Abstract
The development of novel intervention strategies for the control of zoonoses caused by bacteria such as Salmonella spp. in livestock requires appropriate experimental models to assess their suitability. Here, a novel porcine intestinal in vitro organ culture (IVOC) model utilizing cell crown (CC) technology (CCIVOC) (Scaffdex) was developed. The CCIVOC model was employed to investigate the characteristics of association of S. enterica serovar Typhimurium strain SL1344 with porcine intestinal tissue following exposure to a Lactobacillus plantarum strain. The association of bacteria to host cells was examined by light microscopy and electron microscopy (EM) after appropriate treatments and staining, while changes in the proteome of porcine jejunal tissues were investigated using quantitative label-free proteomics. Exposure of porcine intestinal mucosal tissues to L. plantarum JC1 did not reduce the numbers of S. Typhimurium bacteria associating to the tissues but was associated with significant (P < 0.005) reductions in the percentages of areas of intestinal IVOC tissues giving positive staining results for acidic mucins. Conversely, the quantity of neutrally charged mucins present within the goblet cells of the IVOC tissues increased significantly (P < 0.05). In addition, tubulin-α was expressed at high levels following inoculation of jejunal IVOC tissues with L. plantarum. Although L. plantarum JC1 did not reduce the association of S. Typhimurium strain SL1344 to the jejunal IVOC tissues, detection of increased acidic mucin secretion, host cytoskeletal rearrangements, and proteins involved in the porcine immune response demonstrated that this strain of L. plantarum may contribute to protecting the pig from infections by S. Typhimurium or other pathogens.
Worldwide nontyphoidal Salmonella (NTS) bacterial strains are responsible for between 200 million and 1.3 billion human infections each year, resulting in an estimated 3 million deaths (13). Salmonella enterica serovar Typhimurium is the second most common serovar among those that are isolated from humans in the United Kingdom, and infections by that serovar represent a major public health issue (25). S. Typhimurium is prevalent in pig herds in the United Kingdom and in 2007 was responsible for 69.7% of all reported infections by that species (15).
S. Typhimurium is commonly resistant to therapeutically important antibiotics such as fluoroquinolones (11, 37). Until recently, enteropathogens such as Salmonella spp. and Escherichia coli were controlled in pigs by the addition of in-feed antimicrobial growth promoters (AGPs) (8, 28), but this practice was banned in the European Union in 2006 (12), and alternative approaches to reduce food-borne pathogens in pigs are needed (28).
Probiotics have been advocated as potential alternatives to AGPs in treatment of pigs, and there is some evidence for their efficacy as growth promoters in vivo (10, 28). Probiotics are defined as live microorganisms that, when administered in adequate quantities, confer a health benefit to the host (18). In recent years, many significant advances have been made toward understanding the mechanisms by which probiotics act against food-borne pathogens (45). Probiotics are typically lactic acid bacteria (LAB) such as lactobacilli, bifidobacteria, or enterococci that can contribute to the control of E. coli, S. Typhimurium, and Campylobacter jejuni in livestock (6, 10, 50). The mechanisms by which they confer protection are largely unknown.
Investigations into the interaction between the probiotic, host intestinal epithelium, and pathogen have been limited by the paucity of relevant in vitro and in vivo models (3). Traditionally, assays using cultured mammalian cell lines, gut explants, or whole-animal models have been used to study the adhesion of probiotics to host epithelial cells and how they may affect the association of pathogens with the intestinal mucosa (46). However, in vitro models of infection that use cultured epithelial cells do not accurately replicate the response of epithelial tissues to bacterial colonization (22, 44). Suitable alternative assays that accurately mimic the host intestinal epithelium are required. One such model is in vitro organ culture (IVOC), which permits studies of host-pathogen interactions under physiologically relevant conditions, where tissues retain their natural architecture and mucin layers.
To date, the conventional sponge biopsy IVOC method has been used to study E. coli and Salmonella pathogenesis (24, 44) but not for evaluating probiotics. Here we describe a novel IVOC model to study the interaction of S. Typhimurium with a porcine L. plantarum strain on porcine jejunal and colonic tissues. Additionally, we assessed the modulation of host tissue responses to probiotics (14, 16, 52) in this IVOC system using a proteomic approach, MultiDimensional Protein Identification Technology (MuDPIT).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. Typhimurium strain SL1344 and L. plantarum JC1 (B2028) were obtained from the Veterinary Laboratories Agency Culture Collection (Weybridge, Surrey). SL1344 is an isolate that is virulent in calves and has been extensively used to study Salmonella pathogenicity both in vitro and in vivo (43, 56). L. plantarum JC1 is a putative probiotic strain isolated from the feces of a 6-week-old male crossbred pig that was confirmed to be free from Salmonella colonization by the use of standard microbiological techniques (55). Furthermore, this L. plantarum strain has demonstrated strong antimicrobial activity against a panel of S. Typhimurium field isolates and was antagonistic for S. Typhimurium invasion in vitro (13a). All stock cultures were maintained in heart infusion broth (HIB)-30% (vol/vol) glycerol (Oxoid) at −80°C. Lactobacilli were cultured statically in 20 ml of Mann, Rogosa, Sharpe (MRS) broth (Difco, United Kingdom) in an anaerobic cabinet (Don Whitley, Yorkshire, United Kingdom) (H2-CO2-N2 at 10:10:80) at 37°C for 24 h. S. Typhimurium strains were cultured aerobically in broth composed of Luria-Bertani without glucose (LB-G broth) at 37°C with gentle agitation (225 rpm) for 16 h or on LB-G agar plates.
Porcine in vitro organ culture.
Ten commercially crossbred, 6-week-old pigs were housed for 7 days and fed a commercial ration with water ad libitum. Immediately prior to IVOC studies, pigs were euthanized by stunning followed by exanguination. Pigs were immediately subjected to postmortem examinations and the gastrointestinal tissues exteriorized and aseptically sampled and then placed into precooled complete RPMI 1640 medium (Sigma; Poole, United Kingdom) containing 10% fetal bovine serum (Gibco, NY), 0.25% lactalbumin hydrosylate, 75 mM mercaptoethanol, 0.2 μg ml−1 hydrocortisone (Sigma; Poole, United Kingdom) (1:1 chloroform/ethanol), 0.1 μg ml−1 insulin, and 2 mM l-glutamine and l-aspartate as previously described (23). All chemicals, unless otherwise indicated, were purchased from Sigma (Poole, United Kingdom). Tissues were subsequently aseptically trimmed to remove excess mesenteric adipose tissue. Trimmed tissues were then manipulated so that the mucosal side was uppermost and immobilized in Scaffdex tissue crowns between the cell crown and its base, providing a fixed surface area and limiting the bacterial inocula to the mucosal side of the explant tissue. Immobilized tissues were then placed into a 24-well plate (BD Biosciences) with the mucosal side facing upwards. Tissues were submerged in a predetermined volume of complete RPMI 1640 medium as described above, and IVOC tissues were inoculated differently depending upon the experiment as indicated below. For all studies, the 24-well plates containing IVOC tissues were incubated at 37°C on a rocker in a 95% O2 and 5% CO2 atmosphere. Uninfected control IVOC tissues were tested for the presence of Salmonella bacteria to confirm the absence of preexisting infection. Following infection, tissues were washed thoroughly using Hanks balanced salt solution, homogenized in phosphate-buffered saline (PBS), and serially diluted to facilitate enumeration (36). For this, dilutions from 10−1 to 10−4 were plated onto brilliant green agar (BGA; Oxoid, Basingstoke) supplemented with 1 μg/ml novobiocin in triplicate (Sigma; Poole, United Kingdom) and incubated aerobically for 16 h at 37°C to determine numbers of associated Salmonella bacteria. All IVOC experiments enumerating viable bacteria were repeated in quadruplicate on two separate occasions.
Porcine in vitro organ culture competitive exclusion assays.
For in vitro competitive exclusion assays (Table 1), 100 μl of a 24-h L. plantarum JC1 culture (∼1 × 108 CFU) was used to inoculate IVOC tissues as described previously (23). The S. Typhimurium inoculum (0.1 ml) was prepared by centrifugation of a 16-h S. Typhimurium culture by the use of a Sigma Howie centrifuge at 2,447 × g for 10 min, the supernatant was discarded, and the bacterial pellet was resuspended in 0.1 M PBS (pH 7.2) at an optical density (OD) of 1.2 absorbance units, which equated to ∼1 × 109 CFU/ml. Ten porcine S. Typhimurium isolates (see Table S1 in the supplemental material) were evaluated for their characteristics of association to porcine epithelial tissue; no significant differences were observed with S. Typhimurium strain SL1344 or any of the nine S. Typhimurium isolates, following 30 min of exposure to either porcine jejunal or colonic IVOC tissues (see Fig. S1 in the supplemental material). Therefore, S. Typhimurium strain SL1344 was used to model S. Typhimurium association in all further assays.
TABLE 1.
Association of S. Typhimurium with porcine jejunal and colonic IVOC tissues following probiotic competitive exclusion assaysa
| Tissue type | Assay (duration) | Mean CFU/ml | P value |
|---|---|---|---|
| Jejunum | S. Typhimurium (30 min) | 2.58E+06 | NAb |
| Competition | 2.18E+06 | 0.5837 | |
| Protection | 3.11E+06 | 0.8935 | |
| S. Typhimurium (1 h) | 2.41E+06 | 0.0043c | |
| Displacement | 2.14E+06 | 0.2845d | |
| Colon | S. Typhimurium (30 min) | 3.14E+06 | NA |
| Competition | 1.05E+07 | 0.7045 | |
| Protection | 7.81E+06 | 0.9274 | |
| S. Typhimurium (1 h) | 6.50E+06 | 0.1790 | |
| Displacement | 5.86E+06 | 0.6402d |
Each assay was repeated in quadruplicate with tissues pooled from two pigs on two separate occasions (n = 8).
