Abstract
To obtain genetic information about Campylobacter jejuni and Campylobacter coli from broilers and carcasses at slaughterhouses, we analyzed and compared 340 isolates that were collected in 2008 from the cecum right after slaughter or from the neck skin after processing. We performed rpoB sequence-based identification, multilocus sequence typing (MLST), and flaB sequence-based typing; we additionally analyzed mutations within the 23S rRNA and gyrA genes that confer resistance to macrolide and quinolone antibiotics, respectively. The rpoB-based identification resulted in a distribution of 72.0% C. jejuni and 28.0% C. coli. The MLST analysis revealed that there were 59 known sequence types (STs) and 6 newly defined STs. Most of the STs were grouped into 4 clonal complexes (CC) that are typical for poultry (CC21, CC45, CC257, and CC828), and these represented 61.8% of all of the investigated isolates. The analysis of 95 isolates from the cecum and from the corresponding carcass neck skin covered 44 different STs, and 54.7% of the pairs had matching genotypes. The data indicate that cross-contamination from various sources during slaughter may occur, although the majority of Campylobacter contamination on carcasses appeared to originate from the slaughtered flock itself. Mutations in the 23S rRNA gene were found in 3.1% of C. coli isolates, although no mutations were found in C. jejuni isolates. Mutations in the gyrA gene were observed in 18.9% of C. jejuni and 26.8% of C. coli isolates, which included two C. coli strains that carried mutations conferring resistance to both classes of antibiotics. A relationship between specific genotypes and antibiotic resistance/susceptibility was observed.
Campylobacteriosis is the leading food-borne bacterial gastroenteritis worldwide (12, 15). In Switzerland, the number of registered campylobacteriosis cases has rapidly increased to more than 100 per 100,000 inhabitants in the past few years (14), and this trend has also been observed in the European Union (EU) (12). However, the real number of cases is likely higher, because not all cases are reported due to the self-limiting nature of the disease and its potentially mild symptoms.
Campylobacter jejuni and Campylobacter coli are commonly associated with human infection, and they can be detected in up to 85% and 15% of cases, respectively (33). Despite the important role that C. jejuni and C. coli play as zoonotic pathogens worldwide, there is little information regarding the route(s) of transmission (17). Numerous case-control and modeling studies on the infection sources of C. jejuni and C. coli have suggested that handling and consumption of contaminated poultry meat are associated with a risk of human campylobacteriosis (17, 45, 47, 49, 51). Initial meat contamination with C. jejuni or C. coli from the chicken intestine may occur during commonly used automated slaughter processing through several routes, such as the air, water, previously slaughtered flocks, or machinery (19, 36, 37).
Precise genotyping and continuous comparison of the strains obtained from, e.g., the production site, flocks, slaughterhouse, retail meat, and infected humans would help to trace the source of infection and might indicate possible intervention strategies for the contaminated site.
DNA sequence-based typing methods, such as multilocus sequence typing (MLST), are well suited for this purpose (28), and MLST has become the method of choice for genotyping of Campylobacter (6, 8). Moreover, extension of the classical MLST technique for C. jejuni and C. coli with sequencing of the short variable region (SVR) within the flagellin-encoding gene flaB allows a more precise differentiation among strains that have the same MLST sequence type (ST) (9, 29). An extended MLST work flow was recently developed that reduces the associated time and cost (24). In addition, the new approach allows genetic determination of antibiotic resistance to quinolones and macrolides. Resistance to these antibiotics is a worldwide issue of concern, as an increasing number of Campylobacter isolates are resistant to them. Strikingly, a number of strains are resistant to ciprofloxacin (a quinolone) and, to a lesser extent, erythromycin (a macrolide), which is problematic, because these drugs are typically used to treat campylobacteriosis. Resistance to quinolones is mainly associated with a point mutation in the DNA gyrase gene (gyrA) at position C257T, and a transition in the 23S rRNA gene at position A2075G is commonly responsible for macrolide resistance (1). Simple sequence-based analysis of these common mutational positions can therefore provide information about the antibiotic susceptibility or resistance of a strain. Besides the prudent use of antibiotics, knowledge about the genetic composition of the infectious agent can be helpful to both treat the disease and prevent the spread of resistant strains.
In the current study, MLST, flaB typing, and sequence-based determination of quinolone and macrolide resistances were used to investigate the genetic background of C. jejuni and C. coli isolates collected from Swiss broilers in a spatiotemporal study in 2008. We addressed the following three aspects: (i) the diversity of Campylobacter isolates that were recovered from pooled cecum samples and the carcass neck skin, (ii) the possible impact of cross- and self-contamination during slaughter, and (iii) the antibiotic resistance of Campylobacter strains from the broiler flocks and chicken carcasses. All of the data, including the strain information and trace files, were entered into a commercial Web-based Campylobacter MLST database (SmartGene, Zug, Switzerland). This database allows users to retrieve and compare information for any analyzed strain for monitoring purposes (24).
MATERIALS AND METHODS
Sampling and template preparation.
Broiler cecal and neck skin samples were collected during the EU baseline study on the prevalence of Campylobacter in broiler flocks (11), in which Switzerland took part within the framework of the bilateral approach. Samples were obtained from 411 different flocks at 5 different suppliers in Switzerland (Table 1), which together cover approximately 80% of poultry meat production in the country. Each sample consisted of intact ceca from 10 birds and neck skin from one bird from the same slaughter batch. The samples were collected at the slaughterhouses once a week for at least 44 weeks during 2008. The cecal samples were taken at the time of evisceration by careful manual traction at the junction with the intestine. The neck skins were collected directly after chilling but before further processing, such as freezing, cutting, or packaging (11). The samples were processed in the Center for Zoonosis, Bacterial Animal Diseases and Antibiotic Resistance in Bern, Switzerland (ZOBA), according to the EU recommendations (11). The neck skin batches were processed as previously described for qualitative detection of thermotolerant Campylobacter (43). For the cecal samples, the cecal contents of 10 birds were aseptically removed and pooled. For direct cultivation, a loopful of the material was streaked onto a modified charcoal-cefoperazone-desoxycholate agar (mCCDA) (Oxoid, Pratteln, Switzerland), as well as a campylosel (Oxoid) medium. The plates were incubated under microaerobic conditions at 41.5°C for 48 h. After initial cultivation of bacteria from the neck skin and the cecal batches, at least one presumptive Campylobacter sp. colony from each plate was subcultured on tryptic soy agar (TSA) medium (Oxoid) at 41.5°C for 24 to 48 h under microaerobic conditions. Isolates that belonged to the genus Campylobacter were identified by microscopy, a positive oxidase test, and the absence of growth on TSA after 24 to 48 h under aerobic conditions at 41.5°C, as well as microaerobic incubation at 25°C. Phenotypic species identification was performed as previously reported by Schnider et al. (43). Hydrolysis of indoxyl acetate was used to differentiate the Campylobacter spp. from Helicobacter pullorum, as it was negative for the latter. A cell lysate was prepared from each pure culture as described previously (24).
