Abstract
A total of 560 Legionella species were isolated from environmental water sources from public facilities from June to September 2008 throughout South Korea. The distribution of Legionella isolates was investigated according to geographical region, facility type, and sample type. The genetic diversity of 104 isolates of Legionella pneumophila serogroup 1 (sg 1) was analyzed by sequence-based typing (SBT). L. pneumophila was distributed broadly throughout Korea, accounting for 85.0% of the isolates, and L. pneumophila sg 1 predominated in all of the public facilities except for the springs. Legionella anisa and Legionella bozemanii predominated among non-L. pneumophila species (48.1% and 21.0%, respectively). The second most dominant strain differed depending on the facility type: L. anisa was the second most dominant strain in the buildings (10.8%), L. pneumophila sg 5 in public baths (21.6%), L. pneumophila sg 6 in factories (12.0%), and L. pneumophila sg 7 in hospitals (13.1%). In the SBT analysis, 104 L. pneumophila sg 1 isolates were differentiated into 26 sequence types (STs) and categorized into 3 clonal groups (CGs) and 10 singleton STs via the eBURST V3 program. ST1, a potential founder of major CG1, was commonly distributed (48.1%). The dominant ST in hot water was ST-K1 (7, 12, 17, 3, 35, 11, 11), which was designated in this study (36.1%). The second most dominant strain differed depending on the type of facility from which the samples were obtained. The unique allelic profile of ST-K1, obtained from hot water, was not found in the European Working Group for Legionella Infections (EWGLI) SBT database.
Legionella species, ubiquitous Gram-negative bacteria, are found in a variety of artificial water systems, natural freshwaters, and soils. Currently, the Legionella genus includes 52 species and more than 70 different serogroups, and more than 20 species have been proven to be causative agents of Legionnaires' disease (LD). The species Legionella pneumophila accounts for approximately 90% of confirmed cases of legionellosis, and L. pneumophila serogroup 1 (sg 1) has been recognized as the most important agent in this regard, as that specific strain was initially implicated as the pathogen causative of LD in 1977 (15; http://www.bacterio.cict.fr/l/legionellaceae.html). The other non-L. pneumophila sg 1 strains, sg 2 to 15, accounted for 7.4% of cases, and Legionella longbeachae (3.9%) and Legionella bozemanii (2.4%) have also been associated with the pathogen of LD. In particular, L. longbeachae has been recognized as accounting for 30.4% of community-acquired Legionella isolates in Australia and New Zealand (53).
The most common transmission mechanism of legionellosis is the inhalation of aerosols from the water systems of artificial facilities, including large buildings, hotels, hospitals, public baths, spas, or decorative fountains contaminated by Legionella species (1). Therefore, hot water and water from cooling towers have been perceived as sources of infection in cases of community-acquired, nosocomially acquired, or travel-associated LD (15, 26, 31, 37, 38, 39, 41, 43). Thus, it is important from a public health perspective to continually survey environmental water systems for the presence of Legionella species (2, 34, 35). In particular, hot-water systems used as public baths, such as springs, spas, or tubs, have become a popular means of recreation in a lot of countries, including South Korea. The contamination of hot-water systems has gradually become recognized as an important risk factor all over the world (4, 12, 18, 23, 42, 50), as sources of legionellosis have been detected increasingly since 1982 (52) and many cases of nosocomially acquired (32, 51) and community-acquired (6, 7, 48) LD have been detected in Legionella-contaminated hot-water systems or hot springs.
In South Korea, several cases of nosocomial infection and community-acquired pneumonia have occasionally been reported (9, 45) since the first recognized outbreak in South Korea in 1984, which was associated with Legionella gormanii (27). Since 2006, the Korean National Infectious Disease Surveillance (NIDS) program (http://dis.cdc.go.kr/) has reported an average of 20 cases of LD per year (29). In South Korea, surveys of Legionella acquired from environmental water in public facilities such as hot springs and public baths has been gradually enhanced since 2007. An annual training program for the detection of Legionella species from environmental water systems and clinical specimens is currently conducted for the personnel of 16 Provincial Institute of Health and Environment locations (PIHEs) throughout South Korea. Recently, the rate of detection of environmental Legionella bacteria has been gradually increasing (8.1% in 2006, 9.4% in 2007, and 10.3% in 2008).
The principal objectives of this study were to assess the current distribution of Legionella species from environmental water sources from public facilities such as buildings, hotels, public baths, springs, hospitals, or factories throughout South Korea. Additionally, the molecular typing of L. pneumophila sg 1 isolates was conducted using sequence-based typing (SBT) to assess the genetic diversity among the isolates.
MATERIALS AND METHODS
Sampling and bacterial isolates.
Sixteen PIHEs (in Seoul, Incheon, Gyeonggi, Gangwon, Daejeon, Chungbuk, Chungnam, Jeonbuk, Jeonnam, Gwangju, Gyeongbuk, Gyeongnam, Daegu, Ulsan, Busan, and Jeju) participated in a survey of Legionella in public facilities throughout South Korea from June to September 2008.
A total of 4,938 environmental water samples were collected largely from cooling towers and hot-water systems in large buildings, hospitals, or public baths in each region. The sampling was conducted via identical sampling protocols in all geographic regions. One liter of water samples from cooling towers, faucets, or showers was collected in sterile specimen bottles. The numbers of samples and isolates depended on the number of facilities located in each region. A total of 560 Legionella isolates from 13 out of 16 PIHEs were then sent to our laboratory in order to confirm the serological identification.
Identification of Legionella species.
