Abstract
The protozoan parasite Giardia lamblia undergoes stage differentiation in the small intestine of the host to an environmentally resistant and infectious cyst. Encystation involves the secretion of an extracellular matrix comprised of cyst wall proteins (CWPs) and a β(1-3)-GalNAc homopolymer. Upon the induction of encystation, genes coding for CWPs are switched on, and mRNAs coding for a Myb transcription factor and enzymes involved in cyst wall glycan synthesis are upregulated. Encystation in vitro is triggered by several protocols, which call for changes in bile concentrations or availability of lipids, and elevated pH. However, the conditions for induction are not standardized and we predicted significant protocol-specific side effects. This makes reliable identification of encystation factors difficult. Here, we exploited the possibility of inducing encystation with two different protocols, which we show to be equally effective, for a comparative mRNA profile analysis. The standard encystation protocol induced a bipartite transcriptional response with surprisingly minor involvement of stress genes. A comparative analysis revealed a core set of only 18 encystation genes and showed that a majority of genes was indeed upregulated as a side effect of inducing conditions. We also established a Myb binding sequence as a signature motif in encystation promoters, suggesting coordinated regulation of these factors.
The differentiation of Giardia lamblia cells to cysts is a key step in the simple life cycle of this ubiquitous intestinal parasite. Induced trophozoites, the flagellated, motile, and attachment-competent stage, exit the cell cycle at the G2-M transition (1) and begin to synthesize the components of an extracellular matrix, the cyst wall (CW). Synthesis and export of the cyst wall proteins (CWPs) to specialized organelles, termed encystation-specific vesicles (ESVs), is completed between 8 and 10 h postinduction (p.i.). After proteolytic cleavage of the CWP2 C terminus, the CWPs are sorted into two fractions which are deposited sequentially and polymerize on the surface of the cyst (18). The first layer of the CW, which eventually completely encloses the parasite (19, 24), is secreted in the last minutes of differentiation simultaneously with nuclear division and morphological transformation of the cell (cyst formation). In vitro, the entire encystation process typically takes 20 to 24 h in our hands and can be induced by modifying medium components and/or the concentrations of bile, fatty acids (cholesterol), or lactic acid (3, 10, 15, 22). Because the condition(s) which induce encystation in vivo are not known, all factors and components mentioned above are implicated since their concentrations vary in the small intestine, as does the pH, which increases from ∼6 in the duodenum to 7.5 to 8.0 in the distal ileum. Although all available data indicate that differentiation is induced by one or several environmental signals, the mechanism(s) for reception and transduction have not been characterized. Upregulation of the genes coding for the three major structural proteins and the enzymes involved in the synthesis of the glycan component of the cyst wall has been documented by various small- and large-scale analysis methods (16, 29, 31, 36, 40, 42). Considering how effectively the cyst wall protects the parasite in the environment, the exported cyst wall matrix (CWM) appears to have a surprisingly low complexity. The three cyst wall proteins (CWP1 to CWP3 [CWP1-3]) have been identified as the major protein components of the structure (21, 40). In addition, a simple β(1-3)-GalNAc homopolymer glycan (9, 13) contributes ∼60% of the CWM, but its manner of integration with CWPs in this extracellular matrix is unknown. A membrane-anchored, cysteine-rich protein (high-Cys nonvariant cyst protein [HCNC protein]) which localizes to the endoplasmic reticulum and the periphery of cysts (5) could function as a possible link between the CW proper and the cell surface. The three CWPs are paralogous members of a protein family (40), and their corresponding genes are not transcribed in trophozoites. Their transcript levels during in vitro encystation peak around 7 h p.i. and appear to be sharply downregulated thereafter (6, 11). The inducible expression of a reporter under the control of a CWP1 promoter showed that downregulation of the reporter mRNA after peak induction was dependent on a short downstream region flanking the CWP1 open reading frame (ORF) (11). Another category of upregulated genes, in addition to those coding for structural proteins, are factors needed for the synthesis of the cyst wall glycan. The transcription of five enzymes involved in the synthesis of UDP-GalNAc from glucose was found to be induced relatively late during encystation (20). At least one of those enzymes, UDP-N-acetylglucosamine phosphorylase, is also allosterically regulated (4).
Several investigations have addressed how the expression of CWPs and the enzymes involved in cyst wall biosynthesis may be stage regulated. A Myb2-like protein (Giardia Myb) is one of the few transcription factors identified in Giardia (12, 39, 42). Recently, a plant-like WRKY protein was also shown to bind to promoter sequences and influence transcription levels (30). Both trans-acting factors have binding sites within 100 bp upstream of the ORFs of CWP1 and CWP2 and genes coding for several other factors involved in biosynthesis of the cyst wall. While it seems clear that the Giardia Myb plays a role in regulating the transcription of CWPs and of itself by binding to short sequence motifs upstream of these ORFs, the literature does not quite agree on the nature of these motifs. The results of random site selection experiments suggested that a 5′-GTTT(G/T)(G/T)-3′ motif and Myb were involved, the latter binding to both CWP1 and Myb promoters (42). Gillin and coworkers described a binding motif, 5′-C(T/A)ACAG-3′, in the upstream regions of the Myb, CWP1-3, and G6PI-B genes to which this putative transcription factor binds (39). Interestingly, three Myb binding sequences (MBSs) inserted upstream of a constitutively expressed gene confer only weak stage regulation (2.6-fold), but insertion upstream of the regulated G6PI-B gene promoter boosts induction >10-fold in encysting cells. This suggests that Giardia Myb is able to enhance transcriptional upregulation. A binding motif, 5′-(C/T)TGAC(C/T)-3′, for a giardial WRKY protein has been identified in the upstream region of the stage-regulated WRKY, CWP1, CWP2, and Myb genes but also in a number of nonregulated genes (30). The WRKY binding motif appears to act as a positive regulatory element of transcription in vegetative and in differentiating cells. An interesting aspect of encystation-specific promoter function was revealed by a deletion study of the CWP2 upstream sequence. Davis-Hayman and coworkers (6) showed that deletions in a 64-nucleotide (nt) region upstream of the CWP2 gene led to the expression of a reporter in proliferating trophozoites also, effectively abolishing stage regulation. Thus, while Myb and other transcription factors may play a role in modulating the expression of encystation genes, this evidence suggests that stage-specific control of CWP transcription is mediated by gene silencing in trophozoites and derepressed upon the induction of encystation.
