Abstract
Extracellular enzymes synthesized by soil microbes play a central role in the biogeochemical cycling of nutrients in the environment. The pH optima of eight hydrolytic enzymes involved in the cycles of carbon, nitrogen, phosphorus, and sulfur, were assessed in a series of tropical forest soils of contrasting pH values from the Republic of Panama. Assays were conducted using 4-methylumbelliferone-linked fluorogenic substrates in modified universal buffer. Optimum pH values differed markedly among enzymes and soils. Enzymes were grouped into three classes based on their pH optima: (i) enzymes with acidic pH optima that were consistent among soils (cellobiohydrolase, β-xylanase, and arylsulfatase), (ii) enzymes with acidic pH optima that varied systematically with soil pH, with the most acidic pH optima in the most acidic soils (α-glucosidase, β-glucosidase, and N-acetyl-β-glucosaminidase), and (iii) enzymes with an optimum pH in either the acid range or the alkaline range depending on soil pH (phosphomonoesterase and phosphodiesterase). The optimum pH values of phosphomonoesterase were consistent among soils, being 4 to 5 for acid phosphomonoesterase and 10 to 11 for alkaline phosphomonoesterase. In contrast, the optimum pH for phosphodiesterase activity varied systematically with soil pH, with the most acidic pH optima (3.0) in the most acidic soils and the most alkaline pH optima (pH 10) in near-neutral soils. Arylsulfatase activity had a very acidic optimum pH in all soils (pH ≤3.0) irrespective of soil pH. The differences in pH optima may be linked to the origins of the enzymes and/or the degree of stabilization on solid surfaces. The results have important implications for the interpretation of hydrolytic enzyme assays using fluorogenic substrates.
Measurements of the activities of extracellular enzymes involved in the turnover of nutrients from organic compounds provide important information on biogeochemical cycles in tropical soils (13, 59, 63). In particular, they are the primary mechanism by which microbes decompose organic matter and can provide key information on the nutrient status of the ecosystem (45, 60). For example, changes in the activities of phosphatase and N-acetyl-β-glucosaminidase in soil chronosequences reflect long-term changes in nitrogen and phosphorus availability during pedogenesis in both tropical and temperate rain forests (3, 34).
The pH of the soil solution exerts a strong control on enzyme activity, because it influences the conformation of the enzyme, its adsorption on solid surfaces, and the ionization and solubility of substrates and cofactors (38, 51). Although some studies have determined enzyme activity at the soil pH (e.g., references 19 and 63), assays are usually conducted at the optimum pH for the enzyme, which yields a measure of its maximum potential activity at a given temperature (7, 27). For example, the assay of acid phosphomonoesterase activity using the chromogenic substrate para-nitrophenyl phosphate is typically performed at pH 6.5 (51), based on the determination of the pH optima of this enzyme in a series of temperate agricultural soils (15, 21, 50).
Once the optimum pH for a given enzyme has been determined, it is usually assumed that this will apply broadly to other soils, allowing the recommendation of a single buffer pH in standardized procedures (44, 51). However, the pH optima of some enzymes can vary markedly among soils. For example, Niemi and Vepsäläinen (33) reported soil-specific pH optima for three hydrolytic enzymes (acid phosphomonoesterase, phosphodiesterase, and N-acetyl-β-glucosaminidase) in six soils under contrasting land uses in Finland. Such soil-specific pH optima led Malcolm (27) to recommend, in a critique of soil enzyme assays, that the pH optimum of the enzyme under study should be determined for each soil.
Differences among soils in relation to the pH optima of an individual enzyme might be due to a variety of factors, including the composition of the soil microbial community (i.e., if isoenzymes originating from different organisms have different pH optima) and the location of the enzyme in the soil matrix (e.g., intracellular, free in solution, or adsorbed on solid surfaces, etc.) (4). For example, the sorption of an enzyme on a clay surface can increase its optimum pH by one or two pH units relative to that of the same enzyme in solution (31, 40). This is due to “unfolding” of enzymes on solid surfaces, which is most likely to occur at soil pH values below the isoelectric point of the enzyme (26, 38).
Information on the optimum pH of enzyme activity is of particular importance for studies that use fluorogenic substrates in multiwell plates to assay several enzymes simultaneously. Such studies are usually simplified by assaying all enzymes in a single buffer, such as acetate at pH 5.5 (57) or 2-(N-morpholino)ethanesulfonic acid (MES) at pH 6.1 (30). However, this may not coincide with the pH optima of all the enzymes involved, especially if the optimum pH for a given enzyme varies among soils. Further, most studies of the pH optima of hydrolytic enzymes have been conducted using chromogenic substrates linked to para-nitrophenol, and it is not clear whether the values correspond to the pH optima for fluorogenic substrates linked to 4-methylumbelliferone.
Here I report the pH optima for eight hydrolytic enzymes involved in the cycles of carbon, nitrogen, phosphorus, and sulfur in a series of soils under a lowland tropical rain forest in the Republic of Panama. The aim was to determine the extent to which the pH optima of activity varied among enzymes and soils, in order to develop a method suitable for the measurement of enzyme activities in a broad range of tropical rain forest soils.
MATERIALS AND METHODS
Site description and soil properties.
Soils were sampled from seven sites under lowland moist tropical forest in central Panama (Table 1). The long-term mean annual air temperature on Barro Colorado Island, close to several of the sites, is 27.1°C, and the mean monthly temperature varies by <1°C throughout the year (61). Annual rainfall varies from 1,730 mm at Cerro Galera to 2,159 mm at Rio Paja (Table 1), and there is a strong 4-month dry season from January to April. Samples were taken during the wet season to a depth of 10 cm at various times between 2006 and 2008. Upon return to the laboratory, soils were screened (<9-mm mesh) to break up large aggregates, and visible stones and roots were removed by hand. Samples were then further sieved (<2 mm) to isolate the fine earth fraction, stored at 4°C in plastic bags, and analyzed within 1 month. Extracellular enzyme activities in tropical forest soils appear relatively insensitive to short-term storage (55), so it is considered unlikely, although possible, that the pH optima were influenced by storage (11).
TABLE 1.
Site characteristics and properties of seven soils under lowland moist tropical forest in the Republic of Panama
| Parameter | Value for soil |
||||||
|---|---|---|---|---|---|---|---|
| Cardenas | Albrook | Rio Paja | Gamboa | Las Cruces | Cerro Galera | Caritas | |
| Plot codea | P25 | P24 | P27 | ||||
| Latitude | 08°59′26″N | 08°58′37″N | 09°04′43″N | 09°07′25″N | 09°04′50″N | 08°55′36″N | 09°03′58″N |
| Longitude | 79°34′03″W | 79°33′50″W | 79°47′56″W | 79°40′36″W | 79°38′42″W | 79°37′24″W | 79°34′06″W |
| Elevation (m) | 78 | 53 | 110 | 50 | 180 | 300 | 110 |
| Rainfall (mm yr−1) | —b | 1,860 | 2,159 | 2,153 | 2,033 | 1,730 | 2,027 |
| Soil pH (H2O) | 3.3 | 3.4 | 3.6 | 5.4 | 5.5 | 6.3 | 6.4 |
| Topsoil texture | Clay loam | Loam | Silty-clay loam | Clay | Clay loam | Clay loam | Clay |
| Clay (%) | 28 | 23 | 31 | 49 | 38 | 33 | 41 |
| Total C (g C kg−1) | 43.7 | 60.6 | 33.9 | 73.4 | 36.6 | 37.5 | 56.6 |
| Total N (g N kg−1) | 2.77 | 4.45 | 2.62 | 6.29 | 3.07 | 2.85 | 4.54 |
| Total P (mg P kg−1) | 345 | 376 | 91 | 794 | 532 | 761 | 505 |
From reference 36 (Pyke et al.).
Rainfall is similar to that of the nearby Albrook plot.
