Abstract
Linalool production was evaluated in different Saccharomyces cerevisiae strains expressing the Clarkia breweri linalool synthase gene (LIS). The wine strain T73 was shown to produce higher levels of linalool than conventional laboratory strains (i.e., almost three times the amount). The performance of this strain was further enhanced by manipulating the endogenous mevalonate (MVA) pathway: deregulated overexpression of the rate-limiting 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMG-CoA reductase) doubled linalool production. In a haploid laboratory strain, engineering of this key step also improved linalool yield.
Monoterpenes are a class of isoprenoids of increasing industrial and clinical interest usually produced by plants. They are used as aromatic additives in the food and cosmetics industries and are also important components in wine aroma. Moreover, certain monoterpenes display antimicrobial, antiparasitic, and antiviral properties as well as a plethora of promising health benefits (for recent reviews, see references 2, 7, 15, 28, and 30 and references cited therein). To date, many studies have focused on plant metabolic engineering of monoterpene production (for selected reviews, see references 1, 14, 19, 29, and 35 and references cited therein), and few studies have been carried out on microorganisms (9, 21, 22, 34, 38). Efficient microbial production of these metabolites could provide an alternative to the current methods of chemical synthesis or extraction from natural sources. In this regard, a considerable number of studies have shown the utility of Saccharomyces cerevisiae as a valuable platform for sesquiterpene, diterpene, triterpene, and carotene production (references 5, 10, 23, 26, 30, 31, 32, and 33 and references cited therein). However, all the efforts dedicated to the improvement of isoprenoid yields in S. cerevisiae have been performed using conventional laboratory strains, and there are no studies concerning natural or industrially relevant isolates.
In recent years, many genes that encode plant monoterpene synthases (MTS), a family of enzymes which specifically catalyze the conversion of the ubiquitous C10 intermediate of isoprenoid biosynthesis geranyl pyrophosphate (GPP) to monoterpenes, have been characterized. Such is the case with the LIS gene (codes for S-linalool synthase) of Clarkia breweri, the first MTS-encoding gene to be isolated (13). In contrast to plants, S. cerevisiae cannot produce monoterpenes efficiently, mainly due to the lack of specific pathways involving MTS. However, GPP is formed as a transitory intermediate in the two-step synthesis of farnesyl pyrophosphate (FPP), catalyzed by FPP synthase (FPPS) (Fig. 1), and some natural S. cerevisiae strains have been shown to possess the ability to produce small amounts of monoterpenes (8). Whether this occurs through unspecific dephosphorylation of a more available endogenous pool of GPP and subsequent bioconversions is not known. In addition, it has recently been established that S. cerevisiae has enough free GPP to be used by exogenous monoterpene synthases to produce monoterpenes under laboratory and vinification conditions (22, 34).
FIG. 1.
Simplified isoprenoid pathway in S. cerevisiae, including the branch point to linalool. Dotted arrows indicate that more than one reaction is required to convert the substrate to the product indicated. Dashed arrows indicate the engineered steps. Abbreviations: HMG-CoA, 3-hydroxy-3-methylglutaryl coenzyme A; IPP, isopentenyl pyrophosphate; GPP, geranyl pyrophosphate; FPP, farnesyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; HMGR, HMG-CoA reductase; FPPS, FPP synthase; LIS, linalool synthase.
Here we present the process for selecting and optimizing yeast strains for foreign monoterpene production. We have chosen the C. breweri LIS gene as a prototype because, when heterologously expressed in S. cerevisiae, it specifically results in the production of linalool (3,7-dimethyl-1,6-octadien-3-ol; a floral scent and bioactive acyclic monoterpene identified in numerous fruits and flowers) and no other by-products (22). Two S. cerevisiae strains of different origins have been selected and their endogenous mevalonate (MVA) pathways engineered to enhance the production of linalool. These strategies might be employed to produce any other recombinant monoterpene in S. cerevisiae by expressing the appropriate monoterpene synthase.
MATERIALS AND METHODS
Strains, culture media, and growth conditions.
