Abstract
Development of the strategy known as consolidated bioprocessing (CBP) involves the use of a single microorganism to convert pretreated lignocellulosic biomass to ethanol through the simultaneous production of saccharolytic enzymes and fermentation of the liberated monomeric sugars. In this report, the initial steps toward achieving this goal in the fermentation host Zymomonas mobilis were investigated by expressing heterologous cellulases and subsequently examining the potential to secrete these cellulases extracellularly. Numerous strains of Z. mobilis were found to possess endogenous extracellular activities against carboxymethyl cellulose, suggesting that this microorganism may harbor a favorable environment for the production of additional cellulolytic enzymes. The heterologous expression of two cellulolytic enzymes, E1 and GH12 from Acidothermus cellulolyticus, was examined. Both proteins were successfully expressed as soluble, active enzymes in Z. mobilis although to different levels. While the E1 enzyme was less abundantly expressed, the GH12 enzyme comprised as much as 4.6% of the total cell protein. Additionally, fusing predicted secretion signals native to Z. mobilis to the N termini of E1 and GH12 was found to direct the extracellular secretion of significant levels of active E1 and GH12 enzymes. The subcellular localization of the intracellular pools of cellulases revealed that a significant portion of both the E1 and GH12 secretion constructs resided in the periplasmic space. Our results strongly suggest that Z. mobilis is capable of supporting the expression and secretion of high levels of cellulases relevant to biofuel production, thereby serving as a foundation for developing Z. mobilis into a CBP platform organism.
The biological conversion of lignocellulosic biomass to ethanol represents a potential major source of future domestic transportation fuels, but the current cost of converting biomass to fermentable sugars still needs to be reduced further (12). Most current strategies for ethanol production via biochemical conversion of lignocellulosic feedstocks utilize simultaneous saccharification and fermentation (SSF) or simultaneous saccharification and cofermentation (SSCF) processes (8, 21, 22). The process configuration known as consolidated bioprocessing (CBP) (20) would alleviate the financial strain of producing saccharolytic enzyme cocktails by combining the necessary steps for ethanol production as the action of one microorganism.
A particularly attractive microbial candidate for the development of a CBP microorganism is the Gram-negative fermentative bacterium Zymomonas mobilis. Z. mobilis has been studied for its exceptionally high ethanol production rate, yield, and tolerance to the toxicity of the final product (15-17, 20, 31-33, 35, 43). In addition, Z. mobilis has the ability to ferment sugars at low pH and has a naturally high tolerance to many of the inhibitory compounds found in hydrolysates derived from lignocellulosic biomass (45, 46) Furthermore, the use of the Entner-Doudoroff pathway (37) allows Z. mobilis to achieve the near-theoretical maximum ethanol yields during fermentation while achieving relatively low biomass formation. Accordingly, Z. mobilis has been used successfully in SSF and SSCF processes (14, 24, 36). Additionally, Z. mobilis has been successfully engineered to ferment the pentose (C5) sugars, xylose (45) and arabinose (10).
A necessary prerequisite to establishing Z. mobilis as a CBP host is the ability to achieve high levels of cellulolytic enzyme expression. However, there is not yet a strong consensus on how to achieve maximal heterologous protein expression in Z. mobilis. Multiple groups have attempted heterologous expression of numerous genes, including cellulolytic enzymes in Z. mobilis with various degrees of success (6, 7, 9, 19, 27, 42, 44). Unfortunately, there are no obvious correlations between the expression strategies employed compared to the results obtained. Intriguingly, however, when researchers used the tac promoter (Ptac) to drive expression of native Z. mobilis genes, they were able to express several genes to extremely high levels (2). The results from this study (2) suggest that while the potential to achieve high levels of heterologous cellulase expression in Z. mobilis certainly exists, the ability to do so on a consistent basis will need further investigation.
While achieving high-level expression of cellulases is an important hurdle to overcome in the development of the CBP technology, it is imperative that these enzymes additionally be translocated to the extracellular medium in order to directly contact the lignocellulosic substrate. The most obvious means by which to achieve this translocation is by harnessing the host cell's protein secretion apparatus. There is, however, in general, very little fundamental knowledge regarding the capacity of Z. mobilis to secrete proteins. There is only one account to our knowledge of fusing secretion signals native to Z. mobilis onto proteins from exogenous sources, where extracellular secretion of a recombinant β-glucosidase reached only 11% of the total amount of enzyme synthesized (43).
We initially report the finding that several Z. mobilis strains natively produce an endogenous activity against carboxymethyl cellulose (CMC) and that this activity can be detected extracellularly. Together, these results suggest that Z. mobilis may be adept at producing and secreting cellulolytic enzymes, and as this attribute is essential for a CBP organism, Z. mobilis serves as an ideal candidate for further investigation.
We next describe the expression of two cellulolytic enzymes (E1 and GH12) in both Escherichia coli and Z. mobilis. E1 (locus tag Acel_0614) and GH12 (locus tag Acel_0619) are both from the acidothermophile Acidothermus cellulolyticus and are representative of glycoside hydrolase families 5 and 12, respectively (10a, 38a). E1 is an endo-1,4-β-glucanase, and GH12 is an uncharacterized enzyme that has a very high sequence identity to the GH12 domain of GuxA (Acel_0615) from A. cellulolyticus. GuxA has activities against a wide variety of substrates, including carboxymethyl cellulose, arabinoxylan, xylan, and xyloglucan (W. Adney, unpublished results). While GH12 has yet to be fully characterized, we used homology modeling (3) to predict the enzyme class of GH12 and found that it strongly resembles an endo-1,4-β-glucanase. These enzymes were chosen because of their relatively low molecular weight, high stability, and activity over a broad temperature and pH range using only the catalytic domains (Adney, unpublished). We report the successful expression of both enzymes in Z. mobilis by addressing several variables related to gene expression. Additionally, the use of codon optimization was explored as a way of enhancing heterologous expression in Z. mobilis. After successfully demonstrating the intracellular expression of E1 and GH12 in Z. mobilis, we further show that Z. mobilis is capable of secreting these proteins extracellularly through the use of native secretion signals predicted to utilize two separate protein translocation pathways in Z. mobilis, the SecB-dependent and twin arginine translocation (TAT) pathways. This finding should prove valuable beyond the production of cellulases and could include all classes of recombinant proteins.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Z. mobilis strains ATCC 39676, ZM4 (ATCC 31821), and CP4 (41) were routinely grown in RMG medium (1% [wt/vol] yeast extract, 0.2% [wt/vol] KH2PO4, 2% [wt/vol] glucose, and for plates, 1.5% [wt/vol] Bacto agar) at 30°C and shaken at 120 rpm. Where applicable, tetracycline was added to a final concentration of 20 μg/ml for plates and 10 μg/ml for liquid culture.
Z. mobilis transformations.
The transformation protocol was adapted from the method of Deanda et al. (10). Three to five micrograms of plasmid DNA was used to transform approximately 1010 cells per ml of Z. mobilis in 100 μl of 10% glycerol. A Bio-Rad gene pulser was used with the following conditions: 200 Ω, 25 μF, and 1.6 kV in a 0.1-cm cuvette. Following electroporation, 1 ml of mating medium [50 g/liter glucose, 10 g/liter yeast extract, 5 g/liter tryptone, 2.5 g/liter (NH4)2SO4, 0.2 g/liter K2HPO4, and 1 mM MgSO4] was added to the cells, and the cells were then incubated at 30°C for at least 6 h. One hundred to 200 μl of cells were then plated onto mating medium agar plates (without MgSO4) supplemented with 20 μg/ml tetracycline and incubated at 30°C for 2 or 3 days anaerobically. Positive transformants were identified by NotI restriction digestion of purified plasmid DNA.
