Abstract
Capsule polysaccharide (CPS) plays an important role in the virulence of Streptococcus pneumoniae and is usually used as the pneumococcal vaccine target. Glycerol-2-phosphate is found in the CPS of S. pneumoniae types 15A and 23F and is rarely found in the polysaccharides of other bacteria. The biosynthetic pathway of the nucleotide-activated form of glycerol-2-phosphate (NDP-2-glycerol) has never been identified. In this study, three genes (gtp1, gtp2, and gtp3) from S. pneumoniae 23F that have been proposed to be involved in the synthesis of NDP-2-glycerol were cloned and the enzyme products were expressed, purified, and assayed for their respective activities. Capillary electrophoresis was used to detect novel products from the enzyme-substrate reactions, and the structure of the product was elucidated using electrospray ionization mass spectrometry and nuclear magnetic resonance spectroscopy. Gtp1 was identified as a reductase that catalyzes the conversion of 1,3-dihydroxyacetone to glycerol, Gtp3 was identified as a glycerol-2-phosphotransferase that catalyzes the conversion of glycerol to glycerol-2-phosphate, and Gtp2 was identified as a cytidylyltransferase that transfers CTP to glycerol-2-phosphate to form CDP-2-glycerol as the final product. The kinetic parameters of Gtp1 and Gtp2 were characterized in depth, and the effects of temperature, pH, and cations on these two enzymes were analyzed. This is the first time that the biosynthetic pathway of CDP-2-glycerol has been identified biochemically; this pathway provides a method to enzymatically synthesize this compound.
Capsule polysaccharide (CPS) of Gram-positive bacteria, external to the cell wall, provides resistance to phagocytosis. CPS in Streptococcus pneumoniae is the most important virulence factor and the target of pneumococcal vaccines (2). Ninety individual CPS serotypes have been recognized so far by immunological and chemical techniques (9). Each has a structurally distinct CPS, composed of repeating oligosaccharide units joined by glycosidic linkages. The components of the repeat units are transferred from nucleoside diphosphate (NDP) derivatives. Among the 54 identified CPS structures, several sugars and related compounds have been found. Seven NDP-monosaccharide precursors (glucopyranose, N-acetylglucosamine, galactopyranose, N-acetylgalactosamine, 2-acetamido-4-amino-2,4,6-trideoxy-d-galactopyranose, ribitol-phosphate, and phosphorylcholine) are available from housekeeping metabolic pathways, and the biosynthetic genes for 14 NDP-monosaccharide precursors were found in the pneumococcal cps loci. Among the 14 components, the pathways of five (NDP-d-mannitol, NDP-d-arabinitol, NDP-ribofuranose [Rib], CDP-glycerol [CDP-Gro], and NDP-2-glycerol) are putative and have not yet been identified (1, 4).
Glycerol-2-phosphate is rarely present in bacteria and has been found in S. pneumoniae types 15A and 23F. The NDP-2-glycerol biosynthetic pathway has been proposed to include three enzymes: Gtp1, Gtp2, and Gtp3. Gtp3 has been proposed to be a glyceraldehyde-2-phosphotransferase and to be involved in the synthesis of glyceraldehyde-2-phosphate from glyceraldehyde. Gtp1, a putative dehydrogenase, has been proposed to be responsible for the conversion of glyceraldehyde-2-phosphate to glycerol-2-phosphate. The last step of the synthesis of CDP-2-glycerol is catalyzed by the putative glycerol-2-phosphate cytidyltransferase Gtp2 (14). The three genes, gtp1, gtp2, and gtp3, have also been found to be present in the cps loci of S. pneumoniae serotypes 15B, 15C, 15F, 23A, 23B, 28A, and 28F (4). However, the biosynthetic pathway for NDP-2-glycerol has never been identified by molecular and biochemical methods.
In this study, we found that the enzymes were not reactive by the previously proposed CDP-2-glycerol biosynthetic pathway. Therefore, a new pathway was proposed, and the three enzymes, Gtp1, Gtp2, and Gtp3, were identified and confirmed biochemically as 1,3-dihydroxyacetone/glyceraldehyde reductase, glycerol-2-phosphate cytidylyltransferase, and glycerol-2-phosphotransferase, respectively. This is the first report on the characterization of the CDP-2-glycerol biosynthetic pathway.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1 .
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or sequence | Sourcea |
|---|---|---|
| Bacterial strains | ||
| G1934 | S. pneumoniae 23F type strain | CIDM |
| E. coli BL21 | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| E. coli DH5α | F− φ80lacZM15 endA recA1 hsdR(rK−mK−) supE44 thi-1 gyrA96 relA1 Δ(lacZYA-argF)U169 | TBC |
| Plasmids | ||
| pET28a+ | T7 expression vector, Kanr | Novagen |
| pLW1282 | pET28a+ containing N-terminally six-histidine-tagged S. pneumoniae 23F gtp1 at the NdeI/XhoI site | This work |
| pLW1207 | pET28a+ containing C-terminally six-histidine-tagged S. pneumoniae 23F gtp3 at the NcoI/XhoI site | This work |
| pLW1263 | pET28a+ containing C-terminally six-histidine-tagged S. pneumoniae 23F gtp2 at the NcoI/XhoI site | This work |
| Primers | ||
| wl-5886 | 5′-GGGAATTCCATATGTTGAAAAATAATGATTTAAAGATA-3′ | |
| wl-5887 | 5′-CCGCTCGAGCTACAACTCGCTTATGAGTTC-3′ | |
| wl-9071 | 5′-CATGCCATGGGCATGAAATTGACAAATAGAGTTGA-3′ | |
| wl-9072 | 5′-CCGCTCGAGGACAATTCCTTTCCACATT-3′ | |
| wl-5892 | 5′-CATGCCATGGGCATGAAAGCACTTATTTTAGCA-3′ | |
| wl-5929 | 5′-CCGCTCGAGAGCAAATAGTTTTTCTGCAG-3′ |
CIDM, Centre for Infectious Diseases and Microbiology, Westmead Hospital, New South Wales, Australia; TBC, Tianjin Biochip Corporation, Tianjin, China.
