Abstract
Chitin, an insoluble polymer of N-acetyl-d-glucosamine (GlcNAc), is one of the most abundant carbohydrate polymers in marine and terrestrial environments. Chitin hydrolysis by Listeria monocytogenes depends on two chitinase-encoding genes, chiA and chiB, and the aim of this study was to investigate their regulation. Chitin induces the expression of both chitinases in late exponential growth phase, and chiA but not chiB is furthermore induced by the monomer GlcNAc. Furthermore, their expression is subjected to catabolite control. Chitinases expressed by bacterial pathogens have proven to be important not only for nutrient acquisition and environmental survival but also for infecting animals and humans. Interestingly, the central L. monocytogenes virulence gene regulator, PrfA, is required for the chitinolytic phenotype, as chitinase activity was significantly reduced in prfA mutant cells compared to its level in wild-type cells. In agreement with this, Northern blot analysis showed that the amounts of chiA and chiB transcripts upon induction by chitin were significantly lower in the prfA mutant than in the wild type. The chitinolytic activity and chiA and chiB expression were reduced in the absence of the sigB gene, indicating that σB is also important for the production of chitinases. The chiA, chiB, and chiA chiB mutants were not impaired for in vitro adhesion and invasion in epithelial cell lines, but the chiA chiB double mutant showed less survival ability in a chitin-enriched medium. The regulation of chitinolytic activity in L. monocytogenes is complex, and taken together, the results indicate that the biological role of this activity may not be limited to the external environment.
Listeria monocytogenes is a Gram-positive, facultative intracellular pathogen primarily causing severe infections in elderly and immunocompromised individuals. Although L. monocytogenes is widely distributed in nature and though food and feed transmission is critical for human and animal infections, the transmission pathways and natural reservoirs are not well defined (44). Recently, we observed that Listeria spp., including L. monocytogenes, encode two chitinases, ChiA (Lmo1883) and ChiB (Lmo0105), that are able to hydrolyze chitin, a highly insoluble carbohydrate polymer widely distributed in nature (28).
Many bacteria from terrestrial and marine environments utilize chitin as a source of carbon and nitrogen by expressing chitinase activities, resulting in the formation of the dimer chitobiose (GlcNAc)2 and the monomer N-acetylglucosamine (GlcNAc). The majority of bacterial chitinases belong to glycosyl hydrolase family 18, including the Listeria enzymes (50). Chitinolytic bacteria, e.g., species of the genera Bacillus, Listeria, Serratia, and Vibrio, often produce multiple chitinases, and the synergy between these is assumed to be necessary for effective chitin degradation (2, 38, 49, 57, 58). The expression of chitinases is regulated by substrates; however, there is great diversity among various chitinolytic bacteria regarding regulatory mechanisms. Substrates such as (GlcNAc)2-6 and chitin induce chitinase gene expression, whereas GlcNAc can act both as a repressor and an inducer (4, 30, 31, 51, 54, 58). In addition, a number of regulatory parameters are known to play a role in the expression of chitinases in various Gram-negative bacteria, including a two-component regulatory system (chiS) (30), quorum-sensing signaling (12, 56), and the histone-like nucleoid-structuring proteins H-NS and StpA (13, 23). Also, environmental factors, such as temperature, pH, salinity, and the concentration of nitrogen, play a role in the expression of chitinases (4, 11).
The genetic regulation of chitinolytic activity is not as well understood for Gram-positive bacteria, but in silico and microarray analyses have identified some regulatory parameters in L. monocytogenes. Thus, it was found by microarray analysis that the stress-responsive alternative sigma factor σB and the motility gene repressor MogR are involved in the regulation of chiA expression (20, 46). Furthermore, in silico analysis of the L. monocytogenes genome revealed that chiB is preceded by a completely conserved σ54 promoter and, therefore, is putatively σ54 regulated (1).
Chitinases expressed by bacterial pathogens have proven to be important not only for nutrient acquisition and environmental survival but also for infecting humans and animals. Recently, it was shown that a chiA mutant of Legionella pneumophila has a reduced ability to persist in the mouse lung (10) and that a chitin-binding protein of Vibrio cholerae is involved in attachment to intestinal epithelia (25). Furthermore, the expression of chitinase genes of non-cholerae Vibrio species and of chiA in L. monocytogenes is upregulated in the rabbit and mouse intestine, respectively (4, 53). Like Legionella and pathogenic vibrios, L. monocytogenes is an environmental pathogen capable of switching from a saprophytic to a pathogenic lifestyle (10, 19, 53). In L. monocytogenes, this transformation appears to be mediated through complex regulatory pathways that modulate the expression of virulence factors in response to environmental conditions and include σB and central virulence regulatory factor PrfA (24, 26, 37, 47). PrfA positively regulates the expression of virulence genes that are essential for the intracellular survival of L. monocytogenes. The activity of PrfA is influenced by environmental conditions like temperature, pH, fermentable carbohydrates that are taken up by the phosphoenolpyruvate-dependent phosphotransferase (PTS) system, and bacterial growth phase (26, 33), ensuring that virulence factors are downregulated outside a host.
In this study, we examined regulatory aspects of the expression of chiA and chiB in L. monocytogenes EGD, and we show that the expression of chiA and chiB is subject to catabolite control and is induced by chitin in the late exponential growth phase. Furthermore, we find that the chitinolytic activity and the expression of chiA and chiB of L. monocytogenes are stimulated by PrfA and σB. These results are discussed in the context of the diverse habitats of L. monocytogenes.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Wild-type strain L. monocytogenes EGD and prfA mutant strains were obtained from Werner Goebel (Biozentrum, University of Würzburg, Germany). The isogenic EGD ΔchiA and ΔchiB deletion mutants and the chiA chiB double mutant (26) were obtained from our laboratory collection. The ΔsigB mutant was obtained from Lone Brøndsted within our department. An in-frame deletion in the coding region of lmo0106 of L. monocytogenes EGD was constructed as described previously (27) by the gene splicing by overlap extension method (16) using primers lmo106-a, lmo106-b, lmo106-c, and lmo106-d (Table 1).
