Abstract
The production of selenomethionine (SeMet) derivatives of recombinant proteins allows phase determination by single-wavelength or multiwavelength anomalous dispersion phasing in X-ray crystallography, and this popular approach has permitted the crystal structures of numerous proteins to be determined. Although yeast is an ideal host for the production of large amounts of eukaryotic proteins that require posttranslational modification, the toxic effects of SeMet often interfere with the preparation of protein derivatives containing this compound. We previously isolated a mutant strain (SMR-94) of the methylotrophic yeast Pichia pastoris that is resistant to both SeMet and selenate and demonstrated its applicability for the production of proteins suitable for X-ray crystallographic analysis. However, the molecular basis for resistance to SeMet by the SMR-94 strain remains unclear. Here, we report the characterization of SeMet-resistant mutants of Saccharomyces cerevisiae and the identification of a mutant allele of the MUP1 gene encoding high-affinity methionine permease, which confers SeMet resistance. Although the total methionine uptake by the mup1 mutant (the SRY5-7 strain) decreased to 47% of the wild-type level, it was able to incorporate SeMet into the overexpressed epidermal growth factor peptide with 73% occupancy, indicating the importance of the moderate uptake of SeMet by amino acid permeases other than Mup1p for the alleviation of SeMet toxicity. In addition, under standard culture conditions, the mup1 mutant showed higher productivity of the SeMet derivative relative to other SeMet-resistant mutants. Based on these results, we conclude that the mup1 mutant would be useful for the preparation of selenomethionyl proteins for X-ray crystallography.
Structural analyses of proteins have provided meaningful insights into the relationship between protein conformation and biological function. Different approaches, including X-ray crystallographic analysis, nuclear magnetic resonance (NMR) analysis, and electron microscopy analysis, are applicable to determine protein structures. Although the principal method for determining three-dimensional structures of purified proteins is X-ray crystallography, substantial efforts are required to determine protein structures using this method, such as the expression and purification of recombinant proteins, optimization of crystallization conditions, and solving phase problems. Recent advances in structural biology have resulted from the substitution of Met residues for selenomethionine (SeMet) for the phase determination of proteins, using single-wavelength anomalous dispersion (SAD) and multiwavelength anomalous dispersion (MAD) phasing methods (9, 22). In addition, the use of SeMet derivatives for solving phase problems is indispensable for high-throughput determination of protein structure for structural genomic studies that aim to understand biological phenomena in whole-cell systems at the atomic level (10, 26).
The use of SeMet-incorporated proteins for X-ray crystallography was originally reported in the 1990s (9). At that time, the majority of tertiary structures were determined by SAD or MAD phasing using SeMet-containing crystals that were routinely prepared in Escherichia coli cells cultured with SeMet. However, it is considered more difficult to incorporate SeMet into proteins expressed in eukaryotic systems than in E. coli cells, and eukaryotic proteins which require posttranslational modification often fail to be expressed in E. coli cells. Therefore, the incorporation of SeMet into eukaryotic proteins is limited to those proteins that can be successfully expressed in E. coli. Although there are a few reports on the production of recombinant proteins labeled with SeMet in mammalian and insect cells, these reports emphasize mainly the practical use of the specified host cells and did not examine the mechanisms by which SeMet toxicity is overcome (1, 8). Yeast is an attractive host for the production of eukaryotic proteins of interest, as cells are capable of rapid growth under simple culture conditions and production of large amounts of recombinant proteins at low cost. In addition, the potential exists to minimize or eliminate SeMet toxicity through the isolation of a SeMet-resistant mutant of yeast.
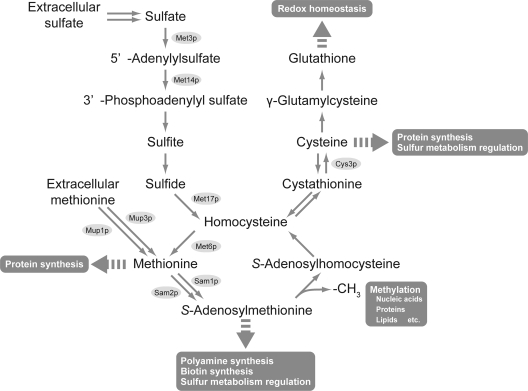
The first report of SeMet-resistant mutants in the budding yeast Saccharomyces cerevisiae suggested that the observed resistance of the eth10 and eth2 mutants was dependent upon the increase of intracellular Met concentrations as a result of enhanced sulfate assimilation during biosynthesis (6). Subsequent genetic and biochemical analyses identified that the eth10 and eth2 mutant cells possess a single, recessive mutation in the unlinked genes SAM1 and SAM2, which encode isomers of S-adenosylmethionine (AdoMet) synthetase (5). A recent study demonstrated that the deletion of both SAM1 and SAM2 confers increased SeMet resistance and allows the production of recombinant proteins with 95% of SeMet occupancy (18). In a different approach, Bockhorn et al. screened a collection of single-gene deletion mutants of S. cerevisiae for resistance to SeMet and demonstrated that a mutant lacking cystathionine γ-lyase activity (cys3Δ) showed the highest resistance to SeMet and has an ability to incorporate SeMet that is equal to or slightly higher than that of sam1Δ sam2Δ cells (2). However, the extracellular supply of expensive AdoMet or Cys, which are involved in a wide range of important biological phenomena, is required to support cellular growth of these mutants and thus limits their use in practical applications (Fig. 1).
FIG. 1.
Metabolic pathways of sulfur compounds in S. cerevisiae. The main sulfur compounds are methionine, S-adenosylmethionine, and cysteine, which are involved in protein synthesis and sulfur metabolism regulation. The S-adenosylmethionine also participates in the methylation of nucleic acids, proteins, and lipids as a methyl group donor and in the biosynthesis of biotin and polyamines. Glutathione plays a pivotal role in redox homeostasis.
Previously, we isolated a SeMet-resistant mutant of the methylotrophic yeast Pichia pastoris (SMR-94 strain) that also showed resistance to selenate (13). The mutant cells were able to produce recombinant human lysozyme containing a sufficient amount of SeMet to allow determination of its crystal structure by the SAD phasing method without the need for supplementation of AdoMet and Cys. However, the mutation sites of the P. pastoris SMR-94 strain responsible for SeMet resistance remain unclear because unlike S. cerevisiae, there is a lack of established genetic approaches and techniques for P. pastoris. Here, in an attempt to reveal the molecular basis for SeMet resistance and generate a suitable host for the production of SeMet derivatives of eukaryotic proteins, we isolated SeMet-resistant mutants of S. cerevisiae. Two obtained mutants (SRY5-3 and SRY5-7) were characterized genetically and biochemically. Furthermore, we examined the ability of these mutants to produce SeMet derivatives of epidermal growth factor (EGF) peptide.
