Abstract
Marine hydrocarbon seeps supply oil and gas to microorganisms in sediments and overlying water. We used stable isotope probing (SIP) to identify aerobic bacteria oxidizing gaseous hydrocarbons in surface sediment from the Coal Oil Point seep field located offshore of Santa Barbara, California. After incubating sediment with 13C-labeled methane, ethane, or propane, we confirmed the incorporation of 13C into fatty acids and DNA. Terminal restriction fragment length polymorphism (T-RFLP) analysis and sequencing of the 16S rRNA and particulate methane monooxygenase (pmoA) genes in 13C-DNA revealed groups of microbes not previously thought to contribute to methane, ethane, or propane oxidation. First, 13C methane was primarily assimilated by Gammaproteobacteria species from the family Methylococcaceae, Gammaproteobacteria related to Methylophaga, and Betaproteobacteria from the family Methylophilaceae. Species of the latter two genera have not been previously shown to oxidize methane and may have been cross-feeding on methanol, but species of both genera were heavily labeled after just 3 days. pmoA sequences were affiliated with species of Methylococcaceae, but most were not closely related to cultured methanotrophs. Second, 13C ethane was consumed by members of a novel group of Methylococcaceae. Growth with ethane as the major carbon source has not previously been observed in members of the Methylococcaceae; a highly divergent pmoA-like gene detected in the 13C-labeled DNA may encode an ethane monooxygenase. Third, 13C propane was consumed by members of a group of unclassified Gammaproteobacteria species not previously linked to propane oxidation. This study identifies several bacterial lineages as participants in the oxidation of gaseous hydrocarbons in marine seeps and supports the idea of an alternate function for some pmoA-like genes.
Hydrocarbon seeps are widespread along continental margins and emit large amounts of oil and gas into the surrounding environment. This gas is primarily composed of methane, a powerful greenhouse gas, and marine hydrocarbon seeps are estimated to contribute 20 Tg year−1 methane to the atmosphere, representing about 5% of the total atmospheric flux (21, 39). Seeps of thermogenic gas also release an estimated 0.45 Tg year−1 ethane and 0.09 Tg year−1 propane to the atmosphere (20). Each of these three fluxes would be substantially larger if not for microbial oxidation in the sediments and water column (68). Methane, ethane, and propane are subject to anaerobic oxidation in anoxic sediments and water columns (44, 53, 68) or to aerobic oxidation in oxic and suboxic water columns and oxygenated surface sediment (10, 47, 53, 80). We focus here on aerobic oxidation.
The majority of known aerobic methane-oxidizing bacteria are members of either Gammaproteobacteria (type I) or Alphaproteobacteria (type II) (29), though several strains of highly acidophilic methanotrophic Verrucomicrobia have also been recently isolated (63). Most methanotrophs are capable of growth only on methane or other one-carbon compounds (17, 29), using a methane monooxygenase (MMO) enzyme to oxidize methane to methanol. There are two known forms of this enzyme: soluble methane monooxygenase (sMMO) and particulate methane monooxygenase (pMMO). sMMO is a soluble, di-iron-containing monooxygenase found only in certain methanotrophs and typically expressed only under low-copper conditions (57). In contrast, pMMO is a membrane-bound enzyme believed to contain copper and iron (26). It is found in all known methanotrophs, with the exception of species of the genus Methylocella (16). pmoA, the gene encoding the α subunit of pMMO, is often used to identify methanotrophic bacteria (54). Very few methanotrophs from marine environments have been cultured (22, 49, 72, 74), but several previous studies of marine methanotrophs (35, 62, 77, 82, 85) have been performed with culture-independent methods and have almost exclusively detected type I methanotrophs. Many of the pmoA sequences from methane seep sites are quite different from those of cultured organisms, suggesting that these environments may contain many novel methanotrophs (77, 82, 85).
Even less is known about the organisms that oxidize ethane or propane in marine environments. The number of such isolates, which primarily represent high G+C Gram-positive bacteria (Nocardia, Pseudonocardia, Gordonia, Mycobacterium, and Rhodococcus) or Pseudomonas species, is limited (70). Nearly all of these strains were isolated from soil and selected for their ability to grow on propane or n-butane as the sole carbon source. Most propane-oxidizing strains can oxidize butane, as well as a range of longer chain n-alkanes, but differ in the ability to oxidize ethane. These strains show little, if any, ability to oxidize methane, and none have been shown to grow with methane as the sole carbon source (13, 27, 38, 45, 65). As with methane metabolism, the first step in aerobic ethane and propane metabolism is the oxidation of the alkane to an alcohol (70). Several different enzymes are known to catalyze this step. Thauera butanivorans uses a soluble di-iron butane monooxygenase related to sMMO to oxidize C2 through C9 n-alkanes (18, 73). Gordonia sp. strain TY-5, Mycobacterium sp. strain TY-6, and Pseudonocardia sp. strain TY-7 contain soluble di-iron propane monooxygenases that are capable of both terminal and subterminal propane oxidation and differ in their substrate ranges (45, 46). Nocardioides sp. strain CF8 is believed to possess a copper-containing monooxygenase similar to pMMO and ammonia monooxygenase (27, 28). An alkane hydroxylase typically used to oxidize longer-chain n-alkanes has also shown some ability to oxidize propane and butane but not ethane (38). The variety of enzymes and their substrate ranges make it difficult to identify ethane or propane oxidizers with a single functional gene.
In order to identify the organisms responsible for methane, ethane, and propane oxidation at hydrocarbon seeps, we used stable isotope probing (SIP). SIP allows the identification of organisms actively consuming a 13C-labeled substrate of interest, based on the incorporation of 13C into biomass, including DNA and lipids (67). We collected sediment from the Coal Oil Point seep field and incubated sediment-seawater slurries with 13C methane, ethane, or propane. Samples were removed at three time points, chosen to ensure sufficient 13C incorporation into DNA while minimizing the spread of 13C through the community as a result of cross-feeding on metabolic byproducts. 13C-DNA was separated from 12C-DNA by CsCl density gradient ultracentrifugation, and we used the fractionated DNA for terminal restriction fragment length polymorphism (T-RFLP) and clone library analysis. We also measured 13C incorporation into fatty acids in order to confirm significant 13C enrichment in membrane lipids, to determine the carbon labeling pattern for each substrate and lipid, and to further characterize the composition of the microbial community.
MATERIALS AND METHODS
Study site and sample collection.
Sediment for SIP incubations was collected from Shane Seep in the Coal Oil Point seep field, offshore of Santa Barbara, CA (34°24.370′N, 119°53.428′W; 21 m depth). Coal Oil Point is one of the world's most prolific areas of hydrocarbon seepage, emitting 2 × 1010 g year−1 methane, 1.9 × 109 g year−1 ethane, 1.4 × 109 g year−1 propane, and 5.8 × 106 liters year−1 oil (9, 31). The shallow, nearshore seeps are highly dynamic environments, with variable gas flux levels and seasonal changes in sediment deposition (43). A neighboring seep, Brian Seep, has been described in detail by Kinnaman et al. (43) and Treude and Ziebis (78); Shane Seep is very similar, though the sediment contains more oil than at Brian Seep. At Brian Seep, convective pore water transport allows oxygen penetration through the top centimeter of sediment; the high methane oxidation rates observed in this layer have been attributed to aerobic methane oxidation (78). The distribution of chemicals in sediment pore fluids at Shane Seep suggests such that convective processes are also active (42). For this study, divers collected sediment from the top 1 to 2 cm in areas of active bubbling at Shane Seep; this sediment had no detectable sulfide odor. Sediment and the overlying seawater were collected in sterile polypropylene tubes, and samples were kept on ice until return to the laboratory. SIP incubations were prepared immediately.
SIP incubations.
SIP incubations were carried out with 13C-labeled methane, ethane, or propane (Isotec) (99% 13C). Parallel incubations used natural abundance levels of 13C for each hydrocarbon substrate (referred to as 12C controls). No hydrocarbon was added to an additional control sample. Sediment and overlying seawater from six 50-ml collection tubes were pooled and mixed to form a slurry, of which ∼25 g was added to each 125-ml serum bottle. Bottles were sealed with butyl rubber stoppers and aluminum crimp caps. A 5-ml volume of hydrocarbon gas was injected into each bottle, and air was added for overpressure to allow removal of samples for gas analysis. Samples were incubated at room temperature in the dark, with shaking at 75 rpm. Every 24 to 72 h, headspace concentrations of hydrocarbons, oxygen, and carbon dioxide were measured with a 3000A MicroGC system (Agilent) equipped with a thermal conductivity detector. When the hydrocarbon gas had nearly been consumed, the headspace was purged with air and an additional 5 ml of hydrocarbon gas was added. Oxygen concentrations in the headspace remained above 10% at all times. Sediment samples (3 ml) were removed at 3 days and 6 days and after the second 5-ml aliquot of hydrocarbon had been consumed (9 days for methane and ethane, 14 days for propane). Sediment was stored frozen until analysis.