To reconfirm the identification of the Legionella species, all of the collected strains were subcultured onto BCYE agar without l-cysteine or BCYE agar with l-cysteine. After the colonies growing only on BCYE agar were selected, Legionella species were confirmed via PCR using primers for Legionella genus-specific 16S rRNA and L. pneumophila species-specific mip genes (10, 24). Serological identification was then performed using a latex agglutination test (Oxoid, England), an antiserum kit (Denka, Japan), or a direct fluorescent-antibody assay (DFA) kit (m-Tech) for Legionella spp. Isolates not identified by serological methods were confirmed via 16S rRNA, mip, or rpoB gene amplification and comparative sequence analyses using the NCBI database (24, 25, 28).
SBT and allelic diversity analyses.
Genotyping was conducted via the standard sequence-based typing (SBT) method of the European Working Group for Legionella Infections (EWGLI) using 7 genes (flaA, pilE, asd, mip, mompS, proA, and neuA) (16, 40). Nucleotide analyses used the SBT database available on the EWGLI website (http://www.ewgli.org/), and the sequences were compared with those in the SBT database, which are also available on the website (http://www.hpa-bioinformatics.org.uk/legionella/legionella_sbt/php/sbt_homepage.php). Additionally, sequence types (STs) that were not available in the EWGLI SBT database were represented alphanumerically in this study (e.g., ST-K1).
The clonal complexes were analyzed using eBURST V3 (http://eburst.mlst.net), and clusters of related STs that descended from a common ancestor were defined as clonal groups (CGs). Single genotypes corresponding to no CG were defined as singletons.
Statistical analyses.
All calculations were conducted using SPSS 12.0 software (SPSS, Inc., Chicago, IL). The chi-square test was used to compare the proportional distributions of Legionella species according to environmental water source type.
RESULTS
Collected samples and isolates.
A total of 560 isolates were classified into 7 regions based on geographic location (Table 1), and the number of isolates was proportional to the number of collected samples. In particular, samples collected from the Gyeongsang region accounted for 34.4% of the overall samples, and the isolates from this region accounted for 44.1% of the total. For this reason, more institutions (Gyeongbuk, Gyeongnam, Daegu, Ulsan, and Busan) in this region than in other regions participated in this study. Although the isolates analyzed in this study were not selected in one region, the possibility that a bias was induced by regional differences in the number of strains could not be dismissed.
TABLE 1.
Distribution of samples and Legionella isolates from 7 geographical regions
| Geographical region | Cooling tower water |
Hot water |
Total |
|||
|---|---|---|---|---|---|---|
| No. of samples (%) | No. of isolates (%) | No. of samples (%) | No. of isolates (%) | No. of samples (%) | No. of isolates (%) | |
| Seoul | 316 (8.5) | 4 (1.3) | 187 (15.6) | 51 (20.2) | 503 (10.2) | 55 (9.8) |
| Gyeonggi | 784 (21.0) | 21 (6.8) | 157 (13.1) | 12 (4.8) | 941 (19.1) | 33 (5.9) |
| Gangwon | 367 (9.8) | 3 (1.0) | 264 (22.0) | 13 (5.2) | 631 (12.8) | 16 (2.9) |
| Chungcheong | 664 (17.8) | 120 (39.0) | 102 (8.5) | 33 (13.1) | 766 (15.5) | 153 (27.3) |
| Gyeongsang | 1,239 (33.2) | 124 (40.3) | 459 (38.2) | 123 (48.8) | 1,698 (34.4) | 247 (44.1) |
| Jeolla | 373 (10) | 34 (11) | 29 (2.4) | 20 (7.9) | 402 (8.1) | 54 (9.6) |
| Jeju | 93 (2.5) | 2 (0.6) | 4 (0.3) | 0 (0.0) | 97 (2.0) | 2 (0.4) |
| Total | 3,736 (100) | 308 (100) | 1,202 (100) | 252 (100) | 4,938 (100) | 560 (100) |
During sample collection, water characteristics such as temperature, pH, and residual chlorine concentration were usually not measured in this study.
Serological distribution of Legionella species.
A total of 560 isolates of Legionella were isolated from buildings (36.3%), public baths (28.9%), hospitals (25.9%), factories (4.5%), springs (2.5%), and hotels (2.0%). Fifty-five percent of the total samples were collected from cooling tower water, and the rest of the samples were collected from hot water. Among the 560 isolates, the 479 L. pneumophila isolates statistically predominated (85.5%), whereas Legionella species other than L. pneumophila accounted for 14.5% of the total. Among the 479 L. pneumophila species, the sg 1 strain accounted for 262 (54.7%), whereas strains sg 5, sg 6, sg 7, sg 3, and sg 10 accounted for 11.9%, 11.5%, 6.3%, 4.8%, and 2.7%, respectively (Fig. 1A). Thirty-nine Legionella anisa isolates were isolated, making L. anisa the dominant species (48.1%) among 81 non-L. pneumophila species, followed by L. bozemanii, which accounted for 21.0%. A total of 23.5% of the total isolates remained unidentified, despite genetic analyses using the 16S rRNA, mip, or rpoB genes (Fig. 1B).
FIG. 1.
Distribution of Legionella species from environmental water sources in South Korea for 2008. (A) L. pneumophila; (B) Non-L. pneumophila species. “Non-serogroup*” comprised L. pneumophila isolates not identified as members of any serogroup, and “other Legionella species**” comprised Legionella isolates not identified as members of any known species. sg 11, sg 13, sg 14, and sg 15 accounted for 0.2% each, and sg 12 accounted for 0.8%.
Analysis of geographic distribution of Legionella.