The soils contained contrasting physical and chemical properties, including different pH values (3.3 to 6.5 in deionized water), textures (loam to clay), organic matter (33.9 to 73.4 g C kg−1 soil), and total phosphorus concentrations (91 to 794 mg P kg−1 soil). To determine soil properties, subsamples of each soil were air dried (22°C for 10 days) and then ground in a ball mill. Total carbon and nitrogen were determined by combustion and gas chromatography using a Thermo Flash NC1112 soil analyzer (CE Elantech, Lakewood, NJ). Total phosphorus was determined by ignition (550°C for 1 h) and extraction in 1 M H2SO4 (1:50 soil-to-solution ratio for 16 h), with phosphate detection by automated molybdate colorimetry using a Lachat Quickchem 8500 system (Hach Ltd., Loveland, CO). Texture was determined by the pipette method on the <2-mm fraction (i.e., not milled) after pretreatment to remove organic matter (H2O2 oxidation) and iron oxides (dithionite reduction). Soil pH was determined in a 1:2 soil-to-solution ratio in deionized water by use of a glass electrode.
Enzyme assays.
The activities of eight hydrolytic enzymes were determined using fluorogenic substrates (30, 55). The enzymes, the substrates, and their functions in soil are described in Table 2, although it should be noted that not all enzymes were assayed in all soils. Substrates were purchased from Glycosynth Ltd. (Warrington, United Kingdom) and were dissolved in 0.4% methylcellosolve (2-methoxyethanol; 0.1% final concentration in the assay), except for 4-methylumbelliferyl sulfate (a potassium salt rather than a free acid), which was dissolved in deionized water.
TABLE 2.
Enzymes and substrates used in the study and recommended buffer pHs
| Enzyme | General function | Substrate(s) | EC no. | Recommended assay pH |
|---|---|---|---|---|
| Phosphomonoesterase | Release of inorganic phosphate from simple phosphomonoesters, including organic and condensed inorganic phosphates | 4-Methylumbelliferyl phosphate | 3.1.3.2 (acid) | 6.5a |
| 3.1.3.1 (alkaline) | 11.0a | |||
| Phosphodiesterase | Release of a phosphate monoester from phosphodiesters | Bis-(4-methylumbelliferyl) phosphate | 3.1.4.1 | 8.0a |
| Cellobiohydrolase (cellulose 1,4-β-cellobiosidase) | Hydrolysis of cellulose from plant cell walls | 4-Methylumbelliferyl β-d-cellobiopyranoside | 3.2.1.91 | 5.5b |
| β-Xylanase (xylan 1,4-β-xylosidase) | Hydrolysis of hemicellulose from plant cell walls | 4-Methylumbelliferyl β-d-xylopyranoside | 3.2.1.37 | |
| α-Glucosidase | Hydrolysis of α-glucosyl residues to release α-d-glucose | 4-Methylumbelliferyl α-d-glucopyranoside | 3.2.1.20 | 6.0c |
| β-Glucosidase | Hydrolysis of β-glucosyl residues to release β-d-glucose; the final step in cellulose hydrolysis | 4-Methylumbelliferyl β-d-glucopyranoside | 3.2.1.21 | 6.0a |
| N-Acetyl-β-glucosaminide (β-N-acetylhexosaminidase) | Hydrolysis of glycosidic (N-acetyl-β-glucosaminide) bonds in chitin | 4-Methylumbelliferyl N-acetyl-β-d-glucosaminide | 3.2.1.52 | 5.5d |
| Arylsulfatase | Release of inorganic sulfate from organic sulfate esters | 4-Methylumbelliferyl sulfate, potassium salt | 3.1.6.1 | 5.8a |
Recommended by Tabatabai (51), with use of chromogenic substrates.
Recommended by Deng and Tabatabai (12) for cellulase, with use of a carboxymethyl cellulose substrate.
Recommended by Eivazi and Tabatabai (16), with use of a chromogenic substrate.
Recommended by Parham and Deng (35), with use of a chromogenic substrate.
For each sample, soil suspensions were prepared in a 1:100 soil-to-water ratio (containing 1 mM NaN3 to inhibit microbial activity) by stirring on a magnetic stir plate for 15 min. Soil suspension (50 μl) was then pipetted into wells on a microwell plate (eight wells for each pH) containing 100 μl of substrate and 50 μl of modified universal buffer (47). Use of modified universal buffer allows the assessment of pH profiles over a wide pH range in the same chemical solution, which is important because the ionic composition of the buffer can have a marked effect on enzyme activity (15, 21). Modified universal buffer was made by dissolving 6.3 g of boric acid, 14.0 g of citric acid, 11.6 g of maleic acid, 12.1 g of Trizma base, and 19.5 g of NaOH in 500 ml of deionized water. Aliquots (50 ml) were adjusted to the required pH with 1.0 M HCl or 1.0 M NaOH and diluted to 100 ml with deionized water. Buffer was stored at 4°C for up to 2 weeks.
Note that the buffer described above is stronger than the normal-strength modified universal buffer used in standard methods (51), but it was diluted 4-fold in the final assay solution. Thus, the final concentration of each chemical in the final assay mixture was 25 mM, compared to 20 mM in the normal-strength modified universal buffer. This was necessary to maintain the required pH during the assays (determined by measurement of assay pH before and after incubation), despite the solution-to-soil ratio in the final assays solution being 400:1 (200 μl solution and 0.5 mg soil), which is 80-fold greater than that in standard assays with a 5:1 solution-to-soil ratio (51). The pH range was 3.0 to 11.5, although pHs of 2.0 and 2.5 were used for arylsulfatase in one soil (Caritas). For all enzymes, the final concentrations of substrate in the assay were 100 μM for the Albrook, Cardenas, and Cerro Galera soils and 250 μM for the other four soils.
Plates were incubated for either 30 min (phosphatases, β-glucosidase, N-acetyl-β-glucosaminidase, and arylsulfatase) or 2 h (cellulase, β-xylanase, and α-glucosidase) at 26°C to approximate the daytime temperature in the upper 10 cm of soil in lowland forests in central Panama (29). The incubation times were based on preliminary assays to assess the linearity of the reaction over time. The reaction was terminated by adding 50 μl of 0.5 M NaOH (final pH of solution, >11) and the fluorescence determined immediately on a FLUOstar Optima multidetection plate reader (BMG Labtech, Offenburg, Germany), with excitation at 360 nm and emission at 460 nm. Control wells were prepared for each substrate and contained substrate, buffer, and 1 mM NaN3 (no soil suspension). Blank wells contained soil suspension and buffer only (no substrate). Standard wells contained buffer, 1 nmol methylumbelliferone (MU), and either soil suspension or 1 mM NaN3 to account for the reduction of fluorescence in the presence of soil (quenching) (22). Standard curves showed that fluorescence was linear to at least 2 nmol MU under these assay conditions. All enzyme activities are expressed as nmol MU g−1 soil (dry weight) min−1.
RESULTS
Phosphorus enzymes. (i) Phosphomonoesterase.