Gene synthesis, codon optimization, and plasmid construction.
The coding sequences of the catalytic domains of E1 and GH12 from A. cellulolyticus were codon optimized using Gene Designer (39) based on the codon bias of Z. mobilis strain ZM4 and synthesized by DNA 2.0 (Menlo Park, CA).
Plasmids.
Schematic representations of all plasmids used in this study are shown in Table 1. The sequences of all plasmids have been verified, and the GenBank accession numbers associated with novel sequences are listed below. Using standard overlap PCR techniques, the gap promoter region from Z. mobilis genomic DNA (ATCC 39676) was fused to the T7 terminator region from plasmid pET101/D-topo (Invitrogen, Carlsbad CA) separated by a NotI restriction site to create plasmid pJL100.
TABLE 1.
Plasmids used in this study
a The plasmid names and relevant expression elements are shown graphically. For E1 and GH12 plasmids, the amino acids (AA's) that are encoded are shown in parentheses.
b N/A, not available.
To create plasmids pJL101 and pJL103, PCR products representing coding sequences for E1 and GH12 were cloned into pFLAG-CTC (Sigma-Aldrich, St. Louis, MO). A. cellulolyticus genomic DNA was used as the PCR template for the E1 and GH12 coding sequences for plasmids pJL101 and pJL103. pJL101 carries the gene coding for the first 274 amino acid residues of GH12, while pJL103 carries the gene coding for amino acid residues 42 to 404 of E1.
Plasmids p25143 and p25144 were codon optimized, synthesized, and subcloned by DNA 2.0 (Menlo Park, CA) and carry genes coding for amino acid residues 42 to 422 of E1 and amino acid residues 35 to 274 of GH12, both lacking the predicted native signal peptide. These sequences entail the catalytic domains and are lacking the cellulose binding domains (CBM). These sequences were then subcloned into the NotI site of vector pZB188 (45) to create plasmids p25143 and p25144.
Plasmid pJL110 was created by PCR amplifying the Z. mobilis ATCC 39676 strain pdc gene including extended 5′ (75-bp) and 3′ (122-bp) noncoding sequences with flanking NotI sequences incorporated. This PCR product was Topo-TA cloned into pYES2.1 (Invitrogen, Carlsbad, CA) to create plasmid pYes2.1-PDC. The E1 gene was PCR amplified using primers with homology to the 5′ and 3′ noncoding sequences of Z. mobilis pdc. The primer sequences were as follows with E1-specific nucleotides shown in lowercase type and Z. mobilis pdc homologous nucleotides shown as uppercase type: Forward, CCTGATTCAGACATAGTGTTTTGAATATATGGAGTAAGCAatgtgtggaattgtgagcgg; Reverse, GGACGGGCTTTTCGCCTTAAGCTCTAAGTTTATTTAAAAAttagcttggactgggactgg. Saccharomyces cerevisiae strain w303 was transformed with the resulting PCR product as well as KpnI-linearized pYES2.1-E1. Through endogenous homologous recombination in yeast, the Z. mobilis pdc open reading frame (ORF) was exchanged with the E1 ORF on plasmid pYes2.1-E1 to create plasmid pYES2.1-PDC-E1-PDC. The E1 fragment containing the 5′ and 3′ sequences of pdc was excised with NotI and ligated into pZB188 to create plasmid pJL110.
To create plasmids pJL111 and pJL116, the sequence representing the predicted secretion signal of the phoC gene (ZM0130, ATGATAAAAGTCCCGCGGTTCATCTGTATGATCGCGCTTACATCCAGCGTTCTGGCAAGCGGCCTTTCTCAAAGCGTTTCAGCTCAT) was fused to the 5′ termini of E1 and GH12 genes, respectively. For plasmid pJL112, the predicted secretion signal of the predicted Z. mobilis ORF ZM0331 (ATGAAAAGAAAGCTTGGTCGTCGCCAGTTATTAACTGGCTTTGTTGCCCTTGGCGGTATGGCGATTACAGCTGGTAAGGCGCAGGCTTCT) was fused to the 5′ terminus of the E1 gene sequence.
Immunoblots and CMC zymogram analysis.
Following SDS-PAGE, cellular proteins were transferred to a polyvinylidene fluoride (PVDF) membrane at a constant 200 V for 1 h. A mouse monoclonal anti-E1 antibody diluted 1:4,000 in 3% milk in Tris-buffered saline containing Tween 20 (TBST) was added to the PVDF membranes, which were then incubated for 2 h at room temperature. After the membranes were washed with TBST, they were incubated in TBST containing 3% milk and an alkaline phosphatase-conjugated goat anti-mouse secondary antibody (diluted 1:4,000) for 1 h at room temperature. We prepared the carboxymethyl cellulose (CMC) zymograms by the protocol of Taylor et al. (38), except that the reaction buffer used was 50 mM sodium citrate buffer (pH 7.0).
Total and extracellular fraction preparations.
For each strain, an equal volume of cell culture was moved into two separate microcentrifuge tubes. In one of the replicates, the cells were removed by two rounds of centrifugation (15,000 × g for 5 min each time) to create the extracellular fraction. Equal volumes of the culture with cells (total) and the extracellular fraction were treated with 10× BugBuster lysis buffer (Novagen) to create a 1× concentration of lysis buffer. Lysis buffer was added to the extracellular fraction to ensure that the presence or absence of lysis buffer had no effect on the enzyme activity assays performed on these fractions. The samples were vortexed and incubated at room temperature for 20 min. The samples were centrifuged (15,000 × g for 5 min), and the supernatants were moved to a new tube.
Subcellular fractionations.
Cultures of Z. mobilis (50 ml) were grown to the beginning of stationary phase, and the cells were centrifuged at 4,000 × g for 10 min. The cells were resuspended in periplasting buffer at a volume corresponding to 4 ml/g (wet weight) of the cell pellet. Periplasting buffer consisted of 200 mM Tris-HCl (pH 7.5), 20% sucrose, 1 mM EDTA, and 2.5 million units of lysozyme (0.00625 g/5 ml; catalog no. L6876 [Sigma-Aldrich]). Multiple incubation times were tested empirically to find the longest time point where no more than 5% of the alcohol dehydrogenase (ADH) activity was found within the periplasmic fraction (typically 5 min; ADH assay detailed below). This was done to ensure maximal release of periplasmic contents, while minimizing contamination with cytoplasmic contents. Following the incubation in periplasting buffer, ice-cold H2O was added to a volume corresponding to 6 ml/g (wet weight) of the original cell pellet. The cells were incubated on ice for 10 min and then centrifuged at 4°C for 10 min at 4,000 × g. The periplasmic fraction (supernatant) was transferred to a new tube, and BugBuster HT lysis buffer (Novagen) was added to the cell pellet at a volume corresponding to 10 ml/g (wet weight) of the original cell pellet. Following vortexing to ensure complete resuspension, this lysing reaction mixture was incubated for 20 min at room temperature and centrifuged for 5 min at 15,000 × g. The resulting supernatant represented the cytoplasmic fraction.
