Abstract
Bacillus subtilis is able to utilize arabinopolysaccharides derived from plant biomass. Here, by combining genetic and physiological analyses we characterize the AraNPQ importer and identify primary and secondary transporters of B. subtilis involved in the uptake of arabinosaccharides. We show that the ABC-type importer AraNPQ is involved in the uptake of α-1,5-arabinooligosaccharides, at least up to four l-arabinosyl units. Although this system is the key transporter for α-1,5-arabinotriose and α-1,5-arabinotetraose, the results indicate that α-1,5-arabinobiose also is translocated by the secondary transporter AraE. This broad-specificity proton symporter is the major transporter for arabinose and also is accountable for the uptake of xylose and galactose. In addition, MsmX is shown to be the ATPase that energizes the incomplete AraNPQ importer. Furthermore, the results suggest the existence of at least one more unidentified MsmX-dependent ABC importer responsible for the uptake of nonlinear α-1,2- and α-1,3-arabinooligosaccharides. This study assigns MsmX as a multipurpose B. subtilis ATPase required to energize different saccharide transporters, the arabinooligosaccharide-specific AraNPQ-MsmX system, a putative MsmX-dependent ABC transporter specific for nonlinear arabinooligosaccharides, and the previously characterized maltodextrin-specific MdxEFG-MsmX system.
Transport across biological membranes is a fundamental process for life and is accomplished by channels, primary and secondary transporters, and group translocators (19). ATP-binding cassette (ABC) transporters constitute one of the largest families of translocation facilitators and are distributed across all domains of life. Although they are involved in a variety of distinct processes, such as nutrient uptake, resistance to antibiotics and other drugs, lipid trafficking, cell division, sporulation, immune response, and pathogenesis, all ABC transporters, regardless of the polarity of transport (exporters and importers), share a structure and a general mechanism (reviewed in references 3 and 8). Their organization comprises two transmembrane domains (TMDs) coupled to two cytosolic nucleotide-binding domains (NBDs), or ATP-binding cassettes, responsible for ATP binding and hydrolysis-driven conformational changes.
The majority of the eukaryotic ABC transporters are exporters facilitating translocation from the cytoplasm. In contrast, prokaryotic ABC permeases are involved mainly in the import of nutrients, vitamins, and trace elements (3, 6, 8). Canonical bacterial ABC importers are dependent predominantly on high-affinity substrate-binding proteins (BPDs) that capture the substrate and deliver it to the transporter. The canonical maltose/maltodextrin importer MalEFGK2 of Escherichia coli/Salmonella enterica serovar Typhimurium (5) is one of the most-characterized members of this superfamily of translocation facilitators, serving as model for ABC importers in general (14, 15). In Gram-negative bacteria, BPDs are proteins located in the periplasmic space between the inner and outer membranes. In Gram-positive organisms, which are devoid of this type of cellular compartment, BPDs often are lipoproteins that are anchored to the extracellular side of the cytoplasmic membrane via its N-terminal domain (3 and 8 and references therein).
In nature, the major source of carbohydrates for microorganisms to utilize is plant biomass. Thus, in their natural habitat, such as soil, aquatic environments, or animal digestive tracts, bacteria secrete a vast number of polysaccharolytic enzymes for the degradation of plant-derived polysaccharides. The resulting mono-, di-, and oligosaccharides enter the cell mainly through specific ABC transporters. The number of well-characterized ABC transporters devoted to the uptake of products resulting from the degradation of hemicellulose is very scarce (6), and in Gram-positive organisms only two systems, BxlEFG of Streptomyces thermoviolaceus (31) and XynEFG (29) of Geobacillus staerothermophilus, both dedicated to the transport of xylodextrins, have been characterized in detail.
An in silico analysis of the Bacillus subtilis genome estimated the existence of at least 78 ABC transporters based on the identification of 86 NBDs in 78 proteins, 103 MSD proteins, and 37 BPD proteins, which account for about 5% of the protein-coding genes of this model organism (17). At least 10 ABC systems are predicted to be involved in the uptake of sugars (20). One of these ABC importers, AraNPQ, is clustered together with genes encoding enzymes involved in arabinose catabolism and the degradation of arabinooligosaccharides in a large operon, araABDLMNPQ-abfA (23). AraN is the BPD, and AraP and AraQ are the TMDs. This transporter, which lacks the NBD protein partner, was proposed to be involved in the uptake of arabinose oligomers mainly by genomic context and in silico analysis (10, 11, 23).
Here, by combining genetic and physiological analyses, we characterize the AraNPQ importer and identify primary and secondary transporters of B. subtilis involved in the uptake of arabinosaccharides. Furthermore, this study assigns the role of MsmX as a multipurpose B. subtilis ATPase required to energize different saccharide transporters.
MATERIALS AND METHODS
Substrates.
α-1,5-Linked arabinooligosaccharides (arabinobiose, arabinotriose, and arabinotetraose), arabinan, and debranched arabinan (sugar beet, purity 95%) were purchased from Megazyme International Ireland Ltd.; arabinose, maltotriose, maltotetraose, and maltopentaose were from Sigma-Aldrich Co.; and glucose was from BDH.
DNA manipulation and sequencing.