TABLE 1.
Summary of MLST data, supplier, and number of Swiss broiler isolates analyzed
| Origin | Bacterial species | CCa | STb | No. of isolates from slaughterhouse: |
Total | ||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | |||||
| Cecum | C. jejuni | 21 | 19 | 0 | 0 | 0 | 0 | 1 | 1 |
| 21 | 0 | 4 | 1 | 1 | 1 | 7 | |||
| 50 | 0 | 1 | 4 | 0 | 1 | 6 | |||
| 262 | 0 | 1 | 0 | 0 | 0 | 1 | |||
| 883 | 0 | 1 | 0 | 0 | 0 | 1 | |||
| 917 | 0 | 1 | 0 | 0 | 0 | 1 | |||
| 22 | 22 | 1 | 0 | 2 | 0 | 0 | 3 | ||
| 45 | 45 | 2 | 7 | 2 | 0 | 1 | 12 | ||
| 137 | 0 | 2 | 1 | 0 | 0 | 3 | |||
| 1964 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| 48 | 48 | 1 | 5 | 2 | 1 | 1 | 10 | ||
| 61 | 61 | 0 | 1 | 0 | 0 | 0 | 1 | ||
| 206 | 122 | 0 | 2 | 1 | 0 | 0 | 3 | ||
| 257 | 257 | 0 | 1 | 4 | 1 | 1 | 7 | ||
| 283 | 267 | 0 | 1 | 0 | 0 | 0 | 1 | ||
| 383 | 0 | 1 | 0 | 0 | 0 | 1 | |||
| 354 | 354 | 0 | 1 | 0 | 0 | 0 | 1 | ||
| 1073 | 0 | 0 | 0 | 0 | 1 | 1 | |||
| 443 | 51 | 0 | 1 | 0 | 0 | 0 | 1 | ||
| 460 | 2952 | 0 | 2 | 0 | 0 | 0 | 2 | ||
| 607 | 3963 | 0 | 0 | 0 | 2 | 0 | 2 | ||
| ND | 464 | 0 | 2 | 0 | 0 | 3 | 5 | ||
| ND | 586 | 0 | 2 | 1 | 0 | 1 | 4 | ||
| ND | 2655 | 0 | 1 | 0 | 0 | 0 | 1 | ||
| Total | 4 | 37 | 19 | 5 | 11 | 76 | |||
| C. coli | 828 | 825 | 0 | 2 | 1 | 2 | 0 | 5 | |
| 827 | 1 | 4 | 6 | 0 | 0 | 11 | |||
| 829 | 0 | 0 | 0 | 0 | 1 | 1 | |||
| 854 | 0 | 1 | 3 | 0 | 1 | 5 | |||
| 1096 | 0 | 2 | 1 | 0 | 0 | 3 | |||
| 1545 | 0 | 1 | 0 | 0 | 0 | 1 | |||
| 1556 | 0 | 0 | 0 | 1 | 0 | 1 | |||
| 1563 | 0 | 1 | 0 | 0 | 0 | 1 | |||
| 1614 | 0 | 0 | 0 | 0 | 1 | 1 | |||
| 2142 | 0 | 2 | 2 | 0 | 1 | 5 | |||
| 3336 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| ND | 1049 | 0 | 0 | 0 | 0 | 1 | 1 | ||
| ND | 1680 | 0 | 0 | 1 | 0 | 0 | 1 | ||
| ND | 3989 | 0 | 1 | 0 | 0 | 0 | 1 | ||
| Total | 1 | 14 | 15 | 3 | 5 | 38 | |||
| Neck skin | C. jejuni | 21 | 19 | 0 | 1 | 0 | 0 | 0 | 1 |
| 21 | 0 | 5 | 3 | 2 | 0 | 10 | |||
| 50 | 0 | 4 | 7 | 0 | 1 | 12 | |||
| 262 | 0 | 1 | 1 | 0 | 0 | 2 | |||
| 883 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| 917 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| 3988 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| 22 | 22 | 1 | 3 | 6 | 0 | 0 | 10 | ||
| 42 | 42 | 0 | 2 | 0 | 0 | 0 | 2 | ||
| 45 | 11 | 0 | 1 | 0 | 0 | 0 | 1 | ||
| 45 | 1 | 8 | 11 | 0 | 2 | 22 | |||
| 137 | 0 | 2 | 0 | 0 | 0 | 2 | |||
| 418 | 0 | 1 | 0 | 0 | 0 | 1 | |||
| 782 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| 1964 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| 2197 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| 2219 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| 48 | 48 | 0 | 4 | 3 | 0 | 0 | 7 | ||
| 52 | 52 | 0 | 0 | 1 | 0 | 1 | 2 | ||
| 61 | 61 | 0 | 2 | 1 | 0 | 0 | 3 | ||
| 177 | 1388 | 0 | 1 | 0 | 0 | 0 | 1 | ||
| 206 | 122 | 0 | 3 | 0 | 0 | 0 | 3 | ||
| 227 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| 572 | 0 | 1 | 0 | 0 | 0 | 1 | |||
| 257 | 257 | 0 | 1 | 15 | 0 | 4 | 20 | ||
| 824 | 0 | 0 | 0 | 0 | 1 | 1 | |||
| 283 | 267 | 0 | 2 | 1 | 0 | 2 | 5 | ||
| 383 | 0 | 1 | 0 | 0 | 0 | 1 | |||
| 353 | 353 | 0 | 2 | 0 | 0 | 0 | 2 | ||
| 354 | 878 | 1 | 3 | 4 | 0 | 0 | 8 | ||
| 1073 | 0 | 2 | 1 | 0 | 2 | 5 | |||
| 362 | 587 | 0 | 0 | 1 | 0 | 0 | 1 | ||
| 443 | 51 | 0 | 1 | 1 | 0 | 0 | 2 | ||
| 460 | 2952 | 0 | 1 | 0 | 0 | 0 | 1 | ||
| 607 | 607 | 0 | 1 | 1 | 0 | 0 | 2 | ||
| 3963 | 0 | 1 | 2 | 7 | 0 | 10 | |||
| 677 | 677 | 0 | 0 | 1 | 0 | 0 | 1 | ||
| 1034 | 1956 | 1 | 0 | 0 | 0 | 0 | 1 | ||
| ND | 441 | 0 | 0 | 1 | 1 | 0 | 2 | ||
| ND | 464 | 0 | 3 | 0 | 0 | 2 | 5 | ||
| ND | 586 | 0 | 5 | 3 | 0 | 2 | 10 | ||
| ND | 1962 | 0 | 0 | 1 | 0 | 0 | 1 | ||
| ND | 3964 | 0 | 0 | 0 | 0 | 1 | 1 | ||
| 362 | 587 | 0 | 0 | 1 | 0 | 0 | 1 | ||
| Total | 4 | 62 | 73 | 10 | 18 | 167 | |||
| C. coli | 828 | 825 | 0 | 1 | 2 | 4 | 0 | 7 | |
| 827 | 3 | 10 | 7 | 0 | 1 | 21 | |||
| 829 | 0 | 3 | 0 | 0 | 1 | 4 | |||
| 854 | 0 | 1 | 2 | 1 | 0 | 4 | |||
| 1016 | 0 | 2 | 0 | 0 | 0 | 2 | |||
| 1096 | 0 | 2 | 0 | 0 | 1 | 3 | |||
| 1413 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| 1614 | 0 | 0 | 1 | 0 | 1 | 2 | |||
| 2142 | 0 | 5 | 4 | 0 | 0 | 9 | |||
| 3023 | 0 | 0 | 0 | 1 | 0 | 1 | |||
| 3990 | 0 | 0 | 1 | 0 | 0 | 1 | |||
| 3991 | 0 | 1 | 0 | 0 | 0 | 1 | |||
| ND | 1584 | 0 | 0 | 0 | 2 | 0 | 2 | ||
| ND | 3989 | 0 | 1 | 0 | 0 | 0 | 1 | ||
| Total | 3 | 26 | 18 | 8 | 4 | 59 | |||
ND, not defined.
New STs are indicated by boldface.
A total of 149 isolates from the cecum and 248 isolates from the neck skin were obtained after cultivation and phenotypic identification. Isolate species identification through phenotypic testing was verified by sequence analysis of the rpoB gene fragment. Only the isolates confirmed as C. jejuni or C. coli were investigated further. The strain set allowed a comparison of 95 isolate pairs in which there was cultivation of Campylobacter spp. from the ceca and neck skin of the same slaughtered flock.