TABLE 1.
PCR primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| chiA-fw | CTTTTGGCACTAGGT |
| chiA-re | CATCCACATTGGCTG |
| chiB-fw | CCACGCCTTGTTTAT |
| chiB-re | TTACGGTGATTGGTC |
| lmo106-fw | TTATTTCTGCGACAG |
| lmo106-re | CAGCTTTAGCGGAAC |
| lmo106-a | ATATAAAGTGTACCGAAATGGGACCGAAGT |
| lmo106-b | TCCACCAATATCGTATACAAAATACATTCCTCT |
| lmo106-c | TGTATACGATATTGGTGGACAAAATAAAAGGTAACT |
| lmo106-d | AAGCTAGATGCCACGGAAGAAGAAATTATT |
The bacteria were cultured on brain heart infusion agar (BHI agar; Oxoid) or BHI broth (Oxoid) that contained 2 g liter−1 glucose or in Luria broth (LB) containing tryptone (10.0 g liter−1), yeast extract (5.0 g liter−1), NaCl (10.0 g liter−1), agar (15.0 g liter−1), and phosphate buffer, pH 6.9 (Oxoid). The effect of chitin, glucose, or N-acetyl-d-glucosamine (GlcNAc) (catalog no. A8625; Sigma-Aldrich) on gene expression was examined by supplementing LB with acid-hydrolyzed chitin (2.5 g liter−1) (catalog no. C9213; Sigma-Aldrich) or GlcNAc (2 g liter−1) and/or glucose (2 g liter−1) (Merck). Acid-hydrolyzed/colloidal chitin was prepared as described previously (17), omitting the filtration and freeze-drying steps. All cultures were incubated under shaken aerobic conditions at 30°C, except for one experiment that was done at 37°C. Samples for RNA extraction of bacteria in late exponential and early stationary phases were taken at optical densities at 600 nm (OD600) of 0.3 and 0.7, respectively, for bacteria grown in LB and at OD600 of 0.6 and 1.5, respectively, for bacteria grown in LB supplemented with either glucose or GlcNAc and in BHI. The higher OD values in the latter cases were due to the higher final OD value of Listeria grown in LB supplemented with carbohydrates and in BHI compared to the OD value after growth in LB alone. The numbers of CFU ml−1 at the time of sampling were determined by plating serial dilutions in 0.9% saline on BHI agar plates.
Survival during long-term starvation.
To determine survival during long-term starvation, wild-type and mutant bacteria were grown in LB supplemented with glucose (0.5 g liter−1) and with or without chitin (2.5 g liter−1) at 30°C. The numbers of CFU ml−1 were determined after serial dilutions in 0.9% saline on BHI agar plates.
Examination of chitinase activity.
Examination of the chitinase activities of L. monocytogenes cultures was done as previously described (28), except that acid-hydrolyzed/colloidal chitin (see above) was used as the substrate. The plates were incubated under aerobic conditions at 30°C and scored for hydrolytic ability (clearing zones) for up to 10 days.
RNA extraction and Northern hybridization.
Cells of L. monocytogenes wild-type and mutant strains were grown to mid-logarithmic growth phase and late exponential phase in LB or in LB supplemented with one or more of the following additional carbohydrates: chitin, glucose, or GlcNAc in a concentration of 2.5, 2, or 2 g liter−1, respectively. The bacterial cells were lysed using a FastPrep FP120 instrument (BIO101; ThermoSavant) for 45 s at speed setting 6.0. Total RNA was extracted from the cells using an RNeasy mini kit (Qiagen, Denmark) according to the manufacturer's directions. Analysis of transcripts was done as previously described (27). Hybridization probes were generated by PCR from chromosomal DNA of L. monocytogenes EGD using primers specific for the chiB gene (chiB-fw and chiB-re), the chiA gene (chiA-fw and chiA-re), and the lmo0106 gene (lmo106-fw and lmo106-re) (Table 1). RNA extracted from at least two independent experiments was analyzed.
Adhesion and invasion.
Invasion was studied in the murine macrophage cell line J774A.1 (ATCC TIB-67), the mouse fibroblast cell line L929 (European Collection of Animal Cell Cultures [ECACC] no. 85011425), and in enterocyte-like Caco-2 cells (ATCC HTB-37; Promochem). Adhesion was only studied in Caco-2 cells. J774A.1 and L929 were propagated in Dulbecco's modified Eagle's medium (DMEM, catalog no. BE12-604F; Lonza) supplemented with 10% heat-inactivated (30 min at 56°C) fetal bovine serum (FBS) (catalog no. DE14-830; Lonza) and 25 μg/ml gentamicin (catalog no. 15750-037; Gibco). Caco-2 cells were cultured in MEM enriched with Glutamax and HEPES, supplemented with 20% heat-inactivated FBS, 0.1 mM nonessential amino acids, and 0.5 ml of gentamicin (50 mg/ml). All chemicals were from Invitrogen. Cells were incubated with 5% CO2 at 37°C. All cell assay experiments were carried out in duplicate in two independent trials with the gentamicin protection assay as previously described (18). Briefly, the monolayers of cells were infected with approximately 1 × 107 CFU ml−1 of bacteria, and after 1 h of infection, the monolayers were washed. For determination of adhesion, 0.1% Triton X-100 was added to loosen and lyse the Caco-2 cells. For invasion, the washed cells were overlaid with medium containing 25 μg ml−1 gentamicin to kill extracellular bacteria before the cells, after washing, were lysed with 0.1% Triton X-100. The numbers of adhered and intracellular bacteria were determined by plating appropriate dilutions on BHI agar plates. The adhesion and intracellular indices were calculated as the number of adhered or invaded bacteria divided by the number of bacteria in the inoculum times 100%.
RESULTS
Chitin induces the expression of chitinase genes chiA and chiB, but only chiA is induced by the monomer GlcNAc.