MATERIALS AND METHODS
Yeast strains, media, and genetic manipulations.
S. cerevisiae strains used in this work are listed in Table 1. Yeast cells were routinely cultured at 30°C in YPAD medium containing 1% Bacto yeast extract (Difco), 2% Bacto peptone (Difco), and 0.01% adenine hemisulfate or in synthetic complete (SC) medium containing 0.67% Bacto yeast nitrogen base without amino acids (Difco), 2% glucose, 0.01% adenine hemisulfate, 0.002% uracil, and amino acids. SDCA medium lacking uracil (SDCA-Ura) was composed of 0.67% Bacto yeast nitrogen base without amino acids, 2% glucose, 0.01% adenine hemisulfate, 0.002% tryptophan, and 0.5% Casamino Acids (Difco). For the Met uptake assay, a minimal drop-in medium was used as described by Isnard et al. (11). For the SeMet resistance assay, l-selenomethionine (Nacalai Tesque) was added to SC medium lacking Met (SC-Met) at a final concentration of 0.025 to 0.5 mM. For the selenate resistance assay, sodium selenate (Wako) was added to YPAD medium at a final concentration of 10 mM.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| W303-1A | MATaleu2-3,112 his3-11 ade2-1 ura3-1 trp1-1 can1-100 | 27 |
| W303-1B | MATα leu2-3,112 his3-11 ade2-1 ura3-1 trp1-1 can1-100 | 27 |
| SRY5-3 | MATasam1-224 W303 | This study |
| SRY5-7 | MATamup1-100 W303 | This study |
| SRY5-3d | MATa/MATα SAM1/sam1-224 W303 | This study |
| SRY5-7d | MATa/MATα MUP1/mup1-100 W303 | This study |
| TKY357 | MATamet6Δ::His3MX SRY5-7 | This study |
| TKY358 | MATamet17Δ::His3MX6 SRY5-7 | This study |
| TKY386 | MATamup1Δ::His3MX6 W303 | This study |
| TKY387 | MATamup3Δ::His3MX6 W303 | This study |
| TKY388 | MATamup1Δ::TRP1 mup3Δ::His3MX6 W303 | This study |
| TKY371-4A | MATamup1-100 sam1-224 W303 | This study |
| TKY340 | MATasam1Δ::KanMX sam2Δ::His3MX6 W303 | This study |
| TKY417 | MATacys3Δ::His3MX6 W303 | This study |
Genetic crosses, sporulation, and dissection of the yeast strains were performed as described by Sherman (24). Gene disruption was performed using a one-step method as described previously (17).
Plasmid construction.
The primers used in this study to construct expression plasmids are listed in Table S1 in the supplemental material. For the expression of SAM1, a DNA fragment which contained the SAM1 open reading frame (ORF) and the 1.0-kbp upstream and 0.6-kbp downstream regions of the SAM1 ORF was amplified by PCR using the primers SAM1-FW and SAM1-RV. The amplified fragment was digested with SmaI, purified using QIAquick gel extraction kit (Qiagen), and then cloned into the corresponding site of pRS316 (25). The DNA sequence of the resulting plasmid (pRS316-SAM1) was confirmed using an automated DNA sequencer (Applied Biosystems). For the expression of MUP1, a DNA fragment which contained the MUP1 ORF and the 0.9-kbp upstream and 0.6-kbp downstream regions was amplified by PCR using the primers MUP1-FW and MUP1-RV. The amplified fragment was digested with SmaI, purified, and cloned into the corresponding sites of pRS315 and pRS316, and the DNA sequences of the resulting plasmids (pRS315-MUP1 and pRS316-MUP1) were confirmed as described above. To express the EGF peptide derivative containing a Met substitution at Leu-123 (L123M), the site-directed mutation was introduced as follows. The plasmid pJJ-αEGFVII (7), which contains an expression cassette comprised of the α-factor pre-pro sequence and the 6×His-EGF domain, was used as a template to substitute Leu-123 for a Met residue. The mutation was introduced into pJJ-αEGFVII using the QuikChange site-directed mutagenesis kit (Stratagene) with the primers L123M-FW and L123M-RV. The generated plasmid, pJJ-αEGFVII-L123M, was digested with BamHI, and then a DNA fragment containing the TDH3 promoter, the ORF encoding the L123M EGF peptide, and the TDH3 terminator (1.4 kbp) was subcloned into the corresponding site of pBluescript II SK(+) (Stratagene). The resulting plasmid, pBS-L123M, was confirmed by sequencing. To express the L123M EGF peptide under the control of the GAL10 promoter, a DNA fragment containing the ORF encoding the L123M EGF peptide and the TDH3 terminator was amplified by PCR using the primer aFactor-ER and the M13 primer M3 (TaKaRa), with pBS-L123M as a template. The amplified fragment was digested with EcoRI and BamHI, the resulting 0.6-kbp DNA fragment was cloned into EcoRI- and BglII-digested pHIG126 derived from pESC-URA (Stratagene), generating pGAL-L123M (PGAL10-L123M, 2μm, URA3), and its DNA sequence was confirmed as described above.
Isolation of SeMet-resistant mutants of S. cerevisiae.
S. cerevisiae W303-1A was cultivated overnight in 5 ml of liquid YPAD medium. The cells were harvested by centrifugation at 1,000 × g for 5 min and then washed and resuspended in 10 ml of sterile water. One hundred microliters of cell suspension (approximately 2 × 106 cells) was plated on SC-Met medium containing 0.025 mM SeMet, followed by incubation at 30°C. After 4 days, all growing colonies were replica plated on SC-Met medium containing 0.5 mM SeMet. After further incubation at 30°C for 4 days, the strains SRY5-3 and SRY5-7 were isolated as representative clones showing a SeMet-resistant phenotype.
Identification of the genes responsible for SeMet resistance.
