Abstract
The acclimatization of methanogens to two-phase olive mill wastes (TPOMW) was investigated in pilot fermenters started up with cattle excreta (37°C) and after changing their feed to excreta plus TPOMW (37°C or 55°C) or TPOMW alone (37°C) until a steady state was reached (28 days). Methanogenic diversity was screened using a phylogenetic microarray (AnaeroChip), and positive targets were quantified by real-time PCR. Results revealed high phylogenetic richness, with representatives of three out of the four taxonomic orders found in digesters. Methanosarcina dominated in the starting excreta (>96% of total 16S rRNA gene copies; over 45 times more abundant than any other methanogen) at high acetate (0.21 g liter−1) and ammonia N concentrations (1.3 g liter−1). Codigestion at 37°C induced a 6-fold increase of Methanosarcina numbers, correlated with CH4 production (rPearson = 0.94; P = 0.02). At 55°C, the rise in temperature and H2 partial pressure induced a burst of Methanobacterium, Methanoculleus, Methanothermobacter, and a group of uncultured archaea. The digestion of excreta alone resulted in low but constant biogas production despite certain oscillations in the methanogenic biomass. Unsuccessful digestion of TPOMW alone was attributed to high Cu levels inducing inhibition of methanogenic activity. In conclusion, the versatile Methanosarcina immediately adapted to the shift from excreta to excreta plus TPOMW and was responsible for the stimulated CH4 production at 37°C. Higher temperatures (55°C) fostered methanogenic diversity by promoting some H2 scavengers while yielding the highest CH4 production. Further testing is needed to find out whether there is a link between increased methanogenic diversity and reactor productivity.
Turning residues into energy is a societal and scientific priority due to climate change, fossil fuel exhaustion, and waste accumulation. In 2006, in Europe (EU27), less than 3% of electricity production came from biomass and wastes (11). Biogas plants, which anaerobically treat organic wastes to produce energy, are increasingly promoted in Europe, but their distribution is highly biased (35). While thousands of full- and farm-scale biogas plants are spread over central and northern Europe, anaerobic digestion technology in Mediterranean countries—Portugal, Spain, Italy, Greece, and Turkey—is in its early stages (35). These nations and other circum-Mediterranean countries lead in the production of olive oil and thus in olive mill wastes and wastewaters, which have a huge biogas production potential due to their lipid composition (1). Spain alone generates one-third of the world's oil production and millions of tons of two-phase olive mill wastes (TPOMW) per year. TPOMW are mostly burned or composted (28), hence releasing methane into the atmosphere. This compels a change in strategy: methane production from TPOMW should be optimized in engineered environments and transformed into energy.
TPOMW is a humid residue containing the olive pulp and stone. Its anaerobic digestibility is hampered by its low pH, low ammonia N, and high content in antimicrobial substances (1). However, it has been successfully fermented under laboratory conditions by supplementing it with nutrients and increasing the reactor organic loading rate stepwise (2) or by codigesting it with residues with a high buffering capacity, e.g., cattle excreta (17). These approaches seem to facilitate the adaptation of the methane-producing anaerobic community to the environmental conditions that TPOMW impose.
Methanogenic archaea—microbes clustered within five orders of the Euryarchaeota—constitute the last step in the trophic chain of decomposers degrading organic matter in oxygen-free environments (36). Methanogenesis is often the rate-limiting step of anaerobic digestion of organic wastes (3) due to the fast duplication times of bacteria, which generate all substrates for the slow-growing methane-producing archaea. It is also the most sensitive step in processing imbalances (4), likely due to the lack of functional redundancy among methanogens (8). High concentrations of volatile fatty acids, salts, ammonia, and heavy metals can be inhibitory for methanogens (5, 22) and are the most common reasons for reactor failure (3). Our objective was to understand the adaptation of methanogenic communities to TPOMW. We investigated methanogenic diversity and abundance in pilot digesters fed with cattle excreta and after changing their feed to TPOMW or TPOMW plus excreta. We expected that mixing both residues would allow a faster adaptation and more efficient performance of the methanogenic communities in digesting TPOMW. The cofermentation was evaluated at 37°C and 55°C. During an acclimatization period of 28 days, we screened the methanogenic diversity using an in-house-devised phylogenetic microarray, the AnaeroChip (13), and quantified dominant genera by real-time quantitative PCR (qPCR). We have taken primers from the literature, and we present four new sets of genus-specific primers and SYBR green I-optimized assays for quantifying methanogens in anaerobic environments.
MATERIALS AND METHODS
Reactor operation and performance.
Four 75-liter (working volume) anaerobic continuously stirred tank reactors were completely loaded with cattle excreta (mixed excrements and urine), progressively heated up to 37°C, and fed daily with 3.5 liters day−1 of cattle excreta, corresponding to a hydraulic retention time (HRT) of 21.4 days. Similar steady conditions were achieved in all reactors, that is, stable biogas production of 241 ± 18 ml biogas liter−1 sludge day−1 with 68.7% ± 0.4% (vol/vol) CH4 for three consecutive days and similar physical and chemical properties of the effluent sludge (average data are given in Table 1). This initial step was performed in order to set up common starting conditions for all reactors and prove reproducibility of reactor performance under equal operating conditions. On day 4, cattle excreta were replaced with the following specific feed solutions at a flow rate of 3.5 liter day−1. Reactors EX_OL37 and EX_OL55 were fed with a mixture of 3:1 excreta/TPOMW and operated at 37°C or 55°C. Both showed an immediate and sharp increase in CH4 production that stabilized as soon as 4 and 14 days, respectively, after changing of the feed. Biogas production averaged 1,096 and 1,289 ml biogas liter−1 sludge day−1 at 37°C and 55°C after the complete turnover of the reactor load (21.4 days). The other two reactors were loaded with either cattle excreta/water (EX37) or water/TPOMW (OL37) at a ratio of 3:1 and operated at 37°C to assess the contribution of each residue to the mesophilic codigestion. Reactor EX37 performed constantly, and after one HRT, it produced 251 ml biogas liter−1 sludge day−1. Reactor OL37 acidified, its volatile fatty acid/alkalinity ratio exceeding 0.8 after 21 days, and CH4 production ceased completely on day 30. A detailed analysis of all operational parameters can be found elsewhere (17).
