Abstract
One clone encoding glycoside hydrolases was identified through functional screening of a rumen bacterial artificial chromosome (BAC) library. Of the 68 open reading frames (ORFs) predicted, one ORF encodes a novel endo-β-1,4-xylanase with two catalytic domains of family GH43 and two cellulose-binding modules (CBMs) of family IV. Partial characterization showed that this endo-xylanase has a greater specific activity than a number of other xylanases over a wide temperature range at neutral pH and could be useful in some industrial applications.
The ruminal microbes possess a repertoire of hydrolases, including glycosyl hydrolases and esterases, which mediate hydrolysis and subsequent fermentation of the diets (mainly cellulose, xylan, amylopectin, amylose, pectin) (10, 18). Thus, the ruminal microbiome has been recognized as a rich source of enzymes important not only for feed and animal industries but also for the bioenergy industry (20, 26). However, this enzyme source is largely untapped because the majority of ruminal microbes remain uncultured (17). Activity-based metagenomics enables direct identification of genes and enzymes of interest by screening metagenomic libraries for desired heterologous phenotypic traits expressed in a surrogate bacterial host. Collectively, previous studies have identified 12 esterases, 10 endoglucanases, two lipases, one cyclodextrinase, one polyphenol oxidase, and one unique multifunctional glycosyl hydrolase from ruminal metagenomic libraries (2, 6, 8-9, 21, 23). These studies provided further evidence that the ruminal microbiome contains a rich source of glycosyl hydrolases possessed by as-yet-uncultured microbes. The objective of this study was to examine a bacterial artificial chromosome (BAC) library constructed previously from the ruminal samples collected from Chinese Holstein cows for hydrolases.
Functional screening and bioinformatic analysis.
The BAC library was constructed from metagenomic DNA extracted from ruminal content collected from Chinese Holstein cows using the CopyControl pCClBAC vector and Escherichia coli TransforMax EPI300 (Epicentre Biotechnologies, Madison, WI) and has been reported elsewhere (31). Amylase- and xylanase-positive clones were screened for by plating the BAC clones on LB agar plates containing 1.0% starch and xylan, respectively (15, 30). From screening approximately 15,360 BAC colonies, we identified 10 clones with amylase activity and 18 clones with xylanase. One BAC clone (designated U4) was found to have both amylase and xylanase activities in the screening. A shotgun library was constructed for this clone by subcloning mechanically sheared DNA (1.5 to 3 kb) into the pUC18 vector. The entire insert of clone U4 (designated URE4) was sequenced to 6× coverage using the Sanger DNA sequencing technology. The assembled insert URE4 sequence is 85,006 bp long and has 61% G+C content (GenBank accession no. FJ529691). A total of 68 open reading frames (ORFs) that code for 50 or more amino acid (aa) residues were identified using the GeneMark program (see Table S1 in the supplemental material). Based on manual comparisons to the proteins archived in the Clusters of Orthologous Groups (COG) database in NCBI, 15% of the predicted URE4 genes encode proteins involved in carbohydrate transport and metabolism (see Fig. S1 in the supplemental material). It is likely that insert URE4 was recovered from a bacterium that is involved in digestion and utilization of polysaccharides, at least starch and xylan.
The host origin of insert URE4 was inferred by comparing all the ORFs to GenBank (http://www.ncbi.nlm.nih.gov/) and FibRumBa database (http://blast.jcvi.org/rumenomics/) sequences using the BLASTP algorithm. The best hit was selected based on its low E value. The majority (70%) of the predicted genes exhibited best matches to genes of Bacteroidetes, with the remaining genes appearing to be related to Firmicutes, Proteobacteria, Verrucomicrobia, and Spirochaetes (see Table S1 in the supplemental material). When all the ORFs were analyzed using the MEGAN 3 program against the NCBI taxonomy database, 25 (37%) ORFs matched genes of Bacteroidetes, 13 (19%) ORFs matched genes of other bacteria, and 30 ORFs were not assigned to any existing bacterial phyla. To further determine the possible bacterial origin of URE4, we compared all the ORFs to the Signature gene database of known phylogeny using the “Signature search” algorithm (http://www.cmbi.ru.nl/signature) (7). The Signature genes contain a wealth of gene content-based clade-specific information and thus can serve as a strong evolutionary signal that could be used to phylogenetically characterize DNA sequences, such as metagenomic sequences, in a manner that is complementary to sequence-based analyses (7). The two signature orthologous groups (OGs) (NOG0647 and NOG19098) we identified (data not shown) suggest that URE4 was recovered from a bacterium of Bacteroidetes. Based on the results of the above three types of analysis, we concluded that URE4 was recovered from a bacterium of the phylum Bacteroidetes, a predominant bacterial phylum present in the rumen.