Phospholipid fatty acid extraction, quantification, and isotopic analysis.
Samples were lyophilized and then extracted in dichloromethane (DCM)/methanol (9:1) by the use of a microwave-assisted extraction system (CEM Mars5) at 100°C for 20 min with stirring. The total lipid extract was collected and filtered using a GFF-A filter, and the solvent was removed under dry N2 conditions. The lipid extract was resuspended in 0.5 ml of hexane, and fatty acid fractions were purified by solid-phase extraction (Phenomenex Sepra NH2) (0.5 g), with elution by the use of 8.0 ml of 2% formic acid in DCM. Fatty acids were converted to their methyl esters by reaction with BF3-methanol at 70°C for 20 min and were extracted into 10 ml of 3× hexane.
Fatty acid structure and abundance were quantified by gas chromatography/mass spectrometry/flame ionization detection (GC/MS/FID) using a Thermo Finnigan Trace/DSQ system at Caltech. GC separation employed a ZB-5ms column (Phenomenex) (30 m by 0.25 mm; 0.25 μm pore size) with a programmable-temperature vaporization (PTV) injector. The effluent from the GC column was split 80/20 between the MS and FID, allowing for simultaneous structure identification by MS and quantitation using the FID signal by comparison to an internal standard (palmitic acid isopropyl ester). No correction for relative response factors in the FID was made.
Stable carbon isotope ratios (δ13C values) were measured using a Thermo Finnigan GC-isotope ratio mass spectrometer (IRMS) at the Marine Science Institute Analytical Lab, University of California (UC), Santa Barbara, CA. This system used a Trace GC sytem with an Omegawax 250 capillary column (Supelco) (30 m by 0.25 mm; 0.25 μm pore size) with a splitless injector, a GC combustion III interface, and a Delta Plus XP mass spectrometer. The GC combustion III unit catalyzed the oxidation of organic analytes to CO2 over Cu/Ni/Pt wire heated to 950°C, and water was removed through a selectively permeable membrane. The carrier gas was helium, with a flow rate of 2.4 ml min−1. Peaks eluted from the GC column were converted into CO2 and analyzed by IRMS. Carbon isotope ratios for each compound were measured relative to a CO2 working standard with a nominal δ13C of −32.6‰. All δ13C values were normalized to a decanoic acid methyl ester (C10:0) isotope standard obtained from Arndt Shimmelmann at Indiana University and then corrected for the addition of the methyl group from methanol, assuming δ13Cmethanol = −25‰. Each sample was analyzed twice; in all cases, the atomic %13C was within ±0.5% 13C between duplicates for 13C-labeled samples and within ±0.01% 13C for 12C controls.
CsCl gradient fractionation.
Sediment DNA was extracted with a FastDNA spin kit for soil (MP Biomedicals). 12C-DNA and 13C-DNA were separated by ultracentrifugation and density gradient fractionation as described by Neufeld et al. (60). Briefly, ∼5 to 7 μg of DNA was added to a CsCl solution and centrifuged in a VTi 65.2 rotor (Beckman) at 177,000 × g for 48 to 60 h at 20°C. Gradients were separated into 12 425-μl fractions, where fraction 1 was the heaviest and fraction 12 was the lightest. The density of selected fractions was checked with a refractometer (Reichert). DNA was precipitated with polyethylene glycol and glycogen, washed with 70% ethanol, and eluted in 30 μl of Tris-EDTA buffer. The amount of DNA in each fraction was quantified with a PicoGreen double-stranded DNA (dsDNA) assay kit (Invitrogen).
T-RFLP.
T-RFLP analysis was used for initial characterization of the gradient DNA. For fractions 2, 4, 6, 8, 10, and 12 of each sample, the 16S rRNA gene was PCR amplified using primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′; labeled with 6-carboxyfluorescein) and 1392R (5′-ACGGGCGGTGTGTAC-3′). Each 50-μl reaction mixture contained 1× buffer, 1.5 mM MgCl2, 200 μM each dNTP (deoxynucleoside triphosphate), 0.5 μM each primer, 0.2 mg/ml bovine serum albumin (BSA), 1.25 U of Taq (Qiagen), and 2 or 4 μl of DNA (2 μl for the three “light” fractions and 4 μl for the three “heavy” fractions). Thermal cycling conditions were as follows: initial denaturation at 95°C for 2 min; 30 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 90 s; and a final extension at 72°C for 5 min. Duplicate PCRs were pooled and cleaned with a Wizard SV DNA purification kit (Promega). DNA (100 ng) was digested with MspI or HhaI for 3 h at 37°C, followed by heat inactivation for 20 min at 65°C. Digests were purified using a Montage PCR centrifugal filter device kit (Millipore) and analyzed at the UC Berkeley DNA Sequencing Facility. T-REX (10) was employed for data processing, using a clustering threshold of 1 bp. Relative abundances were calculated using peak areas. Preliminary samples were run in duplicate. The relative abundances of all peaks that were >1% of the total peak area were within ±2% of the total peak area of the duplicates.
16S rRNA and pmoA gene clone libraries.
16S rRNA gene clone libraries were constructed from the initial sediment and at least one fraction of “heavy” DNA (fraction 4 or 6; 1.73 or 1.75 g ml−1) from each substrate at each time point. Clone libraries were also created from selected controls: “light” DNA (fraction 10; 1.68 g ml−1) from 13C incubations and “heavy” DNA from 12C control incubations. PCR conditions were the same as those for T-RFLP, except that an unlabeled forward primer was used.
pmoA clone libraries were constructed from the initial sediment and heavy DNA from day 3 of the methane incubation using the primers pmoA189f and mb661r (14). For the ethane incubations, we used additional degenerate primers designed to amplify the highly divergent pmoA sequences listed in GenBank for Methylococcaceae ET-HIRO and ET-SHO (AB453960 to AB453963) as well as other Methylococcaceae species. Heavy DNA from day 6 of the ethane incubation was amplified with the following pairs of primers: pmoA189f and mb661r, ethmo189f (5′-GGYGAYTGGGAYTTYTGG-3′) and ethmo661r (5′-CTGGAGCMACGTCTTTAC-3′), and ethmo189f and ethmo649r (5′-GTWCCMCGCTCNAYCATBC-3′). Reaction concentrations were the same as those described above, and thermal cycling conditions were as follows: initial denaturation at 94°C for 3 min; 30 cycles of 94°C for 60 s, 54°C for 90 s, and 72°C for 60 s; and a final extension at 72°C for 5 min. For the ethane sample, the template volume was limited and yields were low in all three reactions, so the PCR products were pooled.
For both 16S rRNA and pmoA, PCR products were cleaned with a Wizard SV DNA purification kit (Promega) and cloned with a PCR cloning kit (Qiagen). Plasmid DNA was isolated from randomly selected clones with an UltraClean 6-minute Mini Plasmid Prep kit (Mo Bio) and sequenced at the UC Berkeley DNA Sequencing Facility.
Phylogenetic analysis and assignment of T-RFs.
Sequences were edited and assembled with Sequencher (Gene Codes Corp.). 16S rRNA gene sequences were screened for potential chimeras with Bellerophon (32), CHIMERA CHECK (12), and Pintail (1) software; suspected chimeras were not considered in further analysis. Phylogenetic affiliations of 16S rRNA genes and their relative abundances were determined with the RDP Classifier tool (81). Representative 16S rRNA and pmoA sequences and related reference sequences were aligned using CLUSTALW, and neighbor-joining phylogenetic trees were constructed with MEGA software (76).
Sequences were further classified by in silico digestions performed with MspI and HhaI to predict terminal restriction fragment (T-RF) lengths. These were used to assign identities to T-RFs from the T-RFLP analysis. 16S rRNA gene sequences from both 13C-DNA and controls were used to create a list of sequences with each predicted T-RF length. Predicted and observed T-RFs correlated closely, provided that each observed T-RF length was corrected by 4 bp. Differences between observed and predicted T-RF lengths have often been observed in T-RFLP studies (64, 69), and can differ based on purine content or fragment length (41), but in this case appeared to take the form of a constant 4-bp offset. The T-RF lengths shown represent the observed T-RF length plus 4 bp. T-RFs were then combined to form six major groups, based on the clone library sequences with these predicted T-RF lengths.