NA, not applicable.
Significant result (P < 0.05).
P value calculated from data obtained with experimental control S. Typhimurium assayed for 1 h but not S. Typhimurium assayed for 30 min.
Histopathological analysis of IVOC tissues. (i) Sample fixation and preparation.
Following the appropriate infection period as outlined in Table 1, IVOC tissues were collected into 10% (vol/vol) neutral buffered formalin held at ambient temperature and allowed to fix for a minimum of 24 h. Trimmed tissues were processed routinely in paraffin wax, and 4-μm-thick sections were cut from the blocks and subsequently stained with hematoxylin and eosin (HE), as described previously (32). Tissues were then examined microscopically using a Leica DMLB light microscope (Leica Microsystems).
(ii) PAS/Alcian blue staining and subsequent quantification.
Porcine tissues were infected and fixed as described above. Subsequently, representative 4-μm-thick sections were stained with periodic acid-Schiff (PAS) stain for neutral mucin detection or Alcian blue (AB) for acidic mucins as described previously (7). For quantification of PAS/AB staining, the examined thresholds were predefined by selecting representative examples across the normal expected spectrum for each stain. All microscopic evaluation studies were carried out under blinded conditions by a veterinary pathologist. Twelve nonoverlapping consecutive fields containing all the mucosal layers for duplicate pieces of IVOC tissue were examined, and any area expressing positive staining within that threshold and within the selected measurement field was expressed as a percentage of that field. Thus, results were quantified as percentages of areas of positive staining.
SEM.
Porcine tissues were infected as described above. Subsequently, IVOC tissues were fixed in 3% gluteraldehyde prepared in a 0.1 M phosphate buffer. Processing of the samples for scanning electron microscopy (SEM) was carried out as described previously (23). Briefly, the specimens were cut into 2-cm2 pieces, washed in 0.1 M phosphate buffer, and postfixed in 1% (wt/vol) osmium tetroxide. Subsequently, the specimens were rinsed in several changes of 0.1 M PBS, dehydrated through the use of a graded ethanol series (up to 100%), and placed into acetone. The specimens were then subjected to critical point drying with liquid carbon dioxide and the fixed tissues attached to aluminum stubs by the use of silver conductive paint, sputter-coated with gold, and examined by SEM using a Zeiss Evo LS10 SEM at 10 to 20 kV.
Quantitative label-free proteomics of jejunal IVOC tissues.
Porcine jejunal IVOC tissues from two pigs were used for these studies and infected as described above. This process was repeated on three separate occasions with pigs obtained from the same litter, fed on the same diet, and housed under identical conditions (n = 6). Postinfection IVOC tissues were stored at 4°C in 75 mM HEPES containing protease and phosphate inhibitors (Sigma; Poole, United Kingdom) prior to cellular lysis. Subsequently, tissues were homogenized using an Omni tissue homogenizer (Camlab; Cambridge, United Kingdom) until the IVOC tissues had been totally disrupted. To prevent further microbial contamination of the homogenate, samples were boiled for 1 h to kill the majority of the associated microflora and samples were stored at 4°C until required. Subsequently, 200 μl of the concentrated IVOC homogenate was sampled and mixed with 2 ml of lysis buffer containing urea (5 M), thiourea (2 M), CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate)} (2% [wt/vol]), SB3-10 (2% [wt/vol]), pharmalytes (0.5% [vol/vol]), dithiothreitol (DTT) (100 mM), and Tris (40 mM). The protein extract was desalted by dilution with ammonium bicarbonate (50 mM [pH 8.0]) and concentrated to ∼0.5 ml by centrifugation in a 5-kDa molecular-cutoff (MWCO) Vivaspin centrifugal filter (Sigma; Poole, United Kingdom). Protein concentrations were estimated using the Bradford assay as described previously (4).
Following protein quantification, 0.05 mg of the protein extracts was digested with sequence-grade trypsin and then subjected to one-dimensional (1D) chromatography using an Agilent series 1100 high-pressure liquid chromatography (HPLC) system and a Biobasic SCX column (ThermoHypersil [part no. 73205-102130]) (100 by 2.1 mm; 5 μm pore size). Tryptic peptides were subjected to chromatography at a flow rate of 0.25 μl/min with mobile phases comprising 2.5 mM ammonium acetate/ acetonitrile (75:25) (pH 4.5) (A) and 250 mM ammonium acetate/acetonitrile (75:25) (pH 4.5) (B) (Fischer, United Kingdom), with a 39-min binary gradient (0% B for 0 min, 0% B for 5 min, 35% B for 18 min, 100% B for 20 min, 0% B for 22 min, 0% B for 32 min, and 0% B for 39 min). The optical density of the HPLC eluent was recorded at 280 nm, and 15 sequential 1-min fractions were collected using an automated fraction collector (Agilent Technologies). Each of the fractions was concentrated in a centrifugal evaporator (Eppendorf), resuspended in 20 μl of 0.1% formic acid and separated using HPLC-coupled mass spectroscopy (MS), with the exception that an Agilent 6520 Acurate-Mass Q-TOF LC/MS system with an HPLC Chip Cube source (Agilent Technologies, West Lothian, United Kingdom) was used. The chip consisted of a 40-nanoliter enrichment column and a 150-mm separation column (part no. G4240-62002; Agilent Technologies) driven by an Agilent Technologies 1100 series nano/capillary liquid chromatography system, and both systems were controlled by MassHunter workstation data acquisition for Q-TOF (version B.01.03; Agilent Technologies). Tryptic peptides were subjected to chromatography at a flow rate of 0.6 μl/min with a 47-min binary gradient of 0.1% formic acid and acetonitrile (Fischer; United Kingdom) as follows: 5% acetonitrile for 0 min, 5% acetonitrile for 5 min, 40% acetonitrile for 40 min, 80% acetonitrile for 41 min, 80% acetonitrile for 45 min, and 5% for 47 min. This series was followed by the next injection, with the injection volume set at 4 μl and the subsequent eluent running straight into the mass spectrometer. The mass spectrometer was run in positive-ion mode, and MS precursor scans were collected over an m/z range of 250 to 3,000 and at 5 spectra per s. Precursor ions were selected for tandem MS (MS/MS) at an absolute threshold of 2,000 and a relative threshold of 0.01, with a maximum of five data-dependent MS/MS sequence scans per cycle and active exclusion set at 1 spectrum and released after 3 min. Precursor charge-state selection preferences were set to 2 and then 3 positively charged ions.
Bioinformatics.
Tryptic peptides were identified and assigned to proteins using Spectrum Mill software (Agilent Technologies) by searching a range of appropriate databases. The spectrum count method was used to calculate the relative abundances of proteins, and results are representative of the number of peptide counts detected for each protein, following published guidelines (21, 31, 57). The porcine genome sequence was incomplete at the time of writing, and it was therefore necessary to interrogate the MS/MS data against multiple databases to accurately identify proteins of interest from the HPLC-MS. Previous studies by Hornshøj et al. utilized the trEST database, which is based on a library of porcine expressed sequence tag (EST) sequences (26) (ftp://ftp.isrec.isb-sib.ch/pub/databases/trest); however, we were unable to convert this database to a format compatible with Spectrum Mill software (Agilent Technologies). Agilent provides a mammalian NCBInr database built into the Spectrum Mill software (Agilent Technologies), and many porcine proteomics studies to date have successfully utilized mammalian databases to identify porcine proteins (14). For this study, a database was created based upon the Sus scrofa build1 FASTA file, which is downloadable from NCBI (ftp://ftp.ncbi.nih.gov/genomes/Sus_scrofa/protein/). This database contains the current build of the porcine genome project and sequences from chromosomes 1, 4, 5, 7, 11, 13, 14, 15, 17, MT, and X. The NCBI Sus scrofa build 1 database was evaluated against the NCBInr mammalian database for use in this study, and although the NCBInr database yielded significantly (P < 0.001) more protein identifications (see Table S2 in the supplemental material), only 20.51% with a porcine origin were identified. Therefore, we adopted the NCBI Sus scrofa build 1 database for this study, demonstrating a consistent identification of only porcine proteins across all test groups (see Table S2 in the supplemental material). Masshunter data acquisition files were extracted using Agilent Spectrum Mill software (Agilent; United Kingdom), and the subsequent MS data files were processed against the Sus scrofa database (see Table S3 in the supplemental material). Following the database searches, results were compared using Microsoft Access; only proteins present in all three of the replicates were processed. The false-discovery rate (FDR) was determined by searching one data set using a combined forward-reverse decoy data set created from Sus scrofa build 1 and the freeware DBToolkit (34), which is available for downloading (http://code.google.com/p/dbtoolkit/). Proteins that contained fewer than two distinct tryptic peptides after the database search were discarded, 92 spectra and 12 proteins were identified from the reverse database, and the FDR was calculated at 6.5% according to the methods of Wang et al. (54).
Spectrum count normalization was applied to each sample replicate, enabling accurate comparisons between test groups to be made. Spectrum counts were normalized as described previously (58), and statistical analyses were performed using the log of the normalized spectrum abundance factor [ln(NSAF)] for spectrum counts, with changes in protein expression determined using Student's t test (58). Briefly, the NSAF for any protein k was calculated by determining the spectrum count (SpC) divided by the protein length (L [in numbers of amino acids]) and dividing by the sum of SpC/L for all the proteins (N) which were present in all three biological replicates in the experiment, as follows:
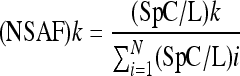 |
(1) |
Statistical analysis.
