Abstract
Despite a century of research and increasing environmental and human health concerns, the mechanistic basis of the toxicity of derivatives of the metalloid tellurium, Te, in particular the oxyanion tellurite, Te(IV), remains unsolved. Here, we provide an unbiased view of the mechanisms of tellurium metabolism in the yeast Saccharomyces cerevisiae by measuring deviations in Te-related traits of a complete collection of gene knockout mutants. Reduction of Te(IV) and intracellular accumulation as metallic tellurium strongly correlated with loss of cellular fitness, suggesting that Te(IV) reduction and toxicity are causally linked. The sulfate assimilation pathway upstream of Met17, in particular, the sulfite reductase and its cofactor siroheme, was shown to be central to tellurite toxicity and its reduction to elemental tellurium. Gene knockout mutants with altered Te(IV) tolerance also showed a similar deviation in tolerance to both selenite and, interestingly, selenomethionine, suggesting that the toxicity of these agents stems from a common mechanism. We also show that Te(IV) reduction and toxicity in yeast is partially mediated via a mitochondrial respiratory mechanism that does not encompass the generation of substantial oxidative stress. The results reported here represent a robust base from which to attack the mechanistic details of Te(IV) toxicity and reduction in a eukaryotic organism.
The metalloid tellurium (Te) is one of the rarest elements in the Earth's crust, and its derivatives, particularly the oxyanion tellurite, TeO32− or Te(IV), are also highly toxic to living organisms. Despite more than 100 years of physiological and molecular research, the mechanism by which tellurium species exert their toxicity still constitutes a scientific conundrum (7). This toxicity is of concern because the expanding use of tellurium in electronic appliances, optics, and batteries, together with its natural occurrence in sulfide-rich ores, gives rise to high local concentrations in connection with waste dumps and metallurgical plants, with detrimental effects on the environment and human health. In fact, although tellurium is currently not known to be an essential element, it is nevertheless one of the most abundant trace elements in human bone, superseded only by iron, zinc, and rubidium (45). An intriguing twist is that Te(IV) is reduced by living cells to volatile methylated forms such as dimethyltellurium or to elemental tellurium, Te(0) (21, 24, 52). In bacteria, the latter is typically deposited as black aggregates with strain-specific nanostructures resulting in a distinct darkening of cells and tissues (30, 55), a phenomenon that is exploitable in environmental viability screens (31).
It has long been known that different bacterial species vary in their tolerance to Te(IV), which has prompted the inclusion of TeO32− in a variety of selective bacteriological tests (48). However, the mechanistic basis of this variance remains elusive. In general, Te(IV) resistance in bacteria has been thought to be associated with accumulation of tellurium (53), although Te(IV) resistance without tellurium accumulation has been observed in phototrophic bacteria (72). For some bacterial species, the capacity to grow at high Te(IV) concentrations has been shown to depend on the presence of Te(IV) resistance-encoding genetic determinants carried on IncHI, IncHII, and IncP plasmids (65). In addition, chromosomal genes important for growth in the presence of Te(IV) have been identified in a few species, but the physiological basis of their involvement has not been clearly determined (28, 54, 58). A favorite culprit of metal toxicity is oxidative damage of proteins, DNA, and lipids. It has been proposed that this toxicity results from an ability of Te(IV) to act as a strong oxidizing agent (33), as Te(IV) directly interacts with and oxidizes cellular thiol groups (63, 64). Te(IV) has also been implicated in redox reactions involving the respiratory chain (59). Recent reports favor an oxidative mode of action; Te(IV) increases the production of reactive oxygen species (ROS) in vitro as well as in vivo, increases protein carbonylation, a common sign of oxidative damage, and inactivates oxidative stress-sensitive Fe-S enzymes (41). Furthermore, the transcription, as well as the activity, of the superoxide dismutases, critical components of the oxidative stress defense, is elevated in Te(IV)-stressed bacteria (4, 41). Nevertheless, the absence of genome-wide data on tellurium exposure leaves open the question of whether oxidative damage constitutes the dominant source of Te(IV) toxicity. Alternative toxicity mechanisms for Te(IV) and the chemophysically related selenium derivative Se(IV) have been suggested, such as the inactivation of proteins by the replacement of sulfhydryl groups or the direct quenching of respiratory chain components (30, 59, 61, 62).
To provide an unbiased and system-wide view of the mechanisms of Te(IV) toxicity and reduction in eukaryotes, we here quantified the tolerance of the Saccharomyces cerevisiae gene knockout collection to Te(IV), as well as measured Te(0) accumulation. Our results demonstrated that Te(IV) toxicity and reduction to tellurium are inversely correlated; low Te(0) accumulation coincided with high Te(IV) tolerance and vice versa, suggesting a causal relationship between accumulation and toxicity. In addition, we show that two cellular routes mediate the bulk of Te(IV) reduction and toxicity in yeast, the sulfate assimilation pathway and a mitochondrial mechanism that encompasses respiration but not the generation of oxidative damage. We also found that Te(IV) toxicity and reduction strongly overlapped with Se(IV) toxicity and reduction and with selenomethionine toxicity, suggesting a common mode of action.
MATERIALS AND METHODS
Strains and media.
Throughout this study, the diploid S. cerevisiae type strain S288c and diploid (BY4743 [MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met17Δ0/MET17 LYS2/lys2Δ0 ura3Δ0/ura3Δ0]) or haploid (BY4741 [MATa his3Δ1 leu2Δ0 met17Δ0 ura3Δ0] and BY4742 [MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0]) derivatives thereof were used (5). A genome-wide deletion strain screen was carried out using the haploid BY4741 deletion collection (as described above, ORF::kanMX4) and strains with significant deviations were confirmed by rescreening using the diploid homozygote BY4743 collection (as described above, ORF::kanMX4/ORF::kanMX4). For Schizosaccharomyces pombe, the standard wild-type (WT) strain 972h− was used. S. cerevisiae strains were cultivated in synthetic complete medium (0.14% yeast nitrogen base, 0.5% ammonium sulfur, 1% succinic acid, 2% [wt/vol] glucose, and 0.077% complete supplement mixture supplemented with 20 mg/liter cysteine, pH 5.8) with and without K2TeO3, Na2SeO3, and selenomethionine (all chemicals were from Sigma Aldrich). For the study of respiratory growth, glucose was replaced with a mixture of 2% (wt/vol) ethanol and 3% (wt/vol) glycerol. Where indicated, methionine-free medium was also used. S. pombe strains were cultivated in YES medium (0.5% yeast extract, 3% [wt/vol] glucose, 225 mg/liter histidine, leucine, lysine, arginine, adenine, and uracil) where indicated. Glutathione suppression experiments were performed with synthetic complete medium supplemented with 5 mM reduced glutathione (GSH). Agar at 20 g/liter was added to solid medium for growth experiments. All strains were stored in 20% (wt/vol) glycerol solution at −80°C and cultivated at 30°C.
Liquid medium microcultivation.
Liquid medium growth curves of yeast strains growing for 72 h in the presence and or absence of the indicated concentrations of K2TeO3 were obtained using a high-resolution microcultivation approach as previously described (66, 67). Strains were tested in duplicate (n = 2).
Optical microscopy of Te(IV)-stressed cells.
Te(0) plaque formation in individual exponentially growing cells was monitored by time-lapse microscopy. S. cerevisiae and S. pombe cells were precultivated overnight in liquid medium and resuspended to an optical density at 610 nm (OD610) of ∼0.1. When an OD610 of ∼0.2 was reached, 0.5 mM K2TeO3 was added and cells were placed in an enclosed 160-μl chamber, heated to 30°C, and mounted in an inverted microscope (Nikon TE2000 PFS). Except where otherwise stated, bright-field images were acquired every 5 min for 20 h using a 60×/numerical aperture (NA) 1.4 or a 100×/NA 1.45 oil immersion objective (Nikon) and a charge-coupled device camera (iXon; Andor). Te(0) plaque formation in S. cerevisiae cells grown on agar plates containing 0.5 or 2 mM K2TeO3 at 30°C was followed by imaging of the cells every 24 h for 5 days. Prior to imaging, cells were resuspended in liquid medium, placed between two cover glasses, and imaged in bright-field mode (as described above). To follow Te(0) plaque localization with regard to the vacuole, time-lapse measurements were performed with S. cerevisiae and S. pombe cells fluorescently stained with the vacuolar membrane styryl dye FM 4-64 (T-3166; Invitrogen). Cells were precultivated in medium (as described above), resuspended to an OD610 of ∼0.3 in 0.5 mM K2TeO3 medium, and cultivated for 4 h (S. pombe) or ∼18 h (S. cerevisiae). Cultivated cells were concentrated ∼10× by centrifugation (10,000 rpm, 2 min) and resuspension in 100 to 200 μl medium, and 1 to 4 μl of dye was added. Following 20 to 30 min of preincubation at 30°C to facilitate uptake of the dye, cells were pelleted (10,000 rpm, 2 min), resuspended in 1 ml of 0.5 mM K2TeO3 medium, cultivated for ∼2 h at 30°C, and concentrated ∼5× (as described above) for imaging. A 160-μl volume of culture was placed between cover glasses precoated with immobilizing concanavalin A and heated to 30°C. Cells were imaged every 30 s (as described above). Fluorescence images were acquired using excitation filter D540/25x, dichroic mirror 565DCLP, and emission filter D605/55m (Chroma Corporation).