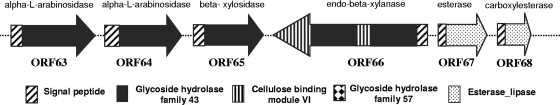
Sequence annotation revealed an α-amylase gene (ORF6) and a cluster of xylanase genes (ORF63 to ORF68) carried by URE4 (Fig. 1; see also Table S1 in the supplemental material). As calculated using the Compute pI/Mw tool (http://www.expasy.ch/tools/pi_tool.html), the amylase gene encodes a protein of 49.70 kDa with a theoretical pI of 5.87. Based on BLAST searches against GenBank and SwissProt databases, the putative amylase (designated UA6) is 61% identical in amino acid sequence (E value = 3e−150) to an α-amylase found in Algoriphagus sp. PR1. Phylogenetic analysis of UA6 and related proteins using the Mega program (http://www.megasoftware.net/) also suggests that UA6 was probably derived from a bacterium of Bacteroidetes. Amino acid sequence comparisons with databases indicated that the putative α-amylase belongs to glycosyl hydrolase family GH57. Amylases are important industrial enzymes and have broad applications in different industries, such as food, textile, and paper industries (24). Most of these industrial amylases belong to family GH13 and are categorized as α-amylases. Only a few α-amylases belong to family GH14 or GH57, whose members have a catalytic nucleophile of glutamate (13). Despite belonging to family GH57, the UA6 α-amylase is not very closely related to any known α-amylases. Future studies are needed to further characterize this new amylase.
FIG. 1.
Organization of the xylanase system. Arrows indicate the location and direction of the transcription of individual ORFs.
The cluster of xylanolytic genes (ORF63 to ORF68) encodes four putative xylanases and two esterases (Fig. 1; see also Table S1 in the supplemental material). All four xylanases belong to family GH43. ORF63 and ORF64 are predicted to encode α-l-arabinosidases (605 and 537 aa residues, respectively), both of which are most similar to an α-l-arabinosidase of Bacteroides thetaiotaomicron VPI 5482. ORF65 encodes a β-xylosidase of 535 aa residues that shares 35% aa sequence identity with a β-xylosidase of Bacteroides fragilis YCH45. ORF66 was predicted to encode a relatively large protein (designated UX66), while ORF67 and ORF68 encode an esterase and a small carboxylesterase, respectively. The UX66 protein was predicted to contain 868 aa residues with a calculated molecular mass of 97.70 kDa and a pI of 5.05. When expressed in E. coli, UX66 was a single peptide (data not shown). Interestingly, the UX66 protein seems to contain a fusion of two endo-β-1,4-xylanases (Fig. 1), which were designated UX66a (nucleotides 80940 to 82166; 409 aa residues, including a 19-aa signal peptide) and UX66b (nucleotides 82179 to 83543; 455 aa residues). UX66a and UX66b each have one catalytic domain of family GH43 and one cellulose-binding module (CBM) of class VI (CBM_6). UX66a and UX66b share 35% aa sequence identity and are linked by 4 aa residues (HLNM). Based on a BLASTn search, UX66a showed 46% aa sequence identity to a known xylanase from Bacteroides vulgatus ATCC 8482 (E value = 6e−100), and UX66b showed an identity of 66% to a known xylanase from Bacteroides ovatus ATCC 8483 (E value = 7e−164). Alignment of the amino acid sequences of UX66a and UX66b with some endo-xylanases in family GH43 revealed that both UX66a and UX66b have the catalytic residues of glutamate and aspartate (Fig. 2).
FIG. 2.
An alignment of the amino acid sequences of the endo-xylanases UX66a and UX66b with those in family GH43. Sequences (GenBank accession no.) are for Bacteroides thetaiotaomicron (NP812573), Bacteroides fragilis (YP101639), and Cellvibrio japonicus (YP001981313). Conserved regions are indicated as follows: *, identical in all species; :, similar in all species; and •, similarities exist. The active site and catalytic residues are indicated by ▴.