Nucleotide sequence accession numbers.
These sequence data have been submitted to the GenBank database under accession numbers GU584298 to GU584873 (16S rRNA gene) and GU584229 to GU584297 (pmoA).
RESULTS
13C enrichment in fatty acids.
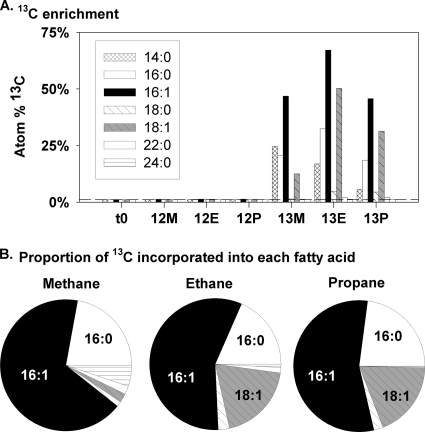
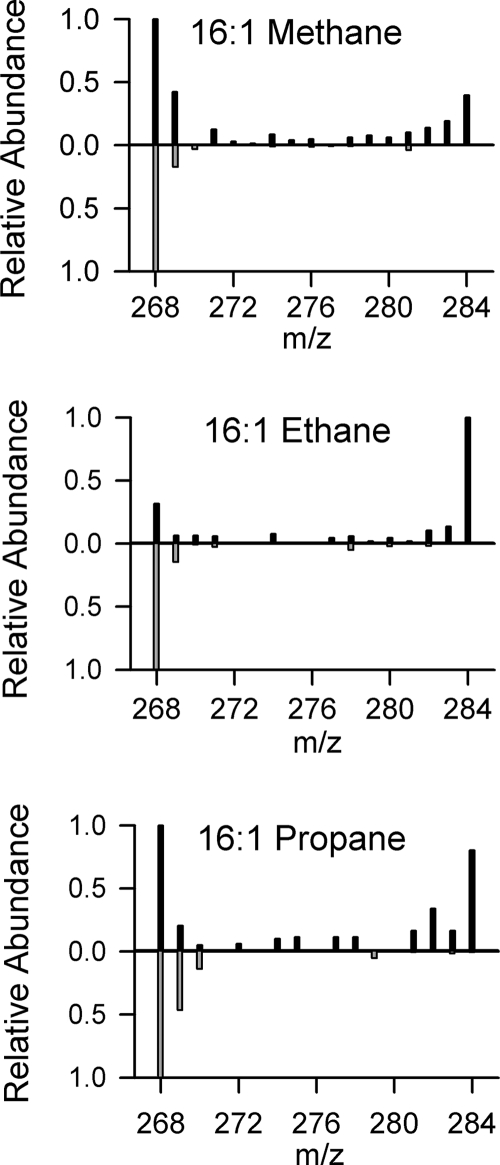
Fatty acids were extracted from the initial sediment used for inoculation and from the sediment remaining after the final time point of each SIP incubation. In all cases, the most abundant fatty acids were the 16- and 18-carbon-saturated fatty acids (16:0 and 18:0) and monounsaturated fatty acids (16:1 and 18:1; multiple positional isomers were present for each but were not differentiated) (data not shown). In all cases, the 13C incubations showed very high levels of 13C incorporation into individual fatty acids (up to 63%), while the 12C controls showed no significant 13C enrichment (<1.25% 13C) (Fig. 1 A). The 16:1, 16:0, and 18:1 fatty acids were heavily enriched with 13C in all samples, while the 18:0 and 14:0 fatty acids showed much less 13C incorporation. In all samples, >50% of the 13C incorporated into fatty acids was found in the 16:1 fatty acid, ∼20% in the 16:0 fatty acid, and <3% in the 18:0 fatty acid. The 18:1 fatty acid accounted for ∼20% of the 13C in the ethane and propane incubations but for only 4% in the methane incubation (Fig. 1B). Mass spectra for the highly labeled fatty acids, particularly for the 16:1 fatty acid, indicated a bimodal pattern of isotopic distribution, where most molecules either contained a natural abundance of 13C (no 13C or one 13C per molecule) or were fully labeled with 13C (Fig. 2). Mass spectra from unlabeled (12C) controls showed no appreciable incorporation of 13C, which was in agreement with the δ13C values measured by GC/IRMS.
FIG. 1.
(A) 13C enrichment of phospholipid fatty acids of 13C methane (M), ethane (E), and propane (P) incubations and 12C controls in initial sediment (t0) and at final time points. The 16:1 and 18:1 isomers were not differentiated. The dashed line indicates natural abundance levels of 13C. (B) Proportion of 13C incorporated into each fatty acid.
FIG. 2.
Mass spectra for the 16:1 fatty acid molecular ion peak, showing the extent of 13C labeling at the final time point of the methane, ethane, and propane incubations. The m/z values for the spectra span the full range from no 13C incorporation (m/z = 268) to full 13C labeling (m/z = 268 + 16 = 284). The spectra oriented upward correspond to incubations with 13C labeling, whereas the inverted spectra correspond to control incubations with substrate lacking 13C labeling. Each spectrum is normalized to the height of its tallest peak.
16S rRNA gene clone libraries.
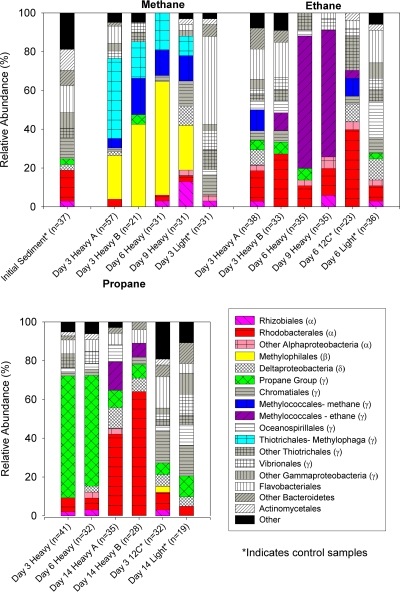
DNAs from each 13C sample and its 12C control were separated by density gradient ultracentrifugation into 12 fractions, with the buoyant densities ranging from 1.66 g ml−1 to 1.78 g ml−1). 16S rRNA gene clone libraries were constructed from at least one heavy DNA fraction (1.73 or 1.75 g ml−1) from each gas at each time point, as well as from a heavy DNA fraction from selected 12C controls and a light DNA fraction (1.69 g ml−1) from selected 13C samples. Because we were primarily interested in the most abundant phylotypes, we sequenced 24 to 48 clones from most fractions, for a total of 595 sequences. The phylogenetic affiliations of these sequences are shown in Fig. 3 and 4.
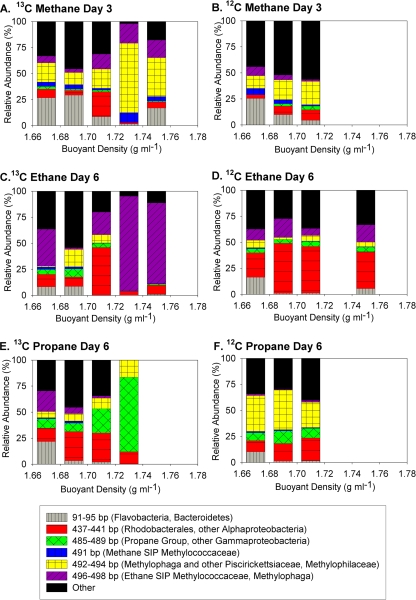
FIG. 3.
Relative abundances of 16S rRNA sequences in clone libraries from heavy DNA (fraction 4 or 6 [noted as Heavy A or Heavy B] or both) from the three time points of the 13C methane, ethane, and propane incubations, selected controls (light DNA from the 13C incubations and heavy DNA from the 12C controls), and the initial sediment. Sequences were grouped using the RDP Classifier tool; “other” combines sequences that, classified at the order level, represented less than 5% of the clones in any individual clone library. Representative sequences from each of the groups indicated in methane, ethane, or propane oxidation are included in the phylogenetic tree in Fig. 4.