Cloning and plasmid construction.
Genes gtp1, gtp2, and gtp3 from S. pneumoniae 23F (G1934) were amplified by PCR using the primers listed in Table 1 (wl-5886 and wl-5887 for gtp1, wl-9071 and wl-9072 for gtp2, wl-5892 and wl-5929 for gtp3). A total of 30 cycles were performed using the following conditions: denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min (final volume of 20 μl). The amplified genes were cloned into pET28a+ to construct pLW1282 (containing gtp1), pLW1207 (containing gtp2), and pLW1263 (containing gtp3), and the presence of the inserts was confirmed by sequencing using an ABI 3730 Sequencer.
Protein expression and purification.
Escherichia coli BL21 carrying each of the recombinant plasmids was grown in LB medium containing 50 μg ml−1 kanamycin overnight at 37°C. The overnight culture (5 ml) was inoculated into 500 ml of fresh LB medium and grown at 37°C until the A600 reached 0.6. The expression of Gtp1 and Gtp2 was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 25°C (0.1 mM FeCl2 and 0.05 mM ascorbate were also needed for Gtp1 expression), and Gtp3 was induced with 0.1 mM IPTG for 4 h at 37°C. After IPTG induction, the cells were harvested by centrifugation, washed with binding buffer (50 mM Tris-HCl, pH 7.4, 300 mM NaCl, and 10 mM imidazole; for Gtp1, NaCl was not in the buffer), resuspended in 5 ml of the same buffer containing 1 mM phenylmethanesulfonyl fluoride (PMSF) and 1 mg of lysozyme ml−1, and sonicated. The cell debris was removed by centrifugation, and total soluble proteins in the supernatant were collected. The His6-tagged fusion proteins in the supernatant were purified by nickel ion affinity chromatography with a chelating Sepharose fast-flow column (GE Healthcare), according to the manufacturer's instructions. Unbound proteins were washed out with 100 ml of wash buffer (50 mM Tris-HCl, pH 7.4, 300 mM NaCl, and 25 mM imidazole). The fusion proteins were eluted with 3 ml of elution buffer (50 mM Tris-HCl, pH 7.4, 300 mM NaCl, and 250 mM imidazole; for Gtp1, NaCl was not in the buffer) and dialyzed overnight against 50 mM Tris-HCl buffer (pH 7.4) at 4°C. Protein concentration was determined by the Bradford method.
Enzyme activity assays.
For the Gtp1 assay, a reaction mixture containing 10 mM 1,3-dihydroxyacetone, 1 mM NADH, 50 mM K2HPO4-KH2PO4 (pH 6.5), and 2.97 μM purified Gtp1 protein in a total volume of 20 μl was used and the reaction was carried out at 37°C for 0.5 h. A converse reaction mixture for Gtp1 contains 10 mM glycerol, 1 mM NAD+, 50 mM K2HPO4-KH2PO4 (pH 6.5), and 2.97 μM purified Gtp1 protein in a total volume of 20 μl. For the Gtp2 assay, a reaction mixture containing 20 mM glycerol-2-phosphate, 5 mM CTP, 0.05-U ml−1 inorganic pyrophosphatase (IP), 10 mM MgCl2, 50 mM K2HPO4-KH2PO4 (pH 8.0), and 0.013 mM purified Gtp2 protein was added to the mixture (20 μl), and the reaction was carried out at 37°C for 20 min. For the Gtp2 and Gtp3 assays, a reaction mixture containing 10 mM glycerol, 5 mM ATP, 5 mM CTP, 0.05-U ml−1 IP, 50 mM K2HPO4-KH2PO4 (pH 8.0), 0.071 mM purified Gtp3, and 0.026 mM Gtp2 protein in a total volume of 20 μl was used, and the reaction was carried out at 37°C for 1 h. Products from each of the reactions were analyzed by capillary electrophoresis (CE). In addition, products from the reaction catalyzed by Gtp2 were analyzed by electrospray ionization mass spectrometry (ESI MS) and nuclear magnetic resonance (NMR) spectroscopy. Enzyme activities were indicated by the conversion of substrates into products.
CE analysis.
CE was performed using a Beckman Coulter P/ACE MDQ capillary electrophoresis system with a photo-diode array (PDA) detector (Beckman Coulter, CA). The capillary was bare silica (75 μm [internal diameter] by 57 cm), with the detector at 50 cm. The capillary was conditioned before each run by washing it with 0.1 M NaOH for 2 min, deionized water for 2 min, and 25 mM borate-NaOH (pH 9.6) (used as the mobile phase) for 2 min. Samples were loaded by pressure injection at 0.5 lb/in2 for 10 s, and separation was carried out at 20 kV. Peak integration and trace alignment were done with Beckman P/ACE station software (32 Karat version 5.0). The conversion ratio was calculated by comparing the peak areas of substrate and product.
