Abstract
The intestinal microbiota of broiler chickens and the microbiota in the litter have been well studied, but the interactions between these two microbiotas remain to be determined. Therefore, we examined their reciprocal effects by analyzing the intestinal microbiotas of broilers reared on fresh pine shavings versus reused litter, as well as the litter microbiota over a 6-week cycle. Composite ileal mucosal and cecal luminal samples from birds (n = 10) reared with both litter conditions (fresh versus reused) were collected at 7, 14, 21, and 42 days of age. Litter samples were also collected at days 7, 14, 21, and 42. The microbiotas were profiled and compared within sample types based on litter condition using PCR and denaturing gradient gel electrophoresis (PCR-DGGE). The microbiotas were further analyzed using 16S rRNA gene clone libraries constructed from microbiota DNA extracted from both chick intestinal and litter samples collected at day 7. Results showed significant reciprocal effects between the microbiotas present in the litter and those in the intestines of broilers. Fresh litter had more environmental bacteria, while reused litter contained more bacteria of intestinal origin. Lactobacillus spp. dominated the ileal mucosal microbiota of fresh-litter chicks, while a group of bacteria yet to be classified within Clostridiales dominated in the ileal mucosal microbiota in the reused-litter chicks. The Litter condition (fresh versus reused) seemed to have a more profound impact on the ileal microbiota than on the cecal microbiota. The data suggest that the influence of fresh litter on ileal microbiota decreased as broilers grew, compared with temporal changes observed under reused-litter rearing conditions.
The intestines of broiler chickens harbor a complex microbiota that plays an important role in the growth and health of the bird. Nurmi and Rantala (27) provided the first reported evidence that a healthy gut microbiota could protect broiler chicks against a challenge by enteric pathogens. That study led to the concept of “competitive exclusion.” Since then, the colonization of the intestines by beneficial bacteria has been shown to promote epithelial cell turnover (34), increase mucous production (26), upregulate expression of genes involved in several important intestinal functions (14), and help with reinforcement of the mucosal barrier, modulation of the immune system, and metabolism of nutrients by the host (46). Although studies conducted using either cultivation-based or molecular biology analyses have documented that the intestinal microbiota of mature broiler chickens is relatively stable (10, 24, 45), this microbiota is still dynamic and can be manipulated to a large extent (8, 31, 37).
Commercial broiler flocks in the United States are primarily floor raised in enclosed, environmentally controlled facilities. Within these commercial broiler houses, poultry litter can be reused for a year or longer if managed well and maintained in a relatively dry state (6). At an estimated accumulation rate of 1.45 metric tons per 1,000 broilers, the U.S. poultry industry, which produces >8.5 billion broilers per year (39), produces >12.3 million metric tons of poultry litter annually (4). Management and disposal of such a large quantity of poultry litter are two of the greatest challenges faced by U.S. broiler producers (6). Reuse of poultry litter can help alleviate this challenge, but there is concern that the reused-litter environment, both biotic and abiotic, may negatively affect the intestinal microbiota of the bird, potentially resulting in poor health and reduced production efficiency.
Poultry litter is primarily a mixture of bedding materials and bird excreta. In addition to variation in the physiochemical properties, reused poultry litter harbors microbes of typically intestinal origin that are not commonly present in fresh litter (23, 35). During a broiler growth cycle, a constant influx of nutrients and intestinal microbes results in a complex litter microbiota. With continued reuse, the litter environment becomes more complex, which may have a profound impact on flock growth performance and health. Recognizing the importance of the litter environment, especially in commercial broiler houses with high stocking densities, several research groups have investigated how the physiochemical properties of broiler litter affect the growth and health of the birds (15) and the microbiota in the litter, using either cultivation-based or molecular biology techniques (22, 23, 33). Early studies focused on the effect of litter on enteric pathogens, including Salmonella (35, 12, 28) and cellulitis-causing Escherichia coli (38). From analyzing 16S rRNA gene clone libraries, Lu et al. (23) showed that in broiler litter at various stages of reuse, low-G+C Gram-positive bacteria predominated. Recent studies have focused on the interplay between the unique physiochemical conditions within the litter and the endogenous microbiota (22, 29), as well as the occurrence and persistence of antibiotic resistance in poultry litter (13, 17), providing some insight into the litter microbiota and the intestinal microbiota in broiler chickens. However, the relationship between litter and intestinal microbiotas has not been investigated. Since chickens consume some litter materials, we hypothesized that the intestinal and litter microbiotas may affect each other with respect to their composition and diversity. Using PCR with denaturing gradient gel electrophoresis (PCR-DGGE) and 16S rRNA gene clone library analysis, we tested this hypothesis by examining the effect of the litter microbiota on the ontogeny of the intestinal microbiota in broilers over a 6-week growth cycle.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Two flocks of newly hatched chicks were used in this study. The eggs from which the chicks were hatched were not disinfected. One flock was placed in broiler pens (30 birds/pen) with fresh pine shavings (referred to as fresh-litter chicks or birds), while the other flock was placed in a broiler house with litter that had been used for 2 years prior to the current study (referred to as reused-litter chicks or birds). Both flocks of chicks were fed a commercial all-vegetable, antibiotic-free diet. In the hatchery, all chicks were administered the anticoccidial spray vaccine Coccivac-B (Schering-Plough Animal Health) at 87% of the recommended spray dose (21 ml per 1,000 chicks). Intestinal mucosa and cecal luminal samples were collected from multiple birds (see below) from each flock at 7, 14, 21, and 42 days of age. On each sampling day, a group of 30 birds from each litter condition was randomly selected and immediately transported to the Poultry Research Center at the Ohio Agricultural Research and Development Center (OARDC), where they were randomly divided into three sets of 10 and humanely euthanized by means of cervical dislocation. The ileum (the region between Meckel's diverticulum and the ileocecal junction) was excised, slit lengthwise, and rinsed gently with sterile saline to remove digesta particles. The mucosa was scraped off with the blunt side of a scalpel to collect the ileal mucosa and adherent bacteria. The mucosal samples from each set of 10 birds were pooled into one composite sample and frozen at −80°C immediately after collection. The ceca from each set of 10 birds were excised and milked, and the cecal contents were pooled into one composite sample and frozen at −80°C. A total of three replicate composite samples were prepared per age group per litter condition per site (ileal and cecal). Subsamples of litter were collected and pooled on each sampling day. For the birds reared in the broiler house containing the reused litter, 6 subsamples of litter were collected from each of the following four locations: the brood area, along the water and feed lines, and along the walls, for a total of 24 litter subsamples. For the birds reared in pens containing the fresh pine shavings, 6 subsamples of litter were collected per pen. Fewer fresh-litter subsamples were collected because the smaller pen dimensions resulted in the brood area, water and feed lines, and walls being in closer proximity to one another, greatly reducing litter variation among sampling locations. Subsamples were pooled, mixed thoroughly, and frozen at −80°C.
Microbial community DNA was extracted from the mucosal, cecal, and litter samples using the RBB+C method (44). The quality of the DNA extracts was confirmed using agarose gel (0.8%) electrophoresis, and concentrations were quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE). The concentration of an aliquot of the extracts was adjusted to 50 ng/μl using Tris-EDTA (TE) buffer prior to PCR amplification.