E1 and GH12 activity assays.
Protein lysates (10 μl of whole-cell lysates and periplasmic and cytoplasmic fractions) were added to 90 μl of reaction buffer (50 mM sodium citrate [pH 7.0] and 0.00125 g/5 ml 4-methylumbelliferyl β-d-cellobiopyranoside [Sigma-Aldrich]) in a 96-well microtiter plate. Reaction mixtures were incubated at 50°C for 30 to 60 min and analyzed for fluorescence using a BMG Labtech FLUOstar Omega with an excitation wavelength of 355 nm and an emission wavelength of 460 nm. Measurements were taken at multiple time points to ensure that fluorescence detection was not saturated. Each lysate was created from individual cultures independently at least three times, and each of those was run in triplicate on the same microtiter plate. Assays were blanked against the average of three independent mock reactions. For each experiment, the highest raw value average was set at 1, and the remaining samples were normalized as a fraction of 1. This was done for both the whole-cell lysates and the individual subcellular or extracellular fractions.
ADH assays.
Forty-four microliters of 50 mM sodium pyrophosphate (pH 8.8) was added to each well of a 96-well microtiter plate, followed by 2 μl of either the periplasmic or cytoplasmic fractions prepared as described above. Then, 50.6 μl of 15 mM β-NAD hydrate (catalog no. N7004; Sigma-Aldrich) and 3.4 μl of 95% ethanol were added to the wells. The absorbance at 340 nm was measured immediately and subsequently every 2 min for 14 min using a BMG Labtech FLUOstar Omega. The ΔA340 was calculated, and the relative contribution of the periplasmic and cytoplasmic fraction to the total rate was calculated to determine the percent localization of ADH activity.
Growth curve analysis and doubling time calculations.
The protocol used to analyze growth data was adapted from the method of Franden et al. (11). Logarithmically growing cultures were used to inoculate 200 μl of RMG medium to an optical density at 600 nm (OD600) of 0.05 in a 100-well honeycomb plate. Using a Bioscreen C MBR analyzer (Growth Curves USA, Piscataway, NJ), turbidometric measurements were made using a wide band filter (420 to 580 nm) every 15 min for 24 h with no agitation. Growth rate constant measurements were taken from the early portion of logarithmic growth (from OD420-580 [OD420 to OD580] of ∼0.1 to 0.3), using the following equation: Y = Xo − eμt where Y is the absorbance at time t, Xo is the initial absorbance (near OD420-580 of 0.1), and μ is the growth rate. Doubling times shown in the Results section were calculated as μ divided by the natural log of 2. Means ± 1 standard deviation are shown.
DNA sequence accession numbers.
The sequences for the following expression cassettes were deposited in GenBank: p25143 (E1-codon optimized; GenBank accession no. HM595415), p25144 (GH12-codon optimized; GenBank accession no. HM595416), pJL111 (pTac-Z130-E1-T1T2; GenBank accession no. HM595412), pJL112 (pTac-Z331-E1-T1T2; GenBank accession no. HM595413), and pJL116 (pTac-Z130-GH12-T1T2; GenBank accession no. HM595414).
RESULTS
Zymomonas mobilis has endogenous cellulolytic activities.
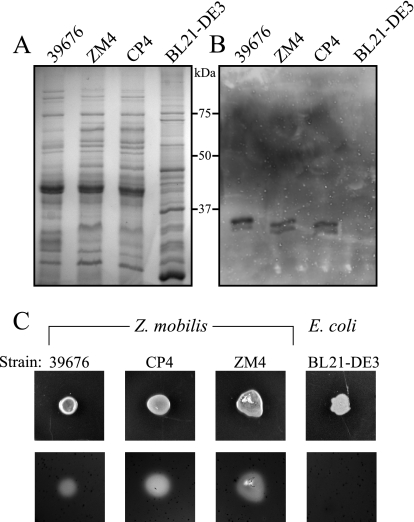
During the process of investigating cell lysates of Z. mobilis for carboxymethyl cellulose (CMC) activity, it became evident that all of the Z. mobilis strains used in this study (i.e., ATCC 39676, ZM4, and CP4) demonstrated hydrolytic activity against the CMC substrate. This activity manifested itself as two, closely spaced, yet distinct molecular mass bands on zymograms (Fig. 1 A and B). The apparent masses of these bands (Fig. 1B) are consistent with the predicted molecular mass of the Z. mobilis CelA protein (37 kDa [30]), suggesting that one (or both) of the bands may represent CelA. Additionally, when grown on agar plates containing CMC, Z. mobilis but not E. coli can clearly hydrolyze the CMC as can be seen by the zones of clearing (Fig. 1C). Importantly, celA is the only annotated cellulolytic gene found within the published genome of Z. mobilis (http://cmr.jcvi.org), providing further support that the CM-cellulolytic activity seen in Fig. 1 is likely attributable to CelA. The finding that many Z. mobilis strains naturally express and secrete at least one endogenous cellulase provides support that Z. mobilis might prove adept at expressing and secreting heterologous cellulases.
FIG. 1.
Activity of the native cellulolytic proteins in Z. mobilis and E. coli strains. Z. mobilis strains ATCC 39676, ZM4, and CP4 and E. coli strain BL21(DE3) were studied. (A) Coomassie blue-stained polyacrylamide gel. (B) Carboxymethyl cellulose (CMC) zymogram. (C) Patched colonies of Z. mobilis and E. coli growing on an RMG-CMC plate, and the same plate with the cells removed and stained with Congo red to reveal areas of CMC degradation.
Intracellular expression of heterologous cellulolytic enzymes in E. coli and Z. mobilis.
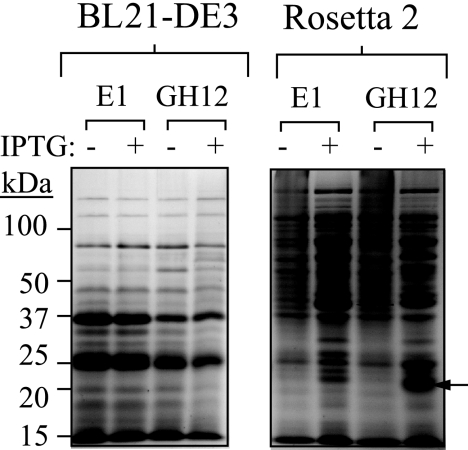
An altered codon bias between A. cellulolyticus and Z. mobilis could hinder the successful expression of E1 and GH12 in Z. mobilis. Since there are no commercially available codon-enhanced strains of Z. mobilis, the issue of codon bias was initially addressed by using a codon-enhanced strain of E. coli. Separate plasmids containing Ptac-driven genes encoding the E1 and GH12 endoglucanases (Acel-0614 and Acel-0619, respectively) from A. cellulolyticus were introduced into a strain of E. coli designed to enhance the expression of heterologous proteins that contain codons rarely used in E. coli. Figure 2 shows that while expression of E1 and GH12 was not detected upon isopropyl-β-d-thiogalactopyranoside (IPTG) induction in E. coli BL21(DE3) cells, both proteins were strongly induced in the codon-enhanced Rosetta 2 strain of E. coli (EMD, Madison, WI). These data strongly suggest that the differential codon bias between the host organism (A. cellulolyticus) and the expression organism (E. coli) could be a major barrier to heterologous expression in E. coli. By inference, this may also suggest that differential codon usage between A. cellulolyticus and Z. mobilis may have a detrimental effect on the expression of E1 and GH12 in Z. mobilis.