Routine DNA manipulations were performed as described by Sambrook et al. (22). All restriction enzymes were purchased from MBI Fermentas or New England Biolabs and used according to the manufacturers' recommendations. PCR amplifications were carried out using Phusion high-fidelity DNA polymerase (Finnzymes) or NZYDNAChange DNA polymerase (NZYTech). DNA from agarose gels and PCR products were purified with the GFX PCR DNA gel band purification kit (GE Healthcare). All DNA ligations were performed using T4 DNA ligase (MBI Fermentas). Plasmids were purified using the Qiagen plasmid midi kit (Qiagen) or QIAprep spin Miniprep kit (Qiagen). All constructs described below were confirmed by DNA sequencing performed with the ABI PRISM BigDye terminator ready reaction cycle sequencing kit (Applied Biosystems).
Construction of plasmids and strains.
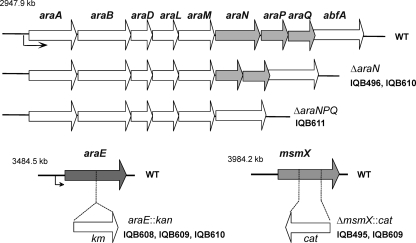
To construct a B. subtilis msmX insertion-deletion mutation by the insertion of a chloramphenicol resistance (Cmr) cassette, msmX was amplified by PCR from chromosomal DNA of B. subtilis 168T+ using primers ARA422 and ARA423, and the amplification product was digested with SacI and KpnI. The resulting 1,360-bp DNA fragment was cloned in the phagemid pBluescript II KS(+) (Table 1), also digested with SacI and KpnI, yielding pMJ1. A Cmr cassette was obtained by the digestion of pMS38 (Table 1) with HindIII and EcoRI and was subcloned in pMJ1 digested with the same restriction enzymes, resulting in pMJ2. This plasmid was linearized with ScaI and used to transform B. subtilis 168T+, yielding strain IQB495 (Table 1).
TABLE 1.
Plasmids, B. subtilis strains, and oligonucleotides used in this study
| Plasmid, strain, or oligonucleotide | Relevant construction, genotype, or sequence (5′-3′) | Source or referenceb |
|---|---|---|
| Plasmids | ||
| pBluescript II KS(+) | Multipurpose cloning/in vitro transcription phagemid vector, bla | Stratagene |
| pMAD | Plasmid used for allelic replacement in Gram-positive bacteria, bla, erm | 2 |
| pLitmus29 | Multipurpose cloning/in vitro transcription phagemid vector, bla | New England Biolabs |
| pMS38 | pLitmus29 derivative used as a source of chloramphenicol resistance cassette, bla, cat | 35 |
| pJL3 | pLitmus29 derivative used as a source of kanamycin resistance cassette, bla, kan | This work |
| pMJ1 | pBluescript II KS(+) derivative harboring the msmX coding region, bla | This work |
| pMJ2 | pMJ1 derivative used for the creation of msmX-null mutations, bla, cat | This work |
| pMJ6 | pMAD derivative used for the creation of araN in-frame deletion mutants, bla, erm | This work |
| pMJ7 | pBluescript II KS(+) derivative harboring the promoterless araE coding region, bla | This work |
| pMJ10 | Integrative plasmid used for the creation of area-null mutations, bla, kan | This work |
| pMJ11 | Integrative plasmid used for the creation of araNPQ in-frame deletion mutants, bla, erm | This work |
| B. subtilis strains | ||
| 168T+ | Prototroph | F. E. Young |
| IQB206 | ΔaraL-abfA::spc | 23 |
| IQB495 | ΔmsmX::cat | pMJ2a→168T+ |
| IQB496 | ΔaraN | pMJ6→168T+ |
| IQB608 | araE::kan | pMJ10a→168T+ |
| IQB609 | ΔmsmX::cat araE::kan | pMJ10a→IQB495 |
| IQB610 | ΔaraN araE::kan | pMJ10a→IQB496 |
| IQB611 | ΔaraNPQ | pMJ11→168T+ |
| Oligonucleotides | ||
| ARA422 | TATTGAGCTCTCAGGGATAGATATCAAATCG | |
| ARA423 | CATCGGTACCATTCTGAGATTTTCAATAGC | |
| ARA426 | GCGGATCCCTTTGGTGACATGCTCGG | |
| ARA427 | CGCTTTCACTTTTTGAATGGGCTGCGTTACCCCTTCCATTCGAGGTGCGGG | |
| ARA428 | CCCGCACCTCGAATGGAAGGGGTAACGCAGCCCATTCAAAAAGTGAAAGCG | |
| ARA429 | GCTTAGTACTGCTCTTTCGGCACATTTTGC | |
| ARA448 | GATAAAGTACTTTTCGAAAAAAGTCATTTTTTTCATCTGCGTTACCCCTTC | |
| ARA449 | GAAGGGGTAACGCAGATGAAAAAAATGACTTTTTTCCAAAAGTACTTTATC | |
| ARA450 | GGCATGCGCATGTTTGAGCTGCCGCAGGC | |
| ARA467 | CCAGCAATATTTTATCC | |
| ARA488 | CAAATTTCGGTACCTCACAGCG |
Transformation was carried out with linearized DNA. The restriction sites used are underlined.
The arrows indicate transformation and point from donor DNA to the recipient strain.