Genotyping and determination of antibiotic resistance.
The rpoB gene fragment was amplified with the broad-range primers CamrpoB-L, Pasrpob-L, and Rpob-R and was subsequently sequenced as the first stage of genetic characterization for species identification (22, 23). The genotyping and antibiotic resistance analyses were performed as previously described by Korczak et al. (24), with a few adaptations concerning the combination of target genes in amplification groups. The 10 target genes were amplified in 3 multiplex PCRs: the first multiplex PCR amplified the glmM, aspA, and 23S rRNA gene fragments; the second PCR amplified the glnA, tkt, flaB, and gyrA gene fragments; and the third PCR amplified the glyA, atpA, and gltA gene fragments.
Data analysis.
Data analysis was performed with the commercial Web-based application for Campylobacter identification, typing, and antibiotic resistance determination (IDNS Campylobacter; SmartGene, Zug, Switzerland). In addition to the data entry and sequence trace file editing, the ST and clonal complex (CC) were assigned with an integrated automated link to the public PubMLST data bank (http://pubmlst.org/campylobacter). The CCs are defined as a group of independent isolates that share at least four alleles (8). The genotypes of flaB were determined with a tool that is provided by the PubMLST database (http://pubmlst.org/campylobacter). All of the new genotypes were submitted directly to the curator of the PubMLST database. The BioNumerics software, version 5.1 (Applied Maths NV, Sint-Martens-Latem, Belgium), was used for cluster analysis of concatenated sequences, whereas BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to compare the rpoB sequences and to identify species.
Statistical analysis.
The binomial probability function of Statistix 9.0 for Windows (Analytical Software, Tallahassee, FL) was used to calculate the probability values for matching STs in the paired cecum and neck skin samples. The association of certain genotypes with the presence of quinolone resistance and their local distribution were tested by Fisher's exact two-tailed test. A P value of ≤0.05 was used to indicate statistically significant results.
The Simpson's index of diversity was calculated as previously described (18).
The frequency distributions of STs that were obtained from the different suppliers were compared by calculating the proportional similarity index (PSI), also known as the Czekanowski index (42). The frequency distributions of the different sources were estimated by calculating their similarity using the following equation: PSI = 1 − 0.5 Σi|pi − qi|, where pi and qi represent the proportions of strains that belong to ST i out of all of the strains that belong to the sources P and Q, respectively. A value of 1 indicates identical frequency distributions, and a value of 0 indicates no common types.
To investigate the population structures of the isolates from different suppliers, the correlated allele frequency model of STRUCTURE 2.3 was used (13, 40). The population number (K) was set as 5. The parameters were 100,000 burn-in iterations, followed by 100,000 sampling iterations using the admixture model to determine the FST values as a measure of genetic variance among populations, with high FST values indicative of separate populations.
Nucleotide sequence accession numbers.
The new rpoB sequences were submitted to GenBank under the accession numbers listed in Table 2.
TABLE 2.
Accession numbers and numbers of isolates that represent identical rpoB gene fragment sequences
| Species | Accession no.a | No. of identical isolates |
|---|---|---|
| C. jejuni | HM486850 | 55 |
| HM486851 | 1 | |
| HM486852 | 1 | |
| HM486853 | 5 | |
| HM486854 | 11 | |
| CP000538 | 18 | |
| HM486855 | 14 | |
| HM486856 | 1 | |
| DQ174198 | 87 | |
| CP000814 | 49 | |
| HM486857 | 1 | |
| HM486858 | 1 | |
| HM486859 | 1 | |
| C. coli | HM486860 | 7 |
| HM486861 | 2 | |
| HM486862 | 6 | |
| AF372098 | 78 | |
| DQ174193 | 2 | |
| H. pullorum | HM486863 | 2 |
| HM486864 | 1 | |
| HM486865 | 5 | |
| HM486866 | 5 | |
| HM486867 | 2 | |
| HM486868 | 1 | |
| HM486869 | 9 | |
| HM486870 | 2 | |
| HM486871 | 1 | |
| HM486872 | 2 | |
| HM486873 | 1 | |
| HM486874 | 1 | |
| HM486875 | 2 | |
| HM486876 | 1 | |
| HM486877 | 1 | |
| HM486878 | 2 | |
| HM486879 | 4 | |
| HM486880 | 1 | |
| HM486881 | 1 | |
| HM486882 | 1 | |
| HM486883 | 2 | |
| HM486884 | 1 | |
| HM486885 | 1 | |
| HM486886 | 1 | |
| HM486887 | 2 | |
| HM486888 | 2 | |
| A. butzleri | HM486889 | 1 |
| HM486890 | 1 | |
| HM486891 | 1 |
The accession numbers obtained in this study are indicated by boldface.
