Abstract
The bacterium Ralstonia eutropha H16 synthesizes polyhydroxybutyrate (PHB) from acetyl coenzyme A (acetyl-CoA) through reactions catalyzed by a β-ketothiolase (PhaA), an acetoacetyl-CoA reductase (PhaB), and a polyhydroxyalkanoate synthase (PhaC). An operon of three genes encoding these enzymatic steps was discovered in R. eutropha and has been well studied. Sequencing and analysis of the R. eutropha genome revealed putative isologs for each of the PHB biosynthetic genes, many of which had never been characterized. In addition to the previously identified phaB1 gene, the genome contains the isologs phaB2 and phaB3 as well as 15 other potential acetoacetyl-CoA reductases. We have investigated the roles of the three phaB isologs by deleting them from the genome individually and in combination. It was discovered that the gene products of both phaB1 and phaB3 contribute to PHB biosynthesis in fructose minimal medium but that in plant oil minimal medium and rich medium, phaB3 seems to be unexpressed. This raises interesting questions concerning the regulation of phaB3 expression. Deletion of the gene phaB2 did not result in an observable phenotype under the conditions tested, although this gene does encode an active reductase. Addition of the individual reductase genes to the genome of the ΔphaB1 ΔphaB2 ΔphaB3 strain restored PHB production, and in the course of our complementation experiments, we serendipitously created a PHB-hyperproducing mutant. Measurement of the PhaB and PhaA activities of the mutant strains indicated that the thiolase reaction is the limiting step in PHB biosynthesis in R. eutropha H16 during nitrogen-limited growth on fructose.
Polyhydroxyalkanoates (PHAs) are natural polyesters synthesized by a wide range of bacteria as carbon and energy reserves. PHAs are typically stored when organisms are in an environment in which carbon is plentiful but the lack of another nutrient limits normal cell growth. It has been found that in environments with fluctuating carbon levels, PHA producers have crucial advantages over rival species (14). In addition to their importance in the microbial world, these polymers have been studied for their potential uses in biodegradable consumer goods (12) and medical products (22) and as chemical precursors (4). Although many PHA monomers have been discovered, the most common are 3-hydroxyalkanoates (32). Common PHAs are typically characterized by their constituent monomers as short-chain-length polymers (SCL-PHA; C4 and C5 monomers) or medium-chain-length polymers (MCL-PHA; C6 and longer monomers).
The model organism used to study PHA biosynthesis is the Gram-negative bacterium Ralstonia eutropha. This organism accumulates a high percentage of its cell dry weight (CDW) as SCL-PHA under nutrient limitation. When grown on sugars or plant oils, R. eutropha makes poly(3-hydroxybutyrate) (PHB) almost exclusively, although the addition of precursors such as propionate to the growth medium can lead to incorporation of 3-hydroxyvalerate into the polymer chain as well (2). An operon of biosynthetic genes from R. eutropha encoding enzymes sufficient for synthesis of PHB from acetyl coenzyme A (acetyl-CoA), which consisted of phaC-phaA-phaB, was discovered in the late 1980s (25, 26, 36). In this pathway, two molecules of acetyl-CoA are condensed by a β-ketothiolase (PhaA) and the resulting acetoacetyl-CoA is reduced by a reductase (PhaB) to form (R)-3-hydroxybutyryl-CoA (HB-CoA), which is the substrate for the PHA synthase (PhaC). Sequencing and analysis of the R. eutropha genome revealed the existence of putative isologs for each of the PHA synthetic genes (29). While the existence of alternate β-ketothiolases was already known (39), most of the potential isologs identified had never been characterized.