With the exception of the proteomics data, all results were presented as means of the standard deviations of the mean. A one-way analysis of variance (ANOVA) was performed with commercially available software (Microsoft Excel); a P value of <0.05 was taken to be significant.
RESULTS
Establishment of a polarized IVOC system using CC technology.
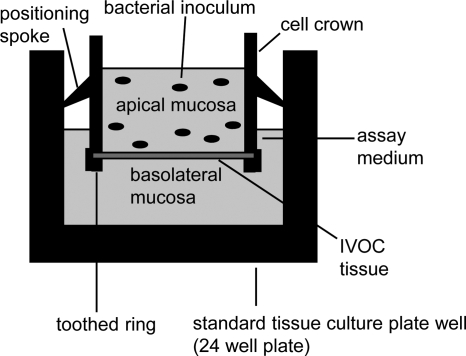
A polarized IVOC system was generated using the CC system from Scaffdex (Fig. 1). Porcine intestinal explants from the jejunum and colon were immobilized between the cell crown and toothed ring, resulting in the separation of the apical (mucosal) and basolateral sides of the tissue explant (Fig. 2). Subsequently, the apical mucosal side of the explant was inoculated with S. Typhimurium or L. plantarum or a combination of the two, as outlined in Table 2. The use of the CC technology (Scaffdex, Tampere, Finland) facilitated the in vitro culture of large porcine intestinal explants with a fixed surface area. In addition to this, the bacterial inoculum was limited to the mucosal (apical) surface of the intestinal explants, thus generating more physiologically relevant bacterial association values and limiting artifacts commonly associated with IVOC, as described previously (44). Furthermore, the IVOC tissues generated using the CCIVOC system remained well preserved following relatively short incubation periods of up to an hour (data not shown).
FIG. 1.
Cell crown insert from Scaffdex.
FIG. 2.
Inoculation of a polarized in vitro organ culture system. Intestinal explants are immobilized between the cell crown and toothed ring, with the mucosal side of the tissue facing the apical part of the cell crown. Bacterial inocula are added to the apical chamber of the cell crown, thus ensuring that the bacterial inoculum is limited to the mucosal surface of the tissue and a fixed surface area is provided for infection.
TABLE 2.
Experimental strategy for IVOC infection studies
| Assay type | Experimental strategy |
|---|---|
| Competition assay | Inoculation with 108 CFU/ml L. plantarum and 108 CFU/ml S. Typhimurium (SL1344) simultaneously for 30 min |
| Protection (exclusion) assay | Preincubation with 108 CFU/ml L. plantarum for 30 min and then inoculation with 108 CFU/ml S. Typhimurium for 30 min |
| Displacement assay | Preincubation with 108 CFU/ml S. Typhimurium for 30 min and then inoculation with 108 CFU/ml L. plantarum for 30 min |
| S. Typhimurium (SL1344)a | Inoculation with 108 CFU/ml S. Typhimurium for 30 min |
| L. plantarum JC1a | Inoculation with 108 CFU/ml L. plantarum JC1 for 30 min |
| S. Typhimurium (SL1344)b | Inoculation with 108 CFU/ml S. Typhimurium for 1 h |
| L. plantarum JC1b | Inoculation with 108 CFU/ml L. plantarum JC1 for 1 h |
| Uninfected tissuea,b | No bacterial inoculums were added |
Control conditions.
Displacement assay control.
L. plantarum JC1 did not reduce the association of S. Typhimurium with porcine IVOC tissues.
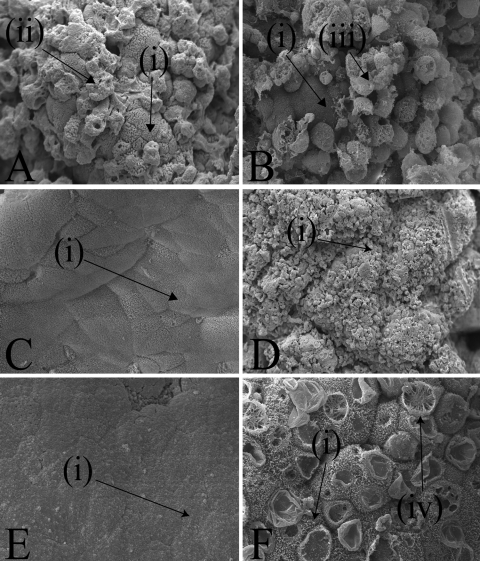
To determine whether L. plantarum JC1 could reduce the association of S. Typhimurium to jejunal and colonic IVOC tissue in vitro, competition, protection, and displacement assays were performed as outlined in Table 2. No reductions in S. Typhimurium association to porcine IVOC tissues were observed, and there appeared be no change in the interaction of S. Typhimurium with host tissues, as typical membrane ruffles on the jejunal IVOC tissues were induced within 30 min regardless of the presence or absence of L. plantarum (Fig. 3 A or B, respectively).
FIG. 3.
Scanning electron microscopy of jejunal IVOC tissue following the addition of S. Typhimurium SL1344 (magnification, ×2.0 k) (A), following the addition of L. plantarum JC1 to the jejunal IVOC tissue before S. Typhimurium in a probiotic protection assay (magnification, ×1.1 k) (B), following the addition of heat-killed L. plantarum JC1 (magnification, ×1.0 k) (C), or following the addition of viable L. plantarum JC1 (magnification, ×0.15 k) to jejunal IVOC tissues (D) or control uninfected jejunal IVOC tissue (magnification, ×0.6 k) (E) and uninfected jejunal IVOC tissue maintained in assay medium at pH 4.5 (magnification, ×0.6 k) (F). Arrows (i) indicate the presence of a brush border; arrow (ii) indicates membrane “pitting” and the loss of the brush border associated with bacterial invasion, arrow (iii) indicates a damaged epithelial cell with characteristic rounded edges, and arrow (iv) demonstrates an unidentified cytoskeletal rearrangement.
Culture of jejunal tissue with L. plantarum JC1 was associated with damage to the brush border in the form of microvillus blunting and membrane blebbing, which is associated with cell death and membrane repair following sublethal cell damage (Fig. 3D) (51). Tissue culture medium adjusted with inorganic acid at pH 4.5 demonstrated similar damage to the epithelium of the IVOC tissues (Fig. 3F), whereas heat-killed (HK) L. plantarum caused no damage to the epithelial cells (Fig. 3C). These data indicated metabolic activity, and possibly a reduced pH was responsible for the extensive damage to the intestinal epithelium of the porcine tissues (Fig. 3C and F). In addition to this, all IVOC tissues that were examined by SEM (Fig. 3) were also evaluated by HE staining; however, there was no evidence of deep tissue damage or disruption of the intestinal epithelial layer under any of the sets of assay conditions outlined in Table 2 (data not shown).
Differential modulation of acidic and neutral intestinal mucin levels following probiotic exposure. (i) Modulation of acidic intestinal mucins.
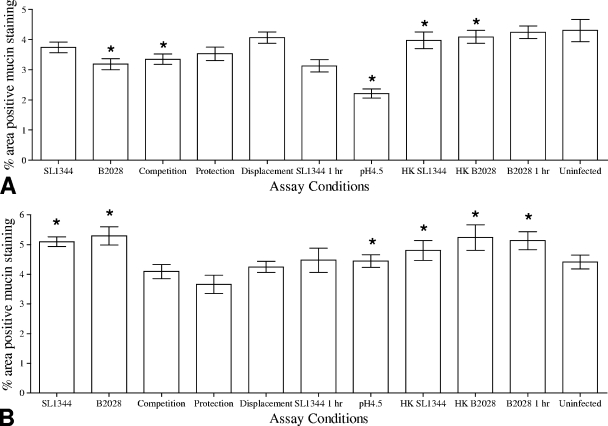
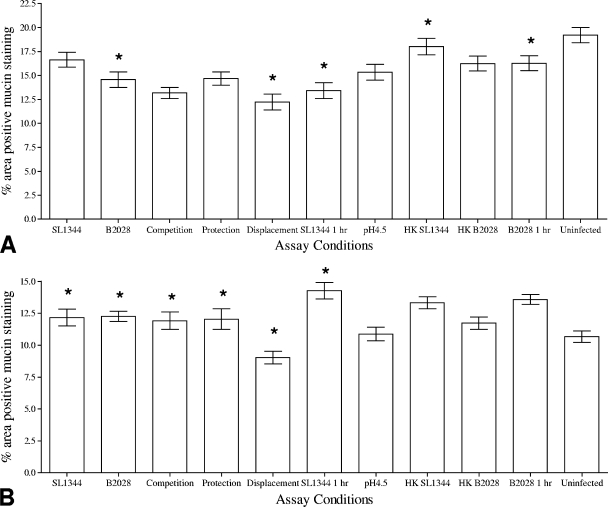
Exposure of jejunal tissues to L. plantarum JC1 for 30 min led to a significant (P < 0.05) reduction in the percentage of the area of the IVOC tissue that exhibited positive Alcian blue staining. Furthermore, porcine jejunal tissues exposed to heat-killed L. plantarum for 30 min demonstrated no significant alterations to the percentage of the IVOC tissues that exhibited positive Alcian blue staining, compared to porcine jejunal tissues exposed to viable L. plantarum JC1 for 30 min. Interestingly, exposure of IVOC tissues to pH 4.5 for 30 min (Fig. 4 A) led to a significant (P < 0.05) 2% reduction in Alcian blue staining across the whole IVOC tissue, although this effect was observed only for jejunal tissues. Unexpectedly, viable bacteria in the colonic tissue, but not heat-killed bacteria, induced a significant (P < 0.05) reduction in the amount of Alcian blue staining (Fig. 5 A). Moreover, combinations of L. plantarum and S. Typhimurium used for CE assays as outlined in Table 2 indicated that S. Typhimurium did not affect the ability of L. plantarum JC1 to reduce the amount of Alcian blue staining (Fig. 4A and 5A).