RESULTS
Genetic identification.
In most cases, phylogenetic rpoB-based analysis (23) confirmed the results from the phenotypic identification of the isolates. There was a total of 245 C. jejuni, 95 C. coli, 54 H. pullorum, and 3 Arcobacter butzleri isolates recognized. The rpoB sequence similarities between the C. jejuni and C. coli, C. jejuni and H. pullorum, and C. jejuni and A. butzleri isolates varied from 83.17 to 98.97%, 71.79 to 74.07%, and 71.14 to 72.58%, respectively. The C. coli isolates showed 71.79 to 75.73% rpoB sequence similarity to H. pullorum and 71.35 to 73.83% sequence similarity to A. butzleri, whereas the H. pullorum and A. butzleri isolates demonstrated 71.85 to 73.5% rpoB sequence similarity. Twenty-one of the isolates (5.3%) had conflicting genotype and phenotype data. Nine strains were phenotypically identified as C. coli but were later classified as C. jejuni because of their rpoB gene sequence identities (sequences identical to C. jejuni accession no. CP000538, CP000814, or DQ174198). Five phenotypically identified C. jejuni isolates were later classified as C. coli (sequences identical to C. coli accession no. AF372098, HM486861, or HM486862). Two strains phenotypically identified as H. pullorum were later identified as C. coli (sequence identical to accession no. AF372098) and C. jejuni (sequence identical to accession no. CP000538) by rpoB gene analysis. The opposite was observed with one C. jejuni and one C. coli isolate, where rpoB gene sequencing identified the strains as H. pullorum (sequences identical to accession no. HM486886 and HM486887). Finally, one C. jejuni and two C. coli isolates were genetically determined to be A. butzleri (Table 2). Interestingly, rpoB-based identification of two indistinguishable paired isolates (i.e., the same strain) did not agree with the results from further genetic analysis. These isolates were identified by rpoB sequencing as C. jejuni (rpoB sequence identical to accession no. DQ174198), but all of the other target genes used for MLST and detection of antibiotic resistance were specific for C. coli.
The prevalences of C. jejuni and C. coli isolated from the cecum and neck skin samples differed slightly, but this difference was not significant. Within 114 cecal strains, 76 (66.7%) were C. jejuni and 38 (33.3%) were C. coli; of the 226 neck skin samples, 167 (73.9%) were C. jejuni and 59 (26.1%) were C. coli.
Typing.
Of the 340 Campylobacter isolates that were analyzed, a total of 65 different STs were identified (Table 1). Six of these were novel STs that comprised 18 (5.3%) isolates. These STs have been submitted to the PubMLST database for appropriate number designations and may be the result of new combinations of previously described alleles. However, for ST-3991, a new aspA allele sequence was found and was designated 248. Twenty of the STs were derived from C. coli isolates, and 45 of the STs were observed within the C. jejuni isolates. Eight STs were found only in isolates from the cecum, and 26 STs were found only in the isolates that were obtained from neck skin; all of the other STs were present in both sample groups. Some of the isolates that belonged to the same ST could be further distinguished when the entire amplicon sequence was compared. Differences within the regions that flank the gene region that is defined as an allele by PubMLST were observed for ST-45, ST-122, ST-267, and ST-1096.
The most frequent STs (number of isolates [n] ≥ 14) within C. jejuni were ST-45 (n = 34/14.0%), ST-257 (n = 27/11.1%), ST-50 (n = 18/7.4%), ST-21 (n = 17/7.0%), ST-48 (n = 17/7.0%), and ST-586 (n = 14/5.8%). The new ST-3963 was observed within several of the C. jejuni isolates (n = 12/4.9%). The predominant STs within C. coli were ST-827 (n = 32/33.0%) and ST-2142 (n = 14/14.4%).
Fifty-five STs, which were present in 305 (89.7%) isolates, belonged to 20 previously defined clonal complexes. The remaining 35 isolates were distributed among 10 STs, which could not be assigned to any of the known lineages. The predominant clonal complexes were CC21 (18.5%), CC45 (18.9%), and CC257 (11.5%) for C. jejuni and CC828 (93.8%) for C. coli.
Investigation of the distribution of STs resulted in recognition of 7 different STs in 13 isolates obtained from slaughterhouse A, 42 STs in 138 isolates obtained from slaughterhouse B, 40 STs in 125 isolates from slaughterhouse C, 10 STs in 26 isolates from slaughterhouse D, and 20 STs in 38 isolates obtained from slaughterhouse E (Table 1). There was a local association of ST-257 and the new ST-3963 that was statistically significant. ST-257 was the most prominent ST in the sample set from slaughterhouse C (15.2%) compared to the others, especially slaughterhouse B, which had a similar sample size but had only a 1.4% representation of ST-257. ST-3963 was dominant in the samples that were obtained from slaughterhouse D (34.6%).
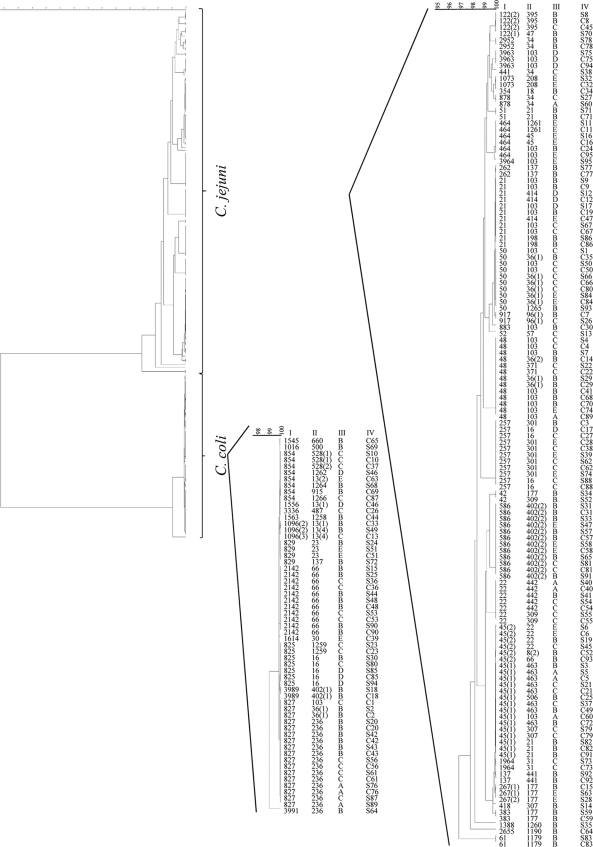
The analysis of 95 paired samples identified 44 different STs. Whereas matching STs were found in 52 (54.7%) of these samples, 43 (45.3%) of the paired samples did not have matching STs (Fig. 1). Given the various STs that were observed, the probability of obtaining this high number of matches by chance is P ≃ 0 based on statistical analysis using binomial probability.