Taken together, the current in vitro data indicate that the rapid induction of gene expression during encystation is controlled on multiple levels, which encompass basic transcription activity, enhancement, and derepression. However, with the exception of a putative repressor for CWP genes, no single factor or cis-acting motif appears to have a dominant role. The results of a recent study show that, ultimately, trophozoite-to-cyst differentiation is controlled on an epigenetic level and depends on histone acetylation (35). The protocol for the induction of encystation in vitro is key. None of the current experimental approaches to identify encystation genes, including those used to generate the serial analysis of gene expression (SAGE) data (2, 29) that are available on the GiardiaDB website, account for the possibility that side effects of encystation protocols might lead to an overestimation of the number of candidates. Analysis of encysting parasites isolated from the gut of a host could serve as a benchmark. However, this would require synchronization of induction, which is not feasible in vivo.
Here, we implement two distinct but equally effective encystation protocols to test the hypothesis that the induction of encystation in vitro activates the transcription of a very limited number of genes coding for products that are directly required for differentiation. We predict that the transcriptional induction of many other factors is due to protocol-specific off-target effects, which are neither (directly) linked to nor required for differentiation. To address this issue, we focused our analysis on the first 7 h of encystation. During this time, the bulk of CWPs is synthesized and exported from the endoplasmic reticulum to specialized organelles, the ESVs, for maturation. Comparative analysis of 7-h-p.i. microarray data sets generated with the two encystation protocols confirmed the upregulation of a small core set of genes and underscored the importance of the Myb binding sequence as a signature motif in promoters of encystation genes.
MATERIALS AND METHODS
Biochemical reagents.
Unless otherwise stated, all chemicals were purchased from Sigma and cell culture reagents were from Gibco BRL.
Cell culture and transfection.
Trophozoites of the G. lamblia strain WBC6 (ATCC catalog number 50803) were grown axenically in 11-ml culture tubes (Nunc, Roskilde, Denmark) containing Diamond's TYI-S-33 medium supplemented with 10% adult bovine serum and bovine bile at 37°C. Parasites were harvested by chilling the culture tubes on ice for 30 min to detach adherent cells, and cells were collected by centrifugation at 1,000 × g for 10 min. The cells were then resuspended/washed in ice-cold phosphate-buffered saline (PBS). Construction of expression plasmids and transfection were performed as described in reference 35.
Induction of encystation.
For protocol A, two-step encystation was induced as described previously (3, 10). Briefly, cells were cultivated for 44 h to confluence in bile-free medium (preencysting medium) and subsequently in prewarmed encystation medium (preencysting medium with an increase in pH to pH 7.85 and the addition of porcine bile [B8631, 0.25 mg/ml; Sigma], as well as lactic acid [L-200, 0.545 mg/ml; Sigma]) to induce encystation.
For protocol B, lipid starvation was induced as described in reference 22. Briefly, bile-free medium containing delipidated fetal calf serum (FCS) (PHM-L Lipsorb; Calbiochem) at a pH of 7.85 was used to induce differentiation of cells grown to confluence in standard trophozoite medium.
Protein analysis.
Protein analysis was performed as described in reference 37. Giardia parasites were harvested for gel electrophoresis by chilling culture tubes in ice and centrifugation at 1,000 × g. The cell pellet was washed once in ice-cold PBS and counted in a Neubauer chamber. The cell pellet was dissolved in SDS sample buffer to obtain 2 × 105 cells in 50 μl and boiled for 3 min. Dithiothreitol (DTT) was added at 7.75 μg/ml before boiling. SDS-PAGE on 12% polyacrylamide gels and transfer to nitrocellulose membranes were done according to standard techniques. The nitrocellulose membranes were blocked in 5% dry milk, 0.05% Tween 20 in PBS and incubated with primary antibody, a biotinylated antihemagglutinin (anti-HA) (high affinity, 3Flo; Roche) at 1:500 dilution, in blocking solution. Bound antibodies were detected with horseradish peroxidase-conjugated streptavidin antibody at 1:1,000 dilution (protein research products; Thermo Scientific Pierce) and developed using Western Lightning chemiluminescence reagent (PerkinElmer Life Sciences, Boston, MA). Data collection was done in a MultiImage light cabinet with AlphaEaseFC software (Alpha Innotech, San Leonardo, CA) using the appropriate settings.
Immunofluorescence analysis.
Cells were harvested as described above. Fixation and preparation for fluorescence microscopy were done as described previously (25). Briefly, cells were washed with cold PBS and fixed with 3% formaldehyde in PBS for 40 min at room temperature (20°C), followed by a 5-min incubation with 0.1 M glycine in PBS. Cells were permeabilized with 0.2% Triton X-100 in PBS for 20 min at room temperature and blocked overnight in 2% bovine serum albumin (BSA) in PBS. Incubation of all antibodies was done in 2% BSA, 0.2% Triton X-100 in PBS. Mouse monoclonal Alexa Fluor 488-conjugated anti-HA (1:30 dilution; Roche Diagnostics, Manheim, Germany) antibody or Cy3-conjugated anti-CWP1 (1:80 dilution; Waterborne, New Orleans, LA) was incubated for 50 min on ice. Washes between incubations were done with 0.5% BSA, 0.05% Triton X-100 in PBS. Labeled cells were embedded in Vectashield (Vector Laboratories, Burlingame, CA) containing the DNA-intercalating agent 4-6-diamidino-2-phenylindole (DAPI) for the detection of nuclear DNA. Immunofluorescence analysis was performed on a Leica DM IRBE fluorescence microscope with a 63× oil immersion objective.
FACS analysis.
For quantification of cells expressing CWP1 and the cyst yield, parasites were induced to encyst with either protocol A or protocol B for the required amount of time. Cysts and detached trophozoites were collected as described above, pelleted by centrifugation at 1,000 × g for 10 min, and washed with PBS. Fixed and permeabilized cells were fluorescently labeled for 50 min on ice using monoclonal Cy3-conjugated anti-CWP1 antibody (Waterborne, New Orleans, LA; dilution 1:80). Before fluorescence-activated cell sorting (FACS) analysis, cells were washed twice in PBS and resuspended in ice-cold PBS. Unlabeled samples were used to determine the background fluorescence and, subsequently, fluorescently labeled cysts were analyzed in triplicate on a FACSCalibur flow cytometer (Becton & Dickinson, Basel, Switzerland). All samples were analyzed in parallel by immunofluorescence assay (IFA) to assess the encystation efficiency and labeling and permeabilization quality.
Bioinformatic analysis. (i) MEME/MAST.
Protocols for the Motif Discovery and Search algorithms used are available at meme.sdsc.edu/.
(ii) Microarray.
Microarray experiments were performed as described in reference 35. Briefly, for RNA extraction, trophozoites were encysted if required and harvested as described above. Total RNA isolation was performed using an RNeasy kit (Qiagen, Stanford, CA), following the animal cell spin protocol. Residual genomic DNA was removed with DNase I digestion according to the manufacturer's protocol. The integrity of the RNA was assessed using a Bioanalyzer (Agilent Technologies, Inc., Palo Alto, CA) with eukaryote total RNA nano series II settings.