Escherichia coli DH5α [endA1 hsdR17 gyrA96 thi-1 relA1 supE44 recA1 ΔlacU169 (φ80 lacZΔM15)] was used for cloning experiments and plasmid propagation. S. cerevisiae strains used in this study are listed in Table S1 in the supplemental material. E. coli was maintained in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl) with or without 100-μg ml−1 ampicillin. S. cerevisiae strains were maintained in YPD rich medium (1% yeast extract, 2% bacteriological peptone, 2% glucose) or SD minimal medium (0.17% yeast nitrogen base without amino acids [Difco Laboratories, Detroit, MI], 2% glucose, 0.5% ammonium sulfate) with or without 20-mg liter−1 uracil. When necessary, SD media were supplemented with 20-mg liter−1 adenine, 20-mg liter−1 histidine, 20-mg liter−1 tryptophan, and 30-mg liter−1 leucine. For solid media, 1.5% agar was added. To determine linalool yields of recombinant yeasts, aliquots from overnight cultures of selected transformants grown in SD medium lacking uracil were transferred to 250-ml flasks containing 50 ml of YPD medium at an initial optical density at 600 nm (OD600) of 0.05. Yeast cultures were grown with continuous shaking (200 rpm) at 30°C, and aliquots of the cultures were taken at different times. For plasmid stability analyses, transformants were grown under both selective (SD medium) and nonselective (YPD medium) conditions, and the colonies growing under each condition were counted.
DNA manipulations, transformations, and Northern analyses.
General DNA manipulations were performed according to standard protocols (41). Transformations of yeast strains were done using lithium acetate to permeabilize the cells as previously described (18, 37). Transformants were selected and maintained on SD medium plates without uracil. S. cerevisiae total RNA was isolated as described previously (3). Northern analyses were carried out essentially as described previously (41). Agarose gels were loaded with 12.5 μg of total glyoxylated RNA per track. Membranes (Hybond-N+; Amersham Biosciences) were stained with 0.03% methylene blue as a loading control, washed, and hybridized. Hybridizations were done using a digoxigenin (DIG)-dUTP-labeled fragment generated by PCR (oligonucleotides LIS-A and LIS-B [see Table S2 in the supplemental material]) according to the manufacturer's instructions (Roche).
Construction of plasmids.
The TDH3p::LIS::PGK1t expression cassette containing the C. breweri LIS cDNA (GenBank accession no. U58314 [13]) was isolated from pG1-Lis (21) as a 3.8-kb PstI-NaeI fragment, which was subsequently subcloned into YEplac195 (17) that had been previously digested with PstI and SmaI, thus creating YEplac195-Lis. To construct the PGK1p::catHMG1::CYC1t expression cassette (catHMG1 codes for the catalytic domain of the isozyme 3-hydroxy-3-methylglutaryl coenzyme A [HMG-CoA] reductase 1, or catHmg1), we first synthesized by PCR each of the three components of the expression unit using genomic DNA from a yeast strain derived from S288c and the primers indicated in Table S2 in the supplemental material. The three PCR fragments were individually subcloned in pGEM-T Easy. A 1.6-kb NcoI-BamHI fragment containing the catHMG1 allele and a 1.49-kb SalI-NcoI fragment containing the PGK1 promoter (PGK1p) were ligated into YEplac195, which had previously been digested with SalI and BamHI, and the intermediate plasmid YEplac195-Hmg18 was thus generated. To construct YEplac195-Hmg1531-1054, the CYC1 terminator obtained as a 0.3-kb BamHI-EcoRI fragment (CYC1t) was subcloned into YEplac195-Hmg18, which had previously been digested with BamHI and EcoRI. In order to transfer the LIS expression cassette into the YEplac195-Hmg1531-1054 vector, the 3.8-kb PstI-NaeI fragment from pG1-Lis was treated with T4 DNA polymerase and ligated into the YEplac195-Hmg1531-1054 vector, which had previously been digested with SalI and blunt ended using T4 DNA polymerase. Plasmid YEplac195-LisHmg1531-1054 was thus generated. Fragments obtained by PCR were checked by sequencing to ensure the absence of PCR-induced mutations. PCR amplifications were carried out using the Expand high-fidelity system (Roche). Plasmids are indicated in Table S1 in the supplemental material.
Monoterpene analysis.
