Abstract
The most common class B biosolids in the United States are generated by mesophilic anaerobic digestion (MAD), and MAD biosolids have been used for land application. However, the pathogen levels in MAD biosolids are still unclear, especially with respect to enteric viruses. In this study, we determined the occurrence and the quantitative levels of enteric viruses and indicators in 12 MAD biosolid samples and of Salmonella enterica in 6 MAD biosolid samples. Three dewatered biosolid samples were also included in this study for purposes of comparison. Human adenoviruses (HAdV) had the highest gene levels and were detected more frequently than other enteric viruses. The gene levels of noroviruses (NV) reported were comparable to those of enteroviruses (EV) and human polyomaviruses (HPyV). The occurrence percentages of HAdV, HAdV species F, EV, NV GI, NV GII, and HPyV in MAD samples were 83, 83, 42, 50, 75, and 58%, respectively. No hepatitis A virus was detected. Infectious HAdV was detected more frequently than infectious EV, and all infectious HAdV were detected when samples were propagated in A549 cells. Based on most-probable-number (MPN) analysis, A549 cells were more susceptible to biosolid-associated viruses than BGM cells. All indicator levels in MAD biosolids were approximately 104 MPN or PFU per gram (dry), and the dewatered biosolids had significantly higher indicator levels than the MAD biosolids. Only two MAD samples tested positive for Salmonella enterica, where the concentration was below 1.0 MPN/4 g. This study provides a broad comparison of the prevalence of different enteric viruses in MAD biosolids and reports the first detection of noroviruses in class B biosolids. The observed high quantitative and infectivity levels of adenoviruses in MAD biosolids indicate that adenovirus is a good indicator for the evaluation of sludge treatment efficiency.
Over the last decade, thousands of people in the United States have been infected with waterborne diseases, a large number of whom were hospitalized. Most of the waterborne disease outbreaks in the United States that occurred between 1991 and 2004 were related to microbial agents, i.e., viruses, bacteria, and parasites (5, 30). The majority of the outbreaks involved unidentified agents, and the Environmental Protection Agency (EPA) suspects that many of the outbreaks due to unidentified sources were caused by enteric viruses (46). Indeed, viruses have a high potential for groundwater pollution due to their small size and low die-off rates. The occurrence of enteric viruses in groundwater has been reported (1, 7, 12, 17). In the United States, approximately 5.6 million dry tons of biosolids are generated annually and 60% of the biosolids are applied to land (36).
Several studies have reported the occurrence of enteric viruses in biosolids (18, 35, 47); however, information on the quantity and infectivity of enteric viruses in biosolids is still limited, and most studies focused solely on enteroviruses (41). Few studies have reported the levels of human adenoviruses (HAdV) in biosolids (6, 47), and no quantitative results have yet been reported on some of the emerging viruses, such as hepatitis A virus (HAV) and noroviruses (NV). Also, only one or two types of enteric viruses were quantified in the previous studies; therefore, it is hard to determine and compare the prevalence of different types of enteric virus in biosolids, since the samples and sample processing methods varied from study to study. A few studies focused on the viral infectivity of biosolids, and the results showed that infectious astrovirus and enteroviruses were still present in the treated biosolids (9, 18, 42). However, no results on the occurrence of adenoviruses in biosolids have been reported.
PCR techniques have been used in most of the recent environmental virology studies. In comparing these techniques to cell culture, the main advantages of PCR methods for virus detection are fast results, less labor intensiveness, high specificity and sensitivity, and the capability of detecting difficult-to-culture or nonculturable viruses (for examples, human noroviruses and adenovirus 40/41). Quantitative real-time PCR (qPCR), which is considered the latest advancement in PCR technology, can provide both qualitative and quantitative results. However, PCR results may not reflect the infectivity of the samples since PCR only detects the genes of the pathogens; therefore, integrated cell culture-PCR (ICC-PCR) was developed to identify the specific infectious enteric viruses. ICC-PCR has been used to detect infectious enteric viruses in river water, tap water, beach water, and wastewater effluent samples (28, 29, 39, 50). However, Buffalo green monkey (BGM) cell culture, currently recommended by the EPA, has been compared with other cell lines, such as A549 and PLC/PRC/5 (28, 39), and the results showed that enteric viruses were propagated better with these cell lines than with BGM cells.
The main objective of this work was to investigate the occurrence and the quantitative levels of the enteric viruses in class B mesophilic anaerobically digested (MAD) biosolid samples by molecular and cell culture methods. These results can be used for risk assessment at biosolid application sites. Also, enteric viruses have been suggested as fecal source tracking indicators (21, 32), and the levels of enteric viruses in biosolids reported in this study would be useful for the determination of which enteric virus is a better fecal source tracking indicator at biosolid application sites. MAD biosolids were chosen since they are the most common type of class B biosolid produced in the United States (47). Three dewatered biosolid samples were also included for comparison purposes. The levels of human adenovirus (HAdV), adenovirus type 40/41 (HAdV 40/41), enterovirus (EV), norovirus GI (NV GI) and NV GII, human polyomavirus (HPyV), and hepatitis A virus (HAV) were quantified by qPCR methods. BGM and A549 cell lines were used to quantify the infectious viruses in the biosolids, and the effectiveness of these two cell lines' ability to propagate infectious viruses was compared. The occurrence of infectious EV and HAdV in biosolids was determined by ICC-PCR, and the serotypes of the infectious adenoviruses propagated on A549 were further determined. The levels of pathogen indicators and Salmonella enterica are also presented in this study.
MATERIALS AND METHODS
Sampling.
Biosolid samples were collected from five different wastewater treatment plants (WWTPs) in Michigan, with three different sampling events at each plant. Four of the plants produced class B MAD biosolids, and one of the plants produced nondigested (dewatered) biosolids. A total of 12 MAD and three dewatered biosolid samples were collected from December 2008 to September 2009. Approximately one or two samples were collected each month during the study period. The class B biosolids are applied in agricultural plots, and the dewatered biosolids are disposed to the local landfill. Table 1 displays the stabilization temperatures, solid retention time, and dewatering processes of each plant for producing biosolids. Two-liter grab samples of each of the anaerobically digested samples were collected from the postdigestion holding tanks and then transferred or shipped to the laboratory on ice overnight. The dewatered samples were collected from the exiting conveyor belt in the loading bay. Upon acquisition of the sample, all indicator tests, Salmonella enterica tests, gravimetric analysis for determination of the solid contents, and viral elution/concentration of each sample were performed immediately.
TABLE 1.
