Abstract
Thermostable phytases, which are active over broad pH ranges, may be useful as feed additives, since they can resist the temperatures used in the feed-pelleting process. We designed new beta-propeller phytases, using a structure-guided consensus approach, from a set of amino acid sequences from Bacillus phytases and engineered Pichia pastoris strains to overproduce the enzymes. The recombinant phytases were N-glycosylated, had the correct amino-terminal sequence, showed activity over a pH range of 2.5 to 9, showed a high residual activity after 10 min of heat treatment at 80°C and pH 5.5 or 7.5, and were more thermostable at pH 7.5 than a recombinant form of phytase C from Bacillus subtilis (GenBank accession no. AAC31775). A structural analysis suggested that the higher thermostability may be due to a larger number of hydrogen bonds and to the presence of P257 in a surface loop. In addition, D336 likely plays an important role in the thermostability of the phytases at pH 7.5. The recombinant phytases showed higher thermostability at pH 5.5 than at pH 7.5. This difference was likely due to a different protein total charge at pH 5.5 from that at pH 7.5. The recombinant beta-propeller phytases described here may have potential as feed additives and in the pretreatment of vegetable flours used as ingredients in animal diets.
Phytases (myo-inositol hexakisphosphate phosphohydrolases; EC 3.1.3.8, EC 3.1.3.26, and EC 3.1.3.72) hydrolyze phytate (myo-inositol hexakisphosphate), the major storage form of phosphorus in feeds of plant origin (27). Monogastric and agastric animals, such as pigs, poultry, and fish, cannot utilize dietary phosphorus because their gastrointestinal tracts are deficient in enzymes with phytase activity (27, 28, 30). Therefore, these enzymes have significant value as animal feed additives.
Based on the presence of a specific consensus motif and their three-dimensional structures, phytases are classified into four major classes: histidine acid, beta-propeller, cysteine, and purple acid phytases (28, 30). Most of the commercially available phytases are histidine acid phytases derived from fungi (of the genus Aspergillus) and possess catalytic activity in the pH range of 2.5 to 6. On the other hand, bacterial phytases from the genus Bacillus are beta-propeller phytases. These phytases are structurally different from the fungal phytases, possess a pH optimum close to 7, and exhibit activity within a range of pHs that is broader than that of the fungal phytases (16, 22, 23, 35). Because animal feeds are commonly pelleted, a useful phytase additive should resist the temperatures of the pelleting process.
Among the protein-engineering strategies described for improving protein thermostability, data-driven protein design uses available sequences and structures to predict potential stabilizing amino acids as targets for mutation. Specifically, stabilizing amino acids can be predicted from the consensus amino acid sequence for homologous proteins, thus reducing the number of candidates to be tested experimentally. This approach has been applied successfully to engineer protein thermostability (25, 26). A further improvement is the structure-guided consensus approach, which uses structural information to further reduce the number of protein candidates to be tested for thermostability (37).
The methylotrophic yeast Pichia pastoris has been developed as a host for the efficient production and secretion of foreign proteins (20). Protein-engineering strategies that use P. pastoris as the host can improve both protein thermostability and protein overproduction. Therefore, we designed new beta-propeller phytases with a high probability of being thermostable and with a broad range of pH activities. We used a structure-guided consensus approach and a set of amino acid sequences from Bacillus phytases. We engineered P. pastoris strains to introduce phytase-encoding sequences that harbor P. pastoris-preferred codons to overproduce the designed phytases. In addition, the produced phytases were characterized biochemically, and their thermostabilities were correlated with protein structures.
MATERIALS AND METHODS
Strains, plasmids, medium composition, chemicals, and enzymes.
Pichia pastoris KM71 (his4), plasmid pPIC9, and all oligonucleotides were purchased from Invitrogen (San Diego, CA). Escherichia coli XL10-GOLD, used as a host for subcloning, and a QuikChange multisite-directed mutagenesis kit were from Stratagene (La Jolla, CA). Plasmid pUC57, used for cloning, mutagenesis, and sequencing, was from GenScript Corp. (Piscataway, NJ). Luria-Bertani (LB) agar plates, LB broth, regeneration dextrose base (RDB), yeast extract-peptone-dextrose (YPD), buffered minimal glycerol (BMG), and buffered minimal methanol (BMM) media were prepared according to the instructions for a Pichia expression kit (Invitrogen). For induction, BMM was supplemented with 0.1% (wt/vol) CaCl2 (BMM-CaCl2), and 0.75% (vol/vol) methanol was used instead of 0.5% (vol/vol) methanol, as recommended by the Invitrogen protocols. Yeast nitrogen base without amino acids was purchased from Difco (Detroit, MI). Taq DNA polymerase, SalI restriction endonuclease, and T4 DNA ligase were purchased from Promega (Madison, WI). XhoI and AvrII restriction endonucleases and endo Hf glycosidase were from New England Biolabs (Beverly, MA). All chemicals were analytical grade and were purchased from Sigma-Aldrich Co. (St. Louis, MO) or from Productos Químicos Monterrey (Monterrey, Nuevo León, México).
Amino acid sequence design.
Phytase amino acid sequences (FTE and FTEII) were designed as beta-propeller phytase sequences by a structure-guided consensus approach using a combination of two methods: (i) the consensus concept for thermostability engineering of proteins (25, 26) from 15 known amino acid sequences of phytases from the genus Bacillus; and (ii) a homology modeling analysis to determine the stabilizing amino acids, using a Bacillus amyloliquefaciens phytase structure (Protein Data Bank [PDB] code 2POO) and Molecular Operating Environment software (Chemical Computing Group Inc., Montreal, Quebec, Canada). The amino acid sequences used were as follows: four B. amyloliquefaciens sequences (GenBank accession no. AAL59320, AAW28542, AAL25193, and AAC38573), four Bacillus licheniformis sequences (GenBank accession no. AAM74021, AAT73627, YP_077686, and YP_090097), six Bacillus subtilis sequences (GenBank accession no. CAB13871, CAE48281, AAK97047, AAG17903, AAC31775, and AAO43434), and one Bacillus sp. sequence (GenBank accession no. AAR99083). The alignment of the 15 amino acid sequences and the consensus sequence determination were performed with the BioEdit v7.0.8.0 program (17). In addition, phenylalanine was considered an artificial first residue of the mature protein to enhance signal peptide cleavage by the Kex2 protease (21, 31, 32).