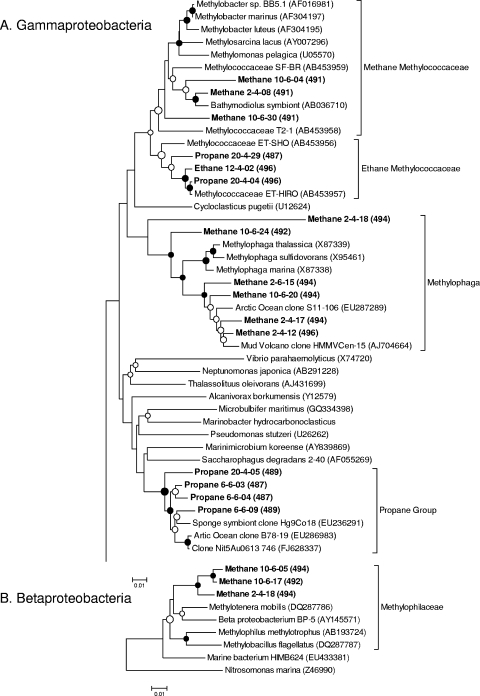
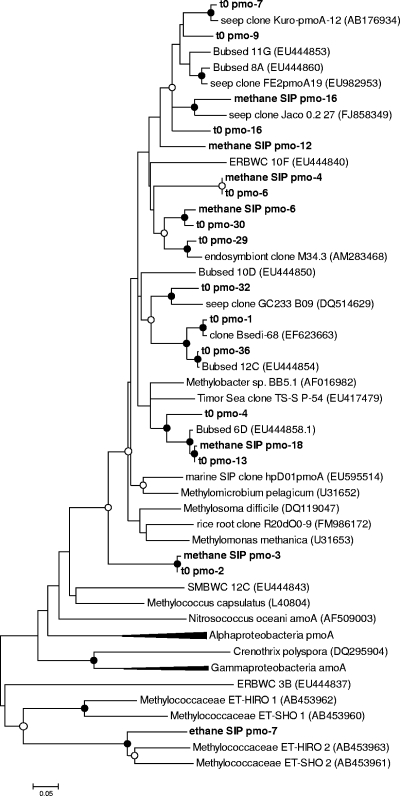
FIG. 4.
Neighbor-joining phylogenetic trees of the 16S rRNA gene sequences from groups involved in methane, ethane, or propane oxidation, based on their abundance in both the heavy DNA clone libraries and the heavy T-RFLP fractions, relative to light DNA and 12C controls. (A) Gammaproteobacteria; (B) Betaproteobacteria. Sequences from this study are shown in bold, with predicted MspI T-RF lengths in parentheses. Reference sequences from GenBank are shown with accession numbers in parentheses. Filled circles indicate bootstrap values above 90% and open circles bootstrap values above 50% (2,000 replicates).
Methane.
The most abundant sequences in the heavy DNA from all three time points of the methane incubation were from species of a group of Gammaproteobacteria related to Methylophaga and a group of Betaproteobacteria in the family Methylophilaceae, followed by a group of Gammaproteobacteria in the family Methylococcaceae. These three groups accounted for 81% of sequences in the heavy fraction 6 from day 3, 91% in the heavy fraction from day 6, and 45% in the heavy fraction from day 9, with sequences of Methylophaga and the Methylophilaceae bacteria making up the majority at all time points. By day 9, two additional groups comprised at least 10% of the heavy DNA sequences: Alphaproteobacteria in the order Rhizobiales and Gammaproteobacteria in the order Chromatiales. In comparison, the most common sequences in the light DNA clone library were Flavobacteriales (45%), followed by Thiotrichales, Vibrionales, and Chromatiales; among the 31 clones, there were no sequences affiliated with the Methylococcaceae, Methylophilaceae, or Methylophaga.
Ethane.
At day 3, the heavy DNA sequences from the 13C ethane incubations were similar to those of the 12C control heavy DNA and the light DNA from the 13C sample, predominately representing Rhodobacterales, Oceanospirillales, Thiotrichales, and Flavobacteriales species. By days 6 and 9, though, the majority of heavy DNA sequences were from a group of Gammaproteobacteria bacteria in the family Methylococcaceae. Like the methane Methylococcaceae sequences, their closest cultured relatives were Methylobacter sp. BB5.1, Methylobacter luteus NCIMB 11914, and Methylobacter marinus strain A45. However, the ethane Methylococcaceae sequences all clustered separately from the methane Methylococcaceae sequences and could be distinguished by their predicted MspI T-RF (496 bp for ethane and 491 bp for methane). The most similar sequences in GenBank were from “Methylococcaceae bacterium ET-HIRO” (AB453957.1) and “Methylococcaceae bacterium ET-SHO” (AB453956.1).
Propane.
The majority of sequences in the heavy DNA clone libraries from days 3 and 6 of the propane incubation belonged to species from a group of unclassified Gammaproteobacteria most closely related to the genera Marinimicrobium, Microbulbifer, and Saccharophagus. This group accounted for 61% and 56% of heavy DNA sequences at days 3 and 6, respectively, but only 6% of heavy DNA from the 12C control. By day 14, however, they made up less than 10% of the sequences in fractions 4 and 6, and Rhodobacterales sequences were most abundant instead (43% and 65%, respectively). Rhodobacterales sequences made up only 5% of the light DNA sequences from the last time point.
pmoA and pmoA-like gene clone libraries.
In order to further characterize the Methylococcaceae species in the ethane incubations, we created small pmoA clone libraries by the use of degenerate primers designed to amplify highly divergent pmoA sequences. The clone library with heavy DNA from the ethane incubation contained several highly divergent pmoA sequences, which grouped with the ET-HIRO and ET-SHO sequences (Fig. 5). We also created small pmoA clone libraries from the initial sediment (46 clones) and heavy DNA from the methane incubations (10 clones) by employing the commonly used primers pmoA189f and mb661r. Representative sequences are also shown in Fig. 5. These sequences all grouped with the gammaproteobacterial pmoA sequences and were most closely related to those from other uncultured marine methanotrophs.
FIG. 5.
Neighbor-joining phylogenetic tree of pmoA gene sequences from seep sediment (t0) and methane and ethane heavy DNA sequences, plus reference sequences from GenBank (accession numbers in parentheses). Only the divergent sequences from the ethane SIP sample are shown; others were identical to those in the t0 and methane SIP samples. Filled circles indicate bootstrap values above 90% and open circles bootstrap values above 50% (2,000 replicates). The alphaproteobacterial pmoA and gammaproteobacterial amoA sequences are condensed for clarity.
T-RFLP fingerprinting of density gradient fractions and correlation with clone libraries.
Though clone libraries provide a more detailed representation of the 16S rRNA genes present in each fraction, T-RFLP is useful for characterizing a broader range of gradient fractions from both 13C incubations and 12C controls. From six fractions of each sample, 16S rRNA genes were amplified and digested with the restriction enzymes MspI and HhaI. The two restriction enzymes produced similar results; only those from MspI are shown here (Fig. 6; see also Fig. S1 to S3 in the supplemental material). It was possible to amplify the 16S rRNA gene from nearly every fraction, but the amount of PCR product was insufficient for further analysis in some of the heavy fractions.
FIG. 6.
T-RFLP fingerprinting of the 16S rRNA gene in density gradient fractions from day 3 of the 13C (A) and 12C (B) methane, day 6 of the 13C (C) and 12C (D) ethane, and day 6 of the 13C (E) and 12C (F) propane incubations. Major T-RFs were identified by in silico digestions of clone library sequences with MspI. Less-abundant T-RFs (<5% of the total peak area in any of the heavy fractions) were combined and are shown as “other.” Fractions with insufficient DNA for analysis are left blank.
In order to correlate T-RFLP and clone library data, we performed in silico digestions of all suitable 16S rRNA sequences from the clone libraries and grouped them by T-RF length. The MspI T-RF lengths clearly distinguished the species of Rhodobacterales and other Alphaproteobacteria (439 or 441 bp) from those of the Flavobacteriales and other Bacteroidetes (91, 93, or 95 bp). All members of the “propane group” of unclassified Gammaproteobacteria species had a predicted T-RF of 485, 487, or 489 bp, though other high-abundance Gammproteobacteria (Oceanospirillales) sequences shared these T-RF lengths. All of the Methylococcaceae sequences from the methane incubation had a predicted T-RF of 491 bp, while all of the Methylococcaceae sequences from the ethane incubation had a T-RF of 496 bp. The 496 bp T-RF was also predicted for some Methylophaga sequences, though the rest of the Methylophaga sequences had T-RFs of 492 or 494 bp. It was not possible to differentiate Methylophaga from Methylophilaceae; these sequences also all had a predicted T-RF of 492 or 494 bp. Sequences of several other groups of Gammaproteobacteria (predominately Thiotrichales and Chromatiales) also had a predicted T-RF of 492, 494, or 496, but these sequences never represented more than 10% of the sequences in the heavy DNA clone libraries (Fig. 3). They were, however, more abundant in the clone libraries from the control samples, so the presence of these fragment lengths in T-RFLP samples from the light fractions was also expected, and these groups of Gammaproteobacteria and the Methylophilaceae could not be clearly differentiated. Digestion with HhaI led to better differentiation of a few groups of Gammaproteobacteria but more poorly resolved others and still did not differentiate between sequences of the methylotrophic Piscirickettsiaceae (Gammaproteobacteria) and Methylophilaceae (Betaproteobacteria) species.
