Abstract
Three GH-6 family cellobiohydrolases are expected in the genome of Magnaporthe grisea based on the complete genome sequence. Here, we demonstrate the properties, kinetics, and substrate specificities of a Magnaporthe oryzae GH-6 family cellobiohydrolase (MoCel6A). In addition, the effect of cellobiose on MoCel6A activity was also investigated. MoCel6A contiguously fused to a histidine tag was overexpressed in M. oryzae and purified by affinity chromatography. MoCel6A showed higher hydrolytic activities on phosphoric acid-swollen cellulose (PSC), β-glucan, and cellooligosaccharide derivatives than on cellulose, of which the best substrates were cellooligosaccharides. A tandemly aligned cellulose binding domain (CBD) at the N terminus caused increased activity on cellulose and PSC, whereas deletion of the CBD (catalytic domain only) showed decreased activity on cellulose. MoCel6A hydrolysis of cellooligosaccharides and sulforhodamine-conjugated cellooligosaccharides was not inhibited by exogenously adding cellobiose up to 438 mM, which, rather, enhanced activity, whereas a GH-7 family cellobiohydrolase from M. oryzae (MoCel7A) was severely inhibited by more than 29 mM cellobiose. Furthermore, we assessed the effects of cellobiose on hydrolytic activities using MoCel6A and Trichoderma reesei cellobiohydrolase (TrCel6A), which were prepared in Aspergillus oryzae. MoCel6A showed increased hydrolysis of cellopentaose used as a substrate in the presence of 292 mM cellobiose at pH 4.5 and pH 6.0, and enhanced activity disappeared at pH 9.0. In contrast, TrCel6A exhibited slightly increased hydrolysis at pH 4.5, and hydrolysis was severely inhibited at pH 9.0. These results suggest that enhancement or inhibition of hydrolytic activities by cellobiose is dependent on the reaction mixture pH.
Cellulose, composed of β-1,4-linked glucosyl units, is the most abundant naturally produced biopolymer on earth and can be utilized as a sustainable and renewable energy resource in place of fossil fuel. Establishing conditions for the efficient degradation of cellulose will contribute to the enhanced use of bioethanol, a biobased alternative to gasoline, which will increase biomass recycling and reduce carbon dioxide emissions (21, 30). Hence, efficient degradation of cellulose is an issue of great importance today.
Bacteria and fungi produce cellulases that catalyze the hydrolysis of β-1,4-glycosidic bonds and are involved in the degradation of cellulose. Cellulases are divided into three major types according to their substrate specificities and the mode of hydrolysis: endoglucanases (EC 3.2.1.4), cellobiohydrolases (EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21). The most efficient hydrolysis of cellulose is thought to result from the combined synergistic actions of cellulases, whereby the enzymatic activity of an enzyme mixture is substantially higher than the sum of the activities of the individual enzymes. Several types of synergy have been described as the cooperative actions of endo- and exo-acting enzymes (20, 25, 32, 33, 45). Such cellulose-degrading enzymes are routinely used in the manufacture of beverages and industrial products, e.g., beer and wine, animal feed, paper, textiles, laundry detergents, and food ingredients (5). Hence, reducing cellulase manufacturing costs by increasing the productivity of cellulases with high specific activities through biotechnological modification is a desired research goal.
Fungal cellobiohydrolases belong to glycosyl hydrolase families 6 and 7 (GH-6 and -7) and act most efficiently on highly ordered crystalline cellulose, hydrolyzing from either the reducing or the nonreducing terminus to liberate predominantly cellobiose (C2) with a minor amount of cellotriose (C3) (6, 39, 40). Also, Trichoderma reesei Cel6A can hydrolyze 1,3-1,4-β-glucan (1, 18), but it is unclear whether in vivo it is hydrolysis of 1,3-1,4-β-glucan that occurs mainly or hydrolysis of cellulose derivatives. The resulting accumulation of cellobiose inhibits the activity of cellobiohydrolase (13, 15, 28, 31, 36, 37, 44). Some microorganisms possess cellulosomes, multienzyme complexes that contribute to the efficient degradation of cellulose. Cellobiohydrolase is a documented component of cellulosomes in Clostridium thermocellum (2, 31).
The three-dimensional (3D) structures of two GH-6 family members have been elucidated, including the cellobiohydrolase of T. reesei and that of Humicola insolens in complex with glucose, cellooligosaccharide, and a nonhydrolyzable substrate analogue (35, 41-43). The proposed structures have identified the significant amino acids associated with the catalytic core domain, where the catalytic site is buried inside a tunnel-shaped cavity and an enzyme-cellooligosaccharide hydrogen bond network. The structure suggests that the mode of action proceeds in a processive manner as cellobiohydrolase progresses along the cellulose chain (7, 19, 34, 40).
The ascomycete fungus Magnaporthe grisea is the pathogen that causes rice blast, the most devastating fungal disease of rice. Since the complete genome sequence of M. grisea has been published (10), mining the database for candidate genes involved in pathogen-plant interactions, cell wall degradation, etc., is quite feasible. The cell wall-degrading enzymes of the genus Magnaporthe that are involved in the infection process have been of particular interest (22-24). Based on the complete genome sequence, M. grisea has three putative GH-6 family cellobiohydrolases and four GH-7 family cellobiohydrolases. Using primers designed from the database of M. grisea cellobiohydrolases, we cloned putative GH-6 and GH-7 family cellobiohydrolases, designated MoCel6A and MoCel7A, respectively, from Magnaporthe oryzae by PCR. The cloned MoCel6A and MoCel7A from M. oryzae were completely identical to those of M. grisea. In this paper, we demonstrate the properties of MoCel6A prepared by homologous overexpression in M. oryzae and examine the effects of cellobiose on the hydrolytic activity of MoCel6A. Furthermore, the effects of cellobiose on the activities of both MoCel6A and a T. reesei GH family 6 cellobiohydrolase (TrCel6A, formerly referred to as CBH II), which were overexpressed in Aspergillus oryzae, were also examined.
MATERIALS AND METHODS
Inoculation of M. oryzae into rice leaves.
M. oryzae (strain Ina72), grown on potato-dextrose agar plates at 28°C for 4 days under dark-blue light, and a suspension of spores (3 × 105 spores/ml) in double-distilled water (ddH2O) were sprayed on leaves of the rice cultivar Shin-2 at the four- to five-leaf stage. The leaves were harvested 1 to 5 day after inoculation (see Fig. S1 in the supplemental material). For cDNA cloning of GH-6 and GH-7 family cellobiohydrolases (MoCel6A and MoCel7A), the leaf at 4 days after infection with M. oryzae was used. Determination of transcript levels was performed by reverse transcription (RT)-PCR using the leaves at various time points after inoculation with M. oryzae.
Cloning of cellobiohydrolase genes.
To extract cellobiohydrolases belonging to GH-6 and GH-7 from M. oryzae, the deduced amino acid sequences of T. reesei (accession no. AF302657), Aspergillus fumigatus (accession no. XP_748511), Chaetomium globosum (accession no. XM_001226028), and H. insolens (accession no. AB048710) Cel6As and T. reesei (accession no. AY928809), A. fumigatus (accession no. XM_745951), and C. thermophilum (accession no. AM711862) Cel7As were used for BLASTP searches against the M. grisea database (http://www.broadinstitute.org/annotation/genome/magnaporthe_grisea/MultiHome.html). Other GH-6 and GH-7 cellobiohydrolases were used for BLASTP searches, but no cellobiohydrolase from M. grisea has been found other than three GH-6 and four GH-7 family cellobiohydrolases.