To create B. subtilis mutant strains with in-frame deletions of araN and araNPQ plasmids, pMJ6 and pMJ11 were constructed. These plasmids were generated using the pMAD plasmid (2) (Table 1). Regions immediately upstream and downstream of araN were amplified by two independent PCR experiments from chromosomal DNA of B. subtilis 168T+ using primers ARA426 and ARA427 (PCR1) and ARA428 and ARA429 (PCR2). The products were joined by overlapping PCR with primers ARA426 and ARA429 (Table 1), and the resulting fragment was digested with ScaI and BamHI and cloned into pMAD SmaI-BamHI, yielding pMJ6. Using an identical technique, regions immediately upstream of araN and downstream of araQ were amplified with primers ARA426 and ARA448 (PCR3) and ARA449 and ARA450 (PCR4) and were joined by overlapping PCR with primers ARA426 and ARA450 (Table 1). The resulting product was phosphorylated by T4 polynucleotide kinase (MBI Fermentas) and then cloned into pMAD digested with SmaI, yielding pMJ11. These two plasmids harboring in-frame deletions of araN and araNPQ, respectively, were used in separate experiments for the integration and generation of clean deletions in the B. subtilis chromosome by following the published procedure described by Arnaud et al. (2). Both in-frame deletions were confirmed by DNA sequencing, and the resulting strains were named IQB496 and IQB611, respectively (Table 1).
To construct an araE-null mutant by disruption with the insertion of a kanamycin resistance (Kmr) cassette, the araE gene was amplified by PCR from chromosomal DNA of B. subtilis 168T+ using primers ARA467 and ARA488. The amplification product was digested with SspI and XmnI, and the resulting 1,445-bp DNA fragment was cloned into pBluescript II KS(+) digested with SmaI, yielding pMJ7. A Kmr cassette was obtained by the digestion of pJL3 with XbaI and EcoRI. Plasmid pJL3 is a pLitmus29 derivative carrying the Kmr cassette from pAH248 (A. O. Henriques, unpublished data) cloned between the BamHI and PstI sites. The cassette was filled in using Klenow fragment (MBI Fermentas), and the resulting 1,504-bp DNA fragment was subcloned into pMJ7 digested with Eco47III, creating pMJ10. This plasmid was linearized with ScaI and used to transform B. subtilis strains 168T+, IQB495, and IQB496, yielding strains IQB608 (araE::kan), IQB609 (ΔmsmX::cat araE::kan), and IQB610 (ΔaraN araE::kan), respectively (Table 1).
The transformation of B. subtilis was performed according to the method described by Anagnostopoulos and Spizizen (1).
Growth conditions.
E. coli DH5α (Gibco-BRL) was used for the construction of all plasmids. All E. coli strains were grown in liquid Luria-Bertani (LB) medium (13) and on LB solidified with 1.6% (wt/vol) agar, and ampicillin (100 μg ml−1), chloramphenicol (25 μg ml−1), kanamycin (20 μg ml−1), or isopropyl-β-d-thiogalactopyranoside (IPTG) (0.1 mM) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg ml−1) were added as appropriate. B. subtilis was grown in liquid LB medium and LB medium solidified with 1.6% (wt/vol) agar, with chloramphenicol (5 μg ml−1), kanamycin (10 μg ml−1), and spectinomycin (60 μg ml−1) being added as appropriate.
Growth kinetic parameters of the wild-type and mutant B. subtilis strains were determined in liquid minimal medium. Cells from freshly streaked B. subtilis strains were grown overnight (37°C, 150 rpm) in C minimal medium (16) supplemented with l-tryptophan (100 μg ml−1), potassium glutamate (8 μg ml−1), and potassium succinate (6 μg ml−1) (CSK medium) (4). The cell cultures were washed and resuspended to an initial optical density at 600 nm (OD600) of 0.05 in 1.5 ml of CSK medium without potassium succinate and supplemented with different carbon and energy sources (glucose, arabinose, arabinobiose, arabinotriose, arabinotetraose, arabinan, debranched arabinan, maltotriose, maltotetraose, and maltopentaose) at a final concentration of 0.1% (wt/vol). The cultures were grown at 37°C and 180 rpm in an Aquatron water bath rotary shaker, and the OD600 was read periodically with an Ultrospec 2100 pro UV/visible spectrophotometer.
RESULTS
Uptake of arabinose oligomers by AraNPQ.
The AraNPQ proteins are the BPD and MSD components of an ABC-type importer (23). The three proteins, which are encoded by the arabinose metabolic operon araABDLMNPQ-abfA, previously were shown to be dispensable for arabinose utilization in a strain bearing a large deletion comprising all genes downstream from araD (23). However, growth experiments indicated a possible involvement in the utilization of arabinose oligomers (10). To confirm this hypothesis, an in-frame deletion mutation in the araN gene was generated by allelic replacement to minimize the polar effect on the genes of the araABDLMNPQ-abfA operon, which is located downstream of araN (Fig. 1). The physiological effect of this knockout mutation in B. subtilis (strain IQB496 ΔaraN) (Table 1) was assessed by the determination of the growth kinetic parameters using various saccharides as the sole carbon and energy source (Table 2). In the presence of glucose and arabinose, the doubling time of the mutant is comparable to that of the wild-type strain, indicating both the stability of the strain bearing the in-frame deletion and that the AraNPQ system is not responsible for the uptake of arabinose. In contrast, the absence of AraN has a negative effect on the ability of strain IQB496 (ΔaraN) to grow on the α-1,5-arabinose oligomers α-1,5-arabinobiose, α-1,5-arabinotriose, and α-1,5-arabinotetraose (Table 2). Similar results were obtained in the growth experiments (Table 2) with a strain harboring an in-frame deletion of the three genes araNPQ (IQB611 ΔaraNPQ) (Table 1), which were constructed by allelic replacement as describe above for the in-frame deletion of araN (Fig. 1). Taken together, the data implicate the AraNPQ system in the uptake of arabinose oligomers.