Raman measurements.
Raman spectra were collected using a Raman spectrometer (Renishaw 2000) in combination with an inverted microscope (TE2000E; Nikon). The microscope was equipped with a 60×/NA 0.7 objective (Nikon) and an argon laser (Spectra Physics 2060) tuned to 514.5 nm for excitation. The integration time was 30 s with a power of ∼3.5 mW at the sample. An angle-tuned holographic notch filter was used to block the Rayleigh scattered light. Raman spectra were collected from WT S. cerevisiae cells cultivated without K2TeO3 or with 2 mM K2TeO3, oxidized tellurium TeO2, or elemental tellurium Te(0). Cells were grown on synthetic complete medium plates (20 g/liter agar) for 11 days and resuspended in phosphate-buffered saline, and 5 μl was withdrawn and put between two cover glasses, of which one was precoated with concanavalin A to immobilize the cells. Te(0)and TeO2 spectra were recorded from the interior and the oxidized surface of freshly cleaved Te(0) grains (Sigma Aldrich), respectively.
Segregation analysis of tellurium traits in the cross BY4741 × BY4742.
To follow the potential cosegregation of tellurium traits and the Met17Δ0 and lys2Δ0 markers in a cross between BY4741 and BY4742, strains were cospotted onto a fresh YPD plate and allowed to mate for 1 day at 30°C. Following suspension in water, cells were streaked onto sporulation plates (1% potassium acetate), sporulated for 7 days at 22°C, and suspended in Lyticase solution (5,000 U/ml; Sigma Aldrich) for 30 min at 37°C. Lysed asci were streaked onto YPD plates, and 10 tetrads were dissected using a micromanipulator (Singer Instruments, United Kingdom). Following incubation for 2 days at 30°C, tetrads were replica plated onto synthetic complete medium plates lacking methionine or lysine but containing 0.1 mM K2TeO3. Plates were incubated for 1 day at 30°C and imaged (as described above), and the potential cosegregation of Met17Δ0 and lys2Δ0 (data not shown) with tellurium traits was evaluated visually.
A genome-wide screen for Te(IV) toxicity and tellurium accumulation.
A genome-wide deletion strain screen for Te(IV) toxicity and tellurium accumulation was carried out using the haploid BY4741 deletion collection (as described above, ORF::kanMX4). Strains were pinned in 1536 format on media containing 0, 0.1, 0.2, 0.3, 0.4, and 0.5 mM K2TeO3 using a benchtop RoToR HDA robot (Singer Instruments, United Kingdom) with default settings. After 1, 2, 3, and 6 days, colonies were imaged using a 10-megapixel digital camera (Powershot A640; Canon) and stored as JPEG images. Images were evaluated by scoring of colony size and coloration using a manual evaluation system with 10 grades, colony size reflecting Te(IV) tolerance and colony darkening reflecting tellurium accumulation (n = 8, replicate colonies evaluated in aggregate). Absolute values of colony size and color were normalized to the strain average for that concentration and day, resulting in relative measurements of tellurium accumulation and Te(IV) tolerance. Compensating for general growth defects, colony size in the presence of Te(IV) was further normalized to colony size in the absence of Te(IV). Gene knockout mutants showing very slow growth in the absence of stress were excluded from evaluation in order to avoid scoring artifacts; a total of 4,311 gene knockout mutants were retained. Gene knockout mutants with similar Te(IV)-specific aberrations in colony size or color in at least three out of six independent evaluations in both screens were rescreened using the diploid homozygote BY4743 deletion collection (as described above, ORF::kanMX4/ORF::kanMX4). Rescreening was performed in 384 pinning format using 0, 0.1, and 0.4 mM K2TeO3; colony size and color were evaluated (as described above) after 1, 2, 3, and 6 days of growth (n = 4). BY4743 gene knockout mutants with confirmed K2TeO3 phenotypes were further tested for Na2SeO3 and selenomethionine phenotypes by pinning onto agar plates containing 0, 0.2, and 0.8 mM Na2SeO3 or 0, 0.03, and 0.06 mM selenomethionine.
RESULTS
Tellurium accumulation in S. cerevisiae and S. pombe.
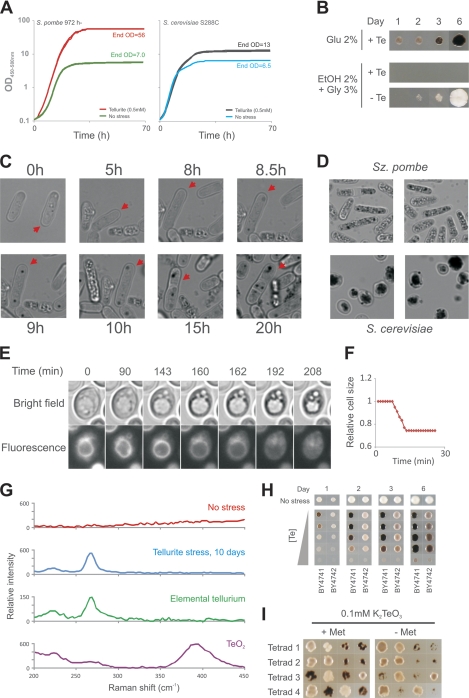
Cultivation of commonly used laboratory strains of the model yeasts S. cerevisiae and S. pombe in the presence of TeO32− resulted in the distinct cell darkening that is a hallmark of tellurium accumulation in bacteria. For batch-cultivated S. pombe cells, this Te(IV)-induced cell darkening was reflected in an OD/turbidity increase in stationary phase of about 8-fold compared to that of cells cultivated in the absence of Te(IV) (Fig. 1A). Only marginal increases were observed during exponential fermentative growth, suggesting that Te(0) accumulation is coupled primarily to respiration. In the presence of high concentrations of Te(IV), yeast also failed to utilize the respiratory carbon sources ethanol and glycerol, confirming a tight link between Te(IV) toxicity and respiration (Fig. 1B). To time the appearance of intracellular darkening following Te(IV) exposure, individual Te(IV)-stressed cells were monitored by time-lapse microscopy. In the average cell, darkening occurred as localized plaque formation with a very distinct onset at 8 to 8.5 h of Te(IV) exposure in S. pombe and 14 to 18 h of Te(IV) exposure in S. cerevisiae; however, following long exposure times, plaque formation spread throughout the cell until it was filled completely (Fig. 1C and D). By staining Te(IV)-exposed S. cerevisiae and S. pombe cells with a vacuolar membrane dye, we found that the intracellular plaques were absent from the vacuolar lumen but were formed in close proximity to the vacuolar membrane (Fig. 1E; see videos S1 to S3 in the supplemental material). After prolonged Te(IV) exposure, the integrity of the vacuole was compromised and the vacuole subsequently fractionated and disintegrated. This was followed by rapid cell shrinkage to about 75% of the initial size (Fig. 1F). In Te(IV)-exposed bacteria, cellular darkening corresponds to the accumulation of elemental tellurium aggregates with little complex binding to any organic matter (60). To confirm that cellular darkening is due to elemental tellurium accumulation also in yeast, we performed Raman spectroscopy of Te(IV)-stressed cells. Raman spectroscopy is a powerful method to obtain vibration “fingerprint” spectra of molecules and solids via inelastic scattering of laser light. No distinct Raman signatures were observed in the sample without Te(IV), but a distinct Raman active vibration peak was observed at around 270 cm−1 in Te(IV)-stressed cells. The peak position is in excellent agreement with what is expected for elemental tellurium (peak position, 268 cm−1, Fig. 1G). It should be noted that elemental tellurium can be oxidized to TeO2 in the presence of oxygen and that TeO2 thus is an alternative reaction product following cellular metabolization of Te(IV). However, the 390- to 395-cm−1 band characterizing TeO2 is absent from Raman spectra measured inside Te(IV)-stressed cells. We therefore conclude that Te(IV)-induced cellular darkening in yeast indeed results from intracellular accumulation of elemental tellurium.