To identify the genes involved in the SeMet resistance of SRY5-3, cells were transformed with a centromeric S. cerevisiae genomic DNA library (URA3 marker) and plated on SDCA-Ura medium containing 10 mM selenate. Plasmids were recovered from the transformants that could grow in the presence of 10 mM selenate. To identify the mutation sites of SRY5-7, TKY358 cells in which the MET17 gene was further disrupted in a SRY5-7 background were transformed with a centromeric S. cerevisiae genomic DNA library and plated on SC-Ura containing 5 μg/ml Met and 10 mM selenate. All transformants were replica plated onto SC-Ura and LA-agar (0.3% Bacto peptone, 0.5% Bacto yeast extract, 4% glucose, 0.02% ammonium sulfate, 0.1% lead acetate, 0.01% adenine sulfate, and 2% agar) plates. Plasmids that appeared as dark brown colonies on LA-agar plates were recovered from the transformants.
Met uptake assay.
The permease activity of Mup1p was measured as described by Isnard et al., with minor modifications (11). Briefly, overnight-cultured cells were inoculated into minimal drop-in medium and cultured until reaching mid-log phase. The assay was started by mixing 495 μl of cell culture with 5 μl of a Met solution containing 30.2 kBq of [methyl-3H]Met (PerkinElmer) to give a final concentration of 20 μM. Aliquots (95 μl) of the resulting mixtures were withdrawn, and cells were then filtered onto a GF/C glass fiber filter (Whatman) and washed twice with 5 ml of ice-cold water. The radioactivity on the glass fiber filter was quantified using a liquid scintillation counter (Aloka). The data were normalized for the differences in cell density (1 optical density at 600 nm (OD600) unit = 1 × 107 cells). To evaluate the Met uptake in whole cells, the cells were precultured in SC-Met medium until they reached mid-log phase. The assay was started by the addition of 5 μl of radiolabeled Met (30.2 kBq) to 495 μl of cell cultures to give a final concentration of 0.25 mM. The amount of radioactive [methyl-3H]Met incorporated into the cells was analyzed as described above.
Northern blot analysis.
Total RNA was isolated from 25 ml of cell cultures using an RNeasy minikit (Qiagen) by following the manufacturer's protocol. The obtained RNA (8 μg) was separated on agarose gels containing formaldehyde and transferred onto a Hybond N+ membrane (GE Healthcare) by capillary transfer. The membranes were baked at 80°C for 2 h, and hybridization was performed according to the standard protocol supplied by the manufacturer of the digoxigenin (DIG) system (Roche). Intragenic fragments of MET3, MET14, MET17, and ACT1 were amplified with specific primers (listed in Table S1 in the supplemental material) using the PCR DIG probe synthesis kit (Roche) and were used as DIG-labeled DNA probes. Hybridized probes were visualized by using anti-DIG-alkaline phosphatase Fab fragments (Roche) and CDP-Star reagents (GE Healthcare).
Preparation of a SeMet derivative of the L123M EGF peptide.
Yeast transformants harboring pGAL-L123M were grown overnight at 30°C in SDCA-Ura medium. After the cells were harvested by centrifugation, they were washed once with sterile water, and 1 OD600 unit of cells was used to inoculate 100 ml of SC-Met/Ura containing 2% raffinose and 0.1 M potassium phosphate buffer (pH 6.0). The cells were then cultured overnight at 30°C until an OD600 of approximately 2 was reached. To induce the expression of the SeMet derivative of the L123M EGF peptide, 2% galactose and various concentrations of SeMet ranging from 0.1 to 0.5 mM as a final concentration were added to the culture medium. After a 24-h induction at 30°C, the culture supernatants were collected by centrifugation at 3,890 × g for 5 min. The L123M EGF peptide was purified from the culture supernatant using a HisTrap column (1 ml; GE Healthcare). The pH of the crude samples was adjusted to 8.0 to 8.2 with 5 M potassium hydroxide before they were loaded on the column, which was equilibrated with buffer A (20 mM sodium phosphate, 300 mM NaCl at pH 7.4). The column was washed with 10 ml of buffer A containing 10 mM imidazole, and the peptide was eluted from the column with buffer A containing 500 mM imidazole. The eluate was monitored at 280 nm, and the fractions (1 ml) containing the L123M EGF peptide were collected and stored at −80°C until needed for the quantification of SeMet.
Quantification of the SeMet content in the L123M EGF peptide using HPLC.
High-performance liquid chromatography (HPLC) analysis was conducted using a Cosmosil 5C4-AR-300 column (4.6 mm by 150 mm; Nacalai Tesque). The elution was performed using two solvents at a flow rate of 1.0 ml/min; solvent A was an aqueous solution of 0.1% trifluoroacetic acid (TFA), and solvent B was a solution of acetonitrile containing 0.1% TFA. After the L123M EGF peptide solutions were loaded, the peptide was eluted with a linear gradient of solvent B (48% for 40 min). The peptide was monitored by UV absorbance at 215 nm. To confirm the molecular weight of the L123M EGF peptide, matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry was performed. A portion of the eluates from reverse-phase chromatography were mixed with α-cyano-4-hydroxycinamic acid (Nacalai Tesque) in 50/50 (vol/vol) acetonitrile-water containing 0.5% TFA, followed by analysis using an Ettan MALDI-TOF Pro instrument (GE Healthcare).
RESULTS
Genetic analysis of SeMet-resistant mutants.
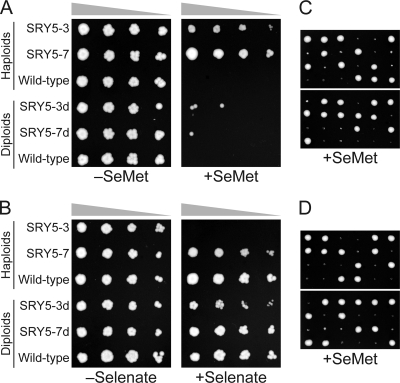
We initially isolated two spontaneous mutants (SRY5-3 and SRY5-7) of S. cerevisiae showing resistance to SeMet (Fig. 2 A). Although both mutants were capable of growing in the presence of 0.05 mM SeMet, the SRY5-7 mutant also showed selenate resistance (Fig. 2B), which is a phenotype similar to that observed for the P. pastoris SMR-94 mutant (13). Interestingly, unlike the wild-type and SRY5-7 cells, the SRY5-3 mutant could not grow in the presence of 10 mM selenate. The diploid cells generated from a cross between SRY5-7 and W303-1B (SRY5-7d) showed a SeMet-sensitive phenotype, similar to that of wild-type haploid cells (Fig. 2A). In the case of SRY5-3, diploid SRY5-3d cells showed not only SeMet sensitivity but also a selenate-resistant phenotype. The dissection of the diploids revealed that both SRY5-7d and SRY5-3d exhibited a 2:2 segregation of viable and nonviable spores in the presence of SeMet (Fig. 2C and D), indicating that the SRY5-7 and SRY5-3 mutant cells acquired SeMet resistance due to a single-gene recessive mutation.