RP HPLC and ESI MS analysis.
The Gtp2 reaction mixture was separated by reversed-phase high-performance liquid chromatography (RP HPLC) using an LC-20AT HPLC (Shimadzu, Japan) with a Venusil MP-C18 column (5-μm particle, 4.6 by 250 mm) (Agela Technologies, Inc.). The mobile phase was composed of 70% acetonitrile and 30% water, and the flow rate was 0.6 ml min−1. Fractions containing the expected products were collected, lyophilized, and redissolved in 50% methanol before being injected into a Finnigan LCQ Advantage MAX ion trap mass spectrometer (Thermo Electron, CA) in negative mode (4.5 kV, 250°C) for ESI MS analysis. For MS/MS analysis, nitrogen was used as the collision gas and helium was used as the auxiliary gas, and collision energies used were typically 20 to 30 eV.
NMR spectroscopy.
A sample of CDP-2-Gro (0.3 mg) was deuterium exchanged by freeze-drying it from D2O, dissolved in 99.96% D2O (200 μl), and examined using a Shigemi (Japan) microtube. NMR spectra were recorded on a Bruker DRX-500 spectrometer (Germany) at 30°C, using internal sodium trimethylsilyl-[2,2,3,3-2H4]propanoate (δH, 0.00) and external aqueous 85% H3PO4 (δP, 0) as references. Two-dimensional NMR spectra were obtained using standard pulse sequences from the manufacturer, and the XWinNMR 2.6 program (Bruker) was used to acquire and process the NMR data.
Kinetic parameter measurements.
To measure the Gtp1 Km and maximum rate of metabolism (Vmax) values, reactions were carried out with 0.128 μM (at 37°C) Gtp1 in a final volume of 20 μl at various concentrations of 1,3-dihydroxyacetone (0.05 to 1 mM) or glyceraldehyde (0.1 to 2 mM) and a constant concentration of NADH (1 mM) or in various concentrations of NADH (0.1 to 2.5 mM) or NADPH (0.3 to 8 mM) and a constant concentration of 1,3-dihydroxyacetone (10 mM). To measure the Gtp2 Km and Vmax values, reactions were carried out with 0.0278 μM Gtp2 in a final volume of 20 μl at various concentrations of glycerol-2-phosphate (0.5 to 4 mM) or glycerol-1-phosphate (5 to 40 mM) and a constant concentration of CTP (5 mM) or with various concentrations of CTP (0.125 to 2 mM) or dCTP (0.5 to 5 mM) and a constant concentration of glycerol-2-phosphate (20 mM). The reactions were terminated by adding an equal volume of chloroform. Conversion of NADH to NAD+ and CTP and glycerol-2-phosphate or glycerol-1-phosphate to CDP-2-glycerol or CDP-glycerol was monitored by CE. Km and Vmax values were calculated based on the Michaelis-Menten equation. The reported data were the averages of results from three independent experiments.
Determination of temperature, pH optima, and divalent cation requirements.
For parameter characterization, 1.48 μM Gtp1 and 5.28 μM Gtp2 were used and the reactions were carried out at 37°C for 30 min (Gtp1) or 10 min (Gtp2). To determine the temperature optimum for Gtp1 and Gtp2, reactions were carried out at 4, 15, 25, 37, 50, 55, 60, 65, and 75°C, respectively. To determine the pH optimum for Gtp1 and Gtp2, reactions were carried out at pHs 5.5, 6, 6.5, 7, 7.5, 8, 8.5, 9, and 9.5. To test the effects of different cations on activity, reactions were carried out in the presence of 10 mM NH4Cl, NiSO4, MgCl2, MnCl2, FeSO4, CuCl2, CaCl2, CoCl2, ZnCl2, and FeCl3. The effect of these cations (5 mM) in the presence of 5 mM MgCl2 for Gtp2 activity alone was also examined. All enzyme activities were determined by CE.
RESULTS
Overexpression and purification of Gtp1, Gtp2, and Gtp3.
Genes gtp1, gtp2, and gtp3 from S. pneumoniae 23F were cloned, which constructed the plasmids pLW1282, pLW1263, and pLW1207, respectively, and were expressed in E. coli BL21 induced by IPTG. The majority of each protein was found in the soluble fraction as estimated by SDS-PAGE analysis (data not shown). The three proteins were purified to near homogeneity as His6-tagged fusion proteins (see Fig. S1 in the supplemental material). The estimated molecular masses were 39.1 kDa for Gtp1, 27.5 kDa for Gtp2, and 31.7 kDa for Gtp3, corresponding well to their calculated masses (38.3, 28.3, and 33.7 kDa, respectively).
Characterization of Gtp1, Gtp2, and Gtp3 activities by CE.