Our group wanted to better understand how acetoacetyl-CoA reduction occurs in R. eutropha. In addition to the earlier-identified phaB gene, now referred to as phaB1 (GeneID, 4249784), the genes phaB2 (GeneID, 4249785) and phaB3 (GeneID, 4250155) were discovered on R. eutropha chromosome 1. Fifteen other potential isologs were also found to encode amino acid sequences that could potentially indicate acetoacetyl-CoA reductase activity (29). The roles of the newly discovered genes in PHB biosynthesis were unclear, especially given the results of an earlier biochemical study that suggested there was a single NADPH-dependent acetoacetyl-CoA reductase in R. eutropha (10). In order to determine the roles of the reductase genes in R. eutropha, we deleted phaB1, phaB2, and phaB3 from the genome both individually and in combination. In addition to characterizing these newly discovered genes, we also hoped to eliminate or diminish formation of HB-CoA by stopping the reduction reaction. Efforts to purify the PHA synthase from R. eutropha have been complicated by the high levels of PHB made by this organism (7). Studying formation and growth of PHB granules is difficult because PHB accumulates at a high rate, causing individual granules to coalesce and become indistinct (44). We therefore believed that an R. eutropha strain with decreased HB-CoA synthesis would be a useful experimental tool and could also serve as a platform for engineering new PHA synthesis pathways into R. eutropha.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
All experiments were performed with Ralstonia eutropha H16 (ATCC 17699) and mutants derived from this strain (Table 1). R. eutropha strains were grown aerobically at 30°C in both rich and minimal media. The rich medium was dextrose-free tryptic soy broth (TSB) medium (Becton Dickinson, Sparks, MD). The minimal medium had an initial pH of 6.8 and was composed of 4.0 g/liter NaH2PO4, 4.6 g/liter Na2HPO4, 0.45 g/liter K2SO4, 0.39 g/liter MgSO4, 62 mg/liter CaCl2, and 1 ml per liter of a trace element solution. The trace element solution consisted of 15 g/liter FeSO4·7H2O, 2.4 g/liter MnSO4·H2O, 2.4 g/liter ZnSO4·7H2O, and 0.48 g/liter CuSO4·5H2O dissolved in 0.1 M hydrochloric acid. Carbon and nitrogen sources were added to this defined medium as described in the text. All media contained 10 μg/ml gentamicin sulfate. Medium components were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise specified.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| R. eutropha strains | ||
| H16 | Wild-type strain; Gm resistant | ATCC 17699 |
| Re2106 | ΔphaB2; made from H16 with pCB28 | This study |
| Re2107 | ΔphaB3; made from H16 with pCB29 | This study |
| Re2111 | ΔphaB1; made from H16 with pCB42 | This study |
| Re2112 | ΔphaB1 ΔphaB2; made from Re2106 with pCB42 | This study |
| Re2113 | ΔphaB1 ΔphaB3; made from Re2107 with pCB42 | This study |
| Re2114 | ΔphaB2 ΔphaB3; made from Re2106 with pCB29 | This study |
| Re2115 | ΔphaB1 ΔphaB2 ΔphaB3; made from Re2112 with pCB29 | This study |
| Re2139 | phaB1 inserted into Re2115 genome; made with pCB65 | This study |
| Re2140 | phaB2 inserted into Re2115 genome; made with pCB66 | This study |
| Re2141 | phaB3 inserted into Re2115 genome; made with pCB67 | This study |
| Re2142 | fabG inserted into Re2115 genome; made with pCB68 | This study |
| Re2143 | phaB3 inserted into Re2115 genome; made with pCB76 | This study |
| E. coli strains | ||
| DH5α | General cloning strain | Invitrogen |
| S17-1 | Strain for conjugative transfer of plasmids to R. eutropha | 38 |
| Tuner(DE3) | Strain for inducible protein expression | Novagen |
| Plasmids | ||
| pCR2.1-TOPO | Vector for cloning and sequencing PCR products, confers Km resistance | Invitrogen |
| pGY46 | Plasmid for deletion of R. eutropha phaC1; carries sacB, oriV, oriT, and traJ; confers Km resistance; based on pJQ200mp18; all other deletion plasmids use backbone from this plasmid | 31, 46 |
| pCB28 | Plasmid for deletion of R. eutropha phaB2 | This study |
| pCB29 | Plasmid for deletion of R. eutropha phaB3 | This study |
| pCB42 | Plasmid for deletion of R. eutropha phaB1 | This study |
| pCB65 | Plasmid for insertion of phaB1 into R. eutropha genome at phaB1 locus; based on pCB42 | This study |
| pCB66 | Plasmid for insertion of phaB2 into R. eutropha genome at phaB1 locus; based on pCB42 | This study |
| pCB67 | Plasmid for insertion of phaB3 (annotated start codon) into R. eutropha genome at phaB1 locus; based on pCB42 | This study |
| pCB68 | Plasmid for insertion of fabG into R. eutropha genome at phaB1 locus; based on pCB42 | This study |
| pCB76 | Plasmid for insertion of phaB3 (upstream start codon) into R. eutropha genome at phaB1 locus; based on pCB42 | This study |
| pET-15b | Plasmid expression of His-tagged proteins in E. coli; confers Ap resistance | Novagen |
| pRARE2 | Plasmid supplying tRNAs for seven rare E. coli codons; confers Cm resistance | Novagen |
| pET-15b-phaB1 | Plasmid for expression of PhaB1 with N-terminal His tag | This study |
| pET-15b-phaB2 | Plasmid for expression of PhaB2 with N-terminal His tag | This study |
| pET-15b-phaB3 | Plasmid for expression of PhaB3 with N-terminal His tag | This study |
| pET-15b-fabG | Plasmid for expression of FabG with N-terminal His tag | This study |
Abbreviations: Gm, gentamicin; Km, kanamycin; Ap, ampicillin; Cm, chloramphenicol.
Growth in rich medium was carried out by inoculating 6 ml of TSB with a single colony from a TSB agar plate. These cultures were incubated for 24 h on a roller drum and then used to inoculate 100-ml cultures of TSB to an initial optical density at 600 nm (OD600) of 0.05. The 100-ml cultures were grown in 500-ml baffled flasks and shaken at 200 rpm. Samples were taken from the flask cultures at various time points for analysis.
