Abstract
The genotyping of numerous isolates of Cryptosporidium parasites has led to the definition of new species and a better understanding of the epidemiology of cryptosporidiosis. A single-locus genotyping method based on the partial sequence of a polymorphic sporozoite surface glycoprotein gene (GP60) has been favored by many for surveying Cryptosporidium parvum and C. hominis populations. Since genetically distinct Cryptosporidium parasites recombine in nature, it is unclear whether single-locus classifications can adequately represent intraspecies diversity. To address this question, we investigated whether multilocus genotypes of C. parvum and C. hominis cluster according to the GP60 genotype. C. hominis multilocus genotypes did not segregate according to this marker, indicating that for this species the GP60 sequence is not a valid surrogate for multilocus typing methods. In contrast, in C. parvum the previously described “anthroponotic” genotype was confirmed as a genetically distinct subspecies cluster characterized by a diagnostic GP60 allele. However, as in C. hominis, several C. parvum GP60 alleles did not correlate with distinct subpopulations. Given the rarity of some C. parvum GP60 alleles in our sample, the existence of additional C. parvum subgroups with unique GP60 alleles cannot be ruled out. We conclude that with the exception of genotypically distinct C. parvum subgroups, multilocus genotyping methods are needed to characterize C. parvum and C. hominis populations. Unless parasite virulence is controlled at the GP60 locus, attempts to find associations within species or subspecies between GP60 and phenotype are unlikely to be successful.
The lack of variable morphological traits to identify oocysts from different Cryptosporidium species has driven the development of numerous genotyping methods to survey the diversity in this genus. Genetic markers such as single-nucleotide polymorphisms (24), restriction fragment length polymorphisms (7, 34), random amplification methods (17, 20), conformational polymorphisms (11), simple sequence repeats (3, 10), and DNA sequence polymorphisms (6, 36) have been used to type Cryptosporidium oocysts excreted by humans and animals and oocysts recovered from the environment. This effort has led to a deeper understanding of the taxonomy of the genus Cryptosporidium and the epidemiology of cryptosporidiosis in humans and livestock. As a result of this work, two species responsible for a majority of human infections, Cryptosporidium parvum and C. hominis, were identified (21) and our understanding of the taxonomy of the genus was refined (35).
The application of genetic markers to define species, i.e., reproductively isolated populations, is straightforward. At this taxonomic level, all genotypes cosegregate and the choice of marker will have little impact on the outcome, provided that the marker, or combination thereof, is sufficiently polymorphic. The classical example is the variable region of the small-subunit rRNA gene which has been used, as in other taxa, to define many Cryptosporidium species. For studying intraspecies polymorphism, the choice of genotyping methods needs to take into consideration the potential for genetic recombination. This is clearly the case for species such as those belonging to the genus Cryptosporidium, which are known to undergo an obligatory sexual cycle during which genetically dissimilar haplotypes can recombine (28).
Among the many markers that have been applied in epidemiological surveys of C. parvum and C. hominis, a variable fragment of the gene encoding a sporozoite surface glycoprotein (8, 26) has been particularly popular. As a result of the widespread adoption of this marker, variously named GP60, cpgp40/15, or gp40, numerous alleles have been identified and deposited in GenBank. The analysis of this continuously growing collection of GP60 sequences has led to the identification of groups of related sequences (18, 27, 32, 33). In an attempt to simplify the comparison of GP60 genotypes among different laboratories, a GP60 nomenclature distinguishing the main groups of alleles has been created (26) and later refined (27).
The desire to streamline the genotyping of large numbers of Cryptosporidium isolates collected during surveys has led to the widespread adoption of the GP60 genotype as the only marker for defining intraspecies groups. Since this approach is not compatible with the reassortment of unlinked loci, the classification of isolates on the basis of the GP60 genotype, or any other single marker, needs to be evaluated. Within a recombining population, no single genetic marker can a priori be expected to serve as a surrogate for other loci or multilocus genotypes (MLGs), and any apparent clustering of isolates is dependent on the marker. To investigate the validity of the GP60 genotyping method as commonly applied to the classification of C. parvum and C. hominis isolates, the GP60 genotype was added to a previously described 9-locus genotype (29) and a diversified collection of 10-locus genotypes was examined for intraspecies clusters. We show that, with the exception of some GP60 alleles apparently restricted to human C. parvum, neither C. parvum nor C. hominis GP60 alleles define subspecies genotypes. These results are discussed in the context of ongoing research to better understand the population structure of these parasites and identify genotypes associated with virulence traits.
