Abstract
Several microorganisms have been isolated that can transform hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), a cyclic nitramine explosive. To better characterize the microbial genes that facilitate this transformation, we sequenced and annotated a 182-kb plasmid, pGKT2, from the RDX-degrading strain Gordonia sp. KTR9. This plasmid carries xplA, encoding a protein sharing up to 99% amino acid sequence identity with characterized RDX-degrading cytochromes P450. Other genes that cluster with xplA are predicted to encode a glutamine synthase-XplB fusion protein, a second cytochrome P450, Cyp151C, and XplR, a GntR-type regulator. Rhodococcus jostii RHA1 expressing xplA from KTR9 degraded RDX but did not utilize RDX as a nitrogen source. Moreover, an Escherichia coli strain producing XplA degraded RDX but a strain producing Cyp151C did not. KTR9 strains cured of pGKT2 did not transform RDX. Physiological studies examining the effects of exogenous nitrogen sources on RDX degradation in strain KTR9 revealed that ammonium, nitrite, and nitrate each inhibited RDX degradation by up to 79%. Quantitative real-time PCR analysis of glnA-xplB, xplA, and xplR showed that transcript levels were 3.7-fold higher during growth on RDX than during growth on ammonium and that this upregulation was repressed in the presence of various inorganic nitrogen sources. Overall, the results indicate that RDX degradation by KTR9 is integrated with central nitrogen metabolism and that the uptake of RDX by bacterial cells does not require a dedicated transporter.
Many military ranges in the United States and Canada are contaminated with explosive and propellant residues (28, 38, 54). Of the nitroaromatic and nitramine explosives that exist on these sites, hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) has a low sorption capacity (48) and can migrate through soil into surface water and groundwater (15). RDX-contaminated groundwater also has been found at sites affected by manufacturing, storage, or disposal facilities (19). Mobile explosive contaminants, like RDX, threaten overall sustainability of Department of Defense (DoD) ranges by migrating away from the source areas and contaminating surface and subsurface soils at relatively low but potentially toxic levels. The potential for bioremediation of RDX in groundwater and soils via the addition of nutrients (19, 23) or via combined abiotic/biotic processes (49) has been established. While in situ bioremediation is an attractive remedial action for these sites, the capability to predict the efficacy of in situ bioremediation of RDX is generally lacking. Nucleic acid-based assays that can detect and quantify appropriate genetic biomarkers would be valuable tools toward demonstrating the genetic potential of a contaminated site for nitramine biodegradation. The development and application of such tools have led to an increased understanding of biomarker dynamics for contaminants such as chlorinated ethenes and polycyclic aromatic hydrocarbons (32, 37). Similarly, successful monitoring of biomarkers for support of in situ bioremediation of explosives will require a detailed understanding of the microorganisms, biochemical pathways, and genes used to degrade RDX in the environment.
Thus far, microbially mediated RDX biodegradation has been reported under a number of conditions, including aerobic (16, 46, 51), nitrate-reducing (12, 22), sulfate-reducing (11), manganese-reducing (12), acetogenic (1, 9, 47), anaerobic (24), and methanogenic (2) conditions. While a number of RDX-degrading bacteria have been isolated from contaminated soils and sediments (16, 17, 46, 55), only a few genes have been implicated in RDX degradation (14, 23, 46). Of these, the cytochrome P450 system encoded by xplAB is perhaps the best characterized (27, 43). XplA is unusual for a microbial cytochrome P450 in that it comprises a flavodoxin domain fused to a C-terminal cytochrome P450 domain. XplB serves as a partner NADH-utilizing flavodoxin reductase. Together these enzymes catalyze the denitration of RDX to yield 4-nitro-2,4-diazabutanal (NDAB), nitrous oxide, ammonium, formaldehyde, and carbon dioxide (21, 27). The xplAB genes have been introduced into various plants for phytoremediation (42). Despite these advances, the overall contribution of xplAB to degradation of RDX in the environment remains to be established. While xplA has been detected in enrichments subjected to 15N-stable isotope probing derived from RDX-contaminated groundwater (41), studies establishing a correlation between the presence of xplA and RDX contamination are lacking. Such a linkage is critical to establishing xplA as a biomarker of RDX biodegradation.
Previously, we isolated two RDX-degrading strains, Gordonia sp. KTR9 and Williamsia sp. KTR4, from a semiarid soil at an explosives manufacturing facility (51). These strains are able to grow on RDX as a sole nitrogen source, similarly to strains Rhodococcus rhodochrous 11Y (46) and Rhodococcus sp. DN22 (16, 21), and produced RDX degradation products consistent with the involvement of a cytochrome P450 (10, 51). Based on the strong cross-reactivity of Gordonia sp. KTR9 and Williamsia sp. KTR4 with a TaqMan PCR assay targeting the xplA gene, we concluded that an XplA homolog was responsible for RDX degradation in these strains (25).
The actinomycete genus Gordonia is a metabolically diverse group of bacteria known for their ability to degrade xenobiotics, environmental pollutants, and recalcitrant natural polymers (4). Despite the recent interest in this genus, the genome sequence for only one organism, Gordonia bronchialis DSM 43247, is publicly available in the genome database at this time (26). In addition, two plasmid sequences, from Gordonia bronchialis DSM 43247 and Gordonia westfalica DSM 44215, respectively, are available in the genome database. Here, we present the complete sequence of a 182-kb plasmid, pGKT2, from Gordonia sp. KTR9. The plasmid contains xplAB gene homologs as well as surrounding genes which may have a role in the metabolism of RDX as a nitrogen source in strain KTR9. In order to ascertain the role of this plasmid, KTR9 strains cured of this plasmid were evaluated for the ability to degrade RDX. In addition, the effects of inorganic nitrogen compounds on the degradation of RDX and the expression of xplA and cotranscribed genes were evaluated in strain KTR9.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains, plasmids, and primers used in this study are listed in Table 1. Stationary-phase cultures of Gordonia sp. KTR9 were obtained from single colonies taken off tryptic soy broth (TSB) or LBP (1% Bacto peptone, 0.5% yeast extract, 1% NaCl) plates and inoculated into a mineral salts medium (51) containing 5 mM succinate, 5 mM glucose, 10 mM glycerol, and 180 μM RDX or 4 mM (NH4)2SO4 as the sole nitrogen source. For those studies evaluating the ability of KTR9 wild-type and derived mutant strains to degrade RDX, KTR9 stationary-phase cells pregrown on mineral salts medium with (NH4)2SO4 were washed twice with mineral salts medium without a carbon and nitrogen source and then transferred at an initial optical density at 600 nm (OD600) of 0.02 into LBP broth or mineral salts medium, each supplemented with 180 μM RDX. For those studies examining the impacts of exogenous nitrogen sources on RDX degradation, KTR9 cells pregrown on RDX as a sole nitrogen source were transferred at an initial OD600 of 0.05 into mineral salts medium supplemented with 180 μM RDX and individual competing nitrogen sources, i.e., (NH4)2SO4, KNO2, or KNO3 (all 4 mM). All KTR9 cultures were incubated at 30°C with moderate shaking. Rhodococcus jostii RHA1 was grown either in M9 mineral medium (33) supplemented with Goodies solution (7) or in PTYG broth (6). RHA1 cultures were incubated at 30°C with shaking at 200 rpm.
TABLE 1.