Construction of Pichia pastoris recombinant strains.
The FTE-encoding sequence (fte) was designed based on P. pastoris-preferred codons (36). AT-rich stretches of more than six nucleotides were removed to introduce silent mutations. Twelve nucleotides from the 3′ terminus of the alpha-factor prepro-secretion signal sequence from Saccharomyces cerevisiae, including the XhoI site, and an AvrII site were introduced at the 5′ and 3′ ends, respectively. The designed nucleotide sequence, with a full length of 1,094 bp, was synthesized (GenBank accession no. HM755449), cloned into vector pUC57, and sequenced by GenScript Corp. (Piscataway, NJ), generating the plasmid pUC57fte.
The fte gene was mutated at the corresponding nucleotides to generate the N336D mutation, thus synthesizing the fteII gene. In the same way, the fteII gene was mutated with F29K, V81A, and K292Q changes to give the fba gene, encoding the B. amyloliquefaciens phytase (GenBank accession no. AAC38573). Both mutagenesis assays were performed with a QuikChange multisite-directed mutagenesis kit according to the manufacturer's instructions. The nucleotide sequences of the fte, fteII, and fba genes were confirmed with a DNA sequencer (ABI Prism 310 genetic analyzer; Applied Biosystems, Foster City, CA) at the Unidad de Biología Molecular of the Instituto de Fisiología Celular of the Universidad Nacional Autónoma de México (UNAM).
DNA fragments harboring the fte, fteII, and fba genes were obtained by digestion of pUC57fte, pUC57fteII, and pUC57fba, respectively, with the XhoI and AvrII restriction enzymes and then were ligated to vector pPIC9 that had previously been digested with the same restriction enzymes to produce the new expression vectors: pPIC9fte, pPIC9fteII, and pPIC9fba. These vectors harbor the fte, fteII, and fba gene sequences in frame with the S. cerevisiae alpha-factor prepro-secretion signal and between the promoter and transcriptional terminator of the AOX1 gene. The correct construction of the vectors was confirmed by XhoI-AvrII double-digested restriction analysis and by a PCR using 5′ and 3′ AOX1 primers, directed to the AOX1 promoter and the transcription terminator, as previously described (6). All DNA manipulations were performed according to standard methods (33).
The P. pastoris host strain, KM71 (his4), was transformed separately with SalI-digested pPIC9fte, pPIC9fteII, and pPIC9fba DNAs by use of a Pichia EasyComp transformation kit (Invitrogen) according to the manufacturer's instructions. The transformants were selected for the ability to grow on histidine-deficient medium (RDB-agar plates) at 30°C until colonies appeared (His+ selection). His+ colonies from each transformation were randomly selected, and the integration of the expression cassette into the genomes of the selected strains was verified by a PCR using the 5′ and 3′ AOX1 primers, as previously described (6).
Production of recombinant phytases FTE, FTEII, and FBA.
Pichia pastoris recombinant strains were tested to select an overproducer strain for each phytase. Twenty His+ colonies for each recombinant strain were reactivated in YPD medium at 30°C and 250 rpm for 12 h and then used to inoculate 100 ml of BMG medium to an initial optical density at 600 nm (OD600) of 0.3. Further incubation was performed at 30°C and 250 rpm for 12 h. Cells were harvested by centrifugation (1,500 × g, 5 min) and used to inoculate 20 ml of BMM-CaCl2 medium to an initial OD600 of 50. Further incubation was carried out at 30°C for 48 h with continuous shaking at 250 rpm. Methanol was added to a final concentration of 0.75% every 24 h. Cell-free culture medium was recovered by centrifugation, CaCl2 was added to a final concentration of 1 mM, and volumetric phytase activity (U/ml) was determined. The final cell concentrations were estimated based on 1 OD600 unit approximately corresponding to 107 cells/ml (2). The amount of extracellular phytase production per cell was calculated as the ratio of volumetric phytase activity of the cell-free culture medium to the final cell concentration. The strains that yielded the highest phytase production levels per cell were considered the overproducer strains.
The effect of using P. pastoris-preferred codons in fteII and fba was evaluated by comparing the expression levels of each overproducer strain with the expression levels under the same culture conditions for P. pastoris strains GS115 Mut+ (GS115PhyC) and KM71 Muts (KM71PhyC), which are overproducers of recombinant Bacillus subtilis phytase C (PhyC-R), previously constructed in our laboratory (12, 13). In addition, the influence of an F (FTEII) or K (FBA) residue at position P1′ of the Kex2 site for the α-mating factor prepro-secretion signal processing was evaluated.
Each overproducer strain was used to produce every recombinant phytase by growing cells in 600 ml of BMG for 12 h, using harvested cells to inoculate 120 ml of BMM-CaCl2 medium, and incubating the cells at 30°C for 48 h. Methanol was added to a final concentration of 0.75% every 24 h, and cell-free culture medium was recovered as described above.
Further phytase purification steps were carried out at 4°C. Each cell-free culture medium was concentrated 20-fold and filtrated by ultrafiltration, using 10-kDa Centricon Plus 70 filters (Millipore, MA) and 100 mM Tris-HCl (pH 8.5) buffer with 100 mM NaCl, 2% glycerol, and 5 mM CaCl2. Aliquots of enzyme concentrates were stored at −20°C until they were analyzed. With each enzyme concentrate, additional phytase purification was performed using a Biologic LP chromatography system (Bio-Rad, Hercules, CA) and a HiTrap Q FF anion-exchange column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) equilibrated with buffer A (25 mM Tris-HCl, 5 mM CaCl2, pH 8.5). Proteins were eluted with buffer B (25 mM Tris-HCl, 5 mM CaCl2, 0.5 M NaCl, pH 8.5), using a stepwise gradient elution process (0 to 10 min, 100% buffer A; 10 to 11 min, 0 to 5% buffer B; 11 to 17 min, 5% buffer B; 17 to 18 min, 5 to 35% buffer B; 18 to 24 min, 35% buffer B; 24 to 25 min, 35 to 60% buffer B; 25 to 31 min, 60% buffer B; 31 to 33 min, 60 to 100% buffer B; and 33 to 43 min, 100% buffer B) at a flow rate of 1 ml/min. Fractions of 1.4 ml were collected, and the protein concentration and phytase activity were determined. The ratio of phytase activity to the protein concentration of the fraction with the maximum phytase activity was considered the specific activity of purified phytase.