FIG. 1.
Organization of the araABDLMNPQ-abfA, araE, and msmX loci of the wild-type B. subtilis chromosome (WT). The location of the three regions is indicated in kilobase pairs. The genes are represented by arrows pointing in the direction of transcription. Below each region are displayed the constructs bearing the mutations used in this work, and the strains harboring each mutation are indicated in front of the construct. Below the araABDLMNPQ-abfA loci are depicted the two in-frame deletions generated by allelic replacement, ΔaraN and ΔaraNPQ. the mutation generated by the insertion of a kanamycin resistance cassette (kan) is represented in the araE locus. Below the msmX locus is presented the insertion-deletion mutation created by a deletion of msmX followed by the insertion of a chloramphenicol resistance cassette (cat) as described in Materials and Methods.
TABLE 2.
Effect of distinct mutations in the growth kinetics of B. subtilis strains in the presence of different carbohydrates
| Carbon source | 168T+ (WT) | IQB496 (ΔaraN) | IQB611 (ΔaraNPQ) | IQB 608 (araE::kan) | IQB610 (ΔaraN araE::kan) | IQB495 (ΔmsmX::cat) | IQB609 (ΔmsmX::cat araE::kan) |
|---|---|---|---|---|---|---|---|
| Glucose | 55.5 ± 1.3 | 53.4 ± 1.0 | 52.5 ± 0.8 | 57.3 ± 0.3 | 58.6 ± 1.5 | 57.7 ± 2.7 | 54.1 ± 1.1 |
| Arabinose | 67.0 ± 0.2 | 66.6 ± 0.9 | 63.7 ± 2.9 | NG | NG | 68.2 ± 2.3 | NG |
| Arabinobiose | 112.4 ± 10.8 | 263.8 ± 37.2 | 258.2 ± 46.1 | 178.6 ± 19.9 | NG | 286.7 ± 33.1 | NG |
| Arabinotriose | 98.2 ± 10.0 | NG | NG | - | - | NG | - |
| Arabinotetraose | 92.4 ± 4.5 | NG | NG | - | - | NG | - |
| Arabinan (sugar beet) | 149.0 ± 6.8 | 177.0 ± 21.6 | 188.8 ± 15.3 | - | - | NG | - |
| Debranched arabinan | 84.0 ± 4.6 | 347.6 ± 23.7 | 330.6 ± 30.8 | - | - | 332.5 ± 6.8 | - |
| Maltoriose | 77.7 ± 2.3 | - | - | - | - | 83.1 ± 2.1 | - |
| Maltotetraose | 76.1 ± 2.2 | - | - | - | - | 125.8 ± 4.5 | - |
| Maltopentaose | 75.0 ± 1.2 | - | - | - | - | 105.4 ± 4.2 | - |
The B. subtilis wild-type (WT) and different mutant strains were grown in minimal CSK liquid medium in the presence of the following sugars (0.1%, wt/vol): glucose, arabinose, α-1,5-arabinobiose, α-1,5-arabinotriose, α-1,5-arabinotetraose, arabinan (sugar beet), debranched arabinan, maltotriose, maltotetraose, and maltopentaose. The kinetic growth parameters were determined, and the results correspond to the averages from three independent experiments and respective standard deviations. NG, no growth; -, not determined.
Interestingly, the absence of a functional AraNPQ system (strains IQB496 ΔaraN and IQB611 ΔaraNPQ) impaired growth in the presence of α-1,5-arabinotriose and α-1,5-arabinotetraose; however, in the presence of α-1,5-arabinobiose, the growth rate decreased only 2.3-fold compared to that of the wild-type strain (Table 2). This observation suggested the existence of an additional transporter for this substrate. In B. subtilis, arabinose enters the cell mainly through the AraE transporter, a proton symport-type permease (24) with broad substrate specificity that also is responsible for the uptake of xylose and galactose (12). Thus, AraE, due to its capacity to transport different substrates, was considered a potential candidate for the transport of α-1,5-arabinobiose. To test this hypothesis, the araE gene was disrupted by an insertion mutation (Fig. 1) as described in Materials and Methods. The resulting strain IQB608 (araE::kan) was unable to grow in the presence of arabinose (Table 2), although it displayed normal growth in the presence of glucose. On the other hand, in the presence of α-1,5-arabinobiose it showed a 1.6-fold decrease of its growth rate compared to that of the wild-type strain (Table 2). This effect is similar to that observed with strain IQB496 (ΔaraN), suggesting the involvement of AraE in the uptake of α-1,5-arabinobiose. To further support this evidence, a double mutant, ΔaraN araE::kan, was constructed (strain IQB610) and shown to be unable to grow in the presence of α-1,5-arabinobiose (Table 2). These data revealed that both AraE and AraNPQ are able to transport the disaccharide α-1,5-arabinobiose and represent the only transport systems accountable for its uptake in B. subtilis.