Summary of operation parameters and biosolid characteristicsa
| Location | Treatment method | Capacity (MGD) | Average flow (MGD) | Sludge treatment | Dewatering process | MAD temp | Solids retention time | % Solids | Disposal of biosolids |
|---|---|---|---|---|---|---|---|---|---|
| East Lansing | Activated sludge | 18.8 | 13.4 | Dewatering | Bell press | NA | NA | 18.30 ± 1.57 | Landfill |
| Traverse City | Activated sludge + membrane bioreactor | 17 | 8.5 | MAD | Gravity thickened | 35°C | ∼45 days | 5.04 ± 1.28 | Agricultural land |
| Romeo | Trickling filter/rotating biological contactors | 2.2 | 0.8 | MAD | Gravity thickened | 35°C | ∼7 days | 7.95 ± 3.63 | Agricultural land |
| Plainwell | Rotating biological contactors | 1.3 | 0.5 | MAD | Gravity thickened | 37°C | ∼7.8 days | 5.19 ± 2.56 | Agricultural land |
| St. Clair | Trickling filter | 4.2 | 1.3 | MAD | Gravity thickened | 37°C | ∼7 days | 6.47 ± 1.24 | Agricultural land |
MGD, million gallons per day; NA, not applicable.
Virus elution process.
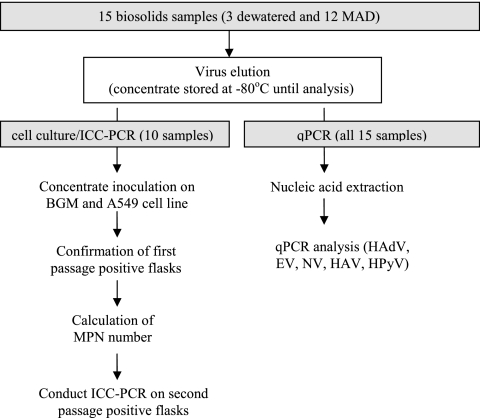
Figure 1 shows a flow chart describing the methodology applied to virus determination. The virus elution and concentration were performed according to ASTM D 4994-89 (3). Briefly, beef extract was added to 10 to 20 g (dry) biosolids and stirred for 30 min to elute the viruses. Then, the solids were spun down by centrifugation and the supernatant was kept for further concentration. The supernatant was flocculated by adjusting the pH to 3.5 and spun again to form a pellet. The pellet was then dissolved in phosphate-buffered saline (PBS) and filtered through a 0.22-μm filter. The final eluent was kept in a −80°C freezer for further analysis.
FIG. 1.
The methodology of enteric virus determination by cell culture, ICC-PCR, and qPCR.
Indicators and Salmonella enterica analysis.
All of the samples were analyzed immediately after they were delivered to the laboratory. The indicators included in this study were fecal coliform (FC) bacteria, Escherichia coli, enterococci, and somatic phages. FC analysis was performed according to U.S. EPA method 1680 (44). E. coli and enterococci were analyzed by IDEXX methods (2, 4). Somatic phages were analyzed by the double layer agar method (43). Each of the indicator measurements were run in triplicate for each sample. Salmonella enterica was measured according to EPA method 1682 (45). All dilutions were made with sterilized phosphate-buffered water.
Nucleic acid extraction.
The virus eluent and cell culture supernatant were extracted by using a MagNA Pure automatic extraction machine (Roche), and the extraction kits used were MagNA Pure compact nucleic acid isolation kits, large volume (Roche). One milliliter of the sample was extracted, and the final elution volume was 100 μl. The nucleic acid (NA) eluents were stored in a −80°C freezer for molecular analysis.
The NA extraction efficiency was evaluated, and the method used was adapted from previous studies (38, 47). NA extraction efficiencies for dewatered and MAD biosolids were evaluated for all the biosolid samples. Amounts of 6.2 × 102 PFU of bovine enteroviruses (BEV) (ATCC VR-754) were spiked into the biosolid eluent and NA-free water. After extraction, the levels of BEV were determined by qPCR. The extraction efficiency was calculated by dividing the BEV RNA recovered from the biosolid matrix by the BEV RNA recovered from the NA-free water. BEV was chosen since no samples tested positive for BEV. The extraction efficiency was incorporated into the calculation of virus concentration in the biosolid samples.
qPCR standards.
The concentrations of the target pathogens in the biosolid samples were quantified by using the standard curves generated from ATCC viruses or environmental isolates. Human adenovirus 40 (ATCC VR-930), Coxsackie virus B5 (ATCC VR-1036AS/MK), hepatitis A virus HM175 (ATCC VR-1402), polyomavirus JC (JCPyV) (ATCC VR-1583), Salmonella enterica (ATCC 14028), and norovirus isolates provided by Ingham County Health Department were used to generate the standard curves.
PCR amplicons of the target gene from HAdV, HPyV, and Salmonella enterica and cDNA of the target gene from EV, BEV, NV, and HAV were cloned into plasmid vectors according to the one-shot chemical transformation described in the manufacturer's instructions (TOPO TA cloning kit for sequencing; Invitrogen, Carlsbad, CA). Plasmids carrying the cloned target gene were purified using a Wizard Plus SV minipreps DNA purification system (Promega, Madison, WI) and sent for sequencing at the Research Technology Support Facility at Michigan State University. The target gene sequences were compared with those published in the National Center for Biotechnology Information (NCBI) database by using the Basic Local Alignment Search Tool (BLAST).
The gene equivalent copies (GEC) of the standard stocks were quantified using a NanoDrop 1000 spectrophotometer and then 10-fold serially diluted. The dilutions ranged from 101 to 108 GEC/real-time PCR (qPCR) reaction mixture volume and were used to calibrate the concentration of the target gene detected in the qPCR assays. The efficiency for each standard curve is illustrated in Table 2.
TABLE 2.