PCR and DGGE.
The V3 hypervariable region of the prokaryotic 16S rRNA gene was amplified using a universal primer set (GC-357f [5′-CCTACGGGAGGCAGCAG-3′] and 519r [5′-ATTACCGCGGCKGCTG-3′]) and resolved using DGGE as reported previously (43). To eliminate artifact DGGE bands, the PCR was ended with a final 30-min extension step at 72°C to reduce the formation of heteroduplex PCR products (16). Due to the limited number of lanes on a DDGE gel, two of the three replicate composite samples from all sampling locations and at each time point were randomly chosen for DGGE analyses and run side by side on a given gel. DGGE gels for each sampling location (ileum, cecum, and litter) were analyzed individually, and no statistical analysis was performed on the DGGE profiles of the replicated composite samples. The DGGE gel images were analyzed using the BioNumerics software program (v.5.1; Applied Maths, Inc., Austin, TX). Bands were detected using both the band-searching algorithm and manual selection or deletion when necessary to ensure accuracy. External reference markers (100-bp ladder; Invitrogen Inc., Carlsbad, CA), and internal reference bands were used to normalize individual gels, allowing for adjustment of migration differences across lanes within a gel. Position tolerance for band matching was set at 1% to help correct for slight migratory variation. Migratory distance for each band was determined by the BioNumerics software and treated as an independent variable. The DGGE banding patterns were transformed into a binary (presence or absence of bands) correlation cross-products matrix. Principal component analysis (PCA) was performed using the PC-ORD software program (V4.01; MJM Software, Gleneden Beach, OR) on each binary matrix to provide visual assessment of the DGGE profiles. The DGGE banding patterns were also analyzed using the Proc GLM procedure of the SAS software program (v.9.1.3; SAS Institute Inc., Cary, NC) with significance declared at a P value of ≤0.05. The influence of the litter condition on banding patterns within a sampling day was analyzed with litter condition, DGGE profile, and litter condition-DGGE profile interaction as independent variables. A significant litter condition-DGGE profile interaction suggests that the DGGE profile (number or pattern of bands) differed between litter conditions for samples collected on the same day. The influence of bird age on banding patterns within a given litter condition (fresh or reused) was analyzed with bird age (sampling day), DGGE profile, and age-DGGE profile interaction as the independent variables. A significant age-DGGE profile interaction suggests that the DGGE profiles varied due to the sampling day within a given litter condition.
Construction of 16S rRNA gene libraries.
The microbiota diversity and composition were further examined using 16S rRNA gene clone libraries that were constructed from samples collected at day 7. Purified DNA was pooled from the two replicate composite samples of ileal mucosa, cecal lumen, and litter that were used in the DGGE analysis for both fresh and reused litter. PCR was carried out using the universal bacterial primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1525r (5′-AAGGAGGTGWTCCARCC-3′) (19) to amplify the near-full-length 16S rRNA gene. For each DNA sample, 1 μl of microbiota DNA template (50 ng/μl) was added to a 49-μl master mix (40.1 μl distilled water [dH2O], 5 μl 10× PCR buffer, 1.75 μl 50 mM MgCl2, 1 μl 3.36% bovine serum albumin, 0.4 μl 100 μM deoxynucleoside triphosphate [dNTP], 0.25 μl [each] 100 μM primer, and 0.25 μl 5-U/μl Platinum Taq high-fidelity polymerase). The PCR mix was first subjected to an initial 5-min denaturation step at 94°C. Subsequent cycles consisted of a 30-s denaturation step at 95°C, a 45-s annealing step at 55°C, and a 90-s elongation step at 72°C. After 35 cycles, there was a final 7-min extension at 72°C before a 4°C hold. The PCR products (∼1,500 bp) were confirmed using agarose gel (1%) electrophoresis.
The PCR product was cloned using a Topo TA cloning kit for sequencing (Invitrogen Inc., Carlsbad, CA). For each of the three samples (ileal mucosa, cecal lumen, and litter), per litter condition, 288 randomly selected clones were cultured in 1 ml LB broth supplemented with 50 ng/ml ampicillin in three 96-well culture blocks. Following incubation at 200 rpm at 37°C for 36 to 48 h, 240 μl/well of the broth culture was transferred to PCR plates. The plates were centrifuged at room temperature for 10 min to pellet the E. coli cells. The cell pellet was resuspended in 50 μl TE, and 1 μl thereof was used as a PCR template to screen for the presence of recombinant plasmids containing the cloned 16S rRNA gene using the primers M13f and M13r as done previously (21). Following confirmation of the PCR products (∼1,650 bp), the clones were grouped by restriction fragment length polymorphism (RFLP) using HaeIII and AluI to reduce sequencing redundancy (21). Clones exhibiting the same RFLP banding pattern were assumed to contain the same 16S rRNA gene. One representative clone was selected from each RFLP group and consolidated into one 96-well culture block for each of the three composite samples per litter condition. These clones were propagated in 96-well culture blocks and prepared as glycerol stocks. From the glycerol stock, a 150-μl aliquot of each clone was sent to the University of Washington High-Throughput Genomics Unit for DNA sequencing using the primer 907r (5′-CCGTCAATTCMTTTRAGTTT-3′) (19).
Sequence analysis.
Ambiguous nucleotides at both ends of each sequence read were trimmed off, and the remaining high-quality sequences were manually edited using the FinchTV software program, v.1.4.0 (Geospiza, Inc., Seattle, WA). Each of the sequences (>500 bp) were checked for errors and chimera formation using the Mallard (3) and Pintail (2) programs. Multiple sequence alignment was performed using the ClustalW software program (20). Distance matrices were constructed using DNADIST with Jukes-Cantor correction implemented within the PHYLIP V3.6 software package (Department of Genome Sciences, University of Washington, Seattle, WA). Distance matrices were processed using the DOTUR software program (36), which generates rarefaction curves and diversity indices. Sequences were classified using the Classifier program, and libraries were compared using the Lib Compare program implemented in the Ribosomal Database Project (41). All the sequences were also aligned against the Greengenes database using the NAST aligner (7). Sequences representing operational taxonomical units (OTUs) were added to “tree_all” of the Greengenes ARB database environment (25) (http://greengenes.lbl.gov/Download/Sequence_Data/Arb_databases/greengenes.arb.gz), using parsimony insertion. Twelve reference sequences from the database were kept, while all other sequences were removed.
Diversity indices, including OTU richness, maximum number of OTUs likely present in each of the libraries, evenness, and maximal Shannon-Wiener index (Hmax), were calculated as described previously (21). The Shannon-Wiener (H′) index was calculated using DOTUR (36).
Nucleotide sequence accession numbers.
Sequences identified in the present study were deposited in the GenBank database under accession numbers HM574695 to HM575172.
RESULTS
DGGE profiles of litter microbiota.