In L. monocytogenes, chitin hydrolysis depends on two genes encoding chitinases, chiB (lmo0105) and chiA (lmo1883) (28). Examination of the L. monocytogenes EGD-e genome sequence revealed putative transcriptional terminators downstream of both chiA and chiB, and a recent experimental analysis of the L. monocytogenes operon structure showed that chiA and chiB are expressed as monocistronic transcripts (53).
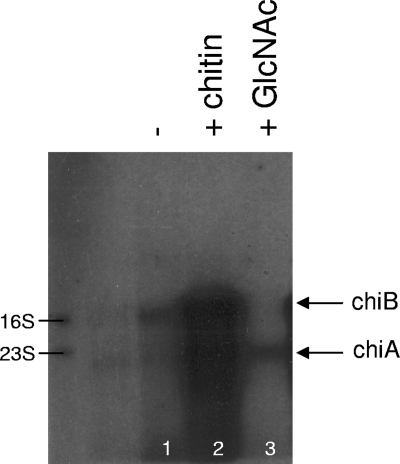
To identify inducers of chiA and chiB expression in L. monocytogenes EGD, we grew the bacteria in the absence and presence of chitin and GlcNAc and measured the expression of chiA and chiB by Northern blot analysis. We have previously suggested that chitinolytic activity in L. monocytogenes is subjected to catabolite repression mediated by easily fermentable carbohydrates (28). Therefore, the identification of inducible substrates was performed in Luria broth, which is a poor growth medium for L. monocytogenes due to low concentrations of fermentable carbohydrates (3, 45). When chitin was added to the growth medium of L. monocytogenes EGD, the amounts of both chiA and chiB mRNA were dramatically increased (Fig. 1). Probing with chiA- and chiB-specific probes identified major bands of approximately 1,100 and 2,300 bp, respectively, indicating that chiA and chiB are expressed as monocistronic transcripts, in agreement with the findings of Toledo-Arana et al. (53) (Fig. 1). When GlcNAc was added to the growth medium, a weak increase in the amount of chiA transcripts was detected (Fig. 1). Thus, GlcNAc is sufficient to induce the expression of chiA in L. monocytogenes. Interestingly, we were unable to detect any transcripts of chiB in medium with GlcNAc alone, indicating a variation in the inducing capacity of this compound (Fig. 1).
FIG. 1.
The expression of chiA and chiB of Listeria monocytogenes EGD is induced by chitin, whereas only chiA is induced by GlcNAc. Northern blot analysis of chiA and chiB transcripts. RNA was isolated from wild-type cells grown in LB (−; lane 1) or in LB supplemented with 0.25% (wt/vol) chitin (lane 2) or with 0.2% GlcNAc (lane 3) to early stationary phase. The positions of the 16S and 23S rRNA bands are indicated. The arrows indicate the chiA and chiB transcripts. The results presented are representative of three independent experiments.
lmo0106 is not a transcriptional regulator of chiA and chiB.
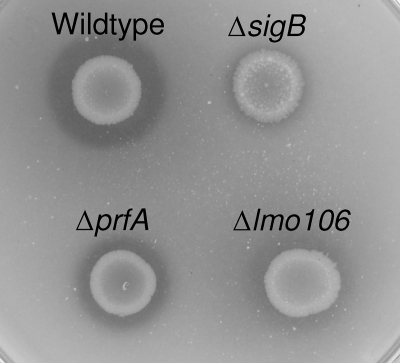
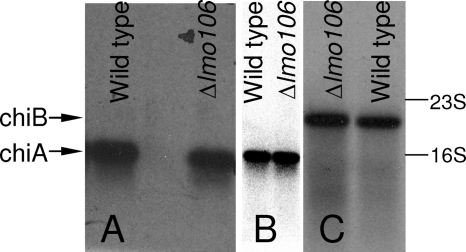
The chiB gene is located upstream of lmo0106 that encodes a gene annotated as a transcriptional regulator/sugar kinase. In order to investigate whether Lmo0106 modulates chitinolytic activity, we constructed an in-frame deletion of lmo0106 and observed that the chitinolytic activity of the resulting mutant was markedly reduced compared to that in the wild type when examined on chitin agar plates (Fig. 2). To examine whether Lmo0106 is a transcriptional regulator of chiA or chiB, we compared the amount of chiA and chiB mRNA produced in the wild type and the lmo0106 deletion mutant in chitinase-inducing growth medium. The expression of chiA and chiB was similar in both strains after growth in LB supplemented with either chitin or GlcNAc (Fig. 3). Thus, Lmo0106 is not a transcriptional regulator of chiA and chiB, and we propose a posttranscriptional role in modulating chitin hydrolysis.
FIG. 2.
The chitinolytic activity of Listeria monocytogenes depends on σB, PrfA, and Lmo0106. Wild-type and prfA, sigB, and lmo0106 mutant strains were spotted on chitin agar plates, and the plates were incubated for 5 days at 30°C. The photograph presented is representative of 3 independent experiments.
FIG. 3.
The expression of chiA (A, B) and chiB (C) of Listeria monocytogenes is not affected by lmo0106. Northern blot analysis of chiA and chiB transcripts. RNA was isolated from wild-type and lmo0106 mutant strains grown in LB supplemented with 0.2% GlcNAc (A) or grown in LB with 0.25% (wt/vol) chitin (B, C) to late exponential phase. The positions of the 16S and 23S rRNA bands are indicated. The arrows indicate the chiA and chiB transcripts. The results presented are representative of two independent experiments.
chiA and chiB expression is growth-phase dependent and subject to catabolite repression.