Primers and probes for enteric virus detection
| Virus | Primer or probe | Concn (μM) per reaction mixture volume | Sequence (5′-3′) | Reaction conditions (temp [°C], time [s]; purpose) | Amplification efficiency (%) | Reference |
|---|---|---|---|---|---|---|
| Total HAdV | Forward | 0.9 | C(A/T)TACATGCACATC(G/T)C(C/G)GG | 95, 15; denaturation | 95.4 | 16 |
| Reverse | 0.9 | C(A/G)CGGGC(G/A)AA(C/T)TGCACCAG | 60, 60; annealing | |||
| Probe-1 | 0.45 | CCGGGCTCAGGTACTCCGAGGCGTCCT | ||||
| Probe-2 | 0.45 | CCGGACTCAGGTACTCCGAAGCATCCT | ||||
| HAdV40/41 | Forward | 0.4 | ACCCACGATGTAACCACAGAC | 95, 10; denaturation | 107.2 | 50 |
| Reverse-1 | 0.2 | ACTTTGTAAGAGTAGGCGGTTTC | 60, 30; annealing | |||
| Reverse-2 | 0.2 | CACTTTGTAAGAATAAGCGGTGTC | 72, 12; extension | |||
| Probe | 0.3 | CGACKGGCACGAAKCGCAGCGT | ||||
| EV | Forward | 1.0 | ACATGGTGTGAAGAGTCTATTGAGCT | 95, 15; denaturation | 112.8 | 14 |
| Reverse | 1.0 | CCAAAGTAGTCGGTTCCGC | 60, 60; annealing | |||
| Probe | 0.6 | TCCGGCCCCTGAATGCGGCTAAT | ||||
| NV-G1 | Forward | 0.2 | CGCTGGATGCGNTTCCAT | 95, 15; denaturation | 93.9 | 10 |
| Reverse | 0.2 | CCTTAGACGCCATCATCATTTAC | 60, 60; annealing | |||
| Probe | 0.2 | TGGACAGGAGAYCGCRATCT | ||||
| NV GII | Forward | 0.4 | CARGARBCNATGTTYAGRTGGATGAG | 95, 15; denaturation | 98.2 | 25 |
| Reverse | 0.4 | TCGACGCCATCTTCATTCACA | 56, 60; annealing | |||
| Probe | 0.25 | TGGGAGGGCGATCGCAATCT | ||||
| HPyV | Forward | 0.5 | AGTCTTTAGGGTCTTCTACCT TT | 95, 15; denaturation | 96.6 | 32 |
| Reverse | 0.5 | GGTGC AACCTATGGAACAG | 55, 15; annealing | |||
| Probe | 0.15 | TCATCA CTGGCA AACAT | 60, 60; extension | |||
| HAV | Forward | 0.25 | GGTAGGCTACGGGTGAAAC | 95, 10; denaturation | 92.3 | 24 |
| Reverse | 0.25 | AACAACTCACCAATATCCGC | 55, 20; annealing | |||
| Probe | 0.15 | CTTAGGCTAATACTTCTATGAAGAGATGC | 72, 15; extension | |||
| BEV | Forward | 0.5 | GCCGTGAATGCTGCTAATCC | 95, 15; denaturation | 99.1 | 23 |
| Reverse | 0.5 | GTAGTCTGTTCCGCCTCCACCT | 60, 60; annealing | |||
| Probe | 0.25 | CGCACAATCCAGTGTTGCTACGTCGTAAC | ||||
| Salmonella | Forward | 0.25 | GCGTTCTGAACCTTTGGTAATAA | 95, 15; denaturation | 94.5 | 37 |
| Reverse | 0.25 | CGTTCGGGCAATTCGTTA | 62, 60; annealing | |||
| Probe | 0.4 | TGGCGGTGGGTTTTGTTGTCTTCT | 72, 10; extension |
qPCR assays.
All qPCR assays were performed in a Roche LightCycler 1.5 instrument (Roche Applied Sciences, Indianapolis, IN). Each target in each sample was run in triplicate qPCRs for determination. All PCR runs included a negative control (PCR-grade H2O without template) and a positive-control reaction mixture. The crossing point of each PCR was automatically determined by the LightCycler software, version 4.0, and used to calculate the genomic copies. All of the primer and probe sequences are summarized in Table 2. Each qPCR mixture included 10 μl of 2× LightCycler 480 TaqMan master mix, an appropriate volume of primers and probes to obtain the concentration described in Table 2, 5 μl of DNA or cDNA sample, and an appropriate volume of PCR-grade water to make up a final reaction mixture volume of 20 μl. The real-time PCR running program (all thermocycles were performed at a temperature transition rate of 20°C/s) was 95°C for 15 min followed by 45 cycles of denaturation, annealing, and extension (temperatures and times are listed in Table 2). All reactions ended with a final cooling step at 40°C for 30 s.
Reverse transcription was required before the qPCR on EV, BEV, HAV, and NV. Each reverse transcription reaction mixture included 2.5 μl of 10 μM reverse primer, 1 μl of reverse transcriptase (Promega), 4 μl of 5× Transcriptor reaction buffer (Roche), 20 U of Protector RNase inhibitor (Roche), and 2 μl of 10 mM deoxynucleotide (Roche). The reaction conditions for all three RNA viruses were the same; the reaction mixture was incubated at 55°C for 30 min and then at 85°C for 5 min to inactivate the enzyme.
To evaluate the presence of inhibition in the biosolid extracts, 104 copies of BEV RNA were spiked into each biosolid NA extract at different dilutions (no dilution, 1:5 dilution, and blank control consisting of NA-free water). The BEV threshold cycle values of each biosolid NA extract with no dilution and with the 1:5 dilution were compared with the one in the blank control. If the threshold cycle value of the blank control was 5% lower than that of the biosolid NA extract with no dilution, it indicated that inhibition was present, and dilution of the biosolid NA extract was performed until no inhibition was observed.
The limit of quantification was defined previously as the lowest concentration of the target gene remaining within the linear range of quantification (38). Ten copies of the virus gene was the limit of quantification for all qPCR assays used in this study. Even though <10 copies of the virus gene were detected in some assays, the signals were not consistent. Therefore, we set 10 copies as our qPCR detection limit, and the gene levels of the samples that tested negative were calculated using 10 copies per qPCR.
Cell culture and ICC-PCR assay.
Cell culture was performed on 10 biosolid samples (two samples from each WWTP) to determine their infectivity levels. Two cell lines, BGM (passage 140 to 170, obtained from Shay Fout at the EPA) and A549 (passage 100 to 130, obtained from ATCC [CCL 185]), were used to culture the viruses in biosolids. Briefly, the cells were grown in flasks until reaching at least 80 to 90% confluence. Virus eluents with different serial 10-fold dilutions were added to multiple culture flasks at each dilution and incubated at 36.5 ± 1°C for 1 h with rocking every 15 min to ensure complete contact between the cells and viral particles. Cells were maintained with minimum essential medium (MEM) supplemented with l-glutamine, Earle's salts, and 2% fetal bovine serum. Cytopathic effects (CPE; indicative of a viral infection) in the cell cultures were monitored for up to 14 days. All flasks that displayed CPE were frozen at −80°C for confirmation (second passage). The confirmation was done by inoculating aliquots of 1 ml of the supernatant into new 80-to-90%-confluent flasks and incubating for 7 days at 36.5 ± 1°C. The CPE in each flask was recorded, and the mean viral concentrations of the samples were estimated with free most-probable-number (MPN) software downloaded from http://www.i2workout.com/mcuriale/mpn/index.html. The results were expressed as MPN/4 g (dry), and only one MPN value was obtained for each sample due to the high labor intensiveness of replication of MPN cell culture experiments. The positive flasks were then frozen at −80°C for ICC-PCR assay. The primers/probes of the total HAdV and EV assays described in Table 2 were used for the ICC-PCR assay to determine the occurrence of infectious HAdV and EV in the biosolid samples. ICC-PCR assays were carried out on all flasks for the samples that had no flasks displaying CPE after first passage.