MATERIALS AND METHODS
Laboratory methods.
The C. parvum and C. hominis isolates included in the analyses are listed in Table 1. These isolates were selected from a larger study examining the geographic structure of numerous C. parvum and C. hominis 9-locus genotypes (29). The isolates included here were selected from this larger collection based on the availability of DNA needed to amplify and sequence a portion of the GP60 gene or the availability of the diagnostic GP60 fragment in GenBank. This sequence was amplified using a nested PCR protocol with external primers gp15ATG and gp15STOP (26) and internal primers 15A and 15E (19). Amplicons were purified using a MiniElute PCR purification kit (Qiagen) and sequenced in both directions using the internal primers.
TABLE 1.
C. parvum and C. hominis isolates
| Isolatea | Species | GP60 allele | MLGb | Referencec |
|---|---|---|---|---|
| UG996 | C. hominis | Ia | 7 | 29 |
| UG1481 | C. hominis | Ia | 16 | 29 |
| UG285 | C. hominis | Ia | 57 | 29 |
| UG94 | C. hominis | Ia | 76 | 29 |
| UK3004ft | C. hominis | Ia | 78 | 29 |
| UK3334ft | C. hominis | Ia | 79 | 29 |
| UK3555ft | C. hominis | Ia | 80 | 29 |
| UK3351 | C. hominis | Ia | 81 | 29 |
| UGTU502 | C. hominis | Ia | 52 | 29 |
| UK3000 | C. hominis | Ib | 22 | 29 |
| UK2696 | C. hominis | Ib | 23 | 29 |
| UK3812 | C. hominis | Ib | 24 | 29 |
| UK4498 | C. hominis | Ib | 27 | 29 |
| UK3871 | C. hominis | Ib | 30 | 29 |
| UK4691 | C. hominis | Ib | 25 | 29 |
| UK4733 | C. hominis | Ib | 33 | 29 |
| UK4771 | C. hominis | Ib | 26 | 29 |
| UK11766 | C. hominis | Ib | 1 | 29 |
| USNEMC1 | C. hominis | Ib | 52 | 26 |
| US9897 | C. hominis | Ib | 73 | 26 |
| USECHIV | C. hominis | Ib | 75 | Unpublished |
| UG918 | C. hominis | Ie | 5 | 29 |
| UG1467 | C. hominis | Ie | 6 | 29 |
| UK5139 | C. hominis | Ie | 32 | 29 |
| USTU728 | C. hominis | Ie | 19 | Unpublished |
| UK4938 | C. hominis | If | 37 | 29 |
| UK4776 | C. hominis | If | 36 | 29 |
| UK3010 | C. hominis | Ig | 3 | 29 |
| UK3957 | C. hominis | Ig | 2 | 29 |
| UG26 | C. hominis | IIc | 43 | 29 |
| TR13 | C. parvum | IIa | 21 | 30 |
| IL-7 | C. parvum | IIa | 32 | 30 |
| TR39 | C. parvum | IIa | 36 | 30 |
| IL-3 | C. parvum | IIa | 44 | 30 |
| IL-8 | C. parvum | IIa | 47 | 30 |
| IL-13 | C. parvum | IIa | 49 | 30 |
| IL-15 | C. parvum | IIa | 51 | 30 |
| IL-1 | C. parvum | IIa | 37 | 30 |
| NZHhu1 | C. parvum | IIa | 40 | 29 |
| NZHhu6 | C. parvum | IIa | 41 | 29 |
| NZHbo2 | C. parvum | IIa | 42 | 29 |
| SRB6 | C. parvum | IIa | 56 | 29 |
| SRB80 | C. parvum | IIa | 58 | 29 |
| SRB112 | C. parvum | IIa | 64 | 29 |
| USiowa | C. parvum | IIa | 50 | 1 |
| UKMD | C. parvum | IIa | 39 | 6 |
| UG1224 | C. parvum | IIc | 1 | 29 |
| UG2428 | C. parvum | IIc | 7 | 29 |
| UG30 | C. parvum | IIc | 13 | 29 |
| UG2553 | C. parvum | IIc | 18 | 29 |
| UG1504 | C. parvum | IIc | 54 | 29 |
| UGtu114 | C. parvum | IIc | 2 | 29 |
| UG1601 | C. parvum | IIc | 15 | 29 |
| UG9 | C. parvum | IIc | 8 | 29 |
| SRB42 | C. parvum | IId | 66 | 29 |
| UG1862 | C. parvum | IIe | 25 | 29 |
| UG216 | C. parvum | IIi | 23 | 29 |
| UGtu154 | C. parvum | IIi | 22 | 29 |
| SRB24 | C. parvum | IIj | 68 | 29 |
| SRB58 | C. parvum | IIj | 74 | 29 |
The first 2 or 3 letters indicate the country of origin: IL, Israel; NZ, New Zealand; SRB, Serbia; UG, Uganda; UK, United Kingdom; US, United States; TR, Turkey. ft, sample originated from person reporting recent travel outside the United Kingdom.