Growth in minimal media was carried out by inoculating 3 ml of TSB with a single colony from a TSB agar plate. These cultures were incubated 24 h on a roller drum and then used to inoculate 5 ml of minimal medium containing 2% fructose and 0.1% NH4Cl. Aliquots from the minimal medium precultures were used to inoculate minimal medium flask cultures to an initial OD600 of 0.05. The minimal medium generally contained 2% fructose and 0.05% NH4Cl. This fructose concentration was sufficiently high that carbon limitation never occurred in these cultures. In one set of experiments, 1% emulsified palm oil (Wilderness Family Naturals, Silver Bay, MN) was used as the sole carbon source in place of fructose. Gum arabic (0.3%; Sigma-Aldrich) was included in the palm oil medium as the emulsifying agent. Gum arabic is a natural glycoprotein (8) that is not metabolized by R. eutropha. The oil was emulsified by mixing the medium with a Sorvall Omni-Mixer for 1 min. Flask cultures had volumes of either 50 ml (in 250-ml baffled flasks) or 100 ml (in 500-ml baffled flasks) and were shaken at 200 rpm. Samples were taken from the flask cultures at various time points for analysis.
Plasmid and strain construction.
The method for deletion and insertion of genes in the R. eutropha genome was in accordance with the procedure described by York et al. (46). Standard techniques were used to amplify, manipulate, and prepare DNA (35). DNA was routinely amplified using high-fidelity Taq polymerase (Qiagen, Valencia, CA) and digested using restriction enzymes from New England BioLabs (Ipswich, MA). All oligonucleotide sequences used in this study are provided in Table S1 in the supplemental material.
Plasmids used to make markerless deletions in the R. eutropha chromosome were created by first constructing stretches of DNA in which the regions upstream and downstream of a given gene were connected. This was done by first amplifying ∼500 bp of sequence upstream and downstream of the gene. Primers were designed such that the two fragments had identical 16-bp sequences at the ends that were to be connected. The 16-bp sequence included a SwaI restriction site for future cloning applications. A single, connected DNA fragment was created by overlap extension PCR. The primers used in the overlap PCR were designed so that the final product had BamHI restriction sites at each end. The product of the overlap PCR was cloned into a TOPO vector (Invitrogen, Carlsbad, CA) and sequenced. The TOPO vector was digested with BamHI, and the fragment for making the deletion was isolated and then ligated into the backbone of pGY46 digested with the same enzyme. The plasmid pGY46 has previously been used to delete the R. eutropha phaC1 gene (46). Gene deletion plasmids were transformed into Escherichia coli S17-1 and introduced into R. eutropha via conjugative transfer. R. eutropha strains with potential deletions were assessed by diagnostic PCR.
Complementation experiments were carried out by integrating potential reductase genes into the genome of strain Re2115. The genes phaB1, phaB2, phaB3, and fabG (GeneID, 4246984) were amplified from the R. eutropha genome by PCR, cloned into TOPO vectors, and sequenced. The gene phaB3 was cloned twice, once using the sequence as annotated in the published genome (29) and once using an alternate start codon 30 bp upstream of the annotated start. The primers were designed such that all genes had an AscI restriction site upstream of the gene and a PacI site downstream of the gene. Additionally, the primers that hybridized to the 5′ end of each gene were designed so that the 11 bp immediately upstream of the start codon of each gene were AGGAGATCTCC, which ensured that each gene had an identical ribosome binding site (RBS) in the complemented strains. The TOPO vectors containing each gene were digested with AscI and PacI, and then the DNA fragment with the gene was isolated and blunted using a New England BioLabs quick-blunting kit. Finally, each gene was cloned into the SwaI site of pCB42 (the plasmid used to delete phaB1), creating plasmids for integrating each gene into the Re2115 genome at the ΔphaB1 locus. Gene integration was carried out as previously described (46). All strains and plasmids used in this study are described in Table 1.
Polymer analysis.
PHB content and cell dry weight (CDW) were measured by transferring 4 to 9 ml of culture to preweighed glass test tubes at various time points. Cells were pelleted, washed with 5 ml cold water, pelleted again, and dried under a vacuum at 80°C. Samples from palm oil cultures were prepared using the same protocol, except the washing was performed with a mixture of 4 ml cold water and 2 ml cold hexane. The hexane was added to remove unused oil from the samples. PHB content and CDW were determined from the dried samples using established methods (16, 47). The residual cell dry weight (RCDW) was calculated as the CDW minus the mass of PHB.
PHB molecular weight was measured for polymer extracted from strains grown in fructose minimal medium. After 48 and 72 h of growth, 40 ml of culture was harvested, pelleted, washed with 25 ml cold water, pelleted again, resuspended in 5 ml water, frozen at −80°C, and lyophilized. At each time point, samples were also taken for quantification of PHB content. Freeze-dried cells were weighed into glass test tubes, and sufficient chloroform was added so that the dissolved PHB would have a concentration of 3 mg/ml. The test tubes were incubated in a Reacti-Therm heating/stirring module (Pierce, Rockford, IL) at 50 to 55°C with refluxing for 48 h, with stirring provided by magnetic stir bars. Chloroform lost due to evaporation was replaced over the course of the extraction. At the end of the extractions, the samples were cooled and cellular debris was removed by filtering the PHB solutions using 0.2 μm polyvinylidene difluoride (PVDF) syringe filters (Pall, Port Washington, NY).