FIG. 2.
Expression of E1 and GH12 in E. coli strains BL21(DE3) and Rosetta 2. Coomassie blue-stained polyacrylamide gel of protein lysates from E. coli strains BL21(DE3) and Rosetta 2 harboring plasmids pJL101 (E1 lanes), and pJL103 (GH12 lanes) with (+) or without (−) protein induction with 1 mM IPTG.
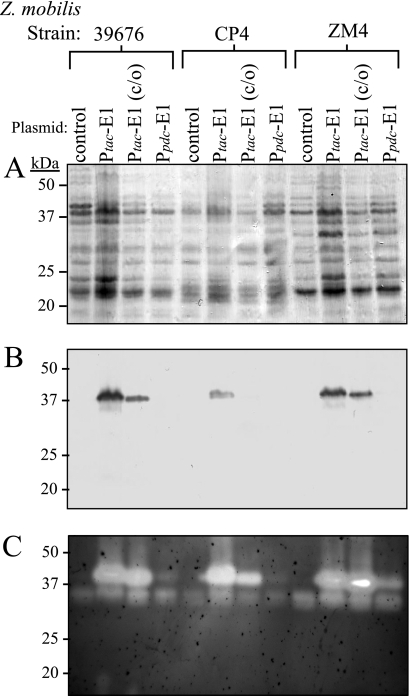
To test the effect that differential codon bias might have on gene expression, the gene sequence encoding the E1 catalytic domain was codon optimized for Z. mobilis (DNA 2.0, Menlo Park, CA), and the synthesized gene was tested for expression in Z. mobilis. The expression of the native and codon-optimized (E1-c/o) gene sequence of the E1 catalytic domain (E1) was examined in multiple strains of Z. mobilis. Both E1 and E1-c/o can be detected using immunoblot and zymogram gel analysis (Fig. 3). Surprisingly, both constructs were expressed nearly equally as well in Z. mobilis, though the native E1 protein might have been expressed slightly better than E1-c/o (Fig. 3B and 3C), when total protein loading is taken into account (Fig. 3A). The fact that the codon-optimized version of E1 was not expressed significantly better than the native E1 coding sequence suggests that either the codon optimization strategy undertaken was suboptimal or that differential codon usage bias was not a major obstacle (or at least not the sole obstacle) to achieving higher levels of expression.
FIG. 3.
Expression of various E1 constructs in multiple strains of Z. mobilis. Protein lysates from Z. mobilis strains ATCC 39676, CP4, and ZM4 transformed with the plasmids pZB188 (control lanes), pJL113 (Ptac-E1 lanes), p25143 [Ptac-E1 (c/o) lanes], and pJL110 (Ppdc-E1 lanes) were run identically on two independent 12% polyacrylamide gels supplemented with 0.12% carboxymethyl cellulose (CMC). (A) A PVDF membrane stained with amido black to show total protein. (B) Immunoblot probed with an anti-E1 antibody. (C) A CMC zymogram performed on the second of two duplicate polyacrylamide gels to show cellulolytic activity. E1 activity is represented by the top band, and cellulolytic activity endogenous to Z. mobilis can be seen by the bottom band.
Expression of the codon-optimized E1 gene sequence was also examined using the promoter and the 5′ and 3′ untranslated regions (UTRs) of the Z. mobilis pdc gene, which is a highly transcribed gene with very stable mRNA (25). It was reasoned that this strategy might increase E1 transcription levels and its mRNA stability, thus increasing E1 protein expression. We compared the expression of this new construct with expression of the Ptac-driven constructs and surprisingly found very low levels of E1 expression using the pdc transcriptional unit (Fig. 3). It should be noted that while Ppdc-driven E1 is difficult to visualize in Fig. 3B, it was, in fact, clearly visible on the immunoblot. To ensure that the low level of E1 expression detected was not a strain-specific phenomenon, the expression of the various E1 constructs was examined using multiple Z. mobilis strains. While both Z. mobilis strains ATCC 39676 and ZM4 showed similar expression levels of the E1 constructs, strain CP4 repeatedly showed a reduced level of expression for all of the constructs, suggesting that differences in gene expression can also be strain specific. The possibility that the E1 protein was being expressed to a high level but was escaping our detection by either being released into the supernatant or forming SDS-insoluble aggregates was ruled out by immunoblot analysis of the culture supernatant and using the agarose gel electrophoresis to resolve aggregates (AGERA) technique, respectively (40; data not shown).
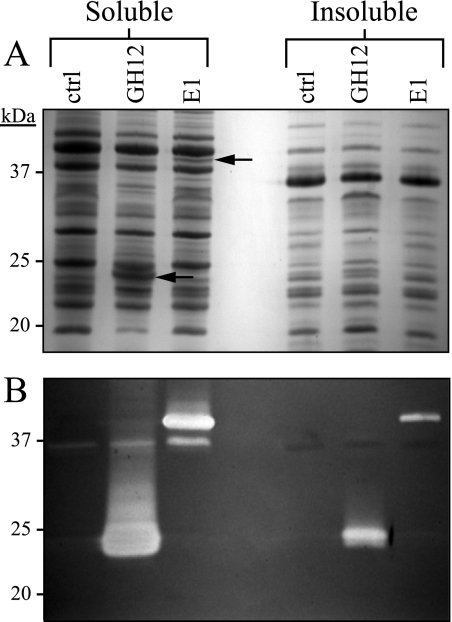
The intracellular expression of GH12 from A. cellulolyticus in Z. mobilis (strain ATCC 39676) was also examined. Using an identical approach to E1, the GH12 catalytic domain was codon optimized and subcloned into a plasmid under the control of Ptac. Surprisingly, the level of GH12 expression observed in Z. mobilis stood in stark contrast to that of E1. Figure 4 A clearly shows that GH12 can be detected by Coomassie blue-stained SDS-PAGE (Fig. 4A). The GH12 protein represented approximately 4.6% (standard deviation, 1.3%; n = 6) of the total cellular protein in logarithmically growing cells (as measured by densitometry analysis of Coomassie blue-stained polyacrylamide gels). This stands in contrast to the low levels of recombinant E1 protein produced which represented 1.7% (standard deviation, 0.2%; n = 3) of the total protein. Furthermore, a significant portion of the GH12 protein expressed in Z. mobilis was soluble (Fig. 4A). In addition, the protein was enzymatically active as evident by the zones of substrate clearing on a CMC zymogram (Fig. 4B).
FIG. 4.
Expression, solubility analysis, and activity of E1 and GH12 in Z. mobilis strain ATCC 39676. Protein lysates from Z. mobilis strain ATCC 39676 transformed with pZB188 (control [ctrl]), p25144 (GH12 lanes), and p25143 (E1 lanes) were run identically on two independent 12% polyacrylamide gels supplemented with 0.12% carboxymethyl cellulose (CMC). (A) One of the duplicate polyacrylamide gels was stained with Coomassie blue to show total protein. The locations of the GH12 and E1 proteins are indicated by black arrows in the gel. (B) A CMC zymogram performed on the second of two duplicate polyacrylamide gels designed to show cellulolytic activity.
Extracellular secretion of heterologously expressed cellulases in Z. mobilis.