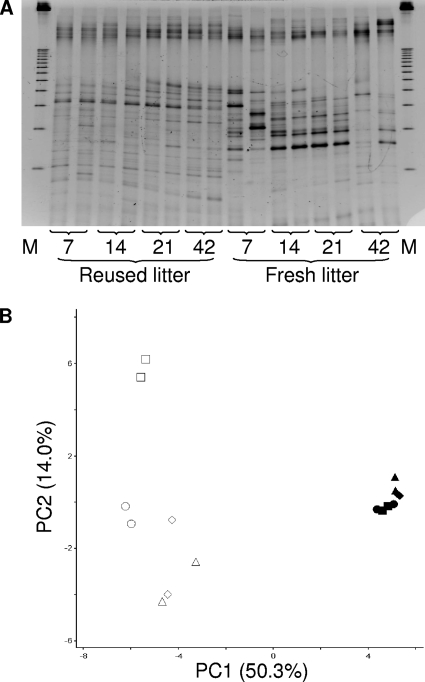
The reused-litter samples had significantly more DGGE bands per lane than the fresh-litter samples on all sampling days (Fig. 1 A). All the fresh litter samples, with the exception of the day 42 samples, had at least two intense bands, while the reused-litter samples had bands of relatively similar intensities. Principal component analysis (PCA) revealed clusters and separation between the fresh and the reused litter (Fig. 1B). Reused-litter samples formed a tight cluster and were separated from the fresh-litter samples along the first principal component (PC1) axis, which accounted for 50.3% of the variation. The second principal component (PC2) accounted for 14.0% of the variation, with the fresh-litter samples being separated by age along this axis. Fresh-litter samples collected at day 42 clustered apart from the fresh-litter samples collected at earlier time points. The third principal component (PC3) accounted for 8.0% of the variation (data not shown), with no separation trend. The results indicated a significant litter condition-DGGE profile interaction over the entire study period. With the exception of the fresh-litter samples collected at days 7 and 14 (P < 0.80), significant age-DGGE profile interactions were also observed for both litter conditions, indicating temporal successions in the litter microbiota as the birds grew.
FIG. 1.
DGGE banding profiles (A) and corresponding PCA analysis (B) of the litter microbiotas collected at days 7, 14, 21, and 42. In figure A, “M” represents the electrophoresis marker. In figure B, symbols are as follows: triangle, day 7; diamond, day 14; circle, day 21; square, day 42; open symbols, fresh litter samples; closed symbols, reused litter samples.
DGGE profiles of ileal mucosal microbiota.
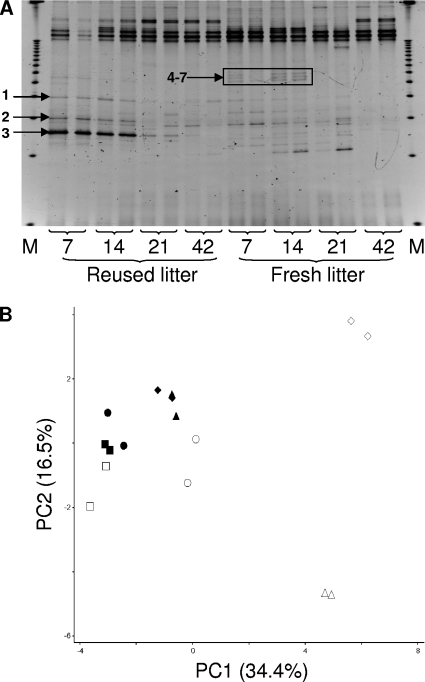
For the ileal mucosal samples, fresh-litter birds had more DGGE bands than their reused-litter counterparts at all the sampling days (Fig. 2 A). It was also noted that several bands decreased in intensity and then disappeared (e.g., bands 1 to 3 of the day 7 and 14 samples of the reused-litter birds), while a few bands appeared and then disappeared (e.g., the quadruplet bands 4 to 7 of the fresh-litter birds) over the same sampling period. For both litter conditions, a trend was observed in which the number of discernible bands per sample increased from day 7 to day 14 and then decreased thereafter, reflecting temporal succession in the ileal mucosal microbiota. Day 42 samples displayed the lowest number of bands per lane for both litter conditions. All lanes exhibited high-intensity bands in the upper portion of the gel (low denaturant), reflecting the possible presence of low-G+C bacteria at greater abundance in all the samples analyzed.
FIG. 2.
DGGE banding profiles (A) and corresponding PCA analysis (B) of the ileal mucosal microbiotas collected at days 7, 14, 21, and 42. In figure A, “M” represents the electrophoresis marker. Bands 1 to 3 and 4 to 7 changed intensity and disappeared over time, respectively. In figure B, symbols are as follows: triangle, day 7; diamond, day 14; circle, day 21; square, day 42; open symbols, fresh-litter birds; closed symbols, reused-litter birds.
The PC1 of the PCA on the ileal mucosal samples (Fig. 2B) accounted for 34.4% of the variation in band presence/absence and clearly identified separate DGGE profiles between fresh-litter and reused-litter chicks (7 and 14 days). Along the PC1, the day 21 samples from the fresh-litter birds tended to cluster with the samples from the young reused-litter chicks (7 and 14 days), and the day 42 samples from the fresh-litter birds clustered with the day 21 and 42 samples from the reused-litter birds. These results suggest that the ileal mucosal microbiota in the older fresh-litter birds became similar to that of the reused-litter birds in a delayed fashion. The PC2 accounted for 16.5% of the variation and mainly indicated separation between samples collected at days 7 and 14 of the fresh-litter chicks. The PC3 accounted for 15.6% of the variation (data not shown) with no separation trend.
The statistical analysis revealed significant litter condition-DGGE profile and age-DGGE profile interactions over the entire study period, indicating that DGGE profiles varied as a function of litter condition and bird age, suggesting that the ileal mucosal microbiota was affected by both litter condition and bird age.
DGGE profiles of cecal luminal microbiota.
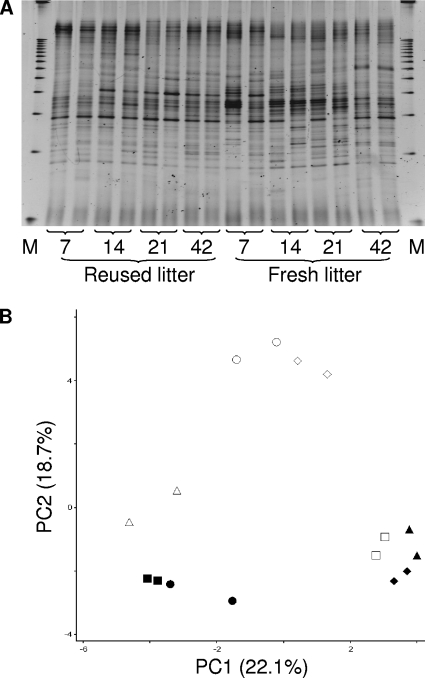
The cecal luminal samples had a large number of bands that were distributed along the entire gradient (Fig. 3 A). Relative to the litter and ileal mucosal samples, substantially more bands were resolved from the cecal luminal samples, as expected, irrespective of litter condition. A greater number of bands were resolved in the middle of the gel than in the top and bottom ends of the gel, and the number of bands per sample increased as the birds aged.
FIG. 3.
DGGE banding profiles (A) and corresponding PCA analysis (B) of the cecal luminal microbiotas collected at days 7, 14, 21, and 42. In figure A, “M” represents the electrophoresis marker. In figure B, symbols are as follows: triangle, day 7; diamond, day 14; circle, day 21; square, day 42; open symbols, fresh-litter birds; closed symbols, reused-litter birds.