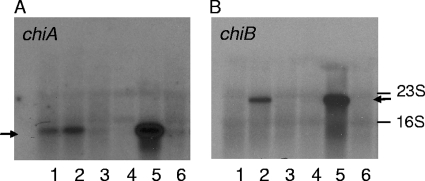
We examined the expression of the chitinase genes chiA and chiB in response to growth phase by Northern blot analysis. L. monocytogenes cells were grown to late exponential (1.4 ×108 ± 0.4 ×108 CFU ml−1 [mean ± standard deviation]) and early stationary (3.9 ×108 ± 0.5 ×108 CFU ml−1) growth phase, respectively, in growth medium with and without chitin. The induction of chiA and chiB by chitin was clearly stronger at early stationary phase than at late exponential growth phase (Fig. 4). This was not a pH effect caused by the fermentative activity of L. monocytogenes, as the pH of the growth medium was similar at both growth phases (data not shown). chiB was not expressed in the absence of chitin at either growth phase, whereas there seems to be detectable transcription of chiA in the absence of chitin by cells in late exponential phase (Fig. 4). Previously, we found that the chitinolytic activity of L. monocytogenes EGD is repressed by glucose, suggesting that chitinolytic activity is subjected to catabolite repression (28). This conclusion was confirmed in the present study, as the amounts of chiA and chiB transcripts were reduced when 0.2% glucose was included in the chitin-containing growth medium (Fig. 4). We also examined the expression of chiA and chiB after growth in BHI (without chitin) at 37°C, since these growth conditions are generally used for culturing L. monocytogenes, and found that the amounts of transcripts were below the level of detection in early stationary phase (see Fig. S1 in the supplemental material). In contrast, chiA and chiB transcripts were detected at 37°C in LB supplemented with chitin but at reduced levels compared to the levels at 30°C (see Fig. S1 in the supplemental material).
FIG. 4.
The expression of chiA and chiB of Listeria monocytogenes is growth phase dependent and repressed by glucose. Northern blot analysis of chiA (A) and chiB (B) transcripts. RNA was isolated from wild-type cells grown in LB without chitin (lanes 1 and 4), with 2.5 g liter−1 (wt/vol) chitin (lanes 2 and 5), or with 2.5 g liter−1 chitin and 2 g liter−1 glucose (lanes 3 and 6) to an OD600 of 0.3 (lanes 1 to 3) or an OD600 of 0.7 (lanes 4 to 6). The positions of the 16S and 23S rRNA bands are indicated. The arrows indicate the chiA and chiB transcripts. The results presented are representative of two independent experiments.
ChiA and ChiB contribute to long-term survival in chitin-containing medium.
Chitin has previously been shown to support the survival of L. monocytogenes in minimal medium with glucose (39). We therefore determined whether ChiA and ChiB are important for the survival of L. monocytogenes during starvation in chitin-containing medium. Wild-type and mutant strains were inoculated into LB supplemented with 0.5 g liter−1 glucose and with or without 2.5 g liter−1 chitin and incubated at 30°C for up to 6 weeks. No differences between wild-type and chitinase mutant strains were detected during the first 11 days of starvation (Table 2). However, after prolonged starvation, the number of CFU ml−1 of the chiA chiB double mutant decreased slightly more than the number of CFU ml−1 of the wild type, and the number was approximately 5 to 10 times lower than the number of the wild type (Table 2 and data not shown). The numbers of the chiA and the chiB mutants were between the numbers of the wild type and of the chiA chiB double mutant (Table 2).
TABLE 2.
Survival of Listeria monocytogenes EGD during starvation in growth medium with 0.5 g liter−1 glucose and 2.5 g liter−1 chitin
| Day | No. in viable population (log10CFU ml−1) ofa: |
|||
|---|---|---|---|---|
| Wild type | chiA mutant | chiB mutant | chiA chiB mutant | |
| 0 | 8.9 | 8.9 | 9.0 | 9.0 |
| 11 | 8.6 | 8.6 | 8.6 | 8.5 |
| 18 | 8.6 | 8.1 | 8.3 | 7.5 |
| 26 | 7.9 | 7.3 | 7.2 | 6.8 |
Values are the means of duplicate samples. Numbers were determined after plating on BHI agar plates. The experiment was repeated three times with similar results.
prfA and σB are important for the expression of chiA and chiB, but the chitinases are not important for infection of epithelial cell lines.
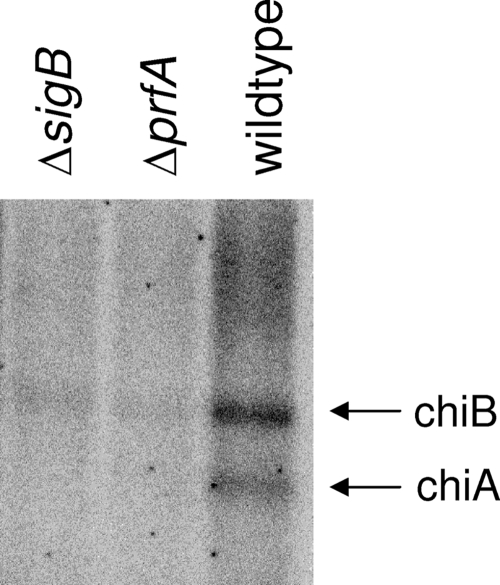
In some pathogens, chitinases may be important for virulence, as shown for L. pneumophila (10). Therefore, we set out to determine whether PrfA was involved in the chitinolytic activity of L. monocytogenes EGD. Comparison of the chitinase activity of the EGD wild type with that of a prfA mutant showed that the mutant had reduced chitinolytic activity on chitin agar plates (Fig. 2). In agreement with this, we found by Northern blot analysis that the amounts of chiA and chiB transcripts were significantly smaller upon induction by chitin in the prfA mutant than in the wild type (Fig. 5). The alternative sigma factor σB contributes to virulence in L. monocytogenes and coregulates a subset of virulence genes with positive regulatory factor A (35, 37). Therefore, we determined the role of σB in the expression of chiA and chiB and in the chitinolytic activity of L. monocytogenes. The results showed that the expression of chiA and chiB (Fig. 5) and chitinolytic activity (Fig. 2) were reduced in the absence of σB, indicating that σB is important for the production of chitinases. Thus, in addition to the issue of the presence of carbohydrates, virulence-associated parameters affect the expression of the chitinolytic phenotype of L. monocytogenes. Therefore, we investigated the role of the chitinases in virulence in vitro by examining adhesion and invasion of epithelial cells. The results showed that there were no differences between the wild type and the chiA, the chiB, and the chiA chiB mutants in their ability to adhere or invade to Caco-2 cells (data not shown). Likewise, the chitinase mutants were not impaired in invasion of the murine macrophage cell line J774A.1 or the mouse fibroblast cell line L929 (data not shown).