Linalool was analyzed by headspace solid-phase microextraction (HS-SPME) coupled with gas chromatography (GC) and GC-mass spectrometry (MS) as described previously (22). 2-Octanol (0.2 μg) was used as an internal standard. Linalool concentrations were calculated using standard solutions and are given as the averages of results from at least three independent cultures. Calibration curves were determined using the same protocol by exposure of the polydimethylsiloxane (PDMS) filament to the headspace of YPD spiked with standards at final concentrations of 1, 10, 100, 1,000, and 10,000 μg liter−1. Compounds were identified by comparing retention times and mass spectra with those of standard compounds (Sigma, St. Louis, MO) using a Hewlett-Packard (HP) capillary gas chromatograph, model 5890, series II, equipped with an HPInnowax column (length, 15 m; inside diameter, 0.25 mm; film thickness, 0.25 mm) and an Agilent 5973N mass selective detector coupled to an Agilent 6890 gas chromatograph (Agilent Technologies, Waldbronn, Germany), under the same chromatographic conditions.
LIS enzyme assays.
To determine linalool synthase activities, we adapted the protocol described for Lactococcus lactis (21). Yeast cells were grown for 24 h as described above. Twenty-milliliter aliquots were centrifuged (4,000 rpm, 15 min, 4°C), and the resulting pellets were maintained frozen until use. To make cell extracts, pellets were resuspended in 1 ml ice-cold linalool assay buffer (LAF) containing 50 mM Tris-HCl, pH 7.5, 10 mM dithiothreitol (DTT), 10% (vol/vol) glycerol, 20 mM MgCl2, 5 mM MnCl2, 1 mM sodium ascorbate, and protease inhibitor cocktail (Roche). Cells were disrupted by bead beating (4 times for 30 s each cycle) using 1 ml glass beads (diameter, 0.5 mm), and the resulting supernatant was recovered after centrifugation (12,000 rpm, 5 min, 4°C). The protein concentrations of extracts were measured using the Bio-Rad/Bradford assay employing bovine serum albumin (BSA) as the standard. Supernatant volumes corresponding to protein contents of 2 and 4 mg were adjusted with LAF to yield final volumes of 1 ml and transferred to 10-ml Pyrex tubes. Twenty microliters of nonradioactive GPP (from a 2.6 mM solution; Sigma) was added, vials were closed with Teflon septa, and the reaction mixtures were incubated at 30°C with gentle shaking (100 rpm) for 30 min. Subsequently, 0.2 g NaCl was added and the reaction products were collected with a PDMS fiber and measured as described above. Linalool (∼83 μg liter−1) was detected in the control vial (GPP alone). This background amount was subtracted from the linalool produced in the enzymatic assays. Linalool was not detected as a product of GPP derived from the endogenous phosphatase activity of the microbial host, since the yeast strains transformed with empty plasmid (no LIS gene) produce linalool levels similar to that of the control. Under the assay conditions used, linalool production was proportional to the amount of protein present in the assays. Enzyme assays were performed in duplicate.
RESULTS AND DISCUSSION
Expression of the C. breweri LIS gene in different laboratory strains of S. cerevisiae results in similar linalool yields.
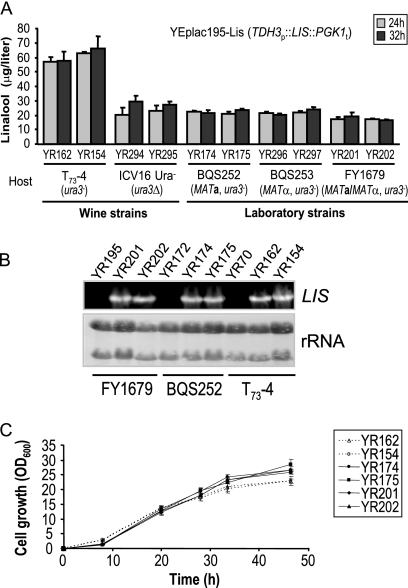
As the genetic background of the host may have dramatic consequences with regard to recombinant monoterpene production, we first evaluated the performance of six S. cerevisiae strains of different genotypes and ploidies. It has been suggested (24) that MATa strains might synthesize more FPP, as this compound is needed for prenylation of the mating pheromone (4). Indeed, the yields of the sesquiterpene epicedrol (synthesized in one step from FPP) in MATa strains are twice those in MATα strains (24). We therefore studied the effect of mating type on linalool production. The LIS expression cassette (TDH3p::LIS::PGK1t) from plasmid pG1-Lis (22) was subcloned into the 2μm shuttle vector YEplac195 (17) and used to transform ura3-auxotrophic strains (derived from S288) of opposite mating types, BQS252 (MATa ura3-52) (16) and BQS253 (MATα ura3-52), using the LiAc/ss-DNA/PEG method (18). Two uracil-prototrophic transformants of each genetic background were thus isolated, YR174 and YR175 (MATa) and YR296 and YR297 (MATα), and their abilities to excrete linalool were compared. Yields of linalool were similar in both mating types (average, 21.92 ± 1.75 μg liter−1) (Fig. 2 A), suggesting that the potential diversification of the MVA pathway to provide prenyl groups for the mating pheromone does not improve GPP availability. We then used the a mating type for further analyses. As expected, linalool was not detected in the complemented control strains YR172 and YR304 (BQS252 and BQS253 transformed with YEplac195).