FIG. 4.
Analysis of the percentages of areas of jejunal IVOC tissues staining positive for AB (A) or PAS (B) following in vitro competitive exclusion assays using L. plantarum JC1 (B2028). Uninfected tissue, heat-killed L. plantarum and S. Typhimurium, and assay medium at pH 4.5 were used as control conditions for both tissues. Results are presented as the means ± standard deviations of the means. Asterisks indicate conditions where the percentages of areas of the IVOC tissue exhibiting positive mucin staining were significantly (P < 0.05) different from the uninfected control results.
FIG. 5.
Analysis of the percentages of areas of colonic IVOC tissues staining positive for AB (A) or PAS (B) following in vitro competitive exclusion assays using L. plantarum JC1 (B2028). Uninfected tissue, heat-killed L. plantarum and S. Typhimurium, and assay medium at pH 4.5 were used as control conditions for both tissues. Results are presented as the means ± standard deviations of the means. Asterisks indicate conditions where the percentages of areas of the IVOC tissue exhibiting positive mucin staining were significantly (P < 0.05) different from the uninfected control results.
(ii) Modulation of porcine neutral intestinal mucins.
Treatment of jejunal IVOC tissues with either heat-killed or live S. Typhimurium or L. plantarum resulted in a significant (P < 0.05) reduction in the percentage of the IVOC tissue exhibiting positive PAS staining (Fig. 4B), suggesting that modulation of neutral mucins in porcine jejunal tissues is a generic host response to bacteria. Furthermore, both S. Typhimuirum and L. plantarum induced significant reductions in the quantity of negatively charged mucins present in the colonic tissues, as indicated by the percentage of the tissue demonstrating positive PAS staining (P = 0.05 and P = 0.001, respectively) (Fig. 5B).
Host proteome changes in response to the exposure of ex vivo jejunal tissues by L. plantarum and S. Typhimurium. (i) S. Typhimurium-induced host proteome changes.
Following infection of IVOC tissues with viable S. Typhimurium for 30 min, significant (P < 0.05) changes were induced in the relative levels of expression of 6 host proteins (Table 3). Of these, the relative abundances of four proteins increased, while those of two decreased compared to uninfected tissue protein (Table 3). The levels of cytoskeletal proteins (tropomyosin and actin) were below the detection limit in the uninfected control tissue, although following infection of IVOC tissues with viable S. Typhimurium for 30 min, those proteins were detected with extremely high ln(NSAF) values of −3.21 and −3.41, respectively, and were the most abundant proteins detected by MuDPIT within the tissue (Table 4 ). Moreover, SEM of Salmonella-infected IVOC tissues confirmed the presence of membrane ruffles, significant loss of microvilli, and disruption to the brush border integrity of cells, which are all characteristic of the induced cellular pathology of S. Typhimurium-infected cells (Fig. 3A) (35, 43). Additionally, inoculation of IVOC tissues with S. Typhimurium significantly (P < 0.029) reduced the expression of a 90-kDa heat shock protein that is associated with the protection and trafficking of eukaryotic signaling complexes and microtubule stability (Table 3) (30, 38).
TABLE 3.
Differentially expressed proteins in jejunal IVOC tissues following a probiotic protection assay or the monoassociation of S. Typhimurium or L. plantarum
| Treatment group | Accession no.b | Protein identificationc | Mean NSAF (uninfected tissue) | Mean ln(NSAFUninfected tissue)a | Mean NSAF (test condition) | Mean ln (NSAFTest condition) | P value |
|---|---|---|---|---|---|---|---|
| Salmonella | 194044822 | (Predicted) similar to peroxiredoxin 4 | 5.66E-03 | −5.17 | 8.96E-03 | −4.71 | 0.0079 |
| 194039391 | (Predicted) heat shock 90-kDa protein 1, beta | 8.51E-03 | −4.77 | 6.12E-03 | −5.1 | 0.0129 | |
| 194042660 | (Predicted) similar to heterogeneous nuclear ribonucleoprotein F isoform 2 | 2.95E-03 | −5.83 | 5.35E-03 | −5.23 | 0.0188 | |
| 47523680 | Heat shock 10-kDa protein 1 | 2.09E-02 | −3.87 | 3.37E-02 | −3.39 | 0.0277 | |
| 194043710 | (Predicted) collagen, type VI, alpha 3 isoform 1 | 2.07E-03 | −6.18 | 3.12E-03 | −5.77 | 0.0280 | |
| 194040685 | (Predicted) importin 5 | 2.85E-03 | −5.86 | 1.86E-03 | −6.29 | 0.0286 | |
| L. plantarum | 194044029 | (Predicted) heat shock 60-kDa protein 1 (chaperonin) isoform 1 | 1.02E-02 | −4.59 | 2.13E-02 | −3.85 | 0.0072 |
| 194041795 | (Predicted) similar to phosphoglycerate mutase 1 (phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1) isoform 2 | 7.13E-03 | −4.94 | 1.30E-02 | −4.34 | 0.0092 | |
| 178056753 | Phosphorylase, glycogen (liver) | 3.15E-03 | −5.76 | 1.43E-03 | −6.55 | 0.0111 | |
| 47522618 | Vinculin | 6.14E-03 | −5.09 | 1.10E-02 | −4.51 | 0.0184 | |
| 47523680 | Heat shock 10-kDa protein 1 | 2.09E-02 | −3.87 | 3.11E-02 | −3.47 | 0.0311 | |
| 194040685 | (Predicted) importin 5 | 2.85E-03 | −5.86 | 1.63E-03 | −6.42 | 0.0336 | |
| 194035835 | (Predicted) coatomer protein complex, subunit alpha isoform 1 | 2.25E-03 | −6.1 | 1.54E-03 | −6.47 | 0.0413 | |
| Protection assay | 194044029 | (Predicted) heat shock 60-kDa protein 1 (chaperonin) isoform 1 | 1.02E-02 | −4.59 | 2.03E-02 | −3.9 | 0.0009 |
| 194040622 | (Predicted) lymphocyte cytosolic protein 1 (L-plastin) isoform 2 | 7.16E-03 | −4.94 | 1.16E-02 | −4.45 | 0.0045 | |
| 194035835 | (Predicted) coatomer protein complex, subunit alpha isoform 1 | 2.25E-03 | −6.1 | 1.63E-03 | −6.42 | 0.0064 | |
| 194037554 | (Predicted) similar to ATP synthase subunit beta, mitochondrial | 2.46E-02 | −3.71 | 1.63E-02 | −4.12 | 0.0075 | |
| 194043617 | (Predicted) ubiquitin C isoform 1 | 4.01E-03 | −5.52 | 8.19E-03 | −4.8 | 0.0165 | |
| 194039820 | (Predicted) similar to histone cluster 1, H2bd | 6.28E-02 | −2.77 | 3.08E-02 | −3.48 | 0.0166 | |
| 194037683 | (Predicted) similar to lumican | 7.26E-03 | −4.92 | 1.12E-02 | −4.49 | 0.0236 | |
| 194037527 | (Predicted) nonmuscle myosin light chain | 5.85E-02 | −2.84 | 3.04E-02 | −3.49 | 0.0261 | |
| 194041795 | (Predicted) similar to phosphoglycerate mutase 1 (phosphoglycerate mutase isozyme B) (PGAM-B) (BPG-dependent PGAM 1) isoform 2 | 7.13E-03 | −4.94 | 2.55E-03 | −5.97 | 0.0426 | |
| 194036973 | (Predicted) tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 2.72E-02 | −3.6 | 1.56E-02 | −4.16 | 0.0480 | |
| 194037380 | (Predicted) similar to integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | 1.02E-03 | −6.89 | 3.78E-04 | −7.88 | 0.0497 | |
| 194034970 | (Predicted) similar to ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | 1.08E-02 | −4.53 | 1.38E-02 | −4.28 | 0.0505 |
ln(NSAF) is the natural log of the normalized spectrum abundance factor (NSAF). The higher the ln(NSAF) value, the greater the protein abundance within a sample.
NCBI GI numbers.
BPG, 2,3-bisphosphoglycerate.
TABLE 4.