All protein concentrations were determined by the Bradford protein assay, using bovine serum albumin as the standard.
Phytase activity assays.
Phytase activity was determined for all of the cell-free culture media, for protein concentrates obtained by ultrafiltration, and for the chromatographic fractions. For enzyme assays of cell-free culture media, samples were previously desalted by passage through a PD-10 column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) into a 100 mM Tris-HCl (pH 8.5) buffer with 100 mM NaCl, 2% glycerol, and 5 mM CaCl2. All samples were diluted in a suitable volume of the above Tris-HCl buffer. Phytase activities were measured in reaction mixtures containing the enzyme preparation, 1.6 mM sodium phytate, 100 mM Tris-HCl buffer (pH 7.5), and 1 mM CaCl2, with incubation at 37°C for 30 min. The reaction mixtures were stopped by adding an equal volume of 15% trichloroacetic acid, and the released inorganic phosphate was measured using the ascorbic acid method (8). One unit of phytase activity was defined as the amount of enzyme required to liberate 1 μmol of phosphate per min from sodium phytate under the assay conditions.
Biochemical characterization of phytases.
The biochemical characterization of phytases was performed with each enzyme concentrate by ultrafiltration from the cell-free culture medium.
N-glycosylation was evaluated by assessing the migration shift of endo Hf-treated proteins in a Coomassie blue-stained 12% SDS-polyacrylamide gel. Reactions were carried out by incubating phytase concentrates with endo Hf for 1 h at 37°C, according to the manufacturer's instructions.
Amino-terminal amino acid analyses were performed with the deglycosylated and electrophoretically resolved bands, which were then transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA), cut out, and analyzed at the Protein Facility of the Iowa State University Office of Biotechnology by the Edman degradation method, using a model 494 Procise protein/peptide sequencer and an online model 140C PTH amino acid analyzer, both from Perkin Elmer Applied Biosystems.
The effect of pH on enzymatic activity was determined at 37°C, using 250 mM glycine-HCl (pH 2.5), 360 mM sodium acetate (pH 5.5), and 100 mM Tris-HCl (pH 7.5 and 9.0) as buffers. The effect of temperature on enzymatic activity was determined by measuring the phytase activity in each enzyme preparation at various temperatures ranging from 25 to 80°C.
Thermostability of each phytase preparation was evaluated by measuring residual phytase activity after incubation at 25, 37, 60, and 80°C for 10 min in 100 mM Tris-HCl (pH 7.5) or 360 mM sodium acetate (pH 5.5) buffer in the presence of 1 or 5 mM CaCl2. In addition, the thermostability of PhyC-R was tested for comparison with the other three recombinant phytases.
All results were compared between statistical groups, using analysis of variance (ANOVA) and Tukey's multiple comparisons, with a significance cutoff of a P value of <0.05.
Computational analysis.
Among all of the phytases studied (FTE, FTEII, FBA, and PhyC-R), PhyC-R and FBA were the least similar, differing in 28 residues. The effects of these residues on the structural stability of the four phytases were evaluated with Swiss-PdbViewer/DeepView 4.0 (14) and iMolTalk (5), using molecular models of PhyC (GenBank accession no. AAC31775), FTE, and FTEII and information from the crystal structure of B. amyloliquefaciens phytase in its fully calcium-loaded state (PDB code 2POO). Molecular models were constructed by homology modeling using SWISS-MODEL 8.05 (34), with the 2POO structure as a template. In addition, the Ikai aliphatic index (19) was calculated for the four phytases as a positive factor for increases in thermostability, using the ProtParam tool available from the ExPASy server (10). Surface areas were calculated with the GetArea 1.0 server (9). Potential N-linked glycosylation sites were predicted with the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/). Isoelectric points, pKs of ionizable residues, and total charges at pH 7.5 and 5.5 of the recombinant phytases were estimated with the H++ server (11).
The system used for residue position identification for all of the recombinant phytases corresponded to the B. amyloliquefaciens phytase numbering, with the first amino acid residue being the methionine of the native signal peptide. Because all of the recombinant phytases produced were mature proteins, they started at position 29.
RESULTS
Amino acid sequence design.
The 105 pairwise sequence identities of the 15 amino acid sequences of Bacillus phytases ranged from 60 to 100% and were distributed mainly into two groups: 47 (45%) had values of >90%, while 41 (39%) had identities ranging from 60 to 70%. The obtained consensus amino acid sequence differed from the 15 sequences used as input; however, sequence identities of >90% were seen with 10 sequences. In addition, the consensus amino acid sequence presented a sequence identity of 98% with the B. amyloliquefaciens phytase (GenBank accession no. AAC38573), for which five three-dimensional structures have been described (PDB codes 1POO, 2POO, 1QLG, 1CVM, and 1H6L) (16, 35), differing in only seven amino acids (V81, D148, D288, K292, I331, I352, and L377) plus F29, the artificial first residue of the mature protein. The comparative analysis of the 2POO structure with the homology model of the consensus sequence showed that the A81V change is buried in the protein core, while the K29F and Q292K changes are solvent exposed. Also, the three changes could stabilize the protein structure of the consensus sequence. In the A81V change, the valine side chain increases the apolar surface area by 53 Å2 with respect to the original alanine residue. In addition, using the parameterization proposed by Xie and Freire (38), the enthalpic contribution of the A81V change to stability is estimated to be about 0.4 kcal/mol. The electrical charge of the 2POO structure at the K29 side chain is directed toward the solvent, while the aromatic ring at F29 of the consensus sequence could bury the hydrophobic surface area by its own side chain and from L30, a residue that is in contact with the ring. Similarly, the contribution of F29 to the consensus sequence stability is estimated to be about 1.1 kcal/mol, according to the increment of the buried apolar surface area. Therefore, the A81V and K29F changes might reduce the value of the unfolding equilibrium constant by 1 order of magnitude. In the Q292K change, the electrical charge in the amino group of the K292 side chain could be useful for decreasing the aggregation between the unfolded chains, thus favoring folding. Also, K292 might form salt bridges with nearby residues D248 and E288. Lastly, since residue 336 plays an important role in the interaction with a Ca2+ ion—stabilizing the three-dimensional structure of the protein (16) (but a discrepancy exists in the literature about the nature of this position in the five three-dimensional structures described for the B. amyloliquefaciens phytase [the 1CVM, 1QLG, and 1H6L structures have N336, while the 1POO and 2POO structures have D336])—the influence of N336 or D336 on the thermostability of the protein was studied by constructing both variants. Therefore, the newly designed amino acid sequences, called FTE and FTEII, differed in three residues (F29, V81, and K292) from the B. amyloliquefaciens phytase sequence (GenBank accession no. AAC38573), along with N336 for FTE and D336 for FTEII.