TABLE 1.
Physical and chemical sludge properties in influent and effluent solutions and gas quality parameters in all reactors under initial steady-state conditions and after 1 HRTa
| Parameterb | Initial value |
Value after 1 HRT for reactor: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EX_OL37 |
EX_OL55 |
EX37 |
OL37 |
||||||||||||
| IN | EF | Gas quality | IN | EF | Gas quality | IN | EF | Gas quality | IN | EF | Gas quality | IN | EF | Gas quality | |
| TS (% [wt/vol]) | 4.7 | 4.0 (0.1) | 11.0 | 5.6 (0.4) | 11.0 | 5.3 (0.4) | 3.6 | 3.2 (0.0) | 7.5 | 3.7 (0.1) | |||||
| VS (% [wt/wt] of TS) | 71.1 | 66.5 (0.8) | 75.1 | 74.7 (1.0) | 75.1 | 72.3 (0.8) | 71.1 | 63.8 (0.4) | 21.8 | 71.4 (2.1) | |||||
| Particulate COD (g liter−1) | 25.5 | 20.0 (0.4) | 74.9 | 28.8 (0.9) | 74.9 | 26.1 (0.2) | 19.2 | 16.5 (0.05) | 52.7 | 23.0 (0.3) | |||||
| Soluble COD (g liter−1) | 14.7 | 7.0 (0.7) | 43.6 | 12.3 (0.8) | 43.6 | 19.0 (1.7) | 11.0 | 5.7 (0.4) | 28.4 | 13.7 (0.1) | |||||
| pH | 7.4 | 7.80 (0.02) | 6.7 | 7.67 (0.04) | 6.7 | 7.85 (0.06) | 7.4 | 7.60 (0.06) | 4.9 | 7.20 (0.06) | |||||
| EC (mS cm−1) | 17.2 | 16.4 (0.8) | 14.4 | 16.0 (0.1) | 14.4 | 16.6 (0.1) | 17.2 | 15.1 (0.2) | 4.1 | 12.2 (0.4) | |||||
| Acetate (g liter−1) | 3.34 | 0.21 (0.01) | 0.21 (0.09) | 0.20 (0.03) | 0.03 (0.01) | 2.70 (0.4) | |||||||||
| Propionate (g liter−1) | 0.64 | BDLc | 0.55 (0.03) | 0.60 (0.14) | BDL | 1.21 (0.12) | |||||||||
| NH4-N (g liter−1) | 1.20 | 1.32 (0.04) | 0.83 | 0.88 (0.01) | 0.83 | 1.02 (0.01) | 0.90 | 1.07 (0.03) | BDL | 0.58 (0.02) | |||||
| Alkalinity (mmol H+ eq liter−1) | 166 | 188.7 (6.4) | 124 | 188.6 (2.4) | 124 | 191.6 (1.4) | 125 | 172.8 (2.4) | 10 | 110.3 (10.3) | |||||
| CH4 (liters day−1) | 12.9 (0.7) | 54.3 (4.1) | 66.5 (7.5) | 12.9 (1.5) | 2.4 (2.6) | ||||||||||
| H2 (ml day−1) | 0.34 (0.07) | 4.26 (0.63) | 38.38 (7.67) | 0.27 (0.07) | 0.34 (0.55) | ||||||||||
IN, influent; EF, effluent; “Initial,” initial steady-state conditions (excreta at 37°C); one HRT = 21.4 days. Reactor conditions: for EX_OL37, 3:1 excreta/TPOMW at 37°C; for EX_OL55, 3:1 excreta/TPOMW at 55°C; for EX37, 3:1 excreta/water at 37°C; for OL37, 3:1 water/TPOMW at 37°C. “Initial” data are averages for all four reactors. Effluent sludge data after 1 HRT are averages for the last two sampling dates. CH4 and H2 data are averages of seven consecutive daily measurements. Standard deviations are given in parentheses.
TS, total solids; VS, volatile solids; COD, chemical oxygen demand; EC, electrical conductivity; NH4-N, ammonia nitrogen.
BDL, below detection limit.
Sampling of sludge, DNA extraction, and AnaeroChip analyses.
Three sludge samples (ca. 500 ml) were collected every 3.5 days from each reactor. One subsample was used for determination of physical and chemical parameters. Table 1 summarizes the physical and chemical properties of the influent and effluent sludge after one HRT. A second subsample was kept at −20°C until DNA extraction.
Sludge samples (1 ml) were submitted to three freeze-thaw cycles consisting of 30 min at −80°C and 5 min at 65°C. After centrifugation (5 min, 14,000 rpm), DNA was extracted from the pellet using the PowerSoil DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA). Sludge DNA from samples collected on days 4, 14, and 28 was subjected to PCR amplification of the 16S rRNA gene using the universal archaeal primers 109F and 934R (Table 2) to screen the methanogenic diversity in the start, middle, and end stages of the codigestion. PCR amplifications were performed in a Flexcycler instrument (Analytik Jena, Germany) in 50-μl volumes, with each reaction mixture containing a final concentration of 1× reaction buffer [16 mM (NH4)2SO4, 67 mM Tris-HCl {pH 8.8}, 1.5 mM MgCl2, 0.01% Tween 20], 200 μM each deoxynucleoside triphosphate (dNTP), 0.8 μM primer 109F with a Cy5 label in the 5′ end, 0.2 μM primer 934R with a PO4− group in the 5′ end, 0.1 mM MgCl2, 0.4 mg ml−1 bovine serum albumin (BSA), 1.25 U BioTherm DNA polymerase (GeneCraft, Germany), 10 mM tetramethylammonium chloride, and sterile water. Two μl DNA was directly applied to the reaction mix. Thermal cycling, preparation of single-stranded PCR products, array hybridization, washing, and scanning were performed as described in reference 13.