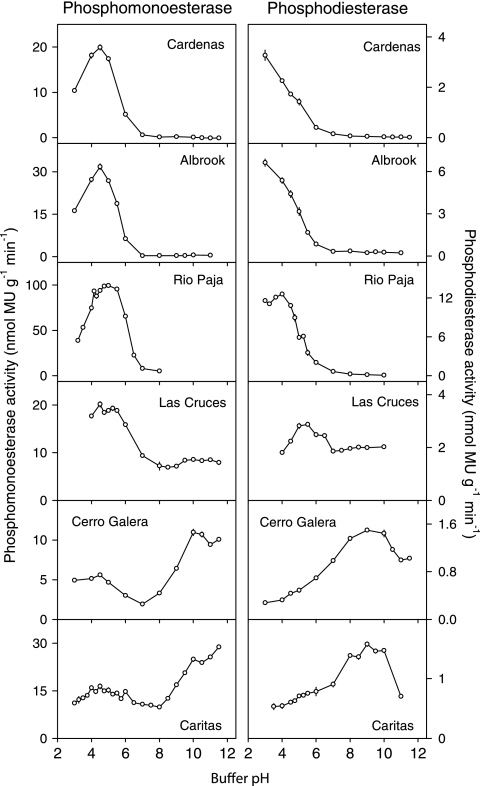
There were marked differences in phosphomonoesterase activities and pH optima among the seven soils assayed (Fig. 1 and Table 3). In particular, soils with a pH of <4 (Albrook, Cardenas, and Rio Paja) had high rates of acid phosphomonoesterase activity but negligible activity in the alkaline pH range. In contrast, soils with a pH of >6 (Cerro Galera and Caritas) had high rates of alkaline phosphomonoesterase activity that were greater than those of acid phosphomonoesterase (Fig. 1). Another soil with pH 5.5 (Las Cruces) had an acidic optimum pH but showed considerable activity across the entire pH range, including the alkaline region (Fig. 1). Overall, the optimum pH for acid phosphomonoesterase activity was between 4.0 and 5.0 (mean 4.5), whereas alkaline phosphomonoesterase activity, detected in three soils, showed broad optima between pH 9.5 and pH 11.5 (mean pH, 10.3). The highest acid phosphomonoesterase activity was detected in the Rio Paja soil, which also contained the lowest total phosphorus concentration, while the lowest acid phosphomonoesterase activity was detected in the Cerro Galera soil (soil pH of 6.5), which contained the highest total phosphorus concentration of the soils analyzed for this enzyme (Table 1).
FIG. 1.
pH optima of phosphomonoesterase activity (left panel) and phosphodiesterase activity (right panel) in six soils under lowland tropical rain forest in the Republic of Panama. Assays were conducted at 26°C using fluorogenic substrates in modified universal buffer. Error bars are the standard errors of the means for eight replicate wells per pH.
TABLE 3.
Optimum pH values and the enzyme activities at those values for 10 enzymes in seven tropical rain forest soils from the Republic of Panama
| Soil | Optimum pH (activity in nmol MU g−1 soil min−1)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phosphorus cycle |
Carbon cycle |
N-Acetyl-β-glucosaminidase (nitrogen cycle) | Arylsulfatase (sulfur cycle) | |||||||
| Acid PMEase | Alkaline PMEase | Acid PDEase | Alkaline PDEase | Cellobiohydrolase | β-Xylanase | α-Glucosidase | β-Glucosidase | |||
| Cardenas | 4.5 (19.9) | 3.0 (3.27) | ND | ND | ND | 3.0 (4.48) | 3.0 (4.37) | 3.0 (0.15) | ||
| Albrook | 4.5 (31.8) | 3.0 (6.62) | ND | ND | ND | 3.0 (6.44) | 4.0 (2.99) | 3.0 (1.57) | ||
| Rio Paja | 5.0 (99.3) | ND | 4.0 (12.6) | ND | 4.0 (0.97) | 4.5 (2.75) | 3.0-3.5 (0.32) | 4.0 (7.97) | 4.0 (9.14) | 3.0 (1.17) |
| Gamboa | ND | ND | ND | ND | 4.5 (3.73) | 5.0 (3.38) | 6.0 (0.50) | ND | 4.5 (17.3) | ND |
| Las Cruces | 4.5 (20.6) | 9.5-11.0 (8.4-8.6) | 5.5 (2.87) | 8.5-10.0 (2.00) | 5.0 (0.87) | 4.5 (0.87) | ND | 4.25 (6.13) | 5.0 (6.67) | 3.0 (2.43) |
| Cerro Galera | 3.0-4.5 (5.60) | 10.0 (11.0) | 9.0 (1.50) | 4.5 (1.22) | 5.5 (1.78) | ND | 4.5 (2.31) | 4.5 (2.20) | 3.0 (0.83) | |
| Caritas | 4.5 (16.4) | 11.5 (28.8) | 9.0 (1.58) | ND | ND | 7.0 (0.93) | 4.75 (10.46) | 4.5 (9.89) | 3.0 (2.65) | |
| Mean pH | 4.5 | 10.3 | 3.8 | 9.1 | 4.5 | 5.0 | 5.3 | 3.9 | 4.2 | 3.0 |
PMEase, phosphomonoesterase; PDEase, phosphodiesterase; ND, not determined.
(ii) Phosphodiesterase.
There were marked differences in the pH optima for phosphodiesterase activity in the seven soils studied, including acid and alkaline phosphodiesterases (Fig. 1 and Table 3). These included (i) a very acidic optimum pH, 3.0 to 4.0, for soils with a pH of <4 (although the optima may have been lower for two soils, as assays were not conducted at a pH of <3), with negligible activity in the alkaline range, (ii) a moderately acid optimum pH around 5.5 for one soil with the same soil pH (Las Cruces; soil pH 5.5), which also had high activity in the alkaline range, and (iii) an alkaline optimum pH at 8.5 to 10.0 (mean pH, 9.1) for two soils with a pH of >6 (Cerro Galera and Caritas). Thus, the optimum pH for phosphodiesterase appeared to vary systematically with soil pH, with clear acid and alkaline optima. Phosphodiesterase activity was between 5- and 10-fold lower than acid phosphomonoesterase activity in all soils, with the highest activity in the Rio Paja soil (Fig. 1 and Table 3).
Carbon enzymes. (i) Cellobiohydrolase.
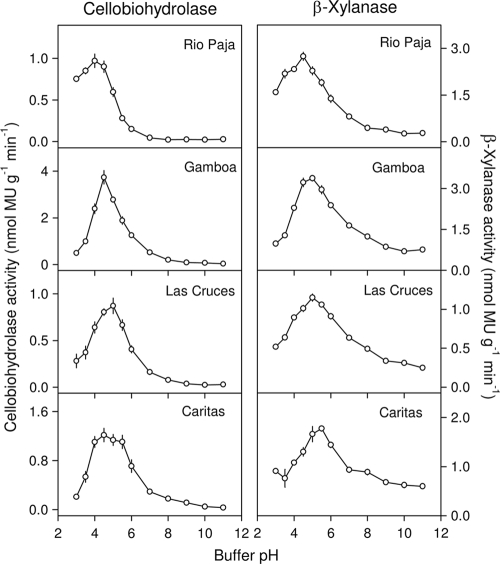
The pH optima of cellobiohydrolase activity were similar in the four soils assayed for this enzyme (Table 3 and Fig. 2), ranging from pH 4.0 for the Rio Paja soil (soil pH, 3.6) to pH 5.0 for the Las Cruces soil (soil pH, 5.5). For the Caritas soil, the optimum pH range was relatively broad (4.0 to 5.5). For the Rio Paja soil, there was considerable activity at a pH of <4, whereas activity declined markedly in this pH range for the other three soils (Fig. 2). There was negligible cellobiohydrolase activity in the alkaline range for all soils. The highest cellobiohydrolase activity was detected in the Gamboa soil, which also contained the highest concentration of total carbon (Table 1).
FIG. 2.
pH optima of cellobiohydrolase (cellulose 1,4-β-cellobiosidase) activity (left panel) and β-xylanase (xylan 1,4-β-xylosidase) activity (right panel) in four soils under lowland tropical rain forest in the Republic of Panama. Assays were conducted at 26°C using fluorogenic substrates in modified universal buffer. Error bars are the standard errors of the means for eight replicate wells per pH.
(ii) β-Xylanase.
The pH optima for β-xylanase activity in the four soils assayed for this enzyme were between 4.5 and 5.5 (mean, 5.0) (Table 3 and Fig. 2). Like the case for cellobiohydrolase activity, the highest β-xylanase activity was detected in the Gamboa soil. Unlike for cellobiohydrolase, there was relatively high activity in the alkaline range for three soils with a pH of >5 (Table 1 and Fig. 2).
(iii) α-Glucosidase.