Fig. 1.
Tellurium is accumulated in yeast following Te(IV) exposure. (A) Growth curves were recorded for S. cerevisiae and S. pombe cells microcultivated with and without 0.1 mM K2TeO3 for 70 h by automated measurements of medium OD/turbidity. (B) S. cerevisiae cells (BY4741) were spotted onto fermentative (glucose [Glu]) and respiratory media with and without 0.5 mM K2TeO3 and cultivated for 6 days. EtOH, ethanol; Gly, glycerol. (C) Cellular darkening and plaque formation in a single S. pombe cell (red arrow) through 20 h of 0.5 mM K2TeO3 stress. (D) Cellular darkening and plaque formation in S. pombe (3 days, liquid cultivation) and S. cerevisiae (11 days, solid medium cultivation) after exposure to 0.5 mM K2TeO3. (E) Tellurium plaque formation with respect to the vacuole was followed by time-lapse microscopy of S. cerevisiae S288c cells stained with a fluorescent vacuolar membrane dye. The time sequence shows a typical cell. Time zero is ∼19 h after the addition of 0.5 mM K2TeO3. See also videos S1 to S3 in the supplemental material. (F) The relative cell size decreases to about 75% of the initial cell size in Te(IV)-exposed cells following collapse of the vacuole. The relative size of a typical S. cerevisiae cell is shown. Time zero is ∼21 h after the addition of 0.5 mM K2TeO3. (G) Raman spectra of S. cerevisiae cells grown for 11 days in the absence or presence of 2 mM K2TeO3, as well as Raman spectra of oxidized (TeO2) and nonoxidized elemental tellurium, Te. (H) Tellurium accumulation and Te(IV) tolerance of WT S. cerevisiae strains BY4741 (MATa met17Δ0) and BY4742 (MATα lys2Δ0), which are isogenic except for their auxotrophic markers and mating types. (I) High tellurium accumulation and Te(IV) sensitivity cosegregate with the met17Δ0 marker in the F1 haploid progeny of the BY4741 × BY4742 cross. Shown is colony growth 2 days after replica plating from rich medium onto a medium supplemented with 0.1 mM K2TeO3. Met, methionine.
Investigating Te(IV) toxicity and tellurium accumulation in two essentially isogenic derivatives of the reference strain, BY4741 (met17Δ0) and BY4742 (lys2Δ0), we noted very strong differences (Fig. 1H). These strains differ only in their mating types and auxotrophic markers, providing a clear indication as to the genetic basis of their trait differences. In fact, controlled mating of the two strains, followed by tetrad dissection and phenotyping and genotyping of the F1 haploid progeny, showed a complete cosegregation of the tellurium accumulation trait and the met17Δ0 marker; absence of Met17 gave rise to high tellurium accumulation and correspondingly low tolerance to Te(IV) (Fig. 1I). MET17 encodes a cysteine and methionine synthase at the root of the divergence of the methionine and cysteine biosynthesis pathways, implicating sulfur assimilation as one determinant of tellurium accumulation in yeast.
Sulfate assimilation mediates tellurium accumulation in yeast.
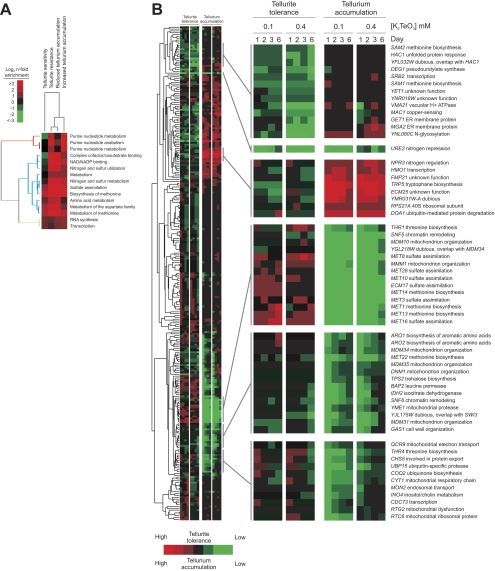
To provide a genome-wide view of the genetic determinants that control the intracellular fate of Te(IV), we screened a collection of nonessential haploid S. cerevisiae gene knockout mutants in the BY4741 background for aberrations in Te(IV) tolerance and Te(0) accumulation. Changes in Te(IV) tolerance were quantified as Te(IV)-specific deviations in colony size on agar, whereas Te(0) accumulation variations were scored as deviations in colony darkening. Evaluating 4,310 gene knockout mutants, we found 3.1% to have deviations in Te(0) accumulation and 6.3% to feature variations in Te(IV) tolerance. We observed very strong links between tolerance and accumulation variations; gene knockout mutants with high tellurium accumulation also tended to show low tolerance to Te(IV) (7.3-fold enrichment [P = 5 × 10−9; Fisher's exact test]) whereas gene knockout mutants with reduced tellurium accumulation tended to have high tolerance to Te(IV) (9.4-fold enrichment [P = 2 × 10−11; Fisher's exact test]). This suggests that reduction of Te(IV) to bioaccumulated tellurium is an important cause of Te(IV) toxicity, in complete contrast to observations from bacterial studies indicating that accumulation of tellurium is frequently associated with Te(IV) resistance (53). To identify cellular functions linked to variations in tellurium accumulation and Te(IV) tolerance, gene ontology functions overrepresented among gene knockout mutants with variations in tellurium phenotypes were identified. Essentially all of the biological processes that were enriched among Te(IV)-tolerant gene knockout mutants were also enriched among gene knockout mutants with low tellurium accumulation, confirming the inverse functional link between tolerance and accumulation (Fig. 2A). To exclude the possibility of recessive secondary site mutations penetrating in the haploid background and also control for the influence of the met17Δ0 marker, gene knockout mutants with tellurium phenotypes were rescreened using homozygous diploid knockout mutants (BY4743 [met17Δ0/MET17 lys2Δ0/LYS2]). In the diploid knockout mutants, 75% of the knockout phenotypes could be stringently verified while the remaining 25% were discarded.
Fig. 2.
A genome-wide screen for gene knockout-caused variations in tellurium accumulation and tolerance in S. cerevisiae. A genome-wide screen for gene knockout-induced variations in tellurium accumulation and tolerance in S. cerevisiae was carried out. About 4,200 gene knockout mutants of haploid reference strain BY4741 were robotically pinned onto K2TeO3 (0.1 or 0.4 mM)-containing solid medium, and colony size and color were recorded after 1, 2, 3, and 6 days of growth. (A) Cellular functions enriched (Fisher's exact test, P < 0.001) among gene knockout mutants with significant aberrations in Te(IV) tolerance and/or tellurium accumulation (colony color) using biological process annotations from MIPS (http://mips.gsf.de/genre/proj/yeast). Heat map colors reflect degrees of enrichment. (B) Gene knockout mutants influencing Te(IV) tolerance and/or tellurium accumulation were confirmed using homozygote diploid knockout mutants in the BY4743 background. Confirmed gene knockout mutants were hierarchically clustered (Pearson correlation coefficients, average-linkage clustering) according to degree of deviation in Te(IV) tolerance and tellurium accumulation in relation to the BY4743 reference strain.
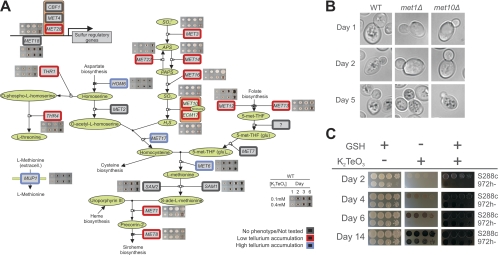
The most striking functional characteristic among gene knockout mutants with low tellurium accumulation was the strong overrepresentation of genes involved in sulfate assimilation (Fig. 2A). Knockout mutants lacking all of the genes in the sulfur assimilation pathway upstream of MET17, i.e., MET3, MET10, MET14, MET16, MET22, and ECM17, consistently showed low tellurium accumulation and correspondingly high Te(IV) tolerance (Fig. 2A and 3A). These effects were observed in both the haploid BY4741 and the diploid BY4743 and were thus independent of met17Δ0. A knockout mutant lacking the key transcriptional regulator of sulfur assimilation genes, MET28, featured phenotypes similar to those of the above-mentioned MET genes, as did met12Δ and met13Δ in the intersecting branch from folate biosynthesis (Fig. 3A). Knockout mutants lacking SAM1, SAM2, and MET6 downstream of Met17 showed normal or strongly increased tellurium accumulation, depending on the absence or presence of MET17, and sam1Δ and sam2Δ also caused low Te(IV) tolerance (Fig. 2B). Interestingly, met1Δ and met8Δ knockout mutants lacking enzymes that act downstream of Sam1, Sam2, and Met6 showed very low tellurium accumulation and high Te(IV) tolerance (Fig. 2B and 3A). This is very likely explained by the fact that they are required for the production of siroheme, an indispensable cofactor of the sulfite reductase encoded by MET10 and ECM17, in the upper parts of the pathway (Fig. 4A). The low tellurium accumulation in met1Δ and met10Δ mutant cells was verified by time-resolved microscopic monitoring of tellurium plaque formation in single cells; the decreased rate of plaque formation was clear in both met1Δ and met10Δ mutant cells, and only after 5 days of Te(IV) exposure did substantial Te(0) accumulation occur (Fig. 3B). Taken together, these observations are consistent with the idea that the activity of the sulfate assimilation pathway upstream of Met17 is a major determinant of Te(IV) toxicity and tellurium accumulation in yeast.