Rice leaves infected with M. oryzae were ground with a pestle in liquid nitrogen, and total RNA was prepared using an RNeasy Plant Mini Kit (Qiagen). A cDNA pool was synthesized from total RNA using an oligo(dT)20 primer and Superscript III reverse transcriptase (Invitrogen). T. reesei (NBRC31326) was grown on potato-dextrose agar plates at 28°C for 4 days. Total RNA extraction and cDNA synthesis were carried out as described above. GH-6 and GH-7 family cellobiohydrolase cDNAs were amplified with PrimeStar GXL DNA polymerase (Takara) from the cDNA pool by PCR using the following primers: 5′-ATGGCTAGCAAGCTGTTCCTCGCCGC-3′ and 5′-CTACAAGGGTGGGTTGGCGTTGGTGAG-3′ for MoCel6A (accession no. XP_360146), 5′-ATGAAGCGCGCGCTCTGTGCCTCCC-3′ and 5′-TTAGTCGACCTGGTAGGTGCTGCCA-3′ for MoCel7A (accession no. XP_367082), and 5′-ATGATTGTCGGCATTCTCACCACGC-3′ and 5′-TTACAGGAACGATGGGTTTGCGTTTG-3′ for T. reesei Cel6 (TrCel6A; accession no. GU724763). These primers were designed from putative GH-6 and GH-7 family cellobiohydrolases found in the M. grisea and T. reesei genome sequences. As a result of sequencing the amplified cDNA fragments, we found that the cloned cDNAs from M. oryzae were completely identical to those of M. grisea.
For mRNA analyses of MoCel6A, corresponding DNA fragments 304 bp in length were amplified from the cDNA pool of M. oryzae-infected rice leaves with PrimeStar GXL DNA polymerase using the specific primers 5′-CTTCGACGAAAAGAAGTACATCTCTG-3′ and 5′-CGCGTCGCTCATGGCGCCGTAGTCCG-3′ for MoCel6A. These primers were designed from the cloned cDNA of M. oryzae. As a control, an actin (accession no. XM_364702) DNA fragment 296 bp in length from M. oryzae was amplified by PCR using the following specific primers: 5′-TCGACGTCCGAAAGGATCTGTACAAC-3′ and 5′-ACTCCTGCTTCGAGATCCACATCTGC-3′. The amplified DNA fragments were subjected to electrophoresis on a 1% agarose gel and stained with ethidium bromide.
Overexpression of recombinant cellobiohydrolases.
To aid protein purification and immunoblot analysis of recombinant proteins, the coding sequence for seven contiguous histidine residues, 5′-TTAGTGATGGTGATGGTGGTGATGGCTAGG-3′, was fused in frame to the 3′ ends of the cDNAs by a PCR extension method and then amplified again to add the restriction nuclease digestion sites of XbaI and HindIII using primers, 5′-CTAGTCTAGAATGGCTAGCAAGCTGTTCCTCGCCGC-3′ and 5′-CCCAAGCTTTAGTGATGGTGATGGTGGTGATG-3′. For overexpression of MoCel6A in M. oryzae, the PCR products were digested with XbaI and HindIII and ligated into the pBAFp expression vector driven by the MPG1 promoter (4). The ligation mixture was transformed into Escherichia coli DH5α by heat shock, and transformants were selected on LB agar plates supplemented with 20 μg/ml chloramphenicol. Prepared pBAFp DNA (5 μg) carrying the cellobiohydrolase gene was transformed into M. oryzae Ina 72 according to the method described by Sweigard et al. (38), and the transformants were screened on oatmeal plates containing bialaphos (250 μg/ml).
For overexpression of MoCel6A and TrCel6A in A. oryzae, DNA fragments obtained by PCR using primers 5′-TCGCCGCTTGCCGTTGAAGAGCGTC-3′ and 5′-TTAGTGATGGTGATGGTGGTGATGGCTAGG-3′ (histidine tag) for MoCel6A and 5′-GTGCCTCTCGAGGAGCGGCAGGCTTG-3′ and 5′-TTAGTGATGGTGATGGTGGTGATGGCTAGG-3′ (histidine tag) for TrCel6A were ligated into the pPPamyBSP expression vector driven by the amyBp promoter (46). The ligation mixture was transformed into E. coli DH5α by heat shock, and transformants were selected on LB agar plates supplemented with 20 μg/ml ampicillin. Prepared pPPamyBSP DNA (5 μg) carrying the cellobiohydrolase gene was transformed into A. oryzae according to the method described by Gomi et al. (14), and the transformants were screened on Czapek-Dox agar (1% glucose, 30 mM Na2CO3, 7 mM KCl, 4 mM K2HPO4, 4 mM MgSO4, 0.04 mM FeSO4) plates containing 0.1 mg/ml pyrithiamine.
Transformants of M. oryzae and A. oryzae were cultured in YG (1% yeast extract and 5% glucose) medium and YPM (1% yeast extract, 2% peptone, and 2% maltose) medium, respectively, at 25°C for 4 days. The culture media were subjected to immunoblot analysis using a horseradish peroxidase-conjugated monoclonal antibody directed against the histidine tag.
Purification of recombinant cellobiohydrolases.
The culture media of M. oryzae and A. oryzae transformants were filtered through cheesecloth, concentrated to 10 to 20 ml by ultrafiltration (Amicon Ultra-4; Millipore), and equilibrated with wash/equilibration buffer (50 mM sodium phosphate, pH 7.0, 50 mM NaCl). The solution was loaded onto a histidine tag-binding resin (Talon metal affinity resin; Clontech) equilibrated with wash/equilibration buffer, and the resin was washed with the same buffer, followed by washing with 0.1× elution buffer (10 mM sodium phosphate, pH 7.0, 50 mM NaCl, 20 mM imidazole). Protein bound to the resin was eluted with 1× elution buffer (50 mM sodium phosphate, pH 7.0, 50 mM NaCl, 200 mM imidazole). The eluate was equilibrated with sodium phosphate buffer (10 mM, pH 6.0) and concentrated to 100 to 150 μl by ultrafiltration. The purity of the proteins was confirmed by electrophoresis on a 12.5% SDS-acrylamide gel, followed by staining with Coomassie brilliant blue.
Cellobiohydrolase activity on polysaccharides.