PCR primers developed by Xu et al. (51) were used to further classify the infectious HAdV species in A549-positive samples. The PCR amplification conditions were 94°C for 4 min, followed by 30 cycles at 94°C for 60 s, 54°C for 45 s, 72°C for 2 min, and finally, 5 min at 72°C. A total reaction mixture volume of 50 μl consisted of 1 μl each of the 10 μM primers, 25 μl of Promega 2× master mix (Promega, WI), 18 μl of molecular-grade water, and 5 μl of DNA sample.
Statistical analysis.
All microbial data were log10 transformed before the statistical analysis since it was determined by the Anderson Darling test that the data were log-normally distributed. To determine significant differences between the concentrations of indicators and enteric viruses, an analysis of variance (ANOVA) single-factor test was performed using SPSS, version 17.0. P values of less than 0.05 indicate a significant difference.
RESULTS
Indicators and Salmonella enterica levels in biosolids.
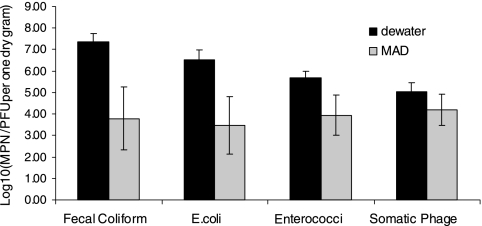
Figure 2 illustrates the indicator levels in the biosolids. All of the indicator levels in the MAD biosolids were around 104 MPN or PFU per gram, and no significant difference between each indicator was observed (P ≥ 0.05). Only 1 out of 12 MAD samples exceeded the U.S. EPA regulatory limits for class B biosolids (2 × 106 CFU/g). The dewatered biosolids had significantly higher levels of all four indicators than the MAD biosolids (P ≤ 0.05). The log differences between dewatered and MAD biosolid samples were 3.56, 3.06, 1.74, and 0.81 for FC bacteria, E. coli, enterococci, and somatic phages, respectively.
FIG. 2.
Indicator levels in biosolid samples (n = 3 dewatered biosolid samples; n = 12 MAD biosolid samples). Error bars represent the standard deviations of the measurement values of samples collected from different sampling events.
Six MAD and one dewatered sample were tested for Salmonella enterica. The only two MAD samples that tested positive were from the St. Clair WWTP and had concentrations of only 0.487 and 0.954 MPN/4 g. However, the dewatered sample had a much higher level of Salmonella enterica, which was 976 MPN/4 g. All seven samples were also tested by qPCR, but none of the samples tested positive.
Nucleic acid extraction efficiency and inhibition control.
No inhibition was observed in any of the 15 biosolid extracts, since the difference between the threshold cycle values of the blank control and the biosolid NA extract with no dilution, as well as the biosolid NA extract with a 1:5 dilution, were less than 5%. The NA extraction efficiencies for dewatered and MAD biosolids were 31.0 ± 10.4% (mean ± standard deviation) and 80.0 ± 35.2%, respectively.
Gene levels of enteric viruses in biosolids.
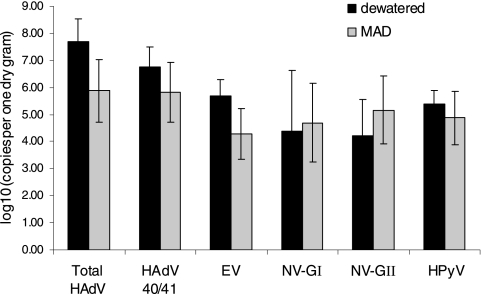
Figure 3 and Table 3 illustrate the average gene levels of enteric viruses and the percentages of qPCR- and cell culture-positive samples in both types of biosolid samples. The levels of HAdV were significantly higher than the levels of other enteric viruses (P ≤ 0.005), and they were detected more frequently than other enteric viruses. The levels of HAdV were at least 0.71 and 2.0 logs higher than the levels of other enteric viruses in the MAD and dewatered biosolids, respectively. The average level and percentage of positive samples of HAdV in the MAD biosolids were 7.5 × 105 copies/g and 83%, respectively. No significant differences were observed among the levels of EV, NV, and HPyV (P ≥ 0.05) in both types of biosolids. No HAV was detected in any of the samples. The levels of NV GI and NV GII were 5.0 × 104 and 1.5 × 105 copies/g in the MAD biosolids, which is comparable to the levels of EV. NV was detected more frequently than EV in the MAD samples. Both HPyV and HAdV are double-stranded DNA viruses, but the levels of HPyV (7.4 × 104 and 2.5 × 105 copies/g for the MAD and dewatered samples, respectively) were lower than the levels of HAdV and HPyV was detected less frequently. Only the levels of HAdV and EV in dewatered samples were significantly higher than their levels in MAD samples (P ≤ 0.05).
FIG. 3.
Enteric virus levels in biosolid samples (n = 3 dewatered biosolid samples; n = 12 MAD biosolid samples). Error bars represent the standard deviations of the measurement values of samples collected from different sampling events.
TABLE 3.
Percentages of qPCR- and cell culture-positive samples
| Assay and virus or cell line | % of indicated type of biosolid samples positive bya: |
|||
|---|---|---|---|---|
| qPCR |
Cell culture |
|||
| Dewatered (n = 3) | MAD (n = 12) | Dewatered (n = 2) | MAD (n = 8) | |
| qPCR | ||||
| Total HAdV | 100 | 83 | NA | NA |
| HAdV 40/41 | 100 | 83 | NA | NA |
| EV | 100 | 42 | NA | NA |
| NV GI | 67 | 50 | NA | NA |
| NV GII | 67 | 75 | NA | NA |
| HPyV | 100 | 58 | NA | NA |
| HAV | 0 | 0 | NA | NA |
| Cell culture | ||||
| BGM | NA | NA | 100 | 50 |
| A549 | NA | NA | 100 | 100 |
NA, not applicable.
Infectivity of enteric viruses.