Although α-glucosidase activity was assayed in only three soils, there were marked differences in pH optima that appeared to vary systematically with soil pH (Table 3 and Fig. 3). Optimum pH values were 3.0 in the Rio Paja soil (soil pH, 3.6), 6.0 in the Gamboa soil (soil pH, 5.4), and 7.0 in the Caritas soil (soil pH, 6.4). The optimum pH range for the Gamboa soil was broad compared to those of the other two soils. The highest α-glucosidase activity was detected in the Caritas soil, although rates were at least an order of magnitude lower than those for β-glucosidase in the same soils (see below).
FIG. 3.
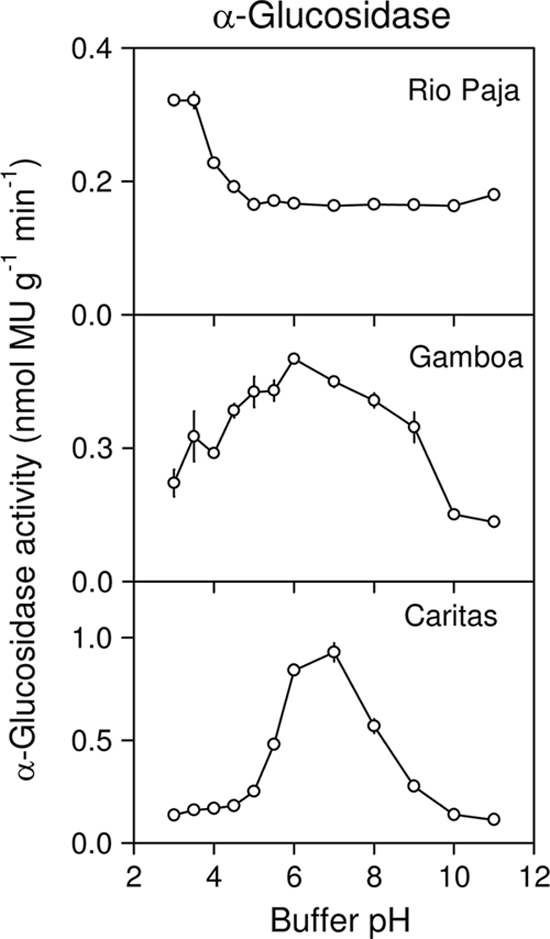
pH optima of α-glucosidase activity in three soils under a lowland tropical rain forest in the Republic of Panama. Assays were conducted at 26°C using a fluorogenic substrate (4-methylumbelliferyl α-d-glucopyranoside) in modified universal buffer. Error bars are the standard errors of the means for eight replicate wells per pH.
(iv) β-Glucosidase.
β-Glucosidase activity was confined to the acidic pH range, with negligible activity in the alkaline range (Fig. 4). However, the pH optima for β-glucosidase differed among soils and appeared to vary systematically with soil pH (Table 3 and Fig. 4). The pH optima were as low as 3.0 for the two most acidic soils (Albrook and Cardenas), 4.5 to 4.75 for two soils with a pH of >6 (Cerro Galera and Caritas), and at intermediate pH values for two other soils (Rio Paja [soil pH, 3.6] and Las Cruces [soil pH, 5.5]). In the Las Cruces soil, β-glucosidase showed a broad optimum pH range between 4.0 and 5.25. These pH optima therefore corresponded to optima for α-glucosidase in the Rio Paja soil (α-glucosidase optimum pH also at 3.0) but not for the Caritas soil (α-glucosidase optimum pH at 7.0). β-Glucosidase activity was highest in the Caritas soil (soil pH, 6.4), lowest in the Cerro Galera soil (soil pH, 6.3), and at similar intermediate values in other soils.
FIG. 4.
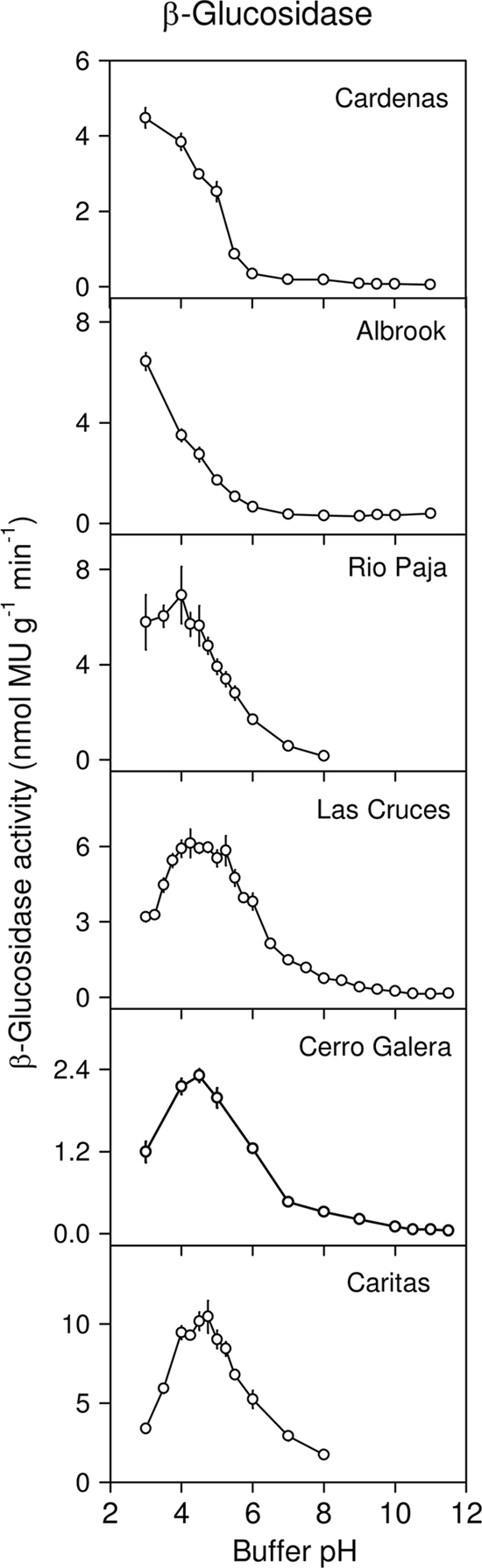
pH optima of β-glucosidase activity in six soils under lowland tropical rain forest in the Republic of Panama. Assays were conducted at 26°C using fluorogenic substrate (4-methylumbelliferyl β-d-glucopyranoside) in modified universal buffer. Error bars are the standard errors of the means for eight replicate wells per pH.
N-Acetyl-β-glucosaminidase.
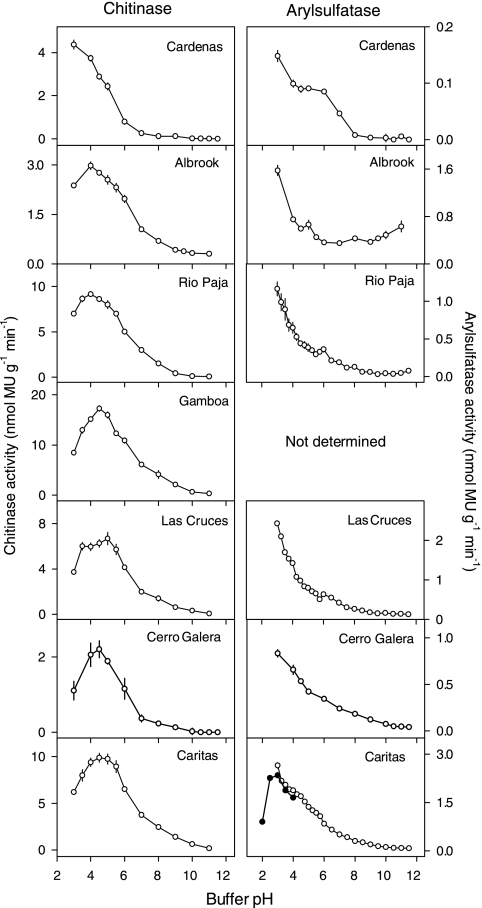
For six soils, the optimum pH for N-acetyl-β-glucosaminidase activity was centered broadly between pH 4.0 and 5.0, with a mean of 4.2 (Fig. 5 and Table 3). However, there was a trend toward more acidic pH optima in the more acidic soils, with pH values of <4.0 for the two most acidic soils. The highest N-acetyl-β-glucosaminidase activity was detected in the Gamboa soil, which also contained the highest concentration of total nitrogen (Table 1). There was little N-acetyl-β-glucosaminidase activity in the alkaline range for any soil (Fig. 5).