To assay for the hydrolytic activity of MoCel6A expressed in M. oryzae, the increase in reducing power was determined by the PAHBAH method (29), as described below. A reaction mixture (100 μl) containing polysaccharide (0.5 to 5 mg), 100 mM sodium phosphate (pH 6.0), and MoCel6A (0.1 μg) was incubated for several hours at 30°C. When water-insoluble polysaccharides were used as substrates, the reaction mixture was vigorously agitated using a microtube mixer. After centrifugation at 22,000 × g for 5 min, the supernatant (50 μl) was transferred to a microcentrifuge tube and mixed with 150 μl of PAHBAH solution (1% [wt/vol] 4-hydroxybenzoic hydrazide). The mixture was heated in a boiling water bath for 5 min, and the absorbance was measured at 410 nm. The polysaccharides tested as substrates were cellulose (Sigmacell 20; Sigma), Avicel (Fluka), phosphoric acid-swollen cellulose (PSC) made from Sigmacell 20, barley 1,3-1,4-β-glucan (here referred to as β-glucan) (Megazyme), carboxymethyl cellulose (CMC) (Sigma), hydroxyethyl cellulose (HEC) (Sigma), oat spelt arabinoxylan (Sigma), and tamarind xyloglucan (Megazyme).
The dependence of pH on the hydrolytic activity of MoCel6A on cellulose (Sigmacell 20) was evaluated by equilibrating the reaction mixture with 100 mM sodium acetate (pH 3.5 to 5.5), sodium phosphate (pH 5.5 to 7.5), Tris-HCl (pH 7.5 to 9.0), or CAPS (N-cyclohexyl-3-aminopropanesulfonic acid; pH 9.0 to 11.0). Determination of the optimum temperature for MoCel6A hydrolysis of cellulose was examined by incubating replicate reaction mixtures for 18 h at 4, 10, 20, 30, 40, 50, or 60°C. The effects of pH and temperature on hydrolytic activity were quantified by the PAHBAH method.
Product analysis by LC/MS.
Hydrolysates produced by MoCel6A were added to methanol containing 0.5 M 3-methyl-1-phenyl-5-pyrazolone (PMP) and 0.3 M NaOH and heated at 70°C for 30 min. The solution was neutralized with 0.3 N HCl and dried. The PMP sugars were dissolved in methanol and passed through a 0.45-μm filter (Minisart RC15; Sartorius AG). Analyses of PMP sugars were performed with an Agilent 1100 series liquid chromatography/mass spectrometry (LC/MS) system (Agilent Technologies, Inc.) equipped with an Agilent Zorbax Eclipse XDB-C18 column (4.6 by 150 mm; Agilent Technologies, Inc.). The mobile phases were 100 mM ammonium acetate and acetonitrile. Elution was conducted with a gradient of 20 to 30% acetonitrile in 20 min at a flow rate of 0.5 ml/min. Quantification of each peak area representing each PMP sugar was calculated based on standard calibration curves. During cellotetraose (C4) hydrolysis by MoCel6A, two cellobioses are released in one cycle of hydrolysis. Therefore,the hydrolytic rate of cellotetraose was estimated from the halved concentration of released cellobiose.
Preparation of sulforhodamine-conjugated cellooligosaccharides.
The reducing termini of cellohexaose (C6), cellopentaose (C5), cellotetraose, cellotriose, and cellobiose were labeled with lissamine rhodamine B sulfonyl chloride (Roche) according to the procedure reported by Fry (12). The labeled cellooligosaccharides were loaded on a Bio-Gel P-2 (Bio-Rad) column (1.5 by 80 cm) in water. Fractions corresponding to sulforhodamine-conjugated cellohexaose (SR-C6), cellopentaose (SR-C5), and cellotetraose (SR-C4) were rechromatographed on the same Bio-Gel P-2 columns in water. Crude preparations of sulforhodamine labeled-cellotriose (SR-C3) and cellobiose (SR-C2) were used as standards.
Cellobiohydrolase activities on celloligosaccharide derivatives.
For assaying the hydrolytic activities of M. oryzae cellobiohydrolases on cellooligosaccharides and sulforhodamine-conjugated cellooligosaccharides, a reaction mixture (20 μl) containing 1% (wt/vol) cellooligosaccharide or sulforhodamine-conjugated cellooligosaccharide, 100 mM sodium phosphate (pH 6.0), and MoCel6A or MoCel7A (0.1 μg) was incubated at 30°C for various times. The reaction products of cellooligosaccharides were developed by thin-layer chromatography (TLC) on silica gel (60 F254; Merck) plates in butan-1-ol-acetic acid-water (2:1:1 [vol/vol]) and stained with 0.5% (wt/vol) thymol in ethanol-H2SO4 (19:1 [vol/vol]). Quantification of reaction products was performed by LC/MS. The reaction products of sulforhodamine-conjugated cellooligosaccharides were separated by silica gel chromatography in butan-1-ol-acetic acid-water (2:1:1 [vol/vol]), and fluorescent spots were observed under UV light (254 nm) in a Luminescent Image Analyzer LAS-3000.
Effect of cellobiose on cellobiohydrolase activities.
A reaction mixture (20 μl) containing either 1% (wt/vol) SR-C6 or -C5, 100 mM sodium phosphate (pH 6.0), and cellobiohydrolase (0.1 μg) was incubated in the presence of 2.9 to 438 mM cellobiose at 30°C for various times. Analysis of the reaction products was conducted by TLC and LC/MS as described above.
RESULTS
Cloning of MoCel6A.
Three putative cellobiohydrolases belonging to the GH-6 family were found in the genome DNA sequence of M. grisea by BLASTP search using the amino acid sequences of other fungal GH-6 family cellobiohydrolases. MoCel6A, one of the three cellobiohydrolases, is composed of a catalytic core domain and a cellulose binding domain (CBD) that are connected by a linker peptide, whereas the others do not contain a CBD. Amino acid alignment revealed that MoCel6A has high similarity to other fungal cellobiohydrolases, for example, 62% similarity to T. reesei Cel6A and 68% similarity to H. insolens Cel6A (see Fig. S2 in the supplemental material). To determine the transcript accumulation of MoCel6A during infection of rice by M. oryzae, RT-PCR was conducted using specific primers. The transcript level of MoCel6A increased during infection (Fig. 1), whereas transcripts from the other cellobiohydrolases were not detected (data not shown).
FIG. 1.
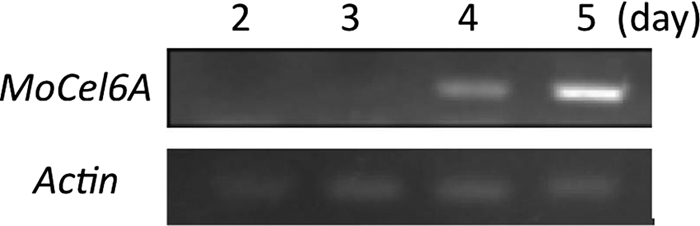
RT-PCR analysis of transcript levels of MoCel6A. cDNA pools were prepared from rice leaves 2 to 5 days after infection with M. oryzae. A DNA fragment of MoCel6A was amplified from the cDNA pools by PCR. M. oryzae actin DNA was amplified as a control.
Dependency of MoCel6A on pH and temperature.
MoCel6A was purified by affinity chromatography using a resin to bind the histidine tag from 4-day culture medium of an M. oryzae transformant overexpressing MoCel6A (see Fig. S3 in the supplemental material). Routinely, about 120 μg MoCel6A could be purified from a 1-liter culture. MoCel6A exhibited a broad pH optimum for cellulose hydrolysis ranging from pH 4.5 to 9.0 at 30 to 50°C (Fig. 2), indicative of an alkaline-stable enzyme. Thereafter, assays for hydrolytic activity of MoCel6A were conducted in sodium phosphate buffer (100 mM, pH 6.0) at 30°C.