Fig. 3.
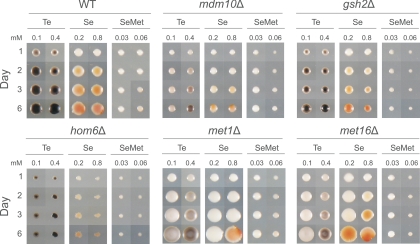
Tellurium accumulation is mediated via the sulfate assimilation pathway. (A) Significant deviations in tellurium accumulation (colony color) of homozygote diploid gene knockout mutants in the sulfur assimilation superpathway after 1, 2, 3, and 6 days of exposure to 0.1 or 0.4 mM K2TeO3. THF = tetrahydrofolate. The phenotype caused by mup1Δ was only observed in the met17Δ mutant background. (B) Time-resolved microscopy of tellurium accumulation in WT and met1Δ and met10Δ mutant cells exposed to 0.5 mM Te(IV) stress. Representative cells are displayed. (C) Growth of S. cerevisiae (S288c) and S. pombe (972h−) cells after 2 to 14 days of growth in the presence or absence of 0.5 mM K2TeO3 and in the presence or absence of 5 mM GSH. Observe that the medium in the presence of both K2TeO3 and GSH immediately turns black because of extracellular conversion of Te(IV) to Te(0).
Fig. 4.
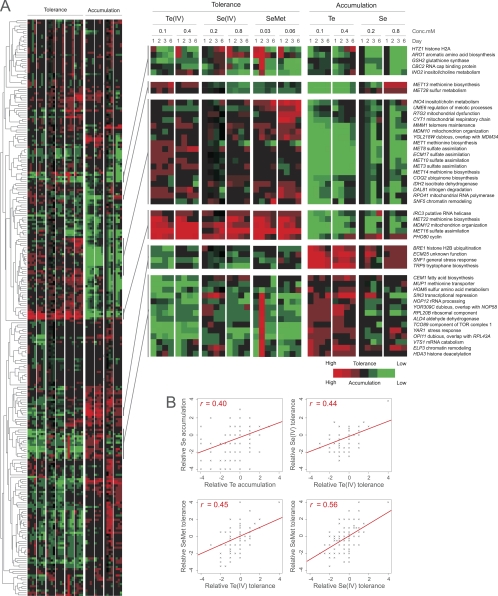
Te(IV), selenite, and selenomethionine have toxicity mechanisms in common. Homozygote diploid gene knockout mutants with confirmed significant deviations in Te phenotypes were pinned onto solid agar medium containing 0.2 or 0.8 mM Na2SeO3 or 0.03 or 0.06 mM selenomethionine (SeMet). Relative selenium accumulation (colony color) and selenite and selenomethionine tolerance (colony size) were quantified by visual scoring after 1, 2, 3, and 6 days of growth on solid agar medium. (A) Relative deviations in Te(IV), selenite, and selenomethionine tolerance and reduction (accumulation) were used to group gene knockout mutants by hierarchical clustering (average-linkage clustering, Pearson correlation coefficients). Heat map colors indicate degrees of tolerance/accumulation in relation to the reference strain (WT). (B) Comparison of accumulation/tolerance phenotypes following growth on selenite, Te(IV), and selenomethionine. Values indicate degrees of tolerance/accumulation in relation to the reference strain (WT). Linear correlations are indicated.
Our screens were performed in the presence of >1 mM external methionine, repressing the activity of the endogenous sulfur assimilation system to less than 10% of its maximal capacity (9). Not surprisingly, loss of MUP1, encoding the high-affinity methionine permease in yeast (19), and the accompanying activation of the sulfate assimilation activity resulted in strongly reduced tolerance of Te(IV) (Fig. 2B and 3A) and, in the met17Δ0 background, high tellurium accumulation. Methionine removal also strongly increased Te(IV) toxicity: in the absence of methionine, WT cells failed to grow in high (>1 mM) concentrations of Te(IV) (data not shown). Hence, both the rate of Te(0) accumulation and the toxicity of Te(IV) were linked to the activity of the sulfur assimilation pathway. MET gene transcription is largely controlled by the Met28-Met31-Met32 and Met28-Cbf1-Met4 complexes. MET4 is essential, Met 31/Met32 is functionally redundant, and Cbf1 is not known to lead to defects in sulfate assimilation; hence, it is not surprising that met28Δ is alone among the corresponding knockout mutants in showing strong tellurium phenotypes. However, many MET gene promoters also contain inositol/choline-responsive elements which are typically bound by the Ino2p/Ino4p transcription activation complex (1). Consequently, the dependence of MET gene expression on Ino2/4 promoter activation may explain the very low tellurium accumulation observed in ino2Δ and ino4Δ mutants (Fig. 2B; see Table S1 in the supplemental material). Loss of THR1 or THR4, converting homoserine to threonine, lowered tellurium accumulation, whereas loss of HOM6, which catalyzes the production of homoserine, conferred increased tellurium accumulation and a corresponding lower tolerance (Fig. 2B and 3A). External cysteine was supplied to allow screening of cysteine-auxotrophic gene knockout mutants which all showed normal tellurium phenotypes. Furthermore, loss of GSH2, which encodes the glutathione synthase acting downstream of cysteine biosynthesis, conferred only a mild increase in Te(IV) toxicity and somewhat lower Te(0) accumulation. Surprisingly, external addition of an excess of GSH caused an immediate and drastic darkening of the growth medium, suggesting glutathione-mediated extracellular conversion of Te(IV) to Te(0) (Fig. 3C). This external reduction of Te(IV) suppressed the growth defects almost completely, in agreement with the lower toxicity of elemental Te(0). Taken together, these observations suggest that GSH is capable of reducing Te(IV) to Te(0) extracellularly in the yeast growth medium but that the GSH generated intracellularly only has a limited effect on Te(IV) tolerance and reduction. Major regulators of the amino acid starvation response, Ure2 mediating nitrogen repression and Npr2-Npr3 mediating nitrogen activation, also featured some of the strongest tellurium phenotypes in the screen, with the npr2Δ and npr3Δ mutants showing high Te(0) accumulation and the ure2Δ mutant featuring low Te(0) accumulation (Fig. 2B; see Table S1 in the supplemental material). Although nitrate reductases possess the ability to reduce Te(IV) to Te(0) in Escherichia coli (2), S. cerevisiae lacks nitrate reductases. Hence, mechanisms other than nitrate reduction must form the basis of the link between nitrogen metabolism and tellurium accumulation in yeast.
Imbalances in intracellular oxidation status and mitochondrial respiration affect tellurium accumulation.
Sulfate assimilation consumes vast amounts of reductive power; the sulfite reductase alone requires three NADPH molecules in the reduction of each sulfite (IV) molecule to sulfide (69). Hence, the enrichment of genes involved in NAD/NADP binding, and consequently in redox status, among gene knockout mutants with low Te(0) accumulation is not surprising (Fig. 2A). Among knockout mutants with low tellurium accumulation, we also see an enrichment of proteins with a known localization in the mitochondria (enrichment = 1.6-fold [P = 0.01; Fisher's exact test]). Mitochondrial respiration is the dominant intracellular source of ROS and has profound effects on cellular redox status. Knockout mutants lacking the key respiratory genes yield petite colonies that are too small to allow stringent evaluation of Te phenotypes; however, loss of nonpetite components such as CYT1, encoding cytochrome c1, Coq1 and Coq2, catalyzing the first steps in ubiquinone biosynthesis, and the cytochrome c reductase subunit Qcr9 conferred low Te(0) accumulation (Fig. 2B). Similarly, a large group of genes involved in mitochondrial import featured very low Te(0) accumulation. Knockout mutants lacking the components of the Mdm10-Mdm12-Mmm1-Mdm34 complex, mediating assembly of mitochondrial import channels (25), or of their genetic interaction partners MDM31 and MDM35 (12) all showed high Te(IV) tolerance and low Te(0) accumulation (Fig. 2B; see Table S1 in the supplemental material). These mitochondrial import mutants all have respiratory deficiencies due to problems in assembling fully functional respiratory chain complexes (11). Respiratory defects may also cause the very low Te(0) accumulation observed following loss of the functionally interdependent SNF5 and SNF6 genes (26) (Fig. 2B); these SWI/SNF chromatin remodeling complex components are absolutely required for respiratory growth because of their role in the transcriptional regulation of genes in the respiratory chain.
