans proteins. Having originally been used to investigate properties of the C. glabrata Epa adhesin family, all fusion proteins expressed from pBC542 carry a short segment of Epa1p toward the C terminus. However, use of a Cwp1p control strain, overexpressing a major S. cerevisiae cell wall protein, demonstrated that this Epa1p segment played no significant role in mediating attachment to Streptococcus gordonii or any of the other receptor substrata tested here. As shown for C. glabrata (50), pBC542 is therefore a viable expression vector for adhesion and biofilm studies with C. albicans.
Utilizing the S. cerevisiae Gateway expression system, C. albicans CWPs Als3p and Hwp1p and, to a lesser extent, Rbt1p were found to promote attachment and biofilm formation on a salivary pellicle. As every surface within the oral cavity is continually bathed in saliva, an ability to bind immobilized salivary components will likely play a significant role in facilitating oral colonization. Als3p and Hwp1p have already been implicated in mediating biofilm formation. To date, however, such studies have focused on in vitro and in vivo catheter models (28, 29, 47). This is the first study to consider biofilm formation upon the salivary pellicle, which approaches conditions that might be anticipated within the oral cavity. Both allelic forms of Als3p exhibited similar phenotypes, implying that the number of tandem repeats within this protein (amino acid residues 434 to 862 or 754 for large and small alleles, respectively) do not notably affect biofilm-forming capabilities. With three of the four heterologously expressed candidal proteins investigated demonstrating various levels of binding to a salivary pellicle, it is clear that redundancy in adhesive function exists across the CWPs. Such a strategy likely ensures that C. albicans can maintain attachment to saliva-coated tissues under a variety of conditions, therefore promoting persistence within the mouth. Interestingly, als1Δ/als1Δ, als3Δ/als3Δ, and hwp1Δ/hwp1Δ mutant cells, which individually are impaired in biofilm formation on catheters, were recently shown to form mixed biofilm communities on silicone when two or more mutant strains were incubated together (30). These observations support the notion of redundancy in C. albicans adhesin functions, as suggested by our results, but also the possibility of complementarity in recognition of surface receptors. It is possible, therefore, that such complementary adhesive function between Hwp1p and Als3p may be applicable to biofilm formation on a range of substrata.
Eap1p has previously been suggested to mediate binding of C. albicans cells to polystyrene (20). Data presented here support these findings and also show that Hwp1p and both allelic forms of Als3p can promote substantial attachment to polystyrene. Likewise, these studies corroborate previous reports that Als3p (47) and Hwp1p (29) mediate attachment to silicone, functions that have potentially important implications for candidal colonization of indwelling medical devices such as catheters and pacemakers. Such a function has also been reported for Eap1p (22), but only low-level binding to silicone, albeit statistically significant, was seen for Eap1p- compared to Als3p- and Hwp1p-expressing cells here. This discrepancy likely reflects problems with cell dispersal, which hindered biofilm studies. It might also relate to the fact that previous work derives from an in vivo rat central venous catheter model (22), for which environmental conditions will have been quite different from those used in these in vitro assays. Importantly, these data collectively serve to confirm that candidal proteins expressed on the surface of S. cerevisiae using the Gateway cloning system are presented in a conformation that maintains previously reported functions. It is also of note that Rbt1p was found to facilitate adhesion to both salivary pellicle and polystyrene, albeit at a lower level than Als3p and Hwp1p. Rbt1p has been implicated in pathogenesis based on evidence that an rbt1Δ/rbt1Δ mutant was found to exhibit significantly reduced virulence in mouse and rabbit infection models compared to that of the parent strain (6). This is the first time, however, that Rbt1p has been shown to function as an adhesin, like Hwp1p, with which it shares 43% amino acid sequence identity.