Bacterial strains, primers, and genetic constructs used in this study
| Bacterial strains, constructs, and primers | Catalog no., sequence, or description | Reference |
|---|---|---|
| Strains | ||
| Gordonia sp. | ||
| KTR9 | 51 | |
| KTR9/pGKT2-1Δ | KTR9 strain with 182-kb plasmid cured | This study |
| KTR9/pGKT2-2Δ | KTR9 strain with 182-kb plasmid cured | This study |
| Rhodococcus jostii RHA1 | 46a | |
| Escherichia coli BL21(DE3) | Stratagene 200131 | |
| Plasmid constructs | ||
| pEMTXplA | 1,668-bp fragment representing xplA from KTR9 synthesized into pET11a | This study |
| pEMT-Cyp151C | 1,197-bp fragment representing a cytochrome P450 from KTR9 synthesized into pET11a | This study |
| pTpXplA | xplA cloned into pTip vector | This study |
| pKTR9-gntR | 3-kb flanking region of gntR from KTR9 synthesized into pCR2.1 and subcloned into BamHI site of pK18mobsacB | This study |
| pKTR9-xplA | 3-kb flanking region of xplA from KTR9 synthesized into pCR2.1 and subcloned into BamHI site of pK18mobsacB | This study |
| Primers | ||
| Real-time PCR | ||
| xplaFm2239 | 5′GATGACCGCTGCGTCCATCGAT3′ | This study |
| xplaR2333 | 5′CCTGTTGCAGTCGCCTATACC3′ | This study |
| xplR518F | 5′CAGAGATGTGCCGGATGCT3′ | This study |
| xplR568R | 5′CGGTCACCGTGCTCATCAC3′ | This study |
| Cyp151C678F | 5′TGATGAGCTGCGTGATGTGA3′ | This study |
| Cyp151C728R | 5′GTGTCGTATCCGCCGAAGAT3′ | This study |
| Glutamine68F | 5′TCGACGCGATCAAGCAGTAC3′ | This study |
| Glutamine11R | 5′TCACGGCCTGGAAGTCGACG3′ | This study |
| xplB175F | 5′CAGGGCACGAAGAACGTGAT3′ | This study |
| xplB225R | 5′CCGATCGTCGAATACCCG3′ | This study |
| Miscellaneous | ||
| xplRF | 5′TGAGTGACGCCATCACGATC3′ | This study |
| xplRR | 5′TCTCTGCCTGAAGAGTGACC3′ | This study |
| xplAF | 5′GAACCCGAGAATCACCCGAACACC3′ | This study |
| xplAR | 5′AGGAAACCGCAGGAAAGAC3′ | This study |
| 182plasmid_gapF(gln-xplB) | 5′AGGTCGAACTCAACTGGATG3′ | This study |
| 182plasmid_gapR(gln-xplB) | 5′AGTCCGATCGTCGAATACCC3′ | This study |
| ktr9plas120565aF | 5′CGAAGGGCTTCTCACTTCAC3′ | This study |
| ktr9plas121248aR | 5′GACGAGACCTCCTTGCAGAC3′ | This study |
| ktr9plas149923bF | 5′TGGCGAACTGAGACCTTCTT3′ | This study |
| ktr9plas150701bR | 5′GCATTCTGTCCTTGCTGTCA3′ | This study |
Rhodococcus jostii RHA1 and Escherichia coli expression studies.
A 1,668-bp fragment representing the KTR9 xplA putative coding region was synthesized (Celtek Biosciences, Nashville, TN) in expression vector pET11a (New England Biolabs, Beverly, MA). The resulting plasmid, pEMTxplA, was cut with NdeI and HindIII, liberating a 1,953-bp DNA fragment containing the xplA gene. The resulting fragment was gel purified and ligated with NdeI/HindIII-restricted pTip vector TypeII, an E. coli-Rhodococcus shuttle vector for heterologous protein production in Rhodococcus (35). The ligation mixture was used to transform E. coli DH5α. The plasmid that carries the xplA gene in the correct orientation was then used to transform R. jostii RHA1 by electroporation. The empty pTip vector was also introduced by electroporation into RHA1 for use as a control. Precultures of both transformed strains were obtained by inoculating single colonies into 2 ml PTYG with 25 μg ml−1 chloramphenicol and incubated overnight at 30°C. Precultures of each strain were used to inoculate triplicate flasks of 40 ml PTYG with 25 μg ml−1 chloramphenicol at a 1% inoculum level. Cultures were induced at an OD of 0.5 by addition of thiostrepton dissolved in dimethyl sulfoxide (DMSO) to a final concentration of 50 μg ml−1. The cells were allowed to grow overnight, washed, resuspended, and concentrated 2-fold in 0.1 M sodium phosphate, pH 7.0, with 60 μg ml−1 RDX. Flasks were incubated at 30°C with shaking. Samples were taken over 5 h and analyzed by high-pressure liquid chromatography (HPLC).
A 1,197-bp fragment containing cyp151C was synthesized (Celtek Biosciences, Nashville, TN) in expression vector pET11a (New England Biolabs, Beverly, MA). The resulting plasmid construct, pEMTCyp151C, along with pEMTxplA, was transformed into E. coli BL21-Gold (DE3) competent cells (Stratagene, La Jolla, CA), and transformants were selected for on LB plates with ampicillin (100 μg ml−1). Overnight cultures of each recombinant E. coli strain were diluted 1/20 in LB supplemented with RDX (40 μg ml−1), incubated at 37°C for 2 h, and induced with the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Final concentrations of RDX were determined after 18 h of incubation at 37°C. E. coli culture samples derived from each recombinant plasmid were mixed with an equal volume of Laemmli sample buffer (33) containing 5% β-mercaptoethanol and heated to 95°C for 5 min. Denatured samples along with a low-range unstained protein standard (Bio-Rad, Hercules, CA) were loaded on a 12.5% Tris-Cl polyacrylamide gel run in 1× TGS buffer (25 mM Tris, 192 mM glycine, 0.1% [wt/vol] SDS, pH 8.3). Samples were visualized with Biosafe Coomassie G250 stain (Bio-Rad, Hercules, CA) according to the manufacturer's instructions.
Gene deletion.
Gene deletion constructs of xplA and xplR were synthesized (Celtek Biosciences) with 1.5-kb flanking sequences to create a 3-kb insert in the pCR2.1 vector (Invitrogen, Carlsbad, CA) based on DNA sequence information of pGKT2. Gene deletion inserts were liberated from the pCR2.1 vector via BamHI restriction enzyme digestion, gel purified with a Wizard SV Gel and PCR Cleanup System kit (Promega, Madison, WI), and subcloned into the BamHI site of the mobilizable plasmid pK18mobsacB (44). The resulting mutagenic plasmids, pKTR9xplA and pKTR9xplR, were used for gene deletion analysis in Gordonia sp. KTR9 based on a conjugation strategy (52) using a sacB counterselectable marker, with the exceptions of using kanamycin and nalidixic acid at 50 μg ml−1. Transconjugants were replica plated on the same medium with and without 10% sucrose. Sucrose-sensitive colonies were propagated overnight at 30°C in 25 ml of LBP medium and then spread on LBP plates supplemented with 10% sucrose. Sucrose-resistant colonies were checked for kanamycin sensitivity and screened for the presence of the deleted gene by PCR amplification using primers listed in Table 1.
PFGE and Southern hybridization.
The Bio-Rad CHEF DRII system (Bio-Rad, Hercules, CA) was employed for pulsed-field gel electrophoresis (PFGE). Cultures of Gordonia sp. KTR9 and derived mutant strains were grown and harvested from late exponential phase in LBP medium. Cell plugs were prepared from approximately 1 × 108 cells per plug in accordance with the manufacturer's instructions, with the addition of 180 U mutanolysin (Sigma-Aldrich, St. Louis, MO) to increase cell lysis. Electrophoresis parameters consisted of 0.5× Tris-borate-EDTA (TBE) at 5.5 V cm−1 with a switch time of 40 to 90 s at a 120° angle for 21 h at a constant temperature of 14°C. The Lambda ladder marker (Bio-Rad) was used as a size standard.