A potential explanation for the strong tellurium phenotypes of mitochondrial deletion mutants is the effect on Fe-S protein assembly due to Te replacement of, or strong binding to, S. Fe-S centers catalyze key reactions in the respiratory chain and are assembled in the mitochondrial matrix in a series of reactions known to involve 19 proteins. Ten of these proteins are essential, precluding the screening of Te phenotypes. However, of the nine viable Fe-S assembly deletion mutants, only those with the above-mentioned qcr9Δ mutation, which indirectly interferes with Fe-S assembly by destabilizing the Rieske Fe-S complex, showed altered Te phenotypes. Furthermore, GAL1-driven individual overexpression (49) of the 19 Fe-S assembly components produced normal Te(IV) tolerance and, at most, marginal deviations in tellurium accumulation (data not shown). Hence, Fe-S assembly does not seem to be directly connected to tellurium toxicity in yeast.
Tellurite, selenite, and selenomethionine share a toxicity mechanism.
A key question is whether Te(IV) exerts its toxicity by mechanisms exclusive to Te(IV) or through mechanisms shared with other chemically related elements. To resolve this issue, we first investigated a potential overlap with the selenite SeO32−, where the chalcogen selenium occurs as Se(IV). Se(IV) is chemophysically related to Te(IV) and is converted inside the cell to Se(0), producing a reddish colony pigmentation, via a set of reactions that is poorly understood in yeast but that in bacteria involves glutathione and the release of ROS (46, 62). Screening gene knockout mutants with significant Te(IV) phenotypes on comparable concentrations of Se(IV), we found substantial overlap between Te(IV) and Se(IV) phenotypes (linear correlation coefficient [r], >0.4) (Fig. 4A); gene knockout mutants with low tellurium accumulation tended to show low accumulation of Se(0) (Fig. 4 and 5). Genes required for both tellurium and selenium accumulation included the components of the Mmm1-Mdm10-Mdm12-Mdm34 and Ino2-Ino4 transcription complexes, the respiratory chain components CYT1 and COQ2, and almost all of the MET genes. Interestingly, gsh2Δ, which only produced somewhat reduced Te(0) accumulation, produced much lower Se(0) accumulation, suggesting a more direct involvement of glutathione in selenite conversion to selenium (Fig. 4A and 5). Gene knockout mutants with high tellurium accumulation, e.g., snf1Δ and snf4Δ mutants lacking the glucose repression switch, produced a similar high accumulation of selenium pigmentation (Fig. 4A). Hence, despite the fact that Te is bulkier than Se and less prone to form stable bonds with carbon and hydrogen, intracellular conversion of Te(IV) to Te(0) appears to proceed largely along the same metabolic routes as conversion of Se(IV) to Se(0). One potential cause for the toxicity of Se(IV) and Te(IV) is the intracellular conversion into selenomethionine and telluromethionine, respectively, which, following replacement of methionine during protein biosynthesis, may cause protein inactivation. To investigate Se-methionine and Te-methionine formation as potential toxicity mechanisms for Se(IV) and Te(IV), we screened gene knockout mutants with aberrations in Te phenotypes following selenomethionine exposure. Telluromethionine is unstable and rapidly decomposes, impeding phenotypic screening. No quantifiable Se(0) accumulation was observed in selenomethionine-exposed cells, suggesting that the conversion of Se(IV) to selenomethionine is unidirectional (Fig. 5). However, comparing gene knockout tolerance traits, we found a strong overlap of Se(IV), Te(IV), and selenomethionine, showing that toxicity and detoxification mechanisms are partially shared by these compounds (Fig. 4). Specifically, loss of MET genes conferred tolerance to Se(IV), Te(IV), and selenomethionine whereas loss of HOM6 or MUP1 resulted in sensitivity, suggesting a similar role for the sulfate assimilation system in Se(IV), Te(IV), and selenomethionine toxicity.
Fig. 5.
Te(IV), selenite, and selenomethionine toxicity and bioaccumulation. Homozygous diploid gene knockout mutants with confirmed significant deviations in Te(IV) phenotypes were pinned onto solid agar medium containing 0.2 or 0.8 mM Na2SeO3 or 0.03 or 0.06 mM selenomethionine (SeMet) and evaluated manually after 1, 2, 3, and 6 days of growth on solid agar medium.
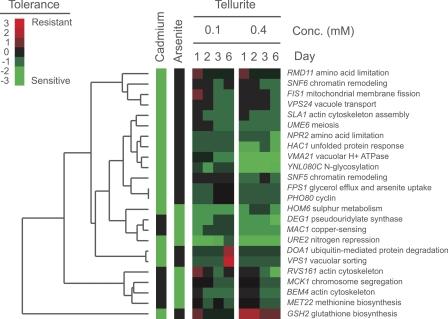
To address whether more general metal/metalloid tolerance systems are involved in Te(IV) detoxification, we also compared genes revealed in this study as important for Te(IV) tolerance to genes previously reported to be important for optimal cadmium (Cd) (20, 43, 47, 57) or arsenite [As(III)] (18, 20, 57) tolerance. Circumventing the issue of a generally small overlap between screens, core sets, as defined by Thorsen et al. (57), corresponding to genes identified as required for resistance in at least two As(III) or two Cd screens were used. We found significant (P < 10−5; Fisher's exact test) enrichments of both Cd-sensitive (enrichment = 3.7-fold) and As(III)-sensitive (enrichment = 4.2-fold) gene knockout mutants among the Te(IV)-sensitive mutants identified here. Hence, the toxicity of Te(IV) partially follows routes that overlap not only with Se(IV) toxicity but apparently also with chemophysically more dissimilar metals. These routes involved actin interaction (RVS161, SLA1, and BEM4), metal homeostasis regulators (MAC1), redox metabolism (GSH2 and HAC1), and nitrogen metabolism (URE2, UME6, and NPR2), as well as MET22 and HOM6, which are involved in the metabolism of sulfur-containing amino acids (Fig. 6). The overlap with cadmium specifically included vacuolar/endosomal transport (VPS1, VMA21, and VPS24) and chromatin remodeling via SNF5 and SNF6 (Fig. 6). However, it should be emphasized that the similarity between the Te(IV) and Cd/As mutant sensitivity profiles, although significant, was less pronounced than the strong overlap of the Te, Se, and selenomethionine phenotypes. Specifically, none of the most prominent Te, Se, and selenomethionine toxicity effects, such as the hypertolerance of sulfate assimilation knockout mutants, were observed for Cd or As.
Fig. 6.
Te(IV) toxicity is partially mediated via general metal toxicity routes. Hierarchical clustering (average-linkage clustering based on Pearson correlation coefficients) of gene knockout mutants with reduced Te(IV) tolerance (reduced colony size) and reduced tolerance to cadmium and/or arsenite. Colors indicate degrees of tolerance in relation to that of the WT; for cadmium and arsenite, an arbitrary intermediate value was used, as the data were qualitative.