The PC1 of the PCA on the cecal luminal samples (Fig. 3B) accounted for 22.1% of the variation in the presence/absence of bands and characterized clustering of samples collected from the young reused-litter chicks (days 7 and 14) with samples from the older fresh-litter birds (day 42). This cluster is separated along the PC1 from a cluster of samples collected at days 21 and 42 from the reused-litter birds and at day 7 from the fresh-litter chicks. A cluster of samples collected from the fresh-litter birds at days 14 and 21 is separated along the PC2 (18.7% of the variation) from all other cecal samples. The PC3 accounted for 12.2% of the variation (data not shown), with no separation trend.
A significant litter condition-DGGE profile interaction was observed at all sampling days, suggesting a significant effect of litter conditions on cecal luminal microbiota. The age-DGGE profile interaction was significant within each litter condition for all the samples, except for the samples collected from the reused-litter birds at days 14 and 21 (P < 0.09), suggesting temporal successions in the cecal luminal microbiota.
Phylogenetic diversity.
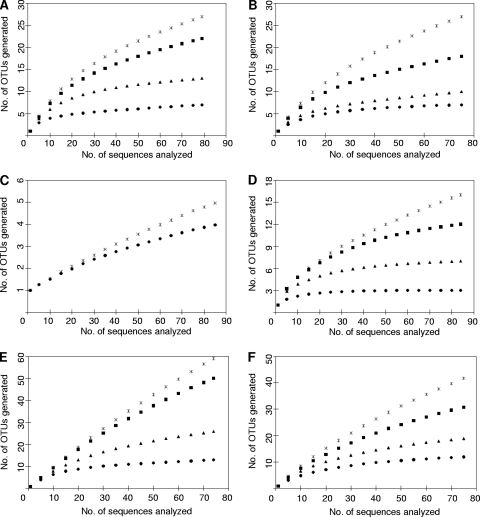
A total of 474 RFLP groups were identified from the 576 random clones selected from the samples collected at day 7. One representative clone from each of the RFLP groups was successfully sequenced. These sequences were free of chimeras and were of high quality and therefore were further analyzed. Rarefaction curves based on OTUs at 97% (equivalent to species), 95% (genus), 90% (family), and 85% (order) sequence similarity levels are presented in Fig. 4. At 90 and 85% similarity levels, nearly complete coverage was achieved for all the samples, except for the ileal mucosa of reused-litter chicks (Fig. 4C). At the 95% similarity level, the curves for the fresh litter (Fig. 4B) and the ileal mucosa of fresh-litter chicks (Fig. 4D) are approaching their asymptotes, suggesting that substantial coverage of the majority of the phylogenetic diversity was achieved for these samples but not for the remaining four samples (Fig. 4A, C, E, and F). Complete coverage was not achieved for any of the samples at the 97% similarity level (Fig. 4 and Table 1).
FIG. 4.
Rarefaction curves of clones sequenced from reused litter (A), fresh litter (B), ileal mucosa of reused-litter chicks (C), or fresh-litter chicks (D) or cecal content of reused-litter chicks (E) or fresh-litter chicks (F). Sequence similarity level: asterisks, 97%; squares, 95%; triangles, 90%; circles, 85%. There is no 95 or 90% similarity curve for the ileal clones of reused-litter chicks (see Fig. 3C) because the few OTUs identified for this sample differed only by a count of 1 between 97 and 85% similarity.
TABLE 1.
OTUs (97% similarity) observed and predicted, estimated percent coverage, and diversity indices for clones from ileal mucosa and cecal contenta
| Sample location | Litter | No. of clones | No. of OTUs observed | No. of OTUs predicted | Estimated coverage (%) | Richness (d) | Shannon-Wiener index (H′) | Maximum Shannon-Wiener index (H′max) | Eveness (e) |
|---|---|---|---|---|---|---|---|---|---|
| Litter | Reused | 79 | 27 | 31 | 87.10 | 13.70 | 2.93 | 4.36 | 2.05 |
| Fresh | 75 | 26 | 35 | 74.26 | 13.33 | 2.78 | 4.31 | 1.96 | |
| Ileum | Reused | 86 | 5 | 12 | 41.67 | 2.07 | 0.30 | 4.45 | 0.43 |
| Fresh | 84 | 16 | 21 | 76.19 | 7.80 | 1.86 | 4.43 | 1.55 | |
| Cecum | Reused | 74 | 58 | 160 | 36.25 | 30.49 | 3.99 | 4.30 | 2.26 |
| Fresh | 76 | 42 | 74 | 56.76 | 21.80 | 3.40 | 4.33 | 2.09 |
From 7-day-old broiler chicks and litter samples collected at day 7.
The fresh litter and the reused litter had very similar numbers of OTUs identified and had similar diversity indices (Table 1). Coverage was quite complete, with approximately 25% (or less) of predicted OTUs remaining to be identified. All but one of the 79 sequenced RFLP groups from the reused litter belonged to the phylum Firmicutes (Table 2 and Fig. 5). The class Bacilli was the most predominant class observed in the reused litter, with Lactobacillus, Staphylococcus, and Jeotgalicoccus representing the predominant genera and accounting for 20.5%, 19.2%, and 17.9% of the sequenced RFLP groups, respectively. Unclassified Bacillaceae-like sequences (about 15% of the sequenced RFLP groups) were also found. The majority of the bacteria found in the fresh litter belong to Firmicutes (77.3%), with Clostridia being the most predominant class and a candidate genus, Lachnospiraceae incertae sedis, representing 71.4% of the RFLP types thereof. Lactobacillus and Enterococcus were represented by 1 and 8 RFLP groups, respectively. Bacteria in the phylum Proteobacteria (22.7%) were also identified in the fresh-litter but not in the reused-litter samples. Within the Proteobacteria, Acinetobacter spp. were predominant, accounting for 58.8% of the RFLP groups, with the remaining RFLP groups belonging to Pseudomonas and enterobacteria.
TABLE 2.