The molecular weight of the extracted PHB was measured using gel permeation chromatography (GPC). Molecular weights were determined relative to a series of low-polydispersity polystyrene standards, with peak molecular weights ranging from 1.1 × 103 to 13.2 × 106 (part no. PL2010-0104; Polymer Laboratories). All molecular weight standards and experimental samples contained isopropanol as an internal standard to normalize retention times. Samples were run on an Agilent 1100 high-performance liquid chromatograph (HPLC; Santa Clara, CA) connected to a computer running Chemstation software. Polymers were separated using a PLgel Olexis guard column and two PLgel Olexis analytical columns, all connected in series and purchased from Polymer Laboratories (part no. PL1110-6400 and PL1110-1400). Chloroform was used as the mobile phase at a flow rate of 1 ml/minute, with the columns maintained at 30°C. One hundred microliters of each sample was injected onto the columns, and the eluted polymer was detected with a refractive index detector. Calibration of the system and analysis of the experimental samples were performed using the Agilent GPC data analysis software package.
Enzymatic assays.
Acetoacetyl-CoA reductase activity was measured for the soluble fraction from cellular lysates of R. eutropha strains. Cultures were grown in fructose minimal medium, harvested after 24 h of growth, pelleted, and stored at −80°C. Lysates were prepared by thawing the pellets on ice and resuspending them in 50 mM potassium phosphate buffer (pH 6) using 5 ml buffer per gram of wet cell mass. One milliliter of suspended cells was transferred to a 2-ml screw top plastic vial containing 0.6 g of 0.1-mm zirconia/silica beads (BioSpec Products, Bartlesville, OK). Vials were loaded onto a FastPrep-24 (MP Biomedicals, Solon, OH) and treated twice at 6.0 m/s for 40 s, with a 5-min break between treatments. After lysis, the samples were centrifuged for 15 min at 4°C. Remaining insoluble debris was removed from the supernatants by filtering it through 0.45-μm low-protein-binding Supor syringe filters (Pall), yielding the soluble lysate fractions that were used for enzymatic assays. Lysates were stored on ice while the experiments were conducted. Reductase activity obtained with the use of either NADPH or NADH as the cofactor was measured at pH 6 and 25°C using the published protocol (10). The assay reaction mixtures contained 50 mM potassium phosphate buffer, 0.1 mM NADPH or NADH, and 32 μM acetoacetyl-CoA. When acetoacetyl-CoA was not included in the assay, addition of R. eutropha lysate did not result in increased NADPH or NADH oxidation relative to the level for the no-lysate control.
The β-ketothiolase activity in the soluble fraction of cellular lysates of R. eutropha strains was measured in the thiolysis direction using acetoacetyl-CoA as the substrate. Lysates were prepared as described above, except cells were harvested after 48 h of growth and the cell pellets were resuspended in 150 mM 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid (EPPS) buffer, pH 8. The enzymatic activity assays were carried out at 25°C as described by Slater et al. (39). The assay reaction mixtures contained 150 mM EPPS (pH 8), 50 mM MgCl2, 40 μM acetoacetyl-CoA, and 0.1 mM CoA. In both the reductase and the β-ketothiolase assays, acetoacetyl-CoA sodium salt was purchased from MP Biomedicals, while all other chemicals were from Sigma-Aldrich. The protein concentrations necessary for specific activity calculations were measured using a modified Bradford assay (48). For both assays, specific activities for pairs of strains were compared using the one-tailed Student t test, with relevant results given in the text.
Purification of His-tagged proteins.
Reductase enzymes from R. eutropha were expressed in E. coli and purified. The expression and purification methods were adapted from a protocol previously used for purification of E. coli FabG (11). The R. eutropha genes phaB1, phaB2, phaB3, and fabG were amplified via PCR with primers that added a BamHI site to the 5′ end of each gene and a BlpI site to the 3′ end of each gene. The PCR fragments were digested with these restriction enzymes and cloned into pET-15b cut with BamHI and BlpI, resulting in genes encoding N-terminal His-tagged versions of each enzyme. Plasmids were transformed into E. coli strain Tuner(DE3), which allowed for inducible protein expression. The R. eutropha genes contain codons that are rare in E. coli; therefore, each Tuner(DE3) strain was also transformed with pRARE2. All strains were grown at 30°C in lysogeny broth (LB) containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. When an OD600 of 0.5 was reached, protein expression was induced by addition of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested 4 h after induction and pelleted. Cell pellets were resuspended in sodium phosphate buffer (20 mM, pH 7.4) and lysed with a French press. Cell debris was removed by centrifuging the lysates and filtering the supernatants through 0.2-μm low-protein-binding filters. Clarified lysate was loaded onto a Ni Sepharose fast-flow column (GE Healthcare, Piscataway, NJ), and fractions were collected with a BioLogic LP chromatography system (Bio-Rad, Hercules, CA). The elution buffer contained 20 mM sodium phosphate and 0.5 M NaCl (pH 7.4) and was run at a flow rate of 5 ml/min. The imidazole concentration in the buffer was increased from 40 mM to 500 mM over the course of each purification. Fractions in which only the protein of interest was visible on an SDS-PAGE gel were collected, concentrated, and dialyzed against storage buffer (20 mM sodium phosphate, 0.2 M NaCl, pH 7.4). The specific activities of the purified enzymes were measured using the reductase assay described above.