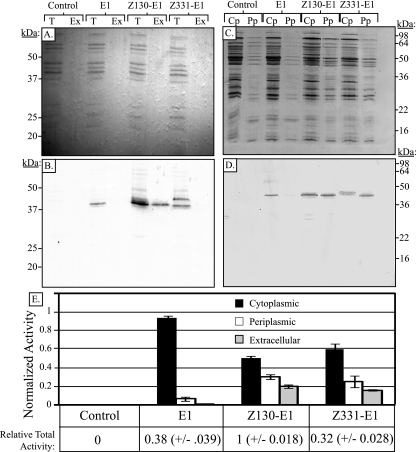
We next examined the capability of Z. mobilis to secrete both E1 and GH12 through the use of predicted secretion signals native to Z. mobilis. We chose two independent secretion signals that are predicted to utilize two separate protein secretion pathways. The first secretion signal was that of the Z. mobilis phoC gene (ZM0130; predicted by Genome Atlas (http://www.cbs.dtu.dk/services/GenomeAtlas/show-subcell.php?KLSO=ASC&KLSK=ORGANISMSORT&kingdom=Bacteria&tableType=Secreted%20Proteins&segmentid=Zmobilis_ZM4_Main&secType=Cytoplasm) which uses the SecB-dependent (type II) pathway (34). The second secretion signal was that belonging to a hypothetical protein (ZM0331) of Z. mobilis predicted by Genome Atlas to utilize the twin arginine translocation (TAT) pathway (18). These signal sequences were fused onto the 5′ end of the coding sequence of the E1 catalytic domain to create the plasmid constructs Z130-E1 (pJL111) and Z331-E1 (pJL112). The resulting plasmids along with the control vector pZB188 and a plasmid containing an intracellularly expressed version of E1 (p25143) were used to examine the cellular localization of E1 in Z. mobilis cultures. Figure 5 A shows an amido black-stained PVDF membrane containing the protein in either the total culture (T lanes; lysate of whole cells plus growth medium) or the extracellular fraction (Ex lanes; mock lysate of the growth medium with the cells removed by centrifugation). Figure 5B shows an anti-E1 immunoblot of the membrane shown in Fig. 5A and reveals the location of E1 in either the total culture or extracellular fractions. This figure clearly shows that a significant portion of Z130-E1 can be found extracellularly, while E1 without a secretion signal and Z331-E1 can be detected only in the total fraction, suggesting they are localized intracellularly. Furthermore, it is interesting to note that the addition of the Z130 secretion signal appears to have increased the overall expression of E1 (Fig. 5).
FIG. 5.
Extracellular secretion and subcellular localization of E1 in Z. mobilis strain ATCC 39676. (A) Amido black-stained PVDF membrane showing total (T) and extracellular medium (Ex) protein lysate fractions to show the total protein load. (B) Anti-E1 immunoblot of the membrane in panel A. (C) Amido black-stained PVDF membrane showing total protein load of protein lysates derived from Z. mobilis expressing multiple versions of E1. Cp, cytoplasmic fraction; Pp, periplasmic fraction. (D) Anti-E1 immunoblot of the membrane in panel C. (E) Relative quantification of E1 activity against methylumbelliferyl cellobiopyranoside (MUC) in periplasmic, cytoplasmic, and extracellular fractions. The relative total activity of equivalent whole-cell lysates from the indicated strains is shown below the bar graph to highlight differential expression between the strains.
To ensure that the E1 enzyme found in the extracellular space was in fact due to protein secretion, and not a by-product of passive release due to increased cell death/lysis in the strains with E1 tagged with secretion signals, we examined the growth medium for alcohol dehydrogenase (ADH), an enzyme activity found exclusively in the cytoplasm and frequently used as a cytoplasmic marker (1, 5, 6, 29). While we were able to detect ADH activity in a lysate of cells plus growth medium, we were unable to detect any ADH activity in the culture medium alone, which suggests that there was not a significant increase in cell lysis (data not shown). In addition there were no significant differences in the cell viability among the strains as measured by the Live/Dead BacLight bacterial viability test (data not shown). Combined, these findings suggest that the E1 protein detected in the extracellular space was a result of protein translocation rather than increased levels of cell death and release of the E1 protein due to cell lysis.
In order to determine how well the Z130 and Z331 signal sequences were functioning to translocate the E1 protein to the periplasmic space, we examined the subcellular localization of E1. To achieve this, periplasmic and cytoplasmic fractions in each of the four strains were isolated and examined for the presence of E1. Figure 5C shows the amido black-stained PVDF membrane and represents the total protein in each subcellular fraction. An anti-E1 immunoblot of the membrane shown in Fig. 5D shows that the periplasmic fractions of the strains harboring the E1 secretion plasmids Z130-E1 and Z331-E1 contain more E1 than either the control or the strain harboring the control E1 construct without a secretion signal. The cellulolytic activity present in the cytoplasmic, periplasmic, and extracellular fractions was determined using the fluorescent substrate 4-methylumbelliferyl-β-d-cellobiopyranoside (MUC). The cellulolytic activity associated with the control E1 construct lacking a secretion signal was found almost exclusively in the cytoplasm (Fig. 5E). In contrast, however, a significant amount of the enzymatic activity in strains containing constructs Z130-E1and Z331-E1 was found in the periplasmic or extracellular spaces. Importantly, while Z331-E1 was undetectable in the extracellular fraction by immunoblotting (Fig. 5B), its activity was clearly detected extracellularly as measured by MUC hydrolysis (Fig. 5E).
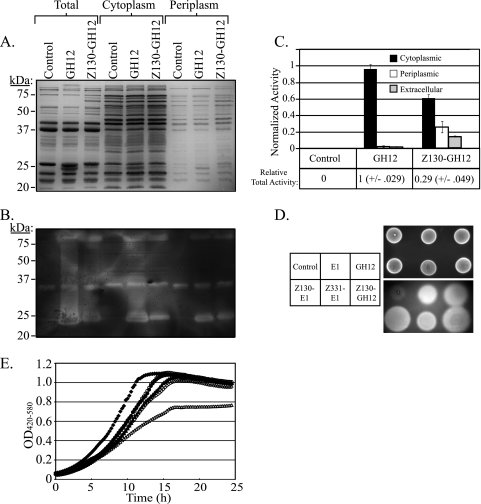
We also examined the subcellular localization of GH12 and GH12 fused with the Z130 secretion signal. An examination of the expression and activity of the subcellular pools of GH12 by Coomassie blue-stained SDS-PAGE (Fig. 6 A) and CMC zymogram analysis (Fig. 6B) reveals several important findings. First, it is clear that the total expression of Z130-GH12 is reduced compared to GH12 without the secretion signal (Fig. 6A, Total, GH12 and A130-GH12 lanes). Second, it appears that while both GH12 and Z130-GH12 show activity in the periplasmic fraction, compared to the activity found in the cytoplasmic or total fraction, Z130-GH12 contributes a relatively higher activity in the periplasmic fraction. As the data presented in Fig. 6A and B are qualitative by nature, we confirmed this result and quantified the activity through the measurement of MUC hydrolysis (Fig. 6C). The lack of a secretion signal resulted in 96% of GH12 activity being localized within the cytoplasm, while the addition of the Z130 signal resulted in a localization of 13% of the GH12 activity in the periplasm and 26% in the extracellular space. Furthermore, no ADH activity was detected in the medium, and cell viability associated with each culture was indistinguishable between the strains, suggesting that the extracellular pool of GH12 was in fact due to secretion, rather than heightened cell lysis. It is also important to note that in contrast to the results obtained with E1 (Fig. 5), the Z130 signal served to decrease the overall expression of GH12 as observed by Coomassie blue-stained polyacrylamide gel electrophoresis, CMC zymogram (Fig. 6A and B), and total activity analysis (Fig. 6C). Figure 6D qualitatively shows the extracellular CMC-degrading activities of all of the strains examined in Fig. 5 and 6. Culture spots shown in the top panel in Fig. 6D were washed from the plate, which was subsequently stained with Congo red to reveal zones of CMC degradation.