Glycoside hydrolases that possess one catalytic domain and one or multiple CBMs are common, but there are only a few glycoside hydrolases that contain multiple catalytic domains (23). Bacteroides cellulosilyticus DSM 14838 produces one hypothetical protein that contains two catalytic domains of different families (GH31 and GH43) and one CBM_6 (GenBank accession number ZP_03677737). The genome of Clostridium nexile DSM 1787 contains a large protein of 1,769 aa residues (GenBank accession number ZP_03289185), which possesses two GH43 domains, one CBM_6, two CBM_4_9, and one β-xylosidase domain (XynB). Among the glycoside hydrolases identified from the ruminal metagenomes by other researchers, only one was found to contain multiple domains: one GH5 domain, one GH26 domain, and two discrete CBMs (23). Unlike the above xylanases, the endo-β-1,4-xylanase UX66 is predicted to contain two catalytic domains and two CBM domains of the same families. To our knowledge, this is the first reported glycoside hydrolase that has such a feature. Additionally, xylanases are generally reported as belonging to families GH10 and GH11, though some xylanases have been found in families GH5, GH7, GH8, GH16, GH26, GH43, GH52, and GH62 (4). The enzymes of GH43 typically have a molecular mass of approximately 64 kDa (12). The dual catalytic domains and CBMs make the UX66 protein twice as large as other GH43 enzymes. If both the catalytic domains of this endo-β-1,4-xylanase are functional, it could have improved xylanase activity. Comparative studies with other xylanases containing one GH domain can help test this hypothesis. Additionally, site-specific mutagenesis should help determine if both catalytic domains are functional.
Characterization of endo-xylanase UX66.
To further characterize protein UX66, we cloned and overexpressed UX66 in E. coli. The gene encoding the endo-β-1,4-xylanase UX66 was amplified by PCR using a sense primer (5′-GGAATTCCATATGGGTGGTGGTATGAAGCGTATCCTCTCTTTTTC-3′) with an NdeI cutting site (underlined) and an antisense primer (5′- CCGCTCGAGCTACCGCGCCATCTTCCACCAG-3′) with an XhoI cutting site (underlined), and then it was cloned and overexpressed using vector pET-30a(+) (Novagen, Madison, WI) and E. coli BL21(DE3) (Invitrogen Corporation, Carlsbad, CA). After induction with IPTG (isopropyl-β-d-thiogalactopyranoside), the UX66 protein was purified using the Ni-NTA kits (Novagen). As estimated by SDS-PAGE analysis, the overexpressed UX66 protein was a single peptide of approximately 99 kDa (data not shown), which is similar to the molecular mass (97.7 kDa) that was predicted from the UX66 DNA sequence. The xylanolytic activity of the overexpressed UX66 protein was assayed with oat spelt xylan (Sigma-Aldrich) as the substrate (14). The reducing sugar released was determined using the dinitrosalicylic acid (DNS) assay according to the standard method, using xylose as the standard. One unit of xylanase was defined as the amount of enzyme that released 1.0 μmol of reducing sugar per minute under the assay conditions.
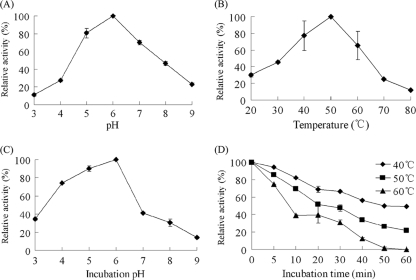
The optimal pH and temperature of the overexpressed UX66 were determined over a pH range of 3 to 9 at temperatures from 20 to 80°C. The pH stability was determined after preincubating the purified UX66 at a pH of 3 to 9 for 120 min. The thermal stability was determined after preincubation for 60 min at 40 to 60°C. The results revealed that the purified UX66 had the optimal pH of 6.0 and temperature of 50°C (Fig. 3 A and B). Following preincubation for 120 min between pH 3.0 and pH 9.0, the UX66 remained active, with no activity being lost at pH 6 (Fig. 3C). Over a period of 60 min of preincubation, the UX66 gradually lost its xylanolytic activity at temperatures between 40 and 60°C, and such a decrease was accelerated at higher temperatures (Fig. 3D). These results indicate that the UX66 was probably derived from a mesophilic neutrophilic bacterium.