FIG. 5.
prfA and sigB are important for the expression of chiA and chiB in Listeria monocytogenes. Northern blot analysis of chiA and chiB transcripts. RNA was isolated from wild-type and sigB and prfA mutant cells grown in LB supplemented with 0.25% (wt/vol) chitin to late log phase (4 × 108 CFU ml−1). The arrows indicate the chiA and chiB transcripts. The results presented are representative of three independent experiments.
DISCUSSION
Many bacterial species possess the ability to catabolize chitin, and not surprisingly, a variety of parameters regulate the expression of this phenotype. One important parameter concerns the presence of the substrate (chitin) and/or its degradation products, GlcNAc and/or (GlcNAc)2, as well as the presence of other potential carbohydrate substrates. V. cholerae represents a particularly well studied organism in this respect. Thus, in the N16961 strain, the expression of three out of five annotated chitinase genes was upregulated by chitin and chitin oligosaccharides [(GlcNAc)2-6]. A fourth gene was induced by both GlcNAc and chitin oligosaccharides, whereas the expression of the fifth gene was unaffected (31). However, species and strain variation is seen, as chitinase genes are constitutively expressed in V. cholerae strains during growth in M9 minimal medium, whereas they are induced by chitin and GlcNAc in non-cholerae Vibrio strains (4). GlcNAc also induce chitinase production by a Pseudoalteromonas strain (51). In contrast, this compound acts as a repressor of chitinase gene expression in Serratia marcescens (54). Also, in Bacillus pabuli, (GlcNAc)2-4 induces the production of chitinases, whereas no chitinases could be detected in the presence of GlcNAc (14).
We show here that the two chitinase genes in L. monocytogenes, chiA and chiB, were induced by different substrates, as only chiA was induced by the monomer GlcNAc, whereas the expression of chiB depended on chitin oligosaccharides present in the chitin suspension. This indicates different roles for the two chitinases. We speculate that GlcNAc and (GlcNAc)2 may be “indicator” molecules to signal the presence of chitin for L. monocytogenes, as hypothesized for marine bacteria (21), as chitin must be present in L. monocytogenes environmental reservoirs such as terrestrial and marine environments in more or less degraded forms (15). It is also of interest to note that chiA appears to be constitutively expressed in small amounts when L. monocytogenes is present in low numbers. Such a steady low-level activity of a presumably secreted protein generates a gradient of soluble chitin degradation products, allowing the chitinolytic organism to home in on available insoluble chitin in the surroundings as outlined for Vibrio furnissii (21). It remains to be demonstrated that L. monocytogenes possesses a chemotactic ability toward chitin.
We observed that the expression of the chitinase genes was repressed in the presence of easily fermentable carbohydrates, similar to other chitinolytic bacteria, such as Bacillus spp. and Streptomyces lividans (14, 36). A microarray study found that lmo0106 and, to a lesser degree, lmo1883 (chiA) are upregulated in two mutants defective in carbon catabolite control, the hprK and ccpA insertion mutants, indicating that lmo0106 presumably is repressed by CcpA/HPr(Ser-P) in the presence of glucose (32). Taken together, these results indicate that one biological role for the chitinolytic phenotype of L. monocytogenes most likely is as a carbon and energy source in the natural environment. This conclusion is supported by the observations that chitin enhances the survival of L. monocytogenes Scott A in a defined minimal medium (39) and that the chiA chiB double mutant does not survive as well as the wild type and the chiA and chiB mutants in medium containing chitin (this study).
The lmo0106 gene encodes a putative product with homology to the ROK protein family (52). The ROK family members are divided into two groups, of which one group carries an N-terminal extension and is composed of transcriptional regulators, whereas the other group is the sugar kinases that only harbor a C-terminal sugar-binding domain (52). When we inactivated lmo0106, we observed reduced chitinolytic activity, although chiA and chiB transcription was not affected. This result agrees with Lmo0106 belonging to the sugar kinase subfamily of ROK and thus, Lmo0106 affects chitinolytic activity at the posttranscriptional level. Interestingly, lmo0106 is conserved in genome-sequenced strains of L. monocytogenes and Listeria welshimeri but not in the genome-sequenced chitinolytic Listeria innocua Clip 11262 strain (www.ncbi.nlm.nih.gov/).
It is also of interest to note that microarray studies have revealed that the chiA gene is upregulated by the alternative sigma factor σB that regulates stress response and virulence functions of L. monocytogenes (20). The growth phase-dependent expression of chiA and chiB, with higher levels of expression in early stationary phase than in late exponential phase, may perhaps be explained by an increased production of σB upon entry into stationary phase (3). We examined the chitinolytic activity of a mutant lacking σB and found that it displayed very low chitinolytic activity, showing that under the growth conditions on the chitin agar plates, σB is also important for chitinase production. Northern blot analysis confirmed that chiA and chiB expression was reduced in the absence of σB, while they were highly expressed in wild-type cells. Chitinolytic activity may also be controlled by quorum sensing, as shown for, e.g., Chromobacterium violaceum (6) and Serratia proteamaculans (7), but further studies are required to clarify this issue.
The expression of chiB is presumably regulated by σ54, since a conserved σ54 promoter was identified upstream of chiB (1). The alternative sigma factor σ54, encoded by rpoN, is involved in nitrogen and carbon utilization in various Gram-negative and Gram-positive bacteria, including L. monocytogenes (1, 48). However, the role of σ54 for chiB expression has not yet been verified in vitro, as microarray analysis of the wild type and mutant cells lacking σ54 revealed no difference in chiB expression in late exponential growth phase in BHI at 42°C, possibly because an unknown cognate activator was not active (1). Accordingly, we were unable to detect any expression of chiA or chiB in BHI without chitin at 37°C.