Construction of P. pastoris recombinant strains KM71FTE, KM71FTEII, and KM71FBA.
DNA sequencing of the pUC57 plasmids confirmed the correct nucleotide sequences of the fte, fteII, and fba genes. We engineered the DNA sequences to produce FTE, FTEII, and FBA phytases as mature polypeptides of 355 amino acids, in accordance with the designed sequences. PCR analyses of the recombinant plasmids by use of AOX1 primers showed 1,537-bp products that confirmed the pPIC9fte, pPIC9fteII, and pPIC9fba constructs.
Transformation of P. pastoris KM71 with the SalI-digested pPIC9fte, pPIC9fteII, and pPIC9fba plasmids gave about 50 His+ transformants each. PCR analysis of the genomic DNAs isolated from P. pastoris KM71FTE, KM71FTEII, and KM71FBA (His+ transformants) showed a 1,537-bp band that corresponds to the expression cassettes (1,065 bp) and fragments of the AOX1 promoter (349 bp) and transcription terminator (123 bp). These results indicate the absence of the AOX1 structural gene and the correct integration of the expression cassettes into the P. pastoris genome, thus confirming the Muts phenotype.
Production of recombinant phytases FTE, FTEII, and FBA.
Cell-free culture media from 48-h methanol-induced recombinant strain cultures showed volumetric phytase activities ranging from 0.12 to 4.97 U/ml (0.39 to 10.74 nU/cell) for the KM71FTE and KM71FTEII strains and from 0.28 to 3.39 U/ml (0.80 to 8.57 nU/cell) for the KM71FBA strain, while the KM71PhyC and GS115PhyC overproducer strains showed volumetric phytase activities of 0.27 and 0.82 U/ml (0.70 and 5.30 nU/cell), respectively. Therefore, the KM71FTEII overproducer strain yielded an extracellular phytase production level per cell that was 15.3-fold and 6.9-fold higher than the production levels per cell yielded by the KM71PhyC and GS115PhyC overproducer strains, respectively. Similarly, the KM71FBA overproducer strain yielded an extracellular phytase production level per cell that was 12.2-fold and 5.5-fold higher than the production levels per cell yielded with the KM71PhyC and GS115PhyC overproducer strains, respectively. Bacillus subtilis phytase C has sequence identities of 93.2 and 92.6% to FTEII and FBA, respectively, but the phyC gene in our constructed recombinant strains has native codons (12, 13), while the fteII and fba genes have P. pastoris-preferred codons. Therefore, the high extracellular phytase production levels per cell for the KM71FTEII and KM71FBA overproducer strains were likely due to the optimal P. pastoris codon bias used by the fteII and fba genes.
The KM71FTEII overproducer strain yielded an extracellular phytase production level per cell that was 1.25-fold higher than the production level per cell yielded by the KM71FBA overproducer strain. This result may be due to the presence of an F in FTEII as the first residue of the mature sequence, while FBA has a K in that position, which would improve the prepro-secretion signal processing by Kex2 endopeptidase (31, 32).
With ultrafiltration processes, we could obtain 6 ml of each phytase concentrate from each cell-free culture medium, with >95% activity recovery.
Biochemical characterization of phytases.
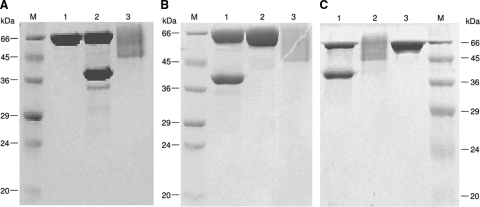
Figure 1 shows the migration shift results in SDS-polyacrylamide gels for the recombinant phytases after treatment with endo Hf. The three phytases (FTE, FTEII, and FBA) without endo Hf treatment showed a smear on the SDS-PAGE gel, ranging from 45 to 66 kDa (Fig. 1A, lane 3, B, lane 3, and C, lane 2). After N-deglycosylation by endo Hf, the three recombinant phytases showed an apparent molecular mass of 39 kDa (Fig. 1A, lane 2, B, lane 1, and C, lane 1). These results clearly indicate that the three recombinant phytases are highly N-glycosylated. These phytases have four potential N-glycosylation sites (121NRSE, 281NSSY, 138NGTL, and 243NGTV), with the last two being more likely to be N-glycosylated since they are at the protein surface.
FIG. 1.
SDS-polyacrylamide gels of FTE (A), FTEII (B), and FBA (C) phytases concentrated by ultrafiltration and treated with or without endo Hf. Lanes M, molecular size marker. (A) Lanes: 1, endo Hf; 2, FTE treated with endo Hf; 3, FTE without endo Hf treatment. (B) Lanes: 1, FTEII treated with endo Hf; 2, endo Hf; 3, FTEII without endo Hf treatment. (C) Lanes: 1, FBA treated with endo Hf; 2, FBA without endo Hf treatment; 3, endo Hf.
The amino-terminal sequences of the 39-kDa bands from the three N-deglycosylated phytases were determined to be FLSDPYHFTV for FTE and FTEII and KLSDPYHFTV for FBA. These sequences are completely identical to their theoretical sequences, demonstrating the identities of the phytases and the correct cleavage of the prepro-secretion signal from the phytases.
Anion-exchange chromatograms for the three phytase concentrates showed two peaks by UV detection at 280 nm, eluted at 0.30 and 0.37 M NaCl. Both peaks showed phytase activity and identical molecular sizes (39 kDa) in SDS-polyacrylamide gels after treatment with endo Hf, indicating that the chromatographic peaks correspond to two different glycosylation forms of each phytase. Specific activities of the purified phytases were 25.7, 13.3, and 18.0 U/mg protein for the FTE, FTEII, and FBA phytases, respectively.