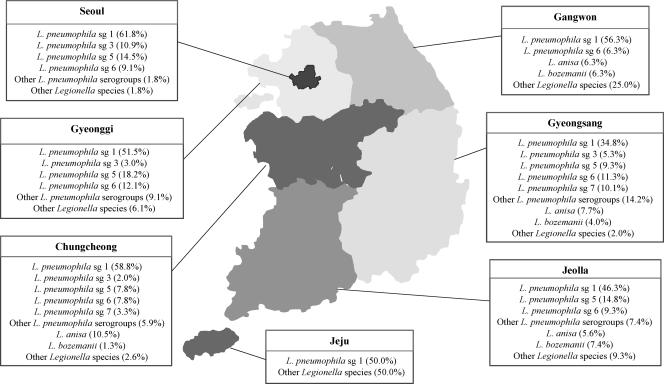
L. pneumophila sg 1 was prevalent in the 7 regions throughout South Korea (Fig. 2), and L. pneumophila sg 5 was represented in Seoul (14.5%), Gyeonggi (18.2%), Chungcheong (7.8%), Gyeongsang (9.3%), and Jeolla (14.8%) but not Gangwon or Jeju. L. pneumophila sg 6 was detected in Seoul (9.1%), Gyeonggi (12.1%), Gangwon (6.3%), Chungcheong (7.8%), Gyeongsang (11.3%), and Jeolla (9.3%) but not Jeju. L. anisa was isolated from 4 regions, all except Seoul, Gyeonggi, and Jeju, and L. bozemanii was also not detected in the Seoul, Gyeonggi, and Jeju regions.
FIG. 2.
Distribution of Legionella species isolated from environmental water sources from 7 geographic regions in South Korea: Seoul (n = 55), Gyeonggi (n = 33), Gangwon (n = 16), Chungcheong (n = 153), Gyeongsang (n = 247), Jeolla (n = 54), and Jeju (n = 2). “Other L. pneumophila serogroups” comprised sg 7 to sg 15, and “other Legionella species” comprised L. dumoffii, L. rubrilucens, L. spiritensis, and Legionella isolates not identified as members of any known species.
Legionella species according to facility type.
L. pneumophila sg 1 accounted for 46.8% (262/560) of the total isolates and predominated in facilities such as buildings, public baths, hospitals, factories, and hotels although not in springs (Fig. 3). The distribution rates of L. pneumophila strains other than sg 1 depended on the facility types; L. pneumophila sg 5 prevailed in public baths (21.6%), L. pneumophila sg 6 in springs and factories (21.4% and 12.0%, respectively), and L. pneumophila sg 7 in hospitals (13.1%). However, it was unreasonable to conclude that L. pneumophila sg 6 predominated in springs, as only 14 isolates were collected from springs. Among other non-L. pneumophila species, L. anisa accounted for 10.8% in buildings.
FIG. 3.
Distribution of Legionella species from environmental water sources according to facility type (n = 560). “Other L. pneumophila serogroups*” comprised sg 4 (2.0% in buildings and 2.8% in hospitals), sg 8 (2.5% in buildings, 1.2% in public baths, and 0.7% in hospitals), sg 9 (0.6% in public baths and 7.1% in springs), sg 11 (0.6% in public baths), sg 12 (0.5% in buildings, 0.6% in public baths, 0.7% in hospitals, and 7.1% in springs), sg 13 and sg 14 (0.5% each in buildings), sg 15 (0.7% in hospitals), and L. pneumophila isolates not identified as members of any serogroup (1.4% in hospitals). “Other Legionella species**” comprised L. dumoffii and L. rubrilucens (0.6% each in public baths) and L. spiritensis (1.5% in buildings and 7.1% in springs). “Non-identifiable Legionella species***” comprised Legionella isolates not identified as members of any known species.
Legionella species in cooling tower water and hot water.
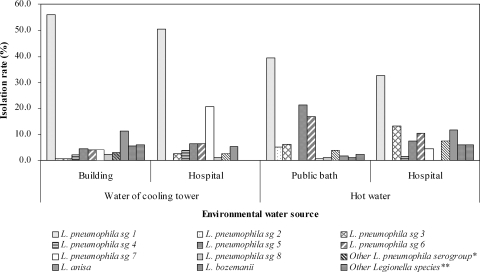
In order to determine whether the distribution of Legionella species depended on the sample type, the species and serogroup distributions were compared between the 276 isolates from the cooling tower water and the 252 isolates from hot water. The predominant strain in the cooling tower water and the hot water was L. pneumophila sg 1 (54.3% and 37.3%, respectively); the secondarily dominant strains depended on the sample type. L. anisa accounted for 9.4% of isolates collected from cooling towers, and L. pneumophila sg 5 accounted for 17.1% of the hot-water samples. In our comparative analysis of distribution between the cooling tower water and the hot water acquired from hospitals, L. pneumophila sg 1 was the predominant strain in both sample types, and the second most dominant strains depended on the sample type: L. pneumophila sg 7 was the second most dominant strain in cooling towers (20.8%), and L. pneumophila sg 3 in hot water (13.2%). L. anisa was the strain of quaternary dominance (5.2%) in the cooling tower samples and of tertiary dominance (11.8%) in the hot-water samples (Fig. 4).
FIG. 4.
Comparison of rates of isolation of Legionella species from cooling tower water (n = 276) and hot water (n = 228) in facilities such as buildings, hospitals, and public baths. “Other L. pneumophila serogroups*” comprised sg 9 to 15 and L. pneumophila isolates not identified as members of any serogroup. “Other Legionella species**” comprised L. dumoffii, L. rubrilucens, L. spiritensis, and Legionella isolates not identified as members of any known species.