The array slides used here were Giardia lamblia microarray version 2 (provided by JCVI free of charge). They are glass slides surface coated with aminosilane with single-stranded oligonucleotides (70-mers), printed with a Genetix arraying machine. The array contains 19,230 elements and covers the whole Giardia lamblia WB strain genome. For further information, check Giardia lamblia microarray version 2 (http://pfgrc.jcvi.org/index.php/microarray/array_description/giardia_lamblia/version2.html).
For dual-channel microarray analysis, extracted total RNA was processed using an amino allyl MessageAmp II RNA amplification kit (Ambion, Austin, TX) and labeled with N-hydroxysuccinimidyl ester-derivatized reactive dye Cy3 or Cy5 according to the manufacturer's protocol. After purification, 2 μg each of Cy3- or Cy5-labeled RNA was denatured, added to SlideHyb buffer I (Ambion), and hybridized to G. lamblia version 2 microarrays (TIGR) in a Tecan HybStation at Functional Genomics Center Zurich, Switzerland. The arrays are aminosilane-surface-coated glass slides with 9,115 oligonucleotides (70-mers) designed to cover the whole G. lamblia WBC6 strain genome.
Prior to hybridization, slides were hydrated and blocked with 150 μl BSA buffer (0.1 mg/ml BSA, 0.1% SDS in 3× SSC buffer [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) for 1 h at 50°C. After washing, samples were injected and hybridized for 16 h at 42°C. Slides were scanned in an Agilent G2565AA scanner, using 543-nm and 633-nm laser lines for excitation of Cy3 and Cy5, respectively. The spatial scanning resolution was 10 μm, single pass. The scanner output files were quantified using Genespotter software (MicroDiscovery GmbH, Berlin, Germany) with the default settings and a 2.5-μm radius. The median spot intensities were evaluated with the Web application MAGMA (32) and normalized using the print-tip-wise loess correction of the LIMMA (Linear Models for Microarray Data) software package (34). Potential gene-specific dye effects were estimated from self-self hybridizations. Differential expression of genes is reported as the fold change compared with the results for control-treated samples, as well as the P value for differential expression as estimated by the empirical Bayes model implemented in limma. All reactions were performed in triplicate.
Expression vector construction and transfection.
Stable chromosomal integration of the described constructs was performed using the pPacV-Integ expression vector (37) using XbaI and PacI restriction sites. The oligonucleotides (5′-to-3′ orientation) used in this study were 3063p_XbaI_s (CGTCTAGATGGGTCGTCGGCTCTACAG), 3063p_HA_EcoRI_as (CGGAATTCCGCGTAGTCTGGGACATCGTATGGGTATCCGTCTCCCGTAGACAG), 3063_EcoRI_s (CGGAATTCAAGCCTCCAGTAGTGCCGTC), 3063_PacI_as (CGTTAATTAATTACAGATTTTTACTAGTAGGGCAG), 32657_s (GCTAGCGGATTGTGACTCTTGTATTAGGTG), and 32657_HA_as (TTAATTAACTACGCGTAGTCTGGGACATCGTATGGGTAATCCATTGTTGCGTACGAG).
For stable integration of plasmid vector DNA into the chromosome, the vectors were linearized at the SwaI restriction enzyme recognition sites adjacent to the TPI homology region (14). Approximately 15 μg of digested plasmid DNA was electroporated (350 V, 960 μF, and 800 Ω) into trophozoites. Integration occurs by homologous recombination, and stable transgenes are selected with the antibiotic puromycin.
RESULTS AND DISCUSSION
Induction of encystation in vitro triggers a bipartite transcriptional response.
The induction levels of CWP mRNA expression reported in the literature range from 100- to 300-fold with low background expression in trophozoites (11, 21, 27, 40). Considering that there is a spontaneous rate of encystation of up to a few percent in standard trophozoite cultures (38), it is more likely that CWP expression is completely silenced at that developmental stage. Peak mRNA levels of the most abundant proteins exported to the cyst wall (CWP1-3) in induced parasites are detected at ∼7 h p.i. (11, 23). To investigate the transcriptional induction of cargo exported to ESVs, we therefore focused our analysis on this initial stage of the differentiation process, which is relatively synchronous. RNA was isolated from encysting trophozoites at 45 min, 3 h, and 7 h p.i. and used for dual-color hybridization to a full-genome microarray with uninduced trophozoite RNA as the reference sample. For this time-dependent analysis, we used the standard two-step method for induction (3, 11), henceforth referred to as protocol A. The three data sets comprise 63 different genes with a signal increase of >2 (Fig. 1A). The results in Fig. 1B show that the early (45 min, 18 hits) and late (7 h, 29 hits) data sets do not overlap. The intermediate data set (3 h, 16 hits) is the smallest and has four unique positions, two overlaps with the early data set, and 10 positions shared with the late data set. Together with the signal increase in all but two of these shared positions at 7 h p.i., this indicates that the transcriptional response is divided into an early phase, which disappears before 3 h p.i., and a late phase, which is sustained through 7 h p.i. Only four genes are unique to the 3-h time point. The mRNAs coding for CWP1-3, an HCNC protein, and the transcription factor Myb are strongly upregulated after 3 h p.i. In addition, several genes associated with synthesis of the CW glycan (20) are upregulated in the late fraction, indicating that this data set also includes genes coding for later steps in CW synthesis. In total, mRNAs from all previously identified regulated genes are included in the combined 3-h and 7-h data sets. Thus, looking at upregulated genes only, we find a qualitatively significant but quantitatively limited, bipartite transcriptional response to the encystation stimulus in this microarray study. This fits with the idea that encystation requires relatively small changes in expression patterns except for the synthesis of the CW components (25). While the late response is easily recognized as encystation related, we currently have no explanation for the transient upregulation of 16 genes only at 45 min p.i. Two annotated but unconfirmed ORFs in this group (ORF 3595 and ORF 16519) code for predicted iron-sulfur proteins. The ORF 3595 product could be involved in DNA damage repair, although the significance of this is not clear. Interestingly, no classical stress genes are upregulated before 3 h p.i., suggesting that the induction of encystation is not associated with a stress response. However, 5 of the 16 genes which are transiently upregulated at 45 min p.i. are hypothetical proteins with unknown function. Thus, we cannot determine whether any of those play a role in an early process of encystation or whether the upregulation of all or some members of this group can be considered an off-target effect.
Fig. 1.