The biosynthetic pathway of CDP-2-glycerol was proposed as shown in Fig. 1. The activities of Gtp1, Gtp2, and Gtp3 were individually confirmed by comparing the enzyme catalyzing reaction products with standard compounds using CE (Fig. 2). In the reaction catalyzed by Gtp1, NADH was converted to NAD+, indicating that the substrate 1,3-dihydroxyacetone was converted to glycerol, which cannot be detected by CE. NADPH can also be used as the cofactor, and glyceraldehyde can also be used as the substrate in this reaction (described in the following sections). Gtp1 did not catalyze the converse reaction. In the reaction catalyzed by Gtp3, ATP was not converted to ADP. After the addition of Gtp2 and CTP, CTP was converted to a new product that eluted at 15.4 min, indicating that Gtp3 was inactive in the absence of Gtp2. In the reaction catalyzed by Gtp2, the substrates glycerol-2-phosphate and CTP were converted to a new product that eluted at 15.4 min. No products were produced when Gtp1, Gtp2, or Gtp3 was heat denatured before addition.
FIG. 1.
Biosynthetic pathway of CDP-2-glycerol.
FIG. 2.
CE chromatographs of the Gtp1, Gtp2, and Gtp3 products. (A) Gtp1 reaction product; (B) Gtp2 and Gtp3 reaction products; (C) Gtp2 reaction products; (D) NADH standard; (E) NAD+ standard; (F) ATP standard; (G) ADP standard; (H) CTP standard. a.u., arbitrary units.
Identification of Gtp2 products by ESI MS and tandem MS.
Products of the Gtp2 reaction were purified by RP HPLC. Fractions containing Gtp1 and Gtp2 products were collected and analyzed by ESI MS (see Fig. S2 in the supplemental material). Ion peaks at m/z 476.08 were obtained, which is in agreement with the expected mass for CDP-2-glycerol (477.25). MS/MS analysis of the product peak at m/z 476.08 resulted in the detection of ion peaks at m/z 432.93, 384.08, 322.05, and 233.13, matching the masses of CDP-2-glycerol minus CO2, CDP minus H2O, CMP (CDP-2-glycerol minus glycerol-2-phosphate), and CDP-2-glycerol minus the N and C heterocycle parts, respectively. Fragments corresponding to each peak are recorded in Table 2 .
TABLE 2.
Interpretations of ion peaks present in ESI MS and MS/MS
| Composition of fragment | Molecular formula | Mol wt | Mass (kDa; negative) |
|---|---|---|---|
| CDP-2-glycerol (full scan) | |||
| CDP-2-glycerol | C12H21O13N3P2 | 477.25 | 476.08 |
| CDP-2-glycerol (MS/MS ion peak 476) | |||
| CDP-2-glycerol minus CO2(N heterocycle opened) | C11H21O11N3P2 | 433.24 | 432.93 |
| CDP minus H2O | C9H13O10N3P2 | 385.16 | 384.08 |
| CDP-2-glycerol minus glycerol-2-phosphate (CMP) | C9H13O8N3P | 322.18 | 322.05 |
| CDP-2-glycerol minus N and C heterocycles | C3H9O8P2 | 235.04 | 233.13 |
Determination of the Gtp2 product by NMR spectroscopy.
A sample of CDP-2-Gro was studied by 1H, 13C, and 31P NMR spectroscopy. A two-dimensional 1H-1H correlation spectroscopy (COSY) experiment demonstrated the following correlations: H1′/H2′ at d 6.01/4.32 for d-ribofuranose (Rib); H1a, H1b/H2 at d 3.74, 3.77/4.32, and H2/H3a, H3b at d 4.32/3.74, 3.77 for glycerol (Gro); and H5/H6 at d 6.14/17.95 for cytosine. The other 1H NMR signals and the 13C NMR signals for Rib and Gro were found using a two-dimensional H-detected 1H-13C heteronuclear single quantum coherence (HSQC) experiment, which showed the following cross-peaks: H2′/C2′, H3′/C3′, H4′/C4′, and H5a′, H5b′/C5′ for Rib and H1a, H1b/C1, H2/C2, and H3a, H3b/C3 for Gro (for chemical shifts, see Table 3). The assigned chemical shifts were in agreement with published data for Rib 5′-phosphate (8) and Gro 2-phosphate (18). The 31P NMR spectrum contained signals for a diphosphate group at d −10.9 and −11.1, which, as expected, showed strong correlations with H5a′5b′ signals of Rib at d −10.9/4.22, 4.27 and H2 signals of Gro at d −11.1/4.32 in a two-dimensional H-detected 1H-31P HMQC spectrum. These data proved the structure of CDP-2-Gro (Fig. 3).
TABLE 3.
1H and 13C NMR data of the CDP-2-Groa
| Ribofuranose |
Glycerol |
Cytosine |
||||||
|---|---|---|---|---|---|---|---|---|
| Atom(s) | δ (ppm) for atom: |
Atom(s) | δ (ppm) for atom: |
Atom | δ (ppm) for atom: |
|||
| 1H | 13C | 1H | 13C | 1H | 13C | |||
| 1′ | 6.01 | 90.7 | 1a, 1b | 3.74, 3.77 | 62.6 | 5 | 6.14 | 98 |
| 2′ | 4.32 | 75.6 | 2 | 4.32 | 79 | 6 | 7.95 | ND |
| 3′ | 4.36 | 70.8 | 3a, 3b | 3.74, 3.77 | 62.6 | |||
| 4′ | 4.28 | 84.3 | ||||||
| 5a′, 5b′ | 4.22, 4.27 | 66.3 | ||||||
ND, not determined.
FIG. 3.