Genetic diversity.
For SBT, among 262 isolates of L. pneumophila sg 1, 104 isolates were selected randomly, and these isolates were differentiated by SBT into 26 different sequence types (STs). ST1 (1, 4, 3, 1, 1, 1, 1), as the predominant type, accounted for 48.1% and was distributed commonly throughout the majority of facilities and regions (Table 2). ST36 (3, 4, 1, 1, 14, 9, 1) of L. pneumophila sg 1 (Philadelphia-1; ATCC 33152) was found only in one of the hot-water samples, and the profile of ST-K1 to -K14 could not be found in the EWGLI SBT database. According to the results of our eBURST analysis, 26 STs belonged to 3 CGs and 10 singleton STs (Table 2). In 3 CGs, CG1, the prevalent clonal group, included ST1, ST-K2, ST296, ST304, ST-K5, ST-K3, ST-K4, and ST-K8; the putative ancestor of CG1 was predicted to be ST1. The reminders were CG2 (ST50, ST159, ST-154, and ST-K14) and CG3 (ST59, ST363, ST-K1, and ST-K11). Among the 10 singleton STs, ST45 was detected in 3 isolates.
TABLE 2.
Distribution of clonal groups from 26 SBT profiles for L. pneumophila sg 1 isolates (n = 104) in South Korea
| Clonal group | ST | Alleic profile |
No. of isolates (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| flaA | pilE | asd | mip | mompS | proA | neuA | |||
| CG1 (n = 61) | 1 | 1 | 4 | 3 | 1 | 1 | 1 | 1 | 50 (48.1) |
| K2 | 1 | 4 | 3 | 1 | 1 | 1 | 4 | 2 (1.9) | |
| 296 | 1 | 4 | 3 | 1 | 1 | 1 | 11 | 2 (1.9) | |
| 304 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 3 (2.9) | |
| K5 | 1 | 12 | 3 | 1 | 1 | 1 | 1 | 1 (1.0) | |
| K3 | 1 | 4 | 3 | 1 | 1 | 3 | 1 | 1 (1.0) | |
| K4 | 1 | 10 | 1 | 1 | 1 | 9 | 1 | 1 (1.0) | |
| K8 | 3 | 10 | 1 | 1 | 1 | 9 | 3 | 1 (1.0) | |
| CG2 (n = 21) | 59 | 7 | 6 | 17 | 3 | 13 | 11 | 11 | 4 (3.8) |
| 363 | 7 | 6 | 3 | 3 | 13 | 11 | 11 | 2 (1.9) | |
| K1 | 7 | 12 | 17 | 3 | 35 | 11 | 11 | 14 (13.5) | |
| K11 | 7 | 10 | 17 | 3 | 13 | 14 | 11 | 1 (1.0) | |
| CG3 (n = 10) | 150 | 11 | 14 | 16 | 1 | 15 | 13 | 1 | 4 (3.8) |
| 159 | 11 | 14 | 16 | 1 | 15 | 13 | 2 | 2 (1.9) | |
| 154 | 11 | 14 | 16 | 16 | 15 | 13 | 2 | 3 (2.9) | |
| K14 | 11 | 14 | 3 | 1 | 15 | 13 | 1 | 1 (1.0) | |
| Singletons (n = 12) | K7 | 2 | 23 | 17 | 3 | 9 | 4 | 6 | 1 (1.0) |
| K6 | 2 | 6 | 14 | 10 | 1 | 4 | 11 | 1 (1.0) | |
| 45 | 5 | 1 | 22 | 26 | 6 | 10 | 12 | 3 (2.9) | |
| K13 | 10 | 10 | 7 | 28 | 8 | 18 | 6 | 1 (1.0) | |
| 42 | 4 | 7 | 11 | 3 | 11 | 12 | 9 | 1 (1.0) | |
| K12 | 10 | 6 | 7 | 21 | 16 | 18 | 9 | 1 (1.0) | |
| 36a | 3 | 4 | 1 | 1 | 14 | 9 | 1 | 1 (1.0) | |
| 22 | 2 | 3 | 6 | 10 | 2 | 1 | 6 | 1 (1.0) | |
| K10 | 6 | 10 | 15 | 14 | 21 | 7 | 11 | 1 (1.0) | |
| K9 | 6 | 10 | 1 | 3 | 19 | 4 | 11 | 1 (1.0) | |
ST36 represents the SBT profile of L. pneumophila sg 1 (Philadelphia-1; ATCC 33152).
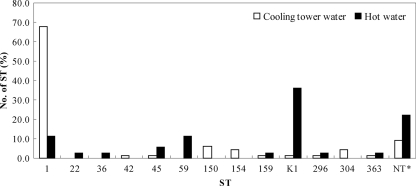
In our comparative analysis of the SBT distribution of the isolates according to sample type (Fig. 5), ST1 was the predominant type in isolates from the cooling tower water (67.6%, 46/68) and ST-K1 (7, 12, 17, 3, 35, 11) was the dominant type in the hot-water samples (36.1%). However, ST-K3, -K4, -K5, -K6, -K7, and -K14 were found only in isolates from the cooling tower water, and ST-K2, -K8, -K9, -K10, -K11, -K12, and -K13 were found only in the hot-water isolates.
FIG. 5.
Comparison of SBT profiles of L. pneumophila sg 1 isolates from cooling tower water (n = 68) and hot water (n = 36). The nontypeable (NT*) STs, which could not be found among the EWBLI SBT data, were ST-K2, ST-K3, ST-K4, ST-K5, ST-K6, ST-K7, and ST-K14 for cooling tower water (1.5% each) and ST-K8, ST-K9, ST-K10, ST-K11, ST-K12, ST-K13, and ST-K14 for hot water (2.8% each).