Whole-genome microarray analysis of encystation induced by protocol A. (A) List of statistically significantly upregulated genes (cutoff, >2-fold signal increase) at 45 min, 3 h, and 7 h postinduction. (B) Graphical representation (Venn diagram) of overlaps in the three data sets. Note that no gene is present in all three data sets.
Two different protocols result in highly comparable induction of encystation.
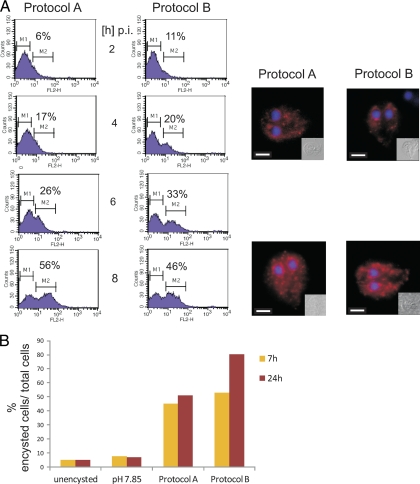
The 7-h data set generated with cells encysted with protocol A contained all previously identified regulated genes. We therefore decided to use this time point for a comparative analysis of encystation protocols with the aim of filtering out off-target effects. Transferring cultured trophozoites to cholesterol (lipid)-free medium with an elevated pH (protocol B) is an efficient (albeit more expensive) method to induce encystation. As a first step, we tested whether the kinetics of induction elicited by protocols A and B were comparable. As criteria, we used the expression of CWP1 protein, ESV formation, and the appearance of cysts. FACS analysis of chemically fixed and detergent-permeabilized cells labeled with anti-CWP1 monoclonal antibody showed that the kinetics of CWP1 production during the first 8 h p.i. was very similar with both protocols (Fig. 2A). Immunofluorescence analysis at 4 h and 8 h p.i. confirmed equal formation of ESVs. Quantitation of encysting cells and cysts at 7 h and 24 h p.i. revealed a slightly higher cyst output with protocol B at 24 h p.i. This suggests that the second half of the encystation process might progress slightly faster in the latter. In these tests, we also confirmed the importance of an elevated pH (7.85) for the induction with protocol B (Fig. 2B). Conversely, an increase of the standard culture medium pH alone is not sufficient for induction.
Fig. 2.
Comparison of the kinetics of the two encystation protocols. (A) Flow cytometry analysis of 105 encysting cells each induced by protocol A or protocol B. Chemically fixed and detergent-permeabilized cells were labeled with an anti-CWP1 antibody coupled to Cy3. Gates for trophozoites (nonlabeled, M1) and cells containing CWP1 in ESVs (M2) at four different time points postinduction are indicated together with calculated percentages of encysting cells. Representative IFA images of labeled cells (CWP1, red; DAPI, blue) are shown for the time points 4 h and 8 h p.i. Insets are bright-field images. Scale bars, 3 μm. Counts, recorded events; FL2-H, fluorescence intensity (Cy3). (B) Quantitation of encystation at different conditions by microscopy (results of representative experiment). The percentages of encysting cells containing ESVs and cysts labeled with an anti-CWP1 antibody coupled to Cy3 at 7 h and 24 h in each field of view were determined.
It is likely that in vivo, an elevated pH in combination with another signal involving lipids is required to induce encystation, indicating that differentiation in vivo is only triggered efficiently in the distal ileum. The combination of two signals suggests that temporal as well as spatial criteria are important. It is tempting to speculate that the latter may provide a way to segregate rapidly proliferating trophozoites from differentiating parasites. Since conditions change significantly between the duodenum and the distal ileum, an interesting question in this respect is how long inducing conditions have to persist for the cells to complete encystation. Preliminary in vitro data indicated that regardless of the protocol used, this time is more than 7 h but less than 20 h p.i. Parasites which are brought back to standard culture conditions before 7 h p.i. do not complete encystation and often become severely damaged (data not shown). This suggests that inducing conditions do not trigger a “fire-and-forget” response but, rather, that parasites need sustained stimulation to complete encystation.
Comparative analysis of induced genes generated with two protocols reveals a small core set of bona fide encystation genes.
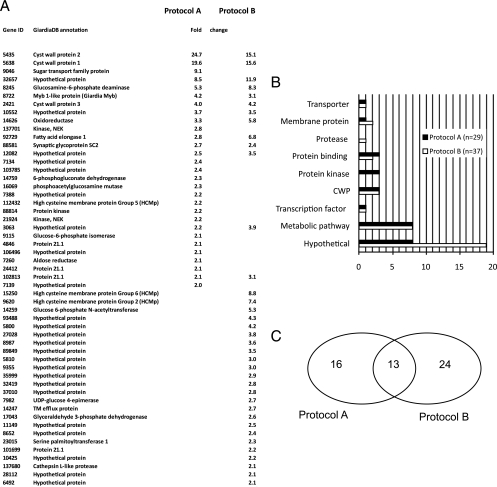
Microarray analysis of cells induced with protocol B yielded a set of 37 genes whose signal was increased >2 fold at 7 h p.i. (Fig. 3A). A categorization of the genes identified in the 7-h data sets indicated clear protocol-specific differences (Fig. 3B). Most notable was the high number of hypothetical proteins and the identification of two different high-Cys membrane proteins (HCM proteins) in the protocol B data set.
Fig. 3.
Comparative analysis of gene regulation induced by protocols A and B. (A) The tabular depiction of ORFs with a statistically significant change of signal of >2.0 in the two data sets shows 13 common ORFs. TM, transmembrane. (B) Categorization of ORFs in the data sets according to function shows that the protocol B data set contains significantly more hypothetical ORFs. (C) Graphical representation (Venn diagram) of the two data sets. The intersecting positions are considered bona fide encystation ORFs.
A detailed comparative analysis of the upregulated genes in the two data sets revealed a greater number of divergent than of intersecting positions (graphical depiction in Fig. 3C). If we follow the definition that only genes significantly induced in both protocols are truly linked to encystation, the result suggests that each protocol produces major off-target effects. However, the intersecting data set of 13 positions contained all confirmed, highly regulated encystation genes, including CWPs and Myb. The HCNC protein identified in a previous study (5) did not appear in any of the data sets, and the three upregulated HCM protein family members are not associated with encystation according to these criteria. Genes in the intersecting data set show a comparable degree of signal increase in both protocols, with the exception of ORF 92729 (annotated as fatty acid elongase 1), which shows an induction difference of >2-fold between data sets. The induction of CWP3 appears to be independent of the protocol used. Conversely, the induction levels of CWP1 and -2 are lower with protocol B. This is consistent with the minor differences in CWP1 expression observed at early time points in the comparative FACS analysis (Fig. 2A) and could reflect differences in expression kinetics rather than the final amount of product.