Part of a two-dimensional 1H,13C HSQC spectrum of CDP-2-Gro. The corresponding part of the 1H NMR spectrum is displayed along the top axis. Arabic numerals refer to cross-peaks in the ribose, glycerol, and cytosine moieties designated R, G, and C, respectively. Signals due to contamination are marked with an asterisk. The structure of CDP-2-Gro is shown within the boxed area. HDO, deuterium protium oxide.
Kinetic parameters for Gtp1 and Gtp2.
Kinetic parameters of Gtp1 for the possible substrate (1,3-dihydroxyacetone or glyceraldehyde) and the cofactor NADH and of Gtp2 for the substrate (glycerol-2-phosphate and CTP) were measured. The initial velocities were measured and used for the kinetic parameter calculations. The kinetics of the reaction catalyzed by Gtp1 and Gtp2 fit reasonably well into the Michaelis-Menten model (see Fig. S3 in the supplemental material), and the detailed kinetic parameters for the two enzymes are listed in Table 4. Gtp1 has a higher kcat/Km ratio for NADPH (401.95 mM−1·min−1) than for NADH (116 mM−1·min−1), indicating a preference for NADPH as the cofactor.
TABLE 4.
Kinetic parameters
| Enzyme | Substrate | Km (mM) | Vmax(mM min−1) | kcat (min−1) |
|---|---|---|---|---|
| Gtp1 | Glyceraldehyde | 0.55 ± 0.08 | 0.024 ± 0.008 | 168.34 ± 70.91 |
| Gtp1 | 1,3-Dihydroxyacetone | 0.19 ± 0.08 | 0.038 ± 0.004 | 296.88 ± 28.16 |
| Gtp1 | NADH | 2.42 ± 0.14 | 0.18 ± 0.009 | 280.73 ± 14.52 |
| Gtp1 | NADPH | 4.6 ± 0.67 | 0.24 ± 0.07 | 1,848.96 ± 565.17 |
| Gtp2 | CTP | 0.15 ± 0.02 | 0.021 ± 0.007 | 755.63 ± 251.89 |
| Gtp2 | dCTP | 1.37 ± 0.06 | 0.051 ± 0.024 | 188.7 ± 88.53 |
| Gtp2 | Glycerol-2-phosphate | 3.03 ± 0.32 | 0.024 ± 0.002 | 875.6 ± 54.99 |
| Gtp2 | Glycerol-1-phosphate | 13.49 ± 0.18 | 0.017 ± 0.004 | 623.73 ± 149.84 |
Determination of physicochemical parameters: optimal temperature and pH for Gtp1 and Gtp2 activities.
Activities of Gtp1 and Gtp2 at temperatures ranging from 4 to 75°C and 4 to 65°C are shown in Fig. 4a. For Gtp1, the conversion ratio increased along with a rise in temperature and reached 86.6% at 65°C, and the cofactor became degraded at temperatures higher than 65°C. The results indicated that Gtp1 is more active at higher temperatures. For Gtp2, activity was detected over a broad range of temperatures from 4 to 75°C, with the highest conversion ratio of 61.2% at 37°C. Gtp1 and Gtp2 had a broad pH range of activities, which is shown in Fig. 4b. For Gtp1, activity was observed for pHs of >5, with an optimum pH between 6 and 9. Gtp2 was active over a wide pH range, with an optimum pH between 6 and 9 and with the highest conversion ratio of 65% at pH 8.
FIG. 4.
Effects of temperature (a), pH (b), and cation (c) on the conversion ratios of Gtp1 and Gtp2. The reactions of Gtp2* were carried out in the presence of Mg2+.
Analysis of cation requirements of Gtp1 and Gtp2.
The effects of cation, including NH4+, Fe2+, Ca2+, Mn2+, Ni2+, Co2+, Cu2+, Zn2+, Mg2+, and Fe3+ on the activities of Gtp1 and Gtp2 are shown in Fig. 4c. The results showed that Fe2+ was favorable for Gtp1 activity and that Cu2+ and Ni2+ can partially inhibit its activity. Gtp2 activity was observed in the presence of Fe2+, Mn2+, Co2+, and Mg2+. Mg2+ was favorable for the reaction, with a conversion ratio of 72.6%, and those of the others (Fe2+, Mn2+, Co2+) were lower, with conversion ratios from 17.1 to 41.2%. The effect of cation was also examined in the presence of Mg2+. Whereas the addition of some cations to the reaction mixture including Mg2+ had no effect, the cations Ni2+ and Cu2+ strongly inhibited Gtp2 activity (Fig. 4c). When EDTA as the cation-chelating agent was added, no Gtp2 activities were detected, indicating that Gtp2 is divalent dependent. EDTA had no effect on Gtp1 activity.
Analysis of substrates for Gtp1 and Gtp2.