FIG. 2.
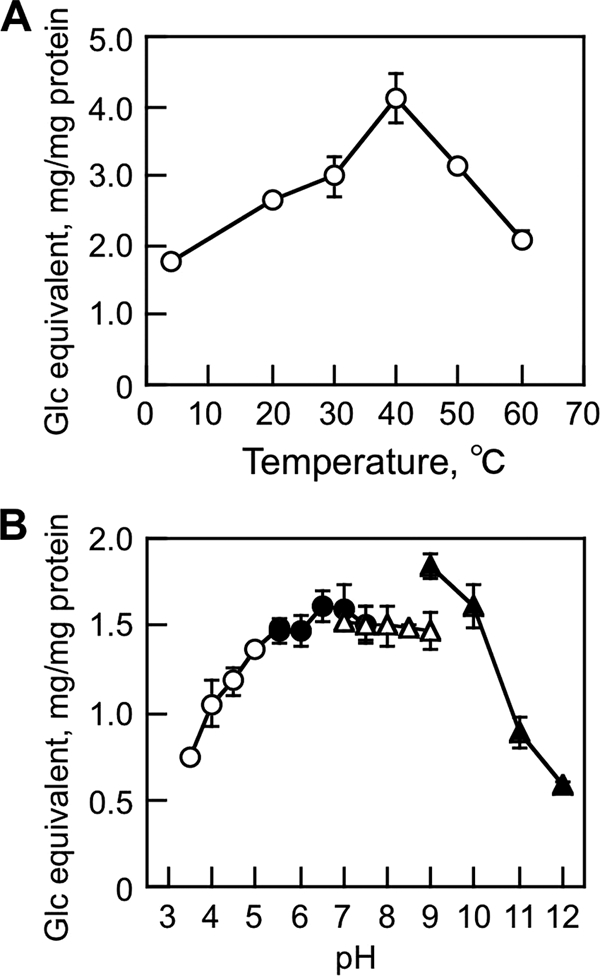
Effects of temperature (A) and pH (B) on cellulose hydrolysis by MoCel6A. The assay mixture (100 μl) containing cellulose (5 mg), sodium phosphate (100 mM; pH 6.0), and MoCel6A (0.1 μg) was incubated for 18 h at various temperatures. To determine the effect of pH on enzyme activity, the following buffers (each at 100 mM) were used: sodium acetate, pH 3.5 to 5.5; sodium phosphate, pH 5.5 to 7.5; Tris-HCl, pH 7.5 to 9.0; and CAPS, pH 9.0 to 11.0. The data are the means of three determinations ± standard error (SE).
Substrate specificity of polysaccharides.
The hydrolytic activity of MoCel6A expressed in M. oryzae was assayed on water-insoluble and water-soluble polysaccharides (Table 1). MoCel6A showed the hydrolysis of not only cellulose, but also PSC and barley β-glucan, whereas CMC, HEC, xylan, and xyloglucan were not hydrolyzed. In comparison with the hydrolysis of cellulose, PSC and barley β-glucan were about 7 times more efficiently hydrolyzed by MoCel6A. The results suggest that MoCel6A catalyzes the hydrolysis of amorphous and water-soluble β-1,4-linked glucans more effectively than cellulose and that chemical modification of the glucose residues in CMC and HEC interferes with substrate recognition.
TABLE 1.
Substrate specificity of MoCel6A on polysaccharides
| Substrate | Relative activity (%)a |
|---|---|
| Cellulose (5 mg) | 100 |
| Cellulose (0.5 mg) | 30 |
| Avicel | 191 |
| PSC | 217 |
| HEC | 0 |
| CMC | 0 |
| β-Glucan | 227 |
| Xyloglucan | 1 |
| Xylan | 0 |
| Mannan | 0 |
Reaction mixtures (100 μl) containing cellulose (5 and 0.5 mg), Avicel (5 mg), or other polysaccharides (0.5 mg) were incubated with MoCel6A (0.1 μg) in sodium phosphate buffer (100 mM; pH 6.0) for 18 h at 30°C, and activity was determined by the PAHBAH method. The values represent the averages of three individual determinations.
Substrate specificity of cellooligosaccharides.
Hydrolysis of naturally occurring cellooligosaccharides and sulforhodamine-conjugated cellooligosaccharides by MoCel6A was examined by LC/MS and TLC analyses (Fig. 3). Production of cellobiose and cellotriose from cellooligosaccharides (C4, C5, and C6) was much greatger than when cellulose was used as the substrate (Fig. 3A). C4 and C5 were hydrolyzed by one reaction into 2 mol of cellobiose and 1 mol each of cellobiose and cellotriose, respectively. In consideration of this result, cellotetraose was the best substrate for MoCel6A, showing a higher Vmax value; however, the Km value for C5 was smaller than that for C4, since formation of an enzyme-substrate complex is easier for C5, with a longer chain of glucose residues. TLC analysis of hydrolyzed products from SR-C6 showed SR-C4 and SR-C3, which were generated by liberating cellobiose and cellotriose, respectively, from the nonreducing ends of SR-C6. When SR-C5 was used as a substrate, only SR-C3 was produced by releasing cellobiose from the nonreducing ends of SR-C5. In this case, cellotriose from the nonreducing ends was not evident because SR-C2 was not produced. SR-C4 was not hydrolyzed after long incubation periods, indicating that SR-C4 was not a substrate for MoCel6A, although C4 was a good substrate. These results indicate that MoCel6A liberates cellobiose and cellotriose from the nonreducing ends and requires at least four contiguous β-1,4-linked glucosyl units for a substrate but did not recognize sulforhodamine-conjugated 1-amino-1-deoxyglucitol.
FIG. 3.