Induced proteins in jejunal IVOC tissues following a probiotic protection assay or the monoassociation of S. Typhimurium or L. plantarum
| Species or assay | Accession no. | Protein identification | Mean NSAF | Mean ln(NSAF)a |
|---|---|---|---|---|
| Salmonella | 194034943 | (Predicted) actin, alpha, cardiac muscle 1 | 4.28E-02 | −3.21 |
| 194034025 | (Predicted) tropomyosin 2 (beta) isoform 1 | 3.34E-02 | −3.41 | |
| 194040330 | (Predicted) similar to 40S ribosomal protein S10 | 1.61E-02 | −4.16 | |
| 194037982 | (Predicted) similar to thioredoxin domain-containing protein 5 precursor (thioredoxin-like protein p46) (endoplasmic reticulum protein ERp46) | 1.13E-02 | −4.49 | |
| 194038246 | (Predicted) similar to calmodulin | 9.66E-03 | −4.67 | |
| 194041070 | (Predicted) similar to myosin light chain kinase | 7.11E-03 | −4.98 | |
| 194044922 | (Predicted) similar to adenine nucleotide translocator 2 | 6.15E-03 | −5.09 | |
| 178056732 | guanine nucleotide binding protein (G protein), alpha-inhibiting activity polypeptide 2 | 5.18E-03 | −5.31 | |
| 55742824 | HLA-B-associated transcript 1 | 4.75E-03 | −5.36 | |
| 47523618 | Citrate synthase | 4.40E-03 | −5.48 | |
| 194034464 | (Predicted) similar to fermitin family homolog 2 | 3.81E-03 | −5.64 | |
| 178056768 | RAD51 homolog | 3.60E-03 | −5.63 | |
| 194041072 | (Predicted) similar to 155-kDa myosin light chain kinase homolog | 3.10E-03 | −5.82 | |
| 194037042 | (Predicted) similar to cadherin 17 | 1.93E-03 | −6.29 | |
| L. plantarum | 194043861 | (Predicted) tubulin, alpha 1a | 1.78E-02 | −4.03 |
| 47523692 | Thioredoxin | 9.84E-03 | −4.68 | |
| 194037982 | (Predicted) similar to thioredoxin domain-containing protein 5 precursor (thioredoxin-like protein p46) (endoplasmic reticulum protein ERp46) | 9.26E-03 | −4.68 | |
| 194041992 | Glutathione S-transferase omega | 7.99E-03 | −4.84 | |
| 194035873 | (Predicted) similar to mucosal pentraxin | 6.53E-03 | −5.05 | |
| 194044922 | (Predicted) similar to adenine nucleotide translocator 2 | 6.07E-03 | −5.13 | |
| 55742824 | HLA-B-associated transcript 1 | 5.41E-03 | −5.4 | |
| 194036006 | (Predicted) similar to CCT3 protein | 4.00E-03 | −5.54 | |
| 194039491 | (Predicted) similar to isocitrate dehydrogenase 3 (NAD+) alpha | 3.36E-03 | −5.7 | |
| 194034464 | (Predicted) similar to fermitin family homolog 2 | 2.41E-03 | −6.04 | |
| 194035771 | (Predicted) similar to guanylate binding protein 4 | 2.41E-03 | −6.07 | |
| 194037042 | (Predicted) similar to cadherin 17 | 1.96E-03 | −6.29 | |
| 194037849 | (Predicted) alpha-2-macroglobulin | 8.13E-04 | −7.12 | |
| Protection | 194034943 | (Predicted) actin, alpha, cardiac muscle 1 | 3.06E-02 | −3.51 |
| 194034025 | (Predicted) tropomyosin 2 (beta) isoform 1 | 2.54E-02 | −3.68 | |
| 113205892 | Beta 5-tubulin | 2.49E-02 | −3.7 | |
| 194043861 | (Predicted) tubulin, alpha 1a | 1.57E-02 | −4.16 | |
| 5835867 | ATP synthase F0 subunit 8 | 1.29E-02 | −4.4 | |
| 47523632 | Proteasome activator subunit 2 | 1.04E-02 | −4.61 | |
| 114326183 | ADP-ribosylation factor family-like protein | 9.96E-03 | −4.66 | |
| 194038246 | (Predicted) similar to calmodulin | 9.82E-03 | −4.66 | |
| 194040330 | (Predicted) similar to 40S ribosomal protein S10 | 7.79E-03 | −4.9 | |
| 194037546 | (Predicted) similar to protein canopy homolog 2 precursor (MIR-interacting saposin-like protein) (transmembrane protein 4) (putative secreted protein ZSIG9) isoform 3 | 5.50E-03 | −5.26 | |
| 194038713 | (Predicted) hypothetical protein | 5.46E-03 | −5.25 | |
| 194041070 | (Predicted) similar to myosin light chain kinase | 5.20E-03 | −5.27 | |
| 5835866 | Cytochrome c oxidase subunit II | 5.19E-03 | −5.26 | |
| 47523692 | Thioredoxin | 4.75E-03 | −5.45 | |
| 194034195 | (Predicted) annexin A1, partial | 4.29E-03 | −5.56 | |
| 194041992 | Glutathione S-transferase omega | 4.15E-03 | −5.54 | |
| 194040559 | (Predicted) similar to periostin precursor (PN) (osteoblast-specific factor 2) (OSF-2) | 4.11E-03 | −5.51 | |
| 194035873 | (Predicted) similar to mucosal pentraxin | 3.94E-03 | −5.58 | |
| 55742824 | HLA-B-associated transcript 1 | 3.63E-03 | −5.62 | |
| 178056732 | Guanine nucleotide binding protein (G protein), alpha-inhibiting activity polypeptide 2 | 3.62E-03 | −5.67 | |
| 194042175 | (Predicted) similar to ribosomal protein S24 isoform 2 | 3.56E-03 | −5.73 | |
| 194037558 | (Predicted) similar to nascent polypeptide-associated complex alpha subunit isoform 3 | 3.39E-03 | −5.7 | |
| 194038973 | (Predicted) similar to purine nucleoside phosphorylase (inosine phosphorylase) (PNP) | 3.32E-03 | −5.72 | |
| 194044922 | (Predicted) similar to adenine nucleotide translocator 2 | 3.22E-03 | −5.76 | |
| 47523786 | Voltage-dependent anion channel 3 | 3.19E-03 | −5.98 | |
| 194040738 | (Predicted) similar to potassium channel tetramerization domain-containing 12 | 3.17E-03 | −5.77 | |
| 194044488 | (Predicted) similar to dynein light chain 2a | 3.01E-03 | −5.82 | |
| 74024903 | Hypoxanthine phosphoribosyltransferase | 2.73E-03 | −5.95 | |
| 194043483 | (Predicted) similar to catechol-O-methyltransferase isoform 1 | 2.55E-03 | −6.02 | |
| 72535188 | GTP-binding protein SAR1a | 2.54E-03 | −6.08 | |
| 113205694 | Protein phosphatase 1 catalytic subunit gamma isoform | 2.42E-03 | −6.11 | |
| 194038763 | (Predicted) similar to proteasome alpha 6 subunit | 2.42E-03 | −6.07 | |
| 194036227 | (Predicted) similar to selenium-binding protein 1 (56-kDa selenium-binding protein) (SP56) | 2.40E-03 | −6.12 | |
| 194041072 | (Predicted) similar to 155-kDa myosin light chain kinase homolog | 2.30E-03 | −6.14 | |
| 194036006 | (Predicted) similar to CCT3 protein | 2.05E-03 | −6.4 | |
| 194034464 | (Predicted) similar to fermitin family homolog 2 | 2.03E-03 | −6.24 | |
| 194042288 | (Predicted) similar to heterogeneous nuclear ribonucleoprotein H3 | 1.92E-03 | −6.36 | |
| 194044393 | (Predicted) similar to CG5862 CG5862-PA | 1.86E-03 | −6.32 | |
| 178056768 | RAD51 homolog | 1.76E-03 | −6.39 | |
| 194040203 | (Predicted) hypothetical protein | 1.72E-03 | −6.38 | |
| 148225750 | Heat shock 105-kDa/110-kDa protein 1 | 1.66E-03 | −6.48 | |
| 194037849 | (Predicted) alpha-2-macroglobulin | 1.65E-03 | −6.42 | |
| 55742742 | Decorin | 1.65E-03 | −6.45 | |
| 194037042 | (Predicted) similar to cadherin 17 | 1.60E-03 | −6.52 | |
| 194039570 | (Predicted) BTB (POZ) domain-containing 1 | 1.51E-03 | −6.51 | |
| 194036441 | (Predicted) putative homeodomain transcription factor 1 | 1.48E-03 | −6.57 | |
| 194042021 | (Predicted) similar to programmed cell death 4 | 1.46E-03 | −6.58 | |
| 194034819 | (Predicted) similar to sorbitol dehydrogenase | 1.40E-03 | −6.67 | |
| 194038870 | (Predicted) phosphoenolpyruvate carboxykinase 2 | 1.34E-03 | −6.79 | |
| 194038294 | (Predicted) chromogranin A (parathyroid secretory protein 1) | 1.32E-03 | −6.67 | |
| 194044577 | (Predicted) similar to tissue transglutaminase homologue (TGH2) | 1.29E-03 | −6.82 | |
| 194037239 | (Predicted) similar to thiosulfate sulfurtransferase (rhodanese) | 1.23E-03 | −6.72 | |
| 194033945 | (Predicted) similar to proteasome (prosome, macropain) 26S subunit, non-ATPase 5 | 1.22E-03 | −6.8 | |
| 194035538 | (Predicted) similar to N-myc downstream regulated gene 1 | 1.20E-03 | −6.74 | |
| 194035771 | (Predicted) similar to guanylate binding protein 4 | 1.13E-03 | −6.83 | |
| 51592133 | H+ ATPase | 1.11E-03 | −6.85 | |
| 194035289 | (Predicted) similar to 5 nucleotidase, ecto | 1.03E-03 | −6.91 | |
| 194042126 | (Predicted) similar to LOC506564 protein | 8.66E-04 | −7.1 | |
| 194034450 | (Predicted) similar to nidogen-2 precursor (NID-2) (osteonidogen) | 6.70E-04 | −7.36 | |
| 194036727 | (Predicted) similar to tyrosine-protein kinase Lyn | 6.29E-04 | −7.39 | |
| 194037681 | (Predicted) similar to early endosome antigen 1 (endosome-associated protein p162) (zinc finger FYVE domain-containing protein 2) | 4.48E-04 | −7.91 | |
| 178056221 | Complement C4 | 3.94E-04 | −7.89 |
ln(NSAF) is the natural log of the normalized spectrum abundance factor (NSAF). The higher the ln(NSAF) value, the greater the protein abundance within a sample.