The results of this comparative analysis underscore the importance of distinguishing between upregulation due to off-target effects and bona fide encystation genes. This is relevant because large data sets from genome-wide transcriptome analyses (SAGE and microarray) of differentiation processes (encystation/excystation) are available to the community via GiardiaDB, and information on stage-regulated expression is included in the data sheet of each giardial gene. From a biological perspective, a direct comparative analysis of encystation with two distinct in vitro protocols is important. This allows narrowing down the region where encystation is triggered in the host's small intestine. Cultured parasites are exposed to complex changes in environmental conditions during in vitro encystation. Similarly, trophozoites, which reside in the duodenal region, experience strong temporal and local fluctuations of bile and pancreatic secretions, in addition to changes induced by dietary factors. Importantly, there is a considerable pH gradient between the proximal and distal small intestine. The most consistent parameter of the two encystation protocols is the absolute requirement of a pH of 7.8 to 7.9. Because this is necessary but not sufficient for strong induction of encystation in vitro, it seems unlikely that encystation in vivo is triggered in the duodenal region, where the pH is 6 to 6.5. The lumen of the jejunum and the ileum, however, is increasingly alkaline and reaches pH 7 to 8 in distal regions. Cholesterol and fatty acids are resorbed by enterocytes in the lower small intestine, and thus, parasites in the distal regions of the ileum likely encounter conditions which trigger differentiation.
A Myb binding sequence is a signature motif for encystation genes.
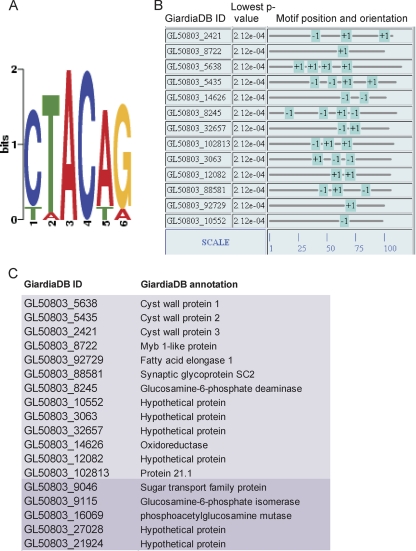
The cis-acting elements for control of Giardia gene expression are included in short, ∼100-bp sequences upstream of transcription start sites. The only element common to many giardial promoters is a short AT-rich sequence (initiator) close to the transcription start site (7). CWP promoters follow this rule, and in addition, contain binding sites for the Giardia Myb transcription factor. A sequence analysis of ∼100 bp upstream of the 13 identified encystation genes revealed that all promoters contained at least one MBS. A MEME analysis using a motif width of between 4 and 12 nt showed that the MBS was the only motif which was overrepresented in the promoter regions of encystation genes (Fig. 4A). We also identified the position and orientation of 33 MBSs in 13 promoter sequences (∼100 bp of upstream sequence) (Fig. 4B) and generated a more refined representation of the MBS 6-mer motif 5′-(C/T–T/A–A–C–A/T–G/A)–3′ with this training set. The result is consistent with the motif identified in the scanning mutagenesis experiment to analyze binding properties of Myb that was performed by Sun and coworkers (39). We tested whether this motif was also overrepresented in the 40 (16 plus 24) nonoverlapping genes in the two 7-h data sets. This analysis revealed one MBS each in the promoter sequences of five additional genes that appeared in either the protocol A or B data set at 7 h p.i. These candidates were not in the intersecting group because they were eliminated for technical reasons (e.g., large variation between technical replicates, a P value of >0.01), or a signal increase just below the 2.0-fold cutoff (see Fig. S1 in the supplemental material). Nevertheless, taking into account additional criteria, such as a statistically significant signal increase of >1.9, detection of the MBS in the MAST analysis, upregulation at the 3 h time point, or mention in the literature (20, 40), we decided to include these five candidates in a final set of 18 encystation genes (Fig. 4C).
Fig. 4.
Representation and distribution of the MBSs in the promoter regions of regulated genes. (A) MEME analysis and sequence logo of the MBS generated with the 13 encystation promoters as a training set. A single change to the alternative nucleotide at position 1, 2, 5, or 6 is tolerated. (B) The MEME analysis also shows the positions of MBSs in the sense (+) and antisense (−) orientation in promoters. Position 1 on the scale indicates the most proximal nucleotide next to the start codon of the ORF. (C) Tabular depiction of the final set of encystation ORFs, including five additional positions (darker shading).
We expanded our analysis and found that none of the promoters of the 18 genes in the 45-min data set (protocol A) contained an MBS. However, in 16 of these promoters, at least one AT-rich motif was identified by a MEME/MAST analysis, predominately in the distal half of the promoter (see Fig. S2A in the supplemental material). This is additional support for the idea that the early response to protocol A is regulated separately and does not involve Myb. In the corresponding analysis of the 3-h data set, a slightly divergent version of the MBS emerges in a MEME motif search (see Fig. S2B in the supplemental material). Six out of nine genes in this group are included in the final set of encystation genes. Finally, a global MAST analysis of the promoter sequences of 4,969 predicted genes in the Giardia genome found the 6-nt MBS sequence in 715 promoter regions (including the 18 bona fide encystation genes). This frequency of occurrence is bracketed by the statistically predicted frequencies of the 6-mer (sense orientation) of 121 if no variability is allowed and of 1,941 if we allow one variation at position 1, 2, 5, or 6 with equal probability for two nucleotides in the motif. Together with the highly significant overrepresentation in the promoter regions of the encystation genes, this strongly suggests an otherwise random frequency of occurrence of the MBS motif in upstream regions of genes.
Myb is constitutively expressed but upregulated during encystation. Thus, since Myb is not the central regulator of encystation gene expression, the presence of the MBS in other promoters is not surprising. Positive autoregulation via an MBS in the Myb promoter (28, 39) is a likely mechanism to ensure a rapid increase of Myb mRNA levels in differentiating cells. Sun et al. (39) showed that three tandem MBS copies confer some upregulation to the constitutive Ran promoter in a reporter construct. Together with data demonstrating a cis-acting sequence in the promoter of CWP2 conferring tight transcriptional repression of the gene in trophozoites (6), this fits with the idea that MBS modulates but does not control stage-specific transcription. Indeed, the promoters of two of our final set of 18 encystation genes contain no MBS on the sense strand. The need for rapid upregulation of Myb during encystation could also be explained by competition for space on the short promoter sequence.