Both 1,3-dihydroxyacetone and glyceraldehyde could be used as the substrate for Gtp1. However, Gtp1 had a higher kcat/Km ratio for 1,3-dihydroxyacetone (1563 mM−1·min−1) than for glyceraldehyde (306 mM−1·min−1), indicating a preference for 1,3-dihydroxyacetone. Both glycerol-1-phosphate and glycerol-2-phosphate were tested as substrates in the reaction catalyzed by Gtp2, and Gtp2 was active when either was used (data not shown). However, Gtp2 had a higher kcat/Km ratio for glycerol-2-phosphate (288.98 mM−1·min−1) than for glycerol-1-phosphate (46.24 mM−1·min−1), indicating a preference for glycerol-2-phosphate. Based on that, the final product of the pathway was determined to be CDP-2-glycerol, and the substrate for Gtp2 in this reaction should be glycerol-2-phosphate. dATP, dTTP, dGTP, dCTP, ATP, TTP, and CTP were tested as the nucleoside triphosphate (NTP) donors for the transformation reaction catalyzed by Gtp2, and only CTP and dCTP were active NTP donors for Gtp2 (data not shown). This analysis showed that the Gtp2 kcat/Km ratio for CTP (5037.53 mM−1·min−1) was higher than for dCTP (137.73 mM−1·min−1), indicating that CTP is the preferred NTP donor in the reaction.
DISCUSSION
This is the first study on the full characterization of the enzymes in the CDP-2-glycerol biosynthetic pathway, which is included in the capsular antigen of S. pneumoniae. The substrate 1,3-dihydroxyacetone or glyceraldehyde in the initial step of the pathway is intermediate of glycerolipid metabolism. The gldA gene (encoding glycerol dehydrogenase (GDH) in S. pneumoniae) responsible for the conversion from glycerol to 1,3-dihydroxyacetone (3) can be found in S. pneumoniae.
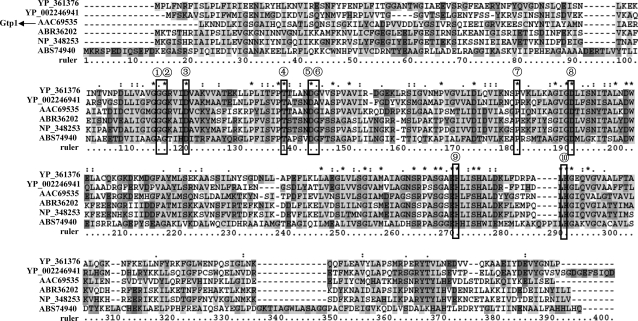
Gtp1 is 16 to 39% identical to glycerol-1-phosphate dehydrogenase (G1PDH), GDH, and 3-dehydroquinate synthase (DHQS) from different species, and all the proteins belong to the same dehydroquinate synthase-like superfamily (CL0224). Alignment of Gtp1 and some proteins from G1PDH, GDH, and DHQS revealed that residues proposed in the binding of NAD+ and the metal cofactor Zn2+ (based on the identified binding sites in the crystal structures of GDH and DHQS) (7) were present in Gtp1 (Fig. 5). Compared to the above dehydrogenases, Gtp1 was identified as a reductase catalyzing the conversion of 1,3-dihydroxyacetone or glyceraldehyde to glycerol and as requiring NADH/NADPH as a cofactor; it was not active for the reverse reaction, and Fe2+ instead of Zn2+ was favorable for Gtp1 activity. Several NADH/NADPH binding motifs, such as GXGXXA (24), GXGXXG (23), and GXGXXP (21), have been reported, and we proposed that the GXGXXR motif near the position of the NAD+ binding site may be the possible motif for Gtp1 to combine with NADH/NADPH. The binding sites of Zn2+ in G1PDH may be responsible for the binding of Fe2+ in Gtp2; however, further experimental evidence is needed. In order to obtain more obviously optimal pHs and temperatures for enzymes, reactions were stopped before the time for the highest rate. Gtp1 seems to be a thermozyme with high activity at 65°C and active over a broad pH range; therefore, it is a good candidate enzyme for industry applications.
FIG. 5.
Alignment of Gtp1 and some G1PDH, GDH, and DHQS proteins. YP 361376, putative glycerol-1-phosphate dehydrogenase from Carboxydothermus hydrogenoformans; YP 002246941, glycerol-1-phosphate dehydrogenase from Coprothermobacter proteolyticus; ABR36202, 3-dehydroquinate synthase from Clostridium beijerinckii; NP 348253, glycerol dehydrogenase from Clostridium acetobutylicum; ABS74940, alcohol dehydrogenase from Bacillus amyloliquefaciens. The seven sites marked with circled 1 to 7 indicate the residues predicted to interact with the coenzyme NADH, and those marked with circled 8 to 10 indicate the residues interacting with the metal cofactor. Asterisks indicate positions which have a single, fully conserved residue. A colon indicates that one of the following “strong” groups is fully conserved: STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, and FYW. A period indicates that one of the following weaker groups is fully conserved: CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, and HFY.