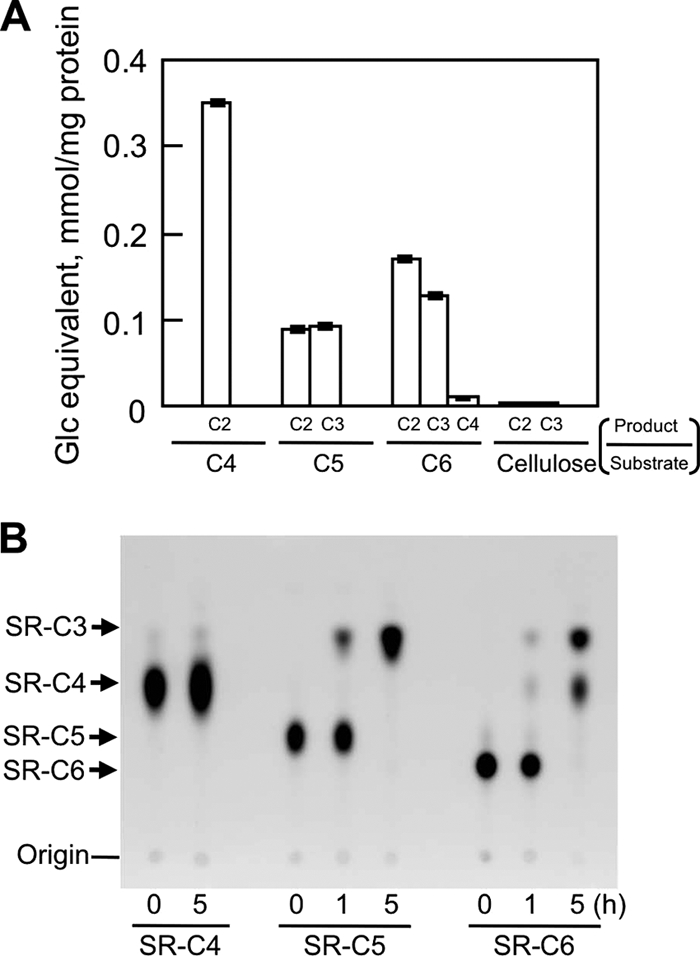
Product analysis of cellooligosaccharides hydrolyzed by MoCel6A. (A) The assay mixtures (10 μl) containing cellooligosaccharides (60 μg) or cellulose (5 mg) and sodium phosphate (100 mM; pH 6.0) were incubated with MoCel6A at 30°C for 2 h, and the hydrolysates were analyzed by LC/MS. From the results of kinetic analyses on initial bond cleavage, the Km and Vmax were 24.3 mM and 454.5 μg/min/mg protein for cellotetraose and 3.3 mM and 63.3 μg/min/mg protein for cellopentaose. The data are the means of three determinations ± SE. (B) Each SR-C4, SR-C5, and SR-C6 was incubated with MoCel6A in sodium phosphate buffer (100 mM; pH 6.0) for 0, 1, and 5 h. After incubation, the reaction mixture was subjected to TLC using butanol-acetic acid-water (2:1:1), and fluorescent spots were visualized under UV light (254 nm).
Effects of cellobiose on hydrolysis of cellooliogsaccharide derivatives.
Cellobiohydrolases mainly release cellobiose from cellulose derivatives, and the presence of cellobiose competitively inhibits the rate of hydrolysis. To investigate the inhibitory effect of the end product, cellobiose, on the activity of MoCel6A compared with MoCel7A, hydrolysis of SR-C6 and -C5 was assayed in the presence of high concentrations of cellobiose (Fig. 4). SR-C6 was hydrolyzed, and SR-C4 and SR-C3 reaction products were generated by MoCel6A hydrolysis, even in the presence of 438 mM cellobiose (Fig. 4A). Conversely, MoCel7A, which was also prepared by homologous overexpression in M. oryzae (see Fig. S3 in the supplemental material), showed decreased hydrolysis of SR-C6 as the cellobiose concentration increased in the reaction mixture (Fig. 4B). The hydrolysis of C5 by MoCel6A to cellotriose slightly increased as the cellobiose concentration increased, whereas MoCel7A hydrolysis of cellopentaose was inhibited when ≥29 mM cellobiose was present in the reaction mixture (Fig. 4C and D). Quantifying the cellotriose produced by MoCel6A by LC/MS resulted in a gradual increase in cellotriose production as the cellobiose concentration increased (Fig. 4E). These results indicate that the hydrolytic activity of MoCel6A on sulforhodamine-conjugated cellooligosaccharide and naturally occurring cellooligosaccharide was not inhibited, but rather, was enhanced slightly in the presence of high concentrations of cellobiose.
FIG. 4.
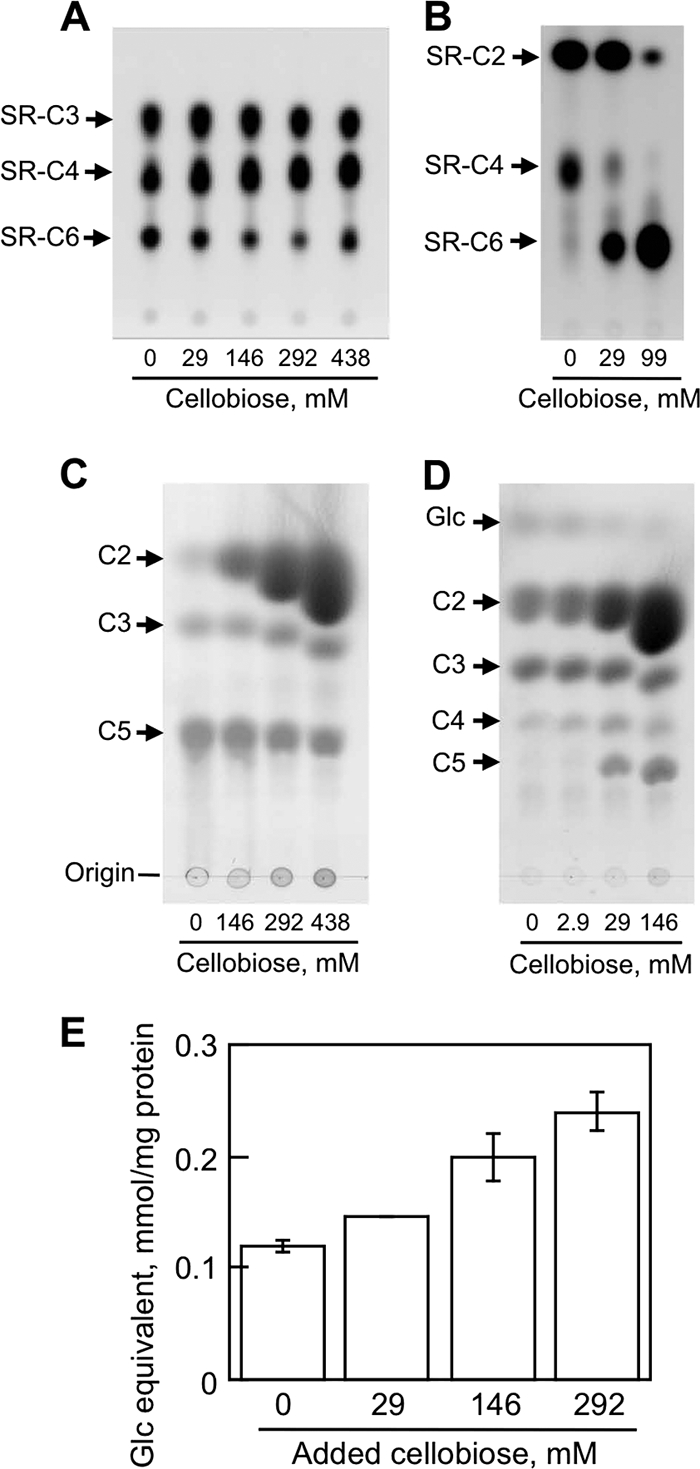
Effects of excess cellobiose on hydrolysis of cellooligosaccharide derivatives. (A) MoCel6A hydrolysates of SR-C6 in the presence of 0 to 438 mM cellobiose were separated by TLC using butanol-acetic acid-water (2:1:1), and fluorescent spots were visualized under UV light (254 nm). (B) Hydrolysis of sulforhodamine-conjugated cellohexaose by MoCel7A in the presence of 0 to 146 mM cellobiose; fluorescent products were observed as described for panel A. (C) MoCel6A hydrolysates of C5 in the presence of 0 to 438 mM cellobiose were separated by TLC in butanol-acetic acid-water (2:1:1) and stained with 0.1% thymol in H2SO4-ethanol (EtOH) (5:95 [vol/vol]). (D) Hydrolysis products of cellopentaose by MoCel7A in the presence of 0 to 146 mM cellobiose were analyzed as described for panel C. (E) Quantification of cellotriose released from cellopentaose by MoCel6A was measured by calculating the cellotriose peak area analyzed by LC/MS. Cellobiose was added to the reaction mixtures at final concentrations of 29 to 292 mM. The data are the means of three determinations ± SE.