RESULTS
Growth and PHB production of R. eutropha reductase mutants in different media.
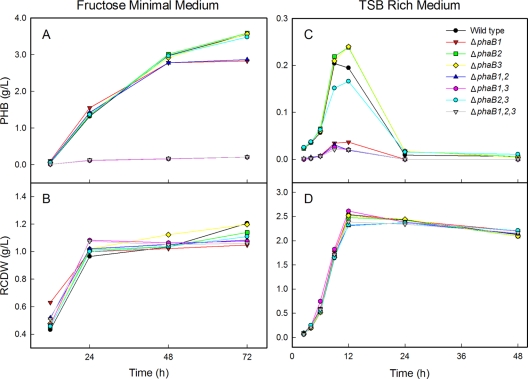
All of the different phaB deletion strains were initially grown in fructose minimal medium and TSB rich medium to determine the influence of the mutations on cell growth and PHB accumulation, as shown in Fig. 1. In minimal medium, the RCDWs of all strains increased until nitrogen in the medium was exhausted, sometime between 12 and 24 h. After nitrogen limitation occurred, RCDW remained constant and all strains reached approximately the same RCDW (Fig. 1B). Similarly, the RCDWs for all strains were nearly identical in TSB (Fig. 1D). This indicates that deletion of the reductase genes did not influence cell growth. In fructose minimal medium, all strains in which phaB1 was present accumulated similar amounts of PHB (Fig. 1A). Strains in which phaB1 was deleted but phaB3 was present showed similar levels of PHB production to H16 from 12 to 48 h, but at the 72 h time point, there was clearly less PHB in these strains than in the wild-type strain. Strains in which both phaB1 and phaB3 were deleted produced markedly less PHB than the other strains in the study. After 72 h in fructose minimal medium, strains Re2113 (ΔphaB1 ΔphaB3) and Re2115 (ΔphaB1 ΔphaB2 ΔphaB3) contained <20% of CDW as PHB, compared to 75% of CDW as PHB for H16. Deletion of phaB2 did not produce an observable phenotype in this medium or under any of the other conditions tested.
FIG. 1.
R. eutropha reductase mutants exhibited different levels of PHB accumulation in both minimal and rich media. Strains were grown in fructose minimal medium, and PHB production (A) and RCDW (B) were measured. For plot A, note that the data points for the low-level-PHB-producing ΔphaB1 ΔphaB3 and ΔphaB1 ΔphaB2 ΔphaB3 strains overlap. Strains were also grown in TSB, and PHB production (C) and RCDW (D) were measured. All data points are averages from duplicate cultures.
It has been shown that when R. eutropha is grown in rich medium, it typically accumulates a relatively low level of PHB early in the culture, which is then metabolized as the cells continue to grow (34). All strains with the phaB1 gene exhibited this pattern of PHB production in TSB, while strains in which phaB1 had been deleted made almost no PHB at any point during the experiment (Fig. 1C). There is some variation in the amount of polymer produced by the strains that make PHB in TSB (H16, Re2106, Re2107, and Re2114), which is most evident at the 9- and 12-h time points. We believe that the observed variations between these strains were due to difficulties in measuring the low levels of PHB and not phenotypic differences between strains.
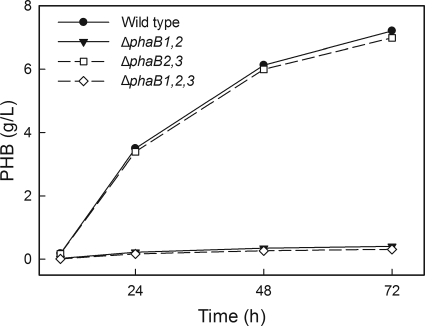
After observing different levels PHB production by some reductase mutant strains in fructose minimal medium and TSB, we decided to test another minimal medium, in which a different carbon source was provided. Plant oils have been proposed as potential feedstocks for industrial PHA production (1). We therefore grew several reductase mutants in minimal medium with palm oil as the sole carbon source and measured PHB production (Fig. 2). The strain with only phaB1 (Re2114) produced the same amount of PHB as H16, as was the case in the fructose medium. The strain with only phaB3 (Re2112), however, accumulated almost no PHB in the palm oil medium, in stark contrast to its behavior in fructose medium.
FIG. 2.
PHB accumulation of Re2112 (ΔphaB1 ΔphaB2) changed when palm oil was used as the sole carbon source instead of fructose. Select strains were grown in palm oil minimal medium, and PHB production was measured. All data points are averages from duplicate cultures.
Expression of reductase genes.