Taken together, the data strongly suggest that the expression of all bona fide encystation genes identified here is modulated by the recruitment of Giardia Myb to their promoter sequences. This alone could be sufficient to increase the levels of gene products which are also expressed in trophozoites. The positive autoregulation of Myb gene expression likely reinforces this effect. However, the CWPs at least, which are silenced in trophozoites, require additional mechanisms (e.g., derepression) for induction. Thus, we postulate that encystation genes can be divided into two groups: encystation-specific genes that are stage-specifically expressed (e.g., structural proteins, such as CWPs), and genes whose expression is upregulated from basal levels in trophozoites after induction. A role of Myb as an amplifier is also consistent with our recently reported data indicating epigenetic control of gene expression upon differentiation (35).
Because the 42 upregulated genes reported in this work (2) are distributed among the 301 hits with sense tags in the three categories encystation (35 hits), differentiation (3 hits), and trophozoites and encyzoites (262 hits), we compared the microarray hits (Fig. 3A and 4C) with the SAGE data only in a unidirectional manner. In addition, we limited our analysis to the SAGE data for the 4-h and 12-h time points. We found a better correlation with hits from protocol A (19/29) than with those from protocol B (18/37) (see Fig. S3A in the supplemental material). The correlation was clearly improved when only the set of 18 encystation genes were considered (14/18). Conversely, the set of nonoverlapping genes from protocols A and B showed the lowest correlation (13/35). This comparison should be taken with a grain of salt because neither the protocol used to induce differentiation nor the time points of the SAGE analysis are directly comparable. However, the relatively high number of encystation genes is consistent with the central prediction of our hypothesis.
The GS strain of Giardia lamblia has been reported to respond less efficiently to induction of encystation in vitro. Sequence differences in the promoter region of CWPs have been discussed as a possible explanation (8). To investigate this further, we analyzed the promoters of 14 GS homologs of encystation genes. We found mutations in one or more MBS motifs in 9 of 14 promoters, which appears to confirm this trend (data not shown). However, the biological significance of this result and a link to less efficient encystation in vitro need to be examined in more detail.
Pharmacologically induced histone hyperacetylation prevents upregulation of encystation genes.
In a previous study (35), we showed that the general level of histone acetylation decreases in encysting trophozoites, suggesting an increase in chromatin condensation in as-yet-undetermined loci as the cells differentiate to a dormant stage. We demonstrated that by preventing hypoacetylation in these parasites with a class I histone deacetylase (HDAC) inhibitor (FR235222), the synthesis and export of cyst wall material was severely compromised, resulting in very low levels of cysts in these cultures. Having identified a set of bona fide encystation genes, we used this existing microarray data to determine whether their induction was affected by FR235222, which would argue further for a common mechanism of regulation. Indeed, inhibition of the giardial HDAC in encysting cells led to decreased (>2-fold) induction of the nine most strongly expressed encystation genes at 7 h p.i. (see Fig. S3B in the supplemental material). Induction of seven additional members of this set was detected as well, but the expression of two of the most weakly induced genes (ORFs 21924 and 9115) did not change in the presence of the drug. The latter result is most likely due to insufficient sensitivity of the microarray analysis. Overall, this is strong support for the idea that, in the first 7 h of p.i., encystation genes are coregulated by derepression in the case of highly expressed genes coding for structural components and/or by positive modulation by binding of Myb to promoters. The combined data also suggest that histone deacetylation during encystation is (directly or indirectly) required for the induction of encystation gene expression.
Downregulated genes.
Ten significantly downregulated genes were identified in the consolidated (i.e., after removal of deprecated genes) protocol A data set at all three time points and only two for the 7-h time point of the protocol B data set (see Fig. S4 in the supplemental material). Although this is much more difficult to interpret, taken together with the comparatively small changes in expression, the data suggest that downregulation may play a smaller but perhaps still significant role in encystation (see below). However, the fact that there is no overlap between the two 7-h data sets casts considerable doubt on the relevance of these genes with respect to differentiation. There is some agreement between the microarray and the SAGE data (2): 5 of 12 identified genes are listed as downregulated. Interestingly, among those is ORF 112103, coding for arginine deiminase (17, 33), an important factor in host-parasite interaction, which was reported to translocate to nuclei and inhibit CWP2 expression (41). Its relatively strong downregulation at 45 min and 3 h p.i. but not at the 7-h time point is consistent with this scenario and warrants further investigation.
Two hypothetical proteins are upregulated and localize to the endoplasmic reticulum.
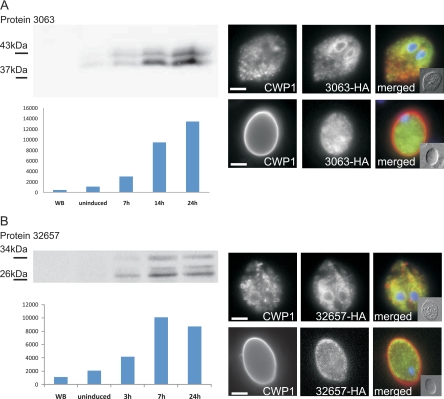
In addition to several known and predicted genes, five newly identified encystation genes coding for hypothetical proteins are included in the final data set. To determine if this group might contain structural proteins of the CW or the CW-plasma membrane interface, we expressed the two predicted secreted proteins encoded by ORFs (ORF 3063 and ORF 32657) as HA epitope-tagged variants under the control of their respective endogenous promoters (Fig. 5; also see Fig. S5 in the supplemental material). Consistent with the microarray data and with previous experiments using CWP sequences, a short, ∼100-bp promoter sequence was sufficient for strong, stage-regulated expression of both tagged proteins. Immunofluorescence analysis showed both tagged products in the endoplasmic reticulum of transgenic encysting parasites (Fig. 5, note the distinct nuclear envelope staining; also see Fig. S5 in the supplemental material). The tagged-ORF 32657 product also localized at the cell periphery, in addition to internal compartments, but not in the cyst wall (Fig. 5A and B; also see Fig. S5B in the supplemental material). The lack of distinct ESV staining makes it unlikely that these candidates are exported to the cyst wall. ORF 32657 encodes a multipass transmembrane protein, while the predicted ORF 3063 product is a type II transmembrane protein. Despite the completely different topologies, both have very similar expression kinetics and localization in trophozoites. Taken together with the domain structure analysis of the final data set, this strongly suggests that no major structural proteins in addition to the three CWP family members are involved in building the cyst wall.
Fig. 5.
Expression of predicted secreted proteins as tagged variants in encysting cells. The expression of HA-tagged ORF 3063 (A) and ORF 32657 (B) is controlled by the respective endogenous promoters. Western analysis of protein extracts separated by SDS-PAGE (left) shows strong induction of the proteins during encystation (0 to 24 h p.i.). Representative IFA images of chemically fixed and detergent-permeabilized cells at 7 h (encysting trophozoites, top rows) and 24 h p.i. (cysts, bottom rows) are shown. For quantitative analysis of colocalization, see Fig. S5 in the supplemental material. WB, untransfected cells (negative control); CWP1, red; 3063-HA, green; DAPI, blue. Scale bars, 3 μm. Insets are bright-field images.