Many kinds of cytidylyltransferases have been identified; examples are glucose-1-phosphate cytidylyltransferase, which is involved in the synthesis of CDP-d-glucose (12, 22), glycerol-1-phosphate cytidylyltransferase, responsible for CDP-glycerol synthesis (15), inositol-1-phosphate cytidylyltransferase, responsible for the synthesis of di-myo-inositol-phosphate (20), and phosphocholine cytidylyltransferase, which catalyzes the pivotal step for phosphatidylcholine synthesis (6). In this study, Gtp2 was the first glycerol-2-phosphate cytidylyltransferase characterized that synthesizes CDP-2-glycerol; however, it was also active when glycerol-1-phosphate was used as the substrate. Gtp2 shares 20 to 30% identity to many glucose-1-phosphate cytidylyltransferases, and they belong to the same protein family: NTP_transferase (Pfam accession no. PF00483). Alignment of Gtp2 and three glucose-1-phosphate cytidylyltransferases revealed a conserved sequence domain (Fig. 6), which is postulated to be responsible for the binding of the nucleotide part or of an allosteric effector molecule (22). Although Gtp2 was active when glycerol-1-phosphate was used as the substrate, it shares very low identity (<10%) with other glycerol-1-phosphate cytidylyltransferases and belongs to a protein family different from those of other known glycerol-1-phosphate cytidylyltransferases (CTP_transf_2 [Pfam accession no. PF01467]). The HWGH motif and RTEGISTT motif in glycerol-1-phosphate cytidylyltransferases, which interact with CTP (16), was not present in Gtp2 (data not shown). The cation requirements, substrate specificities, and kinetic parameters of Gtp2, AscA (the glucose-1-phosphate cytidylyltransferase from Yersinia pseudotuberculosis), and TagD (the glycerol-1-phosphate cytidylyltransferase from Bacillus subtilis) were also compared. The divalent cations Mn2+, Co2+, Fe2+, and Mg2+ were required for the activity of the three enzymes (except that Fe2+ was not tested for AscA), and the divalent cations Cd2+, Hg2+, Sn2+, Cu2+, and Ni2+ inhibited enzyme activity. (The cations Cd2+, Hg2+, and Sn2+ were not tested for AscA and Gtp2 activities, and Ni2+ was not tested for TagD activity) (15, 22). However, the enzymes Gtp1 and Gtp2 were expressed with a His tag, and Cu2+ and Ni2+ may bind them to inhibit their activities. It is noteworthy that Zn2+ inhibited the activity of TagD but did not inhibit that of Gtp2, indicating different origins and functions of the two enzymes (15). Gtp2 and TagD were active when dCTP was used as the substrate (15), and slight AscA activity was also observed when UTP was used as the substrate (22). AscA has an obviously low Km value (81.9 μM) compared to Gtp2 (0.15 mM) and TagD (3.85 mM) for CTP (15, 22), indicating that AscA has the highest affinity toward CTP. TagD has a lower Km value (3.23 mM) for glycerol-1-phosphate than Gtp2 (13.49 mM) (15), showing a higher affinity to glycerol-1-phosphate.
FIG. 6.
Alignment of Gtp2 and three glucose-1-phosphate cytidylyltransferases from Yersinia pseudotuberculosis, Salmonella enterica, and Archaeoglobus fulgidus. The GenBank accession numbers are shown.
Gtp3 is the first characterized glycerol-2-phosphotransferase. It has 32 to 42% identity to many haloacid dehalogenase (HAD) family hydrolases, which includes a diverse range of enzymes that use an Asp carboxylate as a nucleophile (10). Gtp3 contains repeating β-α units, like other HAD family proteins (5). Sequence comparisons showed four conserved motifs in Gtp3 and other HAD family hydrolases, and conserved residues of each motif were found (Fig. 7). The signature DXD in motif I and GDXXXXD in motif IV are required for coordinating the Mg2+ in the active site (11, 13, 17, 19). In our study, Gtp3 was active only in the presence of Gtp2; however, the mechanism is not clear.
FIG. 7.
Alignment of Gtp3 and other HAD family hydrolases from Caldicellulosiruptor saccharolyticus, Fusobacterium nucleatum, Alkaliphilus oremlandii, and Alkaliphilus metalliredigens. The GenBank accession numbers are shown. Four conserved motifs and conserved residues in each motif are marked. The predicted secondary structural elements are indicated with “E” for β-strand regions, “H” for α-helical regions, and - for coil regions.
Rare sugars are potentially useful in the pharmaceutical and chemical industries for drug development. This report provides valuable enzyme sources for the production of CTP-2-glycerol that are not commonly found and are not commercially available.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (30900255, 30788001, and 30870070), the National 863 Program (2010AA10A203), the National Key Programs for Infectious Diseases of China (2008ZX10004-002, 2008ZX10004-009, 2009ZX10004-108, 2008ZX10003, and 2008zx10001-004), and the Tianjin Research Program of the Foundation for the Application of Advanced Technology (10JCYBJC10100).