Arbitrary code assigned to 9-locus MLG; see report by Tanriverdi et al. (29) for actual MLG.
Except for isolates from the United States, all isolates were described by Tanriverdi et al. (29), where the host and geographical origin are described in more detail. The references shown here indicate where the isolate was first described.
The GP60 allele was defined as the number of nucleotides present in the sequence corresponding to nucleotide positions 81 to 357 of the C. parvum GP60 allele from isolate IOWA (cryptoDB, gene identifier cgd_1080). The sequence initiates with a 5′GAG(A/G)G motif and terminates with 3′TGCGG(C/T). The length of the amplicon varies due to the presence of a polymorphic trinucleotide repeat encoding a polyserine tract and an additional polymorphic 13- to 15-bp minisatellite. It is understood that scoring GP60 alleles according to amplicon length ignores nucleotide substitutions, but the approach is consistent with that used for scoring the other 9 markers. The polymorphism of those 9 markers is based exclusively on amplicon length, as previously described (29). Minisatellite MS9 and microsatellite TP14 were developed by Mallon et al. (19). The other 7 markers were part of our previously published multilocus typing method (30).
The GP60 sequences of isolates SRB6, SRB24, SRB42, SRB58, SRB80, SRB112, USIOWA, USNEMC1, US9897, UGTU502, UK3004, UK3010, UK3334, UK3351, UK3555, UK3957, UK4498, UK4691, UK4771, UK4776, and UK11766 were downloaded from GenBank or cryptoDB.org and were not resequenced for this study. The numeric code of the isolates from the United Kingdom was retained, but for consistency the letter “W” was replaced with the country code “UK.” Duplicate isolates displaying the same allele at each of the 10 loci were eliminated to avoid the creation of artifactual clusters of identical multilocus genotypes.
Data analysis.
The GP60 allelic group, sometimes referred to as “subtype family” or “subgenotype,” starting with a roman number indicating the species (I, C. hominis; II, C. parvum), was identified by searching GenBank for the Cryptosporidium sequence with the highest similarity. The length in base pairs (bp) of the above-defined fragment was recorded for each isolate and was used as the only identification of the GP60 allele. Together with the estimated amplicon length of the 9 previously described minisatellite and microsatellite markers (29, 30), this information generated the 10-locus genotype for the 60 isolates shown in Table 1. The GP60 allele nomenclature first proposed by Strong et al. (26) and later amended (27) was adopted.
Pairwise genetic distances were determined using GenAlEx (23). For each pair of 10-locus multilocus genotypes, the haploid and simple sequence repeat (SSR) distances were calculated. The former distance metric is analogous to the Hamming distance (15) and is equal to the number of loci with alleles of unequal length. For the SSR distance, the length difference between pairs of alleles is first determined for each pairwise comparison, and the square of the differences is summed over all loci. A distance matrix was produced for each distance metric and input into the GenAlEx Principal Coordinate Analysis (PCoA) program.