FIG. 2.
Selection and growth phenotypes of SeMet-resistant mutants. The haploid and backcrossed mutants were incubated on SC-Met medium supplemented with or without 0.05 mM SeMet for 3 days at 30°C (A) and on YPAD medium supplemented with or without 10 mM selenate for 2 days at 30°C (B). Twelve tetrads obtained from SRY5-3d (C) and SRY5-7d (D) were incubated on SC-Met medium containing 0.05 mM SeMet for 3 days at 30°C, indicating a 2:2 segregation of viable to nonviable spores in the presence of SeMet.
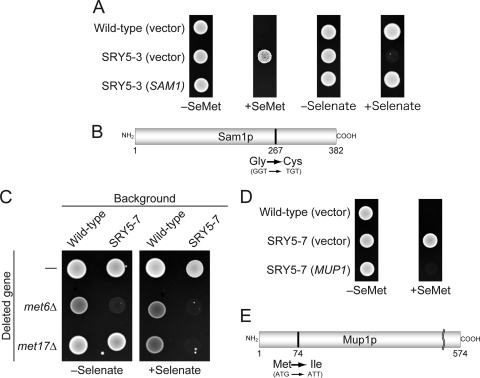
The mutant allele of SRY5-3 was assumed to be involved in the synthesis or regulation of sulfur compounds, as several mutants involved in sulfur metabolism, such as met4Δinh (expressing the mutant Met4p which is not recognized by the ubiquitin ligase) (14), met30-2 (14), and met3Δ (16), are also sensitive to selenate (14, 16). To identify the responsible gene, a library of S. cerevisiae genomic DNA fragments carried on a centromeric plasmid was introduced into SRY5-3 cells, and plasmids were recovered from four independent transformants that were capable of growth in the presence of selenate. DNA sequence analyses revealed that all plasmids contained either SAM1 or SAM2 ORFs encoding isomers of AdoMet synthetase (5). Further sequencing analysis of the PCR fragments of SAM1 and SAM2 ORFs amplified from SRY5-3 genomic DNA identified a point mutation of the Gly-267 residue to Cys (GGT to TGT) for Sam1p (sam1-224) (Fig. 3 A and B), although no mutation in Sam2p was identified.
FIG. 3.
Identification of mutant alleles responsible for SeMet resistance. (A) The SRY5-3 strain was transformed with a centromeric plasmid (URA3 marker) expressing the SAM1 gene under the control of an endogenous promoter. The wild-type and SRY5-3 strains were transformed with empty vector as a control. The transformants were incubated on SC-Met/Ura plates supplemented with or without 0.05 mM SeMet for 3 days at 30°C or on YPAD plates with or without 10 mM selenate for 2day at 30°C. (B) Schematic representation of the mutant protein encoded by sam1-224. The position of the mutation changing Gly-267 to Cys is indicated. (C) Effect of the null mutation of MET6 or MET17 on selenate resistance in the SRY5-7 mutant. Each strain was incubated on SC medium limited in the concentration of Met (5 μg/ml) and supplemented with or without 10 mM selenate for 2 days at 30°C. (D) SRY5-7 was transformed with a centromeric plasmid expressing the MUP1 gene from an endogenous promoter and with the empty vector as a control. The transformants were incubated under the conditions identical to those shown in panel A. (E) Schematic representation of the mutant protein encoded by mup1-100. The position of the mutation changing Met-74 to Ile is indicated.
As the SRY5-7 mutant cells showed selenate resistance (Fig. 2B), the negative screening of the SeMet-sensitive clones by the introduction of a yeast genomic DNA library was required for the identification of the mutated gene in this strain. However, we found that the null mutation of MET6 or MET17 in SRY5-7 resulted in cells which did not grow in the presence of 10 mM selenate on SC medium containing a low concentration of Met (5 μg/ml) (Fig. 3C). Since TKY357 cells (met6Δ in SRY5-7), but not TKY358 cells (met17Δ in SRY5-7), showed a growth defect on SC medium lacking selenate, we introduced the S. cerevisiae genomic DNA library into the TKY358 mutant. To exclude transformants harboring plasmids containing the MET17 ORF, all transformants were replica plated on SC-Ura and LA-agar plates. As MET17-deficient mutants form excess PbS due to the accumulation of H2S, plasmids were recovered from transformants appearing as dark-brown colonies on LA-agar plates (20). Subsequent DNA sequence analyses revealed that these transformants harbored plasmids containing DNA fragments encoding the MUP1, RSC1, and LST7 ORFs. Sequencing of the identical PCR fragment amplified from SRY5-7 genomic DNA revealed a point mutation in the codon encoding the Met-74 residue of Mup1p (mup1-100), resulting in a change to Ile (ATG to ATT) (Fig. 3D and E).
Sulfate assimilation ability of SeMet-resistant mutants.