FIG. 2.
Comparison of levels of linalool production by different host strains. (A) Analysis of linalool yields in different strains of S. cerevisiae. Cultures were sampled at the times indicated, and linalool was quantified. Results are presented as the means and standard deviations from at least three different experiments. (B) Northern blot of total RNA extracted from recombinant yeasts producing linalool and the complemented control strains (24 h). rRNA is shown as a loading control. (C) Growth curves of yeast strains transformed with YEplac195-Lis. These cultures were used to quantify linalool (A).
We also tested linalool production by W303-1A (MATa; laboratory strain from a different origin) (43) transformed with the same replicative LIS expression vector (YEplac195-Lis 2μm URA3) as that used to transform BQS252 and BQS253. Absolute linalool production was around 50% lower in recombinant W303-1ALis strains (YR270 and YR272) than in recombinant BQS252Lis (average, 10.90 ± 2.04 μg liter−1). However, due to the lower level of growth of W303-1A transformed with YEplac195-Lis than that of BQS252Lis and BQS253Lis, when linalool production is measured in reference to OD values, the productivities of these three genetic backgrounds were quite comparable (averages, 1.43 ± 0.27 μg liter−1 OD600−1 and 1.37 ± 0.22 μg liter−1 OD600−1, respectively, at 32 h). These results suggest that haploid laboratory yeast strains have similar abilities to produce monoterpenes. However, in comparison with the linalool levels produced by recombinant wine strains under similar growth conditions (22), the productivity of these laboratory strains is significantly lower.
Wine yeast strain T73-4 has a greater capacity for recombinant monoterpene biosynthesis than other tested strains.
In order to improve wine quality as well as the elaboration processes, wine yeast strains have been selected for a very specialized phenotype and are genetically complex, usually being polyploid or aneuploid (6). To elucidate whether the better ability to convert endogenous GPP into monoterpenes is a common feature in oenological strains of S. cerevisiae, two different wine strains, T73-4 (nearly diploid [37], derived from T73 Lallemand) and ICV16ura3Δ (derived from ICV16 Fermicru Primeur, DSM [25]), as well as the laboratory diploid strain FY1679 (MATa/MATα ura3-52/ura3-52; isogenic to BQS252 and BQS253), were transformed with YEplac195-Lis and two uracil-prototrophic transformants of each genetic background were isolated. Figure 2A shows that linalool production by recombinant FY1679Lis strains (YR201 and YR202) was not greater than that of recombinant BQS252Lis and BQS253Lis, indicating that diploidy alone does not improve linalool levels. In contrast, linalool yields for recombinant wine strains were greater than in their laboratory counterparts, indicating that these genetically complex backgrounds may be more appropriate for producing heterologous monoterpenes. Notably, the inherent capacities of the two industrial strains differ, and production of linalool by recombinant T73-4Lis cells (YR154 and YR162; average, 60.94 ± 6.15 μg liter−1) was about 2-fold greater than that of recombinant ICV16Lis (YR294 and YR295). The reasons for the differing efficacies of yeast hosts are not obvious, and further studies would be necessary to satisfactorily explain this. Differences in transgene transcription are unlikely to be a primary factor, as the LIS gene is similarly expressed in recombinant BQS252Lis, FY1679Lis, and T73-4Lis strains (Fig. 2B). Also, greater linalool production cannot be associated with higher cell densities, as T73-4Lis strains reached slightly lower (around 15%) optical densities than recombinant BQS252 and FY1679 (Fig. 2C). Moreover, as described previously for the wine strain T73-PG1LIS URA (22), both recombinant laboratory (YR175) and wine (YR162 and YR295) yeast strains accumulated linalool during the early and rapid phases of culture growth (see Fig. S1 in the supplemental material). In addition, linalool synthase activities did not differ greatly between recombinant wine strain ICV16Lis or T73-4Lis (588 ± 222 and 617 ± 154 μg liter−1 mg total protein−1, respectively) and laboratory strain BQS252Lis (636 ± 240 μg liter−1 mg total protein−1), despite the different levels of linalool produced by them. Taken together, these results suggest that redirection of the endogenous isoprenoid pathway toward linalool production depends on the genetic background of the hosts, which could affect GPP availability. The low level of linalool accumulation in laboratory strains could reflect other restricting conditions, including limited coupling between the introduced linalool synthase and the endogenous GPP pool or even the limiting availability of a cofactor. Use of the T73 strain as a production host for recombinant monoterpenes and eventually other isoprenoids is an improvement for increasing yield per unit of culture volume.