FIG. 5.
pH optima of chitinase (N-acetyl-β-glucosaminidase) activity (left panel) and arylsulfatase activity (right panel) in soils under lowland tropical rain forest in the Republic of Panama. Assays were conducted at 26°C using fluorogenic substrates in modified universal buffer. Error bars are the standard errors of the means for eight replicate wells per pH.
Arylsulfatase.
Remarkably, the optimum pH for arylsulfatase activity was pH 3.0 (or less) for all soils (Fig. 5 and Table 3). Although the buffering capacity of modified universal buffer is limited below pH 3, arylsulfatase activity for one soil (Caritas) was further tested to pH 2, which confirmed that the optimum pH for this soil, and therefore probably for the other soils, was 2.5 to 3.0. Little arylsulfatase activity was detected in the alkaline range (Fig. 5). The highest arylsulfatase activity was detected in the Caritas and Las Cruces soils of moderately acid pH. The lowest activity was detected in the very acidic Cardenas soil (soil pH, 3.3), although there was evidence from this soil and the other two soils with pH values of <4 for a second arylsulfatase optimum pH between 5.0 and 6.0 (Fig. 5).
DISCUSSION
There is a clear requirement for information on the sensitivity of extracellular enzymes to factors that influence their activity, under both assay conditions and in situ conditions in the environment (60). Of the possible factors involved, assay pH is of central importance, particularly for microwell plate studies that monitor a number of enzymes simultaneously using a single buffer pH. The results presented here demonstrate that optimum pH values can differ markedly among enzymes and, importantly, among soils for the same enzyme.
Patterns of pH optima among enzymes.
The enzymes studied here, including those involved in the cycles of carbon, nitrogen, phosphorus, and sulfur, were classified into three groups based on their optimum pH values and their variation among soils, as follows: (i) enzymes with acidic pH optima that were consistent among soils (cellobiohydrolase, β-xylanase, and arylsulfatase), (ii) enzymes with acidic pH optima that varied systematically among soils, with the lowest pH optima in the most acidic soils and the least acidic pH optima in the least acidic soils (α-glucosidase, β-glucosidase, and N-acetyl-β-glucosaminidase), and (iii) enzymes with both acid and alkaline pH optima, the relative predominance of which depended on the soil pH (phosphomonoesterase and phosphodiesterase). For phosphomonoesterase, the optimum pH values were relatively consistent among soils, while for phosphodiesterase, the pH optima varied systematically with the soil pH.
Phosphatases.
The existence of acid and alkaline pH optima for phosphomonoesterase activity in soil is well known. Typical optimum pH values are 4 to 6 for acid phosphomonoesterase and 8 to 10 for alkaline phosphomonoesterase (49), while a common procedure using para-nitrophenyl phosphate involves determination of acid and alkaline phosphomonoesterases at pH 6.5 and 11.0, respectively (51). A “neutral” phosphomonoesterase with an optimum pH around 7 has also been reported (18, 39). The predominance of either acid or alkaline phosphomonoesterase appears to depend on soil pH. For example, Eivazi and Tabatabai (15) reported pH optima for phosphomonoesterase of 6.5 for four soils with a pH of ≤6.4 and 11.0 for three soils with a pH of ≥7.4. This is broadly consistent with the results reported here, because acid phosphomonoesterase was predominant in the most acidic soils (pH, <4.0), alkaline phosphomonoesterase was predominant in less acidic soils (pH, >6.0), and both acid and alkaline phosphomonoesterases (with the acid type being predominant) were detected in a soil with an intermediate pH (5.4).
The pH optima of acid phosphomonoesterase appear similar for different substrates. For example, Trasar-Cepeda and Gil-Sotres (53) reported pH optima between 4.5 and 6.5, using para-nitrophenyl phosphate, for a range of mainly acidic, low-clay soils under a variety of land uses in Spain, while Niemi and Vepsäläinen (33) reported pH optima between 4.0 and 6.5, using methylumbelliferyl phosphate, for five soils from Finland with contrasting properties and land uses.
The optimum pH for phosphodiesterase activity varied markedly among soils, and this has not been widely reported. Phosphodiesterase is often assumed to have an alkaline optimum pH (6, 15) and is assayed commonly using bis-para-nitrophenyl phosphate substrate at pH 8.0 (51). However, Margesin and Schinner (28) reported pH optima between 4 and 11 for phosphodiesterase with a chromogenic substrate in five Austrian soils, although there did not appear to be a consistent relationship between optimum pH and soil pH. Microwell plate assays that included phosphodiesterase have recently used acidic buffers (3, 30), but there has apparently been no broad assessment of the optimum pH of the enzyme using a fluorogenic substrate. However, Niemi and Vepsäläinen (33) reported pH optima of 4.0 to 4.5, using bis-(4-methylumbelliferyl) phosphate, for four soils with a pH of <7 and 7.5 for one soil with a pH of >7.
Importantly, the optimum pH of phosphodiesterase varied systematically with soil pH, with the most acidic pH optima in the most acidic soils and the least acidic pH optima in the least acidic soils. This suggests that phosphodiesterase assays should be conducted at both acidic and alkaline pH values and that the use of a standardized pH 8.0 buffer may not account for the maximum potential phosphodiesterase activity in acidic soils. The accurate assessment of phosphodiesterase activity is of particular significance given the abundance of phosphate diesters in tropical forest soils (58), wetlands (54), and other low-phosphorus ecosystems (25, 56).
Carbon enzymes.
The two enzymes exhibiting stable acidic pH optima (cellobiohydrolase and β-xylanase) are both involved in the hydrolysis of plant-derived polymeric carbon molecules. This is consistent with the pH optima of cellulase and xylanase reported previously for a variety of soils and substrates (5, 12, 33, 62). Optimum pH values typically range between 4.0 and 5.5, although less acidic pH optima, including pH 5 to 8 for cellulase using carboxymethyl cellulose substrate and pH 5 to 7 for β-xylanase using xylan substrate, were reported for a range of contrasting temperate soils of unspecified land use (43).
In contrast, the pH optima of enzymes involved in the hydrolysis of small carbon molecules (α- and β-glucosidases) varied systematically with soil pH, with the most acidic pH optima in the most acidic soils and the least acidic pH optima in the least acidic soils. In particular, α-glucosidase showed marked differences in pH optima among soils, although its activities were the lowest of all enzyme activities and only three soils were studied. Both α- and β-glucosidases are typically assayed at pH 6.0 when using para-nitrophenol substrates (51). This choice of buffer pH was based largely on results from four soils ranging in pH from 5.1 to 8.0 (16), although similar values were reported in other studies using a chromogenic substrate (1, 5). Using fluorogenic substrates, optimum pH values in a series of Finnish soils were 4.0 to 5.0 for β-glucosidase and 4.0 to 4.5 for α-glucosidase, although the minimum values may have been more acidic, because activity was not assayed at pH <4.0 (33). A second α-glucosidase optimum was also detected at pH 7.0, while high activity at very acidic pH values up to 3.0 was reported for α-glucosidase in one soil with a para-nitrophenol substrate (16).
Nitrogen and sulfur enzymes.
The optimum pH for N-acetyl-β-glucosaminidase activity also varied with soil pH, although only that for the Cardenas soil (optimum pH, 3.0) was outside the range of values reported previously. For example, Parham and Deng (35) reported an optimum pH of 5.5 with para-nitrophenyl-N-acetyl-β-d-glucosaminide in two low-organic-matter arable soils (pH 5.9 and 6.3), while Niemi and Vepsäläinen (33) reported values between 4.0 and 5.5 with a fluorogenic substrate in a series of agricultural and forest soils. Chitinase extracted from beech forest soils exhibited pH optima between 4.5 and 5.5 (62), while values between 5.0 and 5.5 were reported for temperate cultivated soils (42).