In addition to their roles in nutrient acquisition and environmental survival, chitinases may contribute to human and animal infections, as shown for L. pneumophila (10). To investigate the role of ChiA and ChiB of L. monocytogenes in infection, we analyzed the ability of the chiA, the chiB, and the chiA chiB mutants to adhere to and invade epithelial cells. We found that the absence of the chitinases did not influence this ability. Further studies are needed to elucidate whether the chitinases are important for virulence in vivo. Thus, the chiA mutant of L. pneumophila, which is defective in persistence in the lungs of mice, was not impaired in in vitro intracellular infection (10). Our observation that the deletion of prfA, encoding the major L. monocytogenes virulence gene regulator, reduced chitinolytic activity indicates that the biological roles of chitinases are not limited to the external environment, as the activation of prfA in general indicates the activation of a host-associated virulence phenotype (26). This notion is supported by the results of a recent microarray analysis showing that chiA and chiB are upregulated in the mouse intestine compared to their expression in BHI and that the induction of chiA is σB dependent (53). As the mammalian host does not contain chitin, the inducing signal in the host is currently unknown, but it remains a possibility that the absence of catabolite repression plays a role.
An alternative explanation for the induction of chiA and chiB in the mouse intestine would be that they represent bifunctional enzymes with an additional, as-yet-unidentified substrate other than chitin, as suggested for ChiA of L. pneumophila (10). Thus, these enzymes may have other functions in the host, as suggested for ChiA of Escherichia coli (13). Alternative substrates could be postulated to include, e.g., N-linked glycans associated with host proteins, as shown for Streptococcus pyogenes and Enterococcus faecalis (8, 9), or peptidoglycan from the intestinal microflora. We have, however, shown that for at least ChiA, the latter possibility is not valid, as this enzyme lacks lysozymic activity (29). Further research is needed to illuminate the range of potential substrates for the L. monocytogenes chitinases. How PrfA regulates chitinolytic activity is at present unknown, but it is properly not directly linked to the expression of chitinase genes as no PrfA box was identified in the promoter region of chiA or chiB. In addition to the genes preceded by a PrfA box, expression profiles done by Milohanic et al. (34) of L. monocytogenes EGD and its isogenic prfA mutant identified 70 genes differentially transcribed during exponential growth in BHI. These include genes involved in general stress response and the transport and metabolism of carbohydrates. Among these is an operon encoding components of the mannose-specific phosphoenolpyruvate-dependent phosphotransferase system (PTS) and genes encoding ABC transporters (34). PTS and ABC transporters are involved in the transport and metabolism of GlcNAc and (GlcNAc)2 in both Gram-negative and Gram-positive bacteria, and this uptake can be important for the production of chitinases (22, 41, 42, 54). In S. marcescens, for example, (GlcNAc)2 is taken up by the PTS system and mutants defective in (GlcNAc)2 transport are unable to express chitinases (54). Likewise, in Streptomyces coelicolor, a major chitin degrader in soil, (GlcNAc)2 uptake, mediated by ABC transporters, is essential for chitinase production (43). We speculate that the reduced chitinolytic activity in the absence of PrfA is partly caused by a reduced uptake of GlcNAc or (GlcNAc)2. However, other factors linked to carbohydrate uptake and metabolism are presumably involved, and further studies are needed to elucidate how PrfA influences chitinolytic activity.
Recent findings suggest that the mechanism of virulence reflects an adaptive mechanism originating in the environment (25, 40, 55). Thus, toxin-coregulated pili involved in V. cholerae intestinal colonization have a role in biofilm formation on chitin-containing surfaces (40), and a chitin-binding protein mediates adherence to epithelial cells in vitro (25). The present study enhances the understanding of the regulation of the expression of the L. monocytogenes chitinolytic system. This system is regulated by the substrates chitin and GlcNAc, which are presumed to be of importance in the environment. Our finding that PrfA and σB have a role in chitinase expression shows that factors and pathways of L. monocytogenes involved in its pathogenicity in humans and animals may also have roles in its growth in the external environment, as outlined for V. cholerae (55), or that ChiA and/or ChiB potentially play a role in infection. This topic deserves further research into the issue of potential host-related targets of the chitinases, especially for ChiA, as well as the demonstration that they are actual virulence factors by the use of in vivo models. In fact, this was very recently shown, as chiA and chiB mutant cells have a reduced capacity to grow in the liver and spleen after intravenous infection of mice (5).
Supplementary Material
Acknowledgments
The project was funded by the Danish Natural Science Research Council (274-05-0561 and 09-065158). We thank V. P. T. Nguyen and R. L. Jørgensen for expert technical assistance.
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arous, S., C. Buchrieser, P. Folio, P. Glaser, A. Namane, M. Hébraud, and Y. Héchard. 2004. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150:1581-1590. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L., C. Yu, Y. C. Lee, and S. Roseman. 1991. Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266:24276-24286. [PubMed] [Google Scholar]
- 3.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhowmick, R., A. Ghosal, and N. S. Chatterjee. 2007. Effect of environmental factors on expression and activity of chitinase genes of vibrios with special reference to Vibrio cholerae. J. Appl. Microbiol. 103:97-108. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri, S. 2010. Investigation of chitinase as a potential virulence factor in Listeria monocytogenes, abstr. B/P 71, p. 98. ISOPOL XVII Int. Symp. Probl. Listeriosis, 5 to 8 May 2010, Porto, Portugal.