The ability of C. albicans to attach to and colonize mucosal tissues is a risk factor for disseminated candidiasis (25, 42), and three of the candidal proteins tested here have already been associated with mediating interactions with host epithelia. Als3p has been shown to function as an invasin, inducing endocytosis and uptake of C. albicans into endothelial and epithelial cells by targeting N-cadherin and E-cadherin, respectively (35). The N-terminal domain of Hwp1p is structurally like those of small proline-rich proteins expressed on the surface of stratified squamous epithelium. This region is proposed to act as a substrate for mammalian transglutaminases, resulting in cross-linking of C. albicans to the surface of buccal epithelial cells (40). Eap1p has also been shown to promote binding to epithelial cells (20, 22), although no receptor has been identified. Such studies predominantly consider direct binding to epithelial cells, but another potential target for C. albicans in tissue colonization and invasion is the ECM. With the exception of biomass results with fibronectin, which can also be found on epithelial cell surfaces, most ECM components are located within the basement membrane underlying epithelia (19). However, should this layer be perturbed, an ability to bind ECM proteins may promote candidal tissue invasion and subsequent passage into the bloodstream (49). Using microtiter plate assays, both Als3p and Hwp1p were shown to bind fibrinogen, with Hwp1p also exhibiting strong attachment to fibronectin. Sheppard et al. (39) found that expression of Als3p was shown to confer upon S. cerevisiae the ability to bind fibronectin, but in these studies Als3p adhesion to fibronectin was only marginally greater than to the blocking agent gelatin. However, Als3p is not the only fibronectin-binding adhesin expressed by C. albicans, because an als3Δ/als3Δ mutant was not abrogated in adhesion to fibronectin (48). From our studies, Als3p was also shown to mediate attachment to collagen type IV, which constitutes approximately 50% of basement membrane (19). This again demonstrates the multiple substratum specificities of Als3p and Hwp1p. It is also interesting that binding to ECM components was found only for those proteins expressed exclusively on hyphae. Hyphae are the morphological form most associated with candidal invasive disease and invade host tissues by either induction of endocytosis or active penetration through or between cells (9, 33, 35, 43). Consequently, it is this morphology that is most likely to access the underlying basement membrane of mucosal tissues and so utilize ECM proteins as adherence targets.
For each substratum tested to which Hwp1p bound (i.e., saliva, polystyrene, fibronectin, fibrinogen), biofilm formation over 24 h resulted in the highest biomass levels of any of the adhesive heterologous host strains. This likely reflects, at least in part, the fact that expression levels of Hwp1p on the surface of S. cerevisiae were approximately 2-fold greater than those of the other candidal adhesins. Nonetheless, with the exception of biomass results with Eap1p-expressing cells on polystyrene, these differences in biomass were 3-fold or greater, implying that an elevated expression level might not be the only explanation for these observations. All heterologous host strains exhibited similar growth kinetics (data not shown). It is unlikely, therefore, that such variation in biomass resulted from accelerated growth of Hwp1p-expressing cells compared to the other strains. Rather, substantial increases in biomass were seen for Hwp1p-expressing cells from initial attachment (4 h) to subsequent biofilm formation (24 h) compared to those of other heterologous expression strains. This was particularly evident for assays with salivary pellicle, fibronectin, and fibrinogen, and it implies that S. cerevisiae cells expressing Hwp1p might be incorporated more efficiently into a developing biofilm and/or form more stable biofilm structures than those expressing other candidal CWPs. This correlates well with the work of Nobile et al. (29), in which it was shown that an hwp1Δ/hwp1Δ mutant was impaired in retention of cells within an in vitro silicone biofilm model, demonstrated by higher cell numbers in the surrounding medium compared to those of wild-type cells. Likewise, OD600 values for the surrounding supernatants following 24-h biofilm formation on saliva and polystyrene were approximately 2- to 3-fold lower for S. cerevisiae cells expressing Hwp1p than for those expressing other candidal adhesins, with the exception of Eap1p on polystyrene (data not shown). This implies that Hwp1p may function to bind back cells into the developing biofilm from the surrounding environment. Such a role in mediating cell-cell aggregation might also explain why Hwp1p is induced by pheromone during C. albicans mating (3, 46).
Another essential factor in determining the success of colonization is the nature of interactions with other members of the microflora. Pioneer colonizer Streptococcus gordonii mediates attachment to C. albicans via surface protein adhesins SspA, SspB, and CshA and linear cell wall phosphopolysaccharides (13, 14). In addition, it has recently been shown that Streptococcus gordonii promotes hypha formation by C. albicans via a mechanism involving LuxS (1) and that biofilm formation by each species is enhanced by the presence of the other (1). Such a synergistic relationship has potential clinical implications both for the likelihood of long-term C. albicans carriage and for progression to disease. In this study, C. albicans proteins Als3p and Eap1p were found to confer upon S. cerevisiae a strong capacity to bind Streptococcus gordonii. Interactions were also mediated by candidal Hwp1p but to a lesser extent. This is some of the first evidence of specific candidal CWPs facilitating intermicrobial interactions. Als3p (15, 16), Eap1p (20, 21), and Hwp1p (41) share similar structures, with an N-terminal segment containing substratum-binding domains, followed by a central “stalk” of tandem repeat sequences, and terminating in a C-terminal portion with a GPI anchor. It is possible, therefore, that these proteins display a common conformational binding domain that is recognized by streptococci. This hypothesis is currently being tested. Investigations are also under way to determine if these candidal proteins are the receptors targeted directly by streptococcal adhesins SspA and SspB.