After treatment of jejunal tissue with viable L. plantarum JC1 for 30 min, seven proteins were significantly (P < 0.05) differentially expressed (Table 3) compared to uninfected jejunal tissues. Of these, four increased and three decreased in expression compared to the results seen with uninfected tissue. The eukaryotic cytoskeletal protein tubulin-α (alpha 1a) was below the detection limit in the uninfected tissues, whereas after exposure of the IVOC tissues to viable L. plantarum, tubulin-α (alpha 1a) was detected with an ln(NSAF) of −4.03 (Table 4). Moreover, the high ln(NSAF) value for tubulin-α (−4.03) suggests that altered tubulin levels may play a key role in the response of porcine tissue to L. plantarum.
(ii) L. plantarum protection assay-induced host proteome changes.
Following the protection assay, 12 proteins were significantly (P < 0.05) differentially expressed (Table 3). Of these, five proteins were increased and seven decreased in expression compared to uninfected tissue results (Table 3). Interestingly, the majority of the changes in the porcine intestinal proteome following a probiotic protection assay were consistent with the proteome changes exhibited with either bacterial species alone. Additionally, the three most abundant proteins following the protection assay were actin, tropomyosin 2 (beta), and beta-5 tubulin, with ln(NSAF) values of −3.51, −3.68, and −3.7, respectively (Table 4). Moreover, the significant induction of beta-5 tubulin was unique to the probiotic protection assay (Table 4). Tubulin-α was also upregulated, with an ln(NSAF) of −4.16; thus, upregulation of tubulin-α and beta tubulin accentuated the potential modulation of microtubules by lactobacilli in vitro following a probiotic protection assay. In addition to this, a 60-kDa heat shock protein was significantly upregulated following the protection assay and monoassociation with L. plantarum, with ln(NSAF) values of −3.9 and −3.85, respectively. Furthermore, the cytoskeletal proteins KIAA0777, ezrin, nonmuscle myosin, and PDZ domain-containing protein 1 (PDZ-dc1) were detected at high abundance in the uninfected jejunal tissue; however, all of the aforementioned proteins were below the limit of detection in the L. plantarum, S. Typhimurium, and protection assay groups (see Table S4 in the supplemental material).
The major histocompatibility complex class I b molecule (MHC-Ib) HLA-B, an immunoproteasome-associated protease, and an Arf-associated protein were induced following a probiotic protection assay with ln(NSAF) values of −5.62, −4.61, and −4.66, respectively, giving results that were consistent with the start of an adaptive immune response and subsequent antigen presentation by enterocytes (Table 4). Moreover, association of L. plantarum or S. Typhimurium alone with jejunal IVOC tissues induced the expression of HLA-B with ln(NSAF) values of −5.40, −5.36, and −5.62 (Table 4). However, it is unclear whether L. plantarum or S. Typhimurium or a combination of the two in the protection assay is responsible for the induction of proteins involved in an adaptive immune response.
DISCUSSION
Our results demonstrated that L. plantarum JC1 did not competitively exclude S. Typhimurium SL1344 from associating with porcine IVOC jejunal or colonic mucosal tissues (Table 1 [competition, protection, and displacement assays]) and did not affect the cellular pathology (such as ruffle formation) typically observed following early association with S. Typhimurium SL1344 (Table 3 [protection assay] and Fig. 3B). These findings are contrary to observations from previous studies. Servin and others (45, 46) demonstrated that the presence of LAB reduced pathogen association with or invasion into traditionally cultured epithelial cells following in vitro assays. Similarly, we demonstrated that L. plantarum JC1 reduced the association of S. Typhimurium to monolayers and 3D aggregates of the porcine jejunal cell line IPEC-J2 (13a). However, the IPEC-J2 cell line has not been observed to secrete or produce intestinal mucins other than a thin layer of glycocalyx (42). Therefore, the difference in the mucin contents between these in vitro systems may explain in part the observed differences with respect to invasion of S. Typhimurium. Haque et al. demonstrated that intestinal mucin could delay S. Typhimurium invasion into human IVOC tissues (24). These data collectively suggest that the presence of both acidic and neutral mucins represents an important factor affecting the adhesion of S. Typhimurium to the intestinal epithelium. However, Tuomola et al. demonstrated increased in vitro binding of S. Typhimurium to human glycoproteins in the presence of some Lactobacillus strains (53), suggesting that LAB strain differences may also play a role.
To the best of our knowledge, this report describes the first use of IVOC to investigate host cell responses to L. plantarum. The CCIVOC method proved to be a robust and reproducible platform for studying host-pathogen interactions, as the porcine tissues remained well preserved (Fig. 3E). The fixed surface area provided by the cell crown facilitated the reproducible enumeration of bacteria such as S. Typhimurium strain SL1344 by the quantification of viable bacteria associated to the IVOC tissues. Importantly, no bacteria associated to the serosal side of the IVOC tissues were observed by light or electron microscopy (data not shown).
In this study, we demonstrated that L. plantarum JC1 can reduce acidic mucins present in jejunal and colonic goblet cells, suggesting compound exocytosis of the stored mucin granule mass. In contrast, L. plantarum JC1 simultaneously promoted accumulation of PAS-positive neutral mucins within the goblet cells of jejunal and colonic tissues (Fig. 4B and 5B). Previous studies demonstrated that mucin production and secretion can be modulated by L. plantarum and multispecies probiotic preparations (9, 33), but these studies failed to examine the effects of the presence of heat-killed (HK) L. plantarum on mucin secretion. Interestingly, the effect of L. plantarum on mucin secretion requires the presence of HK L. plantarum in the jejunum but viable L. plantarum in the colon (Fig. 4B and 5B, respectively). Moreover, significant (P < 0.05) increases in the levels of neutral mucins were observed in the jejunal tissue by the addition of HK S. Typhimurium and HK L. plantarum to the tissues (Fig. 4B), thus demonstrating a generic host response to bacterial association to the jejunal epithelium. Conversely, modulation of acidic mucin levels in the colon requires viable L. plantarum (Fig. 5A) and is therefore an active process and may indicate the involvement of bacterial proteins acting as a mucin secretagogue. Previous studies demonstrated that L. plantarum 299v secretes a potent heat-stable secretagogue (33), possibly a bacteriocin-like molecule, that modulates the host cell and thus elicits compound exocytosis of the stored mucin granule.
In contrast to the stimulation of mucin secretion by probiotic bacteria, some pathogens (such as Helicobacter pylori and Clostridium difficile) can inhibit mucin secretion to facilitate colonization (5, 47). This study demonstrated that S. Typhimurium increases the amount of neutral mucins stored within goblet cells during the early stages of infection of porcine tissues (Fig. 4B and 5B). While this result needs to be confirmed in vivo, in the jejunal tissues this process was also initiated by both HK L. plantarum and HK S. Typhimurium (Fig. 4B), suggesting that the jejunal tissue is modulated by structural components present on the bacterial cell surface. Interestingly, viable S. Typhimurium strain SL1344 is required to alter the level of neutral colonic mucins (Fig. 5B), thus indicating an active process; however, further study is required to elucidate how S. Typhimurium exerts this effect. Additionally, S. Typhimurium was not able to modulate the levels of acidic mucins in the presence or absence of L. plantarum (Fig. 3). Collectively, these data demonstrate differential characteristics with respect to modulation of acidic and basic mucins in the jejunal and colonic epithelium by S. Typhimurium and L. plantarum. Only L. plantarum could modulate acidic mucin levels (Fig. 4A and 5A), and it is tempting to speculate that the production of excess acidic mucin may facilitate a competitive exclusion effect versus S. Typhimurium. Modulation of neutral mucin was a generic response to the binding or association of bacteria to the epithelium. Finally, the jejunum and colon demonstrated differential mechanisms for the modulation of mucin levels, with colonic tissues requiring live bacteria to elicit changes in the total quantity of secreted mucin and jejunal tissues responding to heat-killed bacteria (Fig. 4 and 5).
The decrease in the quantity of acidic mucin present within the goblet cells of jejunal tissues maintained at pH 4.5 was independent of bacterial association (Fig. 4A). Caballero-Franco et al. demonstrated similar pH-dependent mucin secretion from the LS 174T colonic epithelial cell line, although it was not stated whether these were acidic or basic mucins (9). Furthermore, culture of porcine tissue at pH 4.5 (Fig. 3F) induced membrane perturbations and the formation of apoptotic bodies on epithelial cells, which is consistent with pH-induced damage (19), although it is currently unclear whether this mucin secretion was in response to damage to the epithelium or was merely stimulated by low pH.
Using multidimensional protein identification technology (MuDPIT), we demonstrated significant (P < 0.05) changes in the proteome of porcine jejunal tissues following exposure to L. plantarum JC1. Tubulin-α was expressed at a high relative abundance with an ln(NSAF) of −4.05 in porcine jejunal tissues following exposure to L. plantarum alone compared to uninfected jejunal tissue, where tubulin-α expression was below the detection limit (Table 4). However, the biological significance of this increase in the relative abundance of tubulin-α is unclear.