Transport via the AraNPQ system is energized by MsmX.
The nucleotide-binding domain (NBD) protein partner of the AraNPQ ABC importer is missing in the araABDLMNPQ-abfA transcriptional unit (23). A decade ago, Quentin et al. (17) carried out in silico the inventory and assembly of the ATP binding cassette (ABC) transporter systems in the complete genome of B. subtilis. The authors identified five genes encoding ATPases without any gene coding for TMD protein in their neighborhood. On the basis of similarities, three ATP-binding proteins were proposed to energize 10 incomplete systems. Among these ATPases, MsmX, which is encoded by a monocistronic gene unit, is highly similar (>40% identity) to MalK of E. coli (7) and MsmK of Streptococcus mutans (18), ATPases of transport systems for maltose and multiple sugars, respectively. Thus, the authors proposed that MsmX would be an ATPase involved in the transport of carbohydrates and would be one candidate to energize several incomplete systems in operons that encode only the membrane and solute-binding proteins (17).
To test the possible involvement of MsmX in the transport of AraNPQ substrates, an insertion-deletion mutation was generated in the msmX gene (Fig. 1). The resulting strain, IQB495 (ΔmsmX::cat), was grown as described above, and the physiological response to the presence of different saccharides was determined. The msmX-null mutation does not affect the kinetic growth parameters in the presence of glucose and arabinose. The negative impact of this mutation in the doubling time of strain IQB495 (ΔmsmX::cat) grown in the presence of α-1,5-arabinobiose, α-1,5-arabinotriose, and α-1,5-arabinotetraose was comparable to that observed for strain IQB496 (ΔaraN). This observation clearly indicates that MsmX, like AraN, is essential for the functionality of AraNPQ, playing the role of the ATPase that energizes the unidirectional substrate transport via AraNPQ. Furthermore, since both IQB495 (ΔmsmX::cat) and IQB496 (ΔaraN) exhibit a parallel growth rate in the presence of α-1,5-arabinobiose (Table 2), the additional transporter responsible for the uptake of this substrate must be MsmX independent. Moreover, the phenotype of the double mutant IQB609 (ΔmsmX::cat araE::kan) (Table 2) was very similar to that observed with strain IQB610 (ΔaraN araE::kan) (Table 2). Taken together, these findings corroborate the evidence that AraE is involved in the uptake of α-1,5-arabinobiose.
MsmX contributes to the transport of multiple saccharides.
In addition to its role as an essential partner in the AraNPQ-MsmX system, MsmX previously was shown to be the NBD protein partner of the MdxEFG ABC importer, where MdxE is a maltodextrin-binding protein with high affinities for maltodextrins and a low affinity for maltose, and MdxF and MdxG are the membrane components (27). In this study, we assayed the physiological impact of the mutation ΔmsmX::cat (strain IQB495) on the growth on maltotriose, maltotetraose, and maltopentaose. A negative effect in the doubling time was observed, so the msmX-null mutation does not impair growth in the presence of maltooligosaccharides (Table 2). Studies of transport performed by Schönert et al. (27) using radiolabeled maltotriose showed that the uptake was not prevented in the msmX-null mutation, although in the malP msmX double mutant this transport was drastically reduced (27). Thus, our results correlate with these observations, and the growth observed in the presence of maltooligosaccharides most probably is due to its extracellular degradation and uptake of resulting products, such as maltose and glucose, by other transport systems (27).
Furthermore, mutant and wild-type strains were grown in the presence of two different types of the homopolysaccharide arabinan: branched arabinan, a molecule comprising a backbone of α-1,5-linked l-arabinofuranosyl residues decorated with α-1,2-, and α-1,3-linked l-arabinofuranosyl units, and debranched arabinan, the linear homopolysaccharide α-1,5-l-arabinan. In both mutant strains IQB495 (ΔmsmX::cat) and IQB496 (ΔaraN), a slow but steady growth rate on debranched arabinan was observed compared to that of the wild type (Table 2). Since we showed that a functional AraNPQ-MsmX system is required for the transport of α-1,5-arabinotriose and α-1,5-arabinotetraose and is partially responsible for the uptake of α-1,5-arabinobiose, the slow growth observed most probably is due to the metabolism of some arabinobiose and arabinose residues resulting from the extracellular degradation of linear α-1,5-l-arabinan, which enters the cell through the AraE permease. Interestingly, in the presence of branched arabinan (sugar beet), strains lacking a functional AraNPQ system (ΔaraN and ΔaraNPQ) display only a marginal increase in the doubling time compared to that of the wild-type strain (Table 2); in contrast, the msmX-null mutation (strain IQB495) has a severe impact on growth (Table 2). This fact suggests that most of the arabinooligosaccharides resulting from the extracellular degradation of branched arabinan are nonlinear, i.e., are arabinooligosaccharides with α-1,2- and/or α-1,3-linked arabinosyl residues, and that these products are not transported by the AraNPQ system. The slight decrease of the growth rate observed in strains lacking a functional AraNPQ compared to that of the wild type may represent the reduced amount of linear arabinooligosaccharides resulting from the extracellular degradation of arabinan that, in the wild-type strain, would be transported through AraNPQ and metabolized. Taken together, the growth kinetic parameters of the different strains grown in the presence of two different types of arabinan molecules suggest the existence of an additional transporter for nonlinear arabinooligosaccharides. Furthermore, this putative importer is MsmX dependent due to the lack of growth of the msmX-null mutant in the presence of branched arabinan (sugar beet).