A 683-bp probe specific to pGKT2 was generated by PCR amplification with primers KTR9plas120505aF (5′-CGAAGGGCTTCTCACTTCAC-3′) and KTR9plas121248aR (5′-GACGAGACCTCCTTGCAGAC-3′). Amplification conditions consisted of a single step at 95°C for 5 min followed by 30 cycles at 94°C for 30 s, 63°C for 40 s, and 72°C for 40 s. The resulting PCR amplicon was gel purified and subsequently labeled by random primer extension reaction with the NEBlot Phototope kit (New England Biolabs, Ipswich, MA) following the manufacturer's instructions. The DNA from the PFGE was transferred to a Nytran SuPerCharge nylon membrane using the alkaline transfer method and apparatus from the TurboBlotter kit (Whatman). Hybridization was performed using the Phototope Star detection kit (New England Biolabs). The resultant blot was exposed to Lumi-Film chemiluminescent detection film (Roche).
Quantitative real-time PCR (qPCR) analysis.
Total RNA was isolated from bacterial pellets using Qiagen's RNeasy minikit according to the manufacturer's instructions for Gram-positive bacteria (Qiagen, Valencia, CA). Bacterial Protect reagent (Qiagen) was added to bacterial cell pellets prior to the pellets being stored at −80°C. A 10-min proteinase K treatment (300 μg) step was added to the protocol to enhance RNA recovery. RNA samples were treated with DNase (43 U; Qiagen) for 15 min preceding final purification. RNA samples were run on an Agilent 2100 Bioanalyzer to verify quality and quantity of RNA samples. Total bacterial RNA (up to 2 μg) was reverse transcribed to cDNA using SuperScript III reverse transcriptase (Invitrogen) with random primers (250 ng) according to the manufacturer's instructions. All real-time PCR amplification reactions were performed using the Applied Biosystems 7900HT Fast Real-Time PCR system (Foster City, CA). Amplification reactions were conducted in 20-μl volumes in 384-well reaction plates using 1× SYBR green PCR Master Mix (Applied Biosystems). Primer pairs specific for xplA, the glnA-xplB gene fusion, cyp151C, and xplR (Table 1) were diluted to 500 nM final concentrations for PCR analysis. Template cDNAs were diluted 10-fold prior to PCR analysis. Amplification conditions consisted of a single step at 95°C for 10 min followed by 50 cycles at 95°C for 20 s, 55°C for 20 s, and 72°C for 30 s. PCR data were analyzed using the accompanying Applied Biosystems 7900HT analysis software with threshold determinations automatically performed by the instrument and then manually optimized. Starting mRNA concentrations and PCR efficiencies were calculated using LinRegPCR (40). The mRNA concentrations were normalized to 16S rRNA PCR fluorescent intensity values.
Sequencing and annotation of pGKT2.
The sequence of pGKT2, a 182-kb plasmid, was obtained as part of a whole-genome sequencing of strain KTR9 using the Genome Sequencer FLX system (454 Life Science Corp., Branford, CT). Briefly, 5 μg of genomic DNA (500 ng μl−1) was prepared using Qiagen's DNeasy extraction kits. The 454 reads were assembled into contigs using PHRAP (http://www.phrap.org/).
Open reading frames (ORFs) were identified using the FGENESB program (Softberry Inc., Mount Kisco, NY) and the RAST server (http://rast.nmpdr.org/) (5). Select regions of pGKT2 were analyzed for bacterial promoter elements and Rho-independent terminator sequences using BPROM and FindTerm programs (Softberry Inc.). Identified genes were verified using BLAST searches of the nr database, after which the genes were subjected to a second round of automatic annotation and assignment to a COG functional classification. The Clusters of Orthologous Groups (COG), Kyoto Encyclopedia of Genes and Genomes (KEGG), and RefSeq databases were searched for homologs using BLASTP and an e-value cutoff of 1e−6. The best result for each BLAST search was imported as the gene annotation. The automatic annotation results were then manually checked and adjusted as necessary.
Phylogenetic analysis.
ParA protein sequences were aligned using ClustalW and the program's default settings. Phylogenetic analyses were performed using Proml of Phylip 3.69 (http://www.phylip.com/). Bootstrap analyses were performed using 100 data sets.
HPLC analysis.
Samples (0.6 ml) of pure cultures were mixed with an equal volume of acetonitrile and filtered into autosampler vials with a 0.45-μm polytetrafluoroethylene (PTFE) syringe filter. RDX concentrations were determined with an Agilent 1100 Series HPLC (Palo Alto, CA) equipped with a diode array UV absorbance detector set at 254 nm. An Agilent C18 reversed-phase column (100 × 4.6 mm × 5 μm) was used with an octadecylsilane (ODS)-Hypersil C18 guard column (20 × 4.0 mm × 5 μm). Samples were run under isocratic conditions as described previously (18).
Nucleotide sequence accession number.
The GenBank accession number for the pGKT2 plasmid sequence is CP002112.
RESULTS
Nucleotide sequence and annotation of pGKT2.
To better characterize the basis of the RDX-degrading properties of strain KTR9, its genomic DNA was isolated and subjected to pyrosequencing. A total of 816,115 sequence reads were obtained with an average length of 256 nucleotides. A search of the assembled reads of the KTR9 genome for xplA-specific nucleotide sequences revealed the presence of an xplA homolog on an 182,455-bp sequence contig. Ongoing analysis of the genome sequence revealed that this contig represents one of three circular plasmids of KTR9 (H.-P. Chen, K. J. Indest, and L. D. Eltis, unpublished). This plasmid, named pGKT2, has a G+C content of 63 mol%. Annotation of its nucleotide sequence identified 156 predicted ORFs (accession number CP002112). Those ORFs discussed in this study are listed in Table 2. The functional classes of predicted genes on this plasmid are summarized in Table 3. The largest functional class comprises putative genes involved in replication (recombination and repair), including multiple transposase sequences, an integrase, two topoisomerases, and multiple helicases. Predicted genes involved in general functions encompass the second largest class (13.7%), with the majority categorized as ATPases, including a member of the AAA+ superfamily. Genes involved in biosynthesis of secondary metabolites (10.3%) and transcription (10.3%) constitute perhaps the most significant classes of plasmid genes, with a subset of these being involved in RDX degradation.
TABLE 2.
ORFs with assigned functions from plasmid pGKT2
| ORFa | Gene name | Product sizeb | Closest characterized homologc | % amino acid sequence identityd |
|---|---|---|---|---|
| KTR9_4794 | parA | ParA, Corynebacterium glutamicum ATCC 13032 (BAB98814) | 34 | |
| KTR9_4798 | 635 | Plasmid maintenance protein Orf8 of pKB-1 from G. westfalica Kb1 (NP_954749) | 39 | |
| KTR9_4918 | 679 | Single-stranded DNA-specific exonuclease, C. glutamicum ATCC 13032 (BAB99130) | 43 | |
| KTR9_4921 | xplR | 246 | PipR-, GntR-type transcriptional regulator, Mycobacterium smegmatis mc2155 (YP_890908) | 49 |
| KTR9_4922 | cyp151c | 399 | PipA, cytochrome P450, family 151, Mycobacterium smegmatis mc2155 (YP_890903) | 71 |
| KTR9_4923 | glnA-xplB | 813 | GlnA-XplB fusion protein, glutamine synthetase, flavodoxin reductase, Rhodococcus rhodochrous (AAN27918) | 99 |
| KTR9_4924 | xplA | 552 | XplA, flavodoxin-cytochrome P450, R. rhodochrous (AAN27917) | 99 |
| KTR9_4935 | 145 | Prophage maintenance system killer protein, Nostoc sp. PCC 7120 (NP_478676) | 42 |
Numbered according to the KTR9 genome.
Number of amino acid residues.
Shown as gene product, organism (accession number).
Over length of KTR9 gene product.
TABLE 3.