FIG. 1.
Clustering of the paired sample strains using the entire set of MLST target sequences. The unweighted-pair group method using average linkages (UPGMA) tree was applied in BioNumerics. The ST (I), flaB types (II), and suppliers' designations (III) are indicated. Paired samples (IV) are labeled with corresponding numbers for the cecum (C) and neck skin (S). The numbers in parentheses indicate identical STs that are variants based on differences that were observed within flanking regions of the sequence used for allele definition by PubMLST.
Comparison of the flaB genotypes to the PubMLST database recognized 48 known and 11 new types (Fig. 2). Cluster analysis of the sequenced flaB fragments (446 bp) revealed 67 clusters (Fig. 2). This increase in resolution is due to the differences in flanking regions of the sequence used for allele assignment by the PubMLST database. In some cases, flaB genotyping, as well as sequence analysis, allowed further separation of strains that belonged to the same ST, although different STs can be found with the same flaB sequence. In contrast to the MLST data, there was no clear differentiation between the flaB types from C. jejuni and C. coli, and in some cases, the two species had identical flaB sequences. All 52 of the paired samples that showed identical STs also had matching flaB sequences.
FIG. 2.
Clustering of the strains based on the partial flaB gene sequences. The UPGMA tree was applied in BioNumerics. The flaB type (I), species (II), and ST (III) are indicated. The numbers in parentheses indicate variants of identical flaB types within flanking regions of the sequence used for allele definition by PubMLST. New flaB types described in this study are indicated by boldface.
The Simpson's index of discrimination was calculated to be 0.958 for the MLST analysis alone, 0.953 for the flaB genotype analysis, and 0.978 for the combination of both genotyping methods. Analysis of the concatenated sequences of MLST gene fragments, including the flanking regions of alleles, increased the discriminatory power slightly to 0.963 and increased the combined MLST flaB value to 0.978, whereas the Simpson's index for the flaB sequence-based analysis was 0.955.
Genetic population structure.
The PSI was calculated to assess the similarity of ST distributions between the five different slaughterhouses (Table 3). A relatively high similarity value was observed for ST distribution between suppliers B and C (PSI = 0.579), which are the two major producers in Switzerland. The PSI was also calculated for the cecum and neck skin, and the resulting PSI of 0.697 indicates that there is a high overlap of ST distribution.
TABLE 3.
Proportional similarity indices of ST distribution among the five suppliers
| Supplier | PSI with supplier: |
|||
|---|---|---|---|---|
| A | B | C | D | |
| B | 0.317 | |||
| C | 0.344 | 0.579 | ||
| D | 0.038 | 0.161 | 0.195 | |
| E | 0.132 | 0.426 | 0.467 | 0.117 |
FST values based on the seven MLST alleles were determined for each of the 5 subpopulations with the following values: supplier A = 0.008, supplier B = 0.635, supplier C = 0.511, supplier D = 0.720, and supplier E = 0.671.
Antibiotic resistance.
The point mutation C257T (corresponding to C150T in our fragment) in the gyrA gene, which is associated with quinolone resistance, was observed in 46 C. jejuni (18.9%) and 26 C. coli (26.8%) isolates. Twenty-three cecal isolates (13 C. jejuni and 10 C. coli) and 49 neck skin isolates (33 C. jejuni and 16 C. coli) carried this mutation. The transition A2075G (corresponding to A227G in our fragment) in the 23S rRNA gene, which contributes to macrolide resistance, was observed in 3 C. coli (3.1%) isolates. Two out of these three C. coli strains carried point mutations in both genes, whereas none of the C. jejuni isolates showed macrolide resistance.
Several specific genotypes showed significant association with quinolone resistance. Whereas none of the CC45 (P < 0.01), CC22 (P < 0.01), CC257 (P < 0.01), CC607 (P < 0.05), or ST-586 (P < 0.05) strains had a mutation in the gyrA gene, all isolates of the ST-464 (P < 0.01), ST-829 (P < 0.01), and ST-878 (P < 0.01) strains had the specific mutation that confers quinolone resistance.
DISCUSSION
In this study, we analyzed the genetic diversity and antibiotic resistance of Campylobacter isolates in pooled cecum and neck skin samples from slaughter broilers and possible routes of carcass contamination at various slaughterhouses.
Proper phenotypic identification of Campylobacter isolates, especially differentiation between C. jejuni and C. coli based on the hippurate test, might be difficult and could result in false isolate identification (35). Additionally, phenotypic discrimination between Campylobacter spp. and other phenotypically similar species that are present in the isolate sampling environment, such as H. pullorum and A. butzleri, can be difficult or even impossible (38). rpoB gene analysis allowed individual species identification within the isolated strains; the data we obtained with this analysis corroborated the results from phenotypic characterization (94.7%). Any minor disagreements between the genotype and phenotype data reflect the previously mentioned difficulties with interpretation of the phenotypic tests. The rpoB-based identification of Campylobacter could finally be confirmed by the species-specific STs determined by MLST, as well as by gyrA sequencing results that also allowed phylogenetic separation of the two species (24). The only exception was one case of paired isolates (i.e., the same strain) that possessed an rpoB gene sequence that matched C. jejuni but had an ST and gyrA sequence that were specific for C. coli. This could have been caused by genetic recombination, which is known to occur between these highly related species (3, 46, 50). Thus, even though the rpoB gene sequence can generally be used to discriminate between the two species, false species identification can result from genetic recombination.
Roughly one-third of the isolates were C. coli, which corresponds to previously reported values from isolates in Swiss poultry (24, 53). Thus, both C. jejuni and C. coli are present in poultry samples and may cause campylobacteriosis in humans. Nevertheless, although both of these species are prevalent in poultry samples, less than 15% of C. coli isolates are typically associated with human disease (33). Therefore, C. coli may have a decreased ability to survive on broiler carcasses, there could be a greater number of virulent C. jejuni strains, or other sources may contribute to C. jejuni infection.