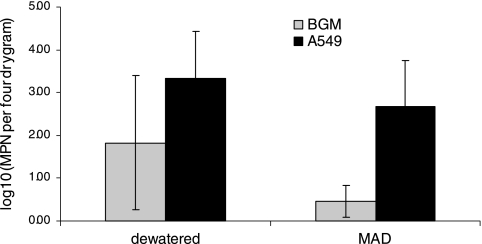
Figure 4 illustrates the average MPN levels of biosolids determined in both cell lines. The results showed that the MPN levels using A549 cells were significantly higher than the levels determined using BGM cells (P ≤ 0.005). The mean levels of infectious viruses in MAD biosolids were 2.9 and 480 MPN/4 g on BGM and A549 cells, respectively. The mean levels in dewatered biosolids were 67 and 2,210 MPN/4 g on BGM and A549, respectively. CPE was observed in all samples on A549 cells, but CPE was observed in only 50% of the MAD samples on BGM cells (Table 3). With the exception of one dewatered sample, all nine samples had higher MPN counts when propagated on A549 cells. There was no significant difference between MPN values of dewatered and MAD samples propagated on both types of cell lines.
FIG. 4.
MPN values for enteric viruses propagated in BGM and A549 cells (n = 2 dewatered biosolid samples; n = 8 MAD biosolid samples). Error bars represent the standard deviations of the measurement values of samples collected from different sampling events.
Table 4 illustrates the ICC-PCR results for HAdV and EV in flasks of BGM and A549 cells. Infectious HAdV and EV were detected in 70% and 10% of the A549 cell-propagated samples, respectively. No infectious HAdV was found in any of the BGM cell-propagated samples, but infectious EV was found in 30% of the BGM cell-propagated samples. Infectious HAdV and EV were found in both dewatered samples. Seventy-five percent and 12.5% of the MAD samples were positive for infectious HAdV and EV, respectively. Interestingly, both HAdV qPCR-negative samples were also ICC-PCR HAdV negative in both cell lines (Table 4). The other ICC-PCR HAdV-negative sample on A549 cells [Plainwell (8/4/2009)] also had a relatively lower HAdV gene concentration. All of the flasks with no CPE tested negative by both HAdV and EV ICC-PCR.
TABLE 4.
Occurrence of HAdV and EV by qPCR and ICC-PCR
| Locationa (mo/day/yr) of sampling | qPCR [mean no. of copies/g (SD)]b |
ICC-PCR using: |
||||
|---|---|---|---|---|---|---|
| A549 cells |
BGM cells |
|||||
| HAdV | EV | HAdV | EV | HAdV | EV | |
| East Lansing (12/1/2008) | 9.4 × 106 (3.3 × 105) | 2.2 × 105 (6.0 × 104) | + | + | − | + |
| East Lansing (6/29/2009) | 3.9 × 108 (7.5 × 106) | 2.4 × 105 (3.1 × 104) | + | − | − | + |
| St. Clair (2/10/2009) | 4.3 × 106 (8.5 × 104) | 2.6 × 104 (1.3 × 103) | + | − | − | − |
| St. Clair (5/4/2009) | 6.9 × 106 (4.2 × 105) | 2.9 × 104 (2.5 × 104) | + | − | − | − |
| Plainwell (4/22/2009) | 1.1 × 106 (2.9 × 105) | ND | + | − | − | − |
| Plainwell (8/4/2009) | 1.5 × 105 (5.8 × 104) | ND | − | − | − | − |
| Romeo (5/29/2009) | 4.1 × 105 (1.4 × 105) | 4.4 × 104 (1.5 × 104) | + | − | − | − |
| Romeo (6/23/2009) | 9.1 × 104 (1.4 × 105) | 7.6 × 104 (7.6 × 103) | + | − | − | − |
| Traverse City (7/16/09) | ND | ND | − | − | − | + |
| Traverse City (8/2/09) | ND | ND | − | − | − | − |
Samples from East Lansing were dewatered biosolids; samples from St. Clair, Plainwell, Romeo, and Traverse City were MAD biosolids.
Numbers inside the parentheses represent the standard deviation of the triplicate qPCR measurement values. ND, none detected.
The species of infectious HAdV detected in positive A549 cell flasks are listed in Table 5. The HAdV species detected were A (f = 4) (f is frequency of detection), B (f = 3), C (f = 3), and D (f = 2). Species E and F were not found.
TABLE 5.
Species of infectious HAdV detected in A549-positive flasks
| Location (mo/day/yr) of sampling | Species of infectious HAdV found |
|---|---|
| East Lansing (12/1/2008) | B |
| East Lansing (6/29/2009) | B, C, D |
| St. Clair (2/10/2009) | A, C |
| St. Clair (5/4/2009) | A, C |
| Plainwell (4/22/2009) | B |
| Romeo (5/29/2009) | A |
| Romeo (6/23/2009) | A, D |
DISCUSSION
Land application of biosolids has been practiced increasingly worldwide since it has the benefit of reducing the environmental contamination by reuse of the biosolids and provides biosolids as an additional source of nutrients to agricultural fields (41). However, there is a growing concern over whether land-applied biosolids would pose a risk of groundwater and/or surface water contamination. Microorganisms generally tend to attach to solid surfaces (31). Therefore, the majority of viruses and other pathogens in wastewater utilities are likely associated with sludge particles and are expected to end up in waste sludge. The most common class B sludge treatment in the United States is mesophilic anaerobic digestion (MAD). The results of a previous study suggested that viruses are resistant to MAD treatment (47). The information provided in this study provides a better understanding of the quantity and infectivity levels of several of the most critical emerging viruses in MAD biosolids measured by both molecular and cell culture methods.
The higher indicator levels in dewatered versus MAD biosolids were expected since the only treatment of dewatered biosolids is to lower the moisture content. The log reduction of FC bacteria between dewatered and MAD samples was greater than the reduction of somatic phages. This observation was similar to previous findings, where FC bacteria had a greater reduction than male-specific phages between class A and B biosolids (47). A similar trend was also observed in our previous study of an anaerobic membrane bioreactor that treats animal waste (49). The log reduction of E. coli by anaerobic digestion was 1.5, but the log reduction for somatic phages was only 0.5. Also, no significant difference was observed between the enteric virus levels in dewatered and MAD samples by both molecular and cell culture measurements (except for total HAdV and EV by qPCR). These findings showed that anaerobic digestion may be effective for the removal of bacterial indicators but not for viruses. Lower levels of Salmonella enterica in the MAD biosolids were expected since previous studies also found that the Salmonella enterica concentrations in the MAD biosolids were several orders of magnitude lower than the indicator concentrations (10, 19). Low-level occurrences of Salmonella enterica in the MAD biosolids were also observed in the previous study, where Gantzer et al. (19) found only 55% of the MAD samples to be positive for Salmonella enterica.