Based on these observations, we combined our T-RFs into six main groups. All remaining T-RFs were combined and are represented as “other”; the relative abundances of the individual T-RFs in this group were never more than 5% in the 13C-labeled heavy fractions. Figure 6 shows T-RFLP profiles from one time point each of the methane, ethane, and propane incubations. The other two time points for each substrate are shown in the supplemental material. In all cases, the T-RFs corresponding to the most abundant sequences in the heavy DNA clone libraries were also most abundant in the heavy fractions of the T-RFLP profiles.
DISCUSSION
Factors to be considered with the SIP technique.
Though SIP can be useful in linking metabolic function to identity, there are challenges in designing these experiments and correctly interpreting the results. A successful SIP study requires sufficient incorporation of 13C into biomass, such that 13C-labeled DNA can be differentiated from the background of unlabeled DNA distributed across all fractions (51, 58). Longer incubations, which increase the extent of labeling, also lead to the spread of 13C throughout the community by cross-feeding involving metabolic byproducts or dead cells (52). Longer incubations and higher substrate concentrations also increase the possibility of selecting organisms adapted for higher substrate concentrations rather than those active in situ (58).
We attempted to minimize these problems in two ways. First, we sampled each incubation at three points in time, increasing the odds of catching the incubations at a point where there was sufficient 13C incorporation but minimal cross-feeding. Second, we ran parallel incubations with 12C methane, ethane, and propane. Though there was DNA present in most of the heavy fractions of the 12C controls, the amount recovered from fractions 6 and 7 was generally smaller than that from the 13C samples (see Fig. S4 in the supplemental material). T-RFLP analysis of 12C samples typically showed similar communities in the heavy and light fractions, while 13C samples showed distinct communities. A distinctive “heavy” community was also observed in the clone libraries, though accompanied by a smaller number of sequences from the most abundant light groups, such as those of Rhodobacterales, Flavobacteriales, Thiotrichales, Vibrionales, and Chromatiales species. While it is possible that these organisms actually incorporated 13C, it is more likely that most of these sequences were from contaminating 12C-DNA. Only groups that were much more abundant in the heavy fractions of the 13C incubations than in the heavy fractions of the 12C incubations and light fractions of the 13C incubations, based on both T-RFLP and clone libraries, were considered 13C labeled. The bimodal distribution of 13C content in fatty acids (Fig. 2) provided additional confirmation of 13C uptake by a subset of the microbial community and supported the results of the clone library and T-RFLP data.
Methane.
From both the T-RFLP data and the clone libraries, it appears that the major consumers of 13C in the methane incubations were affiliated with the Methylophilaceae or Methylophaga, with a smaller contribution from the Methylococcaceae. The Methylophilaceae and Methylophaga are both groups of non-methane-oxidizing methylotrophs, consuming other C1 compounds such as methanol and methylamine (2); previous SIP studies have shown that members of the Methylophaga group were the primary consumers of methanol and methylamine in marine environments (56, 59, 61). Most methane SIP studies have observed some 13C uptake by organisms more closely related to nonmethanotrophic methylotrophs than to known methanotrophs, a results which has been attributed to cross-feeding (8, 33, 50). However, methylotroph sequences in two marine studies were much less abundant than sequences related to known methanotrophs (37, 56), in contrast to the pattern of relative abundance we observed. In a study of rice roots, Qiu et al. (66) found that most of the sequences in a 13C-DNA clone library were affiliated with Methylophilales species, but their long (20-day) incubations make cross-feeding the likely explanation. In our study, the sequences affiliated with Methylophaga and Methylophilales were far more abundant than the Methylococcaceae sequences after just 3 days and remained more abundant over the 9-day incubation period. The early and sustained representation of methylotrophs in the heavy fractions suggests that these Methylophaga and Methylophilales species are either directly incorporating methane or are involved in a closely coupled relationship with methanotrophic Methylococcaceae. In all three of these groups, the most abundant fatty acids are 16:1 and 16:0 (3, 36, 40, 79), which is consistent with the “heavy” labeling of these fatty acids observed at the end of the incubation.
The pmoA data, though limited, show a number of sequences not closely related to cultured methanotrophs. The sequence of the most common phylotype in both the initial sediment and the heavy DNA from the methane incubation was only 81% similar to any other sequence in GenBank. The most closely related sequences came from several uncultured bacteria and two cultured but unclassified Methylococcaceae species. The other pmoA sequences in our methane clone library also clustered with the Methylococcaceae sequences, particularly with those of representatives from methane seep environments like the Eel River Basin (77). We found no sequences affiliated with type II methanotrophs, and very few sequences closely related to cultured type I methanotrophs. Based on the affiliation of these sequences with the gammaproteobacterial pmoA sequences, they likely belong to members of the Methylococcaceae observed in the 16S rRNA clone libraries, though it is also possible that they belong to the Methylophaga-like bacteria, representing a novel group of methanotrophs.
Ethane.
13C ethane was primarily consumed by a group of organisms from the family Methylococcaceae. These organisms have previously been thought to be obligate methanotrophs (3), capable of growing only on methane or a few other C1 compounds. Without pure cultures, we cannot determine whether these ethane-oxidizing Methylococcaceae species are also capable of growth on methane. However, the ethane Methylococcaceae sequences were not detected in any of the heavy DNA clone libraries from the methane incubations, indicating that they were not a major contributor to methane oxidation under the conditions of the SIP incubation. Interestingly, five similar sequences from the last time point of the propane incubation were found in the heavy DNA clone library, raising the possibility that these bacteria also have the ability to consume propane or one of its metabolic byproducts.
Based on the co-occurrence of 16S rRNA genes from members of the family Methylococcaceae and a highly divergent pmoA-like gene in the heavy DNA, we hypothesize that this gene encodes part of a novel ethane monooxygenase enzyme active in some Methylococcaceae species. While molecular genetic and biochemical studies are needed to test this hypothesis, there is some corroboration in existing GenBank sequence entries (AB453956 to -65). These GenBank entries include 16S rRNA and pmoA gene sequences reported to be from the isolated cultures of the “Methylococcaceae bacterium” species ET-HIRO, ET-SHO, T2-1, and SF-BR. They are grouped under the title “Discovery of ethane monooxygenase in marine bacteria belongs to Methylococcaceae that could grow on ethane, but not methane,” but additional information has yet to be published in the peer-reviewed literature. These pmoA-like sequences group outside those of all known pmoA genes from Alphaproteobacteria and Gammaproteobacteria species and amoA genes from Betaproteobacteria and Gammaproteobacteria species. They are also distantly related to the highly divergent sequence observed by Tavormina et al. (77) in the water column above methane seeps in the Eel River Basin. The prevalence and function of both these enzymes are currently unknown, but they may also play a role in the oxidation of other common biogenic molecules such as methane, ammonia, or ethene.
It is difficult to determine the environmental importance of these putative oxidizers of ethane. Both methane- and ammonia-oxidizing bacteria have long been known to oxidize small amounts of ethane (11, 34), but this has only been observed during methane- or ammonia-supported growth. High concentrations of ethane are typically accompanied by propane and even higher concentrations of methane (20, 83), making it unlikely that the bacteria are ever exposed to an environment with ethane as the most abundant carbon source. However, ethane oxidation was detectable in our incubations within 24 h, suggesting that these bacteria required little adaptation to use ethane as the major carbon source. By the final time point, all four of the major fatty acids in the incubations were more enriched in 13C than those in the methane and propane incubations. The ethane incubations also showed the highest levels of 100% 13C-labeled fatty acids—50% of the 16:1 fatty acids were 100% 13C labeled. As mentioned above, Methylococcaceae species typically contain high levels of 16:1 and 16:0 fatty acids (3), which could account for much of the 13C ethane incorporation into fatty acids. In contrast to the methane incubations, though, ∼20% of the 13C was incorporated into 18:1 fatty acids, which are not commonly found at high levels in Methylococcaceae species (4). This result may have been due to different levels of fatty acid content in these Methylococcaceae species or to the presence of cross-feeding bacteria. By the final time point, the second-most-abundant group of T-RFs in the heavy fractions was that assigned to the species of Rhodobacterales, many of which contain predominately 18:1 fatty acids (25, 71, 75). Thus, the most parsimonious explanation for the lipid distributions is that 16:0 and 16:1 fatty acids were primarily produced by ethane-fed Methylococcaceae species throughout the incubation, whereas the 18:1 fatty acid was primarily produced by cross-feeding Rhodobacterales species near the end of the incubation. Elevated levels of intermediate 13C labeling in 18:1 compared to 16:0 and 16:1 fatty acids further support this explanation.