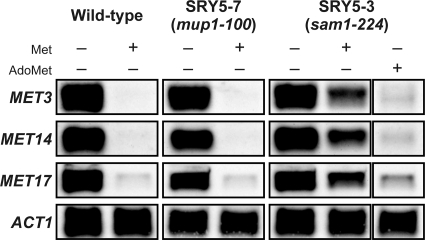
Previously, we isolated a SeMet-resistant P. pastoris mutant (SMR-94 strain) capable of effective production of SeMet derivatives and hypothesized that an increased Met pool resulting from the overexpression of enzymes involved in sulfate assimilation suppressed the toxic effects of SeMet (6, 13). Therefore, we addressed the relationship between SeMet resistance and sulfate assimilation ability in the S. cerevisiae mutants generated in the present study. It has been reported that the MET genes, which encode enzymes involved in sulfate assimilation (Fig. 1), are strictly regulated at the transcriptional level by the transcriptional activator Met4p (23). In response to increases in extracellular and intracellular concentrations of sulfur compounds, the ubiquitin ligase SCFMET30 attaches ubiquitin to Met4p to inactivate its function and prevent transcription (12, 19, 28). Thus, to examine the sulfate assimilation ability of the SRY5-7 and SRY5-3 strains, the mRNA levels of MET genes (MET3, MET14, and MET17) in cells treated with Met and AdoMet were examined by Northern blot hybridization (Fig. 4). The treatment with 0.25 mM Met for 30 min repressed the expression of the MET genes in wild-type and SRY5-7 cells (mup1-100), whereas SRY5-3 cells (sam1-224) expressed MET genes even in the presence of Met, albeit at lower levels than the control cells (no Met). On the other hand, the treatment with 0.25 mM AdoMet, which is generated from Met by Sam1p or Sam2p (Fig. 1), repressed the aberrant expression of the MET genes in SRY5-3 cells. These results indicated that the partial impairment of AdoMet synthesis was caused by a single amino acid change at the Gly-267 residue of Sam1p to Cys.
FIG. 4.
Repression of mRNAs for proteins involved in sulfate assimilation in the presence of Met and AdoMet. The wild-type (left), SRY5-7 (middle), and SRY5-3 (right) strains were grown in SC-Met medium until mid-log phase and then exposed to 0.25 mM Met or AdoMet for 30 min. The expression of MET3, MET14, and MET17 was determined by Northern blot analysis. An ACT1 probe was used as a loading control.
Met and SeMet uptake by SRY5-7 cells.
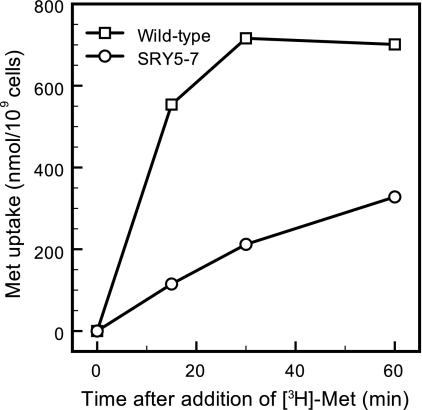
We measured the Met permease activity of SRY5-7 cells. Isnard et al. (11) reported that the apparent affinity constant (Kt) (Km for permease) for Mup1p (13 μM) is considerably lower than that for the Met permease Mup3p (1 mM). In order to evaluate the Mup1p activity alone, the uptake of radiolabeled Met (20 μM) was measured for 2 min in mup1Δ, mup3Δ, and mup1Δ mup3Δ mutants. As shown in Table 2, the uptake rates of Met by TKY386 (mup1Δ) and TKY388 (mup1Δ mup3Δ) cells were very low, ranging from 1.7 to 2.6 nmol/min/109 cells. In contrast, the uptake rate by TKY387 (mup3Δ) cells was 69.7 nmol/min/109 cells, which was 94% of the wild-type Met uptake level, indicating that Met incorporation fully depended on Mup1p function under the assay conditions. In the case of SRY5-7 cells, the rate of Met uptake (13.8 nmol/min/109 cells) was only 19% of the level of wild-type cells (Table 2). These results indicated that Met permease activity of SRY5-7 cells was partially impaired by the single amino acid substitution of the Met-74 residue of Mup1p for Ile. In addition to Met permeases, it is also known that yeast cells have several types of amino acid permeases that exhibit broad substrate specificities (21). We therefore measured the total Met uptake of the SRY5-7 strain after exposure to a high concentration of radiolabeled Met (0.25 mM) in mid-log phase (Fig. 5). Compared to that of wild-type cells, the uptake rate of [3H]Met by SRY5-7 cells was significantly reduced; however, SRY5-7 cells were able to accumulate the extracellular Met to 47% of the level of wild-type cells after 60 min. This result indicates that the partial Met uptake by SRY5-7 cells was a result of compensation by amino acid permeases other than Mup1p.
TABLE 2.
Rate of Met uptake in Met permease mutants of S. cerevisiae
| Strain | Met uptake activity (nmol/min/109 cells)a |
|---|---|
| Wild type | 74.5 ± 15.6 |
| SRY5-7 (mup1-100) | 13.8 ± 1.0 |
| TKY386 (mup1Δ) | 2.6 ± 0.7 |
| TKY387 (mup3Δ) | 69.7 ± 5.5 |
| TKY388 (mup1Δ mup3Δ) | 1.7 ± 0.4 |
Met uptake was measured for 2 min in 20 μM [methyl-3H]Met. Values represent the means ± standard deviations from three independent assays.
FIG. 5.
Total uptake of Met by the wild-type and SRY5-7 (mup1-100) strains. After cells were cultured in SC-Met medium until mid-log phase, 0.25 mM radiolabeled Met (30.2 kBq) was added, and uptake was measured at the indicated time points. The mean values of Met uptakes obtained from two independent experiments are plotted.
As it was speculated that yeast cells incorporate SeMet by a mechanism similar to that used by Met, we measured radiolabeled Met (20 μM) uptake by wild-type cells in the presence of nonradioactive SeMet (200 μM) as a competitor. The uptake of radiolabeled Met decreased to 28% ± 5% in the presence of excess SeMet, which was very similar to the level obtained in the control experiment using excess Met as a competitor (33% ± 2%), indicating that the uptake of SeMet by yeast cells was mediated by the identical permeases involved in Met uptake. Taken together, these results suggest that the SeMet resistance of the SRY5-7 mutant may be due to the partial inhibition of SeMet uptake and indicate the potential usefulness of the mutant to express proteins containing SeMet.
Production of the recombinant EGF peptide by SeMet-resistant mutants in the presence of SeMet.