TABLE 2.
Primers used in this study
| Target | Primers | Sequences (5′-3′) | E. coli positions | Reference |
|---|---|---|---|---|
| Archaea | 109F | ACKGCTCAGTAACACGT | 109-125 | 18 |
| 934R | GTGCTCCCCCGCCAATTCCT | 915-934 | ||
| Methanosarcina | 240F | CCTATCAGGTAGTAGTGGGTGTAAT | 240-264 | 14 |
| 589R | CCCGGAGGACTGACCAAA | 589-606 | ||
| Methanosaeta | MS1b | CCGGCCGGATAAGTCTCTTGA | 585-606 | 30 |
| SAE835R | GACAACGGTCGCACCGTGGCC | 835-851 | ||
| Methanoculleus | 298F | GGAGCAAGAGCCCGGAGT | 298-315 | 14 |
| 586R | CCAAGAGACTTAACAACCCA | 586-606 | ||
| Methanothermobacter | 410F | CTCTTAACGGGGTGGCTTTT | 410-440 | 14 |
| 667R | CCCTGGGAGTACCTCCAGC | 667-686 | ||
| Uncultured group | 195F | AAAACTCCGGTGCCTTAGGATT | 195-230 | 14 |
| 330R | CCCGTAGGGCCTGGACTCA | 330-348 | ||
| Methanobrevibacter | 210F | TTTCGCCTAAGGATGGGTCT | 210-365 | This study |
| 367R | CGATTTCTCACATTGCGGAG | 367-386 | After 32a | |
| Methanobacterium | fMbium | CGTTCGTAGCCGGCYTGA | 576-593 | 32 |
| 748R | TTCGTTACTCACCGTCAGGT | 748-767 | This study | |
| Methanosphaera | 594F | TAAGTCTTTGGTGAAAGCTT | 594-613 | This study |
| 747R | GTTACTCACCGTCAAGAT | 747-764 | ||
| Methanocorpusculum | 193F | TCCTCGAAAGATCCGTC | 193-219 | This study |
| 488R | CTGCCCTTTCTTCACATA | 488-504 |
Modified from reference 32.
Real-time quantitative PCR.
Sludge DNA extracted from samples collected on days 4, 7, 14, 21, and 28 was subjected to real-time quantitative PCR (qPCR) amplification with the genus-specific primers in Table 2. Selection of target organisms was based on positive AnaeroChip results. Primers were either obtained from the literature or designed using the ARB software package (24) as described in reference 13. Standard curves were constructed with PCR-amplified 16S rRNA sequences from pure cultures or environmental clones using the group-specific primers in Table 2 and the source DNA in Table 3. Endpoint PCR amplifications were performed in 25-μl volumes as above with slight modifications (0.2 μM each primer, 0.63 U BioTherm DNA polymerase [GeneCraft, Germany], 1× enhancer [Peqlab, Germany], and no extra MgCl2 added). One μl DNA was applied directly to the PCR mix. Thermal cycling was initiated with 5 min at 95°C, followed by 30 cycles consisting of 45 s at 95°C, 45 s at 51 to 60°C (see specific annealing temperature for endpoint PCR in Table 3), and 45 s at 72°C and terminated with 10 min at 72°C. PCR products were purified (NucleoSpin kit extract II; Macherey-Nagel, Germany), eluted with prewarmed 1× DNase-free Tris-EDTA (TE) buffer in low-DNA binding tubes (Biozym, Austria), quantified in triplicate with the Quant-iT PicoGreen double-stranded DNA kit (Invitrogen, Carlsbad) using low-autofluorescent-microtiter plates (Nunc, VWR International, Austria), and stored at −20°C. The total number of copies of the 16S rRNA gene in the stock DNA was calculated from the length of the amplified fragment and the DNA concentration, assuming that the average weight of a base pair is 650 Da.
TABLE 3.
Source DNA, annealing temperature, and standard curve parameters used for real-time PCR quantification of several groups of methanogens
| Target | Source DNAa | AT, endpoint PCR (°C)b | AT, real-time PCR (°C)b | Standards (gene copies μl−1)c | Slope | Intercept | r2 value |
|---|---|---|---|---|---|---|---|
| Methanosarcina | M. barkeri (DSM 800) | 51 | 64 | 108-104 | −4.12 | 36.50 | 0.998 |
| Methanosaeta | M. concilii (DSM 2139) | 60 | 60 | 104-10 | −3.79 | 31.87 | 0.996 |
| Methanoculleus | Clone F_2FA36 (AM947508) | 58 | 65 | 105-10 | −3.85 | 34.58 | 0.994 |
| Methanothermobacter | M. wolfeii (DSM 2970) | 51 | 61 | 104-1 | −3.58 | 31.86 | 0.996 |
| Uncultured group | Clone F_1FA28 (AM947509) | 58 | 64 | 104-1 | −4.13 | 38.43 | 0.994 |
| Methanobrevibacter | M. smithii (DSM 861) | 54 | 59 | 105-102 | −3.77 | 33.39 | 0.999 |
| Methanobacterium | M. formicicum (DSM 1535) | 54 | 58 | 104-10 | −3.66 | 37.32 | 0.993 |
| Methanosphaera | M. stadtmanae (DSM 3091) | 56 | 61 | 105-10 | −3.63 | 31.58 | 0.994 |
| Methanocorpusculum | M. bavaricum (DSM 4179) | 56 | 58 | 106-102 | −4.02 | 37.72 | 0.999 |
All reference strains from Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). Environmental clones are described in reference 16.
AT, annealing temperature.
Range of standard solutions used for quantification by real-time PCR.