When intermicrobial interactions were visualized, streptococci were often seen to associate with large aggregates of S. cerevisiae cells. Large aggregate formation also occurred in the absence of streptococci for every S. cerevisiae strain expressing a candidal protein. Rauceo et al. (37) reported the propensity of S. cerevisiae expressing Als5p from C. albicans to form large aggregates that could be disrupted by Congo red, a dye that targets amyloid structures and inhibits their formation. This aggregation was later attributed to the presence of a heptapeptide sequence with a high predicted tendency to form β-strand-rich aggregates (31). The heptapeptide sequence is also conserved in Als1p and Als3p (31), providing an explanation for the autoaggregation seen for the Als3p-expressing S. cerevisiae cells. Analysis of the other four proteins expressed via the Gateway system using a statistical algorithm, TANGO, to predict protein aggregation (11) found that Eap1p, Hwp1p, and Rbt1p also carry β-strand-rich aggregate sequences. This is in agreement with recent findings by Ramsook et al. (36). Interestingly, however, no such peptide was found in Cwp1p. This correlates well with the ability of each S. cerevisiae strain expressing these proteins to autoaggregate, and it supports previous reports that Eap1p (21, 22) and Hwp1p (29) are involved in cell-cell interactions. The amyloid-forming sequences listed in Table 3 are characterized by an abundance of β-branched aliphatic amino acids (Ile, Thr, Val). It has been proposed that the hydrophobicity and side chain interactions of these residues serve to stabilize amyloid formation between protein molecules, both on the same cell, leading to adhesin “bundling,” and on adjacent cells, leading to aggregate formation (36). Large aggregates of cells expressing Als3p, Eap1p, or Hwp1p were associated with high levels of streptococcal attachment, possibly related to the provision of large targets for initial bacterial contact. Importantly, however, large-aggregate formation was not essential for streptococcal adherence. Streptococcus gordonii failed to bind strongly to large aggregates of cells expressing Rbt1p. Furthermore, streptococcal adhesion retained its specificity for cells expressing Als3p, Eap1p, and, to a lesser extent, Hwp1p even in the presence of Congo red. Attachment of Streptococcus gordonii to S. cerevisiae cells was therefore dependent upon specific expression of candidal adhesins Als3p, Eap1p, and Hwp1p rather than a nonspecific interaction with large aggregates of these cells. The presence of β-strand-rich aggregate sequences within Hwp1p might function in retaining yeast cells within a developing biofilm. However, Rbt1p contains similar sequences but does not display such a biofilm phenotype, suggesting that these sequences might be necessary but not sufficient for retention of cells within the biofilm.
Taken together, these data confirm the importance of adhesins Als3p and Hwp1p in mediating C. albicans biofilm formation, and they highlight the role of Hwp1p in retaining cells within a developing community. Salivary pellicle and fibrinogen were identified as novel substrata for both adhesins, and Als3p was also found to target collagen type IV, an interaction that might facilitate host tissue penetration. It is clear, therefore, that Als3p and Hwp1p may prove useful as therapeutic targets for C. albicans infections at a range of sites. The next step is to confirm that adhesin substratum specificities demonstrated here also contribute to C. albicans colonization and pathogenesis in vivo. In addition to interactions with host components, Als3p and Eap1p were shown to target receptors on the cell surface of oral bacterium Streptococcus gordonii. This advances our understanding of the molecular mechanisms underpinning the strong synergy seen between these two oral microorganisms (1), identifying for the first time receptors on the surface of C. albicans that are recognized by streptococci. Future studies will seek to identify the adhesive epitopes involved in these adhesin-receptor interactions. Such information could ultimately be applied to the development of novel strategies to control C. albicans colonization and biofilm formation.
ACKNOWLEDGMENTS
We thank Brendan Cormack and Margaret Zupancic (Johns Hopkins University School of Medicine, Baltimore, MD) for provision of destination vector pBC542 and Harriet Turner for technical assistance.
This work was supported by NIH (NIDCR) grant R01 DE016690 awarded to H.F.J.
Footnotes
Published ahead of print on 13 August 2010.