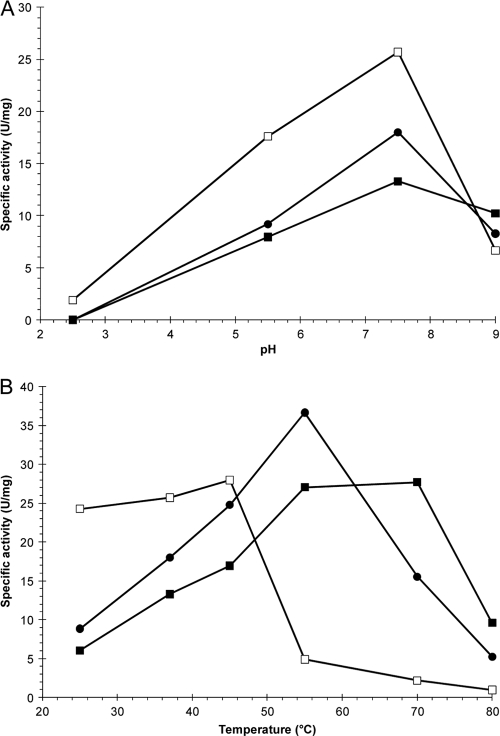
The three recombinant phytases had optimum activity at pH 7.5 (Fig. 2 A). At pH 7.5, FTE showed a higher specific activity than that of FTEII or FBA, while at pH 9.0, FTEII had the highest specific activity of the recombinant phytases. Although the three recombinant phytases had similar pH-activity profiles, the effects of temperature on specific phytase activity were different (Fig. 2B). The optimum temperature was 45°C for FTE and 55°C for FBA, while FTEII showed a broad temperature range for maximum activity (55 to 70°C). FBA showed the highest specific activity at 55°C in comparison with FTE and FTEII; however, FTEII clearly had the highest specific activity at 70°C.
FIG. 2.
Effects of pH (A) and temperature (B) on specific phytase activity at 37°C and pH 7.5 for FTE (□), FTEII (▪), and FBA (•). Points represent the means for at least three independent enzyme assays (coefficient of variation, <5%).
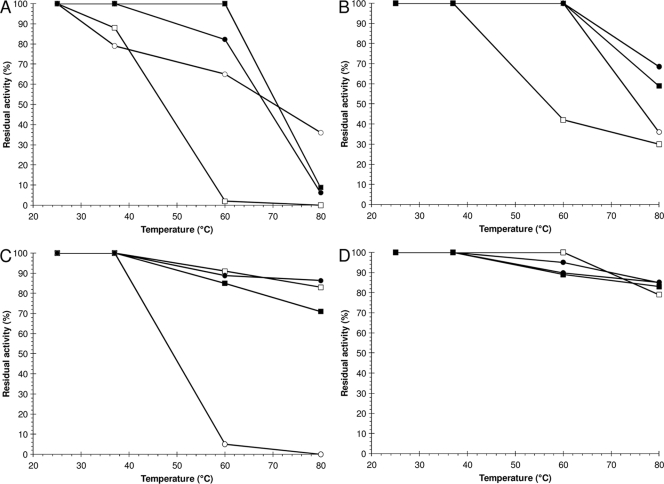
For the three recombinant phytases and PhyC-R, residual activities increased when assays were carried out in the presence of 5 mM instead of 1 mM CaCl2 (Fig. 3), which is typical for Bacillus phytases (22, 23). FTEII and FBA showed the highest thermostability at pH 7.5 and 5 mM CaCl2, followed by PhyC-R and FTE (Fig. 3B), without significant differences between each phytase pair. Nevertheless, the four recombinant phytases showed high thermostability at pH 5.5 (Fig. 3C and D), except in the case of PhyC-R, which had a residual activity that was drastically decreased in the presence of 1 mM CaCl2 (Fig. 3C). Among the four recombinant phytases, FTEII and FBA showed similar thermostability profiles.
FIG. 3.
Residual activities of FTE (□), FTEII (▪), FBA (•), and PhyC-R (○) after 10 min of heat treatment at pH 7.5 (A and B) or 5.5 (C and D) in the presence of 1.0 (A and C) or 5.0 (B and D) mM CaCl2. Points represent the means for at least three independent determinations (coefficient of variation, <5%).
Computational analysis of tridimensional molecular models.
Table 1 shows the results of the analysis of structural factors of the recombinant phytases, evaluated with the 28 residues where PhyC-R differs from FBA. Among the structural factors, differences in hydrogen bonds and salt bridges and the presence of a proline in a surface loop could explain the differences in thermostability at pH 7.5 and 5 mM CaCl2 between FTEII, FBA, and PhyC-R, while the presence of N336 in FTE could explain its reduced thermostability at pH 7.5.
TABLE 1.
Structural factors of recombinant phytases, evaluated with the 28 residues where PhyC differs from FBA
| Structural factor | Value or description for phytase |
|||
|---|---|---|---|---|
| PhyC | FTE | FTEII | FBA | |
| No. (%) of residues | ||||
| In α-helices | 1 (3.6) | 1 (3.6) | 1 (3.6) | 1 (3.6) |
| In β-sheets | 14 (50.0) | 13 (46.4) | 13 (46.4) | 13 (46.4) |
| In coils | 12 (42.9) | 13 (46.4) | 13 (46.4) | 13 (46.4) |
| With >30% accessible surface | 13 (46.4) | 11 (39.4) | 10 (35.7) | 11 (39.3) |
| Buried | 14 (50.0) | 17 (60.7) | 18 (64.3) | 17 (60.7) |
| Ikai aliphatic index | 67.4 | 66.3 | 66.6 | 66.0 |
| No. of H bonds | 37 | 40 | 40 | 40 |
| H bond at N terminus | NPa | K361-T37 | K361-T37 | K361-T37 |
| No. of salt bridges | 3 | 0 | 3 | 3 |
| Proline in surface loop | NP | P257 | P257 | P257 |
NP, not present.
DISCUSSION
Recently, we showed the efficiency of the P. pastoris expression system for synthesizing the B. subtilis VTTE-68013 phytase (PhyC-R) and for secreting the recombinant product into the culture medium with catalytic activity at neutral pH (12, 13). Although this recombinant phytase showed a high residual activity (85%) after 10 min of heat treatment at 80°C and pH 5.5 in the presence of 5 mM CaCl2, it maintained a residual activity of only 36% after heat treatment at pH 7.5. For this reason, we designed new thermostable phytases with catalytic activity at pH 7.5 (FTE and FTEII) from Bacillus phytases by using a structure-guided consensus approach (37). We produced the new recombinant phytases with the P. pastoris expression system by using a synthetic gene with genetic codons biased toward P. pastoris. In addition, we produced the B. amyloliquefaciens phytase with the same expression system to compare its physicochemical properties with those of FTE and FTEII.