Real-time PCR amplifications were conducted using a Rotor-Gene 6000 system (Corbett Life Sciences, Australia) in 20-μl volumes, with each standard reaction mix containing 1× Quantimix Easy SYG kit (Biotools, Spain; 0.8 × for Methanosphaera), 100 nM each primer (150 nM for Methanoculleus and Methanobacterium and 200 nM for Methanosphaera), 0.4 mg ml−1 BSA, and distilled water. Two μl sludge DNA was applied to the PCR mix. Thermocycling was as follows: 95°C for 5 min, 40 cycles of 20 s at 95°C, 20 s at 58 to 65°C (see specific annealing temperature for real-time PCR in Table 3), and 20 s at 72°C. Runs were completed with a melting analysis (65°C to 95°C; ramp, 0.5°C/min) to check for product specificity and primer dimer formation. Each assay included five standard solutions that were freshly prepared from the stock (Table 3). All standards and samples were run in duplicate.
Shannon's diversity index (H′) and evenness (E) were calculated using the equations H′ = −Σ (pi × ln pi) and E = H′/log S (25), where pi denotes the ratio of the corrected 16S rRNA gene copy number of each methanogenic genus to the sum of gene copies of all quantified methanogens, and S denotes the number of methanogenic genera detected at a percentage greater than 0.01. Univariate analysis of variance (ANOVA) was used to test for differences in diversity (Shannon's index) among reactors at the start (day 4) and end (day 28) of the experiment with the SPSS software program, v. 17.0.
Microarray data accession number.
The array design, protocol, and experimental data can be accessed at Array Express (http://www.ebi.ac.uk/arrayexpress/; E-MEXP-2453).
RESULTS AND DISCUSSION
Screening of methanogenic diversity.
Sludge DNA extracted from samples taken from all four reactors on days 4, 14, and 28 was used for hybridization with the AnaeroChip microarray, an oligonucleotide microarray targeting methanogenic genera clustered within Methanomicrobiales, Methanosarcinales, Methanobacteriales, and Methanococcales, which thrive in digesters (13, 20).
Based on the fluorescence intensity data, the signal-to-noise ratio (SNR) was calculated for all probes, and those with an SNR of ≥2 in one or more samples were used for subsequent analyses (see Table S1 in the supplemental material). Microarray data are semiquantitative due to the use of PCR-amplified templates that overrepresent the dominant microbes and the inherent differences in hybridization efficiencies among oligonucleotide probes (29, 37). Still, the SNR values provide an estimate of a target's abundance (7). The facultative acetoclastic Methanosarcina genus dominated all reactors. All six probes targeting this genus fluoresced in most samples, with SNRs up to 271. No clear differences in fluorescence intensity were detected among either samples or sampling dates. Other genera detected with the AnaeroChip microarray in the samples were as follows: the strict acetotrophic genera Methanosaeta, the hydrogenotrophic genera Methanobrevibacter, Methanobacterium, Methanoculleus, Methanothermobacter, Methanocorpusculum, Methanolobus, Methanomicrobium, Methanocalculus, and Methanospirillum, the methanol-consuming genus Methanosphaera, and a group of uncultured archaea. These results indicate a metabolically diverse methanogenic community comprised of organisms capable of methane formation through all known biochemical pathways. Our results also point out an extremely uneven methanogenic community completely dominated by a single genus but highly rich in phylogenetic terms, with representatives of three out of the four taxonomic orders which are commonly found in digesters (20).
Dominance of Methanosarcina has been reported before for many anaerobic environments (23), including manure-digesting reactors (9). This is thought to be due to their tolerance for substances which can be inhibitory to other methanogens at high concentrations, such as total ammonia N, which averaged 1.32 ± 0.04 g liter−1 at the start of our experiment (while all reactors were operated with excreta). Ammonia is presumed to inhibit methanogenesis through diffusion into the cells and subsequent induction of an intracellular pH rise or enzyme inhibition (3). The higher volume-to-surface ratio of the coccoid Methanosarcina and its growth in clusters might result in comparatively lower ammonia diffusion per unit mass and thus increased tolerance to high ammonia levels (5). On the other hand, the relatively high acetate concentration, averaging 212 ± 10 mg liter−1 in the reactors at the start of the experiment, would explain Methanosarcina's ability to outcompete Methanosaeta (39). The former is known to grow at faster specific rates (38) and to prevail at high acetate concentrations (33).
A previous report of methanogenic diversity in TPOMW-fed mesophilic reactors indicated the dominance of Methanosaeta (27). This discrepancy might be due to differences in the following: (i) the start-up strategy, since we initially loaded all reactors with excreta whereas Rincón and coworkers (27) used inocula from an olive mill wastewater-digesting reactor plus a nutrient-trace element solution; (ii) organic loading rates, which were rather low in the work described in reference 27 (0.75 to 3 g chemical oxygen demand [COD] liter−1 day −1), allowing total volatile fatty acid (VFA) levels to be kept at ≤440 mg liter−1. This contrasts with the 3.8- to 5.5-g COD liter−1 day−1 fed in our digesters, which determined total VFA concentrations between 535 and 3760 mg liter−1 in the reactors containing TPOMW (either alone or mixed with excreta) after the period corresponding to one HRT.
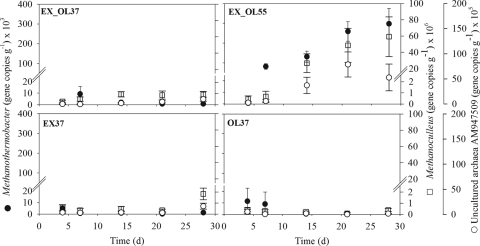
Finally, the codigestion of excreta and TPOMW at 55°C (reactor EX_OL55) induced an increase in Methanoculleus abundance, as indicated by the consistently higher SNR values of the probes Mcul574 and Mcul393 on days 14 and 28 (see Table S1 in the supplemental material).