- 6.Chernin, L. S., M. K. Winson, J. M. Thompson, S. Haran, B. W. Bycroft, I. Chet, P. Williams, and G. S. A. B. Stewart. 1998. Chitinolytic activity in Chromobacterium violaceum: substrate analysis and regulation by quorum sensing. J. Bacteriol. 180:4435-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, A. B., K. Riedel, L. Ebert, L. R. Flodgaard, S. Molin, L. Gram, and M. Givskov. 2003. Quorum-sensing-directed protein expression in Serratia proteamaculans B5a. Microbiology 149:471-483. [DOI] [PubMed] [Google Scholar]
- 8.Collin, M., and V. A. Fischetti. 2004. A novel secreted endoglycosidase from Enterococcus faecalis with activity on human immunoglobulin G and ribonuclease B. J. Biol. Chem. 279:22558-22570. [DOI] [PubMed] [Google Scholar]
- 9.Collin, M., and A. Olsén. 2001. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 20:3046-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DebRoy, S., J. Dao, M. Söderberg, O. Rossier, and N. P. Cianciotto. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. U. S. A. 103:19146-19151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delpin, M. W., and A. E. Goodman. 2009. Nitrogen regulates chitinase gene expression in a marine bacterium. ISME J. 3:1064-1069. [DOI] [PubMed] [Google Scholar]
- 12.Folders, J., J. Algra, M. S. Roelofs, L. C. van Loon, J. Tommassen, and W. Bitter. 2001. Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J. Bacteriol. 183:7044-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francetic, O., C. Badaut, S. Rimsky, and A. P. Pugsley. 2000. The ChiA (YheB) protein of Escherichia coli K-12 is an endochitinase whose gene is negatively controlled by the nucleoid-structuring protein H-NS. Mol. Microbiol. 35:1506-1517. [DOI] [PubMed] [Google Scholar]
- 14.Frändberg, E., and J. Schnürer. 1994. Chitinolytic properties of Bacillus pabuli K1 J. Appl. Bacteriol. 76:361-367. [DOI] [PubMed] [Google Scholar]
- 15.Gooday, G. W. 1990. The ecology of chitin degradation. Adv. Microb. Ecol. 11:387-430. [Google Scholar]
- 16.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528-535. [PubMed] [Google Scholar]
- 17.Ilankovan, P., S. Hein, C.-H. Ng, T. S. Trung, and W. F. Stevens. 2006. Production of N-acetyl chitobiose from various chitin substrates using commercial enzymes. Carbohydrate Polymers. 63:245-250. [Google Scholar]
- 18.Jensen, A., L. E. Thomsen, R. L. Jørgensen, M. H. Larsen, B. B. Roldgaard, B. B. Christensen, B. F. Vogel, L. Gram, and H. Ingmer. 2008. Processing plant persistent strains of Listeria monocytogenes appear to have a lower virulence potential than clinical strains in selected virulence models. Int. J. Food Microbiol. 123:254-261. [DOI] [PubMed] [Google Scholar]
- 19.Jones, D., and H. Seeliger. 1992. The genus Listeria, pp. 1595-1616. In A. Balows, H. Trüper, M. Dworkin, M. Harder, and K. H. Schleifer (ed.), The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd ed. Springer-Verlag, New York, NY.
- 20.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keyhani, N. O., and S. Roseman. 1999. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta 1473:108-122. [DOI] [PubMed] [Google Scholar]
- 22.Keyhani, N. O., L. X. Wang, Y. C. Lee, and S. Roseman. 2000. The chitin disaccharide, N,N′-diacetylchitobiose, is catabolized by Escherichia coli and is transported/phosphorylated by the phosphoenolpyruvate:glycose phosphotransferase system. J. Biol. Chem. 275:33084-33090. [DOI] [PubMed] [Google Scholar]
- 23.Khmel, I. A., M. I. Ovadis, A. V. Mayatskaya, A. M. Veselovskii, I. A. Bass, V. A. Lipasova, A. Bolshoy, I. Chet, and L. S. Chernin. 2005. Activity of Serratia plymuthica IC1270 gene chiA promoter region in Escherichia coli mutants deficient in global regulators of transcription. J. Basic Microbiol. 45:426-437. [DOI] [PubMed] [Google Scholar]
- 24.Kim, H., H. Marquis, and K. J. Boor. 2005. SigmaB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151:3215-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirn, T. J., B. A. Jude, and R. K. Taylor. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863-866. [DOI] [PubMed] [Google Scholar]
- 26.Kreft, J., and J. A. Vázquez-Boland. 2001. Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291:145-157. [DOI] [PubMed] [Google Scholar]
- 27.Larsen, M. H., B. H. Kallipolitis, J. K. Christiansen, J. E. Olsen, and H. Ingmer. 2006. The response regulator ResD modulates virulence gene expression in response to carbohydrates in Listeria monocytogenes. Mol. Microbiol. 61:1622-1635. [DOI] [PubMed] [Google Scholar]
- 28.Leisner, J. J., M. H. Larsen, R. L. Jørgensen, L. Brøndsted, L. E. Thomsen, and H. Ingmer. 2008. Chitin hydrolysis by Listeria spp., including L. monocytogenes. Appl. Environ. Microbiol. 74:3823-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leisner, J. J., M. H. Larsen, H. Ingmer, B. O. Petersen, J. Ø. Duus, and M. M. Palcic. 2009. Cloning and comparison of phylogenetically related chitinases from Listeria monocytogenes EGD and Enterococcus faecalis V583. J. Appl. Microbiol. 107:2080-2087. [DOI] [PubMed] [Google Scholar]
- 30.Li, X., and S. Roseman. 2004. The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc. Natl. Acad. Sci. U. S. A. 101:627-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meibom, K. L., X. B. Li, A. T. Nielsen, C.-Y. Wu, S. Roseman, and G. K. Schoolnik. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U. S. A. 101:2524-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mertins, S., B. Joseph, M. Goetz, R. Ecke, G. Seidel, M. Sprehe, W. Hillen, W. Goebel, and S. Müller-Altrock. 2007. Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes. J. Bacteriol. 189:473-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milenbachs, A. A., D. P. Brown, M. Moors, and P. Youngman. 1997. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol. Microbiol. 23:1075-1085. [DOI] [PubMed] [Google Scholar]