FIG. 3.
Some biochemical features of the overexpressed endo-xylanase UX66. (A) pH optima of UX66 measured at 50°C; (B) temperature optima of UX66 measured at pH 6.0 with incubation for 10 min; (C) stability of UX66 under various pH conditions measured at 50°C; (D) stability of UX66 activity at different temperatures measured at pH 6.0. The values are the average of results from three replicates.
The substrate specificity of endo-xylanase UX66 was evaluated using the optimal conditions determined above and each of the following compounds (wt/vol): 1.0% oat spelt xylan, 1.0% carboxymethyl cellulose (CMC), 1.0% beechwood xylan, 0.1% p-nitrophenyl-α-l-arabinopyranoside, and 0.1% p-nitrophenyl-β-d-xylopyranoside (16). The purified UX66 exhibited the highest specific activity toward oat spelt xylan (1,163 U/mg protein) followed by beechwood xylan (709 U/mg). Additionally, the UX66 also hydrolyzed p-nitrophenyl-α-l-arabinopyranoside (288 U/mg) and CMC (77 U/mg) but to a lesser extent. No significant hydrolysis was observed on p-nitrophenyl-β-d-xylopyranoside. Thus, the UX66 also has some α-l-arabinopyranosidase and cellulase activities.
Complete hydrolysis of natural xylan requires concerted actions of a number of different xylanolytic enzymes (1, 5, 28). Indeed, such multifunctional xylanolytic enzyme systems have been found in Prevotella bryantii B14 (11, 14, 29), Cellulomonas sp. NCIM 2353 (3), Aeromonas caviae W-61 (22), and fungus Melanocarpus albomyces IIS 68 (25). The multiplicity of xylanolytic enzymes (two α-l-arabinofuranosidases, one β-xylosidase, one multidomain β-1,4-endo-xylanase, and two esterases) found in URE4 is also obvious (Fig. 1). Besides the xylanolytic activity, the purified UX66 protein also exhibited considerable cellulase activity, which might contribute to xylan hydrolysis by freeing xylan from embedded celluloses. Taken together, the URE4 xylanolytic system potentially enables the host bacterium to completely and efficiently hydrolyze heteroxylan in the rumen.
Several xylanases have been characterized from other metagenomes (14, 19, 27). In this study, however, we did not experimentally compare the activities of the UX66 and other metagenomic xylanases under the same conditions because they probably have different optimal conditions. Here, we compared the specific activity of the overexpressed UX66 protein to that reported for other metagenomic xylanases. Clearly, UX66 has a greater specific xylanase activity (1,163 U/mg protein) than the overexpressed and purified xylanases recovered from several other metagenomes: Xyn8 (768 U/mg) (19), XynH (118 U/mg) (14), and XynAD1/2 (177 U/mg) (27). Because of its high xylanolytic activity, xylanase UX66 might be useful in some biomass processing industries, such as kraft pulp bleaching, food processing, and animal feed preparation. It should be noted that robust and active xylanases will be increasingly important in utilization of certain bioenergy by-products as animal feed, such as the xylan-rich dried distillers' grains with solubles (DDGS) from the bioethanol industry.
Supplementary Material
Acknowledgments
This work was supported by the National Basic Research Program (973) of China (grant no. 2011CB100805) and the State Key Laboratory of Animal Nutrition [grant no. 2004DA125184(tuan)0801].