Quantification of methanogenic genera detected with the AnaeroChip microarray.
All methanogenic genera found to be more abundant and ubiquitous in the samples according to the AnaeroChip data were targeted with real-time quantitative PCR. Primers designed in this study (Table 2) were specific for their targets after amplification with endpoint and real-time PCR. Prior to the quantification assays, PCR efficiency reduction due to the presence of inhibitors in the sludge matrix was tested by serially diluting the DNA extracts and spiking the standard curves with 2 μl sludge DNA or purified sludge DNA. No differences in amplification efficiencies compared to those for unspiked standards were found (data not shown).
Table 3 summarizes the standard curve parameters (slope, intercept, and r2) developed for all quantification assays. Figures 1 to 4 show the number of 16S rRNA gene copies per gram of sludge, after correction of amplification efficiency, detected in the four reactors on days 4, 7, 14, 21, and 28 for all the methanogenic genera quantified. As expected from the microarray data, Methanosarcina was the most abundant methanogen in all reactors (Fig. 1), with its gene copy numbers representing between 96.2 and 97.5% of the total in the initial methanogenic community (day 4). This was followed by Methanobrevibacter (1.4 to 2.1%), Methanocorpusculum (0.4 to 1.4%), and Methanosaeta (0.3 to 0.6%). All other genera were found to represent <0.2% of all methanogens quantified. However, these figures must be interpreted with caution due to uncertainties in the number of rnn operons in methanogens and differences in amplification efficiency resulting both from the inherent differences among primer pairs and the various abundances of the target organisms, although this is partly solved by data correction.
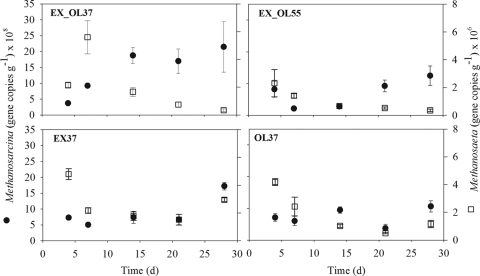
FIG. 1.
16S rRNA gene copy numbers of Methanosarcina and Methanosaeta in reactors EX_OL37 (3:1 excreta/TPOMW at 37°C), EX_OL55 (3:1 excreta/TPOMW at 55°C), EX37 (3:1 excreta/water at 37°C), and OL37 (3:1 water/TPOMW at 37°C). Bars represent standard errors (n = 3).
The change from excreta to excreta plus TPOMW in the mesophilic reactor (EX_OL37) invoked an immediate increase in the 16S rRNA gene copy numbers of Methanosarcina (Fig. 1), which was strongly correlated with the rise in CH4 production (Pearson′s coefficient = 0.94; P = 0.02). The abundance of Methanosarcina in reactor EX_OL37 was correlated with that of total and volatile solids, COD, and with the decrease in ammonia N (for all variables, Pearson's coefficient was ≥|0.92|; P ≤ 0.03). No correlation was found between Methanosarcina 16S rRNA gene copy numbers and acetate or H2, which are two major substrates for most of the species of the genus (23), suggesting that substrates did not limit growth at any stage. Therefore, Methanosarcina easily adapted to the new environmental conditions that resulted from changing the feed from excreta to excreta-TPOMW at 37°C and was responsible for the enhanced methane production because of the codigestion. Methanosarcina's competitive advantage under conditions of system instability has been reported before (26).
The codigestion of excreta and TPOMW at 55°C (EX_OL55) provoked an initial increase of volatile fatty acids, mainly acetate, which averaged 0.21 g liter−1 on day 4 and peaked at 4 g liter−1 on day 7, indicating an imbalance of the microbial guilds involved in anaerobic digestion. This was coincident with a decrease in the gene copy numbers of Methanosarcina (Fig. 1). From day 14 on, concurrent with acetate stabilization to a final concentration of 0.18 g liter−1 on day 28, Methanosarcina grew again in the reactor. Presumably, Methanosarcina thermophila, which is the single thermophilic species known in the genus (15), adapted to the new conditions. This organism has been found to be present in low numbers in mesophilic reactors (39) and to codominate a thermophilic cattle manure-fed reactor together with Methanoculleus thermophilicus (6).
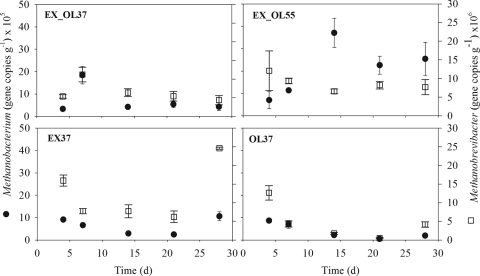
In the same reactor (EX_OL55), setting the temperature at 55°C increased the H2 production 34 times in 3 days, from 0.36 ml day−1 (day 3) to 12.3 ml day−1 (day 6). This increase in the H2 concentration with temperature is consistent with previous observations of methanogenic environments and has been suggested to respond to thermodynamic control (19). From day 6 on, H2 production in EX_OL55 continued to rise progressively, peaking at 42.7 ml day−1 on day 15, which is a 3.5-fold increase during the eight subsequent days. The H2 production in the thermophilic reactor after one HRT (21.4 days) averaged 38.4 ml day−1, multiplying by a factor ranging from 9 to 143 the data corresponding to the three mesophilic reactors (Table 1). The evolution of H2 partial pressure with the fermentation was mirrored in that of the gene copy numbers of several genera of hydrogenotrophic methanogens, which include thermophilic species (15), mostly Methanobacterium, Methanoculleus, and Methanothermobacter, which rose by 1 to 2 orders of magnitude (Fig. 2 and 3). From day 14 on, Methanoculleus became the second most abundant methanogen in the sludge, confirming the AnaeroChip results (see Table S1 in the supplemental material). The immediate growth of these H2 scavengers was probably crucial for equilibrium reestablishment, since an uncontrolled increase in H2 partial pressure might have led otherwise to reactor acidification and instability. Furthermore, a group of uncultured archaea (AM947509), which was found to be abundant in a biowaste-digesting thermophilic full-scale reactor (16), was present and grew rapidly when the temperature had risen to 55°C (Fig. 3). Therefore, this group of uncultured organisms most likely includes hydrogenotrophic thermophilic methanogens. Strong correlations were detected between the mentioned hydrogenotrophic microbes and both reactor H2 partial pressure and temperature (Pearson's coefficient ≥ 0.7; P ≤ 0.001). Previous surveys have also suggested methanogenesis from H2/CO2 under thermophilic conditions in reactors (9, 16) and soils (12). However, a higher H2 partial pressure does not necessarily imply that more energy is available for the hydrogenotrophic methanogens (10). Finally, the evolution of the abundance of Methanoculleus, Methanothermobacter, and the uncultured group in reactor EX_OL55 was highly and positively correlated to the concentration of propionate (Pearson's coefficient ≥ 0.89; P < 0.05). This suggests the involvement of the mentioned H2 scavengers as syntrophic partners of propionate oxidizers, an interaction that is widely known to energetically facilitate propionate degradation (34). Obviously, evidences in this respect would require researching the bacterial community and carrying out experiments to establish the pathways for methanogenesis.