For arylsulfatase, the very acidic optimum pH detected here (≤3.0) contrasts markedly with the values of 5.4 to 6.2 reported previously for studies using para-nitrophenyl sulfate (52) or a pH 5.8 buffer in a standardized procedure (51). This may reflect in part the use of a different substrate, because tests with the same fluorogenic substrate used here for five Finnish soils showed pH optima of ≤4.5 (33). For three soils with a pH of <4.0 in the current study, very acidic pH optima were observed for phosphodiesterase, N-acetyl-β-glucosaminidase, and α- and β-glucosidases but not for phosphomonoesterase, cellobiohydrolase, or β-xylanase. It is perhaps significant that few studies of pH optima have involved very acidic soils (however, see reference 28), because it might be expected that organisms growing under such conditions would synthesize enzymes that function optimally at a very acidic pH. Indeed, some organisms are known to produce enzymes with very acidic pH optima; one such organism is Aspergillus niger, which synthesizes a phosphatase with an optimum pH of 2.5 (24) and a β-glucosidase with high activity at pH 3.0 to 3.5 (37).
Factors influencing differences in optimum pHs among soils.
Several possible mechanisms might explain the observed patterns for pH optima among soils and enzymes observed here, and these include differences in the origin of the enzymes, their location in soil, and methodological artifacts.
(i) Influence of the origin of enzymes on optimum pH.
Soil enzymes are synthesized by a variety of organisms, including plants, bacteria, and fungi (both mycorrhizal and saprotrophic). This might contribute to the differences in pH optima among soils observed here if (i) isoenzymes from the various organisms differ in their pH optima and (ii) there is variation among soils in the relative contribution of enzymes from the different organisms to the total activity. As well as differences in enzymes among groups of organisms (e.g., bacteria, fungi, or plants, etc.), there may be differences within a group of organisms (e.g., among fungal species).
Differences in the relative activities of acid and alkaline phosphatases in soils are likely to be explained by the origin of the enzymes. Tabatabai (51) argued that bacterial phosphatases have higher pH optima than fungal phosphatases, so soils dominated by acid phosphatase are expected to have larger fungal populations than soils dominated by alkaline phosphatase, which are expected to have larger bacterial populations. This should be reflected in the soil pH, because fungi tend to constitute a greater proportion of the microbial biomass in acidic soils, while bacteria and actinomycetes are more abundant in neutral soils (2). The data reported here support this, because acid phosphatase was dominant in soils with a pH of <4.0, alkaline phosphatase was dominant in soils with a pH of >6.0, and there was high activity in both acid and alkaline regions for a soil with an intermediate pH.
Despite this, there can be considerable variation in phosphatases associated with a group of organisms. For example, of nine bacterial species from six genera isolated from a soil (Aridic Argiustoll) under grassland in Colorado (United States), three showed high alkaline phosphomonoesterase activity (pH 8.5), six showed high acid or neutral phosphomonoesterase activity (pH 5.5 or 6.5), one showed high activity at all three pH values, and three showed little activity at any pH value (32). In the same study, of 16 fungal species in 10 genera, 12 showed the highest phosphatase activity at an acidic pH (5.5), while 4 showed the highest activity at an alkaline pH (8.5). The soil from which the organisms were isolated had an optimum pH for phosphomonoesterase activity of 6.0 to 6.5, with a second smaller optimum at pH 11 (all assays conducted using para-nitrophenyl phosphate in modified universal buffer).
For other enzymes, optimum pH values vary among organisms, although they are usually in the acidic range. For example, the pH optima of chitinases from different sources range from 4 to 9 for plants and algae, 4.8 to 7.5 for animals, and 3.5 to 8.0 for microbes, although some of the differences may depend on the substrate used (23). Similar values occur for enzymes involved in the carbon cycle, including cellulase and β-glucosidase (9, 20).
(ii) Influence of enzyme stabilization on optimum pH.
Enzymes occur in a variety of locations in soil, including live or dead cells, free in solution, or stabilized by association with clays or organic matter (8). Although sorption on surfaces usually reduces enzyme activity, it also stabilizes the enzyme to the extent that much of the activity measured in soil enzyme assays may be due to stabilized enzymes (46). Sorption of an enzyme can also change its optimum pH by one or two pH units compared to that of the free enzyme in solution (31, 40), with an increase in the optimum pH expected for enzymes sorbed on negatively charged surfaces, such as clays (26). For example, sorption on kaolinite increased the optimum pH of wheat acid phosphomonoesterase from 5.0 to 5.7 (40) and that of chitinase from 4.7 to 5.7 (48). However, the optimum pH of potato acid phosphomonoesterase became more acidic (from pH 5 to 4) following immobilization on montmorillonite, despite association with tannic acid increasing the pH optimum to 6.0 (41).
The effect of sorption on enzyme pH is best explained by a modification of the enzyme, with proteins “unfolding” on solid surfaces (26). Sorption is more likely in acidic soils or at pH values below the isoelectric point of the enzyme, because negatively charged clay surfaces attract the positively charged proteins. Sorption is reduced at pH values greater than the isoelectric point because negatively charged clay surfaces repel the negatively charged proteins, while at pH values close to the isoelectric point there may be adsorption with little change in conformation of the enzyme (26). Acidic soils are therefore likely to have a larger proportion of their total activity originating from sorbed enzymes, which may in turn influence the optimum pH of enzyme activity. A wide range of isoelectric points can occur for a given class of enzymes; for example, George et al. (17) reported pI values between 3.6 and 7.3 for phytase from a range of sources. Such differences in isoelectric points might contribute to differences in the patterns of pH optima observed for the various enzymes studied here.
(iii) Influence of methodology on optimum pH.
Several methodological artifacts might have influenced the variation in optimum pH among soils and enzymes observed here. Earlier studies typically used chromogenic substrates linked to para-nitrophenol, whereas this study and several recent studies used fluorogenic substrates linked to 4-methylumbelliferone. Although few studies have compared the two types of substrate for soil enzymes, Drouillon and Merckx (14) reported that phosphomonoesterase activities determined in a large number of natural and agricultural soils using both para-nitrophenyl phosphate and 4-methylumbelliferyl phosphate were well correlated. The chromogenic and fluorogenic substrates were compared at the same concentration (10 mM in the final assay solution), although more recent studies using fluorogenic substrates have employed lower substrate concentrations (3, 11, 22, 30, 57). This allows assays to be conducted at substrate concentrations that more closely approximate those in the field, although the possible effects of substrate concentration on pH optima are not known. The choice of buffer can also influence the observed pH optima of enzyme activity (10), although most previous studies that measured optimum pH conducted assays in the same modified universal buffer employed here.
Finally, there might have been an effect of buffer pH on the substrate, influencing its susceptibility to enzyme hydrolysis and, therefore, the apparent pH optimum of activity. For example, Niemi and Vepsäläinen (33) reported that 4-methylumbelliferyl N-acetyl-β-d-glucosaminide was unstable at a high pH, while 4-methylumbelliferyl phosphate was unstable at a low pH. In the current study, chemical hydrolysis was negligible relative to enzymatic hydrolysis and was accounted for by subtracting controls without soil, although increases in fluorescence (>10% compared to that in neutral-pH buffer) were observed at pH 3.0 and pH 11.0 for some substrates. In particular, it was noticeable that control fluorescence for 4-methylumbelliferyl sulfate was greatest in pH 3.0 buffer, suggesting that the high activity at this pH might be due in part to instability of the substrate.
Recommendations.