Our group recently reported whole-cell gene expression microarray data for R. eutropha H16 grown in minimal media with fructose or trioleate as the sole carbon source (3). Trioleate served as a model for plant oils. Transcript levels were measured with each carbon source during the growth phase and PHB storage (i.e., nitrogen-limited) phase of the cultures. We examined these data to determine the expression of the reductase genes under different conditions (Table 2). Gene expression is reported relative to fnr3 expression (GeneID, 4248836), a FNR-like transcriptional regulator. We found that expression of this gene varied <20% across all conditions studied (3), making fnr3 a suitable gene to use for normalization. By this analysis, phaB1 was the most highly expressed reductase. Expression of this gene was approximately constant in fructose cultures but increased in trioleate cultures when nitrogen limitation was reached. Expression levels of phaB2 were very low under all conditions. Expression levels of phaB3 were relatively high during the growth phase of fructose cultures, but expression decreased over 10-fold after nitrogen in the medium was depleted. There was little phaB3 expression at any point in the trioleate cultures. The gene fabG was expressed in the presence of both carbon sources, and in both cases, gene expression levels were lower under nitrogen-limited conditions.
TABLE 2.
Expression of reductase genes in R. eutropha H16 fructose and trioleate minimal medium cultures under growth and PHB storage conditionsa
| Gene | Relative gene expression level for indicated medium and culture condition |
|||
|---|---|---|---|---|
| Fructose |
Trioleate |
|||
| Growth | PHB storage | Growth | PHB storage | |
| phaB1 | 7.48 | 7.32 | 1.31 | 6.55 |
| phaB2 | 0.02 | 0.05 | 0.03 | 0.08 |
| phaB3 | 0.79 | 0.06 | 0.03 | 0.02 |
| fabG | 1.55 | 0.30 | 0.85 | 0.29 |
Expression levels were determined based on data from reference 3. Gene expression is reported relative to fnr3 expression (see Results for details).
Complementation of reductase mutations.
In order to confirm that the deletion of the phaB genes was responsible for the observed decreases in PHB accumulation, we introduced reductase genes into Re2115. Because of reported issues with plasmid stability in R. eutropha (40), we chose to integrate the reductase genes into the Re2115 genome at the original phaB1 locus, which also meant that all reductase genes would then be expressed from the promoter of the phaCAB operon. In addition to the phaB genes, we investigated the ability of R. eutropha fabG to restore PHB production. FabG is a 3-ketoacyl reductase that is part of the fatty acid synthesis pathway (30). This enzyme's normal function is to reduce 3-ketoacyl-[acyl carrier protein] molecules, as opposed to the 3-ketoacyl-CoA substrates used by PhaB1.
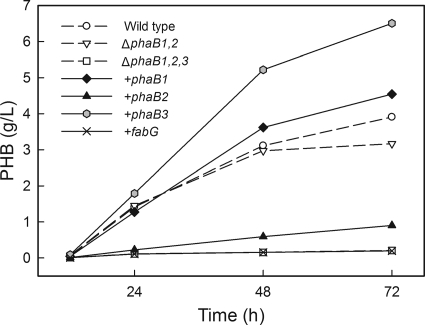
The complemented strains were grown in fructose minimal medium, and PHB production was measured (Fig. 3). We found that the strain in which phaB1 was reintroduced to the Re2115 genome (Re2139) stored slightly more PHB than H16. The strain in which phaB2 was added to Re2115 (Re2140) made more PHB than Re2115 but substantially less than H16. When we initially integrated phaB3 into the Re2115 genome (Re2141), PHB accumulation was the same as that observed for Re2115 (data not shown). This was an unexpected result, as Re2112 made significantly more PHB than Re2115 in fructose cultures. When cloning phaB3 to make Re2141, we used the start codon shown in the annotated genome. After examining the genome sequence, we determined that there was an alternate start codon 30 bp upstream of the annotated start. Addition of these bases to phaB3 results in an open reading frame the same length as phaB1. We cloned the phaB3 gene again using the upstream start codon and created Re2143. Surprisingly, not only was PHB production restored, but Re2143 actually made significantly more polymer than H16. It is unclear why the addition of 10 amino acids (MKKIALVTGG) to the N terminus of PhaB3 was necessary to restore PHB production in this experiment, but protein alignments show that many of these residues are well conserved in both the PhaB sequences and the FabG sequences from several species (5). We therefore concluded that the start codon of phaB3 in the published genome is misannotated. The addition of fabG to the phaB1 locus of Re2115 had no observable impact on PHB production.
FIG. 3.
Addition of reductase genes to the Re2115 genome restored PHB production. H16, Re2112, and Re2115 (dashed lines) were grown at the same time as the complemented strains (solid lines) (genes added to each strain are indicated) in fructose minimal medium, and PHB production was measured. The points labeled “+phaB3” are from strain Re2143. Note that the ΔphaB1 ΔphaB2 ΔphaB3 and “+fabG” points overlap. All data points are averages from duplicate cultures.
PhaB and PhaA activities in mutant strains.