DISCUSSION
The mechanistic causes of the toxicity and intracellular accumulation of Te species are classical enigmas in molecular microbiology. Here we shed light on the Te conundrum by showing that conversion of Te(IV) to Te(0) and the biotoxicity of Te(IV) are mediated largely by the same cellular routes in yeast. A genome-wide screen for tellurium-related traits among yeast gene knockout mutants implicated two main cellular routes, sulfur assimilation and redox-related mitochondrial respiration. From a metal biology perspective, a link between sulfur assimilation and tellurium phenotypes is not unexpected; sulfur metabolism affects the biotoxicity of essentially all well-studied heavy metals and metalloids in yeast, either directly or indirectly via its role in glutathione biosynthesis (68). However, the involvement of sulfur metabolism is typically the reverse of what is observed here; that is, unperturbed sulfur metabolism is a key factor in maintaining optimal heavy metal/metalloid tolerance. Mutants resistant to selenate, Se(VI), which are known to be defective in components of the sulfate assimilation pathway, constitute exceptions (6, 8). Se(VI) is rapidly converted inside the cell to Se(IV), which is chemophysically similar to Te(IV). Hence, the connection between Se- and Te-related traits and sulfate assimilation reported here is not surprising. Sulfate assimilation in S. cerevisiae represents a sequence of redox reactions that are costly in terms of ATP and NADPH (56). The key reduction reaction enabling utilization of sulfur compounds with a high oxidation state is the six-electron reduction of sulfite to sulfide by the two-component sulfite reductase at the expense of three NADPH molecules (69, 70). We identified the sulfate/sulfur assimilation pathway as the most important cellular route mediating Te(IV) toxicity and intracellular accumulation of tellurium in yeast. This directly points to a role for the sulfite reductase in Te(IV) reduction, which is somewhat surprising as there is no evidence from bacterial studies for a sulfite reductase-mediated reduction of either Te(IV) or Se(IV) to H2Te or H2Se, respectively; in addition, reductase component mutations do not produce substantial Se phenotypes in bacteria (14, 62). Instead, bacterial Te metabolism is believed to mimic Se metabolism, which, due to the reactivity of selenium compounds in low oxidation states with sulfur, is largely centered on thiol-mediated reactions (62). In bacteria, glutathione and cysteine are believed to reduce Te(IV) and Se(IV) via the Painter reaction to Te and Se trisulfides which, although stable under acidic conditions, will rapidly decompose to yield Te(0) and Se(0) under neutral conditions (17, 22, 40). This may explain why, in E. coli, Te(IV) tolerance correlates with the relative expression of cysteine biosynthetic genes (16, 64). In yeast, glutathione metabolism and the closely connected cysteine metabolism had a surprisingly limited effect on the intracellular fate of Te species; mutants with cysteine metabolism gene knockout mutants featured normal Te phenotypes, and loss of the glutathione synthase Gsh2 conferred only minor reductions in Te(IV) tolerance and a slight reduction in the conversion of Te(IV) to Te(0). However, external addition of an excess of GSH to Te(IV)-containing growth medium caused an immediate and drastic darkening and suppressed the Te-induced growth defects. This suggests that GSH is capable of reducing Te(IV) to Te(0) extracellularly. It has recently been shown that yeast cells export GSH during As(III) stress in order to complex bind and detoxify the harmful metalloid outside the cell (68); a similar extracellular detoxification mechanism may have a role in Te detoxification.
The inverse correlation observed here between Te(0) accumulation and Te(IV) tolerance suggests that the conversion of Te(IV) to Te(0) is a potent cause of Te(IV) toxicity. Key questions are whether Te(IV), similarly to Se(IV), is bioassimilated; whether the resulting tellurocysteine, telluromethionine, or derivatives thereof are incorporated into peptides; and whether this bioassimilation contributes to Te(IV) toxicity. In contrast to selenium, which is required for the activity of many proteins in higher organisms, no biological functions have yet been discovered for tellurium. The probable reason is the more metallic properties of tellurium, which forms weaker covalent bonds with hydrogen and carbon (34). In bacteria, selenomethionine formation from selenite is believed to pass via reduction of glutathione-bound GS-SeH into H2Se and then further into selenomethionine via the methyl cycle (62). Selenomethionine is then randomly incorporated into proteins due to the editing tolerance of methionyl-tRNA synthetase (10, 35). Although the intracellular fate of telluromethionine is less well known, the existence of telluromethionine, methyltelluromethionine, and, to a lesser extent, tellurocysteine has been reported from the cultivation of yeast and bacteria in the presence of high concentrations of Te(IV) (42, 71). Telluromethionine has also been shown to be able to replace methionine residues in about 40% of the copies of the protein dihydrofolate reductase in E. coli (3). Telluromethionine rapidly decomposes in aqueous solutions (38) and does not permit straightforward tests of toxicity mechanisms. However, we found a remarkable overlap between Te(IV) toxicity and selenomethionine toxicity, suggesting that they stem from shared modes of action. This raises the prospect that the detrimental effect of Te(IV) on yeast growth partially stems from telluromethionine formation. Although it is unclear whether telluromethionine incorporation into peptides would significantly alter protein properties, it has been shown that introduction of tellurocysteine into the active site of a glutathione transferase altered the protein's functionality to a glutathione peroxidase (29). An alternative toxicity mechanism is the incorporation of tellurocysteine into glutathione with concomitant depletion of reducing power. However, the absence of strong Te phenotypes in cysteine and glutathione biosynthesis gene knockout mutants suggests that this is an unlikely toxicity mechanism in yeast. The absence of such phenotypes also fails to support a model where inhibition of glutathione by direct binding of Te to the SH group of glutathione would form the mechanistic basis of Te(IV) toxicity. Such a mechanism has been contemplated for various metals and metalloids (e.g., see reference 51).
Oxidative stress is considered a dominant cause of Te(IV) toxicity in bacterial systems, particularly during respiratory growth (4, 41, 59). In particular, generation of the toxic superoxide radical during Te(IV) reduction is a major contributor to Te(IV) toxicity, in analogy to what has been suggested for selenium oxyanions (46, 50). Surprisingly, we found no support for oxidative damage being an important mechanism of Te(IV) toxicity in yeast. Knockout mutants lacking genes involved in thioredoxin and glutaredoxin reactions, which have been linked to Te(IV) tolerance in bacteria (7), failed to show aberrant Te phenotypes, as did knockout mutants lacking peroxidases, catalases, and the major transcriptional regulators of these systems. The absence of links to oxidative stress protection suggests that ROS generation is not a major source of Te(IV) toxicity in yeast and that routes for Te(IV) toxicity and detoxification may differ substantially between yeast and bacteria. However, genes involved in respiratory functions, as well as transport into the mitochondria, showed low tellurium accumulation and high Te(IV) tolerance. Furthermore, initiation of accumulation of Te(0) coincided with the switch from fermentative to respiratory metabolism in S. cerevisiae and respiring yeast cells were hypersensitive to Te(IV). These observations agree well with observations on Pseudomonas aeruginosa and P. pseudoalcaligenes which show that the rate of Te(IV) reduction is strongly correlated with the rate of respiration, that inhibition of respiration partially inhibits Te(IV) reduction, and that Te(IV) exposure induces changes in multiple components of the respiratory chain (13, 59). Hence, our results identified two cellular routes as key to the intracellular fate of Te species in yeast: sulfate assimilation and a mitochondrial mechanism that encompasses respiration. Interestingly, both sulfur metabolism and mitochondrial genes, including all of the major mitochondrial contributors to Te phenotypes reported here, were recently reported as having low or no rust coloration during bismuth sulfite exposure (23). Bismuth sulfite is converted into bismuth sulfide in yeast in a process dependent on the sulfate assimilation pathway, resulting in a reddish-blackish rust coloration (15, 36). Consequently, bismuth sulfite reduction and Te(IV) reduction proceed along the same cellular routes, both with a strong emphasis on sulfate assimilation and mitochondrial respiration.
Data regarding the toxicity and fate of tellurium compounds in higher eukaryotes, including humans, is scarce. Te species are absorbed in the body, but telluroamino acids have not been found in humans. The present view holds that Te(IV) is metabolized in a manner mimicking that of selenite, implying rapid reduction to telluride followed by methylation to mono-, di-, or trimethylated Te species (37). However, in contrast to selenium, tellurium does not appear to be glycosylated in higher eukaryotes, and trimethyltelluronium, rather than tellurosugars, is the dominating Te species (39). We found no indications that methyltransferases are involved in Te metabolism in yeast, suggesting that Te methylation is of little importance in this organism. In higher eukaryotes, tellurium-induced cell death has been suggested to occur by an oxidative mechanism that at least partially involves oxidation-induced DNA damage (44). Se(IV) exposure is, in fact, well known to lead to DNA double-strand breaks (27, 32). In yeast, however, viable knockout mutants lacking genes involved in DNA repair and replication and in cell cycle checkpoints showed normal Te phenotypes, suggesting that DNA damage contributes little to Te toxicity. Similar to arsenic, which is accumulated as dimethyl arsenical, but in contrast to selenium, tellurium reaches high concentrations in red blood cells as dimethylated Te (37). Hence, arsenic and tellurium may share mechanistic features in mammals. We found a significant overlap between gene knockout sensitivities to Te(IV) and As(III), suggesting that mechanistic similarities exist also in yeast. A common detoxification mechanism of many metals/metalloids, notably, Cd, Sb, As, Hg, and Pb, is sequestration in the vacuole as GSH conjugates (68), a process that is mainly mediated by the ABC transporter Ycf1. However, a ycf1Δ mutant showed normal Te phenotypes, as did knockout mutants lacking the Ycf1 paralogs Bpt1 and Vmr1. Te plaques were also clearly absent from the vacuolar lumen. Intriguingly, however, Te plaques initially emerged in close proximity to the vacuolar membrane, and at later stages of Te stress, the integrity of the vacuole was compromised, followed by vacuole disintegration and rapid cell shrinkage. Several vacuolar mutants also showed substantial Te sensitivity phenotypes. Taken together, these findings suggest that the vacuole may play a role in Te toxicity and detoxification in yeast but that this role does not encompass sequestration.