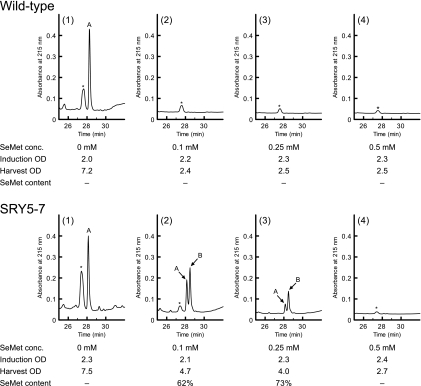
The use of SeMet-resistant mutants as expression hosts to overcome the toxicity of SeMet still results in decreased yields of recombinant proteins (18). We therefore examined whether the SeMet-resistant mutants of S. cerevisiae were able to efficiently express recombinant proteins. The plasmid pGAL-L123M was constructed to overproduce the EGF peptide containing one Met residue (L123M) as a model recombinant protein to confirm SeMet occupancy by HPLC analysis. As an initial trial, the expression of the SeMet-containing L123M peptide was induced by the addition of various concentrations of SeMet (0 to 0.5 mM). After the overexpression and secretion of the L123M EGF peptide by wild-type and SRY5-7 strains, culture supernatant was subjected to affinity purification on a Ni-nitrilotriacetic acid (NTA) column. To evaluate the yield of the L123M peptide and the efficiency of SeMet incorporation simultaneously, the affinity-purified EGF peptides were analyzed by reverse-phase chromatography (Fig. 6). The L123M peptides purified from the cultures of wild-type and SRY5-7 strains without SeMet were eluted as a single peak (Fig. 6, peak A), whereas the peptide from the mutant strain cultured with SeMet was clearly separated into two peaks (peaks A and B). Since SeMet has a higher hydrophobicity than that of Met (3), peak B in Fig. 6 was considered to represent the L123M EGF peptide containing SeMet. Further analysis of the peptide eluates by MALDI-TOF mass spectrometry revealed that the observed masses of peaks A (m/z 5,721.73) and B (m/z 5,768.74) were nearly identical to the theoretical masses of the L123M EGF peptide containing Met (m/z 5,722.20) and SeMet (m/z 5,769.11), respectively (see Fig. S1 in the supplemental material). Based on the HPLC peak area ratio, the SeMet content of the L123M EGF peptide expressed by the mutant was 73% in the presence of 0.25 mM SeMet. The L123M peptide from the SRY5-7 mutant cultured with 0.25 mM SeMet stably contained SeMet residue at 72% ± 1% occupancy using three independent experiments. In contrast, wild-type cells did not grow even in the presence of 0.1 mM SeMet, resulting in the failure of the cells to express the L123M peptide.
FIG. 6.
Evaluation of recombinant protein yield and SeMet incorporation by wild-type and SRY5-7 cells cultured with SeMet. Yeast strains harboring the plasmid pGAL-L123M were cultured with various concentration of SeMet. The L123M EGF peptides in the culture media were analyzed by reverse-phase chromatography after purification by using an Ni-NTA column, as described in Materials and Methods. Peaks A and B were assigned as the L123M peptides containing Met and SeMet, respectively, by mass spectrometry (see Fig. S1 in the supplemental material). Peaks with asterisks show unidentified compounds. SeMet concentration and OD600 values at induction and at harvest are indicated below each chromatogram.
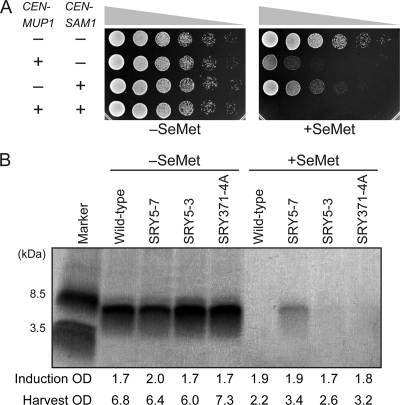
In addition to the SRY5-3 and SRY5-7 mutants, we prepared a double mutant (mup1-100 sam1-224 of TKY371-4A) by tetrad dissection of the diploid cells generated from crosses between each single mutant and confirmed that the introduction of both pRS315-MUP1 and pRS316-SAM1 led to the loss of the SeMet-resistant phenotype of TKY371-4A (Fig. 7 A). The yield of the L123M EGF peptide for each mutant in the absence of SeMet, as determined by silver staining of a Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, was identical to that of wild-type cells (Fig. 7B). However, when the strains were cultured with 0.25 mM SeMet, a peptide band corresponding to the L123M EGF peptide was observed only in the affinity-purified sample prepared from the culture supernatant of SRY5-7, although SRY371-4A grew equivalently to the SRY5-7 mutant in the presence of 0.25 mM SeMet.
FIG. 7.
(A) The double mutation of mup1-100 and sam1-224 increased SeMet resistance of S. cerevisiae. A serial dilution of TKY371-4A (mup1-100 sam1-224) cells harboring various combinations of pRS315-MUP1, pRS316-SAM1, and empty vectors was spotted on SC-Met/Leu/Ura medium with or without 0.05 mM SeMet for 1 day at 30°C. (B) Tricine-SDS-PAGE analysis of the L123M peptide expressed in the presence of 0.25 mM SeMet by SeMet-resistant mutants. The samples from Ni-NTA affinity chromatography were separated on a Tricine gel (15 to 20%; Wako), and protein bands were visualized by silver staining. The OD600 values at induction and harvest are indicated.
We also examined whether the sam1Δ sam2Δ and cys3Δ mutants produce the L123M peptide under the same condition as SRY5-7 (see Fig. S2 in the supplemental material), because these auxotrophic mutants have been reported as being potential hosts to produce SeMet-containing proteins. The sam1Δ sam2Δ mutant showed slow growth when precultured with SC medium containing raffinose rather than glucose and no growth after the addition of galactose and SeMet, although the mutant was isolated as a SeMet-resistant mutant. Moreover, the cys3Δ mutant showed a severe growth defect in the raffinose medium, resulting in the failure to induce expression of the L123M peptide. Thus, we could not obtain the peptide from the cultures of these mutants.
DISCUSSION
In this study, we identified a mutant allele of MUP1 for SeMet resistance and demonstrated the efficient production of the SeMet derivative of the L123M EGF peptide by the SRY5-7 strain (mup1-100). Although the Met and SeMet uptake ability by mup1-100 cells decreased to 47% of the wild-type level, SeMet was contained in the L123M EGF peptide overexpressed by the cells at 73% occupancy. The SeMet-resistant mutant SRY5-3 possessed a sam1-224 mutant allele and was inadequate to produce a SeMet derivative of the L123M EGF peptide. Northern blot analysis revealed that MET genes of SRY5-3 were overexpressed even in the presence of 0.25 mM Met. Since selenite (+4 oxidation state), which could be generated from selenate (+6 oxidation state) through the sulfate assimilation pathway, causes DNA double-strand breaks, the observed selenate sensitivity of the SRY5-3 strain (sam1-224) was thought to be a result of the accumulation of toxic selenite by enhanced sulfate assimilation (15, 16). On the other hand, the aberrant expression of MET genes in SRY5-3 cells was repressed by treatment with 0.25 mM AdoMet, indicating that intracellular AdoMet, which is a key compound for the regulation of the MET genes, decreased in SRY5-3 cells due to the impairment of Sam1p activity (19, 28). A recent report demonstrated that the complete block of AdoMet synthesis also confers a SeMet-resistant phenotype to S. cerevisiae (18). Thus, the observed SeMet resistance of SRY5-3 cells may be due to the reduction of the seleno-compounds derived from SeMet, such as Se-adenosylselenomethionine, rather than the overproduction of Met by enhanced sulfate assimilation (6, 13).