Soil enzymes are usually assayed at the optimum pH of the individual enzyme, although microwell plate assays typically use a common pH that may or may not be optimum for all enzymes involved. This simplifies and standardizes the assay procedure but may confound interpretation of results if there are marked differences in optimum pH values in different soils. If information on maximum potential activity is required, then it seems important to determine the optimum pH of each enzyme in the soils under study, at least for assays involving the three commonly studied enzymes phosphomonoesterase, phosphodiesterase, and β-glucosidase, for which the optimum pH varies markedly with soil pH. Depending on the aims of the study, it may be advisable for microwell plate studies to determine activity at the soil pH and temperature. Although not generally recommended (7, 27), assays at the soil pH provide a measure of the potential activity under field conditions (19, 63). Such studies are required to link the activity measured in laboratory assays to in situ activity in the environment.
Acknowledgments
I thank Bettina Engelbrecht, Didimo Urena, and Julio Rodruigez for assistance with sample collection, Tania Romero, Dianne de la Cruz, Dayana Agudo, and Luis Ramos for laboratory assistance, Rick Condit for providing rainfall data, and Andrew Nottingham for comments on the manuscript.
Footnotes
Published ahead of print on 13 August 2010.
REFERENCES
- 1.Acosta-Martinez, V., and M. A. Tabatabai. 2000. Enzyme activities in a limed agricultural soil. Biol. Fertil. Soils 31:85-91. [Google Scholar]
- 2.Alexander, M. 1964. Introduction to soil microbiology. John Wiley and Sons, New York, NY.
- 3.Allison, V. J., L. M. Condron, D. A. Peltzer, S. J. Richardson, and B. L. Turner. 2007. Changes in enzyme activities and soil microbial community composition along carbon and nutrient gradients at the Franz Josef chronosequence, New Zealand. Soil Biol. Biochem. 39:1770-1781. [Google Scholar]
- 4.Arrieta, J. M., and G. J. Herndl. 2001. Assessing the diversity of marine bacterial β-glucosidases by capillary electrophoresis zymography. Appl. Environ. Microbiol. 67:4896-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batistic, L., J. M. Sarkar, and J. Mayaudon. 1980. Extraction, purification and properties of soil hydrolases. Soil Biol. Biochem. 12:59-63. [Google Scholar]
- 6.Browman, M. G., and M. A. Tabatabai. 1978. Phosphodiesterase activity in soils. Soil Sci. Soc. Am. J. 42:284-290. [Google Scholar]
- 7.Burns, R. G. 1978. Enzyme activity in soil: some theoretical and practical considerations, p. 295-340. In R. G. Burns (ed.), Soil enzymes. Academic Press, London, United Kingdom.
- 8.Burns, R. G. 1982. Enzyme activity in soil: location and a possible role in microbial ecology. Soil Biol. Biochem. 14:423-427. [Google Scholar]
- 9.Busto, M. D., N. Ortega, and M. Perez-Mateos. 1995. Induction of β-glucosidase in fungal and soil bacterial cultures. Soil Biol. Biochem. 27:949-954. [Google Scholar]
- 10.Criquet, S. 2002. Measurement and characterization of cellulase activity in sclerophyllous forest litter. J. Microbiol. Methods 50:165-173. [DOI] [PubMed] [Google Scholar]
- 11.DeForest, J. L. 2009. The influence of time, storage temperature, and substrate age on potential soil enzyme activity in acidic forest soils using MUB-linked substrates and l-DOPA. Soil Biol. Biochem. 41:1180-1186. [Google Scholar]
- 12.Deng, S. P., and M. A. Tabatabai. 1994. Cellulase activity of soils. Soil Biol. Biochem. 26:1347-1354. [Google Scholar]
- 13.Dinesh, R., S. G. Chaudhuri, and T. E. Sheeja. 2004. Soil biochemical and microbial indices in wet tropical forests: effects of deforestation and cultivation. J. Plant Nutr. Soil Sci. 167:24-32. [Google Scholar]
- 14.Drouillon, M., and R. Merckx. 2005. Performance of para-nitrophenyl phosphate and 4-methylumbelliferyl phosphate as substrate analogues for phosphomonoesterase in soils with different organic matter content. Soil Biol. Biochem. 37:1527-1534. [Google Scholar]
- 15.Eivazi, F., and M. A. Tabatabai. 1977. Phosphatases in soils. Soil Biol. Biochem. 9:167-172. [Google Scholar]
- 16.Eivazi, F., and M. A. Tabatabai. 1988. Glucosidases and galactosidases in soils. Soil Biol. Biochem. 20:601-606. [Google Scholar]
- 17.George, T. S., H. Quiquampoix, R. J. Simpson, and A. E. Richardson. 2007. Interactions between phytases and soil constituents: implications for the hydrolysis of inositol phosphates, p. 221-241. In B. L. Turner, A. E. Richardson, and E. J. Mullaney (ed.), Inositol phosphates: linking agriculture and the environment. CAB International, Wallingford, United Kingdom.
- 18.Halstead, R. L. 1964. Phosphatase activity of soils as influenced by lime and other treatments. Can. J. Soil Sci. 44:137-144. [Google Scholar]
- 19.Harrison, A. F., and T. Pearce. 1979. Seasonal variation of phosphatase activity in woodland soils. Soil Biol. Biochem. 11:405-410. [Google Scholar]
- 20.Hayano, K., and K. Tubaki. 1985. Origin and properties of β-glucosidase activity of tomato-field soil. Soil Biol. Biochem. 17:553-557. [Google Scholar]
- 21.Juma, N. G., and M. A. Tabatabai. 1978. Distribution of phosphomonoesterase in soils. Soil Sci. 126:101-108. [Google Scholar]
- 22.Kang, H., and C. Freeman. 1999. Phosphatase and arylsulphatase activities in wetland soils: annual variation and controlling factors. Soil Biol. Biochem. 31:449-454. [Google Scholar]
- 23.Koga, D., M. Mitsutomi, M. Kono, and M. Matsumiya. 1999. Biochemistry of chitinases, p. 111-124. In P. Jollès and R. A. A. Muzzarelli (ed.), Chitin and chitinases. Birkhäuser, Basel, Switzerland. [DOI] [PubMed]
- 24.Komano, T. 1975. Formation of multiple forms of acid and alkaline phosphatases in relation to the culture age of Aspergillus niger. Plant Cell Physiol. 16:643-658. [Google Scholar]
- 25.Leake, J. R., and W. Miles. 1996. Phosphodiesters as mycorrhizal P sources. I. Phosphodiesterase production and the utilization of DNA as a phosphorus source by the ericoid mycorrhizal fungus Hymenoscyphus ericae. New Phytol. 132:435-443. [DOI] [PubMed] [Google Scholar]
- 26.Leprince, F., and H. Quiquampoix. 1996. Extracellular enzyme activity in soil: effect of pH and ionic strength on the interaction with montmorillonite of two acid phosphatases secreted by the ectomycorrhizal fungus Hebeloma cylindrosporum. Eur. J. Soil Sci. 47:511-522. [Google Scholar]
- 27.Malcolm, R. E. 1983. Assessment of phosphatase activity in soils. Soil Biol. Biochem. 15:403-408. [Google Scholar]
- 28.Margesin, R., and F. Schinner. 1994. Phosphomonoesterase, phosphodiesterase, phosphotriesterase, and inorganic pyrophosphatase activities in forest soils in an alpine area: effect of pH on enzyme activity and extractability. Biol. Fertil. Soils 18:320-326. [Google Scholar]
- 29.Marthews, T. R., D. F. R. P. Burslem, S. R. Paton, F. Yangüez, and C. E. Mullins. 2008. Soil drying in a tropical forest: three distinct environments controlled by gap size. Ecol. Model. 216:369-384. [Google Scholar]
- 30.Marx, M.-C., M. Wood, and S. C. Jarvis. 2001. A microplate fluorimetric assay for the study of enzyme diversity in soils. Soil Biol. Biochem. 33:1633-1640. [Google Scholar]
- 31.McLaren, A. D., and E. F. Estermann. 1957. Influence of pH on the activity of chymotrypsin at a solid-liquid interface. Arch. Biochem. Biophys. 68:157-160. [DOI] [PubMed] [Google Scholar]
- 32.Nakas, J. P., W. D. Gould, and D. A. Klein. 1987. Origin and expression of phosphatase activity in a semi-arid grassland soil. Soil Biol. Biochem. 19:13-18. [Google Scholar]
- 33.Niemi, R. M., and M. Vepsäläinen. 2005. Stability of the fluorogenic enzyme substrates and pH optima of enzyme activities in different Finnish soils. J. Microbiol. Methods 60:195-205. [DOI] [PubMed] [Google Scholar]
- 34.Olander, L. P., and P. M. Vitousek. 2000. Regulation of soil phosphatase and chitinase activity by N and P availability. Biogeochemistry 49:175-190. [Google Scholar]
- 35.Parham, J. A., and S. P. Deng. 2000. Detection, quantification and characterization of β-glucosaminidase activity in soil. Soil Biol. Biochem. 32:1183-1190. [Google Scholar]
- 36.Pyke, C. R., R. Condit, S. Aguilar, and S. Lao. 2001. Floristic composition across a climatic gradient in a neotropical lowland forest. J. Veg. Sci. 12:553-566. [Google Scholar]
- 37.Quiquampoix, H., P. Chassin, and R. G. Ratcliffe. 1989. Enzyme activity and cation exchange as tools for the study of confirmation of proteins adsorbed on mineral surfaces. Progr. Colloid Polym. Sci. 79:59-63. [Google Scholar]
- 38.Quiquampoix, H. 2000. Mechanisms of protein adsorption on surfaces and consequences for extracellular enzyme activity in soil, p. 171-206. In J.-M. Bollag and G. Stotzky (ed.), Soil biochemistry, vol. 10. Marcel Dekker, New York, NY.