The interaction of S. Typhimurium with mammalian epithelial cells has previously been well documented (largely using a transcriptomic approach) (2, 17, 27, 40, 41). Shi and others (46a, 46b) used quantitative label-free proteomics to elucidate the response of human macrophages to S. Typhimurium infection. Similarly, in our studies, a marked change in cytoskeletal proteins consistent with the invasion of S. Typhimurium into the porcine epithelial cells was observed after infection of jejunal tissue with S. Typhimurium (Table 3 and Table 4). Moreover, the cytoskeletal proteins actin and tropomyosin were detected at high abundance in jejunal tissues following exposure to S. Typhimurium but were below the detection limit in uninfected tissues (see Table S4 in the supplemental material). In addition to this finding, the actin binding protein actinin and membrane-bound annexin (A2) were highly abundant in uninfected tissues but were below the detection limit in jejunal tissues following infection by S. Typhimurium (see Table S4 in the supplemental material). Additionally, membrane ruffling in jejunal tissue was observed by SEM following S. Typhimurium infection (Fig. 3A), consistent with the observed changes to the proteome of the jejunum following S. Typhimurium infection (20).
To date, little is known with regard to how lactobacilli may mitigate the invasive effects of enteropathogens or how they interact with mammalian epithelial cells. Following a probiotic protection assay (Table 3 and Table 4), the porcine jejunal tissue demonstrated structural proteomic changes indicative of the invasion of S. Typhimurium into host epithelial cells as described above (20). Additionally, the same porcine tissues exhibited proteome changes consistent with the association of L. plantarum to porcine tissue, as described above (Table 3 and Table 4). However, the porcine tissues demonstrated unique proteome changes in response to the probiotic protection assay. These changes were not observed following the association of S. Typhimurium alone or L. plantarum alone to porcine tissues. Specifically, the proteins KIAA0777 (Arg/Abl-interacting protein 2 [ArgBP2]), ezrin (Villin-2), non-muscle-related myosin, and PDZ domain-containing protein 1 (PDZ-dc1) were present in uninfected jejunal tissue but were below the detection limit following the protection assay and indicated the loss of integral membrane proteins that are essential for the normal functioning of a eukaryotic cell (29, 48, 49) (see Table S4 in the supplemental material). Following the probiotic protection assay, the expression of three proteins involved in antigen presentation was induced; these included MHC-Ib (CD 1d in humans), Arf family protein, and a proteasome activator subunit (PA28 in humans) (1, 39) (Table 4). Collectively, these proteins could represent the formation of an immunoproteasome and the subsequent presentation of antigens by the MHC-Ib molecule. It is unclear, however, whether these MHC-Ib molecules are from enterocytes (1), lymphocytes, or other MHC-Ib-expressing cell types already present within the jejunal tissue. MHC-Ib molecules in humans (when epithelium associated) provide a ligand for CD8+ T cells (1). Controls utilizing monoassociated L. plantarum or S. Typhimurium also demonstrated upregulated MHC-Ib molecules (Table 4); thus, it is not possible to determine by this experimental approach which of the aforementioned bacterial species were responsible for initiating an adaptive immune response in jejunal tissue, assuming that this was not the result of combined effects. Furthermore, it is unclear whether intestinal epithelial cells (IECs) or lymphocytes present within the porcine tissue are being primed to present antigens or whether antigen presentation is initiated by L. plantarum or S. Typhimurium association.
Collectively, our data demonstrate that L. plantarum modifies the mucosal barrier function of porcine tissues. Although L. plantarum JC1 was unable to reduce the early association of S. Typhimurium to the porcine intestinal mucosa, increased acidic mucin secretion and the potential modulation of the host cytoskeleton demonstrate that this putative probiotic is a suitable candidate for further in vivo studies in the pig.
Supplementary Material
Acknowledgments
We acknowledge receipt of funding for this study from the Department for Environment, Food, and Rural Affairs, United Kingdom, and the Veterinary Medicines Directorate (project VM2203).
We acknowledge Bentley Crudgington and Ann Long for assisting with the histopathology and subsequent PAS/Alcian blue staining of porcine tissues.
Footnotes
Published ahead of print on 16 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Balk, S. P., S. Burke, J. E. Polischuk, M. E. Frantz, L. Yang, S. Porcelli, S. P. Colgan, and R. S. Blumberg. 1994. Beta 2-microglobulin-independent MHC class Ib molecule expressed by human intestinal epithelium. Science 265:259-262. [DOI] [PubMed] [Google Scholar]
- 2.Boyce, J. D., P. A. Cullen, and B. Adler. 2004. Genomic-scale analysis of bacterial gene and protein expression in the host. Emerg. Infect. Dis. 10:1357-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyen, F., F. Pasmans, F. Van Immerseel, E. Donne, E. Morgan, R. Ducatelle, and F. Haesebrouck. 2009. Porcine in vitro and in vivo models to assess the virulence of Salmonella enterica serovar Typhimurium for pigs. Lab. Anim. 43:46-52. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Branka, J. E., G. Vallette, A. Jarry, C. Bou-Hanna, P. Lemarre, P. N. Van, and C. L. Laboisse. 1997. Early functional effects of Clostridium difficile toxin A on human colonocytes. Gastroenterology 112:1887-1894. [DOI] [PubMed] [Google Scholar]
- 6.Brashears, M. M., M. L. Galyean, G. H. Loneragan, J. E. Mann, and K. Killinger-Mann. 2003. Prevalence of Escherichia coli O157:H7 and performance by beef feedlot cattle given Lactobacillus direct-fed microbials. J. Food Prot. 66:748-754. [DOI] [PubMed] [Google Scholar]
- 7.Brown, P. J., B. G. Miller, C. R. Stokes, N. B. Blazquez, and F. J. Bourne. 1988. Histochemistry of mucins of pig intestinal secretory epithelial cells before and after weaning. J. Comp. Pathol. 98:313-323. [DOI] [PubMed] [Google Scholar]
- 8.Butaye, P., L. A. Devriese, and F. Haesebrouck. 2003. Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on gram-positive bacteria. Clin. Microbiol. Rev. 16:175-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caballero-Franco, C., K. Keller, C. De Simone, and K. Chadee. 2007. The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 292:G315-G322. [DOI] [PubMed] [Google Scholar]
- 10.Casey, P. G., G. E. Gardiner, G. Casey, B. Bradshaw, P. G. Lawlor, P. B. Lynch, F. C. Leonard, C. Stanton, R. P. Ross, G. F. Fitzgerald, and C. Hill. 2007. A five-strain probiotic combination reduces pathogen shedding and alleviates disease signs in pigs challenged with Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 73:1858-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casin, I., J. Breuil, J. P. Darchis, C. Guelpa, and E. Collatz. 2003. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica typhimurium isolates in humans. Emerg. Infect. Dis. 9:1455-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castanon, J. I. 2007. History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. 86:2466-2471. [DOI] [PubMed] [Google Scholar]
- 13.Coburn, B., G. A. Grassl, and B. B. Finlay. 2007. Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 85:112-118. [DOI] [PubMed] [Google Scholar]
- 13a.Collins, J. W., W. Cooley, P. Webb, S. Marsh, E. Waller, R. A. Rastall, G. R. Gibson, M. J. Woodward, and R. M. La Ragione. 2007. Abstr. 161st Meet. Soc. Gen. Microbiol., Edinburgh, United Kingdom, 3 to 6 September 2007, p. 11.
- 14.Danielsen, M., H. Hornshoj, R. H. Siggers, B. B. Jensen, A. G. van Kessel, and E. Bendixen. 2007. Effects of bacterial colonization on the porcine intestinal proteome. J. Proteome Res. 6:2596-2604. [DOI] [PubMed] [Google Scholar]
- 15.DEFRA. 2007. Salmonella in livestock production in GB: 2007 report. Veterinary Laboratories Agency, Surrey, United Kingdom. http://www.defra.gov.uk/vla/reports/rep_salm_rep07.htm.
- 16.Di Caro, S., H. Tao, A. Grillo, C. Elia, G. Gasbarrini, A. R. Sepulveda, and A. Gasbarrini. 2005. Effects of Lactobacillus GG on genes expression pattern in small bowel mucosa. Dig. Liver Dis. 37:320-329. [DOI] [PubMed] [Google Scholar]
- 17.Eckmann, L., J. R. Smith, M. P. Housley, M. B. Dwinell, and M. F. Kagnoff. 2000. Analysis by high density cDNA arrays of altered gene expression in human intestinal epithelial cells in response to infection with the invasive enteric bacteria Salmonella. J. Biol. Chem. 275:14084-14094. [DOI] [PubMed] [Google Scholar]
- 18.Food and Agriculture Organization of the United Nations (FAO). 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. World Health Organization, Geneva, Switzerland. http://www.who.int/foodsafety/publications/fs_management/en/probiotics.pdf.
- 19.Furukawa, O., and S. Okabe. 1997. Effects of growth factors on acid-induced damage to rat gastric epithelial cells. J. Clin. Gastroenterol. 25(Suppl. 1):S79-S83. [DOI] [PubMed] [Google Scholar]
- 20.Galan, J. E., and D. Zhou. 2000. Striking a balance: modulation of the actin cytoskeleton by Salmonella. Proc. Natl. Acad. Sci. U. S. A. 97:8754-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, J., M. S. Friedrichs, A. R. Dongre, and G. J. Opiteck. 2005. Guidelines for the routine application of the peptide hits technique. J. Am. Soc. Mass Spectrom. 16:1231-1238. [DOI] [PubMed] [Google Scholar]
- 22.Garmendia, J., A. D. Phillips, M. F. Carlier, Y. Chong, S. Schuller, O. Marches, S. Dahan, E. Oswald, R. K. Shaw, S. Knutton, and G. Frankel. 2004. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell Microbiol. 6:1167-1183. [DOI] [PubMed] [Google Scholar]
- 23.Girard, F., I. Batisson, G. M. Frankel, J. Harel, and J. M. Fairbrother. 2005. Interaction of enteropathogenic and Shiga toxin-producing Escherichia coli and porcine intestinal mucosa: role of intimin and Tir in adherence. Infect. Immun. 73:6005-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haque, A., F. Bowe, R. J. Fitzhenry, G. Frankel, M. Thomson, R. Heuschkel, S. Murch, M. P. Stevens, T. S. Wallis, A. D. Phillips, and G. Dougan. 2004. Early interactions of Salmonella enterica serovar Typhimurium with human small intestinal epithelial explants. Gut 53:1424-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Health Protection Agency. 2009. Salmonella in humans (excluding S. Typhi & S. Paratyphi). Health Protection Agency, United Kingdom. http://www.hpa.org.uk/webw/HPAweb&HPAwebStandard/HPAweb_C/1195733760280?p=1191942172078.