DISCUSSION
Bacillus subtilis participates in the enzymatic dissolution of plant biomass. Thus, it is able to synthesize a vast variety of glycoside hydrolases capable of the depolymerization of plant cell wall polysaccharides, such as cellulose, hemicellulose, or pectin (28 and 30 and references therein). l-Arabinose, the second most-abundant pentose in plant biomass, next to d-xylose, is found in homopolysaccharides (branched and debranched arabinans) and heteropolysaccharides (arabinoxylans, arabinogalactans, etc.). B. subtilis produces exo- and endo-acting arabinases capable of releasing arabinosyl oligomers and l-arabinose from plant cell walls (10, 11, 33). Although many polysaccharolytic glycoside hydrolases have been purified from bacteria, including several Bacillus spp., information on ABC transporters devoted to the uptake of products resulting from the degradation of hemicellulose is limited (6).
The results presented here lead to the characterization of an ABC primary transporter of B. subtilis involved in the uptake of products of the degradation of arabinans. The AraNPQ system is encoded by the araABDLMNPQ-abfA operon, which is regulated at the transcriptional level by induction in the presence of arabinose and repression by glucose (23). AraN is the high-affinity substrate-binding protein (BPD) that captures the substrate and delivers it to the two transmembrane components (TMDs), AraP and AraQ. The genetic and physiological impact of in-frame deletions on both the araN gene and the entire transporter araNPQ lead us to the conclusion that AraNPQ is involved in the uptake of α-1,5-arabinose oligomers (up to four units) but not arabinose. AraNPQ is the sole transporter for α-1,5-arabinotriose, and α-1,5-arabinotetraose is only partially accountable for the uptake of α-1,5-arabinobiose. The latter also is transported by the AraE permease, the main permease involved in the uptake of arabinose (24), and also is responsible for the transport of xylose and galactose (12). This evidence is supported by the observation that both single mutations in the araN (or araNPQ) and araE genes result in slower growth on α-1,5-arabinobiose; on the other hand, growth on this substrate is impaired by the double mutation araN araE.
In Gram-positive bacteria gene clusters that encode sugar ABC transporters, the BPDs and TMDs frequently lack the nucleotide-binding domains (NBDs). Thus, the NBDs responsible for providing energy for the corresponding transporters is not linked. The B. subtilis genome encodes three ATP-binding proteins without any gene coding for TMD protein in their neighborhood (17). Among these ATPases, MsmX is a strong candidate for supplying energy to the incomplete AraNPQ transporter (17); moreover, the msmX gene is monocistronic and constitutively expressed in both complex and minimal medium (34). MsmX is highly similar to MsmK from Streptococcus mutans (61% identity [18]) and MsiK from Streptomyces lividans (55% identity [9]). In Streptomyces species the MsiK protein is an ATPase involved in several oligosaccharide uptake systems. MsiK energizes the cellobiose, xylobiose, and maltose uptake systems in Streptomyces lividans (9, 26), the trehalose uptake system in Streptomyces reticuli (25), and the N,N′-diacetylchitobiose uptake in Streptomyces coelicolor (21). In Streptococcus mutans, the MsmEFGK system transports raffinose, melibiose, and stachyose, and the MalXFGK transporter is specific for maltodextrins (32). The two ATPase domains, MsmK and MalX, were shown to interact with either transporter system to energize uptake (32). In B. subtilis it was previously shown that MsmX energizes MdxEFG, a transporter specific for maltodextrins, as the cognate ABC domain (27). Here, we show that MsmX is required for the uptake of α-1,5-arabinose oligomers (up to four units) but not arabinose, thus it is the ATPase that supplies energy to AraNPQ. In addition, our findings strongly suggest the existence of an unidentified MsmX-dependent ABC transport system that is involved in the uptake of nonlinear arabinooligosaccharides with α-1,2 and/or α-1,3 linked arabinosyl residues, which are not transported by the AraNPQ system. This hypothesis is supported by two lines of evidence: (i) in the presence of branched arabinan (sugar beet, displaying α-1,2 and/or α-1,3 linked arabinosyl residues to the α-1,5 arabinosyl backbone), strains lacking a functional AraNPQ system (ΔaraN and ΔaraNPQ) display only a minor increase in the doubling time compared to that of the wild-type strain, and (ii) the msmX-null mutation has a severe negative impact on growth on branched arabinan.