FIG. 6.
Extracellular secretion and subcellular localization of GH12 in Z. mobilis strain ATCC 39676. Whole-cell protein lysates and periplasmic and cytoplasmic fractions derived from Z. mobilis strain ATCC 39676 transformed with plasmids pZB188 (Control lanes), p25144 (GH12 lanes), and pJL116 (Z130-GH12 lanes) were run identically on two independent 12% polyacrylamide gels supplemented with 0.12% carboxymethyl cellulose (CMC). (A) Coomassie blue-stained gel to show total protein. (B) A CMC zymogram performed on the second of two duplicate polyacrylamide gels designed to show cellulolytic activity. (C) Relative quantification of GH12 activity against methylumbelliferyl cellobiopyranoside (MUC) in periplasmic, cytoplasmic, and extracellular fractions. The relative total activity of equivalent whole-cell lysates from the indicated strains is shown below the bar graph to highlight differential expression between the strains. (D) Z. mobilis strain ATCC 39676 transformed with plasmids pZB188 (Control), p25143 (E1), p25144 (GH12), pJL111 (Z130-E1), pJL112 (Z331-E1), and pJL116 (Z130-GH12) were spotted onto an agar plate containing 2% glucose and 0.12% CMC. After 18 h of anaerobic growth at 30°C, the plates were photographed (top panel), and the cells were washed off. The plate was subsequently stained with 0.2% Congo red, destained with 1 M NaCl, and photographed again to show CMC degradation (bottom panel). (E) Growth curve analysis of Z. mobilis strain ATCC 39676 in RMG medium harboring the following plasmids: pZB188 (control) (closed diamonds), p25143 (E1) (open diamonds), pJL111 (Z130-E1) (closed triangles), pJL112 (Z331-E1) (open triangles), p25144 (GH12) (closed circles), and pJL116 (Z130-GH12) (open circles).
In order to determine the overall effect of recombinant protein production and secretion, we examined the growth rate of Z. mobilis strain ATCC 39676 harboring the various expression constructs (Fig. 6E). The doubling times (in hours) for strains with the indicated plasmids were as follows in RMG medium: control (pZB188), 2.3 h ± 0.03; E1 (p25143) 2.7 h ± 0.00; Z130-E1 (pJL111), 2.83 h ± 0.51; Z331-E1 (pJL112), 3.06; GH12 (p25144), 2.82 ± 0.04; and Z130-GH12 (pJL116); 2.56 ± 0.07. These numbers indicate that while the expression of E1 and GH12 has a modest effect on the growth rate compared to the control strain, the addition of the Z130 signal peptide does little to inhibit growth in the case of E1 and actually enhances growth rate in the case of GH12. Furthermore, the addition of the Z331 signal peptide to E1 imparts a more detrimental effect on growth rate.
DISCUSSION
The results of our studies first revealed that Z. mobilis has the capacity to degrade carboxymethyl cellulose extracellularly (Fig. 1, 4, and 6). Rajnish et al. (30) identified the Z. mobilis celA gene (ZMO1086) that shows β-1,4-endocellulolytic activity when expressed in E. coli. However, these investigators were unable to detect any cellulolytic activity in Z. mobilis strain ZM4. We independently identified a native CM-cellulolytic activity present in Z. mobilis strains ATCC 39676, CP4, and ZM4. While the data presented in this report cannot definitively attribute the cellulolytic activity we observed to CelA, the bands showing activity had a molecular mass consistent with that predicted for CelA (Fig. 1). While it is possible that one of the bands showing CMC activity may represent an unidentified cellulase, we favor the hypothesis that the higher-molecular-mass band of the two bands (Fig. 1B) represents the full-length version of CelA, while the lower-molecular-mass band most likely represents the mature protein with its signal peptide removed. Our reason for believing this is based on subcellular fractionation experiments detailed in Fig. 6B which show that the higher-molecular-mass band of the two bands is the only band present in the cytoplasmic fractions, while both forms are present in the periplasmic fraction. As signal peptides are typically removed in the periplasm (34), this is exactly what would be expected if in fact the two bands represented the full-length protein (i.e., protein plus signal sequence) and the mature protein. While future identification of these proteins will be required to validate this hypothesis, cellulolytic activity against CMC was detected extracellularly in several Z. mobilis strains (Fig. 1C). This finding is consistent with the fact that CelA contains a predicted secretion signal at the N terminus (4). Together, these findings suggest that Z. mobilis may have evolved as a cellulose-degrading fermentative organism. Given the ability of Z. mobilis to produce at least one cellulase and potentially others (Fig. 1), there may be reason to believe that this organism may harbor an environment conducive to the production of additional heterologous cellulolytic enzymes.
We also demonstrated in this report the exogenous expression and extracellular secretion in Z. mobilis of two endo-1,4-β-glucanases from A. cellulolyticus useful in the degradation of pretreated lignocellulosic biomass. Importantly, both enzymes we heterologously expressed in Z. mobilis (E1 and GH12 from A. cellulolyticus) were found to be enzymatically active, yet any metabolic burden of their expression resulted in minor changes in the growth rate of Z. mobilis. While we show an approximately 20% reduction in the logarithmic growth rate of cells expressing either E1 and GH12 compared to a Z. mobilis control strain, fusing the Z130 secretion signal onto these genes does not seem to impart major additional limitations. Conversely, cultures harboring the Z331-E1 construct have a much larger growth limitation. Our data, taken together with the results of the study by Arfman et al. (2) suggest that Z. mobilis is capable of handling high levels of additional protein expression before major growth rate limitations occur. This is an important finding, as high-level expression of cellulolytic enzymes will be required for the future development of Z. mobilis as a CBP organism, and future efforts should be made to minimize this defect.
The robust levels of expression observed in this study using the GH12 construct certainly warrant further investigation. Not only was Z. mobilis able to produce GH12 at levels approaching 5% of the total cellular protein, but this level of expression was stable throughout the logarithmic and stationary phases of growth. The expression plasmids used in this study containing the Ptac promoter lacked the lacI gene, so the cellulase genes were being expressed constitutively without the need for isopropyl-β-d-thiogalactopyranoside (IPTG) induction. Constitutive cellulase expression will be an important trait for a CBP organism, and the fact that GH12 can be expressed constitutively to a high level in a largely soluble and enzymatically active form provides an optimistic outlook for developing Z. mobilis as a CBP host.