The structure-guided consensus approach led to the production of a phytase (FTEII) which had activity over a broad range of pHs, including pH 7.5, and a thermostability that was higher than that of PhyC-R. In addition, the use of P. pastoris-preferred codons and the presence of an F at the first amino acid residue of FTEII allowed us to obtain high levels of production and secretion into the culture medium for phytases FTE, FTEII, and FBA. Furthermore, the three recombinant phytases had the correct amino-terminal sequences, though we did not include the Glu-Ala repeats after the cleavage site of the alpha-factor signal for peptide release.
The sequence of the synthetic gene used in the present work was designed using the P. pastoris-preferred codons determined from five proximal genes that are highly expressed with methanol as the carbon source (36). The best-expressing P. pastoris strain harboring the fteII gene showed 15-fold increased phytase production in comparison to that of the best-expressing P. pastoris strain harboring the native gene phyC from B. subtilis. The PhyC and FTEII amino acid sequences are very similar (93% identity); however, their corresponding nucleotide sequences are only 72% identical. Similar results have been reported for the production of the Aspergillus niger SK-47 phytase in P. pastoris (39) from a synthetic gene with P. pastoris-preferred codons determined from 28 P. pastoris coding sequences (40). In that report, phytase production increased 14.5 times over the production from the native gene.
Surprisingly, the amino acid sequence obtained by the consensus method in the present work showed a high similarity (98% identity) to the sequence of the B. amyloliquefaciens phytase, which has been reported to be a thermostable phytase (16, 23, 35). This illustrates the power of the consensus concept in designing thermostable beta-propeller phytases, a finding previously reported for the design of thermostable histidine acid phytases from fungal phytases (25, 26). The use of a structure-guided consensus approach allowed for the design of a phytase with physicochemical properties and thermostability similar to those of the B. amyloliquefaciens phytase. In addition, the designed phytase is more thermostable at pH 7.5 than the PhyC-R phytase previously produced in our laboratory (12, 13). Although the four recombinant phytases that were produced were highly similar (>92.6% identity), their specific activities at pH 7.5 and 37°C diverged somewhat, ranging from 11.5 to 25.7 U/mg protein. Similar findings have been observed in the design of thermostable histidine acid phytases from consensus sequences of Aspergillus phytases (25, 26). Since all of the recombinant phytases conserved the same 22 amino acid residues that form phytate and calcium binding sites (except for N336 in FTE and D336 in the other phytases), their different specific activities may be due to different glycosylation percentages in shake-flask cultures. Accordingly, chromatographically purified FTEII produced in a 5-liter bioreactor showed a specific activity of 10.9 U/mg protein (unpublished results), in comparison to 13.3 U/mg protein for FTEII produced in shake flasks.
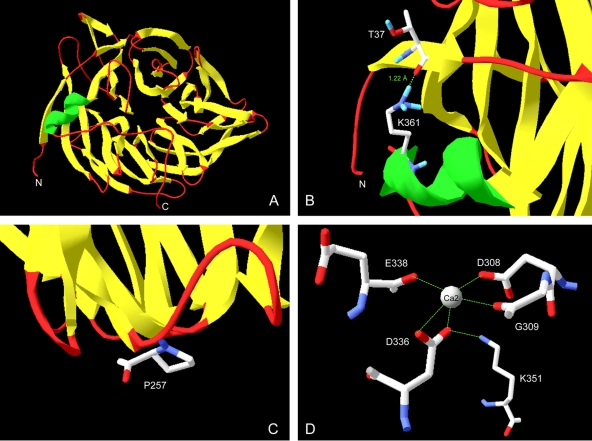
The present work not only deals with the design and production of thermostable phytases with activity at pH 7.5 and 5.5 for use in future biotechnological applications but also contributes to the understanding of the factors involved in the thermostability of enzymes of commercial interest. Numerous structural factors have been proposed to contribute to the thermostability of proteins, such as core and side chain packing, oligomerization, insertions and deletions, proline substitutions, helical content, helical propensities, polar surface area, hydrogen bonds, and salt bridges (24). Our structural analysis with molecular models (Fig. 4 A) suggests that the higher thermostabilities of FTEII and FBA than that of PhyC-R could be due to the larger number of hydrogen bonds in FTEII and FBA, specifically the hydrogen bond between T37 and K361 (Fig. 4B), which binds the ninth residue from the amino-terminal end with the alpha helix of the protein. In addition, the presence of P257, located in a surface loop (Fig. 4C) that does not perturb the three-dimensional structure of the protein, could increase the stability of FTEII and FBA, as suggested in the literature for other proteins (1, 3). A highly thermostable phytase from Bacillus subtilis US417 was recently described (7). The amino acid sequence of this phytase differs from that of PhyC of Bacillus subtilis VTTE-68013 only at position 257 (P versus R, respectively). This observation further supports the stabilizing effect of P257 in the Bacillus phytases. On the other hand, our structural analysis showed that the lower thermostability of FTE at pH 7.5 than those of the other phytases was due to the D336N substitution. This substitution led to the removal of three salt bridges of D336: two with one Ca2+ ion involved in the stability of the enzyme (16) and one with K351 (Fig. 4D). Salt bridges and hydrogen bonds were proposed to be the most frequent factors that confer thermostability to proteins of thermophilic microorganisms in a study that compared sequences of mesophilic and thermophilic homologous proteins (24). Because we designed the FTE and FTEII amino acid sequences by use of homologous phytases from the genus Bacillus, we expect that salt bridges and hydrogen bonds also play important roles in their thermostability.
FIG. 4.
(A) Molecular model of FTEII constructed by homology modeling using SWISS-MODEL 8.05, with the 2POO structure as a template. (B) Hydrogen bond between T37 and K361 in the structure for FTE, FTEII, and FBA, binding the ninth residue from the N-terminal end with the alpha helix of the protein. (C) P257 in a surface loop in FTE, FTEII, and FBA. (D) Salt bridges between D336 and a Ca2+ ion or K351 in FTEII, FBA, and PhyC. Other salt bridges with the Ca2+ ion are also shown.
Although we expected a higher thermostability at pH 7.5 and 5 mM CaCl2 for FTEII than for FBA, due to the theoretical stabilization effect of F29, V81, and K292, no significant differences in thermostability have been found between the two phytases.