The NADPH-dependent acetoacetyl-CoA reductase specific activity measurements for the various strains correspond well to the PHB production data (Table 3). Deletion of phaB3 from H16 resulted in a slight decrease in activity, while deletion of phaB1 led to a dramatic decrease. Both the phaB1 and the phaB3 single mutants have significantly higher levels of reductase activity than Re2115 (P < 0.01 for Re2107 and P < 0.05 for Re2111). Deletion of phaB2 had no significant effect on reductase activity, which agrees with the earlier finding that deletion of this gene did not affect PHB accumulation. Addition of phaB1 and phaB3 to the Re2115 genome led to increases in reductase activity, but neither Re2139 nor Re2143 reached the activity level of Re2114. At the 24-h time point, at which cells were harvested from fructose medium to make these measurements, all strains except Re2113 and Re2115 had approximately the same PHB content, despite their differences in reductase activity. These data suggest that there is a threshold value for reductase activity that allows for wild-type levels of PHB production and that Re2113 and Re2115 fall below this threshold. When NADH was provided as the cofactor, there was no significant difference in the specific activities of the phaB mutants (single-factor analysis of variance [ANOVA]; α = 0.15 and F < Fcrit), indicating that these reductases chiefly use NADPH as the electron donor.
TABLE 3.
Mutant strains of R. eutropha showed different levels of acetoacetyl-CoA reductase specific activity when NADPH was used as the cofactor but similar levels when NADH was the cofactora
| Strain | Genotype | Sp act (U/mg) obtained with: |
|
|---|---|---|---|
| NADPH | NADH | ||
| H16 | Wild type | 4.2 ± 0.3 | 1.12 ± 0.08 |
| Re2106 | ΔphaB2 | 4.0 ± 0.4 | 1.08 ± 0.05 |
| Re2107 | ΔphaB3 | 3.4 ± 0.2 | 1.03 ± 0.03 |
| Re2111 | ΔphaB1 | 0.14 ± 0.03 | 0.97 ± 0.09 |
| Re2112 | ΔphaB1ΔphaB2 | 0.15 ± 0.04 | 1.1 ± 0.2 |
| Re2113 | ΔphaB1ΔphaB3 | 0.09 ± 0.01 | 0.95 ± 0.07 |
| Re2114 | ΔphaB2ΔphaB3 | 3.3 ± 0.4 | 1.2 ± 0.2 |
| Re2115 | ΔphaB1ΔphaB2ΔphaB3 | 0.11 ± 0.01 | 1.2 ± 0.3 |
| Re2139 | Re2115::phaB1 | 0.5 ± 0.1 | 1.0 ± 0.1 |
| Re2143 | Re2115::phaB3 | 0.2 ± 0.1 | 1.1 ± 0.2 |
All strains were grown in duplicate, and each sample was measured twice (n = 4 for each strain). Measurements were made with soluble lysate fractions. Values are reported as means ± standard deviations (SD). One unit of activity is defined as the amount of enzyme needed to convert 1 μmol acetoacetyl-CoA to product per minute at 25°C.
After finding that Re2139 and Re2143 made more PHB than H16, we measured β-ketothiolase activity in some of the mutant strains to determine if mutations at the phaB1 locus influenced thiolase activity, as phaA is located immediately upstream of phaB1 in the genome (Table 4). Re2112 and Re2115 had greater thiolase-specific activity than H16 (P < 0.01), suggesting that deletion of phaB1 increased phaA expression. Addition of genes to the Re2115 genome again altered thiolase activity, with each of the complemented strains showing different results. Although Re2139 made more PHB than H16, there was not a statistically significant difference in the thiolase activities of the two strains. Re2143, which exhibited the highest level of PHB accumulation, had significantly greater thiolase-specific activity than H16 (P < 0.05).
TABLE 4.
The β-ketothiolase specific activity of R. eutropha strains was altered by changes at the phaB1 locusa
| Strain | Genotype | Sp act (U/mg) |
|---|---|---|
| H16 | Wild type | 0.7 ± 0.1 |
| Re2112 | ΔphaB1ΔphaB2 | 2.7 ± 0.2 |
| Re2115 | ΔphaB1ΔphaB2ΔphaB3 | 2.9 ± 0.3 |
| Re2139 | Re2115::phaB1 | 0.7 ± 0.2 |
| Re2140 | Re2115::phaB2 | 2.3 ± 0.2 |
| Re2143 | Re2115::phaB3 | 1.0 ± 0.1 |
All strains were grown in duplicate, and each sample was measured twice (n = 4 for each strain). Measurements were made with soluble lysate fractions. Values are reported as means ± SD. One unit of activity is defined as the amount of enzyme needed to convert 1 μmol acetoacetyl-CoA to product per minute at 25°C.
It has been shown in vitro that copies of all enzymes necessary for PHB synthesis from acetyl-CoA are present on the surfaces of PHB granules isolated from R. eutropha (45). As all enzyme assays in this study were performed using only the soluble fraction of cell lysates, it is possible that some reductase and thiolase activity was not accounted for in our experiments. However, as significant differences in both PhaA and PhaB activities were found in strains making similar amounts of PHB (e.g., H16 and Re2112), we are confident that differences in soluble specific activities reflect actual differences in enzyme levels and are not due to differences in PHB content.
Activities of purified reductases.