COG functional classification of pGKT2 ORFs
| Functional classification | No. of genes | % |
|---|---|---|
| R, general function prediction only | 4 | 13.7 |
| Q, secondary metabolite biosynthesis: transport and catabolism | 3 | 10.3 |
| K, transcription | 3 | 10.3 |
| E, amino acid transport and metabolism | 1 | 3.4 |
| J, translation: ribosomal structure and biogenesis | 1 | 3.4 |
| L, replication: recombination and repair | 13 | 44.8 |
| O, posttranslational modification: protein turnover (chaperones) | 2 | 6.9 |
| D, cell cycle control: cell division (chromosome partitioning) | 1 | 3.4 |
| U, intracellular trafficking: secretion (and vesicular transport) | 1 | 3.4 |
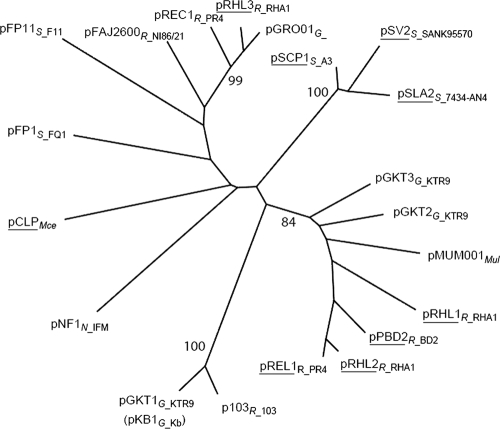
Two predicted plasmid replication proteins of pGKT2 are noteworthy. The first, encoded by KTR9_4798, shares 39% amino acid sequence identity with Orf8 of pKB-1, a 101-kb circular plasmid from Gordonia westfalica Kb1 (13). Orf8 is part of oriV of pKB-1, a region of 2 kb that has been demonstrated to be sufficient for plasmid maintenance. The second predicted plasmid replication protein is a ParA homolog whose gene, KTR9_4794, is located approximately 1.5 kb upstream of KTR9_4798. The encoded protein shares 34% amino acid sequence identity with the functionally characterized ParA of Corynebacterium glutamicum ATCC 13032 (20) and 68% identity with the predicted ParA homolog from Rhodococcus opacus B4. Predicted ParA homologs from actinomycete plasmids, including homologs from each of the three KTR9 plasmids, were aligned using ClustalX. A phylogenetic analysis (Fig. 1) revealed that plasmid-encoded ParAs, including the three from KTR9, are diverse. Even the KTR9 proteins do not cluster according to organismal phylogeny or the topology of the plasmid, consistent with previous findings (34).
FIG. 1.
Phylogenetic relationship of ParA proteins from 20 actinobacterial plasmids. ParAs and their predicted homologs are designated by the name of the plasmid on which they reside. The subscript identifies the genus of the bacterium in which the plasmid resides (first letter) and either the first two letters of the species or the strain number. Linear plasmids are underscored. Bootstrap values of major, stable nodes are indicated as percentages. ParA sequences from the following plasmids were included in the analysis: pGKT1, Gordonia sp. KTR9; pGKT2, Gordonia sp. KTR9; pGKT3, Gordonia sp. KTR9; pKB1, G. westfalica strain DSM 44215T (gi|40217320); pGBRO01, Gordonia bronchialis DSM 43247 (gi|262118119); pSCP1_a2, Streptomyces coelicolor A3(2) (gi|21234217); pCLP, Mycobacterium celatum, (gi|32455743); pMUM001, Mycobacterium ulcerans, (gi|42414723); pNF1, Nocardia farcinica IFM 10152 (gi|54027649); p103, Rhodococcus equi 103 (gi|10657915); pFAJ2600, Rhodococcus erythropolis NI86/21 (gi|2460009); pPBD2, R. erythropolis BD2 (gi|33867067); pREC1, R. erythropolis PR4 (gi|77404542); pREL1, R. erythropolis PR4 (gi|77454578); pRHL1, R. jostii RHA1, (gi|111025387); pRHL2, R. jostii RHA1, (gi|111026081); pRHL3, R. jostii RHA1, (gi|111027090); pSLA2-L, Streptomyces rochei 7434-AN4 (gi|30795070); pFP11, Streptomyces sp. F11 (gi|62184581); pFP1, Streptomyces sp. FQ1 (gi|62184597); pSV2, Streptomyces violaceoruber ANK95570 (gi|32455643). The predicted ParAs of pGKT1 and pKB1 have identical sequences.
Finally, we used IslandPick (31) to search for regions of pGKT2 of probable horizontal origin. Such regions, known as genomic islands (GIs), are frequently associated with particular physiologic adaptation such as virulence, catabolism, or resistance to a toxic compound and can sometimes be identified based on abnormal sequence composition, such as GC mol% and codon bias. The analysis revealed that pGKT2 contained no predicted GIs. Moreover, no insertion sequence (IS) elements were found using IS Finder (http://www-is.biotoul.fr/). However, the plasmid contains five genes predicted to encode transposases or integrases. Of these, the most proximal to xplAB is KTR9_4947, 15 kb away.
xplAB and surrounding genes.
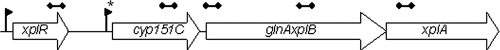
Analysis of the pGKT2 nucleotide sequence revealed that this plasmid encodes an XplA homolog that shares 97% to 99% amino acid sequence identity with characterized XplA cytochromes P450 (3, 45, 46). The gene is the last in a cluster comprising cyp151C, a glnA-xplB gene fusion, and xplA (Fig. 2). Approximately 1.5 kb upstream of this cluster is xplR, encoding a GntR-type transcriptional regulator. Cyp151C shares up to 71% amino acid sequence identity with the mycobacterial pipA/morA-encoded cytochromes P450 that are involved in the utilization of the secondary amines piperidine, pyrrolidine, and morpholine (29, 39, 50). The glnA-xplB fusion is predicted to encode the first 85% of the residues of a glutamine synthetase fused to the entire XplB reductase. PCR analysis of KTR9 DNA genomic template with primers that flank the gene fusion point confirmed that there is no gap between these genes (data not shown). Interestingly, the glutamine synthase portion of the glnA-xplB fusion and XplR share up to 70% and 49% sequence identity, respectively, with the glutamine synthases and GntR-type regulators encoded by genes associated with the mycobacterial cyp151 genes (29, 39, 50).
FIG. 2.
The xpl gene cluster. Putative promoter regions (−10 and −35 regions) were found upstream of the xplR ORF (bp 156809 to 157548) at positions 156741 to 156761 and of the cyp151C ORF (bp 158328 to 159527 bp) at positions 158272 to 158290. A GntR binding site (*) (TnGTnnnACnA) was found 5 bp upstream of the promoter region for cyp151C. Regions targeted by qPCR are shown by barbell lines about the gene boxes.
The organization of the xpl gene cluster suggests that it may constitute an operon that is regulated by XplR. Bioinformatic analysis of this region suggests that putative bacterial promoter elements (−10 and −35 regions) can be found directly upstream of xplR and cyp151C (Fig. 2). A putative GntR binding site (TnGTnnnACnA) of the FadR subtype was also found within 5 bp of the −35 promoter region of cyp151C (53), further suggesting that XplR could regulate transcription of cyp151C. The absence of Rho-independent terminator sequences in cyp151C and the short intergenic space between this gene and glnA-xplB also suggest that cyp151C, glnA-xplB, and xplA are transcribed as a single cistron.
Expression of xplA in R. jostii RHA1 and E. coli.