We further show that Z. mobilis is capable of translocating both E1 and GH12 through the periplasmic space into the extracellular medium when the genes encoding these cellulases are fused with the previously uncharacterized N-terminal secretion signals Z130 and Z331. The Z130 and Z331 signal sequences presumably direct the translocation of proteins via the SecB-dependent and TAT secretory pathways, respectively. As the N-terminal signals used by both secretion pathways function only to translocate the protein to the periplasmic space (18, 34), it is very significant that when fused to E1 and GH12, nearly 50% and 40% of the respective protein translocates through the inner membrane to reside either periplasmically or extracellularly (Fig. 5 and 6). An interesting observation is that while the presence of the Z130 secretion signal resulted in increased expression of the E1 enzyme, it resulted in decreased expression of GH12 (Fig. 5 and 6). The reasons for this are unclear, but one possibility is that perhaps the more oxidizing periplasmic environment (28) favors the proper folding of E1, thus increasing its stability. Regarding GH12, it is possible that since this protein is expressed to such a high level (Fig. 4 and 6), perhaps the incorporation of the secretion signal is overloading the secretion pathway at some level, and GH12 is being degraded at a higher rate. A mechanistic hypothesis supposes that perhaps SecB is binding to the N-terminal signal sequence, keeping GH12 in its unfolded state, yet the overburdening of the translocase complex is slowing the rate of periplasmic translocation, thus leaving GH12 more susceptible to cytoplasmic degradation. Another possibility is that GH12 is being efficiently translocated to the periplasmic space, yet periplasmic proteases are degrading the protein at a higher rate than occurs cytoplasmically. A third possibility is that the addition of the signal sequence itself reduces the translation rate of the GH12 protein.
While the levels of E1 and GH12 secretion demonstrated here certainly represent a significant step toward achieving high-level secretion, the fact remains that in the realm of developing a CBP technology, any cellulolytic enzyme that remains localized within a cell is essentially wasted regarding the degradation of extracellular biomass. Future efforts will be directed to increasing the secretory capacity of Z. mobilis regarding cellulolytic enzymes along three fronts: (i) increasing the translocation through the inner membrane to the periplasm, (ii) increasing the translocation through the outer membrane to the extracellular space, and (iii) exploring the use of alternate secretion pathways, such as the type I secretion pathway which bypasses the two-step secretion mechanism and instead translocates proteins across both membranes (26).
Ultimately, several classes of cellulolytic enzymes will likely need to be coexpressed in order to effectively break down lignocellulosic biomass. At a minimum, we expect that the simultaneous expression of an endoglucanase, an exoglucanase, and a β-glucosidase would be required (13), although the expression of additional enzymes will also quite likely be necessary. We have shown here the effective production and secretion of two endo-1,4-β-glucanases, and future work will need to address the ability of Z. mobilis to express the other two classes of enzymes.
We have shown that Z. mobilis can effectively express and secrete heterologously expressed proteins, and while we personally envision this being useful in developing Z. mobilis into a CBP organism, this could theoretically open up a new pipeline for the production of other classes of valuable proteins. E. coli is one of several organisms that have long been favored for high-level production of recombinant proteins. However, despite extensive knowledge about recombinant protein production in E. coli, not all expression attempts have been successful. Furthermore, E. coli is notoriously poor at secreting heterologous proteins (23), which can be an advantageous route in the expression of recombinant proteins. The ability of Z. mobilis to express and secrete recombinant proteins may represent a complementary pathway to E. coli-based expression systems and may prove valuable to researchers in all disciplines requiring recombinant protein production that is unsuccessful using E. coli.
Z. mobilis has proven to be an extremely valuable organism in the conversion of biomass-derived sugars to ethanol and is currently being developed into a commercial fermentation host as part of an integrated lignocellulosic ethanol process. The data presented in this report show the potential for Z. mobilis to extend its usefulness beyond its fermentative abilities and to play a significant role in the degradation of pretreated lignocellulosic biomass. Given the proven adeptness of Z. mobilis in industrial-scale fermentation, the ability to express high levels of active cellulases shown in this study and the capacity to secrete these enzymes, the foundation has been laid for further investigations in the establishment of Z. mobilis as a CBP organism. While CBP might be considered the ultimate goal, any significant production of cellulolytic enzymes by Z. mobilis could be considered a success, as this would reduce the required loading levels of exogenously produced cellulases during simultaneous saccharification and fermentation or cofermentation, processes in which Z. mobilis has already been tried and tested (14, 24, 36).
Acknowledgments
We gratefully thank Min Zhang, Larry Taylor, Yat-Chen Chou, and Mary Ann Franden for technical advice and invaluable discussions and Philip Pienkos and Min Zhang for a critical review of the manuscript.
Footnotes
Published ahead of print on 6 August 2010.
REFERENCES
- 1.Aldrich, H. C., L. McDowell, M. F. Barbosa, L. P. Yomano, R. K. Scopes, and L. O. Ingram. 1992. Immunocytochemical localization of glycolytic and fermentative enzymes in Zymomonas mobilis. J. Bacteriol. 174:4504-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arfman, N., V. Worrell, and L. O. Ingram. 1992. Use of the tac promoter and lacIq for the controlled expression of Zymomonas mobilis fermentative genes in Escherichia coli and Zymomonas mobilis. J. Bacteriol. 174:7370-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195-201. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Brestic-Goachet, N., P. Gunasekaran, B. Cami, and J. Baratti. 1990. Transfer and expression of a Bacillus licheniformis alpha-amylase gene in Zymomonas mobilis. Arch. Microbiol. 153:219-225. [Google Scholar]
- 6.Brestic-Goachet, N., P. Gunasekaran, B. Cami, and J. C. Baratti. 1989. Transfer and expression of an Erwinia chrysanthemi cellulase gene in Zymomonas mobilis. J. Gen. Microbiol. 135:893-902. [Google Scholar]
- 7.Byun, M. O. K., J. B. Kaper, and L. O. Ingram. 1986. Construction of a new vector for the expression of foreign genes in Zymomonas mobilis. J. Ind. Microbiol. 1:9-15. [Google Scholar]
- 8.Carere, C. R., R. Sparling, N. Cicek, and D. B. Levin. 2008. Third generation biofuels via direct cellulose fermentation. Int. J. Mol. Sci. 9:1342-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carey, V. C., S. K. Walia, and L. O. Ingram. 1983. Expression of a lactose transposon (Tn951) in Zymomonas mobilis. Appl. Environ. Microbiol. 46:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deanda, K., M. Zhang, C. Eddy, and S. Picataggio. 1996. Development of an arabinose-fermenting Zymomonas mobilis strain by metabolic pathway engineering. Appl. Environ. Microbiol. 62:4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Ding, S. Y., W. S. Adney, T. B. Vinzant, M. E. Himmel, and S. R. Decker. June 2006. Thermal tolerant cellulase from Acidothermus cellulolyticus. U.S. patent 7,059,993.