In contrast to the SRY5-3 mutant, the SRY5-7 mutant showed resistance to selenate. Northern blot analysis also demonstrated that MET genes of the SRY5-7 mutant were repressed in the presence of 0.25 mM Met at levels similar to those observed in the wild-type cells (Fig. 4). The uptake of radiolabeled Met also revealed that SeMet is incorporated into S. cerevisiae cells by a mechanism similar to that used by Met. These results indicate that SRY5-7 cells can incorporate extracellular Met and synthesize sulfur compounds involved in the regulation of sulfate assimilation, including AdoMet and Cys, at sufficient levels to repress the expression of MET genes. Although these results also suggest that Se-adenosylselenomethionine and selenocysteine are generated from SeMet, the intracellular levels of seleno-compounds in SRY5-7 cells are considered to be lower than those in wild-type cells, as the total Met uptake by SRY5-7 cells decreased to 47% of the wild-type level (Fig. 5). Based on the insight into the SeMet-resistant mechanism obtained from the analysis of SAM mutants, the SRY5-7 strain likely acquired SeMet resistance by the suppression of toxic seleno-compound generation, resulting from the insufficient uptake of SeMet caused by the mutation of Mup1p. However, SRY5-7 cells still retain the ability to incorporate SeMet and produce a SeMet derivative of the L123M EGF peptide (Fig. 6). It is therefore thought that amino acid permeases with broad substrate specificity, such as Agp1p, Gnp1p, Bap2p, Bap3p, and Lyp1p, compensate for the loss of Mup1p activity and allow SeMet incorporation in this mutant (21). Accordingly, we propose that the moderate incorporation of SeMet by these amino acid permeases allows the SRY5-7 strain to escape the toxic effects of seleno-compounds and efficiently produce SeMet derivatives.
The use of wild-type S. cerevisiae with intact sulfur metabolism has been reported (4). However, a large-scale cultivation of wild-type cells in the presence of SeMet is indispensable for the preparation of protein containing SeMet because crystallization requires a large amount of highly purified protein. SRY5-7 cells gave good yields and stable SeMet incorporation into the L123M peptide grown in shake flasks, whereas we could not obtain peptide from the wild-type strain. Thus, the mutant seems to be a favorable host for the preparation of SeMet derivatives.
Recently, Bockhorn et al. reported that 103 haploid null mutants of S. cerevisiae, including mup1Δ, showed greater SeMet resistance than wild-type cells, and the relative resistance of each mutant was demonstrated (2). According to the report, the SeMet resistance of mup1Δ cells was lower than that of sam1Δ sam2Δ and cys3Δ cells. We also constructed a sam1Δ sam2Δ double mutant and a cys3Δ single mutant in the W303 background; however, cys3Δ did not show any growth in synthetic medium containing raffinose (see Fig. S2 in the supplemental material). Although the sam1Δ sam2Δ mutant could grow in the raffinose medium, the cellular growth of the mutant was much slower than that of the SRY5-7 mutant before the addition of galactose and SeMet. Moreover, the cell density after 24 h of induction was very low (OD600 of approximately 1.5), suggesting a requirement of a large-scale cultivation of sam1Δ sam2Δ mutants to prepare SeMet derivatives for crystallization, even though the mutants are able to incorporate SeMet into proteins by over 90% occupancy. We could not detect a SeMet derivative of the L123M EGF peptide when expressed in sam1-224 mup1-100 cells (TKY371-4A), showing the greatest resistance to SeMet on the SC-Met/Leu/Ura agar medium (Fig. 7A). Taken together, these results indicate that SeMet resistance of the mutant cells is not correlated with the productivity of SeMet derivatives and that the SRY5-7 mutant is superior to the other W303-1A background mutants for the production of proteins with high content levels of SeMet. However, it remains to be verified whether SRY5-7 cells are capable of efficiently producing SeMet derivatives of proteins other than the EGF peptide. With regard to SeMet content, SRY5-7 cells incorporated SeMet into the L123M EGF peptides with 73% occupancy, indicating that the SRY5-7 mutant has the ability to incorporate SeMet and efficiently produce SeMet derivatives, even though a high-affinity Met permease (Mup1p) of the mutant was severely impaired. Although the SeMet replacement was incomplete, it is expected that proteins expressed by SRY5-7cells in the presence of SeMet will be useful for phase determination, as crystals of human lysozyme expressed by the P. pastoris SMR-94 strain containing SeMet with only 65% occupancy were sufficient for SAD phasing (13).
In conclusion, the enhanced production of SeMet derivatives by S. cerevisiae lacking high-affinity Met permease activity was achieved using standard media without the need for supplementation with either Cys or AdoMet. Therefore, the S. cerevisiae mup1 mutant has a clear advantage over the sam1Δ sam2Δ and cys3Δ mutants for the preparation of SeMet derivatives for X-ray crystallography, for which a large amount of highly purified proteins is required.
Supplementary Material
Acknowledgments
We are grateful to Hiroto Hirayama (RIKEN) for providing the plasmid pHIG126. We also thank Yoshiyuki Fujitani (National Research Institute of Brewing) and Takuji Oka (Sojo University) for their helpful discussions.