- 39.Ramírez-Martínez, J. R., and A. D. McLaren. 1966. Determination of soil phosphatase activity by a fluorimetric technique. Enzymologia 30:243-253. [PubMed] [Google Scholar]
- 40.Ramírez-Martínez, J. R., and A. D. McLaren. 1966. Some factors influencing the determination of phosphatase activity in native soils and in soils sterilized by irradiation. Enzymologia 31:23-38. [PubMed] [Google Scholar]
- 41.Rao, M. A., A. Violante, and L. Gianfreda. 2000. Interactions of acid phosphatase with clays, organic molecules and organo-mineral complexes: kinetics and stability. Soil Biol. Biochem. 32:1007-1014. [Google Scholar]
- 42.Rodruigez-Kabana, R., G. Godoy, G. Morgan-Jones, and R. A. Shelby. 1983. The determination of soil chitinase activity: conditions for assay and ecological studies. Plant Soil 75:95-106. [Google Scholar]
- 43.Schinner, F., and W. von Mersi. 1990. Xylanase-, CM-cellulase- and invertase activity in soil: an improved method. Soil Biol. Biochem. 22:511-515. [Google Scholar]
- 44.Sinsabaugh, R. L., M. J. Klug, H. P. Collins, P. E. Yeager, and S. O. Petersen. 1999. Characterizing soil microbial communities, p. 318-348. In G. P. Robertson, D. C. Coleman, C. S. Bledsoe, and P. Sollins (ed.), Standard soil methods for long-term ecological research. Oxford University Press, New York, NY.
- 45.Sinsabaugh, R. L., C. L. Lauber, M. N. Weintraub, B. Ahmed, S. D. Allison, C. Crenshaw, A. R. Contosta, D. Cusack, S. Frey, M. E. Gallo, T. B. Gartner, S. E. Hobbie, K. Holland, B. L. Keeler, J. S. Powers, M. Stursova, C. Takacs-Vesbach, M. P. Waldrop, M. D. Wallenstein, D. R. Zak, and L. H. Zeglin. 2008. Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 11:1252-1264. [DOI] [PubMed] [Google Scholar]
- 46.Skujiņš, J. 1976. Extracellular enzymes in soil. Crit. Rev. Microbiol. 4:383-421. [DOI] [PubMed] [Google Scholar]
- 47.Skujiņš, J. J., L. Braal, and A. D. McLaren. 1962. Characterization of phosphatase in a terrestrial soil sterilized with an electron beam. Enzymologia 25:125-133. [Google Scholar]
- 48.Skujiņš, J., A. Puķite, and A. D. McLaren. 1974. Adsorption and activity of chitinase on kaolinite. Soil Biol. Biochem. 6:179-182. [Google Scholar]
- 49.Speir, T. W., and D. J. Ross. 1978. Chapter 6, Soil phosphatase and sulfatase, p. 197-250. In R. G. Burns (ed.), Soil enzymes. Academic Press, San Diego, CA.
- 50.Tabatabai, M. A., and J. M. Bremner. 1969. Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol. Biochem. 1:301-307. [Google Scholar]
- 51.Tabatabai, M. A. 1994. Soil enzymes, p. 775-833. In R. W. Weaver, J. S. Angle, and P. S. Bottomley (ed.), Methods of soil analysis part 2—microbiological and biological properties. Soil Science Society of America, Madison, WI.
- 52.Tabatabai, M. A., and J. M. Bremner. 1970. Factors affecting soil arylsulfatase activity. Soil Sci. Soc. Am. J. 34:427-429. [Google Scholar]
- 53.Trasar-Cepeda, M. C., and F. Gil-Sotres. 1988. Kinetics of acid phosphatase activity in various soils of Galacia (NW Spain). Soil Biol. Biochem. 20:275-280. [Google Scholar]
- 54.Turner, B. L., and S. Newman. 2005. Phosphorus cycling in wetlands: the importance of phosphate diesters. J. Environ. Qual. 34:1921-1929. [DOI] [PubMed] [Google Scholar]
- 55.Turner, B. L., and T. E. Romero. 2010. Stability of hydrolytic enzyme activity and microbial phosphorus during storage of tropical rain forest soils. Soil Biol. Biochem. 42:459-465. [Google Scholar]
- 56.Turner, B. L., L. M. Condron, S. J. Richardson, D. A. Peltzer, and V. J. Allison. 2007. Soil organic phosphorus transformations during pedogenesis. Ecosystems 10:1166-1181. [Google Scholar]
- 57.Vepsäläinen, M., S. Kukkonen, M. Vestberg, H. Sirviö, and R. M. Niemi. 2001. Application of soil enzyme activity test kit in a field experiment. Soil Biol. Biochem. 33:1665-1672. [Google Scholar]
- 58.Vincent, A. G., B. L. Turner, and E. V. J. Tanner. 2010. Soil organic phosphorus dynamics following perturbation of litter cycling in a tropical moist forest. Eur. J. Soil Sci. 61:48-57. [Google Scholar]
- 59.Waldrop, M. P., T. C. Balser, and M. K. Firestone. 2000. Linking microbial community composition to function in a tropical soil. Soil Biol. Biochem. 32:1837-1846. [Google Scholar]
- 60.Wallenstein, M. D., and M. N. Weintraub. 2008. Emerging tools for measuring and modeling the in situ activity of soil extracellular enzymes. Soil Biol. Biochem. 40:2098-2106. [Google Scholar]
- 61.Windsor, D. M. 1990. Climate and moisture availability in a tropical forest: long term records from Barro Colorado Island, Panama. Smithson. Contrib. Earth Sci. 29:1-145. [Google Scholar]
- 62.Wirth, S. J., and G. A. Wolf. 1992. Micro-plate colourimetric assay for endo-acting cellulase, xylanase, chitinase, 1,3-β-glucanase and amylase extracted from forest soil horizons. Soil Biol. Biochem. 24:511-519. [Google Scholar]
- 63.Yavitt, J. B., S. J. Wright, and R. K. Wieder. 2004. Seasonal drought and dry-season irrigation influence leaf-litter nutrients and soil enzymes in a moist, lowland forest in Panama. Aust. Ecol. 29:177-188. [Google Scholar]