REFERENCES
- 1.Bamford C. V., d'Mello A., Nobbs A. H., Dutton L. C., Vickerman M. M., Jenkinson H. F. 2009. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect. Immun. 77:3696–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Aryeh H., Blumfield E., Szargel R., Laufer D., Berdicevsky I. 1995. Oral Candida carriage and blood group antigen secretor status. Mycoses 38:355–358 [DOI] [PubMed] [Google Scholar]
- 3.Bennett R. J., Uhl M. A., Miller M. G., Johnson A. D. 2003. Identification and characterization of a Candida albicans mating pheromone. Mol. Cell. Biol. 23:8189–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman J., Sudbery P. E. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918–930 [DOI] [PubMed] [Google Scholar]
- 5.Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132 [DOI] [PubMed] [Google Scholar]
- 6.Braun B. R., Head W. S., Wang M. X., Johnson A. D. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaffin W. L. 2008. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72:495–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra J., Kuhn D. M., Mukherjee P. K., Hoyer L. L., McCormick T., Ghannoum M. A. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalle F., Wachtler B., L'Ollivier C., Holland G., Bannert N., Wilson D., Labruere C., Bonnin A., Hube B. 2010. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell. Microbiol. 12:248–271 [DOI] [PubMed] [Google Scholar]
- 10.Egusa H., Soysa N. S., Ellepola A. N., Yatani H., Samaranayake L. P. 2008. Oral candidosis in HIV-infected patients. Curr. HIV Res. 6:485–499 [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Escamilla A. M., Rousseau F., Schymkowitz J., Serrano L. 2004. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22:1302–1306 [DOI] [PubMed] [Google Scholar]
- 12.Gillum A. M., Tsay E. Y., Kirsch D. R. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 13.Holmes A. R., Gopal P. K., Jenkinson H. F. 1995. Adherence of Candida albicans to a cell surface polysaccharide receptor on Streptococcus gordonii. Infect. Immun. 63:1827–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes A. R., McNab R., Jenkinson H. F. 1996. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect. Immun. 64:4680–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyer L. L., Green C. B., Oh S. H., Zhao X. 2008. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med. Mycol. 46:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoyer L. L., Payne T. L., Bell M., Myers A. M., Scherer S. 1998. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr. Genet. 33:451–459 [DOI] [PubMed] [Google Scholar]
- 17.Jenkinson H. F., Douglas L. J. 2002. Interactions between Candida species and bacteria in mixed infections, p. 357–373 InBrogden K. A., Guthmiller J. M. (ed.), Polymicrobial diseases. ASM Press, Washington, DC: [PubMed] [Google Scholar]
- 18.Jenkinson H. F., Lala H. C., Shepherd M. G. 1990. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect. Immun. 58:1429–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeBleu V. S., Macdonald B., Kalluri R. 2007. Structure and function of basement membranes. Exp. Biol. Med. 232:1121–1129 [DOI] [PubMed] [Google Scholar]
- 20.Li F., Palecek S. P. 2003. EAP1, a Candida albicans gene involved in binding human epithelial cells. Eukaryot. Cell 2:1266–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F., Palecek S. P. 2008. Distinct domains of the Candida albicans adhesin Eap1p mediate cell-cell and cell-substrate interactions. Microbiology 154:1193–1203 [DOI] [PubMed] [Google Scholar]
- 22.Li F., Svarovsky M. J., Karlsson A. J., Wagner J. P., Marchillo K., Oshel P., Andes D., Palecek S. P. 2007. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot. Cell 6:931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 24.Makihira S., Nikawa H., Tamagami M., Hamada T., Samaranayake L. P. 2002. Differences in Candida albicans adhesion to intact and denatured type I collagen in vitro. Oral Microbiol. Immunol. 17:129–131 [DOI] [PubMed] [Google Scholar]
- 25.Marr K. A., Seidel K., White T. C., Bowden R. A. 2000. Candidemia in allogeneic blood and marrow transplant recipients: evolution of risk factors after the adoption of prophylactic fluconazole. J. Infect. Dis. 181:309–316 [DOI] [PubMed] [Google Scholar]
- 26.McNab R., Forbes H., Handley P. S., Loach D. M., Tannock G. W., Jenkinson H. F. 1999. Cell wall-anchored CshA polypeptide (259 kilodaltons) in Streptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J. Bacteriol. 181:3087–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNab R., Jenkinson H. F., Loach D. M., Tannock G. W. 1994. Cell-surface-associated polypeptides CshA and CshB of high molecular mass are colonization determinants in the oral bacterium Streptococcus gordonii. Mol. Microbiol. 14:743–754 [DOI] [PubMed] [Google Scholar]