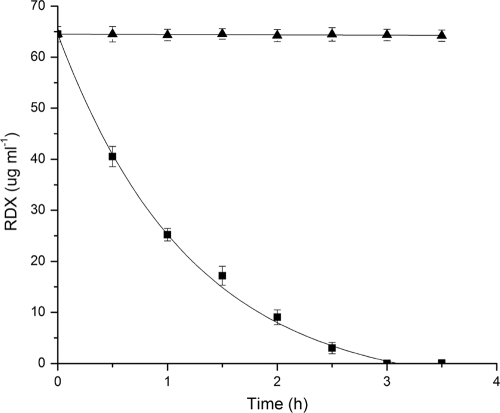
The xplA gene was heterologously expressed in Rhodococcus jostii RHA1 using a pTip vector. Production of the xplA-encoded cytochrome P450 was verified using denaturing acrylamide gel analysis and visible absorption spectroscopy. More particularly, cells carrying pTipXplA but not pTip revealed the characteristic CO-dependent peak at 450 nm, indicating that XplA is both soluble and functional (data not shown). Moreover, RHA1::pXplA rapidly depleted RDX from culture medium within 2.5 h (Fig. 3). In contrast, RHA1::pTip did not detectably deplete RDX in culture media. This indicates that RHA1 efficiently takes up RDX and that the xplA gene alone is sufficient to degrade RDX.
FIG. 3.
Degradation of RDX by Rhodococcus jostii RHA1 containing XplA. Exponentially growing cells containing pTpXplA (▴) or an empty pTip vector (▪) were resuspended to an OD600 of 12 in 0.1 M sodium phosphate, pH 7.5, containing 60 ppm RDX. The fitted curve represents a first-order decay with a half-life (t1/2) of 0.61 ± 0.05 h.
The xplA gene was also heterologously expressed in E. coli BL21 using the pET11a vector. Overnight cultures of the recombinant E. coli strain grown in LB plus RDX (40 μg ml−1) that had been previously induced with the addition of 1 mM IPTG 18 h earlier showed approximately a 71% reduction in RDX compared to control samples (data not shown). In contrast, cyp151C did not elicit degradation of RDX when expressed in E. coli despite the presence of soluble, correctly folded cytochrome P450 as verified by denaturing gel analysis and visible absorption spectroscopy (data not shown).
Generation of a plasmid-free mutant strain of Gordonia sp. KTR9.
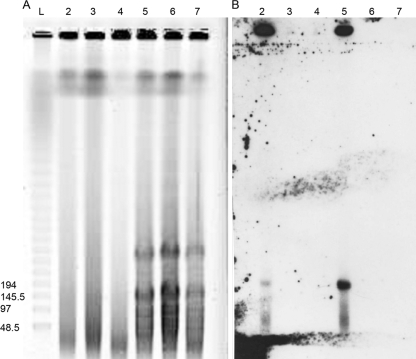
Attempts to independently create unmarked gene deletions in the xplA or xplR loci using a sacB counterselectable marker resulted in curing of pGKT2 from strain KTR9. PCR screening of sucrose-resistant, kanamycin-sensitive derivatives of KTR9 with multiple primer sets targeting different regions of pGKT2 including xplA, xplR, and cyp151C was not amplifiable when tested (data not shown). Loss of the plasmid was further confirmed by PFGE and Southern analysis of uncut and XbaI-digested (one cut site resulting in linearization of the plasmid) DNA samples from both KTR9 and plasmid-cured strains (Fig. 4). Those lanes containing wild-type KTR9 DNA reacted with a biotin-labeled 683-bp probe known to be on the plasmid, revealing the presence of a band in the expected size range for the plasmid. In contrast, neither of the KTR9 plasmid-cured strains produced a signal when probed.
FIG. 4.
PFGE (A) and Southern analysis (B) of KTR9 and derived mutant strains. A biotin-labeled 683-bp DNA fragment, specific for pGKT2, was used to probe a PFGE gel of uncut and XbaI-digested DNA samples from wild-type and mutant KTR9 strains. Lanes: 2, uncut KTR9; 3, uncut deletion mutant 1; 4, uncut deletion mutant 2; 5, cut KTR9; 6, cut deletion mutant 1; 7, cut deletion mutant 1. Lane L shows molecular mass markers, with sizes in kilodaltons at the left.
Plasmid-cured KTR9 strains were evaluated for their ability to degrade RDX and utilize RDX as a sole nitrogen source. KTR9 plasmid-cured strains incubated for up to 67 h in LB broth containing 45 μg ml−1 RDX did not detectably deplete RDX levels despite achieving optical densities at 600 nm of at least 2.0 (Table 4). In contrast, cultures of wild-type KTR9 depleted RDX to trace levels within 67 h. KTR9 plasmid-cured strains incubated in a minimal salts medium containing RDX as the sole nitrogen source did not degrade RDX and as a result did not grow, while the wild-type KTR9 strain grew and completely degraded the RDX within 48 h (Table 4).
TABLE 4.
RDX degradation studies in KTR9 and mutant strains lacking pGKT2a
| Gordonia sp. strains | RDX concn (μg ml−1) |
|
|---|---|---|
| Rich medium | Minimal medium | |
| KTR9 | 1.5 (0.2) | 0.0 (0.0) |
| KTR9/pGKT2-1Δ | 44.6 (0.6) | 44.8 (1.4) |
| KTR9/pGKT2-2Δ | 42.2 (0.2) | 44.2 (1.2) |
Wild-type and plasmid-cured KTR9 strains were grown for up to 67 h in LB medium containing 45 ppm RDX and a minimal medium containing RDX as the sole nitrogen source. Results represent the averages (standard deviations) of triplicate samples.
Effect of nitrogen sources on RDX degradation and xpl gene expression in KTR9.
The effects of ammonium, nitrite, and nitrate on the ability of KTR9 to degrade RDX were investigated. KTR9 degraded RDX most rapidly when this compound was present in liquid cultures as the sole nitrogen source (Fig. 5 A), with approximately 93% of the RDX degraded within 24 h of inoculation. In contrast, only 21% to 34% of the RDX was transformed after 24 h in the presence of a competing nitrogen source. Even after 68 h of incubation, significant concentrations of RDX remained in cultures containing (NH4)2SO4 (63%), KNO2 (42%), or KNO3 (13%).
FIG. 5.
(A) Effects of various inorganic nitrogen sources on the RDX degradation activity (closed symbols) and growth (open symbols) of KTR9. A 4 mM concentration of (NH4)2SO4 (triangles), KNO3 (squares), or KNO2 (diamonds) was included as a competing nitrogen source compared to RDX alone (circles). Error bars are the standard errors of three samples. (B) Quantitative real-time PCR expression analysis using primers for xplR, cyp151C, glnA-xplB, xplA, and a 16S rRNA. RNA was harvested from KTR9 cells grown for 24 h on media containing the indicated nitrogen sources: RDX alone (diagonally striped bars), RDX plus (NH4)2SO4 (white bars), RDX plus KNO2 (black bars), and RDX plus KNO3 (horizontally striped bars). Relative expression of gene targets was normalized to corresponding gene expression levels for cultures in which RDX was the sole nitrogen source.
The expression levels of xplR, cyp151C, glnA-xplB, and xplA were compared in KTR9 cells at the 24-h time point in the growth study described above. RNA samples were subjected to qPCR analysis, and relative gene expression data were normalized to corresponding gene expression levels for cultures in which RDX was the sole nitrogen source. When KTR9 was grown on RDX in the presence of a competing nitrogen source, the expression of all four genes was reduced from 48% to 80% compared to growth on RDX alone (Fig. 5B). These results correlated well with the inhibition of RDX degradation (Fig. 5A). Specifically, expression of xplA was inhibited by 73% in cultures containing ammonium. These same cultures experienced a 79% reduction in the degradation of RDX. Similarly, expression of the xplA gene was inhibited by 75% and 56% in the presence of nitrite and nitrate resulting in a 76% and 66% reduction in the degradation of RDX, respectively.