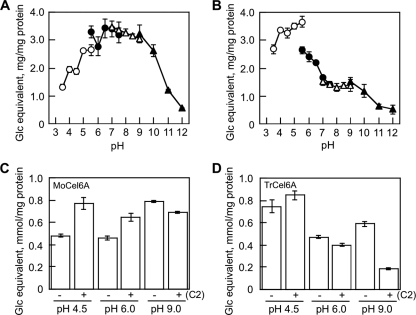
Comparison of cellobiohydrolase activities of MoCel6A and TrCel6A expressed in A. oryzae.
MoCel6A and TrCel6A prepared in A. oryzae were used to compare the effects of cellobiose on hydrolytic activities, since recombinant TrCel6A has not been successfully expressed in M. oryzae. MoCel6A prepared in A. oryzae showed high activity toward cellulose ranging from pH 4.5 to 9.0, similar to that of MoCel6A prepared in M. oryzae (Fig. 5 A). On the other hand, TrCel6A had high activity at pH 4.0 to 6.0, with about 40% of the maximal activity remaining at pH 9.0 (Fig. 5B). Using these enzymes exhibiting different pH dependencies, we measured cellotriose produced by hydrolyzing cellopentaose in the presence of 292 mM cellobiose by LC/MS analysis (Fig. 5C and D). The level of cellotriose produced by MoCel6A increased at pH 4.5 and pH 6.0 with cellobiose in the reaction mixture and decreased at pH 9.0. On the other hand, hydrolysis of cellopentaose by TrCel6A was slightly enhanced at pH 4.5 with cellobiose and severely inhibited at pH 9.0. These results indicate that the hydrolytic activities of MoCel6A and TrCel6A were enhanced by high concentrations of cellobiose at acidic pH and inhibited at basic pH.
FIG. 5.
pH dependency and effects of cellobiose on hydrolytic activities of MoCel6A and TrCel6A. To determine the effect of pH on the activities of MoCel6A (A) and TrCel6A (B), the following buffers (each at 100 mM) were used: sodium acetate, pH 3.5 to 5.5; sodium phosphate, pH 5.5 to 7.5; Tris-HCl, pH 7.5 to 9.0; CAPS, pH 9.0 to 11.0. The hydrolytic activities of cellopentaose in the presence of 292 mM cellobiose by MoCel6A (C) and TrCel6A (D) were determined by LC/MS. The data are the means of three determinations ± SE.
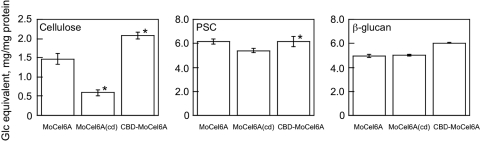
Effects of CBD on the activity of MoCel6A.
MoCel6A is the only cellobiohydrolase with a CBD at the N terminus among the three GH-6 family cellobiohydrolases of M. oryzae. To examine the effect of the CBD on hydrolytic activity, two structurally modified cellobiohydrolases, MoCel6A(cd) and CBD-MoCel6A, were prepared, and their hydrolytic activities were assayed using cellulose, PSC, and β-glucan as substrates (Fig. 5). In comparison with MoCel6A, the hydrolytic activity of MoCel6A(cd) on cellulose was reduced by greater than 50%, and that of CBD-MoCel6A increased. On the other hand, hydrolytic activities on PSC and β-glucan were not influenced by the presence of a CBD, except for a small increase in the activity of CBD-MoCel6A on PSC. Among the carbohydrates tested in our study, the presence of a CBD contributed to the hydrolysis of cellulose and PSC, but not water-soluble β-glucan, indicating that CBD function is dependent on cellulose crystallinity.
DISCUSSION
Cellobiohydrolases belonging to GH families 6 and 7 are thought to be key enzymes that play significant roles in the degradation of cellulose into cellobiose. In this study, we characterized a GH-6 family cellobiohydrolase, MoCel6A from M. oryzae, that was successfully overexpressed and purified by affinity chromatography and compared the hydrolytic activities of MoCel7A and TrCel6A in the presence of high concentrations of cellobiose.
The structure of MoCel6A is composed of a catalytic core domain and a CBD at the N terminus that are connected by a linker peptide. The CBD in some cell wall-degrading enzymes, such as endo-1,4-β-glucanase and -β-mannanase, critically serves in the hydrolysis of their responsible substrates (i.e., substrates the enzymes can hydrolyze), whereas the truncated structure (catalytic core domain only) has reduced activity and the addition of another CBD can enhance activity (16, 17, 26). Similarly, the hydrolytic activity of MoCel6A on cellulose was increased by a tandemly aligned CBD (CBD-MoCel6A) and decreased with the removal of the CBD [MoCel6A(cd)] (Fig. 6). However, influence of the CBD on hydrolysis of PSC and β-glucan was not observed, except for an increase in PSC hydrolysis by CBD-MoCel6A. These observations suggest that the CBD serves to promote the effective hydrolysis of crystalline regions of cellulose.
FIG. 6.
Effects of a CBD on hydrolytic activities. To examine the effects of a CBD on MoCel6A activity, constructs containing a truncated CBD [MoCel6A(cd); catalytic domain only] and a duplicate CBD (CBD-MoCel6A; two CBD motifs attached to a catalytic domain) were made. The hydrolytic activities of the two modified MoCel6A enzymes and the unmodified control enzyme (each 0.1 μg) on cellulose, PSC, and β-glucan were assayed. The data are the means of three determinations ± SE; the asterisks represent significant t test values (P < 0.01) compared with that of MoCel6A.
In comparison with the hydrolysis of cellulose, MoCel6A showed higher activities for PSC, β-glucan, and cellooligosaccharides, which all have β-1,4-linked glucosyl units (Table 1 and Fig. 3A). The ability of MoCel6A to digest β-glucan is advantageous for hydrolyzing monocotyledonous cell walls because β-glucan is one of the main hemicellulosic polysaccharides found in monocots and is proposed to cover the surfaces of cellulose microfibrils (8, 9).
Among the carbohydrates tested, the best substrates for MoCel6A were cellooligosaccharides, such as cellotetraose, cellopentaose, and cellohexaose. To define the mode of action of cellobiohydrolases, cellooligosaccharide derivatives, such as 4-nitrophenyl cellobioside, α- and β-cellobiosyl fluorides, 4-methylumbelliferyl cellobioside, and 1-3H-labeled cellooligosaccharides, have been utilized (3, 44). Fluorescently labeled sulforhodamine-conjugated cellooligosaccharides, such as SR-C5 and SR-C6, used in this study, were also found to be suitable substrates for MoCel6A (Fig. 3B). The use of sulforhodamine-conjugated cellooligosaccharides is beneficial for readily distinguishing the effects of high concentrations of cellooligosaccharides. SR-C4 was not a substrate for MoCel6A but may be useful as a nonhydrolyzable substrate to form enzyme-substrate complexes for 3D structural analysis.