The presence of any significant association of alleles between pairs of loci, i.e., linkage disequilibrium (LD), was tested using Arlequin software (9). For each pair of loci, the program computes a contingency table with all observed combination of alleles (2-locus haplotypes). Similar to a Fisher exact test, the program calculates the probability of obtaining the observed table under the null hypothesis of no association (linkage equilibrium) between loci. The program outputs a table showing pairs of loci with significant linkage disequilibrium, using a P value cutoff of 0.05.
Nucleotide sequence accession numbers.
Newly sequenced GP60 alleles were deposited in GenBank under accession numbers HM365222 to HM365235 and HM370430 to HM370451.
RESULTS
We scored GP60 alleles according to amplicon length, instead of the commonly used alphanumeric code (26), because this scoring method is compatible with the SSR distance calculation, which is based on length differences. Because alleles at the other 9 loci are also defined on the basis of amplicon length, adopting this scoring method for GP60 alleles ensured uniformity across the 10 markers. This coding method reduced diversity to some extent, as mutations which do not affect length are ignored.
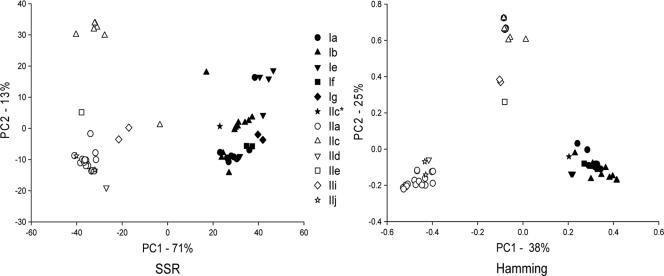
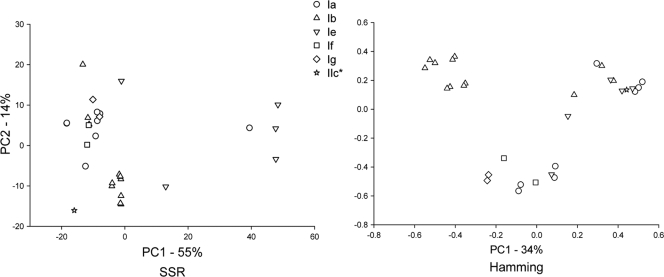
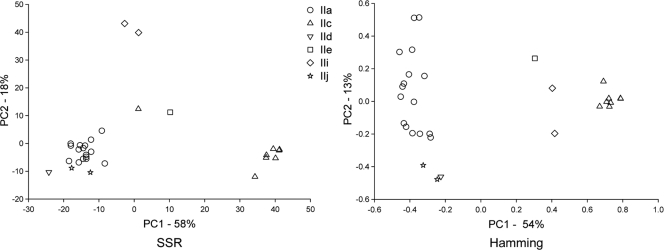
PCoA analysis of 60 multilocus genotypes (MLGs) (30 each for C. hominis and C. parvum) based on 10 polymorphic loci revealed three well-defined clusters (Fig. 1). These clusters correspond to the species C. hominis and two C. parvum subgroups, one comprising only isolates with GP60 allele IIc, the other including isolates with five GP60 alleles. This pattern was not affected by the distance metric, although the relative position of three isolates (one with an IIe GP60 genotype and two with an IIi GP60 genotype) was ambiguous. These isolates appear to cluster with the IIc group when the Hamming distance is used but are closer to the large C. parvum group when the SSR distance was applied. Except for the C. parvum cluster defined by the IIc GP60 allele, no other GP60 genotype appeared to be associated with subspecies clusters. To increase the resolution of the PCoA, each species was analyzed individually. The result of this analysis for C. hominis is shown in Fig. 2. As above, PCoA was performed in duplicate using the SSR and Hamming distance calculations. No GP60-specific clusters were apparent in C. hominis, indicating that in this species the GP60 genotype is not associated with MLG. This finding was not contingent upon the distance metric. In C. parvum, as seen in Fig. 1, the segregation of MLGs in two subgroups was again observed, one subgroup including only MLGs displaying the IIc allele, the other comprising MLGs associated with three GP60 genotypes (Fig. 3). Three isolates with IIe and IIi GP60 genotypes (UG1862_IIe, UG216_IIi, UGtu154_IIi) did not appear to be associated with either cluster. Given the small number of these isolated MLGs, it is unclear if these represent additional C. parvum subgroups, a possibility which is consistent with the apparent absence of these alleles from animals (25, 33). Based on these analyses, we conclude that with the exception of MLGs carrying the IIc allele, none of the C. parvum MLGs cluster according to the GP60 genotype.