DISCUSSION
As part of our efforts to characterize the basis of the RDX-degrading properties of Gordonia sp. KTR9, we describe here the sequence and functional characterization of a circular 182,455-bp plasmid, pGKT2, and the RDX degradation genes that it carries. The plasmid appears to be typical of those of Gordonia based on (i) its G+C content (63 mol%), which is characteristic of DNA from several genera of Corynebacterineae (4), and (ii) the high amino acid sequence identity of key plasmid replication proteins to homologs from Gordonia: ParA and a replication protein similar to Orf8 of pKB-1 from G. westfalica (13). The functional classes of predicted genes on this plasmid, summarized in Table 3, indicated that the majority of the ORFs identified (77%) on this plasmid could not be assigned a function, with 71% having no match in the searched protein databases. In comparison, 76% and 56% of the identified ORFs on pGBRO01 in G. bronchialis and pKB-1 in G. westfalica Kb1, respectively, could not be assigned a function (13, 26).
Functional characterization of xplA confirmed its predicted role in RDX degradation. More specifically, annotation of the pGKT2 ORFs suggested that the RDX catabolic activity of KTR9 is encoded by a four-gene cluster: xplR-cyp151C-glnA-xplB-xplA. Heterologous expression of xplA in E. coli and R. jostii RHA1 demonstrated that xplA is sufficient for the degradation of RDX. Importantly, neither a dedicated transporter nor the xplB-encoded cognate reductase (27, 42) is required in these strains. Moreover, the pGKT2-cured strains of KTR9, generated serendipitously through attempts to delete xplA and xplR, demonstrate that pGKT2 is necessary for RDX degradation and growth on RDX as a sole nitrogen source. Even when grown on rich media, KTR9 plasmid-cured strains did not degrade RDX. This suggests the lack of alternate cometabolic reactions or nitroreductases (30) for RDX degradation by KTR9. Collectively, these results indicate that xplA is necessary and sufficient for RDX degradation in KTR9.
The genomic context of xplAB in KTR9 indicates that these genes were acquired by a recombination event that resulted in their insertion into a locus that may be responsible for the utilization of N-heterocyclic alkanes as growth substrates and nitrogen source. More particularly, xplR, cyp151C, and the truncated glnA domain of pGKT2 are remarkably similar in sequence and order to their homologs in the mor and pip gene clusters responsible for the degradation of morpholine, piperidine, and related compounds in mycobacteria and rhodococci (29, 39, 50). Indeed, a recent comparison of the mor/pip loci in six strains suggests that this locus is subject to a relatively high frequency of recombination events. Where insertions occurred, they were upstream of the truncated glnA. This is in contrast to the case in KTR9, where xplB and xplA occur downstream of glnA. The nucleotide sequence repeats present in the mor/pip loci (29, 39, 50) are absent in the xpl cluster.
The occurrence of xplAB in a locus likely responsible for the utilization of N-heterocyclic alkanes has intriguing functional implications for both sets of genes. Perhaps foremost of these is the possibility that XplR regulates the expression of xplAB in an RDX-dependent fashion and that this regulation is linked to central nitrogen metabolism in KTR9. The qPCR data presented herein indicate that the entire xpl cluster is transcriptionally regulated in such a fashion inasmuch as inorganic nitrogen sources repressed glnA-xplB-xplA transcript levels to a degree that correlated well with inhibition of RDX degradation (Fig. 5). Moreover, a binding site for FadR-like proteins (48), such as XplR, occurs 5 bp from the −35 promoter region of cyp151C (Fig. 2). Finally, transposon mutagenesis studies indicate that the homolog of XplR in Mycobacterium smegmatis mc2155, PipR, is a likely transcriptional repressor, regulating pipA in a piperidine- and pyrrolidine-dependent manner (39). It will be interesting to determine to what extent XplR responds to N-heterocyclic alkanes and RDX, respectively. Similarly, it would be interesting to investigate whether the GlnA-XplB fusion protein is functional and, if so, whether it facilitates the assimilation of nitrogen from both RDX and N-heterocyclic alkanes and provides reducing equivalents to both Cyp151C and XplA. Such functionality in KTR9 may explain why a recombinant strain of RHA1 expressing xplA was able to degrade RDX but was unable to grow on RDX as a sole nitrogen source (data not shown). This finding is surprising since RHA1 grows on nitrite (unpublished observation), one of the products resulting from the xplA-mediated sequential single electron reduction of RDX (27). Alternatively, the inability of RHA1 to grow on RDX as a nitrogen source may reflect a low tolerance of RHA1 to other RDX degradation products such as formaldehyde.
While an analysis of the pGKT2 nucleotide sequence did not reveal the presence of genomic islands, the high sequence identity of the xplAB gene products from different strains (>97%) and the location of these genes on plasmids in strains characterized to date are consistent with their acquisition via horizontal gene transfer. Nevertheless, the genomic context of these genes in KTR9 differs markedly from that in Microbacterium strain MA1, where they occur on pMA1 (3). On this plasmid, xplAB occur immediately downstream from genes predicted to encode a MarR regulator and an amino acid permease, respectively. A dot blot analysis (data not shown) of pGKT2 with the 52.2-kb region from pMA1 showed 4 regions of high sequence identity (97% or greater) ranging from 300 bp to 3,100 bp. Distributed over both plasmids, these corresponded to the xplAB genes and an ORF encoding a putative death-on-curing gene.
Collectively, the current findings suggest that within the past 100 years, some bacteria have evolved an RDX catabolic capability that is integrated and coordinated with the organism's response to nitrogen availability. Our observation along with supporting data from others (36) that expression of xplAB is subjected to repression in the presence of additional nitrogen compounds has implications for the role of xplAB as a real-time biomarker of RDX degradation. RDX bioremediation efforts may need to monitor levels of inorganic nitrogen sources for efficient RDX degradation to take place. Understanding the subtleties that can affect biomarker dynamics has the potential to improve predictability of RDX treatment schemes that incorporate a biomarker monitoring component. Recent studies focused on chlorinated ethenes demonstrate that the expression and hence activity of various functional genes are often variable and may require additional interpretation beyond the potential for degradation (8, 32).
The capacity to accurately quantify signature biomarkers predictive of the breakdown of contaminants in complex environmental samples is viewed as a critical need in the long-term vision for the science and engineering of bioremediation. Accurate estimations of in situ biomarker dynamics in conjunction with other chemical and physical parameters can be used in a weight-of-evidence approach to increase overall predictability of in situ remediation strategies such as bioaugmentation, biostimulation, or monitored natural attenuation. A more in-depth understanding of the microbial ecology of in situ RDX degradation would also be beneficial in this regard, providing additional insights into functional linkages that may exist among the microbial community, the soil geochemistry, and contaminant degradation.
Acknowledgments
This research was funded in part through grants from the Strategic Environmental Research and Development Program (project ER-1609), the U.S. Army Corps of Engineers Environmental Quality Research Program, and Genome BC. The use of RAST was supported in part by National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (NIAID), under contract HHSN266200400042C.
Views, opinions, and/or findings contained herein are those of the authors and should not be construed as an official Department of the Army position or decision unless so designated by other official documentation.
We thank S. Eng and S. J. Hallam for their critical help in running the annotation software.
Footnotes
Published ahead of print on 13 August 2010.