Footnotes
Published ahead of print on 20 August 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aanensen, D. M., A. Mavroidi, S. D. Bentley, P. R. Reeves, and B. G. Spratt. 2007. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189:7856-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso De Velasco, E., A. Verheul, J. Verhoef, and H. Snippe. 1995. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol. Rev. 59:591-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asnis, R. E., and A. F. Brodie. 1953. A glycerol dehydrogenase from Escherichia coli. J. Biol. Chem. 203:153-159. [PubMed] [Google Scholar]
- 4.Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burroughs, A. M., K. N. Allen, D. Dunaway-Mariano, and L. Aravind. 2006. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 361:1003-1034. [DOI] [PubMed] [Google Scholar]
- 6.Chen, B. B., and R. K. Mallampalli. 2007. Calmodulin binds and stabilizes the regulatory enzyme, CTP: phosphocholine cytidylyltransferase. J. Biol. Chem. 282:33494-33506. [DOI] [PubMed] [Google Scholar]
- 7.Daiyasu, H., T. Hiroike, Y. Koga, and H. Toh. 2002. Analysis of membrane stereochemistry with homology modeling of sn-glycerol-1-phosphate dehydrogenase. Protein Eng. 15:987-995. [DOI] [PubMed] [Google Scholar]
- 8.Guijarro, J. I., J. E. Gonzalez-Pastor, F. Baleux, J. L. San Millan, M. A. Castilla, M. Rico, F. Moreno, and M. Delepierre. 1995. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J. Biol. Chem. 270:23520-23532. [DOI] [PubMed] [Google Scholar]
- 9.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hisano, T., Y. Hata, T. Fujii, J. Q. Liu, T. Kurihara, N. Esaki, and K. Soda. 1996. Crystal structure of l-2-haloacid dehalogenase from Pseudomonas sp. YL. An alpha/beta hydrolase structure that is different from the alpha/beta hydrolase fold. J. Biol. Chem. 271:20322-20330. [DOI] [PubMed] [Google Scholar]
- 11.Lahiri, S. D., G. Zhang, D. Dunaway-Mariano, and K. N. Allen. 2002. Caught in the act: the structure of phosphorylated beta-phosphoglucomutase from Lactococcus lactis. Biochemistry 41:8351-8359. [DOI] [PubMed] [Google Scholar]
- 12.Lindqvist, L., R. Kaiser, P. R. Reeves, and A. A. Lindberg. 1994. Purification, characterization, and high performance liquid chromatography assay of Salmonella glucose-1-phosphate cytidylyltransferase from the cloned rfbF gene. J. Biol. Chem. 269:122-126. [PubMed] [Google Scholar]
- 13.Morais, M. C., W. Zhang, A. S. Baker, G. Zhang, D. Dunaway-Mariano, and K. N. Allen. 2000. The crystal structure of bacillus cereus phosphonoacetaldehyde hydrolase: insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily. Biochemistry 39:10385-10396. [DOI] [PubMed] [Google Scholar]
- 14.Morona, J. K., D. C. Miller, T. J. Coffey, C. J. Vindurampulle, B. G. Spratt, R. Morona, and J. C. Paton. 1999. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 23F. Microbiology 145:781-789. [DOI] [PubMed] [Google Scholar]
- 15.Park, Y. S., T. D. Sweitzer, J. E. Dixon, and C. Kent. 1993. Expression, purification, and characterization of CTP: glycerol-3-phosphate cytidylytransferase from Bacillus subtilis. J. Biol. Chem. 268:16648-16654. [PubMed] [Google Scholar]
- 16.Pattridge, K. A., C. H. Weber, J. A. Friesen, S. Sanker, C. Kent, and M. L. Ludwig. 2003. Glycerol-3-phosphate cytidylyltransferase. Structural changes induced by binding of CDP-glycerol and the role of lysine residues in catalysis. J. Biol. Chem. 278:51863-51871. [DOI] [PubMed] [Google Scholar]
- 17.Peisach, E., J. D. Selengut, D. Dunaway-Mariano, and K. N. Allen. 2004. X-ray crystal structure of the hypothetical phosphotyrosine phosphatase MDP-1 of the haloacid dehalogenase superfamily. Biochemistry 43:12770-12779. [DOI] [PubMed] [Google Scholar]
- 18.Perepelov, A. V., B. Liu, S. N. Senchenkova, S. D. Shevelev, W. Wang, A. S. Shashkov, L. Feng, L. Wang, and I. A. Knirel. 2007. The structure of the glycerol phosphate-containing O-specific polysaccharide from Escherichia coli O130. Bioorg. Khim. 33:57-60. [DOI] [PubMed] [Google Scholar]
- 19.Rinaldo-Matthis, A., C. Rampazzo, P. Reichard, V. Bianchi, and P. Nordlund. 2002. Crystal structure of a human mitochondrial deoxyribonucleotidase. Nat. Struct. Biol. 9:779-787. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigues, M. V., N. Borges, M. Henriques, P. Lamosa, R. Ventura, C. Fernandes, N. Empadinhas, C. Maycock, M. S. da Costa, and H. Santos. 2007. Bifunctional CTP:inositol-1-phosphate cytidylyltransferase/CDP-inositol:inositol-1-phosphate transferase, the key enzyme for di-myo-inositol-phosphate synthesis in several (hyper)thermophiles. J. Bacteriol. 189:5405-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roma, G. W., L. J. Crowley, C. A. Davis, and M. J. Barber. 2005. Mutagenesis of glycine 179 modulates both catalytic efficiency and reduced pyridine nucleotide specificity in cytochrome b5 reductase. Biochemistry 44:13467-13476. [DOI] [PubMed] [Google Scholar]
- 22.Thorson, J. S., T. M. Kelly, and H. W. Liu. 1994. Cloning, sequencing, and overexpression in Escherichia coli of the alpha-d-glucose-1-phosphate cytidylyltransferase gene isolated from Yersinia pseudotuberculosis. J. Bacteriol. 176:1840-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang, Q., P. Ding, A. V. Perepelov, Y. Xu, Y. Wang, Y. A. Knirel, L. Wang, and L. Feng. 2008. Characterization of the dTDP-d-fucofuranose biosynthetic pathway in Escherichia coli O52. Mol. Microbiol. 70:1358-1367. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida, Y., Y. Nakano, T. Nezu, Y. Yamashita, and T. Koga. 1999. A novel NDP-6-deoxyhexosyl-4-ulose reductase in the pathway for the synthesis of thymidine diphosphate-d-fucose. J. Biol. Chem. 274:16933-16939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.