Taxonomical hierarchy of 474 sequenced clones from the ileal mucosa and cecal content of 7-day-old broiler chicks and litter samples collected at day 7a
| Taxonomical hierarchy | Name | No. of clones |
|||||
|---|---|---|---|---|---|---|---|
| RL | FL | IR | IF | CR | CF | ||
| Phylum | Actinobacteria | 1 | |||||
| Class | Actinobacteridae | 1 | |||||
| Order | Actinomycetales | 1 | |||||
| Family | Dermabacteraceae | 1 | |||||
| Genus | Brachybacterium | 1 | |||||
| Phylum | Proteobacteria | 17 | 1 | ||||
| Class | Gammaproteobacteria | 17 | 1 | ||||
| Order | Pseudomonadales | 13 | |||||
| Family | Moraxellaceae | 10 | |||||
| Genus | Acinetobacter | 10 | |||||
| Family | Pseudomonadaceae | 3 | |||||
| Genus | Pseudomonas | 3 | |||||
| Order | Enterobacteriales | 4 | 1 | ||||
| Family | Enterobacteriaceae | 4 | 1 | ||||
| Unclassified Enterobacteriaceae | 1 | ||||||
| Genus | Klebsiella | 1 | |||||
| Genus | Shigella | 1 | 1 | ||||
| Genus | Enterobacter | 1 | |||||
| Phylum | Deferribacteres | 1 | |||||
| Class | Deferribacteres | 1 | |||||
| Order | Deferribacterales | 1 | |||||
| Family | Deferribacteraceae | 1 | |||||
| Genus | Mucispirillum | 1 | |||||
| Phylum | Firmicutes | 78 | 58 | 86 | 84 | 74 | 75 |
| Class | Bacilli | 76 | 9 | 3 | 67 | 3 | 1 |
| Unclassified Bacilli | 1 | ||||||
| Order | Lactobacillales | 27 | 9 | 3 | 67 | 2 | 1 |
| Family | Aerococcaceae | 2 | |||||
| Genus | Aerococcus | 2 | |||||
| Family | Lactobacillaceae | 16 | 1 | 3 | 63 | 2 | 1 |
| Genus | Lactobacillus | 16 | 1 | 3 | 63 | 2 | 1 |
| Family | Streptococcaceae | 1 | |||||
| Genus | Streptococcus | 1 | |||||
| Family | Enterococcaceae | 4 | 8 | 0 | 4 | ||
| Genus | Enterococcus | 4 | 8 | 0 | 4 | ||
| Family | Carnobacteriaceae | 4 | |||||
| Genus | Atopostipes | 4 | |||||
| Order | Bacillales | 49 | |||||
| Family | Bacillaceae | 12 | |||||
| Unclassified Bacillaceae | 12 | ||||||
| Family | Staphylococcaceae | 37 | |||||
| Unclassified Staphylococcaceae | 2 | ||||||
| Genus | Jeotgalicoccus | 14 | |||||
| Genus | Salinicoccus | 6 | |||||
| Genus | Staphylococcus | 15 | |||||
| Class | Clostridia | 2 | 49 | 83 | 17 | 70 | 74 |
| Order | Clostridiales | 2 | 49 | 83 | 17 | 70 | 74 |
| Unclassified Clostridiales | 81 | 12 | 6 | ||||
| Family | Lachnospiraceae | 2 | 46 | 13 | 14 | 44 | |
| Unclassified Lachnospiraceae | 1 | 10 | 3 | 3 | 9 | ||
| Genus | Hespellia | 1 | |||||
| Genus | Lachnospiraceaeincertae sedis | 1 | 35 | 10 | 11 | 35 | |
| Family | Ruminococcaceae | 3 | 2 | 4 | 44 | 20 | |
| Unclassified Ruminococcaceae | 3 | 1 | 1 | 25 | 7 | ||
| Genus | Anaerotruncus | 1 | 1 | ||||
| Genus | Ruminococcaceaeincertae sedis | 1 | 1 | 4 | |||
| Genus | Subdoligranulum | 9 | |||||
| Genus | Faecalibacterium | 1 | 6 | 8 | |||
| Genus | Papillibacter | 2 | 2 | ||||
| Family | Veillonellaceae | 2 | |||||
| Genus | Acidaminococcus | 2 | |||||
| Family | Clostridiaceae | 2 | |||||
| Unclassified Clostridiaceae | 2 | ||||||
| Total no. of clones | 79 | 75 | 86 | 84 | 74 | 76 | |
Constructed using the Ribosomal Database Project classifier. LR, reused-litter clones; LF, fresh-litter clones; IR, ileal clones of reused-litter birds; IF, ileal clones of fresh-litter birds; CR, cecal clones of reused-litter birds; CF, cecal clones of fresh-litter birds.
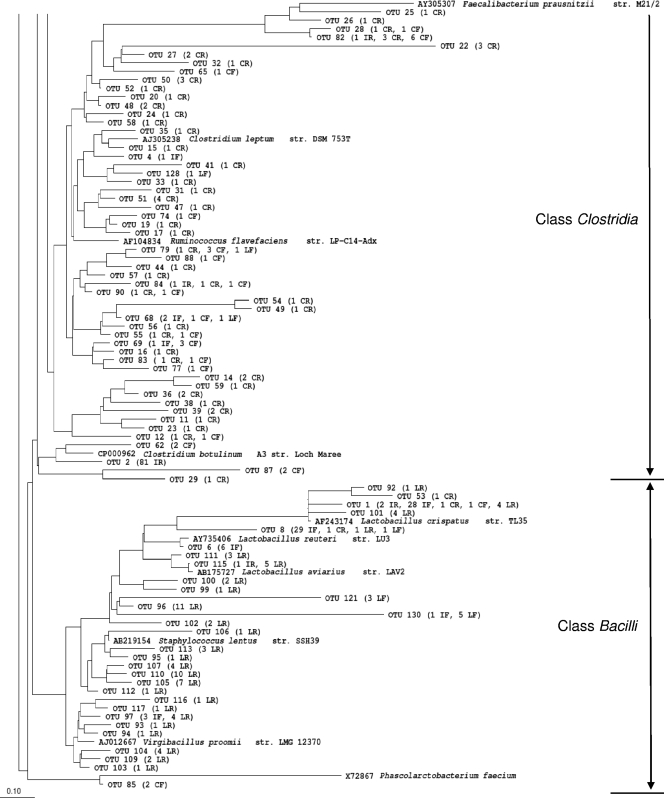
FIG. 5.
Phylogenetic tree of OTUs defined at 97% similarity. LR, reused litter; LF, fresh litter; IR, ileal mucosa of reused-litter chicks; IF, ileal mucosa of fresh-litter chicks; CR, cecal content of reused-litter chicks; CF, cecal content of fresh-litter chicks.
The number of species-level OTUs varied considerably among the intestinal samples, even though similar numbers of RFLP groups were sequenced for each sample (Table 1). Only 5 and 16 OTUs were identified from the ileal mucosal microbiota of the fresh- and reused-litter chicks, respectively. All of these OTUs were assigned to Firmicutes (Table 2). Of the 86 RFLP groups recovered from the ileal mucosal microbiota of the reused-litter chicks, 81 shared ≥97% sequence similarity and represented a novel group of bacteria in the order Clostridiales. The remaining 5 RFLP groups formed four additional species-level OTUs. Consequently, the ileal mucosal microbiota had the lowest species richness, Shannon-Wiener diversity, and evenness indices compared with the cecal luminal and litter samples. This was not surprising, given that only bacteria capable of adhering to the mucosa can colonize the ileal mucosa, and it is consistent with previous reports (9, 10). Compared with the reused-litter chicks, the fresh-litter chicks had more OTUs identified and greater species richness in the ileal mucosal microbiota, resulting in greater Shannon-Wiener diversity and evenness indices. The most dominant OTU was assigned to Lactobacillus (63 of the 84 total RFLP groups), which was a minority (3 of the 86 total RFLP groups) in the reused-litter chicks. Genera identified in the ileal mucosal microbiota of the fresh-litter chicks also included Enterococcus (4 RFLP groups), Papillibacter (2 RFLP groups), and several unclassified genera, including the candidate genus Lachnospiraceae incertae sedis, which was found to be predominant in the fresh-litter samples.