FIG. 2.
16S rRNA gene copy numbers of Methanobacterium and Methanobrevibacter in reactors EX_OL37 (3:1 excreta/TPOMW at 37°C), EX_OL55 (3:1 excreta/TPOMW at 55°C), EX37 (3:1 excreta/water at 37°C), and OL37 (3:1 water/TPOMW at 37°C). Bars represent standard errors (n = 3).
FIG. 3.
16S rRNA gene copy numbers of Methanoculleus, Methanothermobacter, and a group of uncultured archaea (AM947509) in reactors EX_OL37 (3:1 excreta/TPOMW at 37°C), EX_OL55 (3:1 excreta/TPOMW at 55°C), EX37 (3:1 excreta/water at 37°C), and OL37 (3:1 water/TPOMW at 37°C). Bars represent standard errors (n = 3).
All reactors had a similar diversity of quantified methanogens at the start of the experiment (F = 2.32; P = 0.162), but H′ was significantly different after 28 days (F = 30.46; P < 0.001). The reduced abundance of the dominant Methanosarcina genus in the reactor run at 55°C (EX_OL55) and the increased levels of several groups of hydrogenotrophs account for the doubling of evenness and hence diversity of the methanogenic community (Table 4). Karakashev et al. observed that methanogenic communities examined by fluorescence in situ hybridization with family-specific probes were generally richer in digesters operated at 37 to 38°C than in those operated at 51 to 55°C (21). This led them to conclude that mesophilic reactors bear more-diverse methanogenic communities. Our data demonstrate that the shift from 37°C to 55°C can increase the diversity of a highly uneven methanogenic community by imposing growth conditions that are both suboptimal for the dominant population and beneficial for other, less-frequent populations. This might theoretically result in a more efficient use of the resources and thus enhanced system productivity.
TABLE 4.
Richness, evenness, and diversity of methanogens in all reactors at the start (day 4) and end (day 28) of the experimenta
| Reactor | Time (days) | S | E | H′ |
|---|---|---|---|---|
| EX_OL37 | 4 | 8.0 (0.0) | 0.23 (0.04) | 0.20 (0.04) |
| 28 | 7.3 (0.6) | 0.14 (0.01) | 0.12 (0.01) | |
| EX_OL55 | 4 | 8.0 (0.0) | 0.15 (0.03) | 0.14 (0.03) |
| 28 | 9.0 (0.0) | 0.38 (0.04) | 0.36 (0.04) | |
| EX37 | 4 | 8.0 (0.0) | 0.23 (0.04) | 0.21 (0.03) |
| 28 | 8.0 (0.0) | 0.33 (0.03) | 0.30 (0.03) | |
| OL37 | 4 | 8.0 (0.0) | 0.22 (0.06) | 0.20 (0.05) |
| 28 | 8.0 (0.0) | 0.19 (0.05) | 0.17 (0.04) |
Data are averages (n = 3) (standard deviations are given in parentheses). S, richness; E, evenness; H′, diversity.
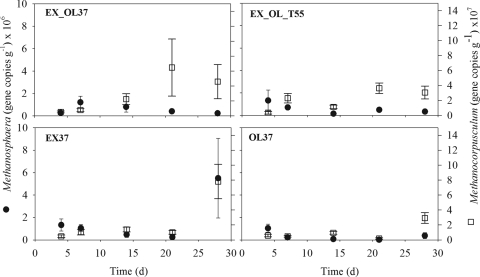
The reactor fed exclusively with excreta had a stable performance in terms of biogas production. It showed small oscillations in the numbers of 16S rRNA gene copies for the methanogenic genera quantified, with no clear temporal trends (EX37 in Fig. 1 to 4). This could simply be a response to various numbers of methanogens being inoculated with the excreta day by day. The magnitude of these differences was not enough to affect the performance of the reactor, which had a constant biogas production throughout the experiment (17). Also, Silvey et al. reported fluctuations in archaeal biomass, estimated from the levels of phospholipid fatty acids, but constant methane production for 14 days of mesophilic digestion of municipal solid waste (31).
FIG. 4.
16S rRNA gene copy numbers of Methanosphaera and Methanocorpusculum in reactors EX_OL37 (3:1 excreta/TPOMW at 37°C), EX_OL55 (3:1 excreta/TPOMW at 55°C), EX37 (3:1 excreta/water at 37°C), and OL37 (3:1 water/TPOMW at 37°C). Bars represent standard errors (n = 3).