- 34.Milohanic, E., P. Glaser, J. Y. Coppée, L. Frangeul, Y. Vega, J. A. Vázquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 35.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen, J., F. Francou, M. J. Virolle, and M. Guérineau. 1997. Amylase and chitinase genes in Streptomyces lividans are regulated by reg1, a pleiotropic regulatory gene. J. Bacteriol. 179:6383-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ollinger, J., B. Bowen, M. Wiedmann, K. J. Boor, and T. M. Bergholz. 2009. Listeria monocytogenes σB modulates PrfA-mediated virulence factor expression. Infect. Immun. 77:2113-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orikoshi, H., S. Nakayama, K. Miyamoto, C. Hanato, M. Yasuda, Y. Inamori, and H. Tsujibo. 2005. Roles of four chitinases (ChiA, ChiB, ChiC, and ChiD) in the chitin degradation system of marine bacterium Alteromonas sp. strain O-7. Appl. Environ. Microbiol. 71:1811-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Premaratne, R. J., W.-J. Lin, and E. A. Johnson. 1991. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57:3046-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reguera, G., and R. Kolter. 2005. Virulence and the environment: a novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J. Bacteriol. 187:3551-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reizer, J., S. Bachem, A. Reizer, M. Arnaud, M. H. Saier, and J. Stülke. 1999. Novel phosphotransferase system genes revealed by genome analysis—the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology 145:3419-3429. [DOI] [PubMed] [Google Scholar]
- 42.Revilla-Nuin, B., A. Reglero, H. Martínez-Blanco, I. G. Bravo, M. A. Ferrero, and L. B. Rodríguez-Aparicio. 2002. Transport of N-acetyl-D-mannosamine and N-acetyl-D-glucosamine in Escherichia coli K1: effect on capsular polysialic acid production. FEBS Lett. 511:97-101. [DOI] [PubMed] [Google Scholar]
- 43.Saito, A., T. Fujii, T. T. Shinya, N. Shibuya, A. Ando, and K. Miyashita. 2008. The msiK gene, encoding the ATP-hydrolysing component of N,N′-diacetylchitobiose ABC transporters, is essential for induction of chitinase production in Streptomyces coelicolor A3(2). Microbiology 154:3358-3365. [DOI] [PubMed] [Google Scholar]
- 44.Sauders, B. D., and M. Wiedmann. 2007. Ecology of Listeria species and L. monocytogenes in the natural environment, pp. 21-53. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety. CRC Press, Boca Raton, FL.
- 45.Sezonov, G., D. Joseleau-Petit, and R. D'Ari. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen, A., and D. E. Higgins. 2006. The MogR transcriptional repressor regulates nonhierarchal expression of flagellar motility genes and virulence in Listeria monocytogenes. PLoS Pathog. 2:e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoll, R., S. Mertins, B. Joseph, S. Müller-Altrock, and W. Goebel. 2008. Modulation of PrfA activity in Listeria monocytogenes upon growth in different culture media. Microbiology 154:3856-3876. [DOI] [PubMed] [Google Scholar]
- 48.Studholme, D. J., and M. Buck. 2000. The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences. FEMS Microbiol. Lett. 186:1-9. [DOI] [PubMed] [Google Scholar]
- 49.Svitil, A. L., S. Chadhain, J. A. Moore, and D. L. Kirchman. 1997. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl. Environ. Microbiol. 63:408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svitil, A. L., and D. L. Kirchman. 1998. A chitin-binding domain in a marine bacterial chitinase and other microbial chitinases: implications for the ecology and evolution of 1,4-beta-glycanases. Microbiology 144:1299-1308. [DOI] [PubMed] [Google Scholar]
- 51.Techkarnjanaruk, S., S. Pongpattanakitshote, and A. E. Goodman. 1997. Use of a promoterless lacZ gene insertion to investigate chitinase gene expression in the marine bacterium Pseudoalteromonas sp. strain S9. Appl. Environ. Microbiol. 63:2989-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Titgemeyer, F., J. Reizer, A. Reizer, and M. H. Saier, Jr. 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140:2349-2354. [DOI] [PubMed] [Google Scholar]
- 53.Toledo-Arana, A., O. Dussurget, G. Nikitas, N. Sesto, H. Guet-Revillet, D. Balestrino, E. Loh, J. Gripenland, T. Tiensuu, K. Vaitkevicius, M. Barthelemy, M. Vergassola, M. A. Nahori, G. Soubigou, B. Régnault, J. Y. Coppée, M. Lecuit, J. Johansson, and P. Cossart. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950-956. [DOI] [PubMed] [Google Scholar]
- 54.Uchiyama, T., R. Kaneko, J. Yamaguchi, A. Inoue, T. Yanagida, N. Nikaidou, M. Regue, and T. Watanabe. 2003. Uptake of N,N′-diacetylchitobiose [(GlcNAc)2] via the phosphotransferase system is essential for chitinase production by Serratia marcescens 2170. J. Bacteriol. 185:1776-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vezzulli, L., A. C. Guzmán, R. R. Colwell, and C. Pruzzo. 2008. Dual role colonization factors connecting Vibrio cholerae's lifestyles in human and aquatic environments open new perspectives for combating infectious diseases. Curr. Opin. Biotechnol. 19:254-259. [DOI] [PubMed] [Google Scholar]
- 56.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe, T., W. Oyanagi, K. Suzuki, and H. Tanaka. 1990. Chitinase system of Bacillus circulans WL-12 and importance of chitinase A1 in chitin degradation. J. Bacteriol. 172:4017-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe, T., K. Kimura, T. Sumiya, N. Nikaidou, K. Suzuki, M. Suzuki, M. Taiyoji, S. Ferrer, and M. Regue. 1997. Genetic analysis of the chitinase system of Serratia marcescens 2170. J. Bacteriol. 179:7111-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.