Supplementary Material
ACKNOWLEDGMENTS
The Giardia lamblia microarrays (version 2) are based on the Giardia (strain WB) genome sequence (26) and were kindly provided through NIAID's Pathogen Functional Genomics Resource Center, managed and funded by the Division of Microbiology and Infectious Diseases, NIAID, NIH, DHHS, and operated by the J. Craig Venter Institute. The Functional Genomics Center Zürich, Switzerland (www.fgcz.uzh.ch) is a joint facility of the ETHZH and the University of Zürich. We are grateful to Therese Michel for technical assistance and to Weihong Qi from the FGCZ for her help with data analysis.
We acknowledge in particular the Stiftung zur Förderung der Wissenschaftlichen Forschung an der Universität Zürich for financial support for this project. C.S. was supported by the Roche and Novartis Foundation and Stiftung für Forschungsförderung, University of Zurich. Research in the Hehl laboratory was supported by the Swiss National Science Foundation (grant no. 31003A-125389).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 6 August 2010.
REFERENCES
- 1.Bernander R., Palm J. E., Svard S. G. 2001. Genome ploidy in different stages of the Giardia lamblia life cycle. Cell. Microbiol. 3:55–62 [DOI] [PubMed] [Google Scholar]
- 2.Birkeland S. R., Preheim S. P., Davids B. J., Cipriano M. J., Palm D., Reiner D. S., Svard S. G., Gillin F. D., McArthur A. G. 2010. Transcriptome analyses of the Giardia lamblia life cycle. Mol. Biochem. Parasitol. 174:62–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher S. E., Gillin F. D. 1990. Excystation of in vitro-derived Giardia lamblia cysts. Infect. Immun. 58:3516–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulik D. A., van Ophem P., Manning J. M., Shen Z., Newburg D. S., Jarroll E. L. 2000. UDP-N-acetylglucosamine pyrophosphorylase, a key enzyme in encysting Giardia, is allosterically regulated. J. Biol. Chem. 275:14722–14728 [DOI] [PubMed] [Google Scholar]
- 5.Davids B. J., Reiner D. S., Birkeland S. R., Preheim S. P., Cipriano M. J., McArthur A. G., Gillin F. D. 2006. A new family of giardial cysteine-rich non-VSP protein genes and a novel cyst protein. PLoS One 1:e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis-Hayman S. R., Hayman J. R., Nash T. E. 2003. Encystation-specific regulation of the cyst wall protein 2 gene in Giardia lamblia by multiple cis-acting elements. Int. J. Parasitol. 33:1005–1012 [DOI] [PubMed] [Google Scholar]
- 7.Elmendorf H. G., Singer S. M., Pierce J., Cowan J., Nash T. E. 2001. Initiator and upstream elements in the alpha2-tubulin promoter of Giardia lamblia. Mol. Biochem. Parasitol. 113:157–169 [DOI] [PubMed] [Google Scholar]
- 8.Franzen O., Jerlstrom-Hultqvist J., Castro E., Sherwood E., Ankarklev J., Reiner D. S., Palm D., Andersson J. O., Andersson B., Svard S. G. 2009. Draft genome sequencing of Giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog. 5:e1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerwig G. J., van Kuik J. A., Leeflang B. R., Kamerling J. P., Vliegenthart J. F., Karr C. D., Jarroll E. L. 2002. The Giardia intestinalis filamentous cyst wall contains a novel beta(1-3)-N-acetyl-D-galactosamine polymer: a structural and conformational study. Glycobiology 12:499–505 [DOI] [PubMed] [Google Scholar]
- 10.Gillin F. D., Boucher S. E., Rossi S. S., Reiner D. S. 1989. Giardia lamblia: the roles of bile, lactic acid, and pH in the completion of the life cycle in vitro. Exp. Parasitol. 69:164–174 [DOI] [PubMed] [Google Scholar]
- 11.Hehl A. B., Marti M., Kohler P. 2000. Stage-specific expression and targeting of cyst wall protein-green fluorescent protein chimeras in Giardia. Mol. Biol. Cell 11:1789–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y. C., Su L. H., Lee G. A., Chiu P. W., Cho C. C., Wu J. Y., Sun C. H. 2008. Regulation of cyst wall protein promoters by Myb2 in Giardia lamblia. J. Biol. Chem. 283:31021–31029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarroll E. L., Manning P., Lindmark D. G., Coggins J. R., Erlandsen S. L. 1989. Giardia cyst wall-specific carbohydrate: evidence for the presence of galactosamine. Mol. Biochem. Parasitol. 32:121–131 [DOI] [PubMed] [Google Scholar]
- 14.Jimenez-Garcia L. F., Zavala G., Chavez-Munguia B., Ramos-Godinez Mdel P., Lopez-Velazquez G., Segura-Valdez Mde L., Montanez C., Hehl A. B., Arguello-Garcia R., Ortega-Pierres G. 2008. Identification of nucleoli in the early branching protist Giardia duodenalis. Int. J. Parasitol. 38:1297–1304 [DOI] [PubMed] [Google Scholar]
- 15.Kane A. V., Ward H. D., Keusch G. T., Pereira M. E. 1991. In vitro encystation of Giardia lamblia: large-scale production of in vitro cysts and strain and clone differences in encystation efficiency. J. Parasitol. 77:974–981 [PubMed] [Google Scholar]
- 16.Kim J., Bae S. S., Sung M. H., Lee K. H., Park S. J. 2009. Comparative proteomic analysis of trophozoites versus cysts of Giardia lamblia. Parasitol. Res. 104:475–479 [DOI] [PubMed] [Google Scholar]
- 17.Knodler L. A., Sekyere E. O., Stewart T. S., Schofield P. J., Edwards M. R. 1998. Cloning and expression of a prokaryotic enzyme, arginine deiminase, from a primitive eukaryote Giardia intestinalis. J. Biol. Chem. 273:4470–4477 [DOI] [PubMed] [Google Scholar]
- 18.Konrad C., Spycher C., Hehl A. B. 2010Selective condensation drives partitioning and sequential secretion of cyst wall proteins in differentiating Giardia lamblia. PLoS Pathog. 6:e1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauwaet T., Davids B. J., Reiner D. S., Gillin F. D. 2007. Encystation of Giardia lamblia: a model for other parasites. Curr. Opin. Microbiol. 10:554–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez A. B., Sener K., Jarroll E. L., van Keulen H. 2003. Transcription regulation is demonstrated for five key enzymes in Giardia intestinalis cyst wall polysaccharide biosynthesis. Mol. Biochem. Parasitol. 128:51–57 [DOI] [PubMed] [Google Scholar]