A different thermostability behavior was seen at pH 5.5 from that at pH 7.5 for the four phytases (Fig. 3), with higher thermostability at pH 5.5 than at pH 7.5. This difference is likely due to a different protein total charge at pH 5.5 from that at pH 7.5 (1 and −5, respectively, for PhyC and 2 and −6, respectively, for FTE, FTEII, and FBA), since the reported isoelectric point of native PhyC is 6.5 (22) and the estimated values for FTE, FTEII, and FBA are 6.66, 6.59, and 6.65, respectively. Among all of the ionizable residues, six residues for PhyC (H91, K232, D258, D312, E338, and H342), eight residues for FTE (H91, H94, D228, D258, D312, D341, H342, and H365), seven residues for FTEII (H91, H94, D258, D312, E338, H342, and H365), eight residues for FBA (H91, H94, E226, D258, D312, E338, H342, and H365), the carboxylate group of the C-terminal end for FTE, and the amino group of the amino-terminal end for FBA have pK values between 5.5 and 7.5 and are the groups responsible for the total charge change. Because native PhyC is more thermostable at pH 7.5 than at pH 5.5 (13), N-linked oligosaccharide chains likely stabilize the change of the total charge, affecting the charge distribution on the surface of the phytase. This phenomenon has been suggested to occur for a glycosylated fungal phytase (15), involving hydrogen bonds between the glycan core and the protein, as described in an X-ray crystallographic investigation of N-glycoprotein linkage region models (29).
Residual activities of PhyC-R and FBA at 80°C, pH 7.5, and 5 mM CaCl2 proved to be 0.5 and 0.8 times lower, respectively, than the residual activities of their native counterparts from B. subtilis, as tested in our laboratory (13), and from B. amyloliquefaciens, as described in the literature (23). This decrease in residual activity for recombinant versus native phytases is likely due to the recombinant phytases being N-glycosylated by P. pastoris, while the native phytases are not N-glycosylated. The presence of N-glycosylation in recombinant proteins has been reported as a stabilizing factor in some cases (15, 18) and a destabilizing factor in others (4). Other explanations could include a kinetic difference in Ca2+ binding in the recombinant phytases versus native phytases due to a steric hindrance of the oligosaccharide chains.
The Pichia pastoris expression system shows significant potential for producing phytases from the genus Bacillus and secreting them into the culture medium. Because high cell densities of P. pastoris can be achieved in a simple bioreactor in a controlled form, a 10- to 100-fold increase in production of the recombinant phytases designed in the present work can be expected. Based on their pH profiles, the designed beta-propeller phytases are more suitable than Aspergillus and E. coli phytases as feed additives for animals with neutral-basic digestive tracts, such as agastric aquaculture species (shrimp and several fishes), since these enzymes have an optimum pH near neutrality and the pHs of the digestive systems of agastric aquaculture species are near pH 7 or higher. In addition, these phytases are quite stable at the high temperatures encountered in the feed-pelleting process. The designed beta-propeller phytases can also be used in industrial food processes under gentle conditions (neutral pH and low temperatures) for phytate hydrolysis.
Acknowledgments
We are thankful for grants CN031-09 (PAICYT) from the Universidad Autónoma de Nuevo León and 59121-06 and 105532 from the Consejo Nacional de Ciencia y Tecnología (CONACYT). J.A.G.-L., J.G.C.-T., and M.C.-G. thank CONACYT for their fellowships.
We also thank Glen D. Wheeler for his stylistic suggestions in the preparation of the manuscript.
Footnotes
Published ahead of print on 6 August 2010.
REFERENCES
- 1.Barzegar, A., A. A. Moosavi-Movahedi, J. Z. Pedersen, and M. Miroliaei. 2009. Comparative thermostability of mesophilic and thermophilic alcohol dehydrogenases: stability-determining roles of proline residues and loop conformations. Enzyme Microb. Technol. 45:73-79. [Google Scholar]
- 2.Byrne, L. J., K. J. O'Callaghan, and M. F. Tuite. 2005. Heterologous gene expression in yeast. Methods Mol. Biol. 308:51-64. [DOI] [PubMed] [Google Scholar]
- 3.Choi, E. J., and S. L. Mayo. 2006. Generation and analysis of proline mutants in protein G. Protein Eng. Des. Sel. 19:285-289. [DOI] [PubMed] [Google Scholar]
- 4.Clark, S. E., E. H. Muslin, and C. H. Henson. 2004. Effect of adding and removing N-glycosylation recognition sites on the thermostability of barley α-glucosidase. Protein Eng. Des. Sel. 17:245-249. [DOI] [PubMed] [Google Scholar]
- 5.Diemand, A. V., and H. Scheib. 2004. iMolTalk: an interactive, Internet-based protein structure analysis server. Nucleic Acids Res. 32:W512-W516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ecamilla-Treviño, L. L., J. M. Viader-Salvadó, H. A. Barrera-Saldaña, and M. Guerrero-Olazarán. 2000. Biosynthesis and secretion of recombinant human growth hormone in Pichia pastoris. Biotechnol. Lett. 22:109-114. [Google Scholar]
- 7.Farhat, A., H. Chouayekh, M. B. Farhat, K. Bouchaala, and S. Bejar. 2008. Gene cloning and characterization of a thermostable phytase from Bacillus subtilis US417 and assessment of its potential as a feed additive in comparison with a commercial enzyme. Mol. Biotechnol. 40:127-135. [DOI] [PubMed] [Google Scholar]
- 8.Fiske, C. H., and Y. Subarrow. 1925. The colorimetric determination of phosphorus. J. Biol. Chem. 66:375-400. [Google Scholar]
- 9.Fraczkiewicz, R., and W. Braun. 1998. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 19:319-333. [Google Scholar]
- 10.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press Inc., Totowa, NJ.