DISCUSSION
The results of this study showed that the ecology of Legionella species differed between the water of cooling towers and the hot-water samples collected from public facilities. With regard to sample type, L. pneumophila sg 1 was identified as a major strain (54.3% in water of cooling towers and 37.3% in the hot-water samples). Among facilities such as buildings, hospitals, public baths, and factories, 66.5% of isolates from cooling tower water were L. pneumophila sg 1, compared to only 41% of the isolates collected from hot water (P < 0.001) (Table 3). Among the hospitals, 94.8% of the isolates collected from cooling tower water were identified as L. pneumophila, compared to only 76.5% of the isolates collected from hot water (P = 0.002) (Table 4). Our results differed from those reported in earlier studies of public facilities. Among the cooling systems and hot-water systems of Spanish health care facilities (42), the predominant species, L. pneumophila, accounted for 85.1% and 91.7% of the total isolates, respectively. With regard specifically to L. pneumophila strains, sg 1 was the most frequently detected strain, at 88.3% and 81.8% in cooling towers and hot-water tanks, respectively. Another study of cooling towers conducted in China (31) reported that L. pneumophila sg 1 accounted for 82% (155/189) of the isolates, followed by Legionella micdadei (23.3% [44/189]). However, in a British analysis of unrelated environmental isolates, L pneumophila sg 1 accounted for 42.7% of the total isolates, followed by 12% and 7.7% for sg 6 and sg 5, respectively (19). In another study, designed to determine the clinical and environmental distribution of Legionella in France, the frequency of L. pneumophila sg 1 isolates from environmental water (75.6%) was lower than that observed for the clinical samples (98.8%), thus suggesting that environmental predominance was unrelated to more-efficient intracellular growth or higher infectivity (11). Some authors have made the point that non-L. pneumophila species with higher prevalence in environmental water samples than in the clinical samples may prove to be less pathogenic than L. pneumophila. This hypothesis has been recognized for the majority of confirmed infections by non-L. pneumophila species occurring in immunosuppressed patients (36, 46). The other studies have identified L. anisa as the cause of Pontiac fever (13, 14) or hospital-acquired LD (5, 14, 43). The detection of L. anisa in water samples should be considered an indication that the water system has been colonized by Legionella species, including L. pneumophila (49, 53). The other Legionella species may prove important in the etiology of community-acquired pneumonia, thus underscoring the need for diagnostic studies, including culture, serology, urinary antigen testing, or gene detection for Legionella species other than L. pneumophila sg 1 (33, 53).
TABLE 3.
Comparative distribution of Legionella species between water in cooling towers and hot water in facilities such as buildings, hospitals, public baths, and factories (n = 560)
| Sample source | No. (%) of Legionella isolates |
No. (%) of L. pneumophila isolates |
||||
|---|---|---|---|---|---|---|
| Total | L. pneumophila | Non-L. pneumophila | Total | sg 1 | Non-sg 1 | |
| Cooling tower water | 308 | 257 (83.4) | 51 (16.6) | 257 | 171 (66.5) | 86 (33.5) |
| Hot water | 252 | 222 (88.1) | 30 (11.9) | 222 | 91 (41.0) | 131 (59.0) |
| Total | 560 | 479 (85.5) | 81 (14.5) | 479 | 262 (54.7) | 64 (51.2) |
TABLE 4.
Comparative distribution of Legionella species between water in cooling towers and hot water in hospitals (n = 145)
| Sample source | No. (%) of Legionella isolates |
No. (%) of L. pneumophila isolates |
||||
|---|---|---|---|---|---|---|
| Total | L. pneumophila | Non-L. pneumophila | Total | sg 1 | Non-sg 1 | |
| Cooling tower water | 125 | 73 (94.8) | 4 (5.2) | 73 | 39 (53.4) | 34 (46.6) |
| Hot water | 20 | 52 (76.5) | 16 (23.5) | 52 | 22 (42.3) | 30 (57.7) |
| Total | 145 | 125 (86.2) | 20 (13.8) | 125 | 61 (64) | 64 (51.2) |
As discussed in a previous publication (20), SBT is a more powerful tool than pulsed-field gel electrophoresis (PFGE) for the subtyping of L. pneumophila strains. The use of SBT data from different countries, then, constitutes a technically uncomplicated and relatively easy method for strain subtyping, especially compared to other contemporary techniques.
ST1 (1, 4, 3, 1, 1, 1) is distributed broadly throughout the world (3, 8, 20, 44) and was the predominant profile in this study. However, the profile of ST-K1 (7, 12, 17, 3, 35, 11) was not detected in the HPA/EWGLI SBT database or in any other studies. ST150 (11, 14, 16, 1, 15, 13) has previously been reported to occur in France (17). Additionally, 13 STs (ST-K2 to -K14) identified as unique types in this study had new allelic profiles.
The 104 isolates of L. pneumophila sg 1 used in SBT were not identified as members of subgroups by use of monoclonal antibodies (MAbs) from the Dresden Panel (21, 22). So, comparison of relationships between subgroups by MAbs and STs, as was the case in other studies, could not be demonstrated in this study.
The 194 clinical isolates of L. pneumophila sg 1 isolated from Ontario, Canada, from 1978 to 2007 comprised 62 STs, and the population of STs was highly diverse. ST36, ST42, ST45, and ST59 were identified as members of cluster II and were also found in the singletons of this study (except for ST59); however, ST150, ST154, ST159, ST296, ST304, and ST363 in this study were not found among the 62 STs (47).