The levels and occurrence of HAdV measured in this study were comparable to the results of 5.0 × 105 copies/g and 88% reported by Viau and Peccia (47). The average HAdV levels of MAD biosolids reported by Bofill-Mas et al. (6) were 103 copies/g, which was approximately 2 logs lower than the HAdV levels observed in this study. The higher levels of HAdV than of other enteric viruses in biosolids could be due to its high resistance to treatment processes and high concentrations in wastewater. Enriquez and Gerba (15) conducted a survival study of HAdV 40/41 in tap water, seawater, and wastewater and concluded that HAdV 40/41 is more stable than poliovirus in tap water and wastewater. Irving and Smith (22) reported that HAdV are more likely to survive the conventional sewage treatment than EV. Katayama et al. (26) found that HAdV had the highest levels during a 1-year survey of NV, EV, and HAdV in six WWTPs.
The average levels of EV in the MAD biosolids were 1.9 × 104 copies/g, which is also comparable to the 1.2 × 104 copies/g reported by Monpoeho et al. (34); however, EV were detected in all of their MAD samples, but only 42% of the samples were detected as positive in this study. The qPCR assay used in this study was adapted from Dierssen et al. (14), and it is different than the one used in the previous study, where their assay was adapted from Monpoeho et al. (33). However, we think the difference in occurrence frequency is likely due to the lower levels of EV in our MAD biosolid samples rather than the use of a different qPCR assay. A comparison study between these two assays that was run in our laboratory for selected samples indicated that the assay developed by Dierssen et al. (14) resulted in higher virus quantities and more frequent detection than the one by Monpoeho et al. (33).
NV is an emerging virus and is one of the causes of gastroenteritis disease worldwide. NV has been detected and quantified in raw sewage and treated effluent (11, 20, 26, 27). However, no quantitative results of NV in biosolids have been reported prior to this study. Even though the levels of NV were not as high as the levels of HAdV, they were comparable to the levels of EV and HPyV. The occurrence levels were also significant, where at least 50% of the MAD samples were found to contain either NV GI or NV GII. NV GII had about half-a-log higher concentrations and 25% more positive samples than NV GI in the MAD biosolids. This observation is similar to the results of previous studies where NV GII was found more abundantly than NV GI in raw sewage (11, 20, 26).
The HPyV levels of MAD biosolids reported by Bofill-Mas et al. (6) were between 103 and 104 copies/g (dry), and all of their biosolid samples were positive for HPyV. The mean HPyV level of our MAD biosolids was 5.91 × 104 copies/g, which is comparable to the previous findings. However, only 58% of our biosolid samples were positive for HPyV. The difference in occurrence frequency between these two studies may be due to our MAD biosolids being collected from several different treatment plants, since HPyV was detected in all of the samples from the St. Clair and Romeo WWTPs but only one sample was positive in all of the Plainwell and Traverse City samples. The positive samples in this study, however, had higher concentrations, ranging from 2.41 × 105 to 1.18 × 106 copies/g. This may be due to the nature of our samples, or it could be due to the qPCR assay used in this study, which could target two main types of HPyV (polyomaviruses JC and BK [JCPyV and BKPyV]), whereas the assay used in the previous study mainly targets JCPyV.
Even though some studies have reported the presence of HAV in environmental water media (8, 13, 40, 48), no HAV was detected in any of our biosolid samples. The occurrence of HAV indicates that the risk of transporting HAV from land-applied biosolids to the natural environment is minimal.
The cell culture MPN results indicated that A549 cells were more susceptible to biosolid-associated viruses than BGM cells. Interestingly, the ICC-PCR results showed that infectious HAdV was present in more samples than infectious EV, and the infectious HAdV was only found in A549 cell-propagated samples. Also, the qPCR results showed a higher quantitative level and occurrence frequency of HAdV than of EV. Previous studies compared BGM with A549 cells and showed that the BGM cell line was not effective in propagating HAdV (30). We have also tested the effectiveness of propagating ATCC HAdV (serotypes 4, 6, 21, 31, 36, 40, and 41) in BGM and A549 cells (data not shown). The results showed that all seven serotypes of HAdV propagated using A549 cells, but only serotype 6 propagated using BGM cells. Based on these facts, we believe that the infectious HAdV levels are higher than the infectious EV levels in the biosolid samples and that BGM cells not being able to propagate the infectious HAdV effectively resulted in a lower infectious unit. Most of the previous studies used BGM cells to evaluate the viral infectivity in biosolids (33, 34, 35, 41). However, most of the recent findings have shown that HAdV are more prevalent than EV in environmental samples. Therefore, HAdV could be a more suitable enteric virus for use as an indicator of human fecal pollution at biosolid application sites.
HAdV species A (HAdV-A), B, C, and D were detected in the positive ICC-PCR samples. In a previous study (28), the infectious HAdV species in river water detected using A549 cells were C (f = 7), D (f = 5), A (f = 4), and F (f = 1). No species B or E was detected. HAdV-A, -C, and -D were detected in the previous study as well as this one. HAdV-F (type 40 and 41) was detected by qPCR in biosolids, and the levels were relatively high compared to the levels of other enteric viruses. As mentioned in the previous paragraph, we have done a study on the propagation of different ATCC HAdV species in A549 cells, and species F (both type 40 and 41) had the least increase in concentration compared to the other HAdV species after 1 week of incubation. Therefore, we think it is likely that there was infectious HAdV-F in the biosolids but that other HAdV species may have outgrown the HAdV-F, which resulted in no detection of HAdV-F during ICC-PCR.
Conclusions.
Currently, monitoring the occurrence of EV in biosolids is suggested by the EPA. However, the results from this study showed that HAdV had the highest gene levels compared to other enteric viruses in biosolids. Infectious HAdV was detected more frequently than infectious EV. Therefore, more studies on the inactivation of HAdV by different sludge treatment processes should be carried out since high levels of HAdV still remained in the MAD biosolids. More cell lines which are susceptible to HAdV should also be investigated. This study provided quantitative levels of NV in biosolids, which has never been reported in the published literature. There is a need to conduct more studies on the occurrence of NV in different types of biosolids since significant levels of NV were found. Finally, low levels of HAV and Salmonella enterica in biosolids may suggest that the risk of water contamination by these pathogens from biosolid application sites would be minimal.
Acknowledgments
This study was funded by Water Environment Research Foundation research grant number SRSK3RO8.
We thank Jim Johnson, Biosolids Coordinator with Michigan DEQ. A special thanks to all the wastewater treatment operators for assisting with sample collection and to Fred Simmons, Arun Kumar, and Matthew Fields for assisting with laboratory analysis.
Footnotes
Published ahead of print on 6 August 2010.