- 11.Franden, M. A., P. T. Pienkos, and M. Zhang. 2009. Development of a high-throughput method to evaluate the impact of inhibitory compounds from lignocellulosic hydrolysates on the growth of Zymomonas mobilis. J. Biotechnol. 144:259-267. [DOI] [PubMed] [Google Scholar]
- 12.Himmel, M. E., S. Y. Ding, D. K. Johnson, W. S. Adney, M. R. Nimlos, J. W. Brady, and T. D. Foust. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804-807. [DOI] [PubMed] [Google Scholar]
- 13.Kumar, R., S. Singh, and O. V. Singh. 2008. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J. Ind. Microbiol. Biotechnol. 35:377-391. [DOI] [PubMed] [Google Scholar]
- 14.Lee, J. H., R. J. Pagan, and P. L. Rogers. 1983. Continuous simultaneous saccharification and fermentation of starch using Zymomonas mobilis. Biotechnol. Bioeng. 25:659-669. [DOI] [PubMed] [Google Scholar]
- 15.Lee, K. J., M. Lefebvre, D. E. Tribe, and P. L. Rogers. 1980. High productivity ethanol fermentations with Zymomonas mobilis using continuous cell recycle. Biotechnol. Lett. 2:487-492. [Google Scholar]
- 16.Lee, K. J., M. L. Skotnicki, D. E. Tribe, and P. L. Rogers. 1980. Kinetic studies on a highly productive strain of Zymomonas mobilis. Biotechnol. Lett. 2:339-344. [Google Scholar]
- 17.Lee, K. J., D. E. Tribe, and P. L. Rogers. 1979. Ethanol production by Zymomonas mobilis in continuous culture at high glucose concentrations. Biotechnol. Lett. 1:421-426. [Google Scholar]
- 18.Lee, P. A., D. Tullman-Ercek, and G. Georgiou. 2006. The bacterial twin-arginine translocation pathway. Annu. Rev. Microbiol. 60:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lejeune, A., D. E. Eveleigh, and C. Colson. 1988. Expression of an endoglucanase gene of Pseudomonas fluorescens var. cellulosa in Zymomonas mobilis. FEMS Microbiol. Lett. 49:363-366. [Google Scholar]
- 20.Lynd, L. R. 1996. Overview and evaluation of fuel ethanol from cellulosic biomass: technology, economics, the environment, and policy. Annu. Rev. Energy Environ. 21:403-465. [Google Scholar]
- 21.Lynd, L. R., W. H. van Zyl, J. E. McBride, and M. Laser. 2005. Consolidated bioprocessing of cellulosic biomass: an update. Curr. Opin. Biotechnol. 16:577-583. [DOI] [PubMed] [Google Scholar]
- 22.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makrides, S. C. 1996. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 60:512-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMillan, J. D., M. M. Newman, D. W. Templeton, and A. Mohagheghi. 1999. Simultaneous saccharification and cofermentation of dilute-acid pretreated yellow poplar hardwood to ethanol using xylose-fermenting Zymomonas mobilis. Appl. Biochem. Biotechnol. 77-79:649-665. [DOI] [PubMed] [Google Scholar]
- 25.Mejia, J. P., M. E. Burnett, H. An, W. O. Barnell, K. F. Keshav, T. Conway, and L. O. Ingram. 1992. Coordination of expression of Zymomonas mobilis glycolytic and fermentative enzymes: a simple hypothesis based on mRNA stability. J. Bacteriol. 174:6438-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mergulhao, F. J., D. K. Summers, and G. A. Monteiro. 2005. Recombinant protein secretion in Escherichia coli. Biotechnol. Adv. 23:177-202. [DOI] [PubMed] [Google Scholar]
- 27.Misawa, N., T. Okamoto, and K. Nakamura. 1988. Expression of a cellulase gene in Zymomonas mobilis. J. Biotechnol. 7:167-178. [Google Scholar]
- 28.Missiakas, D., and S. Raina. 1997. Protein folding in the bacterial periplasm. J. Bacteriol. 179:2465-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Omullan, P. J., T. Chase, and D. E. Eveleigh. 1992. Purification and some properties of extracellular invertase-B from Zymomonas mobilis. Appl. Microbiol. Biotechnol. 38:341-346. [Google Scholar]
- 30.Rajnish, K. N., G. M. K. Choudhary, and P. Gunasekaran. 2008. Functional characterization of a putative endoglucanase gene in the genome of Zymomonas mobilis. Biotechnol. Lett. 30:1461-1467. [DOI] [PubMed] [Google Scholar]
- 31.Rogers, P. L., Y. J. Jeon, K. J. Lee, and H. G. Lawford. 2007. Zymomonas mobilis for fuel ethanol and higher value products. Adv. Biochem. Eng. Biotechnol. 108:263-288. [DOI] [PubMed] [Google Scholar]
- 32.Rogers, P. L., K. J. Lee, and D. E. Tribe. 1980. High productivity ethanol fermentations with Zymomonas mobilis. Process Biochem. 15:7-11. [Google Scholar]
- 33.Rogers, P. L., K. J. Lee, and D. E. Tribe. 1979. Kinetics of alcohol production by Zymomonas mobilis at high sugar concentrations. Biotechnol. Lett. 1:165-170. [Google Scholar]
- 34.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 35.Skotnicki, M. L., K. J. Lee, D. E. Tribe, and P. L. Rogers. 1981. Comparison of ethanol production by different Zymomonas strains. Appl. Environ. Microbiol. 41:889-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spangler, D. J., and G. H. Emert. 1986. Simultaneous saccharification fermentation with Zymomonas mobilis. Biotechnol. Bioeng. 28:115-118. [DOI] [PubMed] [Google Scholar]
- 37.Swings, J., and J. Deley. 1977. Biology of Zymomonas. Bacteriol. Rev. 41:1-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, L. E., II, B. Henrissat, P. M. Coutinho, N. A. Ekborg, S. W. Hutcheson, and R. M. Weiner. 2006. Complete cellulase system in the marine bacterium Saccharophagus degradans strain 2-40T. J. Bacteriol. 188:3849-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Thomas, S. R., R. A. Laymon, and M. E. Himmel. July 1996. Gene coding for the E1 endoglucanase. U.S. patent 5,536,655.
- 39.Villalobos, A., J. E. Ness, C. Gustafsson, J. Minshull, and S. Govindarajan. 2006. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics 7:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss, A., C. Klein, B. Woodman, K. Sathasivam, M. Bibel, E. Regulier, G. P. Bates, and P. Paganetti. 2008. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington's disease. J. Neurochem. 104:846-858. [DOI] [PubMed] [Google Scholar]
- 41.Yablonsky, M. D., A. E. Goodman, N. Stevnsborg, O. G. Delima, J. O. F. Demorais, H. G. Lawford, P. L. Rogers, and D. E. Eveleigh. 1988. Zymomonas mobilis CP4: a clarification of strains via plasmid profiles. J. Biotechnol. 9:71-79. [Google Scholar]
- 42.Yanase, H., J. Kurii, and K. Tonomura. 1986. Construction of a promoter-cloning vector in Zymomonas mobilis. Agric. Biol. Chem. 50:2959-2961. [Google Scholar]
- 43.Yanase, H., K. Nozaki, and K. Okamoto. 2005. Ethanol production from cellulosic materials by genetically engineered Zymomonas mobilis. Biotechnol. Lett. 27:259-263. [DOI] [PubMed] [Google Scholar]
- 44.Yoon, K. H., S. H. Park, and M. Y. Pack. 1988. Transfer of Bacillus subtilis endo-β-1,4-glucanase gene into Zymomonas anaerobia. Biotechnol. Lett. 10:213-216. [Google Scholar]
- 45.Zhang, M., C. Eddy, K. Deanda, M. Finkestein, and S. Picataggio. 1995. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267:240-243. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, M., M. A. Franden, M. Newman, J. Mcmillan, M. Finkelstein, and S. Picataggio. 1995. Promising ethanologens for xylose fermentation. Appl. Biochem. Biotechnol. 51-52:527-536. [Google Scholar]