- 21.Lujan H. D., Mowatt M. R., Conrad J. T., Bowers B., Nash T. E. 1995. Identification of a novel Giardia lamblia cyst wall protein with leucine-rich repeats. Implications for secretory granule formation and protein assembly into the cyst wall. J. Biol. Chem. 270:29307–29313 [DOI] [PubMed] [Google Scholar]
- 22.Lujan H. D., Mowatt M. R., Byrd L. G., Nash T. E. 1996a. Cholesterol starvation induces differentiation of the intestinal parasite Giardia lamblia. Proc. Natl. Acad. Sci. U. S. A. 93:7628–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lujan H. D., Mowatt M. R., Conrad J. T., Nash T. E. 1996b. Increased expression of the molecular chaperone BiP/GRP78 during the differentiation of a primitive eukaryote. Biol. Cell 86:11–18 [DOI] [PubMed] [Google Scholar]
- 24.Marti M., Hehl A. B. 2003. Encystation-specific vesicles in Giardia: a primordial Golgi or just another secretory compartment? Trends Parasitol. 19:440–446 [DOI] [PubMed] [Google Scholar]
- 25.Marti M., Regos A., Li Y., Schraner E. M., Wild P., Muller N., Knopf L. G., Hehl A. B. 2003. An ancestral secretory apparatus in the protozoan parasite Giardia intestinalis. J. Biol. Chem. 278:24837–24848 [DOI] [PubMed] [Google Scholar]
- 26.Morrison H. G., McArthur A. G., Gillin F. D., Aley S. B., Adam R. D., Olsen G. J., Best A. A., Cande W. Z., Chen F., Cipriano M. J., Davids B. J., Dawson S. C., Elmendorf H. G., Hehl A. B., Holder M. E., Huse S. M., Kim U. U., Lasek-Nesselquist E., Manning G., Nigam A., Nixon J. E., Palm D., Passamaneck N. E., Prabhu A., Reich C. I., Reiner D. S., Samuelson J., Svard S. G., Sogin M. L. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921–1926 [DOI] [PubMed] [Google Scholar]
- 27.Mowatt M. R., Lujan H. D., Cotten D. B., Bowers B., Yee J., Nash T. E., Stibbs H. H. 1995. Developmentally regulated expression of a Giardia lamblia cyst wall protein gene. Mol. Microbiol. 15:955–963 [DOI] [PubMed] [Google Scholar]
- 28.Nicolaides N. C., Gualdi R., Casadevall C., Manzella L., Calabretta B. 1991. Positive autoregulation of c-myb expression via Myb binding sites in the 5′ flanking region of the human c-myb gene. Mol. Cell. Biol. 11:6166–6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palm D., Weiland M., McArthur A. G., Winiecka-Krusnell J., Cipriano M. J., Birkeland S. R., Pacocha S. E., Davids B., Gillin F., Linder E., Svard S. 2005. Developmental changes in the adhesive disk during Giardia differentiation. Mol. Biochem. Parasitol. 141:199–207 [DOI] [PubMed] [Google Scholar]
- 30.Pan Y. J., Cho C. C., Kao Y. Y., Sun C. H. 2009. A novel WRKY-like protein involved in transcriptional activation of cyst wall protein genes in Giardia lamblia. J. Biol. Chem. 284:17975–17988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Que X., Svard S. G., Meng T. C., Hetsko M. L., Aley S. B., Gillin F. D. 1996. Developmentally regulated transcripts and evidence of differential mRNA processing in Giardia lamblia. Mol. Biochem. Parasitol. 81:101–110 [DOI] [PubMed] [Google Scholar]
- 32.Rehrauer H., Zoller S., Schlapbach R. 2007. MAGMA: analysis of two-channel microarrays made easy. Nucleic Acids Res. 35:W86–W90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ringqvist E., Palm J. E., Skarin H., Hehl A. B., Weiland M., Davids B. J., Reiner D. S., Griffiths W. J., Eckmann L., Gillin F. D., Svard S. G. 2008. Release of metabolic enzymes by Giardia in response to interaction with intestinal epithelial cells. Mol. Biochem. Parasitol. 159:85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smyth G. K., Michaud J., Scott H. S. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067–2075 [DOI] [PubMed] [Google Scholar]
- 35.Sonda S., Morf L., Bottova I., Baetschmann H., Rehrauer H., Caflisch A., Hakimi M. A., Hehl A. B. 2010. Epigenetic mechanisms regulate stage differentiation in the minimized protozoan Giardia lamblia. Mol. Microbiol. 76:48–67 [DOI] [PubMed] [Google Scholar]
- 36.Stefanic S., Palm D., Svard S. G., Hehl A. B. 2006. Organelle proteomics reveals cargo maturation mechanisms associated with Golgi-like encystation vesicles in the early-diverged protozoan Giardia lamblia. J. Biol. Chem. 281:7595–7604 [DOI] [PubMed] [Google Scholar]
- 37.Stefanic S., Morf L., Kulangara C., Regos A., Sonda S., Schraner E., Spycher C., Wild P., Hehl A. B. 2009. Neogenesis and maturation of transient Golgi-like cisternae in a simple eukaryote. J. Cell Sci. 122:2846–2856 [DOI] [PubMed] [Google Scholar]
- 38.Su L. H., Lee G. A., Huang Y. C., Chen Y. H., Sun C. H. 2007. Neomycin and puromycin affect gene expression in Giardia lamblia stable transfection. Mol. Biochem. Parasitol. 156:124–135 [DOI] [PubMed] [Google Scholar]
- 39.Sun C. H., Palm D., McArthur A. G., Svard S. G., Gillin F. D. 2002. A novel Myb-related protein involved in transcriptional activation of encystation genes in Giardia lamblia. Mol. Microbiol. 46:971–984 [DOI] [PubMed] [Google Scholar]
- 40.Sun C. H., McCaffery J. M., Reiner D. S., Gillin F. D. 2003. Mining the Giardia lamblia genome for new cyst wall proteins. J. Biol. Chem. 278:21701–21708 [DOI] [PubMed] [Google Scholar]
- 41.Touz M. C., Ropolo A. S., Rivero M. R., Vranych C. V., Conrad J. T., Svard S. G., Nash T. E. 2008. Arginine deiminase has multiple regulatory roles in the biology of Giardia lamblia. J. Cell Sci. 121:2930–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang H., Chung H. J., Yong T., Lee B. H., Park S. 2003. Identification of an encystation-specific transcription factor, Myb protein in Giardia lamblia. Mol. Biochem. Parasitol. 128:167–174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.