REFERENCES
- 1.Adrian, N. R., and C. M. Arnett. 2004. Anaerobic biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by Acetobacterium malicum strain HAAP-1 isolated from a methanogenic mixed culture. Curr. Microbiol. 48:332-340. [DOI] [PubMed] [Google Scholar]
- 2.Adrian, N. R., C. M. Arnett, and R. F. Hickey. 2003. Stimulating the anaerobic biodegradation of explosives by the addition of hydrogen or electron donors that produce hydrogen. Water Res. 37:3499-3507. [DOI] [PubMed] [Google Scholar]
- 3.Andeer, P. F., D. A. Stahl, N. C. Bruce, and S. E. Strand. 2009. Lateral transfer of genes for hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) degradation. Appl. Environ. Microbiol. 75:3258-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arenskotter, M., D. Broker, and A. Steinbüchel. 2004. Biology of the metabolically diverse genus Gordonia. Appl. Environ. Microbiol. 70:3195-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aziz, R. K., D. Bartels, A. A. Best, M. DeJongh, T. Disz, R. A. Edwards, K. Formsma, S. Gerdes, E. M. Glass, M. Kubal, F. Meyer, G. J. Olsen, R. Olson, A. L. Osterman, R. A. Overbeek, L. K. McNeil, D. Paarmann, T. Paczian, B. Parrello, G. D. Pusch, C. Reich, R. Stevens, O. Vassieva, V. Vonstein, A. Wilke, and O. Zagnitko. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balkwill, D. L., and W. C. Ghiorse. 1985. Characterization of subsurface bacteria associated with two shallow aquifers in Oklahoma. Appl. Environ. Microbiol. 50:580-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauchop, T., and S. R. Elsden. 1960. The growth of micro-organisms in relation to their energy supply. J. Gen. Microbiol. 23:457-469. [DOI] [PubMed] [Google Scholar]
- 8.Behrens, S., M. F. Azizian, P. J. McMurdie, A. Sabalowsky, M. E. Dolan, L. Semprini, and A. M. Spormann. 2008. Monitoring abundance and expression of “Dehalococcoides” species chloroethene-reductive dehalogenases in a tetrachloroethene-dechlorinating flow column. Appl. Environ. Microbiol. 74:5695-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beller, H. R. 2002. Anaerobic biotransformation of RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine) by aquifer bacteria using hydrogen as the sole electron donor. Water Res. 36:2533-2540. [DOI] [PubMed] [Google Scholar]
- 10.Bhushan, B., S. Trott, J. C. Spain, A. Halasz, L. Paquet, and J. Hawari. 2003. Biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by a rabbit liver cytochrome P450: insight into the mechanism of RDX biodegradation by Rhodococcus sp. strain DN22. Appl. Environ. Microbiol. 69:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boopathy, R., M. Gurgas, J. Ullian, and J. F. Manning. 1998. Metabolism of explosive compounds by sulfate-reducing bacteria. Curr. Microbiol. 37:127-131. [DOI] [PubMed] [Google Scholar]
- 12.Bradley, P. M., and R. S. Dinicola. 2005. RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine) biodegradation in aquifer sediments under manganese-reducing conditions. Bioremediat. J. 9:1-8. [Google Scholar]
- 13.Broker, D., M. Arenskotter, A. Legatzki, D. H. Nies, and A. Steinbüchel. 2004. Characterization of the 101-kilobase-pair megaplasmid pKB1, isolated from the rubber-degrading bacterium Gordonia westfalica Kb1. J. Bacteriol. 186:212-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryant, C., and M. DeLuca. 1991. Purification and characterization of an oxygen-insensitive NAD(P)H nitroreductase from Enterobacter cloacae. J. Biol. Chem. 266:4119-4125. [PubMed] [Google Scholar]
- 15.Clausen, J., J. Robb, D. Curry, and N. Korte. 2004. A case study of contaminants on military ranges: Camp Edwards, Massachusetts, U.S.A. Environ. Pollut. 129:13-21. [DOI] [PubMed] [Google Scholar]
- 16.Coleman, N. V., D. R. Nelson, and T. Duxbury. 1998. Aerobic biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) as a nitrogen source by a Rhodococcus sp., strain DN22. Soil Biol. Biochem. 30:1159-1167. [Google Scholar]
- 17.Crocker, F. H., K. J. Indest, and H. L. Fredrickson. 2006. Biodegradation of the cyclic nitramine explosives RDX, HMX, and CL-20. Appl. Microbiol. Biotechnol. 73:274-290. [DOI] [PubMed] [Google Scholar]
- 18.Crocker, F. H., K. T. Thompson, J. E. Szecsody, and H. L. Fredrickson. 2005. Biotic and abiotic degradation of CL-20 and RDX in soils. J. Environ. Qual. 34:2208-2216. [DOI] [PubMed] [Google Scholar]
- 19.Davis, J. L., A. H. Wani, B. R. O'Neal, and L. D. Hansen. 2004. RDX biodegradation column study: comparison of electron donors for biologically induced reductive transformation in groundwater. J. Hazard. Mater. 112:45-54. [DOI] [PubMed] [Google Scholar]
- 20.Donovan, C., A. Schwaiger, R. Kramer, and M. Bramkamp. 2010. Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum. J. Bacteriol. 192:3441-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fournier, D., A. Halasz, J. Spain, P. Fiurasek, and J. Hawari. 2002. Determination of key metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine with Rhodococcus sp. strain DN22. Appl. Environ. Microbiol. 68:166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freedman, D. L., and K. W. Sutherland. 1998. Biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) under nitrate-reducing conditions. Water Sci. Technol. 38:33-40. [Google Scholar]
- 23.Fuller, M. E., K. McClay, J. Hawari, L. Paquet, T. E. Malone, B. G. Fox, and R. J. Steffan. 2009. Transformation of RDX and other energetic compounds by xenobiotic reductases XenA and XenB. Appl. Microbiol. Biotechnol. 84:535-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawari, J., A. Halasz, T. Sheremata, S. Beaudet, C. Groom, L. Paquet, C. Rhofir, G. Ampleman, and S. Thiboutot. 2000. Characterization of metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) with municipal anaerobic sludge. Appl. Environ. Microbiol. 66:2652-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Indest, K. J., F. H. Crocker, and R. Athow. 2007. A TaqMan polymerase chain reaction method for monitoring RDX-degrading bacteria based on the xplA functional gene. J. Microbiol. Methods 68:267-274. [DOI] [PubMed] [Google Scholar]
- 26.Ivanova, N., J. Sikorski, M. Jando, A. Lapidus, M. Nolan, S. Lucas, T. Glavina Del Rio, H. Tice, A. Copeland, J. Cheng, F. Chen., D. Bruce, L. Goodwin, S. Pitluck, K. Mavromatis, G. Ovchinnikova, A. Pati, A. Chen, K. Palaniappan, M. Land, L. Hauser, Y. Chang, C. Jeffries, P. Chain, E. Saunders, C. Han, J. Detter, T. Brettin, M. Rohde, M. Göker, J. Bristow, J. Eisen, V. Markowit, P. Hugenholtz, H. Klenk, and N. Kyrpides. 2010. Complete genome sequence of Gordonia bronchialis type strain (3410T). Stand. Genomic Sci. 2:1.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson, R. G., E. L. Rylott, D. Fournier, J. Hawari, and N. C. Bruce. 2007. Exploring the biochemical properties and remediation applications of the unusual explosive-degrading P450 system XplA/B. Proc. Natl. Acad. Sci. U. S. A. 104:16822-16827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins, T. F., M. E. Walsh, P. G. Thorne, P. H. Miyares, T. A. Ranney, C. L. Grant, and J. R. Esparza. 1998. Site characterization for explosives contamination at a military firing range impact area. U.S. Army Cold Regions Research and Engineering Laboratory. Special report 98-9. U.S. Army Cold Regions Research and Engineering Laboratory, Hanover, NH.