REFERENCES
- 1.Abbaszadegan, M., M. Lechevallier, and C. Gerba. 2003. Occurrence of viruses in US groundwaters. J. Am. Water Works Assoc. 95:107-120. [Google Scholar]
- 2.APHA. 2000. Chromogenic substrate coliform test, Method 9223B. American Public Health Association, Washington, DC.
- 3.ASTM. 2002. Standard practice for recovery of viruses from wastewater sludges, ASTM D4994-89. ASTM, West Conshohocken, PA.
- 4.ASTM. 2005. Standard test method for enterococci in water using Enterolert, ASTM D6503-99. ASTM, Philadelphia, PA.
- 5.Blackburn, B. G., G. F. Craun, J. S. Yoder, V. Hill, R. L. Calderon, N. Chen, S. H. Lee, D. A. Levy, and M. J. Beach. 2004. Surveillance for waterborne disease outbreaks associated with drinking water—United States, 2001-2002. MMWR Surveill. Summ. 53:23-45. [PubMed] [Google Scholar]
- 6.Bofill-Mas, S., N. Albinana-Gimenez, P. Clemente-Casares, A. Hundesa, J. Rodriguez-Manzano, A. Allard, M. Calvo, and R. Girones. 2006. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 72:7894-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borchardt, M. A., P. D. Bertz, S. K. Spencer, and D. A. Battigelli. 2003. Incidence of enteric viruses in groundwater from household wells in Wisconsin. Appl. Environ. Microbiol. 69:1172-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks, H. A., R. M. Gersberg, and A. K. Dhar. 2005. Detection and quantification of hepatitis A virus in seawater via real-time RT-PCR. J. Virol. Methods 127:109-118. [DOI] [PubMed] [Google Scholar]
- 9.Chapron, C. D., N. A. Ballester, and A. B. Margolin. 2000. The detection of astrovirus in sludge biosolids using an integrated cell culture nested PCR technique. Appl. J. Microbiol. 89:11-15. [DOI] [PubMed] [Google Scholar]
- 10.Dahab, M. F., and R. Y. Surampalli. 2002. Effects of aerobic and anaerobic digestion systems on pathogen and pathogen indicator reduction in municipal sludge. Water Sci. Technol. 46:181-187. [PubMed] [Google Scholar]
- 11.da Silva, A. K., J. C. Le Saux, S. Parnaudeau, M. Pommepuy, M. Elimelech, and F. S. Le Guyader. 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl. Environ. Microbiol. 73:7891-7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, J. V., and E. C. Witt. 1998. Microbiological quality of public water supplies in the Ozark Plateaus Aquifer System Missouri. USGS fact sheet 028-98. U.S. Geological Survey, U.S. Department of the Interior, Rolla, MO.
- 13.De Paula, V. S., L. Diniz-Mendes, L. M. Villar, S. L. B. Luz, L. A. Silva, M. S. Jesus, N. da Silva, and A. M. C. Gaspar. 2007. Hepatitis A virus in environmental water samples from the Amazon Basin. Water Res. 41:1169-1176. [DOI] [PubMed] [Google Scholar]
- 14.Dierssen, U., F. Rehren, C. Henke-Gendo, G. Harste, and A. Heim. 2008. Rapid routine detection of enterovirus RNA in cerebrospinal fluid by a one-step real-time RT-PCR assay. J. Clin. Virol. 42:58-64. [DOI] [PubMed] [Google Scholar]
- 15.Enriquez, C. E., and C. P. Gerba. 1995. Concentration of enteric adenovirus 40 from tap, sea and waste water. Water Res. 29:2554-2560. [Google Scholar]
- 16.Formiga-Cruz, M., G. Tofino-Quesada, S. Bofill-Mas, D. N. Lees, K. Henshilwood, A. K. Allard, A. C. Conden-Hansson, B. E. Hernroth, A. Vantarakis, A. Tsibouxi, M. Papapetropoulou, M. D. Furones, and R. Girones. 2002. Distribution of human virus contamination in shellfish from different growing areas in Greece, Spain, Sweden, and the United Kingdom. Appl. Environ. Microbiol. 68:5990-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fout, G. S., B. C. Martinson, M. W. N. Moyer, and D. R. Dahling. 2003. A multiplex reverse transcription-PCR method for detection of human enteric viruses in groundwater. Appl. Environ. Microbiol. 69:3158-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher, E. M., and A. B. Margolin. 2007. Development of an integrated cell culture-real-time RT-PCR assay for detection of reovirus in biosolids. J. Virol. Methods 139:195-202. [DOI] [PubMed] [Google Scholar]
- 19.Gantzer, C., P. Gaspard, L. Galvez, A. Huyard, N. Dumouthier, and J. Schwartzbrod. 2001. Monitoring of bacterial and parasitological contamination during various treatment of sludge. Water Res. 35:3763-3770. [DOI] [PubMed] [Google Scholar]
- 20.Haramoto, E., H. Katayama, K. Oguma, H. Yamashita, A. Tajima, H. Nakajima, and S. Ohgaki. 2006. Seasonal profiles of human noroviruses and indicator bacteria in a wastewater treatment plant in Tokyo, Japan. Water Sci. Technol. 54:301-308. [DOI] [PubMed] [Google Scholar]
- 21.Harwood, V. J., M. Brownell, S. Wang, J. Lepo, R. D. Ellender, A. Ajidahun, K. N. Hellein, E. Kennedy, X. Y. Ye, and C. Flood. 2009. Validation and field testing of library-independent microbial source tracking methods in the Gulf of Mexico. Water Res. 43:4812-4819. [DOI] [PubMed] [Google Scholar]
- 22.Irving, L. G., and F. A. Smith. 1981. One-year survey of enteroviruses, adenoviruses, and reoviruses isolated from effluent at an activated-sludge purification plant. Appl. Environ. Microbiol. 41:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jimenez-Clavero, M. A., E. Escribano-Romero, C. Mansilla, N. Gomez, L. Cordoba, N. Roblas, F. Ponz, V. Ley, and J. C. Saiz. 2005. Survey of bovine enterovirus in biological and environmental samples by a highly sensitive real-time reverse transcription-PCR. Appl. Environ. Microbiol. 71:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jothikumar, N., T. L. Cromeans, M. D. Sobsey, and B. H. Robertson. 2005. Development and evaluation of a broadly reactive TaqMan assay for rapid detection of hepatitis A virus. Appl. Environ. Microbiol. 71:3359-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katayama, H., E. Haramoto, K. Oguma, H. Yamashita, A. Tajima, H. Nakajima, and S. Ohyaki. 2008. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res. 42:1441-1448. [DOI] [PubMed] [Google Scholar]
- 27.Laverick, M. A., A. P. Wyn-Jones, and M. J. Carter. 2004. Quantitative RT-PCR for the enumeration of noroviruses (Norwalk-like viruses) in water and sewage. Lett. Appl. Microbiol. 39:127-136. [DOI] [PubMed] [Google Scholar]
- 28.Lee, C., S. H. Lee, E. Han, and S. J. Kim. 2004. Use of cell culture-PCR assay based on combination of A549 and BGMK cell lines and molecular identification as a tool to monitor infectious adenoviruses and enteroviruses in river water. Appl. Environ. Microbiol. 70:6695-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, S. H., C. Lee, K. W. Lee, H. B. Cho, and S. J. Kim. 2005. The simultaneous detection of both enteroviruses and adenoviruses in environmental water samples including tap water with an integrated cell culture-multiplex-nested PCR procedure. J. Appl. Microbiol. 98:1020-1029. [DOI] [PubMed] [Google Scholar]
- 30.Liang, J. L., E. J. Dziuban, G. F. Craun, V. Hill, M. R. Moore, R. J. Gelting, R. L. Calderon, M. J. Beach, and S. L. Roy. 2006. Surveillance for waterborne disease and outbreaks associated with drinking water and water not intended for drinking—United States, 2003-2004. MMWR Surveill. Summ. 55:31-65. [PubMed] [Google Scholar]
- 31.Maier, R. M., I. L. Pepper, and C. P. Gerba. 2008. Environmental microbiology. Elsevier Academic Press, San Diego, CA.