Deregulated overexpression of a truncated HMG1 allele increases linalool production in yeasts.
Since another restriction on monoterpene production could be precursor supply, we also wanted to know whether engineering key steps within the MVA pathway affects linalool yields. The major regulatory control point of this pathway (Fig. 1) in many organisms, including yeasts, is the formation of mevalonic acid (MVA) from 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA), a step catalyzed in S. cerevisiae by HMG-CoA reductases 1 and 2 (for reviews, see references 20 and 31). Deregulated overexpression alone of truncated HMG1 alleles (catHMG1 codes for the catalytic domain of the isozyme HMG-CoA reductase 1, or catHmg1) has successfully been employed to increase the rate of flow through the MVA pathway and thus improve the production of squalene (an intermediate of ergosterol synthesis) and amorphadiene (the sesquiterpene precursor of the antimalarial drug artemisin) in S. cerevisiae (11, 36, 39), as well as carotenoids in Candida utilis and Neurospora crassa (42, 44). However, overproduction of catHmg1 alone cannot be considered a precedent for success in improving monoterpene yields, as this strategy failed when employed trying to improve the yields of the sesquiterpene epicedrol (24) and the triterpene β-amyrin (27). Moreover, certain strategies that have been seen to work well in laboratory strains have failed in industrial ones (12). Bearing in mind the above, along with the fact that GPP is a transient intermediate of FPPS in the formation of FPP, it is difficult to predict whether an increase in the flow through of the pathway would result in a concomitant increase of GPP availability.
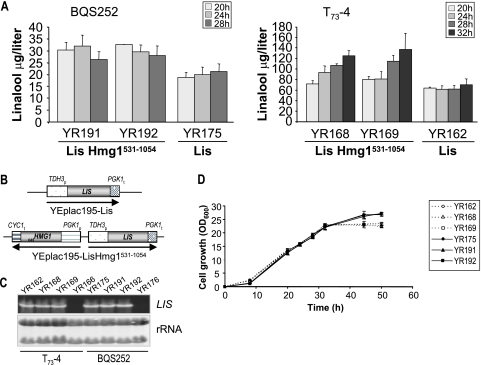
Based on these considerations and looking to increase linalool production, we decided to study the effects of overexpressing the catHMG1 allele (coding for a mutated soluble form of the protein lacking the first 530 amino acid residues and having an engineered Met start codon before Asp531) in S. cerevisiae host cells (BQS252 and T73-4) that produce different levels of linalool. To deregulate the HMG1 gene and avoid recombination between tandemly repeated copies (such as promoter or terminator sequences) that could be looped out by excisional recombination (40), the truncated HMG1 gene was present in YEplac195-LisHmg1531-1054 under the control of the strong constitutive promoter PGK1 and the CYC1 terminator (Fig. 3 B). Plasmid YEplac195-LisHmg1531-1054 was used to transform the selected strains, and the recombinants YR191 and YR192 (BQS252 background) and YR168 and YR169 (T73-4 background) were isolated. Figure 3A shows that coexpression of catHMG1 and LIS in T73-4 was associated with an increase in linalool production to 132.66 ± 24.72 μg liter−1, while in BQS252 the linalool content was 30.19 ± 3.88 μg liter−1. Thus, in comparison with the control strains YR162 (T73-4Lis) and YR175 (BQS252Lis), 2- and 1.5-fold increases in linalool production were achieved. Differences found in the kinetics of linalool production in BQS252LiscatHmg1 and T73-4LiscatHmg1 could explain differences in the final linalool titers. Whereas the highest accumulation of linalool in the engineered laboratory strains (YR191 and