Propane.
Propane was primarily consumed by members of a group of unclassified Gammaproteobacteria whose closest cultured relatives are found in the genera Marinimicrobium, Microbulbifer, and Saccharophagus (16S rRNA genes ∼92% similar). These bacteria are typically known for degradation of complex polysaccharides such as cellulose, chitin, and agar (19, 24), and no members of these genera have previously been linked to propane oxidation or hydrocarbon oxidation in general. However, members of this group also showed ∼90% 16S rRNA gene similarity to some Pseudomonas species, which have been shown to oxidize propane (70), and to obligate hydrocarbon degraders such Alcanivorax and Marinobacter spp. (84).
At the last time point, the clone libraries and the T-RFLP data both showed the Gammaproteobacteria giving way to Rhodobacterales species in the heavy fractions. The Rhodobacterales were not very abundant in the light fractions, so this result appears to have been due to 13C uptake rather than to the presence of contaminating 12C-DNA, perhaps due to a change in community composition over time or, more likely, to cross-feeding. As with ethane, the idea of 13C uptake by this group is supported by the fact that 18:1 fatty acids contained ∼20% of all 13C incorporated into fatty acids; 18:1 fatty acids are abundant in Rhodobacterales species (25, 71, 75). An increase in the abundance of certain members of the Rhodobacterales has often been observed in oil-contaminated environments or oil-amended mesocosm experiments (6, 15, 55). Whether these organisms are actually able to degrade hydrocarbons is unclear (7, 48), though at least one member of this group has been shown to degrade C10 through C13 n-alkanes (30). Rhodobacterales species have not been linked to ethane or propane oxidation, but some species are able to consume ethanol or propanol (23), likely sources of 13C through cross-feeding. It may be that many of the oil-associated Rhodobacterales species are not capable of direct consumption of alkanes but instead feed on the byproducts of alkane metabolism, as appears to have been the case in our incubations.
As in the ethane incubations, the majority of 13C propane was incorporated into 16:1 and 16:0 fatty acids, presumably belonging to the primary propane oxidizers, but the lack of cultured bacteria closely related to these unclassified Gammaproteobacteria species makes comparison impossible. The heavy DNA from the last time point also included several Methylococcaceae sequences similar to those from the ethane incubations, which may account for some of the heavily labeled 16:1 or 16:0 fatty acids. These sequences appeared only in the final samples, suggesting that the ethane oxidizers required an adaptation period before beginning propane oxidation or that they were incorporating a byproduct of propane oxidation. Most previously described ethane oxidizers can oxidize propane, and some propane oxidizers can oxidize ethane (70), so some overlap between the two is not surprising, but ethane and propane oxidation appear to be primarily performed by different communities.
Lipids.
One common and striking feature of the newly produced fatty acids in each experiment is the dominance of monounsaturates, specifically, 16:1 and 18:1. High percentages of these fatty acids in membrane lipids are typical of methanotrophs and are thought to be associated with the extensive network of stacked membranes common in these organisms (5). The gammaproteobacterial (type I) methanotrophs typically contain mostly 16:1 fatty acids, while the alphaproteobacterial (type II) methanotrophs typically contain mostly 18:1 fatty acids (2, 5). The predominant incorporation of 13C-methane into 16:1 fatty acids provides additional evidence that methane was consumed by gammaproteobacterial methanotrophs. The high level of incorporation into 16:1 fatty acids observed in the ethane incubation also supports our interpretation from investigations of 16S rRNA and pmoA clone libraries that the responsible organisms share numerous similarities with methanotrophs, potentially including stacked membranes. A second striking feature determined from these results is the large extent of fully 13C-labeled lipids and low levels of any specific intermediately labeled lipid. These results suggest a high degree of metabolic specificity for all three substrates, though this may include cross-feeding relationships.
Conclusions.
This report demonstrates the ability of SIP to identify previously unknown groups of methane-, ethane-, and propane-oxidizing bacteria at a marine hydrocarbon seep, with a distinct group of bacteria found to be responsible for each process. These results may have implications for our understanding of methane oxidation in marine environments. Novel groups of marine bacteria may contain monooxygenases related to those of terrestrial bacteria but with different substrate affinities, affecting our ability to detect them with common primer sets and to predict biogeochemical function based on the presence or absence of these genes. The multitude of hydrocarbon compounds present in seep environments could support a range of hydrocarbon-oxidizing bacteria, but relatively little is known about which organisms consume which compounds and under what conditions. Further work, with both culturing and culture-independent techniques like SIP, is necessary to improve understanding of these processes and their roles in biogeochemical cycling.
Supplementary Material
Acknowledgments
We thank Michael Friedrich and Bianca Pommerenke for early assistance with the SIP technique. Heather Coleman, Blair Paul, and Cristoph Pierre collected samples, and Lichun Zhang performed the fatty acid extractions and GC/MS analyses.
This work was funded by a National Science Foundation award (OCE-0447395) and a DOE award (DE-NT0005667) to D.L.V. and a National Science Foundation award (EAR-0645502) to A.L.S.
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ashelford, K., N. Chuzhanova, J. Fry, A. Jones, and A. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodelier, P., M. Gillisen, K. Hordijk, J. Damsté, W. Rijpstra, J. Geenevasen, and P. Dunfield. 2009. A reanalysis of phospholipid fatty acids as ecological biomarkers for methanotrophic bacteria. ISME J. 3:606-617. [DOI] [PubMed] [Google Scholar]
- 3.Bowman, J. 2000. The methanotrophs—the families Methylococcaceae and Methylocystaceae, p. 266-289. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community. Springer, New York, NY. http://www.springerlink.com/content/p5120j3327n41805/.
- 4.Bowman, J. 2005. Order VII. Methylococcales ord. nov., p. 248-270. In G. Garrity, D. J. Brenner, N. R. Krieg, and J. R. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2: the proteobacteria, part B-the Gammaproteobacteria. Springer, New York, NY. [Google Scholar]
- 5.Bowman, J., J. Skerratt, P. Nichols, and L. Sly. 1991. Phospholipid fatty acid and lipopolysaccharide fatty acid signature lipids in methane-utilizing bacteria. FEMS Microbiol. Lett. 85:15-22. [Google Scholar]
- 6.Brakstad, O., and A. Lødeng. 2005. Microbial diversity during biodegradation of crude oil in seawater from the North Sea. Microb. Ecol. 49:94-103. [DOI] [PubMed] [Google Scholar]
- 7.Buchan, A., and J. González. 2010. Roseobacter, p. 1335-1344. In K. N. Timmis (ed.), Handbook of hydrocarbon and lipid microbiology, vol. 14. Springer, Berlin, Germany. [Google Scholar]