As expected, the cecal luminal samples had the greatest identified and predicted richness and diversity compared with the ileal mucosal and litter samples (Table 1). The cecal luminal samples of the reused-litter chicks had significantly greater diversity than that of the fresh-litter chicks. Coverage at the species level was rather low, especially for the cecal luminal samples of the reused-litter chicks. All but one RFLP group from the cecal luminal clone libraries belonged to Firmicutes, but only a few OTUs could be classified to known genera (Table 2): Klebsiella, Subdoligranulum, and Papillibacter in the cecum of the reused-litter chicks, Musispirillum and Acidaminococcus in the cecum of the fresh-litter chicks, and Anaerotruncus and Faecalibacterium in all the ceca irrespective of litter conditions. The predominant OTU in the cecal clone library of the reused-litter chicks was related to an unclassified group of Ruminococcaceae, while the predominant OTU from the fresh-litter chicks belonged to the candidate genus Lachnospiraceae incertae sedis.
Collectively, the 474 sequenced RFLP groups from all the samples collected at day 7 were characterized into 136 species-level OTUs (Fig. 5). All the sequences were classified into one of four bacterial phyla. Assigned to Proteobacteria were five OTUs representing clones derived predominantly from the fresh-litter samples and a few clones from the cecal lumen of reused-litter chicks. Deferribacteres and Actinobacteria had one OTU each, representing only one clone, from the cecal lumen of fresh-litter chicks and the reused litter, respectively. The remaining 129 OTUs primarily represent Gram-positive bacteria of either Clostridia or Bacilli within Firmicutes. The reused litter clones were assigned to either Lactobacillaceae or Staphylococcaceae. The majority of fresh-litter clones that did not belong to the Proteobacteria, together with the majority of cecal luminal clones and ileal mucosal clones collected from the reused-litter chicks, were assigned to Clostridia. The vast majority of ileal mucosal clones collected from the fresh litter were classified to Lactobacillaceae.
When the sequences of the 136 OTUs were compared to the RDP database, less than 12% (16 OTUs) were ≥97% identical to the sequences of known bacterial species, while another 4 OTUs (2.9%) were 95 to 96.9% identical. An additional 5 OTUs (3.7%) were 90 to 94.9% identical to known bacterial species. The remaining 111 (81.6%) OTUs were <90% identical to known bacterial species within the database. These results indicate that the microbiotas of poultry intestines and litter have not been sufficiently characterized and that most of the bacteria in these habitats remain to be identified.
DISCUSSION
The reused and the fresh litters differed in DGGE profiles, with the reused litter producing more DGGE bands (Fig. 1). This difference was further revealed by the detection of different bacteria within Proteobacteria and Firmicutes between the two litter conditions. Members of the Proteobacteria were found in the fresh litter (e.g., Acinetobacter, Pseudomonas, and enterobacteria) but not in the reused litter (Table 2 and Fig. 5). Differences were also evident within Firmicutes, the predominant phylum in both litter conditions. Typical intestinal bacteria, such as Lactobacillus, Staphylococcus, Jeotgalicoccus, Salinicoccus, Atopostipes, and a group of unclassified genera of the family Bacillaceae, were the major genera in the reused litter, whereas the candidate genus Lachnospiraceae incertae sedis and Enterococcus were predominant in the fresh litter. One unclassified genus (each) belonging to the family Lachnospiraceae and the family Ruminococcaceae was also present in the fresh litter, but only the unclassified genus belonging to the family Lachnospiraceae was present in the reused litter. Because both unclassified genera were also found to be predominant in the cecal luminal samples of both groups of birds, they are probably of intestinal origin. It remains to be determined why these two genera were more predominant in the fresh litter than in the reused litter. The intestinal bacteria from the excreta should constitute the primary inoculants in the fresh litter, while the more-permissive conditions (e.g., increased nutrients and moisture) in the reused litter than in the fresh litter may explain some of the observed differences. Salinococcus was a predominant genus identified by Lu et al. (23) in reused litter from modern poultry houses located in northeast Georgia, but this genus was not observed in this study. The influence of the geographical region on variation in litter microbiota may be an area for future research.
The microbiota under both litter conditions underwent temporal community succession during the sampling period but to a greater extent in the fresh litter. The deposition of excreta onto the fresh litter might have rapidly altered the biotic and abiotic environments, and thus the microbiota, therein during the study. This hypothesis is consistent with previous studies that demonstrated temporal changes in pH, cellulolytic breakdown, moisture, and nutrient content within poultry litter (5, 22, 30). It should be noted that considerable variation in DGGE profiles was seen between replicate samples of the same litter condition, which indicates heterogeneity in the microbiota among the sampling locations. Indeed, Lovanh et al. (22) reported that the DGGE profiles of litter samples collected along the water and feed lines strongly clustered (cophenetic correlation coefficient ≥ 82%) apart from litter samples collected elsewhere in the same house. New strategies to prepare representative litter samples are needed in future studies.
In this study, we chose to examine the ileal mucosal microbiota because the mucosa serves as the interface between the host and intestinal microbiota, and thus, mucosal microbiota directly interact with and affect the host, as reviewed by Rautava and Walker (32). Both the DGGE and clone library analyses showed clear differences in ileal mucosal microbiota between birds reared under the two litter conditions (Tables 1 and 2 and Fig. 2), suggesting a profound impact of litter on ileal mucosal microbiota. A significant litter condition-DGGE profile interaction also supports this conclusion. As revealed by the greater number of DGGE bands and OTUs observed, the fresh-litter birds had greater bacterial diversity in the ileal mucosa than the reused-litter birds. This intriguing observation cannot be fully explained in the present study, but the presence of environmental microbes in the fresh litter and the greater evenness in the ileal mucosal microbiota might be partially responsible for such a difference. At 7 days of age, the fresh-litter chicks had an ileal mucosal microbiota primarily dominated by Lactobacillus, followed by unclassified Lachnospiraceae and Enterococcus, whereas the ileal mucosa of the reused-litter chicks were primarily colonized by bacteria that cannot be classified within the order Clostridiales. It is interesting to note that species of Lactobacillus were more predominant in the reused litter than in the fresh litter, but it was the ileal mucosal microbiota of the fresh-litter chicks that was dominated by members of this genus. Data from the present study cannot explain this “discrepancy.” It might be speculated that the fresh litter lacked Clostridiales-like bacteria and thus there was less ingestion of these bacteria. Consequently, there was less competition against these bacteria for Lactobacillus to colonize the ileum of the fresh-litter chicks than was the case with the ileum of reused-litter chicks. At the end of the sampling period (day 42), the ileal mucosa of the fresh-litter birds had a microbiota similar to that found in the reused-litter chicks (Fig. 2B). Such a convergence indicates a diminishing effect from the environmental bacteria originally present in the fresh litter, since the intestinal bacteria from the excreta accumulated in the fresh litter. Lu et al. (24) observed an increase from day 3 to day 49 in Clostridium spp. in the broiler ileum. The lower evenness and predominance of clostridia in the ileal mucosal microbiota of the reused-litter chicks than that of the fresh-litter chicks at age 7 days (Tables 1 and 2) suggest that reused litter can aid the acquisition and development of the intestinal microbiota in chicks. Future studies involving sequencing of 16S rRNA gene clones from both younger fresh-litter birds and corresponding litter samples may help verify this notion and identify the participating bacteria.