Within the sample set of 340 isolates, 65 known STs were assigned by MLST. Most of these STs could be assigned to four main CCs (CC21, CC45, CC257, and CC828). CC21 and CC45 are also the most frequently reported C. jejuni genotypes in human disease and can be detected in up to 21.8% and 44.6% of investigated cases, respectively (4, 21, 41). Strains assigned to these lineages are common worldwide in a number of hosts, including poultry, cattle, sheep, wild birds, and the natural environment (7, 21, 44). Therefore, identifying a direct link between a specific carrier and campylobacteriosis may be difficult. In contrast, CC257 can be identified in up to 24.2% of human cases and is mainly found in poultry samples. Thus, human disease caused by C. jejuni isolates assigned to this group probably occurs after handling or consumption of contaminated poultry meat (4, 16, 24, 30, 44, 49).
CC61 is identified frequently in cattle and sheep and the feces of diseased humans (41, 45). Surprisingly, we identified CC61 in one paired sample and two single neck skin samples. Swiss agriculture is based on small mixed-animal farms; thus, strains can be easily transmitted between different hosts (7). Because C. coli is most prevalent in swine, pork products are considered to be the leading source of human infection (39). However, studies of C. coli genetics have shown that only a small number of the genotypes found in swine can be detected in samples from infected humans and other hosts (25, 47). In contrast, a considerable genetic overlap between C. coli isolates from poultry and humans was previously observed, suggesting that infection was most likely due to the consumption of contaminated poultry meat. Interestingly, ST-827, which was the most prevalent genotype of the C. coli isolates (33.0%) that were analyzed in this study, was also predominant (28.1%) in a set of C. coli strains that were isolated from humans (48). Because this C. coli genotype is rare in isolates from hosts other than chickens and humans, ST-827 may have come from chicken meat (6, 26, 31).
In contrast to a recent study in New Zealand, population structure analyses of our set of strains gave only slight indication that specific populations can be discerned from a specific supplier (34). The two major Swiss broiler suppliers analyzed (B and C) had similar ST distributions (PSI = 0.579). An association between a particular sample supplier and several genotypes was observed for two of the STs. ST-257 was the most frequent genotype in the samples that were collected from slaughterhouse C, and the novel genotype ST-3963 was found in 34.6% of isolates from slaughterhouse D. A high FST value of 0.720 for the latter suggests that these genotypes may represent a local clone. Further analyses of Campylobacter bacteria that are present in the delivery areas of both slaughterhouses would be interesting to test for the presence of a local clone and to attempt to track the source of campylobacteriosis.
MLST analysis of Swiss Campylobacter isolates has only recently been utilized. Accumulation of more data about the strain genotypes from various sources will give a more comprehensive picture of the distributions, population structures, and sources of C. jejuni and C. coli contamination in Switzerland.
We compared 95 sample pairs of cultivated Campylobacter isolates that were obtained from the cecum and neck skin from the same flock. More than half of these pairs had matching STs. Statistical analysis indicated that this is a highly significant association and that the majority of samples obtained from the neck skin originated from the slaughtered flock itself. Given the fact that only single colonies were analyzed and that both species and various genotypes can be present in the flock, the true number of self-contaminated carcasses can be quite large (5, 52). The remaining nonmatching isolates that were obtained from the neck skin could have come from other contaminated sources, such as previously slaughtered batches, equipment, working surfaces, or water (19, 36, 49). Additionally, cultivation of Campylobacter from the cecum samples was unsuccessful in some cases. Interestingly, certain STs were found only in the cecum, whereas others were found only on the neck skin. Whether this indicates that environmental factors during the slaughter process lead to carcass contamination remains to be more thoroughly investigated.
The flaB-based genotyping confirmed the results that were obtained with the MLST analysis for the paired samples; pairs that had identical STs also had identical flaB sequences. Generally, the resolution of the flaB sequence-based genotyping was slightly higher than that of MLST. In contrast to the sequence data from the MLST targets, the flaB sequence does not allow phylogenetic analysis of the strains or differentiation between C. jejuni and C. coli. However, it represents a different typing method that is useful for short-term investigations due to the known instability of this marker, wherein that flaB is more stable than the conventionally used flaA (29). Sequence analysis of the entire amplicons of the MLST target genes and flaB increases the discriminatory power only slightly, although this technique may be useful to further differentiate between certain strains.
An increase in the number of C. jejuni and C. coli strains that are resistant to frequently used antibiotics (macrolides and, especially, the quinolones) has been reported worldwide (1, 10, 32). In this study, no C. jejuni isolates and only 3.1% of the C. coli isolates were macrolide resistant, whereas 18.9% of C. jejuni and 26.8% of C. coli isolates were resistant to quinolones. Macrolides and the quinolones are allowed for use in veterinary medicine in Switzerland. A high number of quinolone-resistant isolates could be associated with common therapeutic application of enrofloxacin in broiler flocks; enrofloxacin treatment has been shown to induce Campylobacter resistance and should therefore be used prudently (2, 20). However, the number of antibiotic-resistant C. jejuni and C. coli strains in Switzerland is in the lower range of the European average reported antimicrobial resistance cases in poultry (27).
Remarkably, there was an association between specific genotypes and resistance to quinolone. Because all of the broiler strains that belong to the genotypes ST-464, ST-829, and ST-878 carried the same point mutation within the gyrA gene that is responsible for quinolone resistance, either these Campylobacter genotypes are prone to mutations within this gene or these STs are clonal. On the other hand, strains that belong to the frequently found clonal complexes CC45, CC22, CC257, CC607, and ST-586 appeared to be less susceptible to mutations within the gyrA gene. Previous studies of Belgian and Swiss Campylobacter isolates reported similar results with CC45, where only single strains from this clonal complex were resistant to quinolone (16, 24). Additionally, 30% of C. jejuni CC21 isolates were resistant to quinolone, which was also seen in a previous study (16); this further suggests that there is a correlation between specific genotypes and antibiotic resistance.