In conclusion, our results demonstrated that the proportional populations of environmental isolates of Legionella species isolated from public facilities differed according to the types of facility or sample assessed as well as the geographical locations of the facilities. Additionally, our findings revealed several unique allelic profiles of STs and showed that ST1 of L. pneumophila sg 1 was the prevalent sequence type in South Korea. Routine monitoring of environmental water for Legionella species is expected to prove helpful in efforts to reduce the bacterial contamination of water systems and is also expected to facilitate the development of a more active prevention strategy for LD. Additionally, further study will require that the focus be kept on correlation analysis by clustering between environmental and clinical isolates of Legionella species. Thus, the findings of this study highlight the importance of understanding the epidemiology and ecology of L. pneumophila from public facilities in terms of public health; in this regard, our findings corroborate and reinforce the recommendations made in several previous studies (2, 15, 35).
Acknowledgments
This work was supported by a grant from the Korean National Institute of Health for 2008 (2006-N46002-00).
We express our gratitude to our coworkers in 13 PIHEs who provided useful data and assistance. We also thank Norman Fry for authorizing access to the EWGLI SBT database. Additionally, we thank Kwan Soo Ko, professor, for his advice in our analysis of clonal clustering of STs and Soo Jin Baek for her advice on statistics in comparing the isolates.
Footnotes
Published ahead of print on 6 August 2010.
REFERENCES
- 1.Arnow, P. M., T. Chou, D. Weil, E. N. Shapiro, and C. Kretzschmar. 1982. Nosocomial Legionnaires' disease caused by aerosolized tap water from respiratory devices. J. Infect. Dis. 146:460-467. [DOI] [PubMed] [Google Scholar]
- 2.Boccia, S., P. Laurenti, P. Borella, U. Moscato, G. Capalbo, A. Cambieri, R. Amore, G. Quaranta, F. Boninti, M. Orsini, G. Branca, G. Fadda, V. R. Spica, and G. Ricciardi. 2006. Prospective 3-year surveillance for nosocomial and environmental Legionella pneumophila: implications for infection control. Infect. Control Hosp. Epidemiol. 27:459-465. [DOI] [PubMed] [Google Scholar]
- 3.Borchardt, J., J. K. Helbg, and P. C. Lück. 2008. Occurrence and distribution of sequence types among Legionella pneumophila strains isolated from patients in Germany: common features and differences to other regions of the world. Eur. J. Clin. Microbiol. Infect. Dis. 27:29-36. [DOI] [PubMed] [Google Scholar]
- 4.Borella, P., M. T. Montagna, S. Stami, G. Stancanelli, V. Romano-Spica, M. Triassi, I. Marchesi, A. Bargellini, D. Tatò, C. Napoli, F. Zanetti, E. Leoni, M. Moro, S. Scaltriti, G. R. D'Alcalà, R. Santarpia, and S. Boccia. 2005. Legionella contamination in hot water of Italian hotels. Appl. Environ. Microbiol. 71:5805-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornstein, N., A. Meercatello, D. Martmet, M. Surgot, Y. Deveaux, and J. Fleurette. 1989. Pleural infection caused by Legionella anisa. J. Clin. Microbiol. 27:2100-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bornstein, N., D. Marmet, M. Surgot, M. Nowicki, A. Arslan, J. Esteve, and J. Fleurette. 1989. Exposure to Legionellaceae at hot spring spa: a prospective clinical and serological study. Epidemiol. Infect. 102:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2005. Legionnaires' disease associated with potable water in a hotel—Ocean City, Maryland, October 2003-Februrary 2004. MMWR Morb. Mortal. Wkly. Rep. 54:165-168. [PubMed] [Google Scholar]
- 8.Chasqueira, M. J., L. Rodrigues, M. Nascimento, and T. Marques. 2009. Sequence-based and monoclonal antibody typing of Legionella pneumophila isolated from patients in Portugal during 1987-2008. Euro Surveill. 14(28):pii=19271. [DOI] [PubMed] [Google Scholar]
- 9.Choi, T. Y. 1998. Legionella infection. Korean J. Clin. Microbiol. 1:24-32. [Google Scholar]
- 10.Cloud, J. L., K. C. Darroll, P. Pixton, M. Erali, and D. R. Hilluard. 2000. Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J. Clin. Microbiol. 38:1709-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doleans, A., H. Aurell, M. Reyrolle, G. Lina, J. Freney, F. Vandenesch, J. Etienne, and S. Jarraud. 2004. Clinical and environmental distributions of Legionella strains in France are different. J. Clin. Microbiol. 42:458-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edagawa, A., A. Kimura, H. Doi, H. Tanaka, K. Tomioka, K. Sakabe, C. Nakajima, and Y. Suzuki. 2008. Detection of culturable and nonculturable Legionella species from hot water systems of public buildings in Japan. J. Appl. Microbiol. 105:2104-2114. [DOI] [PubMed] [Google Scholar]
- 13.Fallon, R. J., and N. H. Stack. 1990. Legionnaires' disease due to Legionella anisa. J. Infect. 20:227-229. [DOI] [PubMed] [Google Scholar]
- 14.Fenstersheib, M. D., M. Miller, C. Diggins, S. Liska, L. Detwiler, S. B. Werner, D. Lindquist, W. L. Thacker, and R. F. Benson. 1990. Outbreak of Pontiac fever due to Legionella anisa. Lancet 336:35-37. [DOI] [PubMed] [Google Scholar]