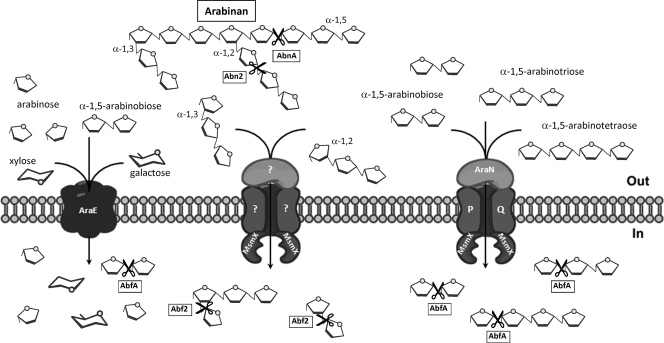
Taken together, these data led to our current model of the transport and utilization of arabinooligosaccharides by B. subtilis (Fig. 2). Arabinan, a homopolymer of arabinose, is extracellularly degraded by two GH43 endo-α-1,5-arabinanases, AbnA and Abn2 (11). The resulting products, mainly arabinooligosaccharides, enter the cell through different transport systems. The ABC-type importer AraNPQ is involved in the uptake of α-1,5-arabinooligosaccharides, at least up to four l-arabinosyl units, and is energized by the ATPase MsmX. The primary AraNPQ-MsmX system is the key transporter for α-1,5-arabinotriose and α-1,5-arabinotetraose. The AraE permease, a proton symporter, the major transporter for arabinose (24), also is responsible for the uptake of xylose and galactose (12) from other hemicellulosic heteropolysaccharides like arabinoxylan, arabinogalactan, or pectin. This broad-specificity secondary transporter also displays capacity for the uptake of α-1,5-arabinobiose, although this sugar enters the cell preferentially through the primary AraNPQ-MsmX importer. At least one more unidentified MsmX-dependent ABC importer is likely to be responsible for the uptake of the nonlinear α-1,2- and α-1,3-arabinooligosaccharides. Once inside the cell, α-1,5-α-1,2- and α-1,3-arabinooligosaccharides are further degraded by the action of two GH51 family α-l-arabinofuranosidases, AbfA and Abf2. The resulting l-arabinose then is metabolized by AraA, AraB, and AraD, which sequentially convert l-arabinose to d-xylulose 5-phosphate, which is further catabolized through the pentose phosphate pathway.
FIG. 2.
Primary and secondary transporters involved in the uptake of arabinooligosaccharides in B. subtilis and the role of MsmX in their transport. Arabinan, a homopolymer of arabinose, is extracellularly degraded by two endo-α-1,5-arabinanases, AbnA and Abn2. The resulting products, mainly arabinooligosaccharides, enter the cell through different transport systems. The ABC-type importer AraNPQ is involved in the uptake of α-1,5-arabinooligosaccharides and is energized by the ATPase MsmX. The AraNPQ-MsmX system is the key transporter for α-1,5-arabinotriose and α-1,5-arabinotetraose. The AraE permease, a proton symporter, the major transporter for arabinose, also is responsible for the uptake of xylose and galactose. This broad-specificity transporter also displays capacity for the uptake of α-1,5-arabinobiose. At least one more unidentified MsmX-dependent ABC importer is likely to be responsible for the uptake of the nonlinear α-1,2- and α-1,3-arabinooligosaccharides. Once inside the cell, arabinooligosaccharides are further degraded to l-arabinose by the action of two α-l-arabinofuranosidases, AbfA and Abf2 (see the text for a detailed explanation).
In this work we provide evidence that MsmX, as a homo- or heterodimer, is responsible for providing energy for at least two, and possibly three, ABC oligosaccharide transport systems. To our knowledge, the capacity of an ATPase to associate and energize distinct transport systems with different specificities in B. subtilis is described here for the first time. These findings support the establishment of this peculiar broad capacity of protein-protein interaction in the general bacterial mechanism of ABC-type sugar import.
Acknowledgments
We thank Michel Débarbouillé for the gift of plasmid pMAD and for advice in the construction of the in-frame deletion mutations and Joana Lima for the construction of pJL3.
This work was supported partially by grant no. PTDC/AGR-AAM/102345/2008 from Fundação para a Ciência e Tecnologia (FCT), FEDER, and POFC 2010, to I.S.-N.