- 8.Cébron, A., L. Bodrossy, N. Stralis-Pavese, A. Singer, I. Thompson, J. Prosser, and J. Murrell. 2007. Nutrient amendments in soil DNA stable isotope probing experiments reduce the observed methanotroph diversity. Appl. Environ. Microbiol. 73:798-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, J., L. Washburn, J. Hornafius, and B. Luyendyk. 2000. Dissolved hydrocarbon flux from natural marine seeps to the southern California Bight. J. Geophys. Res. Oceans 105:11509-11522. [Google Scholar]
- 10.Clayton, C., S. Hay, and S. Baylis. 1997. Alteration of natural gas during leakage from a North Sea salt diapir field. Mar. Geol. 137:69-80. [Google Scholar]
- 11.Colby, J., D. I. Stirling, and H. Dalton. 1977. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem. J. 165:395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole, J., B. Chai, T. Marsh, R. Farris, Q. Wang, S. Kulam, S. Chandra, D. McGarrell, T. Schmidt, and G. Garrity. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooley, R., B. Dubbels, L. Sayavedra-Soto, P. Bottomley, and D. Arp. 2009. Kinetic characterization of the soluble butane monooxygenase from Thauera butanivorans, formerly ‘Pseudomonas butanovora.’ Microbiology 155:2086-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costello, A. M., and M. E. Lidstrom. 1999. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulon, F., B. McKew, A. Osborn, T. McGenity, and K. Timmis. 2007. Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ. Microbiol. 9:177. [DOI] [PubMed] [Google Scholar]
- 16.Dedysh, S., W. Liesack, V. Khmelenina, N. Suzina, Y. Trotsenko, J. Semrau, A. Bares, N. Panikov, and J. Tiedje. 2000. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50:955-969. [DOI] [PubMed] [Google Scholar]
- 17.Dedysh, S. N., C. Knief, and P. F. Dunfield. 2005. Methylocella species are facultatively methanotrophic. J. Bacteriol. 187:4665-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubbels, B., L. Sayavedra-Soto, and D. Arp. 2007. Butane monooxygenase of ‘Pseudomonas butanovora’: purification and biochemical characterization of a terminal-alkane hydroxylating diiron monooxygenase. Microbiology 153:1808-1816. [DOI] [PubMed] [Google Scholar]
- 19.Ekborg, N. A., J. M. Gonzalez, M. B. Howard, L. E. Taylor, S. W. Hutcheson, and R. M. Weiner. 2005. Saccharophagus degradans gen. nov., sp. nov., a versatile marine degrader of complex polysaccharides. Int. J. Syst. Evol. Microbiol. 55:1545-1549. [DOI] [PubMed] [Google Scholar]
- 20.Etiope, G., and P. Ciccioli. 2009. Earth's degassing: a missing ethane and propane source. Science 323:478. [DOI] [PubMed] [Google Scholar]
- 21.Fung, I., M. Prather, J. John, J. Lerner, and E. Matthews. 1991. Three-dimensional model synthesis of the global methane cycle. J. Geophys. Res. 96:13033-13065. [Google Scholar]
- 22.Fuse, H., M. Ohta, O. Takimura, K. Murakami, H. Inoue, Y. Yamaoka, J. Oclarit, and T. Omori. 1998. Oxidation of trichloroethylene and dimethyl sulfide by a marine Methylomicrobium strain containing soluble methane monooxygenase. Biosci. Biotechnol. Biochem. 62:1925-1931. [DOI] [PubMed] [Google Scholar]
- 23.Garrity, G., J. Bell, and T. Lilburn. 2005. Order III. Rhodobacterales ord. nov., p. 161-167. In G. Garrity, D. J. Brenner, N. R. Krieg, and J. R. Staley (ed.), Bergey's manual of systematic bacteriology, vol. 2: the proteobacteria, part C—the Alpha-, Beta-, Delta-, and Epsilonproteobacteria. Springer, New York, NY. [Google Scholar]
- 24.Gonzalez, J., F. Mayer, M. Moran, R. Hodson, and W. Whitman. 1997. Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int. J. Syst. Evol. Microbiol. 47:369-376. [DOI] [PubMed] [Google Scholar]
- 25.Gosnik, J. J. 2005. Genus XI. Octadecabacter, p. 194-197. In G. Garrity, D. J. Brenner, N. R. Krieg, and J. R. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2: the proteobacteria, part C—the Alpha-, Beta-, Delta-, and Epsilonproteobacteria. Springer, New York, NY. [Google Scholar]
- 26.Hakemian, A., and A. Rosenzweig. 2007. The biochemistry of methane oxidation. Annu. Rev. Biochem. 76:223-241. [DOI] [PubMed] [Google Scholar]
- 27.Hamamura, N., and D. J. Arp. 2000. Isolation and characterization of alkane-utilizing Nocardioides sp. strain CF8. FEMS Microbiol. Lett. 186:21-26. [DOI] [PubMed] [Google Scholar]
- 28.Hamamura, N., C. M. Yeager, and D. J. Arp. 2001. Two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8. Appl. Environ. Microbiol. 67:4992-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harwati, T., Y. Kasai, Y. Kodama, D. Susilaningsih, and K. Watanabe. 2008. Tranquillimonas alkanivorans gen. nov., sp. nov., an alkane-degrading bacterium isolated from Semarang Port in Indonesia. Int. J. Syst. Evol. Microbiol. 58:2118-2121. [DOI] [PubMed] [Google Scholar]
- 31.Hornafius, J., D. Quigley, and B. Luyendyk. 1999. The world's most spectacular marine hydrocarbon seeps (Coal Oil Point, Santa Barbara Channel, California): quantification of emissions. J. Geophys. Res. Oceans 104:20703-20711. [Google Scholar]
- 32.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 33.Hutchens, E., S. Radajewski, M. G. Dumont, I. R. McDonald, and J. C. Murrell. 2004. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 6:111-120. [DOI] [PubMed] [Google Scholar]
- 34.Hyman, M. R., I. B. Murton, and D. J. Arp. 1988. Interaction of ammonia monooxygenase from Nitrosomonas europaea with alkanes, alkenes, and alkynes. Appl. Environ. Microbiol. 54:3187-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inagaki, F., U. Tsunogai, M. Suzuki, A. Kosaka, H. Machiyama, K. Takai, T. Nunoura, K. Nealson, and K. Horikoshi. 2004. Characterization of C1-metabolizing prokaryotic communities in methane seep habitats at the Kuroshima Knoll, southern Ryukyu Arc, by analyzing pmoA, mmoX, mxaF, mcrA, and 16S rRNA genes. Appl. Environ. Microbiol. 70:7445-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenkins, O., D. Byrom, and D. Jones. 1987. Methylophilus: a new genus of methanol-utilizing bacteria. Int. J. Syst. Evol. Microbiol. 37:446. [Google Scholar]
- 37.Jensen, S., J. Neufeld, N. Birkeland, M. Hovland, and J. Murrell. 2008. Methane assimilation and trophic interactions with marine Methylomicrobium in deep-water coral reef sediment off the coast of Norway. FEMS Microbiol. Ecol. 66:320-330. [DOI] [PubMed] [Google Scholar]
- 38.Johnson, E. L., and M. R. Hyman. 2006. Propane and n-butane oxidation by Pseudomonas putida GPo1. Appl. Environ. Microbiol. 72:950-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Judd, A. 2004. Natural seabed gas seeps as sources of atmospheric methane. Environ. Geol. 46:988-996. [Google Scholar]
- 40.Kalyuzhnaya, M., S. Bowerman, J. Lara, M. Lidstrom, and L. Chistoserdova. 2006. Methylotenera mobilis gen. nov., sp. nov., an obligately methylamine-utilizing bacterium within the family Methylophilaceae. Int. J. Syst. Evol. Microbiol. 56:2819-2823. [DOI] [PubMed] [Google Scholar]
- 41.Kaplan, C., and C. Kitts. 2003. Variation between observed and true terminal restriction fragment length is dependent on true TRF length and purine content. J. Microbiol. Methods 54:121-125. [DOI] [PubMed] [Google Scholar]
- 42.Kinnaman, F. 2008. Ph.D. thesis. Microbial consumption of natural gas in marine sediments: rates, distributions and isotope effects. University of California, Santa Barbara, CA.