Supplementary Material
ACKNOWLEDGMENTS
We express our gratitude to Lianming Tong, Department of Applied Physics, Chalmers University of Technology, for his assistance with the Raman measurements and to Markus Tamas, University of Gothenburg, for thoughtful comments on the manuscript.
J.W. was supported by the Carl Trygger Research Foundation and the Royal Swedish Academy of Sciences.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Ambroziak J., Henry S. A. 1994. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 269:15344–15349 [PubMed] [Google Scholar]
- 2.Avazéri C., Turner R. J., Pommier J., Weiner J. H., Giordano G., Vermeglio A. 1997. Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology 143(Pt. 4):1181–1189 [DOI] [PubMed] [Google Scholar]
- 3.Boles J. O., Lewinski K., Kunkle M., Odom J. D., Dunlap B., Lebioda L., Hatada M. 1994. Bio-incorporation of telluromethionine into buried residues of dihydrofolate reductase. Nat. Struct. Biol. 1:283–284 [DOI] [PubMed] [Google Scholar]
- 4.Borsetti F., Tremaroli V., Michelacci F., Borghese R., Winterstein C., Daldal F., Zannoni D. 2005. Tellurite effects on Rhodobacter capsulatus cell viability and superoxide dismutase activity under oxidative stress conditions. Res. Microbiol. 156:807–813 [DOI] [PubMed] [Google Scholar]
- 5.Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288c: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132 [DOI] [PubMed] [Google Scholar]
- 6.Breton A., Surdin-Kerjan Y. 1977. Sulfate uptake in Saccharomyces cerevisiae: biochemical and genetic study. J. Bacteriol. 132:224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chasteen T. G., Fuentes D. E., Tantalean J. C., Vásquez C. C. 2009. Tellurite: history, oxidative stress, and molecular mechanisms of resistance. FEMS Microbiol. Rev. 33:820–832 [DOI] [PubMed] [Google Scholar]
- 8.Cherest H., Davidian J. C., Thomas D., Benes V., Ansorge W., Surdin-Kerjan Y. 1997. Molecular characterization of two high affinity sulfate transporters in Saccharomyces cerevisiae. Genetics 145:627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherest H., Eichler F., Robichon-Szulmajster H. 1969. Genetic and regulatory aspects of methionine biosynthesis in Saccharomyces cerevisiae. J. Bacteriol. 97:328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowie D. B., Cohen G. N. 1957. Biosynthesis by Escherichia coli of active altered proteins containing selenium instead of sulfur. Biochim. Biophys. Acta 26:252–261 [DOI] [PubMed] [Google Scholar]
- 11.Dimmer K. S., Fritz S., Fuchs F., Messerschmitt M., Weinbach N., Neupert W., Westermann B. 2002. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimmer K. S., Jakobs S., Vogel F., Altmann K., Westermann B. 2005. Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast. J. Cell Biol. 168:103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Tomaso G., Fedi S., Carnevali M., Manegatti M., Taddei C., Zannoni D. 2002. The membrane-bound respiratory chain of Pseudomonas pseudoalcaligenes KF707 cells grown in the presence or absence of potassium tellurite. Microbiology 148:1699–1708 [DOI] [PubMed] [Google Scholar]
- 14.Fimmel A. L., Loughlin R. E. 1977. Isolation and characterization of cysK mutants of Escherichia coli K12. J. Gen. Microbiol. 103:37–43 [DOI] [PubMed] [Google Scholar]
- 15.Forbes B. A., Sahm D. F., Weissfeld A. S. 1998. Bailey & Scott's diagnostic microbiology, 10th ed.C.V. Mosby Company, St. Louis, MO [Google Scholar]
- 16.Fuentes D. E., Fuentes E. L., Castro M. E., Pérez J. M., Araya M. A., Chasteen T. G., Pichuantes S. E., Vásquez C. C. 2007. Cysteine metabolism-related genes and bacterial resistance to potassium tellurite. J. Bacteriol. 189:8953–8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganther H. E. 1999. Selenium metabolism, selenoproteins and mechanisms of cancer prevention: complexities with thioredoxin reductase. Carcinogenesis 20:1657–1666 [DOI] [PubMed] [Google Scholar]
- 18.Haugen A. C., Kelley R., Collins J. B., Tucker C. J., Deng C., Afshari C. A., Brown J. M., Ideker T., Van Houten B. 2004. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 5:R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isnard A. D., Thomas D., Surdin-Kerjan Y. 1996. The study of methionine uptake in Saccharomyces cerevisiae reveals a new family of amino acid permeases. J. Mol. Biol. 262:473–484 [DOI] [PubMed] [Google Scholar]
- 20.Jin Y. H., Dunlap P. E., McBride S. J., Al-Refai H., Bushel P. R., Freedman J. H. 2008. Global transcriptome and deletome profiles of yeast exposed to transition metals. PLoS Genet. 4:e1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlson U., Frankenberger W. 1993. Biological alkylation of selenium and tellurium, p. 185–227 InSigel H., Sigel A. (ed.), Metal ions in biological systems. Marcel Dekker Inc., New York, NY [Google Scholar]
- 22.Kice J. L., Lee T. W. S., Pan S.-T. 1980. Mechanism of the reaction of thiols with selenite. J. Am. Chem. Soc. 102:4448–4455 [Google Scholar]
- 23.Kim H. S., Huh J., Fay J. C. 2009. Dissecting the pleiotropic consequences of a quantitative trait nucleotide. FEMS Yeast Res. 9:713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konetzka W. A. 1977. Microbiology of metal transformations, p. 317–342 InWeinberg E. D. (ed.), Microorganisms and minerals. Marcel Dekker Inc., New York, NY [Google Scholar]
- 25.Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., Walter P. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325:477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laurent B. C., Treitel M. A., Carlson M. 1991. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc. Natl. Acad. Sci. U. S. A. 88:2687–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letavayová L., Vlasakova D., Spallholz J. E., Brozmanova J., Chovanec M. 2008. Toxicity and mutagenicity of selenium compounds in Saccharomyces cerevisiae. Mutat. Res. 638:1–10 [DOI] [PubMed] [Google Scholar]
- 28.Liu M., Taylor D. E. 1999. Characterization of gram-positive tellurite resistance encoded by the Streptococcus pneumoniae tehB gene. FEMS Microbiol. Lett. 174:385–392 [DOI] [PubMed] [Google Scholar]
- 29.Liu X., Silks L. A., Liu C., Ollivault-Shiflett M., Huang X., Li J., Luo G., Hou Y. M., Liu J., Shen J. 2009. Incorporation of tellurocysteine into glutathione transferase generates high glutathione peroxidase efficiency. Angew. Chem. Int. Ed. Engl. 48:2020–2023 [DOI] [PubMed] [Google Scholar]
- 30.Lloyd-Jones G., Osborn A. M., Ritchie D. A., Strike P., Hobman J. L., Brown N. L., Rouch D. A. 1994. Accumulation and intracellular fate of tellurite in tellurite-resistant Escherichia coli: a model for the mechanism of resistance. FEMS Microbiol. Lett. 118:113–119 [DOI] [PubMed] [Google Scholar]
- 31.Lloyd-Jones G., Williamson W. M., Slootweg T. 2006. The Te-Assay: a black and white method for environmental sample pre-screening exploiting tellurite reduction. J. Microbiol. Methods 67:549–556 [DOI] [PubMed] [Google Scholar]
- 32.Maniková D., Vlasakova D., Loduhova J., Letavayová L., Vigasova D., Krascsenitsova E., Vlckova V., Brozmanova J., Chovanec M. 2010. Investigations on the role of base excision repair and non-homologous end-joining pathways in sodium selenite-induced toxicity and mutagenicity in Saccharomyces cerevisiae. Mutagenesis 25:155–162 [DOI] [PubMed] [Google Scholar]
- 33.Moore M. D., Kaplan S. 1992. Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: characterization of tellurite, selenite, and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J. Bacteriol. 174:1505–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moroder L. 2005. Isosteric replacement of sulfur with other chalcogens in peptides and proteins. J. Pept. Sci. 11:187–214 [DOI] [PubMed] [Google Scholar]