Footnotes
Published ahead of print on 6 August 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barton, W. A., D. Tzvetkova-Robev, H. Erdjument-Bromage, P. Tempst, and D. B. Nikolov. 2006. Highly efficient selenomethionine labeling of recombinant proteins produced in mammalian cells. Protein Sci. 15:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bockhorn, J., B. Balar, D. He, E. Seitomer, P. R. Copeland, and T. G. Kinzy. 2008. Genome-wide screen of Saccharomyces cerevisiae null allele strains identifies genes involved in selenomethionine resistance. Proc. Natl. Acad. Sci. U. S. A. 105:17682-17687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budisa, N., R. Huber, R. Golbik, C. Minks, E. Weyher, and L. Moroder. 1998. Atomic mutations in annexin V—thermodynamic studies of isomorphous protein variants. Eur. J. Biochem. 253:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Bushnell, D. A., P. Cramer, and R. D. Kornberg. 2001. Selenomethionine incorporation in Saccharomyces cerevisiae RNA polymerase II. Structure 9:R11-R14. [DOI] [PubMed] [Google Scholar]
- 5.Cherest, H., and Y. Surdin-Kerjan. 1978. S-Adenosyl methionine requiring mutants in Saccharomyces cerevisiae: evidences for the existence of two methionine adenosyl transferases. Mol. Gen. Genet. 163:153-167. [DOI] [PubMed] [Google Scholar]
- 6.Cherest, H., Y. Surdin-Kerjan, J. Antoniewski, and H. de Robichon-Szulmajster. 1973. Effects of regulatory mutations upon methionine biosynthesis in Saccharomyces cerevisiae: loci eth2-eth3-eth10. J. Bacteriol. 115:1084-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chigira, Y., T. Oka, T. Okajima, and Y. Jigami. 2008. Engineering of a mammalian O-glycosylation pathway in the yeast Saccharomyces cerevisiae: production of O-fucosylated epidermal growth factor domains. Glycobiology 18:303-314. [DOI] [PubMed] [Google Scholar]
- 8.Cronin, C. N., K. B. Lim, and J. Rogers. 2007. Production of selenomethionyl-derivatized proteins in baculovirus-infected insect cells. Protein Sci. 16:2023-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrickson, W. A., J. R. Horton, and D. M. LeMaster. 1990. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iino, H., H. Naitow, Y. Nakamura, N. Nakagawa, Y. Agari, M. Kanagawa, A. Ebihara, A. Shinkai, M. Sugahara, M. Miyano, N. Kamiya, S. Yokoyama, K. Hirotsu, and S. Kuramitsu. 2008. Crystallization screening test for the whole-cell project on Thermus thermophilus HB8. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64:487-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isnard, A. D., D. Thomas, and Y. Surdin-Kerjan. 1996. The study of methionine uptake in Saccharomyces cerevisiae reveals a new family of amino acid permeases. J. Mol. Biol. 262:473-484. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser, P., K. Flick, C. Wittenberg, and S. I. Reed. 2000. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102:303-314. [DOI] [PubMed] [Google Scholar]
- 13.Kitajima, T., E. Yagi, T. Kubota, Y. Chiba, S. Nishikawa, and Y. Jigami. 2009. Use of novel selenomethionine-resistant yeast to produce selenomethionyl protein suitable for structural analysis. FEMS Yeast Res. 9:439-445. [DOI] [PubMed] [Google Scholar]
- 14.Kuras, L., A. Rouillon, T. Lee, R. Barbey, M. Tyers, and D. Thomas. 2002. Dual regulation of the met4 transcription factor by ubiquitin-dependent degradation and inhibition of promoter recruitment. Mol. Cell 10:69-80. [DOI] [PubMed] [Google Scholar]
- 15.Letavayova, L., D. Vlasakova, J. E. Spallholz, J. Brozmanova, and M. Chovanec. 2008. Toxicity and mutagenicity of selenium compounds in Saccharomyces cerevisiae. Mutat. Res. 638:1-10. [DOI] [PubMed] [Google Scholar]
- 16.Logan, H. M., N. Cathala, C. Grignon, and J. C. Davidian. 1996. Cloning of a cDNA encoded by a member of the Arabidopsis thaliana ATP sulfurylase multigene family. Expression studies in yeast and in relation to plant sulfur nutrition. J. Biol. Chem. 271:12227-12233. [DOI] [PubMed] [Google Scholar]
- 17.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 18.Malkowski, M. G., E. Quartley, A. E. Friedman, J. Babulski, Y. Kon, J. Wolfley, M. Said, J. R. Luft, E. M. Phizicky, G. T. DeTitta, and E. J. Grayhack. 2007. Blocking S-adenosylmethionine synthesis in yeast allows selenomethionine incorporation and multiwavelength anomalous dispersion phasing. Proc. Natl. Acad. Sci. U. S. A. 104:6678-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menant, A., P. Baudouin-Cornu, C. Peyraud, M. Tyers, and D. Thomas. 2006. Determinants of the ubiquitin-mediated degradation of the Met4 transcription factor. J. Biol. Chem. 281:11744-11754. [DOI] [PubMed] [Google Scholar]
- 20.Ono, B., N. Ishii, S. Fujino, and I. Aoyama. 1991. Role of hydrosulfide ions (HS−) in methylmercury resistance in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 57:3183-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regenberg, B., L. During-Olsen, M. C. Kielland-Brandt, and S. Holmberg. 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 36:317-328. [DOI] [PubMed] [Google Scholar]
- 22.Rice, L. M., T. N. Earnest, and A. T. Brunger. 2000. Single-wavelength anomalous diffraction phasing revisited. Acta Crystallogr. D Biol. Crystallogr. 56:1413-1420. [DOI] [PubMed] [Google Scholar]
- 23.Rouillon, A., R. Barbey, E. E. Patton, M. Tyers, and D. Thomas. 2000. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCFMet30 complex. EMBO J. 19:282-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 25.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugahara, M., Y. Asada, K. Shimizu, H. Yamamoto, N. K. Lokanath, H. Mizutani, B. Bagautdinov, Y. Matsuura, M. Taketa, Y. Kageyama, N. Ono, Y. Morikawa, Y. Tanaka, H. Shimada, T. Nakamoto, M. Yamamoto, and N. Kunishima. 2008. High-throughput crystallization-to-structure pipeline at RIKEN SPring-8 Center. J. Struct. Funct. Genomics 9:21-28. [DOI] [PubMed] [Google Scholar]
- 27.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 28.Thomas, D., L. Kuras, R. Barbey, H. Cherest, P. L. Blaiseau, and Y. Surdin-Kerjan. 1995. Met30p, a yeast transcriptional inhibitor that responds to S-adenosylmethionine, is an essential protein with WD40 repeats. Mol. Cell. Biol. 15:6526-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.