During the sampling period, the ileal mucosal microbiota also underwent community succession. This conclusion is drawn from the appearance and disappearance of some DGGE bands (Fig. 2A) and the significant age-DGGE profile interaction. Irrespective of litter conditions, ileal mucosal samples showed a similar trend in which the number of DGGE bands increased from day 7 to day 14, corroborating the notion that complexity and diversity of the intestinal microbiota increase as the bird ages. However, a decrease in the number of bands was observed from days 14 to 42, suggesting a subsequent loss of microbial diversity within the ileal mucosa. This trend does not completely agree with previous findings. van der Wielen et al. (40) showed that the number of DGGE bands for the broiler ileum increased from day 1 to day 11, remained unchanged from day 11 to 28, and then increased from day 28 to 39 from chickens fed several antibiotics: nicarbzin and avilamycin from day 1 to 10, avilamycin and salinomycin from day 11 to 33, and avilamycin from day 34 to 40. Gong et al. (11) showed an increase in ileal microbiota richness (an increased number of DGGE bands) from day 3 to 42 in broilers fed an antibiotic-free diet. In both aforementioned studies, the ileal samples analyzed consisted of either digesta or a composite of both digesta and mucosa. An age-dependent increase in ileal microbiota diversity should not be attributed to antibiotic usage or the lack thereof; rather, subtherapeutic antibiotic inclusion in poultry diets has been characterized as having a more profound impact on the intestinal microbiota composition than on the richness at different ages within the broiler ileum and cecum (5, 18, 31, 42). The “loss” of microbial diversity observed in the current study might be a function of the sample type or analysis methods used.
In this study, we chose to examine the cecal luminal microbiota because it is the direct source of the intestinal bacteria found in litter samples. Consistent with previous studies (1, 9, 10, 24, 40), the microbiota in the cecal lumen was observed to be complex (Fig. 3A). Regardless of litter conditions, the complexity of the cecal microbiota also increased with age. These findings are in agreement with those of van der Wielen et al. (40) and Lu et al. (24). Compared to the DGGE profiles of the ileal mucosal microbiota, the DGGE profiles of the cecal luminal samples displayed greater homogeneity at all sampling days irrespective of litter conditions. These findings could be attributed to a less-pronounced impact of the litter on the cecal microbiota than on the ileal microbiota.
Irrespective of litter conditions, most of the OTUs identified in the cecal luminal samples belonged to the class Clostridia (Table 2 and Fig. 5). However, Lachnospiraceae incertae sedis, which was absent in the ileal mucosa of the reused-litter chicks but predominant in the ileal mucosa of the fresh-litter chicks, continued to be the predominant genus found in the cecal lumen of this group of chicks, while Ruminococcaceae, which was minor in the ileal mucosa irrespective of litter conditions, was predominant (particularly Subdoligranulum and the unclassified Ruminococcaceae) in the cecal lumen of the reused-litter chicks. It is also obvious that the group of bacteria yet to be classified within Clostridiales, which was dominant in the ileal mucosa (83 OTUs) of the reused-litter chicks, was also predominant in the cecal lumen (12 OTUs) of this group of chicks. However, this group of bacteria, which was not found in the ileal mucosa of the fresh-litter chicks, was less predominant in the cecal lumen (6 OTUs) of the fresh-litter chicks. It is not certain why rearing birds on reused litter resulted in greater abundance of this group of clostridia while fresh litter favored lachnospira in the cecal lumen. This group of clostridia may be a common bacterial group in the cecum of older chickens, while relatively fresh litter might be the preferred habitat of the lachnospira. Future studies examining both litter conditions over time will help to verify this premise. Unlike the DGGE profiles of the ileal mucosal microbiota, the DGGE profiles of the cecal luminal microbiota did not cluster with respect to litter conditions at the end of the sampling period (Fig. 3A). Future studies are needed to verify if litter impacts on cecal microbiota diminish as excreta accumulate on fresh litter over time.
Taken together, poultry litter conditions can significantly affect the intestinal microbiota, especially in the upper small intestines. The intestinal microbiota of the fresh-litter birds probably harbored more bacteria of litter material origin, while the gut of reused-litter chicks is largely colonized with bacteria of intestinal origin, conceivably excreted from the previous flocks. The cycling of certain bacteria between litter and intestines of poultry is expected. Differences in physiological features, such as tolerance to oxygen, dehydration, and pH, will likely cause different bacteria to persist for various periods of time in the litter and intestines. It should be noted that this study used the traditional Sanger sequencing technology and only a limited number of clones were sequenced. As such, the diversity coverage was limited (Table 1). Future studies using more-comprehensive analyses will help evaluate the scope of the reciprocal effect between litter and intestinal microbiota. Future studies using quantitative analyses are also needed to assess the extent of the effect of exchange of bacteria between litter and intestines, especially with regard to poultry pathogens because of their pragmatic importance to bird health and disease.
Acknowledgments
This work was partially supported by a NIFA grant (award number 2008-35204-18845) to Michael Lilburn and Zhongtang Yu and was partially supported by the Ohio Agriculture Research and Development Center (OARDC) (Wooster, OH).
We thank Normand St. Pierre (Department of Animal Sciences, Ohio State University) for his kind help with statistical analyses. We also thank John Anderson and Keith Patterson of the OARDC for their critical involvement in the care and maintenance of the birds used in this study.
Footnotes
Published ahead of print on 6 August 2010.
REFERENCES
- 1.Amit-Romach, E., D. Sklan, and Z. Uni. 2004. Microflora ecology of the chicken intestine using 16S ribosomal DNA primers. Poult. Sci. 83:1093-1098. [DOI] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamblee, T. N., and R. L. Todd. 2002. Mississippi broiler litter: fertilizer value and quantity produced. Mississippi Agricultural and Forestry Experiment Station research report, vol. 23(5). Mississippi Agricultural and Forestry Experiment Station, Mississippi State, MS.