In conclusion, we found that C. coli represented approximately one-third whereas C. jejuni represented the other two-thirds of the Campylobacter isolates that were obtained from Swiss slaughter broilers. The isolates showed high genetic variability in MLST and flaB genotyping, with most common STs and CCs also described for poultry in other countries. A few genotypes showed association with local suppliers. Comparison of the paired samples indicated that there was mainly “self-contamination” of slaughtered broilers, although cross-contamination of carcasses cannot be excluded. Intervention at the production level could therefore have a most promising effect on the presence of Campylobacter in poultry products. Antibiotic resistance to macrolides and quinolones in C. jejuni and C. coli seems less pronounced in Switzerland than in other European countries. Nevertheless, the presence of antibiotic-resistant strains indicates that such antibiotics should be used carefully, as overuse could lead to selection and spread of resistant bacteria.
Acknowledgments
We thank Elisabeth Lüthi, Stefani Ackermann, Stefanie Kaderli, and Yvonne Schlatter for their technical help. We are grateful to Gerald Heckel for helpful discussions about population genetics.
This research was supported by a Swiss Federal Veterinary Office grant (1.08.12).
Footnotes
Published ahead of print on 13 August 2010.
REFERENCES
- 1.Alfredson, D. A., and V. Korolik. 2007. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 277:123-132. [DOI] [PubMed] [Google Scholar]
- 2.Bruhn, S. 2009. Campylobacter, p. 40-44. In Swiss zoonoses report 2008. Federal Veterinary Office, Bern, Switzerland.
- 3.Caro-Quintero, A., G. P. Rodriguez-Castano, and K. T. Konstantinidis. 2009. Genomic insights into the convergence and pathogenicity factors of Campylobacter jejuni and Campylobacter coli species. J. Bacteriol. 191:5824-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colles, F. M., K. Jones, R. M. Harding, and M. C. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colles, F. M., N. D. McCarthy, S. K. Sheppard, R. Layton, and M. C. Maiden. 2010. Comparison of Campylobacter populations isolated from a free-range broiler flock before and after slaughter. Int. J. Food Microbiol. 137:259-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingle, K. E., F. M. Colles, D. Falush, and M. C. J. Maiden. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingle, K. E., N. D. McCarthy, A. J. Cody, T. E. Peto, and M. C. Maiden. 2008. Extended sequence typing of Campylobacter spp., United Kingdom. Emerg. Infect. Dis. 14:1620-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engberg, J., F. M. Aarestrup, D. E. Taylor, P. Gerner-Smidt, and I. Nachamkin. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Commission. 2007. 2007/516/EC: Commission Decision of 19 July 2007 concerning a financial contribution from the Community towards a survey on the prevalence and antimicrobial resistance of Campylobacter spp. in broiler flocks and on the prevalence of Campylobacter spp. and Salmonella spp. in broiler carcasses to be carried out in the Member States (notified under document number C.2007. 3440). Official J. Eur. Union Legislation L 50:25-37. [Google Scholar]
- 12.European Food Safety Authority. 2010. The community summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in the European Union in 2008, p. 111-137. EFSA J. 8:1496. [Google Scholar]
- 13.Falush, D., M. Stephens, and J. K. Pritchard. 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7:574-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Federal Veterinary Office. 2009. Swiss zoonosis report 2008. Federal Veterinary Office, Bern, Switzerland.
- 15.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. ASM Press, Washington, DC.
- 16.Habib, I., W. G. Miller, M. Uyttendaele, K. Houf, and L. De Zutter. 2009. Clonal population structure and antimicrobial resistance of Campylobacter jejuni in chicken meat from Belgium. Appl. Environ. Microbiol. 75:4264-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphrey, T., S. O'Brien, and M. Madsen. 2007. Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117:237-257. [DOI] [PubMed] [Google Scholar]
- 18.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs-Reitsma, W., U. Lyhs, and J. Wagenaar. 2008. Campylobacter in the food supply, p. 627-644. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter. ASM Press, Washington, DC.
- 20.Jacobs-Reitsma, W. F., C. A. Kan, and N. M. Bolder. 1994. The induction of quinolone resistance in Campylobacter bacteria in broilers by quinolone treatment. Lett. Appl. Microbiol. 19:228-231. [Google Scholar]
- 21.Kärenlampi, R., H. Rautelin, D. Schonberg-Norio, L. Paulin, and M. L. Hanninen. 2007. Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl. Environ. Microbiol. 73:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korczak, B., H. Christensen, S. Emler, J. Frey, and P. Kuhnert. 2004. Phylogeny of the family Pasteurellaceae based on rpoB sequences. Int. J. Syst. Evol. Microbiol. 54:1393-1399. [DOI] [PubMed] [Google Scholar]
- 23.Korczak, B. M., R. Stieber, S. Emler, A. P. Burnens, J. Frey, and P. Kuhnert. 2006. Genetic relatedness within the genus Campylobacter inferred from rpoB sequences. Int. J. Syst. Evol. Microbiol. 56:937-945. [DOI] [PubMed] [Google Scholar]
- 24.Korczak, B. M., M. Zurfluh, S. Emler, J. Kuhn-Oertli, and P. Kuhnert. 2009. Multiplex strategy for MLST, fla typing, and genetic determination of antimicrobial resistance of Swiss Campylobacter jejuni and Campylobacter coli isolates. J. Clin. Microbiol. 47:1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang, P., T. Lefebure, W. Wang, B. P. Pavinski, R. J. Meinersmann, K. Kaya, and M. J. Stanhope. 2010. Expanded multilocus sequence typing and comparative genomic hybridization of Campylobacter coli isolates from multiple hosts. Appl. Environ. Microbiol. 76:1913-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litrup, E., M. Torpdahl, and E. M. Nielsen. 2007. Multilocus sequence typing performed on Campylobacter coli isolates from humans, broilers, pigs and cattle originating in Denmark. J. Appl. Microbiol. 103:210-218. [DOI] [PubMed] [Google Scholar]
- 27.Luangtongkum, T., B. Jeon, J. Han, P. Plummer, C. M. Logue, and Q. Zhang. 2009. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4:189-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellmann, A., J. Mosters, E. Bartelt, P. Roggentin, A. Ammon, A. W. Friedrich, H. Karch, and D. Harmsen. 2004. Sequence-based typing of flaB is a more stable screening tool than typing of flaA for monitoring of Campylobacter populations. J. Clin. Microbiol. 42:4840-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mickan, L., R. Doyle, M. Valcanis, K. E. Dingle, L. Unicomb, and J. Lanser. 2007. Multilocus sequence typing of Campylobacter jejuni isolates from New South Wales, Australia. J. Appl. Microbiol. 102:144-152. [DOI] [PubMed] [Google Scholar]