- 15.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaia, V., N. K. Fry, T. G. Harrison, and R. Peduzzi. 2003. Sequence-based typing of Legionella pneumophila serogroup 1 offers the potential for true portability in legionellosis outbreak investigation. J. Clin. Microbiol. 41:2932-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaia, V., N. K. Fry, B. Afshar, P. C. Lück, H. Meugnier, J. Etienne, R. Peduzzi, and T. G. Harrison. 2005. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 43:2047-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goutziana, G., V. A. Mouchtouri, M. Karanika, A. Kavagias, N. E. Stathakis, K. Gourgoulianis, J. Jremastinou, and C. Hadjichristodoulou. 2008. Legionella species colonization of water distribution systems, pools and air conditioning systems in cruise ships and ferries. BMC Public Health 8:390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison, T. G., N. Doshi, N. K. Fry, and C. A. Joseph. 2007. Comparison of clinical and environmental isolates of Legionella pneumophila obtained in the UK over 19 years. Clin. Microbiol. Infect. 13:78-85. [DOI] [PubMed] [Google Scholar]
- 20.Harrison, T. G., B. Afshar, N. Doshi, N. K. Fry, and J. V. Lee. 2009. Distribution of Legionella pneumophila serogroups, monoclonal antibody subgroups and DNA sequence types in recent clinical and environmental isolates from England and Wales (2000-2008). Eur. J. Clin. Microbiol. Infect. Dis. 28:781-791. [DOI] [PubMed] [Google Scholar]
- 21.Helbig, J., J. Kurtz, P. M. Castellani, C. Pelaz, and P. Luck. 1997. Antigenic lipopolysaccharide components of Legionella pneumophila recognized by monoclonal antibodies: possibilities and limitations for division of the species into serogroups. J. Clin. Microbiol. 35:2841-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helbig, J., S. Brenander, P. M. Castellani, J. Etienne, V. Gaia, S. Lauwer, D. Lindsay, P. C. Lück, T. Marques, S. Mentula, M. F. Peeters, C. Pelaz, M. Struelens, S. A. Uldum, G. Wewalka, and T. G. Harrison. 2002. Pan-European study on culture-proven Legionnaires' disease: distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 21:710-716. [DOI] [PubMed] [Google Scholar]
- 23.Hsu, B. M., C. H. Chen, M. T. Wan, and H. W. Cheng. 2006. Legionella prevalence in hot spring recreation areas of Taiwan. Water Res. 40:3267-3273. [DOI] [PubMed] [Google Scholar]
- 24.Jaulhac, B., M. Nowicki, N. Bornstein, O. Meunier, G. Prevost, Y. Piemont, J. Fleurette, and H. Montiel. 1992. Detection of Legionella spp. in bronchoalveolar lavage fluid by DNA amplification. J. Clin. Microbiol. 30:920-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonas, D., A. Rosenbaum, S. Weyrich, and S. Bhakdi. 1995. Enzyme-linked immunoassay for detection of PCR-amplified DNA of legionellae in bronchoalveolar fluid. J. Clin. Microbiol. 33:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph, C. A., R. Yadav, K. D. Ricketts, et al. 2009. Travel-associated Legionnaires' disease in Europe in 2007. Euro Surveill. 14(18):pii=19196. [DOI] [PubMed] [Google Scholar]
- 27.Kim, J. S., S. W. Lee, H. S. Shim, D. K. Oh, M. K. Cho, H. B. Oh, J. H. Woo, and Y. S. Chong. 1985. Outbreak of legionellosis in ICU of K Hospital, Korea. Korean J. Epidemiol. 7:44-58. [Google Scholar]
- 28.Ko, K. S., H. K. Lee, M. Y. Park, K. H. Lee, Y. J. Yun, S. Y. Woo, H. Miyamoto, and Y. H. Kook. 2002. Application of RNA polymerase β-subunit gene (rpoB) sequences for the molecular differentiation of Legionella species. J. Clin. Microbiol. 40:2653-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korea Centers for Disease Control and Prevention, Division of Bacterial Respiratory Infections. 2009. Diversity of Legionella species isolated from environmental water sources of public facilities throughout Korea in 2009. Public Health Wkly. Rep. 3:5-8. [Google Scholar]
- 30.Kuroki, T., T. Ishihara, K. Ito, and F. Kura. 2009. Bathwater-associated cases of legionellosis in Japan, with a special focus on Legionella concentrations in water. Jpn. J. Infect. Dis. 62:201-205. [PubMed] [Google Scholar]
- 31.Lin, H., B. Xu, Y. Chen, and W. Wang. 2009. Legionella pollution in cooling tower water of air-conditioning systems in Shanghai, China. J. Appl. Microbiol. 106:606-612. [DOI] [PubMed] [Google Scholar]
- 32.Lück, P. C., J. H. Helbig, U. Günter, M. Assmann, R. Blau, H. Koch, and M. Klepp. 1994. Epidemiologic investigation by macrorestriction analysis and by using monoclonal antibodies of nosocomial pneumonia cased by Legionella pneumophila serogroup 10. J. Clin. Microbiol. 32:2692-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNally, C., B. Hackman, B. S. Fields, and J. F. Plouffe. 2000. Potential importance of Legionella species as etiologies in community-acquired pneumonia (CAP). Diagn. Microbiol. Infect. Dis. 38:79-82. [DOI] [PubMed] [Google Scholar]