- 29.Kim, Y. H., I. Kang, H. Bergeron, P. C. Lau, K. H. Engesser, and S. J. Kim. 2006. Physiological, biochemical, and genetic characterization of an alicyclic amine-degrading Mycobacterium sp. strain THO100 isolated from a morpholine-containing culture of activated sewage sludge. Arch. Microbiol. 186:425-434. [DOI] [PubMed] [Google Scholar]
- 30.Kitts, C. L., C. E. Green, R. A. Otley, M. A. Alvarez, and P. J. Unkefer. 2000. Type I nitroreductases in soil enterobacteria reduce TNT (2,4,6,-trinitrotoluene) and RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine). Can. J. Microbiol. 46:278-282. [DOI] [PubMed] [Google Scholar]
- 31.Langille, M. G., W. W. Hsiao, and F. S. Brinkman. 2008. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinformatics 9:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, P. K., T. W. Macbeth, K. S. Sorenson, Jr., R. A. Deeb, and L. Alvarez-Cohen. 2008. Quantifying genes and transcripts to assess the in situ physiology of “Dehalococcoides” spp. in a trichloroethene-contaminated groundwater site. Appl. Environ. Microbiol. 74:2728-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maniatis, T., J. Sambrook, and E. F. Fritsch. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.McLeod, M. P., R. L. Warren, W. W. Hsiao, N. Araki, M. Myhre, C. Fernandes, D. Miyazawa, W. Wong, A. L. Lillquist, D. Wang, M. Dosanjh, H. Hara, A. Petrescu, R. D. Morin, G. Yang, J. M. Stott, J. E. Schein, H. Shin, D. Smailus, A. S. Siddiqui, M. A. Marra, S. J. Jones, R. Holt, F. S. Brinkman, K. Miyauchi, M. Fukuda, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582-15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakashima, N., and T. Tamura. 2004. Isolation and characterization of a rolling-circle-type plasmid from Rhodococcus erythropolis and application of the plasmid to multiple-recombinant-protein expression. Appl. Environ. Microbiol. 70:5557-5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nejidat, A., L. Kafka, Y. Tekoah, and Z. Ronen. 2008. Effect of organic and inorganic nitrogenous compounds on RDX degradation and cytochrome P-450 expression in Rhodococcus strain YH1. Biodegradation 19:313-320. [DOI] [PubMed] [Google Scholar]
- 37.Park, J. W., and D. E. Crowley. 2006. Dynamic changes in nahAc gene copy numbers during degradation of naphthalene in PAH-contaminated soils. Appl. Microbiol. Biotechnol. 72:1322-1329. [DOI] [PubMed] [Google Scholar]
- 38.Pennington, J. C., T. F. Jenkins, S. Thiboutot, G. Ampleman, J. Clausen, A. D. Hewitt, J. Lewis, M. E. Walsh, T. A. Ranney, B. Silverblatt, A. Marois, A. Gagnon, P. Brousseau, J. E. Zufelt, K. Poe, M. Bouchard, R. Martel, D. D. Walker, C. A. Ramsey, C. A. Hayes, S. L. Yost, K. L. Bjella, L. Trepanier, T. E. Berry, D. J. Lambert, P. Dubé, and N. M. Perron. 2005. Distribution and fate of energetics on DoD test and training ranges: report 5. U.S. Army Engineer Research and Development Center. ERDC TR-05-2. U.S. Army Engineer Research and Development Center, Vicksburg, MS.
- 39.Poupin, P., V. Ducrocq, S. Hallier-Soulier, and N. Truffaut. 1999. Cloning and characterization of the genes encoding a cytochrome P450 (PipA) involved in piperidine and pyrrolidine utilization and its regulatory protein (PipR) in Mycobacterium smegmatis mc2155. J. Bacteriol. 181:3419-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramakers, C. J., R. H. Uijter, R. H. Eprez, and A. F. Oorman. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62-66. [DOI] [PubMed] [Google Scholar]
- 41.Roh, H., C. P. Yu, M. E. Fuller, and K. H. Chu. 2009. Identification of hexahydro-1,3,5-trinitro-1,3,5-triazine-degrading microorganisms via 15N-stable isotope probing. Environ. Sci. Technol. 43:2505-2511. [DOI] [PubMed] [Google Scholar]
- 42.Rylott, E. L., R. G. Jackson, J. Edwards, G. L. Womack, H. M. Seth-Smith, D. A. Rathbone, S. E. Strand, and N. C. Bruce. 2006. An explosive-degrading cytochrome P450 activity and its targeted application for the phytoremediation of RDX. Nat. Biotechnol. 24:216-219. [DOI] [PubMed] [Google Scholar]
- 43.Sabbadin, F., R. Jackson, K. Haider, G. Tampi, J. P. Turkenburg, S. Hart, N. C. Bruce, and G. Grogan. 2009. The 1.5-A structure of XplA-heme, an unusual cytochrome P450 heme domain that catalyzes reductive biotransformation of royal demolition explosive. J. Biol. Chem. 284:28467-28475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 45.Seth-Smith, H. M., J. Edwards, S. J. Rosser, D. A. Rathbone, and N. C. Bruce. 2008. The explosive-degrading cytochrome P450 system is highly conserved among strains of Rhodococcus spp. Appl. Environ. Microbiol. 74:4550-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seth-Smith, H. M., S. J. Rosser, A. Basran, E. R. Travis, E. R. Dabbs, S. Nicklin, and N. C. Bruce. 2002. Cloning, sequencing, and characterization of the hexahydro-1,3,5-trinitro-1,3,5-triazine degradation gene cluster from Rhodococcus rhodochrous. Appl. Environ. Microbiol. 68:4764-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Seto, M., K. Kimbara, M. Shimura, T. Hatta, M. Fukuda, and K. Yano. 1995. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:3353-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherburne, L. A., J. D. Shrout, and P. J. Alvarez. 2005. Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) degradation by Acetobacterium paludosum. Biodegradation 16:539-547. [DOI] [PubMed] [Google Scholar]
- 48.Sheremata, T. W., A. Halasz, L. Paquet, S. Thiboutot, G. Ampleman, and J. Hawari. 2001. The fate of the cyclic nitramine explosive RDX in natural soil. Environ. Sci. Technol. 35:1037-1040. [DOI] [PubMed] [Google Scholar]
- 49.Shrout, J. D., P. Larese-Casanova, M. M. Scherer, and P. J. Alvarez. 2005. Sustained and complete hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) degradation in zero-valent iron simulated barriers under different microbial conditions. Environ. Technol. 26:1115-1126. [DOI] [PubMed] [Google Scholar]
- 50.Sielaff, B., and J. R. Andreesen. 2005. Analysis of the nearly identical morpholine monooxygenase-encoding mor genes from different Mycobacterium strains and characterization of the specific NADH:ferredoxin oxidoreductase of this cytochrome P450 system. Microbiology 151:2593-2603. [DOI] [PubMed] [Google Scholar]
- 51.Thompson, K. T., F. H. Crocker, and H. L. Fredrickson. 2005. Mineralization of the cyclic nitramine explosive hexahydro-1,3,5-trinitro-1,3,5-triazine by Gordonia and Williamsia spp. Appl. Environ. Microbiol. 71:8265-8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Geize, R., G. I. Hessels, R. van Gerwen, P. van der Meijden, and L. Dijkhuizen. 2001. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid delta1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 205:197-202. [DOI] [PubMed] [Google Scholar]
- 53.Vindal, V., K. Suma, and A. Ranjan. 2007. GntR family of regulators in Mycobacterium smegmatis: a sequence and structure based characterization. BMC Genomics 8:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walsh, M. E., C. M. Collins, C. H. Racine, T. F. Jenkins, A. B. Gelvin, and T. A. Ranney. 2010. Sampling for explosive residues at Fort Greely, Alaska: reconnaissance visit July 2000. U.S. Army Cold Regions Research and Engineering Laboratory. ERDC/CRREL TR-01-15. U.S. Army Cold Regions Research and Engineering Laboratory, Hanover, NH.
- 55.Zhao, J. S., J. Spain, S. Thiboutot, G. Ampleman, C. Greer, and J. Hawari. 2004. Phylogeny of cyclic nitramine-degrading psychrophilic bacteria in marine sediment and their potential role in the natural attenuation of explosives. FEMS Microbiol. Ecol. 49:349-357. [DOI] [PubMed] [Google Scholar]