- 31.Miller, W. G., M. D. Englen, S. Kathariou, I. V. Wesley, G. Wang, L. Pittenger-Alley, R. M. Siletz, W. Muraoka, P. J. Fedorka-Cray, and R. E. Mandrell. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245-255. [DOI] [PubMed] [Google Scholar]
- 32.Moore, J. E., M. D. Barton, I. S. Blair, D. Corcoran, J. S. Dooley, S. Fanning, I. Kempf, A. J. Lastovica, C. J. Lowery, M. Matsuda, D. A. McDowell, A. McMahon, B. C. Millar, J. R. Rao, P. J. Rooney, B. S. Seal, W. J. Snelling, and O. Tolba. 2006. The epidemiology of antibiotic resistance in Campylobacter. Microbes Infect. 8:1955-1966. [DOI] [PubMed] [Google Scholar]
- 33.Moore, J. E., D. Corcoran, J. S. Dooley, S. Fanning, B. Lucey, M. Matsuda, D. A. McDowell, F. Mégraud, B. C. Millar, R. O'Mahony, L. O'Riordan, M. O'Rourke, J. R. Rao, P. J. Rooney, A. Sails, and P. Whyte. 2005. Campylobacter. Vet. Res. 36:351-382. [DOI] [PubMed] [Google Scholar]
- 34.Müllner, P., J. M. Collins-Emerson, A. C. Midwinter, P. Carter, S. E. Spencer, P. van der Logt, S. Hathaway, and N. P. French. 2010. Molecular epidemiology of Campylobacter jejuni in a geographically isolated country with a uniquely structured poultry industry. Appl. Environ. Microbiol. 76:2145-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakari, U. M., A. Puhakka, and A. Siitonen. 2008. Correct identification and discrimination between Campylobacter jejuni and C. coli by a standardized hippurate test and species-specific polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 27:513-518. [DOI] [PubMed] [Google Scholar]
- 36.Nauta, M. J., W. F. Jacobs-Reitsma, and A. H. Havelaar. 2007. A risk assessment model for Campylobacter in broiler meat. Risk Anal. 27:845-861. [DOI] [PubMed] [Google Scholar]
- 37.Normand, V., M. Boulianne, and S. Quessy. 2008. Evidence of cross-contamination by Campylobacter spp. of broiler carcasses using genetic characterization of isolates. Can. J. Vet. Res. 72:396-402. [PMC free article] [PubMed] [Google Scholar]
- 38.On, S. L. W. 1996. Identification methods for Campylobacters, Helicobacters, and related organisms. Clin. Microbiol. Rev. 9:405-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Payot, S., S. Dridi, M. Laroche, M. Federighi, and C. Magras. 2004. Prevalence and antimicrobial resistance of Campylobacter coli isolated from fattening pigs in France. Vet. Microbiol. 101:91-99. [DOI] [PubMed] [Google Scholar]
- 40.Pritchard, J. K., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ragimbeau, C., F. Schneider, S. Losch, J. Even, and J. Mossong. 2008. Multilocus sequence typing, pulsed-field gel electrophoresis, and fla short variable region typing of clonal complexes of Campylobacter jejuni strains of human, bovine, and poultry origins in Luxembourg. Appl. Environ. Microbiol. 74:7715-7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosef, O., G. Kapperud, S. Lauwers, and B. Gondrosen. 1985. Serotyping of Campylobacter jejuni, Campylobacter coli, and Campylobacter laridis from domestic and wild animals. Appl. Environ. Microbiol. 49:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnider, A., G. Overesch, B. M. Korczak, and P. Kuhnert. 2010. Comparison of real-time PCR assays for detection, quantification, and differentiation of Campylobacter jejuni and Campylobacter coli in broiler neck skin samples. J. Food Prot. 73:1057-1063. [DOI] [PubMed] [Google Scholar]
- 44.Sheppard, S. K., J. F. Dallas, M. MacRae, N. D. McCarthy, E. L. Sproston, F. J. Gormley, N. J. Strachan, I. D. Ogden, M. C. Maiden, and K. J. Forbes. 2009. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int. J. Food Microbiol. 134:96-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheppard, S. K., J. F. Dallas, N. J. Strachan, M. MacRae, N. D. McCarthy, D. J. Wilson, F. J. Gormley, D. Falush, I. D. Ogden, M. C. Maiden, and K. J. Forbes. 2009. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 48:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheppard, S. K., N. D. McCarthy, D. Falush, and M. C. Maiden. 2008. Convergence of Campylobacter species: implications for bacterial evolution. Science 320:237-239. [DOI] [PubMed] [Google Scholar]
- 47.Siemer, B. L., E. M. Nielsen, and S. L. W. On. 2005. Identification and molecular epidemiology of Campylobacter coli isolates from human gastroenteritis, food, and animal sources by amplified fragment length polymorphism analysis and Penner serotyping. Appl. Environ. Microbiol. 71:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sopwith, W., A. Birtles, M. Matthews, A. Fox, S. Gee, S. James, J. Kempster, M. Painter, V. Edwards-Jones, K. Osborn, M. Regan, Q. Syed, and E. Bolton. 2010. Investigation of food and environmental exposures relating to the epidemiology of Campylobacter coli in humans in Northwest England. Appl. Environ. Microbiol. 76:129-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson, D. J., E. Gabriel, A. J. Leatherbarrow, J. Cheesbrough, S. Gee, E. Bolton, A. Fox, P. Fearnhead, C. A. Hart, and P. J. Diggle. 2008. Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson, D. J., E. Gabriel, A. J. Leatherbarrow, J. Cheesbrough, S. Gee, E. Bolton, A. Fox, C. A. Hart, P. J. Diggle, and P. Fearnhead. 2009. Rapid evolution and the importance of recombination to the gastro-enteric pathogen Campylobacter jejuni. Mol. Biol. Evol. 26:385-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wingstrand, A., J. Neimann, J. Engberg, E. M. Nielsen, P. Gerner-Smidt, H. C. Wegener, and K. Molbak. 2006. Fresh chicken as main risk factor for campylobacteriosis, Denmark. Emerg. Infect. Dis. 12:280-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wittwer, M., J. Keller, T. M. Wassenaar, R. Stephan, D. Howald, G. Regula, and B. Bissig-Choisat. 2005. Genetic diversity and antibiotic resistance patterns in a Campylobacter population isolated from poultry farms in Switzerland. Appl. Environ. Microbiol. 71:2840-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zweifel, C., K. D. Scheu, M. Keel, F. Renggli, and R. Stephan. 2008. Occurrence and genotypes of Campylobacter in broiler flocks, other farm animals, and the environment during several rearing periods on selected poultry farms. Int. J. Food Microbiol. 125:182-187. [DOI] [PubMed] [Google Scholar]