- 32.McQuaig, S. M., T. M. Scott, J. O. Lukasik, J. H. Paul, and V. J. Harwood. 2009. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl. Environ. Microbiol. 75:3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monpoeho, S., A. Dehee, B. Mignotte, L. Schwartzbrod, V. Marechal, J. C. Nicolas, S. Billaudel, and V. Ferre. 2000. Quantification of enterovirus RNA in sludge samples using single tube real-time RT-PCR. Biotechniques 29:88-93. [DOI] [PubMed] [Google Scholar]
- 34.Monpoeho, S., A. Maul, C. Bonnin, L. Patria, S. Ranarijaona, S. Billaudel, and V. Ferre. 2004. Clearance of human-pathogenic viruses from sludge: study of four stabilization processes by real-time reverse transcription-PCR and cell culture. Appl. Environ. Microbiol. 70:5434-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monpoeho, S., A. Maul, B. Mignotte-Cadiergues, L. Schwartzbrod, S. Billaudel, and V. Ferre. 2001. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Research Council. 2002. Biosolids applied to land: advancing standards and practices. National Academy Press, Washington, DC.
- 37.Novinscak, A., C. Surette, and M. Filion. 2007. Quantification of Salmonella spp. in composted biosolids using a TaqMan qPCR assay. J. Microbiol. Methods 70:119-126. [DOI] [PubMed] [Google Scholar]
- 38.Rajal, V. B., B. S. McSwain, D. E. Thompson, C. M. Leutenegger, B. J. Kildare, and S. Wuertz. 2007. Validation of hollow fiber ultrafiltration and real-time PCR using bacteriophage PP7 as surrogate for the quantification of viruses from water samples. Water Res. 41:1411-1422. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez, R. A., P. M. Gundy, and C. P. Gerba. 2008. Comparison of BGM and PLC/PRC/5 cell lines for total culturable viral assay of treated sewage. Appl. Environ. Microbiol. 74:2583-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose, M. A., A. K. Dhar, H. A. Brooks, F. Zecchini, and R. M. Gersberg. 2006. Quantitation of hepatitis A virus and enterovirus levels in the lagoon canals and Lido beach of Venice, Italy, using real-time RT-PCR. Water Res. 40:2387-2396. [DOI] [PubMed] [Google Scholar]
- 41.Sidhu, J. P. S., and S. G. Toze. 2009. Human pathogens and their indicators in biosolids: a literature review. Environ. Int. 35:187-201. [DOI] [PubMed] [Google Scholar]
- 42.Soares, A. C., T. M. Straub, I. L. Pepper, and C. P. Gerba. 1994. Effect of anaerobic digestion on the occurrence of enteroviruses and Giardia cysts in sewage sludge. J. Env. Sci. Health A Environ. Sci. Eng. Toxic Hazard. Subst. Control 29:1887-1897. [Google Scholar]
- 43.U.S. Environmental Protection Agency. 2001. Method 1602: male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure. U.S. Environmental Protection Agency, Washington, DC.
- 44.U.S. Environmental Protection Agency. 2006. Method 1680: fecal coliforms in sewage sludge (biosolids) by multiple-tube fermentation using lauryl tryptose broth (LTB) and EC medium. U.S. Environmental Protection Agency, Washington, DC.
- 45.U.S. Environmental Protection Agency. 2006. Method 1682: Salmonella in sewage sludge (biosolids) by modified semisolid Rappaport-Vassiliadis (MSRV) medium. U.S. Environmental Protection Agency, Washington, DC.
- 46.U.S. Environmental Protection Agency. 2006. Prepublication of the Ground Water Rule Federal Register Notice. National primary drinking water regulations: Ground Water Rule. 40 CFR Parts, 9, 141, and 142. EPA-HQ-OW-2002-0061 FRL RIN 2040-AA97. U.S. Environmental Protection Agency, Washington, DC.
- 47.Viau, E., and J. Peccia. 2009. Survey of wastewater indicators and human pathogen genomes in biosolids produced by class A and class B stabilization treatments. Appl. Environ. Microbiol. 75:164-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villar, L. M., V. S. de Paula, L. Diniz-Mendes, F. R. Guimaraes, F. F. M. Ferreira, T. C. Shubo, M. P. Miagostovich, E. Lampe, and A. M. C. Gaspar. 2007. Molecular detection of hepatitis A virus in urban sewage in Rio de Janeiro, Brazil. Lett. Appl. Microbiol. 45:168-173. [DOI] [PubMed] [Google Scholar]
- 49.Wong, K., I. Xagoraraki, J. Wallace, W. Bickert, S. Srinivasan, and J. B. Rose. 2009. Removal of viruses and indicators by anaerobic membrane bioreactor treating animal waste. J. Environ. Qual. 38:1694-1699. [DOI] [PubMed] [Google Scholar]
- 50.Xagoraraki, I., D. H. W. Kuo, K. Wong, M. Wong, and J. B. Rose. 2007. Occurrence of human adenoviruses at two recreational beaches of the Great Lakes. Appl. Environ. Microbiol. 73:7874-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu, W. H., M. C. McDonough, and D. D. Erdman. 2000. Species-specific identification of human adenoviruses by a multiplex PCR assay. J. Clin. Microbiol. 38:4114-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]