FIG. 1.
PCoA of 60 unique C. parvum and C. hominis MLGs. GP60 alleles are colored according to species: white, C. parvum; black, C. hominis. Symbols represent alleles as indicated in the embedded legend. The C. hominis MLG with a IIc allele is shown with a black star and an asterisk in the legend. PCoA is based on SSR genetic distance (left) and Hamming distance (right). The percentages variation explained by the 1st and 2nd axes are indicated.
FIG. 2.
C. hominis PCoA. The symbols represent different GP60 genotypes, as shown in the symbol legend. PCoA is based on SSR genetic distance (left) and Hamming distance (right). Note the overlapping distribution of MLGs with different GP60 genotypes. See the legend to Fig. 1 for additional details.
FIG. 3.
C. parvum PCoA. As in Fig. 1 and 2, PCoA is based on SSR genetic distance (left) and Hamming distance (right). Note the tight cluster of MLGs with the IIc GP60 allele but no apparent segregation of other MLGs. See the legend to Fig. 1 for details.
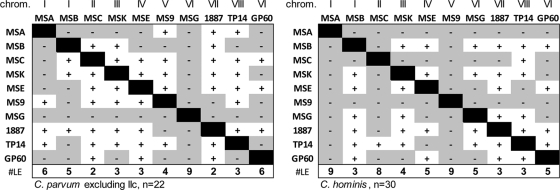
The results from the PCoA imply that, with the exception of IIc, the same GP60 genotype occurs in different MLGs. To verify this finding, we tested for LD among all pairs of loci and compared the numbers of markers in LD across the 10 loci. If GP60 alleles were nonrandomly associated with MLG, the number of loci in LD with GP60 would exceed that for other pairs of loci. In the first LD analysis, all 30 C. parvum isolates were included. Consistent with the segregation of C. parvum into two subgroups, 44/45 pairs of markers were in LD (P < 0.05), with MSA/MSB being the only pair of markers in linkage equilibrium. If the 8 C. parvum isolates with the IIc GP60 genotype were excluded, the number of pairs in LD dropped to 23 (Fig. 4). Significantly, GP60 was in equilibrium with 6 of 9 loci. The number of unlinked pairs of loci for the non-GP60 markers ranged from 2 to 9 (mean, 4.3). The LD analysis for 30 C. hominis MLGs identified 18 of 45 marker pairs in LD. Five of nine markers were in equilibrium with GP60, a number which is close to the mean of 5.4 (range 3 to 9) for this species. Note that the fact that two pairs of markers are located on the same chromosome (MSA/MSB, MSG/GP60) did not increase LD, as expected from the large genomic distance separating these markers (440,000 bp and 140,000 bp, respectively). Consistent with PCoA, LD statistics thus do not indicate that GP60 alleles are linked with MLGs. With the IIc exception noted above, the GP60 genotypes appear in combination with other alleles at other loci, which is consistent with genetic recombination among all loci.
FIG. 4.
Pairwise linkage analysis. Pairs of markers in LD are represented with white squares and +. Linkage equilibrium is indicated with a shaded field and −. Column totals in the bottom row show for each marker the number of markers in linkage equilibrium. The chromosome location of each marker is shown at the top. Two pairs of markers are located on chromosome I and VI. The distance between MSA and MSB is more than 400,000 bp and between MSG and GP60 is over 140,000 bp. Left, C. parvum excluding IIc MLGs (n = 22); right, C. hominis (n = 30).