- 26.Hornshøj, H., E. Bendixen, L. N. Conley, P. K. Andersen, J. Hedegaard, F. Panitz, and C. Bendixen. 2009. Transcriptomic and proteomic profiling of two porcine tissues using high-throughput technologies. BMC Genomics 10:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, B. D. 1997. Host responses to pathogenic Salmonella infection. Genes Dev. 11:679-687. [DOI] [PubMed] [Google Scholar]
- 28.Kritas, S. K., and R. B. Morrison. 2005. Evaluation of probiotics as a substitute for antibiotics in a large pig nursery. Vet. Rec. 156:447-448. [DOI] [PubMed] [Google Scholar]
- 29.Lamprecht, G., and U. Seidler. 2006. The emerging role of PDZ adapter proteins for regulation of intestinal ion transport. Am. J. Physiol. Gastrointest. Liver Physiol. 291:G766-G777. [DOI] [PubMed] [Google Scholar]
- 30.Liang, P., and T. H. MacRae. 1997. Molecular chaperones and the cytoskeleton. J. Cell Sci. 110(Pt. 13):1431-1440. [DOI] [PubMed] [Google Scholar]
- 31.Liu, H., R. G. Sadygov, and J. R. Yates III. 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76:4193-4201. [DOI] [PubMed] [Google Scholar]
- 32.Llewellyn, B. D. 2009. Nuclear staining with alum hematoxylin. Biotech. Histochem. 84:159-177. [DOI] [PubMed] [Google Scholar]
- 33.Mack, D. R., S. Ahrne, L. Hyde, S. Wei, and M. A. Hollingsworth. 2003. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 52:827-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens, L., J. Vandekerckhove, and K. Gevaert. 2005. DBToolkit: processing protein databases for peptide-centric proteomics. Bioinformatics 21:3584-3585. [DOI] [PubMed] [Google Scholar]
- 35.Meyerholz, D. K., T. J. Stabel, M. R. Ackermann, S. A. Carlson, B. D. Jones, and J. Pohlenz. 2002. Early epithelial invasion by Salmonella enterica serovar Typhimurium DT104 in the swine ileum. Vet. Pathol. 39:712-720. [DOI] [PubMed] [Google Scholar]
- 36.Miles, A. A., and S. S. Misra. 1938. The estimation of the bactericidal power of the blood. J. Hyg. (London) 38:732-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parry, C. M. 2003. Antimicrobial drug resistance in Salmonella enterica. Curr. Opin. Infect. Dis. 16:467-472. [DOI] [PubMed] [Google Scholar]
- 38.Pratt, W. B., and D. O. Toft. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 228:111-133. [DOI] [PubMed] [Google Scholar]
- 39.Preckel, T., W. P. Fung-Leung, Z. Cai, A. Vitiello, L. Salter-Cid, O. Winqvist, T. G. Wolfe, M. Von Herrath, A. Angulo, P. Ghazal, J. D. Lee, A. M. Fourie, Y. Wu, J. Pang, K. Ngo, P. A. Peterson, K. Fruh, and Y. Yang. 1999. Impaired immunoproteasome assembly and immune responses in PA28-/- mice. Science 286:2162-2165. [DOI] [PubMed] [Google Scholar]
- 40.Rodenburg, W., I. M. Bovee-Oudenhoven, E. Kramer, R. van der Meer, and J. Keijer. 2007. Gene expression response of the rat small intestine following oral Salmonella infection. Physiol. Genomics 30:123-133. [DOI] [PubMed] [Google Scholar]
- 41.Rodenburg, W., J. Keijer, E. Kramer, S. Roosing, C. Vink, M. B. Katan, R. van der Meer, and I. M. Bovee-Oudenhoven. 2007. Salmonella induces prominent gene expression in the rat colon. BMC Microbiol. 7:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schierack, P., M. Nordhoff, M. Pollmann, K. D. Weyrauch, S. Amasheh, U. Lodemann, J. Jores, B. Tachu, S. Kleta, A. Blikslager, K. Tedin, and L. H. Wieler. 2006. Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem. Cell Biol. 125:293-305. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt, L. D., L. J. Kohrt, and D. R. Brown. 2008. Comparison of growth phase on Salmonella enterica serovar Typhimurium invasion in an epithelial cell line (IPEC J2) and mucosal explants from porcine small intestine. Comp. Immunol. Microbiol. Infect. Dis. 31:63-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schüller, S., M. Lucas, J. B. Kaper, J. A. Giron, and A. D. Phillips. 2009. The ex vivo response of human intestinal mucosa to enteropathogenic Escherichia coli infection. Cell Microbiol. 11:521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Servin, A. L. 2004. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28:405-440. [DOI] [PubMed] [Google Scholar]
- 46.Servin, A. L., and M. H. Coconnier. 2003. Adhesion of probiotic strains to the intestinal mucosa and interaction with pathogens. Best Pract. Res. Clin. Gastroenterol. 17:741-754. [DOI] [PubMed] [Google Scholar]
- 46a.Shi, L., J. N. Adkins, J. R. Coleman, A. A. Schepmoes, A. Dohnkova, H. M. Mottaz, A. D. Norbeck, S. O. Purvine, N. P. Manes, H. S. Smallwood, H. Wang, J. Forbes, P. Gros, S. Uzzau, K. D. Rodland, F. Heffron, R. D. Smith, and T. C. Squier. 2006. Proteomic analysis of Salmonella enterica serovar typhimurium isolated from RAW 264.7 macrophages: identification of a novel protein that contributes to the replication of serovar typhimurium inside macrophages. J. Biol. Chem. 281:29131-29140. [DOI] [PubMed] [Google Scholar]
- 46b.Shi, L., S. M. Chowdhury, H. S. Smallwood, H. Yoon, H. M. Mottaz-Brewer, A. D. Norbeck, J. E, McDermott, T. R. Clauss, F. Heffron, R. D. Smith, and J. N. Adkins. 2009. Proteomic investigation of the time course responses of RAW 264.7 macrophages to infection with Salmonella enterica. Infect. Immun. 77:3227-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slomiany, B. L., Y. H. Liau, R. A. Lopez, J. Piotrowski, A. Czajkowski, and A. Slomiany. 1992. Effect of Helicobacter pylori lipopolysaccharide on the synthesis of sulfated gastric mucin. Biochem. Int. 27:687-697. [PubMed] [Google Scholar]
- 48.Soubeyran, P., A. Barac, I. Szymkiewicz, and I. Dikic. 2003. Cbl-ArgBP2 complex mediates ubiquitination and degradation of c-Abl. Biochem. J. 370:29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spence, H. J., Y. J. Chen, C. L. Batchelor, J. R. Higginson, H. Suila, O. Carpen, and S. J. Winder. 2004. Ezrin-dependent regulation of the actin cytoskeleton by beta-dystroglycan. Hum. Mol. Genet. 13:1657-1668. [DOI] [PubMed] [Google Scholar]
- 50.Stern, N. J., E. A. Svetoch, B. V. Eruslanov, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, V. D. Pokhilenko, V. P. Levchuk, O. E. Svetoch, and B. S. Seal. 2006. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob. Agents Chemother. 50:3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Togo, T., and R. A. Steinhardt. 2004. Nonmuscle myosin IIA and IIB have distinct functions in the exocytosis-dependent process of cell membrane repair. Mol. Biol. Cell 15:688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Troost, F. J., P. van Baarlen, P. Lindsey, A. Kodde, W. M. de Vos, M. Kleerebezem, and R. J. Brummer. 2008. Identification of the transcriptional response of human intestinal mucosa to Lactobacillus plantarum WCFS1 in vivo. BMC Genomics 9:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuomola, E. M., A. C. Ouwehand, and S. J. Salminen. 1999. The effect of probiotic bacteria on the adhesion of pathogens to human intestinal mucus. FEMS Immunol. Med. Microbiol. 26:137-142. [DOI] [PubMed] [Google Scholar]
- 54.Wang, G., W. W. Wu, Z. Zhang, S. Masilamani, and R. F. Shen. 2009. Decoy methods for assessing false positives and false discovery rates in shotgun proteomics. Anal. Chem. 81:146-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woodward, M. J. 2008. Interventions to reduce the carriage of antimicrobial resistance in food producing animals. Veterinary Laboratories Agency, Addlestone, United Kingdom.
- 56.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]
- 57.Zhang, B., N. C. VerBerkmoes, M. A. Langston, E. Uberbacher, R. L. Hettich, and N. F. Samatova. 2006. Detecting differential and correlated protein expression in label-free shotgun proteomics. J. Proteome Res. 5:2909-2918. [DOI] [PubMed] [Google Scholar]
- 58.Zybailov, B., A. L. Mosley, M. E. Sardiu, M. K. Coleman, L. Florens, and M. P. Washburn. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5:2339-2347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.