- 5.Coates, M. E., R. Fuller, G. F. Harrison, M. Lev, and S. F. Suffolk. 1963. A comparison of the growth of chicks in the Gustafsson germ-free apparatus and in a conventional environment, with and without dietary supplements of penicillin. Br. J. Nutr. 17:141-150. [DOI] [PubMed] [Google Scholar]
- 6.Coufal, C. D., C. Chavez, P. R. Niemeyer, and J. B. Carey. 2006. Measurement of broiler litter production rates and nutrient content using recycled litter. Poult. Sci. 85:398-403. [DOI] [PubMed] [Google Scholar]
- 7.DeSantis, T. Z., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engberg, R. M., M. S. Hedemann, S. Steenfeldt, and B. B. Jensen. 2004. Influence of whole wheat and xylanase on broiler performance and microbial composition and activity in the digestive tract. Poult Sci. 83:925-938. [DOI] [PubMed] [Google Scholar]
- 9.Gong, J., R. J. Forster, H. Yu, J. R. Chambers, P. M. Sabour, R. Wheatcroft, and S. Chen. 2002. Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum. FEMS Microbiol. Ecol. 41:171-179. [DOI] [PubMed] [Google Scholar]
- 10.Gong, J., W. Si, R. J. Forster, R. Huang, H. Yu, Y. Yin, C. Yang, and Y. Han. 2007. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol. Ecol. 59:147-157. [DOI] [PubMed] [Google Scholar]
- 11.Gong, J., H. Yu, T. Liu, J. J. Gill, J. R. Chambers, R. Wheatcroft, and P. M. Sabour. 2008. Effects of zinc bacitracin, bird age and access to range on bacterial microbiota in the ileum and caeca of broiler chickens. J. Appl. Microbiol. 104:1372-1382. [DOI] [PubMed] [Google Scholar]
- 12.Hacking, W. C., W. R. Mitchell, and H. C. Carlson. 1978. Sources of salmonellae in broiler chickens in Ontario. Can. J. Comp. Med. 42:392-399. [PMC free article] [PubMed] [Google Scholar]
- 13.Hofacre, C. L., A. R. de Cotret, J. J. Maurer, A. Garritty, and S. G. Thayer. 2000. Presence of fluoroquinolone-resistant coliforms in poultry litter. Avian Dis. 44:963-967. [PubMed] [Google Scholar]
- 14.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 15.Huff, W. E., G. W. Malone, and G. W. Chaloupka. 1984. Effect of litter treatment on broiler performance and certain litter quality parameters. Poult. Sci. 63:2167-2171. [DOI] [PubMed] [Google Scholar]
- 16.Janse, I., J. Bok, and G. Zwart. 2004. A simple remedy against artifactual double bands in denaturing gradient gel electrophoresis. J. Microbiol. Methods 57:279-281. [DOI] [PubMed] [Google Scholar]
- 17.Kelley, T. R., O. C. Pancorbo, W. C. Merka, and H. M. Barnharts. 1998. Antibiotic resistance of bacterial litter isolates. Poult. Sci. 77:243-247. [DOI] [PubMed] [Google Scholar]
- 18.Knarreborg, A., M. A. Simon, R. M. Engberg, B. B. Jensen, and G. W. Tannock. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl. Environ. Microbiol. 68:5918-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. D. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
- 20.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 21.Larue, R., Z. Yu, V. A. Parisi, A. R. Egan, and M. Morrison. 2005. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ. Microbiol. 7:530-543. [DOI] [PubMed] [Google Scholar]
- 22.Lovanh, N., K. L. Cook, M. J. Rothrock, D. M. Miles, and K. Sistani. 2007. Spatial shifts in microbial population structure within poultry litter associated with physiochemical properties. Poult. Sci. 86:1840-1849. [DOI] [PubMed] [Google Scholar]
- 23.Lu, J., S. Sanchez, C. Hofacre, J. J. Maurer, B. G. Harmon, and M. D. Lee. 2003. Evaluation of broiler litter with reference to the microbial composition as assessed by using 16S rRNA and functional gene markers. Appl. Environ. Microbiol. 69:901-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, J., U. Idris, B. Harmon, C. Hofacre, J. J. Maurer, and M. D. Lee. 2003. Diversity and succession of the intestinal bacteria community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattar, A. F., D. H. Teitelbaum, R. A. Drongowski, F. Yongi, C. M. Harmon, and A. G. Coran. 2002. Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediatr. Surg. Int. 18:586-590. [DOI] [PubMed] [Google Scholar]
- 27.Nurmi, E., and M. Rantala. 1973. New aspects of Salmonella infection in broiler production. Nature 241:210-211. [DOI] [PubMed] [Google Scholar]
- 28.Olesiuk, O. M., G. H. Snoeyenbos, and C. F. Smyser. 1971. Inhibitory effect of used litter on Salmonella typhimurium transmission in the chicken. Avian Dis. 15:118-124. [PubMed] [Google Scholar]
- 29.Omeira, N., E. K. Barbour, P. A. Nehme, S. K. Hamadeh, R. Zurayk, and I. Bashour. 2006. Microbiological and chemical properties of litter from different chicken types and production systems. Sci. Total Environ. 367:156-162. [DOI] [PubMed] [Google Scholar]
- 30.Patterson, P. H., E. S. Lorenz, and W. D. Weaver, Jr. 1998. Litter production and nutrient from commercial broiler chickens. J. Appl. Poult. Res. 7:247-252. [Google Scholar]
- 31.Pedroso, A. A., J. F. M. Menten, M. R. Lambais, A. M. C. Racanicci, F. A. Longo, and J. O. B. Sorbara. 2006. Intestinal bacterial community and growth performance of chickens fed diets containing antibiotics. Poult. Sci. 85:747-752. [DOI] [PubMed] [Google Scholar]
- 32.Rautava, S., and W. A. Walker. 2007. Commensal bacteria and epithelial cross talk in the developing intestine. Curr. Gastroenterol. Rep. 9:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothrock, M. J., Jr., K. L. Cook, J. G. Warren, and K. Sistani. 2008. The effect of alum addition on microbial communities in poultry litter. Poult. Sci. 87:1493-1503. [DOI] [PubMed] [Google Scholar]
- 34.Savage, D. C., J. E. Siegel, J. E. Snellen, and D. D. Whitt. 1981. Transit time of epithelial cells in the small intestines of germfree mice and ex-germfree mice associated with indigenous microorganisms. Appl. Environ. Microbiol. 42:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schefferle, H. E. 1965. The microbiology of built up poultry litter. J. Appl. Bacteriol. 28:403-411. [DOI] [PubMed] [Google Scholar]
- 36.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shakouri, M. D., P. A. Iji, L. L. Mikkelsen, and A. J. Cowieson. 2008. Intestinal function and gut microflora of broiler chickens as influenced by cereal grains and microbial enzyme supplementation. J. Anim. Physiol. Anim. Nutr. doi: 10.1111/j.1439-0396.2008.00852.x. [DOI] [PubMed]
- 38.Singer, R. S., J. S. Jeffrey, T. E. Carpenter, C. L. Cooke, E. R. Atwill, W. O. Johnson, and D. C. Hirsh. 2000. Persistence of cellulitis-associated Escherichia coli DNA fingerprints in successive broiler chicken flocks. Vet. Microbiol. 75:59-71. [DOI] [PubMed] [Google Scholar]
- 39.USDA. February 2009. USDA agriculture projections to 2018. http://www.ers.usda.gov/Publications/OCE091/OCE091.pdf. USDA, Washington, DC.
- 40.van der Wielen, P. W. J. J., D. A. Keuzenkamp, L. J. A. Lipman, F. Van Knapen, and S. Biesterveld. 2002. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 44:286-293. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wise, M. G., and G. R. Siragusa. 2007. Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. J. Appl. Microbiol. 102:1138-1149. [DOI] [PubMed] [Google Scholar]
- 43.Yu, Z., and M. Morrison. 2004. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:4800-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, Z., and M. Morrison. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808-812. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, X. Y., T. Zhong, Y. Pandya, and R. D. Joerger. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zocco, M. A., M. E. Ainora, G. Gasbarrini, and A. Gasbarrini. 2007. Bacteroides thetaiotaomicron in the gut: molecular aspects. Dig. Liver Dis. 39:707-712. [DOI] [PubMed] [Google Scholar]