Competitive inhibition by end products is a significant issue for not only cellobiohydrolases, but also other hydrolytic enzymes in efforts to establish conditions for the efficient degradation of carbohydrates. Cellobiose is the main product liberated from cellulose derivatives by the action of cellobiohydrolases and typically decreases the hydrolytic activity of cellobiohydrolases (13, 15, 28, 31, 36, 37, 44). The activity of T. reesei Cel6A (formerly CBH II) was not inhibited by up to 2.5 mM cellobiose, whereas T. reesei and H. insolens Cel7A (formerly CBH I) showed decreased activity in the presence of up to 2.9 mM and 0.5 mM cellobiose, respectively (11, 37). Thus, Cel7A-type enzymes appear to be inhibited by a few millimolar cellobiose, but Cel6A-type enzymes do not. In our results, the activity of MoCel7A, which releases cellobiose from cellulose derivatives as well as MoCel6A, was negatively influenced by more than 29 mM cellobiose, similar to other Cel7A-type enzymes, but MoCel6A was not inhibited, and rather, was enhanced about 2-fold by the addition of 292 mM cellobiose (Fig. 4). The increase in hydrolytic activity was shown not only in MoCel6A, but also in TrCel6A at acidic pH (Fig. 5). Increased activity by high concentrations of cellobiose may be shared in GH family 6 cellobiohydrolases. On the other hand, TrCel6A was strongly inhibited at pH 9.0, but MoCel6A showed only slightly decreased activity. Originally, MoCel6A prepared in both M. oryzae and A. oryzae exhibited favored hydrolysis under alkaline conditions, which may not be susceptible to the inhibitor effect of cellobiose. This property may be caused by the amino acid composition of MoCel6A peptides.
MoCel6A has substitutions at amino acids that are highly conserved among GH family 6 cellobiohydrolases (see Fig. S3 in the supplemental material), some of which are proposed to be associated with substrate binding (41-43). These substitutions may influence pH dependency and the effects of high concentrations of cellobiose on hydrolytic activity. For example, Y169F in T. reesei Cel6A is proposed to play roles in binding to glycosyl residues by a direct hydrogen bond and in pH dependency by forming network interactions, ensuring the protonation of the acid catalyst of D221 (27).
In conclusion, MoCel6A has the ability to hydrolyze cellulose at a broad range of pHs, even under alkaline conditions, and its activity can be enhanced by high concentrations of cellobiose. These properties contribute to the efficient hydrolysis of plant cell walls, because high concentrations of cellobiose and glucose that accumulate during enzymatic hydrolysis are desired in the process of bioethanol production. Experiments are in progress using mutant enzymes to verify how these properties of MoCel6A are determined.
Supplementary Material
Acknowledgments
We are grateful to Takahisa Hayashi for helpful discussions.
Footnotes
Published ahead of print on 13 August 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amano, Y., M. Shiroishi, K. Nisizawa, E. Hoshino, and T. Kanda. 1996. Fine substrate specificities of four exo-type cellulases produced by Aspergillus niger, Trichoderma reesei, and Irpex lacteus on (1→3), (1→4)-β-D-glucans and xyloglucan. J. Biochem. 120:1123-1129. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., R. Lamed, B. A. White, and H. J. Flint. 2008. From cellulosomes to cellulosomics. Chem. Rec. 8:364-377. [DOI] [PubMed] [Google Scholar]
- 3.Becker, D., K. S. H. Johnson, A. Koivula, M. Schulein, and M. L. Sinnott. 2000. Hydrolyses of α- and β-cellobiosyl fluorides by Cel6A (cellobiohydrolase II) of Trichoderma reesei and Humicola insolens. Biochem. J. 345:315-319. [PMC free article] [PubMed] [Google Scholar]
- 4.Beckerman, J. L., and D. J. Ebbole. 1996. MPG1, a gene encoding a fungal hydrophobin of Magnaporthe grisea, is involved in surface recognition. Mol. Plant Microbe Interact. 9:450-456. [DOI] [PubMed] [Google Scholar]
- 5.Bhat, M. K. 2000. Cellulases and related enzymes in biotechnology. Biotechnol. Adv. 18:355-383. [DOI] [PubMed] [Google Scholar]
- 6.Boer, H., T. T. Teeri, and A. Koivula. 2000. Characterization of Trichoderma reesei cellobiohydrolase Cel7A secreted from Pichia pastoris using two different promoters. Biotechnol. Bioeng. 5:486-494. [DOI] [PubMed] [Google Scholar]
- 7.Boisset, C., C. Fraschini, M. Schulein, B. Henrissat, and H. Chanzy. 2000. Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from Humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl. Environ. Microbiol. 66:1444-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpita, N. C., and D. M. Gibeaut. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3:1-30. [DOI] [PubMed] [Google Scholar]
- 9.Carpita, N. C. 1996. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47:445-476. [DOI] [PubMed] [Google Scholar]
- 10.Dean, R. A., et al. 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980-986. [DOI] [PubMed] [Google Scholar]
- 11.Du, F., E. Wolger, L. Wallace, A. Liu, T. Kaper, and B. Kelemen. 2010. Determination of product inhibition of CBH1, CBH2, and EG1 using a novel cellulase activity assay. Appl. Biochem. Biotechnol. 161:313-317. [DOI] [PubMed] [Google Scholar]
- 12.Fry, S. C. 1997. Novel ‘dot-blot’ assays for glycosyltransferases and glycosylhydrolases: optimization for xyloglucan endotransglycosylase (XET) activity. 11:1141-1150. [Google Scholar]
- 13.Gardner, R. M., K. C. Doerner, and B. A. White. 1987. Purification and characterization of an exo-β-glucanase from Ruminococcus flavefaciens FD-1. J. Bacteriol. 169:4581-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomi, K., Y. Imura, and S. Hara. 1987. Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric. Biol. Chem. 51:2549-2555. [Google Scholar]
- 15.Gruno, M., P. Valjamae, G. Pettersson, and G. Johanssen. 2004. Inhibition of the Trichoderma reesei cellulases by cellobiose is strongly dependent on the nature of the substrate. Biotechnol. Bioeng. 86:503-511. [DOI] [PubMed] [Google Scholar]
- 16.Hägglund, P., T. Eriksson, A. Collén, W. Nerinckx, M. Claeyssens, and H. Stalbrand. 2003. A cellulose-binding module of the Trichoderma reesei β-mannanase Man5A increases the mannan-hydrolysis of complex substrates. J. Biotechnol. 101:37-48. [DOI] [PubMed] [Google Scholar]