More overwhelming was the lack of noticeable variability in methanogenic gene copy numbers in the reactor fed with TPOMW (OL37 in Fig. 1 to 4) despite the complete cessation of biogas production after 30 days. We initially hypothesized that methanogenic biomass could have been washed out from the reactor due to the shift from the initial load (75 liters of cattle excreta) to its content after one HRT (18.75 liters of TPOMW plus 56.26 liters of tap water). However, real-time PCR results demonstrated that the amount of methanogenic biomass in reactor OL37 was comparable to that of the other reactors. Therefore, we concluded that methanogens were present in sufficient numbers but their activity stopped. All necessary nutrients were present in the reactor, and 3 times higher copper availability (192 ppm; C. Mondini, personal communication) under the more acidic conditions imposed by TPOMW was hypothesized to be the cause of methanogenesis inhibition (22).
Stepwise forward regression was used to search for variables able to predict methane production in the reactors. The linear combination of four variables (log-transformed 16S rRNA gene copy numbers of Methanoculleus, Methanosarcina, Methanosphaera, and Methanobacterium) significantly (F = 19.91; P < 0.001) predicted 84.2% of CH4 production. All other methanogenic genera quantified in the reactors were excluded from the model, since they did not increase its ability to explain CH4 production.
Conclusions.
A phylogenetically and metabolically rich methanogenic community that stably digested cattle excreta rapidly adapted to the change in feed to excreta plus TPOMW both at 37°C and 55°C but not to the digestion of TPOMW alone. The thermophilic codigestion of the residues was the most productive and held the most diverse methanogenic communities. This is consistent with the theoretical expectation of a more efficient exploitation of resources due to a better occupation of available niches. Linking of the system's diversity and functioning would require further investigating other microbial guilds involved in the complex food web of anaerobic decomposers and experimentally defining the pathways used for methane formation.
Supplementary Material
Acknowledgments
M. Goberna was supported by the Marie Curie Actions (MEIF-CT-2006-041034).
Olive mill waste was obtained from the agricultural cooperative COATO in Totana (Murcia, Spain); thanks go to M. A. Sánchez Monedero, A. Roig, and C. Mondini. Cattle excreta were obtained from the agricultural school in Rotholz (Tirol, Austria). We thank Claudio Mondini for providing nutrient and heavy metal contents of the digestion end products, G. González Barberá for his comments on data analysis, M. Schoen, D. Sperl, and P. Engele for their help in the laboratory, and the K-Regio Centre BIOTREAT, funded by the Tiroler Zukunftsstiftung, for its support. We are grateful to four anonymous reviewers who contributed to improve the final version of the manuscript.
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Angelidaki, I., and B. K. Ahring. 1997. Codigestion of olive oil mill wastewaters with manure, household waste or sewage sludge. Biodegradation 8:221-226. [Google Scholar]
- 2.Borja, R., B. Rincón, F. Raposo, J. Alba, and A. Martín. 2002. A study of anaerobic digestibility of two-phase olive mill solid waste (OMSW) at mesophilic temperature. Process Biochem. 38:733-742. [Google Scholar]
- 3.Braun, R., B. Drosg, G. Bochmann, S. Weiß, and R. Kirchmayr. 2010. Recent developments in bioenergy recovery through fermentation, p. 35-58. In H. Insam, I. H. Franke-Whittle, and M. Goberna (ed.), Microbes at work. From wastes to resources. Springer, Berlin, Germany.
- 4.Briones, A., and L. Raskin. 2003. Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr. Opin. Biotechnol. 14:270-276. [DOI] [PubMed] [Google Scholar]
- 5.Calli, B., B. Mertoglu, B. Inanc, and O. Yenigun. 2005. Community changes during start-up in methanogenic bioreactors exposed to increasing levels of ammonia. Environ. Technol. 26:85-91. [DOI] [PubMed] [Google Scholar]
- 6.Chachkhiani, M., P. Dabert, T. Abzianidze, G. Partskhaladze, L. Tsiklauri, and T. G. J. J. Dudauri. 2004. 16S rDNA characterisation of bacterial and archaeal communities during start-up of anaerobic thermophilic digestion of cattle manure. Bioresour. Technol. 93:227-232. [DOI] [PubMed] [Google Scholar]
- 7.Cho, J.-C., and J. M. Tiedje. 2002. Quantitative detection of microbial genes by using DNA microarrays. Appl. Environ. Microbiol. 68:1425-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis, T. P., and W. T. Sloan. 2004. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 7:221-226. [DOI] [PubMed] [Google Scholar]
- 9.Demirel, B., and P. Scherer. 2008. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion biomass to methane: a review. Rev. Environ. Sci. Biotechnol. 7:173-190. [Google Scholar]
- 10.Dolfing, J., S. R. Larter, and I. M. Head. 2008. Thermodynamic constraints on methanogenic crude oil biodegradation. ISME J. 2:442-452. [DOI] [PubMed] [Google Scholar]
- 11.Eurostat. 2009. Electricity generated from renewable sources. http://epp.eurostat.ec.europa.eu/tgm/table.do?tab=table&init=1&plugin=1&language=en&pcode=tsdcc330. Eurostat, Luxembourg, Luxembourg. Accessed 17 September 2009.