- 35.Munier R., Cohen G. N. 1959. Incorporation of structural analogues of amino acids into bacterial proteins during their synthesis in vivo. Biochim. Biophys. Acta 31:378–390 [DOI] [PubMed] [Google Scholar]
- 36.Nickerson W. J. 1953. Reduction of inorganic substances by yeasts. I. Extracellular reduction of sulfite by species of Candida. J. Infect. Dis. 93:43–56 [DOI] [PubMed] [Google Scholar]
- 37.Ogra Y. 2009. Toxicometallomics for research on the toxicology of exotic metalloids based on speciation studies. Anal. Sci. 25:1189–1195 [DOI] [PubMed] [Google Scholar]
- 38.Ogra Y., Kitaguchi T., Suzuki N., Suzuki K. T. 2008. In vitro translation with [34S]-labeled methionine, selenomethionine, and telluromethionine. Anal. Bioanal. Chem. 390:45–51 [DOI] [PubMed] [Google Scholar]
- 39.Ogra Y., Kobayashi R., Ishiwata K., Suzuki K. T. 2008. Comparison of distribution and metabolism between tellurium and selenium in rats. J. Inorg. Biochem. 102:1507–1513 [DOI] [PubMed] [Google Scholar]
- 40.Painter E. P. 1941. The chemistry and toxicity of selenium compounds which special reference to the selenium problem. Chem. Rev. 28:179–213 [Google Scholar]
- 41.Pérez J. M., Calderon I. L., Arenas F. A., Fuentes D. E., Pradenas G. A., Fuentes E. L., Sandoval J. M., Castro M. E., Elias A. O., Vásquez C. C. 2007. Bacterial toxicity of potassium tellurite: unveiling an ancient enigma. PLoS One 2:e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramadan S. E., Razak A. A., Ragab A. M., el-Meleigy M. 1989. Incorporation of tellurium into amino acids and proteins in a tellurium-tolerant fungi. Biol. Trace Elem. Res. 20:225–232 [DOI] [PubMed] [Google Scholar]
- 43.Ruotolo R., Marchini G., Ottonello S. 2008. Membrane transporters and protein traffic networks differentially affecting metal tolerance: a genomic phenotyping study in yeast. Genome Biol. 9:R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandoval J. M., Leveque P., Gallez B., Vásquez C. C., Buc Calderon P. 2010. Tellurite-induced oxidative stress leads to cell death of murine hepatocarcinoma cells. Biometals 23:623–632 [DOI] [PubMed] [Google Scholar]
- 45.Schroeder H. A., Buckman J., Balassa J. J. 1967. Abnormal trace elements in man: tellurium. J. Chronic Dis. 20:147–161 [DOI] [PubMed] [Google Scholar]
- 46.Seko Y., Saito Y., Kitahara J., Imura N. 1998. Active oxygen generation by the reaction of selenite with reduced glutathione in vitro, p. 70–73 InWendel A. (ed.), Selenium in biology and medicine. Springer-Verlag, Berlin, Germany [Google Scholar]
- 47.Serero A., Lopes J., Nicolas A., Boiteux S. 2008. Yeast genes involved in cadmium tolerance: identification of DNA replication as a target of cadmium toxicity. DNA Repair (Amst.) 7:1262–1275 [DOI] [PubMed] [Google Scholar]
- 48.Smith D. G. 1974. Tellurite reduction in Schizosaccharomyces pombe. J. Gen. Microbiol. 83:389–392 [DOI] [PubMed] [Google Scholar]
- 49.Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S. G., Cyert M., Hughes T. R., Boone C., Andrews B. 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21:319–330 [DOI] [PubMed] [Google Scholar]
- 50.Spallholz J. E. 1994. On the nature of selenium toxicity and carcinostatic activity. Free Radic. Biol. Med. 17:45–64 [DOI] [PubMed] [Google Scholar]
- 51.Stohs S. J., Bagchi D. 1995. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18:321–336 [DOI] [PubMed] [Google Scholar]
- 52.Summers A. O., Silver S. 1978. Microbial transformations of metals. Annu. Rev. Microbiol. 32:637–672 [DOI] [PubMed] [Google Scholar]
- 53.Taylor D. E. 1999. Bacterial tellurite resistance. Trends Microbiol. 7:111–115 [DOI] [PubMed] [Google Scholar]
- 54.Taylor D. E., Hou Y., Turner R. J., Weiner J. H. 1994. Location of a potassium tellurite resistance operon (tehA tehB) within the terminus of Escherichia coli K-12. J. Bacteriol. 176:2740–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor D. E., Walter E. G., Sherburne R., Bazett-Jones D. P. 1988. Structure and location of tellurium deposited in Escherichia coli cells harbouring tellurite resistance plasmids. J. Ultrastruct. Mol. Struct. Res. 99:18–26 [DOI] [PubMed] [Google Scholar]
- 56.Thomas D., Surdin-Kerjan Y. 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61:503–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thorsen M., Perrone G., Kristiansson E., Traini M., Ye T., Dawes I. W., Nerman O., Tamás M. 2009. Genetic basis of arsenite and cadmium tolerance in Saccharomyces cerevisiae. BMC Genomics 10:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toptchieva A., Sisson G., Bryden L. J., Taylor D. E., Hoffman P. S. 2003. An inducible tellurite-resistance operon in Proteus mirabilis. Microbiology 149:1285–1295 [DOI] [PubMed] [Google Scholar]
- 59.Trutko S. M., Akimenko V. K., Suzina N. E., Anisimova L. A., Shlyapnikov M. G., Baskunov B. P., Duda V. I., Boronin A. M. 2000. Involvement of the respiratory chain of gram-negative bacteria in the reduction of tellurite. Arch. Microbiol. 173:178–186 [DOI] [PubMed] [Google Scholar]
- 60.Tucker F. L., Walper J. F., Appleman M. D., Donohue J. 1962. Complete reduction of tellurite to pure tellurium metal by microorganisms. J. Bacteriol. 83:1313–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turner R. J., Aharonowitz Y., Weiner J. H., Taylor D. E. 2001. Glutathione is a target in bacterial tellurite toxicity and is protected by tellurite resistance determinants in Escherichia coli. Can. J. Microbiol. 47:33–40 [PubMed] [Google Scholar]
- 62.Turner R. J., Weiner J. H., Taylor D. E. 1998. Selenium metabolism in Escherichia coli. Biometals 11:223–227 [DOI] [PubMed] [Google Scholar]
- 63.Turner R. J., Weiner J. H., Taylor D. E. 1999. Tellurite-mediated thiol oxidation in Escherichia coli. Microbiology 145(Pt. 9):2549–2557 [DOI] [PubMed] [Google Scholar]
- 64.Vásquez C. C., Saavedra C. P., Loyola C. A., Araya M. A., Pichuantes S. 2001. The product of the cysK gene of Bacillus stearothermophilus V mediates potassium tellurite resistance in Escherichia coli. Curr. Microbiol. 43:418–423 [DOI] [PubMed] [Google Scholar]
- 65.Walter E. G., Taylor D. E. 1992. Plasmid-mediated resistance to tellurite: expressed and cryptic. Plasmid 27:52–64 [DOI] [PubMed] [Google Scholar]
- 66.Warringer J., Anevski D., Liu B., Blomberg A. 2008. Chemogenetic fingerprinting by analysis of cellular growth dynamics. BMC Chem. Biol. 8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Warringer J., Blomberg A. 2003. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast 20:53–67 [DOI] [PubMed] [Google Scholar]
- 68.Wysocki R., Tamas M. J. 2010. How Saccharomyces cerevisiae copes with toxic metals and metalloids. FEMS Microbiol. Rev. [Epub ahead of print.] doi:10.1111/j.1574-6976.2010.00217.x [DOI] [PubMed] [Google Scholar]
- 69.Yoshimoto A., Sato R. 1968. Studies on yeast sulfite reductase. I. Purification and characterization. Biochim. Biophys. Acta 153:555–575 [DOI] [PubMed] [Google Scholar]
- 70.Yoshimoto A., Sato R. 1968. Studies on yeast sulfite reductase. II. Partial purification and properties of genetically incomplete sulfite reductases. Biochim. Biophys. Acta 153:576–588 [DOI] [PubMed] [Google Scholar]
- 71.Yu L., He K., Chai D., Yang C., Zheng O. 1993. Evidence for telluroamino acid in biological materials and some rules of assimilation of inorganic tellurium by yeast. Anal. Biochem. 209:318–322 [DOI] [PubMed] [Google Scholar]
- 72.Yurkov V. V., Beatty J. T. 1998. Aerobic anoxygenic phototrophic bacteria. Microbiol. Mol. Biol. Rev. 62:695–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.