- 17.Hefford, M. A., K. Laderoute, G. E. Willick, M. Yaguchi, and V. L. Seligy. 1992. Bipartite organization of the Bacillus subtilis endo-beta-1,4-glucanase revealed by C-terminal mutations. Protein Eng. 5:433-439. [DOI] [PubMed] [Google Scholar]
- 18.Henriksson, K., A. Teleman, T. Suortti, T. Reinikainen, J. Jaskari, O. Teleman, and K. Poutanen. 1995. Hydrolysis of barley (1→3),(1→4)-β-D-glucan by a cellobiohydrolase II preparation from Trichoderma reesei. Carbohydr. Polymers 26:109-119. [Google Scholar]
- 19.Henrissat, B. 1998. Enzymatic cellulose degradation. Cellulose Commun. 5:84-90. [Google Scholar]
- 20.Henrissat, B., H. Driguez, C. Viet, and M. Schulein. 1985. Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Biotechnology 3:722-726. [Google Scholar]
- 21.Himmel, M. E., S. Ding, D. K. Johnson, W. S. Adney, M. R. Nimlos, J. W. Brady, and T. D. Foust. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804-807. [DOI] [PubMed] [Google Scholar]
- 22.Hirayama, T., H. Nagayama, and K. Matsuda. 1978. Studies on cellulases of a phytopathogenic fungus, Pyricularia oryzae cavara. II. Purification and properties of a beta-glucosidase. J. Biochem. 84:27-37. [DOI] [PubMed] [Google Scholar]
- 23.Hirayama, T., H. Nagayama, and K. Matsuda. 1979. Studies on cellulases of a phytopathogenic fungus, Pyricularia oryzae Cavara. III Multiplicity of beta-glucosidase, and purification and properties of a second component. J. Biochem. 85:591-599. [DOI] [PubMed] [Google Scholar]
- 24.Hirayama, T., H. Nagayama, and K. Matsuda. 1980. Studies on cellulases of a phytopathogenic fungus, Pyricularia oryzae cavara. IV. Kinetic studies on beta-glucosidases. J. Biochem. 87:1203-1208. [PubMed] [Google Scholar]
- 25.Irwin, D. C., M. Spezio, L. P. Walker, and D. B. Wilson. 1993. Activity studies of eight purified cellulases: specificity, synergism and binding domain effects. Biotechnol. Bioeng. 42:1002-1013. [DOI] [PubMed] [Google Scholar]
- 26.Ito, J., Y. Fujita, M. Ueda, H. Fukuda, and A. Kondo. 2004. Improvement of cellulose-degrading ability of a yeast strain displaying Trichoderma reesei endoglucanase II by recombination of cellulose-binding domains. Biotechnol. Prog. 20:688-691. [DOI] [PubMed] [Google Scholar]
- 27.Koivula, A., T. Reinikainen, L. Ruohonen, A. Valkeajärvi, M. Claeyssens, O. Teleman, G. J. Kleywegt, M. Szardenings, J. Rouvinen, T. A. Jones, and T. T. Teeri. 1996. The active site of Trichoderma reesei cellobiohydrolase II: the role of tyrosine 169. Protein Eng. 9:691-699. [DOI] [PubMed] [Google Scholar]
- 28.Lahjouji, K., R. Storms, Z. Xiao, K. Joung, Y. Zheng, J. Powlowski, A. Tsang, and L. Varin. 2007. Biochemical and molecular characterization of a cellobiohydrolase from Trametes versicolor. Appl. Microbiol. Biotechnol. 75:337-346. [DOI] [PubMed] [Google Scholar]
- 29.Lever, M. 1972. A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 47:273-279. [DOI] [PubMed] [Google Scholar]
- 30.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorium. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morag, E., I. Halevy, E. A. Bayer, and R. Lamed. 1991. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J. Bacteriol. 173:4155-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nidetzky, B., W. Steiner, M. Hayn, and M. Claeyssens. 1994. Cellulose hydrolysis by the cellulases from Trichoderma reesei: a new model for synergistic interaction. Biochem. J. 298:705-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reese, E. T., R. G. H. Siu, and H. S. Levinson. 1950. The biological degradation of soluble cellulose derivatives and its relationship to the mechanism of cellulose hydrolysis. J. Bacteriol. 59:485-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reverbel-Leroy, C., S. Page, A. Belaich, J. P. Belaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouvinen, J., T. Bergfors, T. Teeri, J. K. Knowles, and T. A. Jones. 1990. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science 249:380-385. [DOI] [PubMed] [Google Scholar]
- 36.Schmidhalter, D. R., and G. Canevascini. 1993. Purification and characterization of 2 exocellobiohydrolases from the brown rot fungus Coniophora puteana (Schum Ex-Fr) Karst. Archiv. Biochem. Biophys. 300:5591-5598. [DOI] [PubMed] [Google Scholar]
- 37.Schülein, M. 1997. Enzymatic properties of cellulases from Humicola insolens. J. Biotechnol. 57:71-81. [DOI] [PubMed] [Google Scholar]
- 38.Sweigard, J. A., F. Chumley, A. Carroll, L. Farrall, and B. Calent. 1997. A series of vectors for fungal transformation. Fungal Genet. Newsl. 44:52-55. [Google Scholar]
- 39.Teeri, T., A. Koivula, M. Linder, G. Wohlfahrt, C. Divne, and T. A. Jones. 1998. Trichoderma reesei cellobhiohydrolases: why so efficient on crystalline cellulose? Biochem. Soc. Trans. 26:173-178. [DOI] [PubMed] [Google Scholar]
- 40.Tomme, P., E. Kwan, N. R. Gilkes, D. G. Kilburn, and R. A. J. Warren. 1996. Characterization of CenC, an enzyme from Cellulomonas fimi with both endo- and exoglucanase activities. J. Bacteriol. 178:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varrot, A., S. Hastrup, S. Martin, and G. J. Davies. 1999. Crystal structure of the catalytic core domain of the family 6 cellobiohydrolase II, from Humicola insolens, at 1.92 Å resolution. Biochem. J. 337:297-304. [PMC free article] [PubMed] [Google Scholar]
- 42.Varrot, A., T. P. Frandsen, H. Driguez, and G. J. Davies. 2002. Structure of the Humicola insolens cellobiohydrolase Cel6A D416A mutant in complex with a non-hydrolysable substrate analogue, methyl cellobiosyl-4-thio-β-cellobioside, at 1.9 Å. Acta Crystallogr. D Biol. Crystallogr. 58:2201-2204. [DOI] [PubMed] [Google Scholar]
- 43.Varrot, A., T. P. Frandsen, I. von Ossowski, V. Boyer, S. Cottaz, M. Schülein, and G. Davies. 2003. Structure basis for ligand binding and processivity in cellobiohydrolase Cel6A from Humicola insolens. Structure 11:855-864. [DOI] [PubMed] [Google Scholar]
- 44.Vrsanská, M., and P. Biely. 1991. The cellobiohydrolase I from Trichoderma reesei QM 9414: action on cello-oligosaccharides. Carbohydr. Res. 227:19-27. [Google Scholar]
- 45.Wood, T. M. 1992. Fungal cellulases. Biochem. Soc. Trans. 20:46-53. [DOI] [PubMed] [Google Scholar]
- 46.Yano, A., A. Kikuchi, Y. Nakagawa, Y. Sakamoto, and T. Sato. 2009. Secretory expression of the non-secretory-type Lentinula edodes laccase by Aspergillus oryzae. Microbiol. Res. 164:642-649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.