Footnotes
Published ahead of print on 13 August 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Beg, Q. K., M. Kapoor, L. Mahajan, and G. S. Hoondal. 2001. Microbial xylanases and their industrial applications: a review. Appl. Microbiol. Biotechnol. 56:326-338. [DOI] [PubMed] [Google Scholar]
- 2.Beloqui, A., M. Pita, J. Polaina, A. Martinez-Arias, O. V. Golyshina, M. Zumarraga, M. M. Yakimov, H. Garcia-Arellano, M. Alcalde, V. M. Fernandez, K. Elborough, J. M. Andreu, A. Ballesteros, F. J. Plou, K. N. Timmis, M. Ferrer, and P. N. Golyshin. 2006. Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties, structural analysis, and phylogenetic relationships. J. Biol. Chem. 281:22933-22942. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhary, P., and D. N. Deobagkar. 1997. Purification and characterization of xylanase from Cellulomonas sp. NCIM 2353. Biotechnol. Appl. Biochem. 25:127-133. [Google Scholar]
- 4.Collins, T., C. Gerday, and G. Feller. 2005. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 29:3-23. [DOI] [PubMed] [Google Scholar]
- 5.de Vries, R. P., H. C. Kester, C. H. Poulsen, J. A. Benen, and J. Visser. 2000. Synergy between enzymes from Aspergillus involved in the degradation of plant cell wall polysaccharides. Carbohydr. Res. 327:401-410. [DOI] [PubMed] [Google Scholar]
- 6.Duan, C. J., L. Xian, G. C. Zhao, Y. Feng, H. Pang, X. L. Bai, J. L. Tang, Q. S. Ma, and J. X. Feng. 2009. Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J. Appl. Microbiol. 107:245-256. [DOI] [PubMed] [Google Scholar]
- 7.Dutilh, B. E., B. Snel, T. J. Ettema, and M. A. Huynen. 2008. Signature genes as a phylogenomic tool. Mol. Biol. Evol. 25:1659-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrer, M., A. Beloqui, O. V. Golyshina, F. J. Plou, A. Neef, T. N. Chernikova, L. Fernandez-Arrojo, I. Ghazi, A. Ballesteros, K. Elborough, K. N. Timmis, and P. N. Golyshin. 2007. Biochemical and structural features of a novel cyclodextrinase from cow rumen metagenome. Biotechnol. J. 2:207-213. [DOI] [PubMed] [Google Scholar]
- 9.Ferrer, M., O. V. Golyshina, T. N. Chernikova, A. N. Khachane, D. Reyes-Duarte, V. A. Santos, C. Strompl, K. Elborough, G. Jarvis, A. Neef, M. M. Yakimov, K. N. Timmis, and P. N. Golyshin. 2005. Novel hydrolase diversity retrieved from a metagenome library of bovine rumen microflora. Environ. Microbiol. 7:1996-2010. [DOI] [PubMed] [Google Scholar]
- 10.Firkins, J. L., and Z. Yu. 2006. Characterisation and quantification of the microbial populations of the rumen, p. 19-54. In K. Sejrsen, T. Hvelplund, and M. O. Nielsen (ed.), Ruminant physiology: digestion, metabolism and impact of nutrition on gene expression, immunology and stress. Wageningen Academic Publisher, Wageningen, Netherlands.
- 11.Gasparic, A., R. Marinsek-Logar, J. Martin, R. J. Wallace, F. V. Nekrep, and H. J. Flint. 1995. Isolation of genes encoding beta-D-xylanase, beta-D-xylosidase and alpha-L-arabinofuranosidase activities from the rumen bacterium Prevotella ruminicola B1(4). FEMS Microbiol. Lett. 125:135-141. [DOI] [PubMed] [Google Scholar]
- 12.Gosalbes, M. J., J. A. Perez-Gonzalez, R. Gonzalez, and A. Navarro. 1991. Two beta-glycanase genes are clustered in Bacillus polymyxa: molecular cloning, expression, and sequence analysis of genes encoding a xylanase and an endo-beta-(1,3)-(1,4)-glucanase. J. Bacteriol. 173:7705-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta, R., P. Giras, H. Mohapatra, Y. K. Gaswami, and B. Chauhan. 2003. Microbial α-amylases: a biotechnological perspective. Process Biochem. 38:1599-1616. [Google Scholar]
- 14.Hu, Y., G. Zhang, A. Li, J. Chen, and L. Ma. 2008. Cloning and enzymatic characterization of a xylanase gene from a soil-derived metagenomic library with an efficient approach. Appl. Microbiol. Biotechnol. 80:823-830. [DOI] [PubMed] [Google Scholar]