DISCUSSION
PCoA and LD analysis demonstrate that the GP60 genotype is not a substitute for MLGs. As to be expected in sexually reproducing species, genetic recombination generates a multitude of MLGs in which GP60 alleles are associated with different alleles at other loci. This means that single-locus classifications of isolates belonging to the same species, or the same subspecies, are normally not reproducible but a function of the marker. This conclusion also applies to GP60 alleles and is independent of any coding function the marker may have or the level of polymorphism. Terms like “subfamily” or “subgenotype” frequently used in the literature in reference to GP60 alleles could thus be misleading if used as proxies for “multilocus genotype,” particularly when used to classify isolates within a panmictic population. This conclusion does not imply that the GP60 sequence is not useful in molecular epidemiology. The GP60 gene encodes an antigenically important glycoprotein (8, 26), and there is little doubt that this gene is under positive selection. The GP60 glycoprotein by itself, or in association with other proteins, may thus define or contribute to measurable parasite phenotypes, particularly in the context of host-parasite interaction. However, given that the C. parvum and C. hominis genome encodes some 3,800 proteins (1), the likelihood that the GP60 gene by itself will emerge as a marker of a measurable virulence phenotype seems small. Results from Peru showing an association between the GP60 genotype and virulence (4, 5) may originate from local substructuring of the parasite population and may not apply to other geographical areas where GP60 alleles may be found in association with other MLGs. This view is supported by a recent study from India which failed to identify an association between clinical phenotype and the GP60 genotype (2). This reasoning obviously does not apply to the IIc allele or other C. parvum GP60 alleles (IIe and some rare alleles) which apparently occur only in C. parvum found in humans (25, 33).
Although not central to the topic discussed here, the apparent emergence in C. parvum of one or several genetically distinct subpopulations defined by GP60 alleles IIc, IIe, etc., raises interesting questions regarding the evolution of these parasites. We do not wish to speculate here on the origin of this subgroup(s), but it is relevant to note in this context that two isolates originating from the IIc and non-IIc C. parvum subgroups (UKMD and UGtu114) readily recombine in the laboratory (28) and thus belong to the same species. Identifying selective forces that have created and maintained this C. parvum population structure may shed light on the evolution of these parasites and the extent to which host adaptation plays in the emergence of new species. The occurrence of the IIc GP60 genotype in one C. hominis isolate (UG26) is intriguing and raises the possibility of occasional C. parvum-C. hominis hybridization.
The isolates selected for the present analyses originate from different countries (Table 1). However, no assumption of geographic randomness is made, and for the analyses presented here geographic origin is not relevant. The only criterion for inclusion of MLGs in the analyses is that each 10-locus MLG be unique; geographic origin is not a factor.
The PCoA was carried out in duplicate with two distance metrics. The similarity in outcome indicates that the results are robust and not dependent on a specific distance metric. The SSR distance has the advantage of taking into consideration the evolutionary distance between alleles such that alleles which differ in length by multiple repeat units will have more weight than those differing by a single repeat unit. On the other hand, microsatellites with short 2- and 3-bp repeats, specifically markers 1887 and TP14, weigh less in the distance calculation, regardless of the difference in the numbers of repeats. In contrast, the Hamming distance is not affected by this potential pitfall as it only scores equal or different. The drawback of the Hamming distance is that pairs of alleles which differ by a larger number of repeat units and are likely to be evolutionarily distant will contribute the same as those with single-repeat differences to the total distance value. The Hamming distance is also more sensitive to errors in allele scoring than SSR. Small differences in estimated allele size erroneously assigned to identical alleles will have little impact on the SSR distance but contribute to the Hamming distance to the same extent as genuine differences. Given that our set of markers included mini- and microsatellites with repeat units ranging in length from 2 to 19 bp (31) and that visual scoring of alleles at 9 loci may have introduced occasional errors, we did not want to base our analyses on one distance metric only and chose to perform PCoA with SSR and Hamming distances.
The effort and material needed for multilocus genotyping have limited the application of this technique. Many investigators rely instead on the partial sequence of the GP60 gene, which provides a quick method to compare genotypes from different studies and different laboratories. In contrast, different multilocus genotyping methods have been developed (10, 12, 13, 14, 16, 19, 22), making comparisons between different studies more difficult. We have shown here that a single-locus genotype, whether based on the GP60 sequence or another marker, in most cases cannot replace multilocus typing. This outcome is consistent with the observation of genetic recombination in nature (19, 29) and in the laboratory (28). Research to advance our understanding of the epidemiology of cryptosporidiosis would benefit from the application of a common multilocus genotyping method.
Acknowledgments
We thank Eric London for technical assistance, Donna Akiyoshi, Saul Tzipori, James Tumwine, and Rachel Chalmers for DNA samples, and Peter Smouse and Sultan Tanriverdi for helpful comments and suggestions.