- 28.Nobile C. J., Andes D. R., Nett J. E., Smith F. J., Yue F., Phan Q. T., Edwards J. E., Filler S. G., Mitchell A. P. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nobile C. J., Nett J. E., Andes D. R., Mitchell A. P. 2006. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 5:1604–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nobile C. J., Schneider H. A., Nett J. E., Sheppard D. C., Filler S. G., Andes D. R., Mitchell A. P. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 18:1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otoo H. N., Lee K. G., Qiu W., Lipke P. N. 2008. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot. Cell 7:776–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pakula R., Walczak W. 1963. On the nature of competence of transformable streptococci. J. Gen. Microbiol. 31:125–133 [DOI] [PubMed] [Google Scholar]
- 33.Park H., Myers C. L., Sheppard D. C., Phan Q. T., Sanchez A. A., Edwards E., Filler S. G. 2005. Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell. Microbiol. 7:499–510 [DOI] [PubMed] [Google Scholar]
- 34.Perlroth J., Choi B., Spellberg B. 2007. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med. Mycol. 45:321–346 [DOI] [PubMed] [Google Scholar]
- 35.Phan Q. T., Myers C. L., Fu Y., Sheppard D. C., Yeaman M. R., Welch W. H., Ibrahim A. S., Edwards J. E., Jr., Filler S. G. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsook C. B., Tan C., Garcia M. C., Fung R., Soybelman G., Henry R., Litewka A., O'Meally S., Otoo H. N., Khalaf R. A., Dranginis A. M., Gaur N. K., Klotz S. A., Rauceo J. M., Jue C. K., Lipke P. N. 2010. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot. Cell 9:393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauceo J. M., Gaur N. K., Lee K. G., Edwards J. E., Klotz S. A., Lipke P. N. 2004. Global cell surface conformational shift mediated by a Candida albicans adhesin. Infect. Immun. 72:4948–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39.Sheppard D. C., Yeaman M. R., Welch W. H., Phan Q. T., Fu Y., Ibrahim A. S., Filler S. G., Zhang M., Waring A. J., Edwards J. E., Jr 2004. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 279:30480–30489 [DOI] [PubMed] [Google Scholar]
- 40.Staab J. F., Bradway S. D., Fidel P. L., Sundstrom P. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535–1538 [DOI] [PubMed] [Google Scholar]
- 41.Staab J. F., Sundstrom P. 1998. Genetic organization and sequence analysis of the hypha-specific cell wall protein gene HWP1 of Candida albicans. Yeast 14:681–686 [DOI] [PubMed] [Google Scholar]
- 42.Takesue Y., Kakehashi M., Ohge H., Imamura Y., Murakami Y., Sasaki M., Morifuji M., Yokoyama Y., Kouyama M., Yokoyama T., Sueda T. 2004. Combined assessment of beta-D-glucan and degree of Candida colonization before starting empiric therapy for candidiasis in surgical patients. World J. Surg. 28:625–630 [DOI] [PubMed] [Google Scholar]
- 43.Villar C. C., Kashleva H., Nobile C. J., Mitchell A. P., Dongari-Bagtzoglou A. 2007. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect. Immun. 75:2126–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson L. S., Reyes C. M., Stolpman M., Speckman J., Allen K., Beney J. 2002. The direct cost and incidence of systemic fungal infections. Value Health 5:26–34 [DOI] [PubMed] [Google Scholar]
- 45.Zaoutis T. E., Argon J., Chu J., Berlin J. A., Walsh T. J., Feudtner C. 2005. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin. Infect. Dis. 41:1232–1239 [DOI] [PubMed] [Google Scholar]
- 46.Zhao R., Daniels K. J., Lockhart S. R., Yeater K. M., Hoyer L. L., Soll D. R. 2005. Unique aspects of gene expression during Candida albicans mating and possible G1 dependency. Eukaryot. Cell 4:1175–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao X., Daniels K. J., Oh S. H., Green C. B., Yeater K. M., Soll D. R., Hoyer L. L. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152:2287–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X., Oh S. H., Cheng G., Green C. B., Nuessen J. A., Yeater K., Leng R. P., Brown A. J., Hoyer L. L. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150:2415–2428 [DOI] [PubMed] [Google Scholar]
- 49.Zhu W., Filler S. G. 2010. Interactions of Candida albicans with epithelial cells. Cell. Microbiol. 12:273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zupancic M. L., Frieman M., Smith D., Alvarez R. A., Cummings R. D., Cormack B. P. 2008. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol. Microbiol. 68:547–559 [DOI] [PubMed] [Google Scholar]