YR192) was observed at 20 h, as occurs in BQS252Lis and T73-4Lis, the production of linalool in T73-4LiscatHmg1 progressively increased over the whole time course of the experiment (Fig. 3A). Linalool was not detected in strains overexpressing catHMG1. Levels of expression of the LIS transgene were similar in the engineered yeasts (Fig. 3C), indicating that that greater linalool titers are not associated with enhanced expression of the LIS transgene. All transformants reached ODs similar to those of the control strains (YR175 and YR162), and the rates of growth were identical (Fig. 3D), indicating that neither the amount of linalool nor the manipulations introduced within the isoprenoid pathway apparently produce deleterious effects on yeast growth under such conditions. Taken together these results suggest that the pool of GPP increases in both genetic backgrounds as a consequence of overproducing catHmg1 and also that the window in which GPP is available for monoterpene production is wider in the engineered wine T73-4 strain. A previous report showed that overexpression of the catalytic domain of Hmg1 resulted in a dramatic increase in squalene production (11, 36), whereas our study shows a moderate increase in linalool yield. These results may suggest for the enzymes involved in the biosynthesis of ergosterol (i.e., FPPS) a higher affinity for GPP than for LIS.
FIG. 3.
Increase of linalool production by overproducing catHmg1. (A) Histograms showing linalool content in the supernatants of the control and two independent transformants. Cultures were sampled at the times indicated. Values are averages from at least three independent experiments. Error bars indicate standard deviations. (B) Schematic diagram of the gene cassettes in the two plasmids used to transform S. cerevisiae. (C) Expression analysis of the introduced LIS transgene by Northern hybridization using a DIG probe. rRNA is shown as a loading control. (D) Growth curves of yeast strains transformed with YEplac195-LisHmg1531-1054. The same cultures used to build panel A were evaluated.
In conclusion, we describe that pathway optimization and host selection resulted in an increased synthesis of recombinant linalool (up to 6-fold). The S. cerevisiae strains developed in this study can serve, in principle, as platform hosts to make any of the hundreds of known monoterpenes by expressing the appropriate MTS in place of linalool synthase. The levels that we obtained are still low for industrial applications. However, they are sufficient for testing the function of novel monoterpene synthase-encoding genes as well as conducting structural studies on the monoterpenes produced. Moreover, our engineered strains are promising for further development.
Supplementary Material
Acknowledgments
This work was supported by the Spanish Ministerio de Educación y Ciencia/FEDER (grant AGL2005-02542) and grant CSD2007-0063 (Consolider-Ingenio). Juan Rico was the recipient of an FPI predoctoral fellowship from the CICYT.
We thank Eran Pichersky, University of Minesota, for providing us with the C. breweri LIS cDNA clone; Emilia Matallana, Agustín Aranda, José Enrique Pérez-Ortín, and Marce.lí del Olmo, Universitat de València, for the S. cerevisiae strains; and Óscar Herrero for technical advice.