- 43.Kinnaman, F., J. Kimball, L. Busso, D. Birgel, H. Ding, K. Hinrichs, and D. Valentine. 2010. Gas flux and carbonate occurrence at a shallow seep of thermogenic natural gas. Geo-Marine Lett. 30:355-365. [Google Scholar]
- 44.Kniemeyer, O., F. Musat, S. Sievert, K. Knittel, H. Wilkes, M. Blumenberg, W. Michaelis, A. Classen, C. Bolm, S. Joye, and F. Widdel. 2007. Anaerobic oxidation of short-chain hydrocarbons by marine sulphate-reducing bacteria. Nature 449:898-901. [DOI] [PubMed] [Google Scholar]
- 45.Kotani, T., Y. Kawashima, H. Yurimoto, N. Kato, and Y. Sakai. 2006. Gene structure and regulation of alkane monooxygenases in propane-utilizing Mycobacterium sp. TY-6 and Pseudonocardia sp. TY-7. J. Biosci. Bioeng. 102:184-192. [DOI] [PubMed] [Google Scholar]
- 46.Kotani, T., T. Yamamoto, H. Yurimoto, Y. Sakai, and N. Kato. 2003. Propane monooxygenase and NAD+-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY-5. J. Bacteriol. 185:7120-7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krüger, M., T. Treude, H. Wolters, K. Nauhaus, and A. Boetius. 2005. Microbial methane turnover in different marine habitats. Palaeogeog. Palaeoclim. Palaeoecol. 227:6-17. [Google Scholar]
- 48.Kwon, K. K., and S.-J. Kim. 2010. Marine, hydrocarbon-degrading Alphaproteobacteria, p. 1707-1714. In K. N. Timmis (ed.), Handbook of hydrocarbon and lipid microbiology, vol. 19. Springer, Berlin, Germany. [Google Scholar]
- 49.Lidstrom, M. 1988. Isolation and characterization of marine methanotrophs. Antonie Van Leeuwenhoek 54:189-199. [DOI] [PubMed] [Google Scholar]
- 50.Lin, J.-L., S. Radajewski, B. Eshinimaev, Y. Trotsenko, I. McDonald, and J. Murrell. 2004. Molecular diversity of methanotrophs in Transbaikal soda lake sediments and identification of potentially active populations by stable isotope probing. Environ. Microbiol. 6:1049-1060. [DOI] [PubMed] [Google Scholar]
- 51.Lueders, T., M. Manefield, and M. Friedrich. 2004. Enhanced sensitivity of DNA-and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 52.Lueders, T., B. Wagner, P. Claus, and M. Friedrich. 2004. Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ. Microbiol. 6:60-72. [DOI] [PubMed] [Google Scholar]
- 53.Mastalerz, V., G. J. de Langea, and A. Dählmann. 2009. Differential aerobic and anaerobic oxidation of hydrocarbon gases discharged at mud volcanoes in the Nile deep-sea fan. Geochim. Cosmochim. Acta 73:3849-3863. [Google Scholar]
- 54.McDonald, I., L. Bodrossy, Y. Chen, and J. Murrell. 2008. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 74:1305-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKew, B., F. Coulon, A. Osborn, K. Timmis, and T. McGenity. 2007. Determining the identity and roles of oil-metabolizing marine bacteria from the Thames estuary, UK. Environ. Microbiol. 9:165-176. [DOI] [PubMed] [Google Scholar]
- 56.Moussard, H., N. Stralis-Pavese, L. Bodrossy, J. D. Neufeld, and C. J. Murrell. 2009. Identification of active methylotrophic bacteria inhabiting surface sediment of a marine estuary. Environ. Microbiol. Rep. 1:424-433. [DOI] [PubMed] [Google Scholar]
- 57.Murrell, J., I. McDonald, and B. Gilbert. 2000. Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 8:221-225. [DOI] [PubMed] [Google Scholar]
- 58.Neufeld, J., M. Dumont, J. Vohra, and J. Murrell. 2007. Methodological considerations for the use of stable isotope probing in microbial ecology. Microb. Ecol. 53:435-442. [DOI] [PubMed] [Google Scholar]
- 59.Neufeld, J., H. Schäfer, M. Cox, R. Boden, I. McDonald, and J. Murrell. 2007. Stable-isotope probing implicates Methylophaga spp. and novel Gammaproteobacteria in marine methanol and methylamine metabolism. ISME J. 1:480-491. [DOI] [PubMed] [Google Scholar]
- 60.Neufeld, J., J. Vohra, M. Dumont, T. Lueders, M. Manefield, M. Friedrich, and J. Murrell. 2007. DNA stable-isotope probing. Nat. Protoc. 2:860-866. [DOI] [PubMed] [Google Scholar]
- 61.Neufeld, J. D., R. Boden, H. Moussard, H. Schafer, and J. C. Murrell. 2008. Substrate-specific clades of active marine methylotrophs associated with a phytoplankton bloom in a temperate coastal environment. Appl. Environ. Microbiol. 74:7321-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niemann, H., T. Losekann, D. de Beer, M. Elvert, T. Nadalig, K. Knittel, R. Amann, E. Sauter, M. Schluter, M. Klages, J. P. Foucher, and A. Boetius. 2006. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443:854-858. [DOI] [PubMed] [Google Scholar]
- 63.Op den Camp, H. J. M., T. Islam, M. B. Stott, H. R. Harhangi, A. Hynes, S. Schouten, M. S. M. Jetten, N.-K. Birkeland, A. Pol, and P. F. Dunfield. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1:293-306. [DOI] [PubMed] [Google Scholar]
- 64.Osborn, M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 65.Padda, R. S., K. K. Pandey, S. Kaul, V. D. Nair, R. K. Jain, S. K. Basu, and T. Chakrabarti. 2001. A novel gene encoding a 54 kDa polypeptide is essential for butane utilization by Pseudomonas sp. IMT37. Microbiology 147:2479-2491. [DOI] [PubMed] [Google Scholar]
- 66.Qiu, Q., R. Conrad, and Y. Lu. 2009. Cross-feeding of methane carbon among bacteria on rice roots revealed by DNA-stable isotope probing. Environ. Microbiol. Rep. 1:355-361. [DOI] [PubMed] [Google Scholar]
- 67.Radajewski, S., P. Ineson, N. Parekh, and J. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 68.Reeburgh, W. 2007. Oceanic methane biogeochemistry. Chem. Rev. 107:486-513. [DOI] [PubMed] [Google Scholar]
- 69.Schütte, U., Z. Abdo, S. Bent, C. Shyu, C. Williams, J. Pierson, and L. Forney. 2008. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 80:365-380. [DOI] [PubMed] [Google Scholar]
- 70.Shennan, J. 2006. Utilisation of C2-C4 gaseous hydrocarbons and isoprene by microorganisms. J. Chem. Technol. Biotechnol. 81:237-256. [Google Scholar]
- 71.Shibe, T., and J. F. Imhoff. 2005. Genus XVIII. Roseobacter, p. 212-215. In G. Garrity, D. J. Brenner, N. R. Krieg, and J. R. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2: the proteobacteria, part C—the Alpha-, Beta-, Delta-, and Epsilonproteobacteria. Springer, New York, NY. [Google Scholar]
- 72.Sieburth, J., P. Johnson, M. Eberhardt, M. Sieracki, M. Lidstrom, and D. Laux. 1987. The first methane-oxidizing bacterium from the upper mixing layer of the deep ocean: Methylomonas pelagica sp. nov. Curr. Microbiol. 14:285-293. [Google Scholar]
- 73.Sluis, M., L. Sayavedra-Soto, and D. Arp. 2002. Molecular analysis of the soluble butane monooxygenase from ‘Pseudomonas butanovora.’ Microbiology 148:3617-3629. [DOI] [PubMed] [Google Scholar]
- 74.Smith, K., A. Costello, and M. Lidstrom. 1997. Methane and trichloroethylene oxidation by an estuarine methanotroph, Methylobacter sp. strain BB5.1. Appl. Environ. Microbiol. 63:4617-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sorokin, D. Y., F. A. Rainey, R. I. Webb, and J. A. Fuerst. 2005. Genus XXV. Sulfitobacter, p. 224-228. In G. Garrity, D. J. Brenner, N. R. Krieg, and J. R. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2: the Proteobacteria, part C—the Alpha-, Beta-, Delta-, and Epsilonproteobacteria. Springer, New York, NY. [Google Scholar]
- 76.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 77.Tavormina, P., W. Ussler III, and V. Orphan. 2008. Planktonic and sediment-associated aerobic methanotrophs in two seep systems along the North American margin. Appl. Environ. Microbiol. 74:3985-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Treude, T., and W. Ziebis. 2010. Methane oxidation in permeable sediments at hydrocarbon seeps in the Santa Barbara Channel, California. Biogeosciences 7:1905-1933. [Google Scholar]
- 79.Urakami, T., and K. Komagata. 1987. Characterization of species of marine methylotrophs of the genus Methylophaga. Int. J. Syst. Evol. Microbiol. 37:402-406. [Google Scholar]
- 80.Valentine, D., D. Blanton, W. Reeburgh, and M. Kastner. 2001. Water column methane oxidation adjacent to an area of active hydrate dissociation, Eel River Basin. Geochim. Cosmochim. Acta 65:2633-2640. [Google Scholar]
- 81.Wang, Q., G. Garrity, J. Tiedje, and J. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wasmund, K., D. Kurtboke, K. Burns, and D. Bourne. 2009. Microbial diversity in sediments associated with a shallow methane seep in the tropical Timor Sea of Australia reveals a novel aerobic methanotroph diversity. FEMS Microbiol. Ecol. 68:142-151. [DOI] [PubMed] [Google Scholar]
- 83.Xiao, Y., J. Logan, D. Jacob, R. Hudman, R. Yantosca, and D. Blake. 2008. Global budget of ethane and regional constraints on US sources. J. Geophys. Res. 113:D21306. doi: 10.1029/2007JD009415. [DOI] [Google Scholar]
- 84.Yakimov, M., K. Timmis, and P. Golyshin. 2007. Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 18:257-266. [DOI] [PubMed] [Google Scholar]
- 85.Yan, T., Q. Ye, J. Zhou, and C. Zhang. 2006. Diversity of functional genes for methanotrophs in sediments associated with gas hydrates and hydrocarbon seeps in the Gulf of Mexico. FEMS Microbiol. Ecol. 57:251-259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.