Footnotes
Published ahead of print on 6 August 2010.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaud, M., A. Chastanet, and M. Débarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-gc-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson, A. L., E. Dassa, C. Orelle, and J. Chen. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72:317-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Débarbouillé, M., M. Arnaud, A. Fouet, A. Klier, and G. Rapoport. 1990. The sacT gene regulating the sacPA operon in Bacillus subtilis shares strong homology with transcriptional antiterminators. J. Bacteriol. 172:3966-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrmann, M., R. Ehrle, E. Hofmann, W. Boos, and A. Schlösser. 1998. The ABC maltose transporter. Mol. Microbiol. 29:685-694. [DOI] [PubMed] [Google Scholar]
- 6.Eitinger, T., D. A. Rodionov, M. Grote, and E. Schneider. 23 April 2010. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions. FEMS Microbiol. Rev. doi: 10.1111/j.1574-6976.2010.00230.x. [DOI] [PubMed]
- 7.Gilson, E., H. Nikaido, and M. Hofnung. 1982. Sequence of the malK gene in E. coli K12. Nucleic Acids Res. 10:7449-7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 9.Hurtubise, Y., F. Shareck, D. Kluepfel, and R. Morosoli. 1995. A cellulase/xylanase-negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol. Microbiol. 17:367-377. [DOI] [PubMed] [Google Scholar]
- 10.Inácio, J. M., I. L. Correia, and I. Sá-Nogueira. 2008. Two distinct arabinofuranosidases contribute to arabino-oligosaccharide degradation in Bacillus subtilis. Microbiology 154:2719-2729. [DOI] [PubMed] [Google Scholar]
- 11.Inácio, J. M., and I. Sá-Nogueira. 2008. Characterization of abn2 (yxiA), encoding a Bacillus subtilis GH43 arabinanase, Abn2, and its role in arabino-polysaccharide degradation. J. Bacteriol. 190:4272-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krispin, O., and R. Allmansberger. 1998. The Bacillus subtilis AraE protein displays a broad substrate specificity for several different sugars. J. Bacteriol. 180:3250-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Oldham, M. L., A. L. Davidson, and J. Chen. 2008. Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 18:726-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oldham, M. L., D. Khare, F. A. Quiocho, A. L. Davidson, and J. Chen. 2007. Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450:515-521. [DOI] [PubMed] [Google Scholar]
- 16.Pascal, M., F. Kunst, J. A. Lepesant, and R. Dedonder. 1971. Characterization of two sucrase activities in Bacillus subtilis Marburg. Biochimie 53:1059-1066. [DOI] [PubMed] [Google Scholar]
- 17.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287:467-484. [DOI] [PubMed] [Google Scholar]
- 18.Russell, R. R., J. Aduse-Opoku, I. C. Sutcliffe, L. Tao, and J. J. Ferretti. 1992. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J. Biol. Chem. 267:4631-4637. [PubMed] [Google Scholar]
- 19.Saier, M. H., Jr. 2000. Families of proteins forming transmembrane channels. J. Membr. Biol. 175:165-180. [DOI] [PubMed] [Google Scholar]
- 20.Saier, M. H., Jr., S. R. Goldman, R. R. Maile, M. S. Moreno, W. Weyler, N. Yang, and I. T. Paulsen. 2002. Overall transport of capabilities of Bacillus subtilis. .In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives-from genes to cells. American Society for Microbiology, Washington, DC.
- 21.Saito, A., T. Fujii, T. Shinya, N. Shibuya, A. Ando, and K. Miyashita. 2008. The msiK gene, encoding the ATP-hydrolysing component of N, N′-diacetylchitobiose ABC transporters, is essential for induction of chitinase production in Streptomyces coelicolor A3(2). Microbiology 154:3358-3365. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Sá-Nogueira, I., T. V. Nogueira, S. Soares, and H. Lencastre. 1997. The Bacillus subtilis l-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiology 143:957-969. [DOI] [PubMed] [Google Scholar]
- 24.Sá-Nogueira, I., and S. S. Ramos. 1997. Cloning, functional analysis, and transcriptional regulation of the Bacillus subtilis araE gene involved in l-arabinose utilization. J. Bacteriol. 179:7705-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlösser, A. 2000. MsiK-dependent trehalose uptake in Streptomyces reticuli. FEMS Microbiol. Lett. 184:187-192. [DOI] [PubMed] [Google Scholar]
- 26.Schlösser, A., T. Kampers, and H. Schrempf. 1997. The Streptomyces ATP-binding component MsiK assists in cellobiose and maltose transport. J. Bacteriol. 179:2092-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schönert, S., S. Seitz, H. Krafft, E.-A. Feuerbaum, I. Andernach, G. Witz, and M. K. Dahl. 2006. Maltose and maltodextrin utilization by Bacillus subtilis. J. Bacteriol. 188:3911-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shallom, D., and Y. Shoham. 2003. Microbial hemicellulases. Curr. Opin. Microbiol. 6:219-228. [DOI] [PubMed] [Google Scholar]
- 29.Shulami, S., G. Zaide, G. Zolotnitsky, Y. Langut, G. Feld, A. L. Sonenshein, and Y. Shoham. 2007. A two-component system regulates the expression of an ABC transporter for xylo-oligosaccharides in Geobacillus stearothermophilus. J. Bacteriol. 73:874-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stülke, J., and W. Hillen. 2000. Regulation of carbon catabolism in Bacillus subtilis. Annu. Rev. Microbiol. 54:849-880. [DOI] [PubMed] [Google Scholar]
- 31.Tsujibo, H., M. Kosaka, S. Ikenishi, T. Sato, K. Miyamoto, and Y. Inamori. 2004. Molecular characterization of a high-affinity xylobiose transporter of Streptomyces thermoviolaceus OPC-520 and its transcriptional regulation. J. Bacteriol. 186:1029-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb, A. J., K. A. Homer, and A. H. F. Hosie. 2008. Two closely related ABC transporters in Streptococcus mutans are involved in disaccharide and/or oligosaccharide uptake. J. Bacteriol. 190:168-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinstein, L., and P. Albersheim. 1979. Structure of plant cell walls. IX. Purification and partial characterization of a wall-degrading endo-arabanase and an arabinosidase from Bacillus subtilis. Plant Physiol. 63:425-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida, K., I. Ishio, E. Nagakawa, Y. Yamamoto, M. Yamamoto, and Y. Fujita. 2000. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology 146:573-579. [DOI] [PubMed] [Google Scholar]
- 35.Zilhão, R., M. Serrano, R. Isticato, E. Ricca, C. P. Moran, Jr., and A. O. Henriques. 2004. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 186:1110-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]