- 34.Modi, A., J. Gardner, L. Lighton, and N. Coetzee. 2008. Pontiac fever outbreak associated with a spa-pool, United Kingdom, April 2008. Euro Surveill. 13(30):pii=18934. [PubMed] [Google Scholar]
- 35.Mouchtouri, V. A., G. Giutzina, J. Kremastinou, and C. Hadjichristodoulou. 2010. Legionella species colonization in cooling towers: risk factors and assessment of control measures. Am. J. Infect. Control 38:50-55. [DOI] [PubMed] [Google Scholar]
- 36.Muder, R. R., and V. L. Yu. 2002. Infection due to Legionella species other than L. pneumophila. Clin. Infect. Dis. 35:990-998. [DOI] [PubMed] [Google Scholar]
- 37.Neil, K., and R. Berkelman. 2008. Increasing incidence of Legionella in the United States, 1990-2005: changing epidemiologic trends. Clin. Infect. Dis. 47:591-599. [DOI] [PubMed] [Google Scholar]
- 38.Ng, V., P. Tang, F. Jamieson, C. Guyard, D. E. Low, and D. N. Fisman. 2009. Laboratory-based evaluation of legionellosis epidemiology in Ontario, Canada, 1978 to 2006. BMC Infect. Dis. 9:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmore, T. N., F. Stock, M. White, M. Bordner, A. Michelin, J. E. Bennett, P. R. Murray, and K. H. Henderson. 2009. A cluster of cases of nosocomial Legionnaires disease linked to a contaminated hospital decorative water fountain. Infect. Control Hosp. Epidemiol. 30:764-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ratzow, S., V. Gaia, J. H. Heibig, N. K. Fry, and P. C. Lück. 2007. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 45:1965-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ricketts, K. D., and C. A. Joseph on behalf of the European Working Group for Legionella Infections. 2005. Legionnaires' disease in Europe 2003-2004. Euro Surveill. 10:256-259. [DOI] [PubMed] [Google Scholar]
- 42.Rivera, J. M., L. Aguilar, J. J. Granizo, A. Vos-Arenilla, M. J. Gimenez, J. M. Aguiar, and J. Prieto. 2007. Isolation of Legionella species/serogroups from water cooling systems compared with potable water systems in Spanish healthcare facilities. J. Hosp. Infect. 67:360-366. [DOI] [PubMed] [Google Scholar]
- 43.Rosmini, F., M. Castellani-Pastoris, M. F. Mazzotti, F. Forastiere, A. Gavazzoni, D. Greco, G. Ruckdesche, E. Tartagni, A. Zampieri, and W. B. Baine. 1984. Febrile illness in successive cohorts of tourists at a hotel on the Italian Adriatic coast: evidence for a persistent focus of Legionella infection. Am. J. Epidemiol. 119:124-134. [DOI] [PubMed] [Google Scholar]
- 44.Scaturro, M., M. Losardo, G. De Ponte, and M. L. Ricci. 2005. Comparison of three molecular methods used for subtyping of Legionella pneumophila strains isolated during an epidemic of legionellosis in Rome. J. Clin. Microbiol. 43:5348-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seog, W., Y. J. Jung, H. W. Park, H. K. Lee, M. S. Park, M. Y. Park, K. S. Park, M. D. Oh, C. Ahn, E. C. Kim, and K. W. Choe. 1999. A case of community-acquired Legionnaires' disease in a renal transplant recipient. Korean J. Infect. Dis. 31:353-357. [Google Scholar]
- 46.Thacker, W. L., R. F. Benson, L. Hawes, W. R. Mayberry, and D. J. Brenner. 1990. Characterization of a Legionella anisa strain isolated from a patient with pneumonia. J. Clin. Microbiol. 28:122-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tijet, N., P. Tang, M. Romilowych, C. Duncan, V. Ng, D. N. Fisman, F. Jamieson, D. E. Low, and C. Guyard. 2010. New endemic Legionella pneumophila serogroup 1 clones, Ontario, Canada. Emerg. Infect. Dis. 16:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tominaga, M., Y. Aoki, S. Haraguchi, M. Fukuoka, S. Hayashi, M. Tamesada, E. Yabuuchi, and Kohei Nagasawa. 2001. Legionnaires' disease associated with habitual drinking of hot spring water. Intern. Med. 40:1064-1067. [DOI] [PubMed] [Google Scholar]
- 49.van der Mee-Marquet, N., A. S. Domelier, L. Arnault, D. Bloc, P. Laudat, P. Hartemann, and R. Quentin. 2006. Legionella anisa, a possible indicator of water contamination by Legionella pneumophila. J. Clin. Microbiol. 44:56-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veríssimo, A., G. Marrão, F. G. Silva, and M. S. Costa. 1991. Distribution of Legionella spp. in hydrothermal areas in continental Portugal and the island of São Miguel, Azores. Appl. Environ. Microbiol. 57:2921-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visca, P., P. Goldoni, P. C. Lück, J. H. Helbig, L. Cattani, G. Giltri, S. Bramati, and M. C. Pastoris. 1999. Multiple types of Legionella pneumophila serogroup 6 in a hospital heated-water system associated with sporadic infections. J. Clin. Microbiol. 37:2189-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wadowsky, R. M., R. B. Yee, L. Mezmar, E. J. Wing, and J. N. Dowling. 1982. Hot water systems as sources of Legionella pneumophila in hospital and nonhospital plumbing fixtures. Appl. Environ. Microbiol. 43:1104-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, V. L., J. F. Plouffe, M. C. Pastoris, J. E. Stout, M. Schoushoe, A. Widmer, J. Summersgill, T. File, C. M. Heath, D. L. Paterson, and A. Chereshsky. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186:127-128. [DOI] [PubMed] [Google Scholar]