- 12.Fey, A., K. J. Chin, and R. Conrad. 2001. Thermophilic methanogens in rice field soil. Environ. Microbiol. 3:295-303. [DOI] [PubMed] [Google Scholar]
- 13.Franke-Whittle, I. H., M. Goberna, V. Pfister, and H. Insam. 2009. Design and testing of the ANAEROCHIP microarray for investigation of methanogenic communities. J. Microbiol. Methods 79:279-288. [DOI] [PubMed] [Google Scholar]
- 14.Franke-Whittle, I. H., M. Goberna, and H. Insam. 2009. Design, testing and application of real-time PCR primers for the detection of Methanoculleus, Methanosarcina, Methanothermobacter and uncultured methanogens. Can. J. Microbiol. 55:611-616. [DOI] [PubMed] [Google Scholar]
- 15.García, J. L., B. K. C. Patel, and B. Ollivier. 2000. Taxonomic, phylogenetic and ecological diversity of methanogenic Archaea. Anaerobe 6:205-226. [DOI] [PubMed] [Google Scholar]
- 16.Goberna, M., H. Insam, and I. H. Franke-Whittle. 2009. Effect of biowaste sludge maturation on the diversity of thermophilic bacteria and archaea in an anaerobic reactor. Appl. Environ. Microbiol. 75:2566-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goberna, M., M. A. Schoen, D. Sperl, B. Wett, and H. Insam. 2010. Mesophilic and thermophilic cofermentation of cattle excreta and olive mill wastes. Biomass Bioenerg. 34:340-346. [Google Scholar]
- 18.Grosskopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoehler, T. M., M. J. Alperin, D. B. Albert, and C. S. Martens. 1998. Thermodynamic control on hydrogen concentrations in anoxic sediments. Geochim. Cosmochim. Acta 62:1745-1756. [Google Scholar]
- 20.Insam, H., I. H. Franke-Whittle, and M. Goberna. 2010. Microbes in aerobic and anaerobic waste treatment, p. 1-34. In H. Insam, I. H. Franke-Whittle, and M. Goberna (ed.), Microbes at work. From wastes to resources. Springer, Berlin, Germany.
- 21.Karakashev, D., D. J. Batstone, and I. Angelidaki. 2005. Influence of environmental conditions on methanogenic compositions in anaerobic biogas reactors. Appl. Environ. Microbiol. 71:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karri, S., R. Sierra-Alvarez, and J. A. Field. 2006. Toxicity of copper to acetoclastic and hydrogenotrophic activities of methanogens and sulfate reducers in anaerobic sludge. Chemosphere 62:121-127. [DOI] [PubMed] [Google Scholar]
- 23.Kendall, M. M., and D. R. Boone. 2006. The order Methanosarcinales, p. 244-256. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. A handbook on the biology of bacteria, vol. 3. Springer, New York, NY. [Google Scholar]
- 24.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magurran, A. E. 1989. Ecological diversity and its measurement. Vedrà, Barcelona, Spain.
- 26.McMahon, K. D., D. Zheng, A. J. M. Stams, R. I. Mackie, and L. Raskin. 2004. Microbial population dynamics during start-up and overload conditions of anaerobic digesters treating municipal solid waste and sewage sludge. Biotechnol. Bioeng. 87:823-834. [DOI] [PubMed] [Google Scholar]
- 27.Rincón, B., F. Raposo, R. Borja, J. M. González, M. C. Portillo, and C. Saiz-Jiménez. 2006. Performance and microbial communities of a continuous stirred tank anaerobic reactor treating two-phase olive mill solid wastes at low organic loading rates. J. Biotechnol. 121:534-543. [DOI] [PubMed] [Google Scholar]
- 28.Roig, A., M. L. Cayuela, and M. A. Sánchez-Monedero. 2006. An overview on olive mill wastes and their valorisation methods. Waste Manag. 26:960-969. [DOI] [PubMed] [Google Scholar]
- 29.Schadt, C. W., and J. Zhou. 2006. Advances in microarray-based technologies for soil microbial community analyses, p. 189-203. In P. Nannipieri and K. Smalla (ed.), Nucleic acids and proteins in soil. Springer, Berlin, Germany.
- 30.Shigematsu, T., Y. Tang, H. Kawaguchi, K. Ninomiya, J. Kijima, T. Kobayashi, S. Morimura, and K. Kida. 2003. Effect of dilution rate on structure of a mesophilic acetate-degrading methanogenic community during continuous cultivation. J. Biosci. Bioeng. 96:547-558. [DOI] [PubMed] [Google Scholar]
- 31.Silvey, P., P. C. Pullammanappallil, and P. Nichols. 2000. Microbial ecology of the leach bed anaerobic digestion of unsorted municipal solid waste. Water Sci. Technol. 41:9-16. [PubMed] [Google Scholar]
- 32.Skillman, L. C., P. N. Evans, G. E. Naylor, B. Morvan, G. N. Jarvis, and K. N. Joblin. 2004. 16S ribosomal DNA-directed PCR primers for ruminal methanogens and identification of methanogens colonising young lambs. Anaerobe 10:277-285. [DOI] [PubMed] [Google Scholar]
- 33.Smith, K. S., and C. Ingram-Smith. 2007. Methanosaeta, the forgotten methanogen? Trends Microbiol. 15:150-155. [DOI] [PubMed] [Google Scholar]
- 34.Stams, A. J. M., and C. M. Plugge. 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7:568-577. [DOI] [PubMed] [Google Scholar]
- 35.Tabajdi, C. S. 2007. Draft report on sustainable agriculture and biogas: a need for review of EU-legislation (2007/2107 INI). Committee on Agriculture and Rural Development, European Parliament, Brussels, Belgium.
- 36.Thauer, R. K., A.-K. Kaster, H. Seedorf, W. Buckel, and R. Hedderich. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6:579-591. [DOI] [PubMed] [Google Scholar]
- 37.Wagner, M., H. Smidt, A. Loy, and J. Zhou. 2007. Unravelling microbial communities with DNA-microarrays: challenges and future directions. Microb. Ecol. 53:498-506. [DOI] [PubMed] [Google Scholar]
- 38.Yu, Y., J. Kim, and S. Hwang. 2006. Use of real-time PCR for group-specific quantification of aceticlastic methanogens in anaerobic processes: population dynamics and community structures. Biotechnol. Bioeng. 93:424-433. [DOI] [PubMed] [Google Scholar]
- 39.Zheng, D., and L. Raskin. 2000. Quantification of Methanosaeta species in anaerobic bioreactors using genus- and species-specific hybridization probes. Microb. Ecol. 39:246-262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.