- 11.Gordon, J. C., J. B. Myers, T. Folta, V. Shoja, L. S. Heath, and A. Onufriev. 2005. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33:W368-W371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerrero-Olazarán, M., L. Rodríguez-Blanco, J. G. Carreón-Treviño, J. A. Gallegos-López, M. Castillo-Galván, and J. M. Viader-Salvadó. 2007. Bacterial phytase produced in Pichia pastoris. J. Biotechnol. 131:S233-S234. [Google Scholar]
- 13.Guerrero-Olazarán, M., L. Rodríguez-Blanco, J. G. Carreón-Treviño, J. A. Gallegos-López, M. Castillo-Galván, and J. M. Viader-Salvadó. 2010. Expression of a Bacillus phytase C gene in Pichia pastoris and properties of the recombinant enzyme. Appl. Environ. Microbiol. 76:5601-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 15.Guo, M., H. Hang, T. Zhu, Y. Zhuang, J. Chu, and S. Zhang. 2008. Effect of glycosylation on biochemical characterization of recombinant phytase expressed in Pichia pastoris. Enzyme Microb. Technol. 42:340-345. [Google Scholar]
- 16.Ha, N. C., B. C. Oh, S. Shin, H. J. Kim, T. K. Oh, Y. O. Kim, K. Y. Choi, and B. H. Oh. 2000. Crystal structures of a novel thermostable phytase in partially and fully calcium-loaded states. Nat. Struct. Biol. 7:147-153. [DOI] [PubMed] [Google Scholar]
- 17.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 18.Han, Y. M., and X. G. Lei. 1999. Role of glycosylation in the functional expression of an Aspergillus niger phytase (phyA) in Pichia pastoris. Arch. Biochem. Biophys. 364:83-90. [DOI] [PubMed] [Google Scholar]
- 19.Ikai, A. 1980. Thermostability and aliphatic index of globular proteins. J. Biochem. 88:1895-1898. [PubMed] [Google Scholar]
- 20.Ilgen, C., J. Lin-Cereghino, and J. M. Cregg. 2004. Pichia pastoris, p. 143-162. In G. Gellissen (ed.), Production of recombinant proteins—novel microbial and eukaryotic expression systems. Wiley-VCH, Weinheim, Germany.
- 21.Invitrogen. 2009. Pichia expression kit for expression of recombinant proteins in Pichia pastoris. Invitrogen, San Diego, CA. http://tools.invitrogen.com/content/sfs/manuals/pich_man.pdf.
- 22.Kerovuo, J., M. Lauraeus, P. Nurminen, N. Kalkkinen, and J. Apajalahti. 1998. Isolation, characterization, molecular gene cloning, and sequencing of a novel phytase from Bacillus subtilis. Appl. Environ. Microbiol. 64:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, Y. O., H. K. Kim, K. S. Bae, J. H. Yu, and T. K. Oh. 1998. Purification and properties of a thermostable phytase from Bacillus sp. DS11. Enzyme Microb. Technol. 22:2-7. [Google Scholar]
- 24.Kumar, S., C.-J. Tsai, and R. Nussinov. 2000. Factors enhancing protein thermostability. Protein Eng. 13:179-191. [DOI] [PubMed] [Google Scholar]
- 25.Lehmann, M., C. Loch, A. Middendorf, D. Studer, S. F. Lassen, L. Pasamontes, A. P. van Loon, and M. Wyss. 2002. The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Eng. 15:403-411. [DOI] [PubMed] [Google Scholar]
- 26.Lehmann, M., D. Kostrewa, M. Wyss, R. Brugger, A. D'Arcy, L. Pasamontes, and A. P. van Loon. 2000. From DNA sequence to improved functionality: using protein sequence comparisons to rapidly design a thermostable consensus phytase. Protein Eng. 13:49-57. [DOI] [PubMed] [Google Scholar]
- 27.Lei, X. G., and C. H. Stahl. 2001. Biotechnological development of effective phytases for mineral nutrition and environmental protection. Appl. Microbiol. Biotechnol. 57:474-481. [DOI] [PubMed] [Google Scholar]
- 28.Lei, X. G., J. M. Porres, E. J. Mullaney, and H. Brinch-Pedersen. 2007. Phytase: source, structure and application, p. 505-529. In J. Polaina and A. P. MacCabe (ed.), Industrial enzymes: structure, function and applications. Springer, Dordrecht, Netherlands.
- 29.Loganathan, D., and U. Aich. 2006. Observation of a unique pattern of bifurcated hydrogen bonds in the crystal structures of the N-glycoprotein linkage region models. Glycobiology 16:343-348. [DOI] [PubMed] [Google Scholar]
- 30.Mullaney, E. J., and A. H. Ullah. 2007. Phytases: attributes, catalytic mechanisms and applications, p. 97-110. In L. Turner, A. E. Richardson, and E. J. Mullaney (ed.), Inositol phosphates: linking agriculture and the environment. CAB International, Oxfordshire, United Kingdom.
- 31.Ortmann, D., M. Ohuchi, H. Angliker, E. Shaw, W. Garten, and H. D. Klenk. 1994. Proteolytic cleavage of wild type and mutants of the F protein of human parainfluenza virus type 3 by two subtilisin-like endoproteases, furin and Kex2. J. Virol. 68:2772-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rawlings, N. D., F. R. Morton, C. Y. Kok, J. Kong, and A. J. Barrett. 2008. MEROPS: the peptidase database. Nucleic Acids Res. 36:D320-D325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin, S., N. C. Ha, B. C. Oh, T. K. Oh, and B. H. Oh. 2001. Enzyme mechanism and catalytic property of beta propeller phytase. Structure 9:851-858. [DOI] [PubMed] [Google Scholar]
- 36.Sreekrishna, K. 1993. Strategies for optimizing protein expression and secretion in the methylotrophic yeast Pichia pastoris, p. 119-126. In R. H. Baltz, G. D. Hegeman, and P. L. Skatrud (ed.), Industrial microorganisms: basic and applied molecular genetics. American Society for Microbiology, Washington, DC.
- 37.Vázquez-Figueroa, E., J. Chaparro-Riggers, and A. S. Bommarius. 2007. Development of a thermostable glucose dehydrogenase by a structure-guided consensus concept. Chembiochem 8:2295-2301. [DOI] [PubMed] [Google Scholar]
- 38.Xie, D., and E. Freire. 1994. Molecular basis of cooperativity in protein folding. V. Thermodynamic and structural conditions for the stabilization of compact denatured states. Proteins 19:291-301. [DOI] [PubMed] [Google Scholar]
- 39.Xiong, A. S., Q. H. Yao, R. H. Peng, P. L. Han, Z. M. Cheng, and Y. Li. 2005. High level expression of a recombinant acid phytase gene in Pichia pastoris. J. Appl. Microbiol. 98:418-428. [DOI] [PubMed] [Google Scholar]
- 40.Zhao, X., K. K. Huo, and Y. Y. Li. 2000. Synonymous codon usage in Pichia pastoris. Chin. J. Biotechnol. 16:308-311. [PubMed] [Google Scholar]