This study was supported in part by grant AI052781 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 13 August 2010.
REFERENCES
- 1.Abrahamsen, M. S., T. J. Templeton, S. Enomoto, J. E. Abrahante, G. Zhu, C. A. Lancto, M. Deng, C. Liu, G. Widmer, S. Tzipori, G. A. Buck, P. Xu, A. T. Bankier, P. H. Dear, B. A. Konfortov, H. F. Spriggs, L. Iyer, V. Anantharaman, L. Aravind, and V. Kapur. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441-445. [DOI] [PubMed] [Google Scholar]
- 2.Ajjampur, S. S., F. B. Liakath, A. Kannan, P. Rajendran, R. Sarkar, P. D. Moses, A. Simon, I. Agarwal, A. Mathew, R. O'Connor, H. Ward, and G. Kang. 2010. Multisite study of cryptosporidiosis in children with diarrhea in India. J. Clin. Microbiol. 48:2075-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cacciò, S., W. Homan, R. Camilli, G. Traldi, T. Kortbeek, and E. Pozio. 2000. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology 120(Pt. 3):237-244. [DOI] [PubMed] [Google Scholar]
- 4.Cama, V. A., C. Bern, J. Roberts, L. Cabrera, C. R. Sterling, Y. Ortega, R. H. Gilman, and L. Xiao. 2008. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg. Infect. Dis. 14:1567-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cama, V. A., J. M. Ross, S. Crawford, V. Kawai, R. Chavez-Valdez, D. Vargas, A. Vivar, E. Ticona, M. Navincopa, J. Williamson, Y. Ortega, R. H. Gilman, C. Bern, and L. Xiao. 2007. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J. Infect. Dis. 196:684-691. [DOI] [PubMed] [Google Scholar]
- 6.Carraway, M., S. Tzipori, and G. Widmer. 1996. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl. Environ. Microbiol. 62:712-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carraway, M., S. Tzipori, and G. Widmer. 1997. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect. Immun. 65:3958-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cevallos, A. M., X. Zhang, M. K. Waldor, S. Jaison, X. Zhou, S. Tzipori, M. R. Neutra, and H. D. Ward. 2000. Molecular cloning and expression of a gene encoding Cryptosporidium parvum glycoproteins gp40 and gp15. Infect. Immun. 68:4108-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 10.Feng, X., S. M. Rich, D. Akiyoshi, J. K. Tumwine, A. Kekitiinwa, N. Nabukeera, S. Tzipori, and G. Widmer. 2000. Extensive polymorphism in Cryptosporidium parvum identified by multilocus microsatellite analysis. Appl. Environ. Microbiol. 66:3344-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasser, R. B., X. Zhu, S. Cacciò, R. Chalmers, G. Widmer, U. M. Morgan, R. C. Thompson, E. Pozio, and G. F. Browning. 2001. Genotyping Cryptosporidium parvum by single-strand conformation polymorphism analysis of ribosomal and heat shock gene regions. Electrophoresis 22:433-437. [DOI] [PubMed] [Google Scholar]
- 12.Gatei, W., C. A. Hart, R. H. Gilman, P. Das, V. Cama, and L. Xiao. 2006. Development of a multilocus sequence typing tool for Cryptosporidium hominis. J. Eukaryot. Microbiol. 53(Suppl. 1):S43-S48. [DOI] [PubMed] [Google Scholar]
- 13.Gatei, W., P. Das, P. Dutta, A. Sen, V. Cama, A. A. Lal, and L. Xiao. 2007. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect. Genet. Evol. 7:197-205. [DOI] [PubMed] [Google Scholar]
- 14.Grinberg, A., J. Learmonth, E. Kwan, W. Pomroy, N. Lopez Villalobos, I. Gibson, and G. Widmer. 2008. Genetic diversity and zoonotic potential of Cryptosporidium parvum causing foal diarrhea. J. Clin. Microbiol. 46:2396-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamming, R. W. 1950. Error detecting and error correcting codes. Bell Syst. Tech. J. 29:147-160. [Google Scholar]