- 15.Huang, J., G. Wang, and L. Xiao. 2006. Cloning, sequencing and expression of the xylanase gene from a Bacillus subtilis strain B10 in Escherichia coli. Bioresour. Technol. 97:802-808. [DOI] [PubMed] [Google Scholar]
- 16.Iembo, T., M. O. Azevedo, C. Bloch, Jr., and E. X. F. Filho. 2006. Purification and partial characterization οf a new β-xylosidase from Humicola grisea var. thermoidea. World J. Microbiol. Biotechnol. 22:475-479. [Google Scholar]
- 17.Kleessen, B., E. Bezirtzoglou, and J. Matto. 2000. Culture-based knowledge on biodiversity, development and stability of human gastrointestinal microflora. Microb. Ecol. Health Dis. 12(Suppl. 2):53-63. [Google Scholar]
- 18.Krause, D. O., S. E. Denman, R. I. Mackie, M. Morrison, A. L. Rae, G. T. Attwood, and C. S. McSweeney. 2003. Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol. Rev. 27:663-693. [DOI] [PubMed] [Google Scholar]
- 19.Lee, C. C., R. E. Kibblewhite-Accinelli, K. Wagschal, G. H. Robertson, and D. W. Wong. 2006. Cloning and characterization of a cold-active xylanase enzyme from an environmental DNA library. Extremophiles 10:295-300. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S. S., K. J. Shin, W. Y. Kim, J. K. Ha, and I. K. Han. 1999. The rumen ecosystem: as a fountain source of novel enzymes. Asian-Australas. J. Anim. Sci. 12:988-1001. [Google Scholar]
- 21.Liu, K., J. Wang, D. Bu, S. Zhao, C. McSweeney, P. Yu, and D. Li. 2009. Isolation and biochemical characterization of two lipases from a metagenomic library of China Holstein cow rumen. Biochem. Biophys. Res. Commun. 385:605-611. [DOI] [PubMed] [Google Scholar]
- 22.Okai, N., M. Fukasaku, J. Kaneko, T. Tomita, K. Muramoto, and Y. Kamio. 1998. Molecular properties and activity of a carboxyl-terminal truncated form of xylanase 3 from Aeromonas caviae W-61. Biosci. Biotechnol. Biochem. 62:1560-1567. [DOI] [PubMed] [Google Scholar]
- 23.Palackal, N., C. S. Lyon, S. Zaidi, P. Luginbuhl, P. Dupree, F. Goubet, J. L. Macomber, J. M. Short, G. P. Hazlewood, D. E. Robertson, and B. A. Steer. 2007. A multifunctional hybrid glycosyl hydrolase discovered in an uncultured microbial consortium from ruminant gut. Appl. Microbiol. Biotechnol. 74:113-124. [DOI] [PubMed] [Google Scholar]
- 24.Pandey, A., P. Nigam, C. R. Soccol, V. T. Soccol, D. Singh, and R. Mohan. 2000. Advances in microbial amylases. Biotechnol. Appl. Biochem. 31(Pt. 2):135-152. [DOI] [PubMed] [Google Scholar]
- 25.Saraswat, V., and V. S. Bisaria. 1997. Biosynthesis of xylanolytic and xylan-debranching enzymes in Melanocarpus albomyces IIS 68. J. Ferment. Bioeng. 83:352-357. [Google Scholar]
- 26.Selinger, L. B., C. W. Forsberg, and K. J. Cheng. 1996. The rumen: a unique source of enzymes for enhancing livestock production. Anaerobe 2:263-284. [DOI] [PubMed] [Google Scholar]
- 27.Sunna, A., M. D. Gibbs, and P. L. Bergquist. 2000. A novel thermostable multidomain 1,4-beta-xylanase from ‘Caldibacillus cellulovorans’ and effect of its xylan-binding domain on enzyme activity. Microbiology 146(Pt. 11):2947-2955. [DOI] [PubMed] [Google Scholar]
- 28.van Peij, N. N., J. Brinkmann, M. Vrsanska, J. Visser, and L. H. de Graaff. 1997. Beta-xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur. J. Biochem. 245:164-173. [DOI] [PubMed] [Google Scholar]
- 29.Whitehead, T. R., and R. B. Hespell. 1990. The genes for three xylan-degrading activities from Bacteroides ovatus are clustered in a 3.8-kilobase region. J. Bacteriol. 172:2408-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yun, J., S. Kang, S. Park, H. Yoon, M. J. Kim, S. Heu, and S. Ryu. 2004. Characterization of a novel amylolytic enzyme encoded by a gene from a soil-derived metagenomic library. Appl. Environ. Microbiol. 70:7229-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu, Y. X., J. Q. Wang, R. L. Ma, L. Huang, and Z. Y. Dong. 2007. Construction and analysis of rumen bacterial artificial chromosome library from a dairy cow rumen microflora. Wei Sheng Wu Xue Bao 47:213-216. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.