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aharoni, A., M. A. Jongsma, and H. J. Bouwmeester. 2005. Volatile science? Metabolic engineering of terpenoids in plants. Trends Plant Sci. 10:594-602. [DOI] [PubMed] [Google Scholar]
- 2.Ajikumar, P. K., K. Tyo, S. Carlsen, O. Mucha, T. H. Phon, and G. Stephanopoulus. 2008. Terpenoids: oportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol. Pharmaceut. 5:167-190. [DOI] [PubMed] [Google Scholar]
- 3.Amberg, D. C., D. J. Burke, and J. N. Strathern. 2005. Methods in yeasts genetics: a Cold Spring Harbor laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 4.Anderegg, R. J., R. Betz, S. A. Carr, J. W. Crabb, and W. Duntze. 1988. Structure of Saccharomyces cerevisiae mating hormone a-factor. J. Biol. Chem. 263:18236-18240. [PubMed] [Google Scholar]
- 5.Asadollahi, M. A., J. Maury, K. R. Patil, M. Schalk, A. Clark, and J. Nielsen. 2009. Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab. Eng. 11:328-334. [DOI] [PubMed] [Google Scholar]
- 6.Bakalinsky, A. T., and R. Snow. 1990. The chromosomal constitution of wine strains of Saccharomyces cerevisiae. Yeast 6:367-382. [DOI] [PubMed] [Google Scholar]
- 7.Bohlmann, J., and C. I. Keeling. 2008. Terpenoid biomaterials. Plant J. 54:656-669. [DOI] [PubMed] [Google Scholar]
- 8.Carrau, F. M., K. Medina, E. Boido, L. Farina, C. Gaggero, E. Dellacassa, G. Versini, and P. A. Henschke. 2005. De novo synthesis of monoterpenes by Saccharomyces cerevisiae wine yeasts. FEMS Microbiol. Lett. 243:107-115. [DOI] [PubMed] [Google Scholar]
- 9.Carter, O. A., and R. J. Peters. 2003. Monoterpene biosynthesis pathway construction in Escherichia coli. Phytochemistry 64:425-433. [DOI] [PubMed] [Google Scholar]
- 10.Chang, M. C., and J. D. Keasling. 2006. Production of isoprenoid pharmaceuticals by engineered microbes. Nat. Chem. Biol. 2:674-681. [DOI] [PubMed] [Google Scholar]
- 11.Donald, K. A. G., R. Y. Hampton, and I. B. Fritz. 1997. Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 63:3341-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donalies, U. E. B., H. T. T. Nguyen, U. Stahl, and E. Nevoigt. 2008. Improvement of Saccharomyces strains used in brewing, wine making and baking. Adv. Biochem. Eng. Biotechnol. 111:67-98. [DOI] [PubMed] [Google Scholar]
- 13.Dudareva, N., L. Cseke, V. M. Blanc, and E. Pichersky. 1996. Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8:1137-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudareva, N., and E. Pichersky. 2008. Metabolic engineering of plant volatiles. Curr. Opin. Biotechnol. 19:181-189. [DOI] [PubMed] [Google Scholar]
- 15.Edris, A. E. 2007. Pharmaceutical and therapeutic potentials of essential oils and their individual volatile constituents: a review. Phytother. Res. 21:308-323. [DOI] [PubMed] [Google Scholar]
- 16.García-Martínez, J., A. Aranda, and J. E. Pérez-Ortín. 2004. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell 15:303-313. [DOI] [PubMed] [Google Scholar]
- 17.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 18.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 19.Gómez-Galera, S., A. M. Pelacho, A. Gené, T. Capell, and P. Christou. 2007. The genetic manipulation of medicinal and aromatic plants. Plant Cell Rep. 26:1689-1715. [DOI] [PubMed] [Google Scholar]
- 20.Hampton, R., D. Dimster-Denk, and J. Rine. 1996. The biology of HMG-CoA reductase: the pros of contra-regulation. Trends Biochem. Sci. 21:140-145. [PubMed] [Google Scholar]
- 21.Hernández, I., D. Molenaar, J. Beekwilder, H. Bouwmeester, and J. E. T. van Hylckama Vlieg. 2007. Expression of plant flavor genes in Lactococcus lactis. Appl. Environ. Microbiol. 73:1544-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrero, O., D. Ramón, and M. Orejas. 2008. Engineering the Saccharomyces cerevisiae isoprenoid pathway for de novo production of aromatic monoterpenes in wine. Metab. Eng. 10:78-86. [DOI] [PubMed] [Google Scholar]
- 23.Huang, B., J. Guo, B. Yi, X. Yu, L. Sun, and W. Chen. 2008. Heterologous production of secondary metabolites as pharmaceuticals in Saccharomyces cerevisiae. Biotechnol. Lett. 30:1121-1137. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, B. E., E. Hart-Wells, and S. P. T. Matsuda. 2003. Metabolic engineering to produce sesquiterpenes in yeast. Org. Lett. 5:1629-1632. [DOI] [PubMed] [Google Scholar]
- 25.Jiménez-Martí, E., A. Zuzuarregui, I. Ridaura, N. Lozano, and M. del Olmo. 2009. Genetic manipulation of HSP26 and YHR087W stress genes may improve fermentative behaviour in wine yeasts under vinification conditions. Int. J. Food Microbiol. 130:122-130. [DOI] [PubMed] [Google Scholar]
- 26.Kirby, J., and J. D. Keasling. 2008. Metabolic engineering of microorganisms for isoprenoid production. Nat. Prod. Rep. 25:656-661. [DOI] [PubMed] [Google Scholar]
- 27.Kirby, J., D. W. Romanini, E. M. Paradise, and J. Keasling. 2008. Engineering triterpene production in Saccharomyces cerevisiae—β-amyrin synthase from Artemisia annua. FEBS J. 275:1852-1859. [DOI] [PubMed] [Google Scholar]
- 28.Koroch, A. R., J. A. Zygadlo, and H. R. Juliani. 2007. Bioactivity of essential oils and their components, p. 87-115. In R. G. Berger (ed.), Flavors and fragances. Chemistry, bioprocessing and sustainability. Springer-Verlag, Berlin, Germany.