- 16.Hunter, P. R., S. J. Hadfield, D. Wilkinson, I. R. Lake, F. C. Harrison, and R. M. Chalmers. 2007. Subtypes of Cryptosporidium parvum in humans and disease risk. Emerg. Infect. Dis. 13:82-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karanis, P., O. Thekisoe, K. Kiouptsi, J. Ongerth, I. Igarashi, and N. Inoue. 2007. Development and preliminary evaluation of a loop-mediated isothermal amplification procedure for sensitive detection of Cryptosporidium oocysts in fecal and water samples. Appl. Environ. Microbiol. 73:5660-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leav, B. A., M. R. Mackay, A. Anyanwu, R. M. O'Connor, A. M. Cevallos, G. Kindra, N. C. Rollins, M. L. Bennish, R. G. Nelson, and H. D. Ward. 2002. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect. Immun. 70:3881-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallon, M., A. MacLeod, J. Wastling, H. Smith, B. Reilly, and A. Tait. 2003. Population structures and the role of genetic exchange in the zoonotic pathogen Cryptosporidium parvum. J. Mol. Evol. 56:407-417. [DOI] [PubMed] [Google Scholar]
- 20.Morgan, U. M., C. C. Constantine, P. O'Donoghue, B. P. Meloni, P. A. O'Brien, and R. C. Thompson. 1995. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am. J. Trop. Med. Hyg. 52:559-564. [DOI] [PubMed] [Google Scholar]
- 21.Morgan-Ryan, U. M., A. Fall, L. A. Ward, N. Hijjawi, I. Sulaiman, R. Fayer, R. C. Thompson, M. Olson, A. Lal, and L. Xiao. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433-440. [DOI] [PubMed] [Google Scholar]
- 22.Ngouanesavanh, T., K. Guyot, G. Certad, Y. L. Fichoux, C. Chartier, R. I. Verdier, J. C. Cailliez, D. Camus, E. Dei-Cas, and A. L. Banuls. 2006. Cryptosporidium population genetics: evidence of clonality in isolates from France and Haiti. J. Eukaryot. Microbiol. 53:S33-S36. [DOI] [PubMed] [Google Scholar]
- 23.Peakall, R., and P. E. Smouse. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6:288-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. Mac Kenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plutzer, J., and P. Karanis. 2009. Genetic polymorphism in Cryptosporidium species: an update. Vet. Parasitol. 165:187-199. [DOI] [PubMed] [Google Scholar]
- 26.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sulaiman, I. M., P. R. Hira, L. Zhou, F. M. Al-Ali, F. A. Al-Shelahi, H. M. Shweiki, J. Iqbal, N. Khalid, and L. Xiao. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 43:2805-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanriverdi, S., J. C. Blain, B. Deng, M. T. Ferdig, and G. Widmer. 2007. Genetic crosses in the apicomplexan parasite Cryptosporidium parvum define recombination parameters. Mol. Microbiol. 63:1432-1439. [DOI] [PubMed] [Google Scholar]
- 29.Tanriverdi, S., A. Grinberg, R. M. Chalmers, P. R. Hunter, Z. Petrovic, D. E. Akiyoshi, E. London, L. Zhang, S. Tzipori, J. K. Tumwine, and G. Widmer. 2008. Inferences about the global population structures of Cryptosporidium parvum and Cryptosporidium hominis. Appl. Environ. Microbiol. 74:7227-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanriverdi, S., A. Markovics, M. O. Arslan, A. Itik, V. Shkap, and G. Widmer. 2006. Emergence of distinct genotypes of Cryptosporidium parvum in structured host populations. Appl. Environ. Microbiol. 72:2507-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanriverdi, S., and G. Widmer. 2006. Differential evolution of repetitive sequences in Cryptosporidium parvum and Cryptosporidium hominis. Infect. Genet. Evol. 6:113-122. [DOI] [PubMed] [Google Scholar]
- 32.Widmer, G. 2009. Meta-analysis of a polymorphic surface glycoprotein of the parasitic protozoa Cryptosporidium parvum and Cryptosporidium hominis. Epidemiol. Infect. 137:1800-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao, L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80-89. [DOI] [PubMed] [Google Scholar]
- 34.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao, L., R. Fayer, U. Ryan, and S. J. Upton. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]