- 29.Mahmoud, S. S., and R. B. Croteau. 2002. Strategies for transgenic manipulation of monoterpene biosynthesis in plants. Trends Plant Sci. 7:366-373. [DOI] [PubMed] [Google Scholar]
- 30.Marasco, E. K., and C. Schmidt-Dannert. 2007. Biosynthesis of plant natural products and characterization of plant biosynthetic pathways in recombinant microorganisms, p. 1-43. In R. Verpoorte, A. W. Alfermann, and T. S. Johnson (ed.), Applications of plant metabolic engineering. Springer, Dordrecht, Netherlands.
- 31.Maury, J., M. A. Asadollahi, K. Møller, A. Clark, and J. Nielsen. 2005. Microbial isoprenoid production: an example of green chemistry through metabolic engineering. Adv. Biochem. Eng. Biotechnol. 100:19-51. [DOI] [PubMed] [Google Scholar]
- 32.Maury, J., M. A. Asadollahi, K. Moler, M. Schalk, A. Clark, L. R. Formenti, and J. Nielsen. 2008. Reconstruction of a bacterial isoprenoid biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett. 582:4032-4038. [DOI] [PubMed] [Google Scholar]
- 33.Nevoigt, E. 2008. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 72:379-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oswald, M., M. Fischer, N. Dirninger, and F. Karst. 2007. Monoterpenoid biosynthesis in Saccharomyces cerevisiae. FEMS Lett. 7:413-421. [DOI] [PubMed] [Google Scholar]
- 35.Pichersky, E., and N. Dudareva. 2007. Scent engineering: toward the goal of controlling how flowers smell. Trends Biotechnol. 25:105-110. [DOI] [PubMed] [Google Scholar]
- 36.Polakowski, T., U. Stahl, and C. Lang. 1998. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl. Microbiol. Biotechnol. 49:66-71. [DOI] [PubMed] [Google Scholar]
- 37.Puig, S., D. Ramón, and J. E. Pérez-Ortín. 1998. An optimized method to obtain stable food-safe recombinant wine strains. J. Agric. Food Chem. 46:1689-1693. [Google Scholar]
- 38.Reiling, K. K., Y. Yoshikuni, V. J. J. Martin, J. Newman, J. Bohlman, and J. D. Keasling. 2004. Mono and diterpene production in Escherichia coli. Biotechnol. Bioeng. 87:200-212. [DOI] [PubMed] [Google Scholar]
- 39.Ro, D. K., E. M. Paradise, M. Ouelet, K. J. Fischer, K. Newman, J. M. Ndungu, K. A. Ho, R. A. Eachus, T. S. Ham, J. Kirby, M. C. Y. Chang, S. T. Withers, Y. Shibam, R. Sarpong, and J. D. Keasling. 2006. Production of the antimalarial drug precursor artemisic acid in engineered yeast. Nature 440:940-943. [DOI] [PubMed] [Google Scholar]
- 40.Romanos, M. A., C. A. Scores, and J. J. Clare. 1992. Foreign gene expression in yeasts: a review. Yeast 8:423-488. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. E. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Shimada, H., K. Kondo, P. D. Fraser, Y. Miura, T. Saito, and N. Misawa. 1998. Increased carotenoid production by the food yeast Candida utilis through metabolic engineering of the isoprenoid pathway. Appl. Environ. Microbiol. 64:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas, T., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 44.Wang, G. Y., and J. D. Keasling. 2002. Amplification of HMG-CoA reductase production enhances carotenoid accumulation in Neurospora crassa. Metab. Eng. 4:193-201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.