His-tagged versions of PhaB1, PhaB2, PhaB3, and FabG were expressed in E. coli and purified to homogeneity. The upstream start codon was used when phaB3 was PCR amplified for this experiment. The specific activity of each reductase was measured using acetoacetyl-CoA as the substrate and NADPH as the cofactor. Assays were carried out in triplicate, and the average specific activities in U/mg were as follows: for PhaB1, 1,110 ± 50; for PhaB2, 6.3 ± 0.1; for PhaB3, 44 ± 7; and for FabG, 0.64 ± 0.03. The negative control [crude lysate from Tuner(DE3) harboring empty pET-15b] had a specific activity of <0.05 U/mg. It is notable that PhaB1 and PhaB3, the reductases shown to contribute to PHB biosynthesis in our genetic studies, had the highest specific activities.
Molecular weight of PHB from mutant strains.
We next wanted to determine how the sizes of the PHB chains synthesized by some of the mutant strains compared to that of polymer from H16. PHB from H16, Re2112, Re2115, and Re2143 was extracted, and the molecular weights relative to polystyrene standards were measured via GPC (Table 5). We report the number-average (Mn) and the weight-average (Mw) molecular weights for each sample. We observed that for H16, Re2112, and Re2115 PHB, molecular weight decreased with the amount of polymer stored by a given strain. Re2143 deviated from this trend, as PHB from this strain had a lower molecular weight than polymer from H16, despite the fact that Re2143 was more productive. For all strains, polymer extracted from the 72-h samples had lower molecular weight than polymer from the 48-h samples.
TABLE 5.
PHB extracted from different R. eutropha strains had different molecular weightsa
| Strain | Genotype |
Mn (106) at: |
Mw (106) at: |
||
|---|---|---|---|---|---|
| 48 h | 72 h | 48 h | 72 h | ||
| H16 | Wild type | 0.69 ± 0.05 | 0.64 ± 0.06 | 3.5 ± 0.5 | 2.9 ± 0.7 |
| Re2112 | ΔphaB1ΔphaB2 | 0.40 ± 0.02 | 0.29 ± 0.03 | 1.70 ± 0.09 | 1.19 ± 0.02 |
| Re2115 | ΔphaB1ΔphaB2ΔphaB3 | 0.208 ± 0.009 | 0.179 ± 0.008 | 0.528 ± 0.006 | 0.461 ± 0.005 |
| Re2143 | Re2115::phaB3 | 0.53 ± 0.01 | 0.421 ± 0.009 | 1.7 ± 0.2 | 1.2 ± 0.1 |
All strains were grown in triplicate, and samples were taken at 48 and 72 h postinoculation. Mn and Mw values are reported as means ± SD.
DISCUSSION
The number of different acetoacetyl-CoA reductases encoded by the R. eutropha genome and their roles in PHB biosynthesis were previously unclear. Biochemical studies initially suggested that there was one NADPH-dependent reductase and one NADH-dependent reductase expressed in R. eutropha, with only the NADPH-dependent enzyme producing the (R)-HB-CoA necessary for polymerization (10). The gene phaB1 was subsequently discovered using a genetic screen, and the existence of additional phaB genes was proposed (26). Analysis of the R. eutropha genome revealed phaB2 and phaB3, as well as 15 other phaB isologs (29), although it is unclear what cutoff was used in predicting these additional isologs. Phylogenetic analysis of the nucleotide and encoded-amino-acid sequences of phaB1, phaB2, and phaB3 suggests that these genes are paralogs that resulted from gene duplication events (see Fig. S1 in the supplemental material).
The results of this study indicate that under some growth conditions, both PhaB1 and PhaB3 contribute to PHB biosynthesis but that under others, only PhaB1 provides the reductase activity necessary for HB-CoA formation. Interestingly, even strains with the least reductase activity (Re2113 and Re2115) showed a low level of PHB accumulation in minimal media, suggesting either that some reductase activity is provided by other enzymes (see Fig. S1 in the supplemental material) or that there is a secondary route for synthesis of HB-CoA. Our findings are analogous to work done with R. eutropha β-ketothiolases in which multiple enzymes are present but PhaA provides the majority of the activity for PHB synthesis (39), although in the case of the thiolases, the different enzymes exhibit different substrate preferences.
Efforts to purify acetoacetyl-CoA reductase enzymes from a glucose-utilizing R. eutropha mutant, prior to the discovery of the PHB biosynthetic genes, resulted in the suggestion that there was a single NADPH-dependent reductase (10). We were unable to replicate the growth conditions from this study, as wild-type H16 cannot use glucose as a sole carbon source (29). Assuming that both phaB1 and phaB3 were expressed by the mutant during growth on glucose, it is possible that PhaB1 and PhaB3 copurified, as the enzymes have nearly identical molecular weights (26.4 × 103 [PhaB1] and 26.0 × 103 [PhaB3], assuming the upstream start codon) and similar peptide sequences (52% identity according to ClustalW2 alignment [19]). Given that phaB1 is expressed at a much higher level than phaB3 (Table 2), it is also possible that the low level of PhaB3 in the cellular lysate escaped detection.
Interesting questions exist concerning the regulation of phaB3 expression. In fructose minimal medium, mutants with only phaB3 remaining in the genome showed PHB production similar to that observed for the wild type through 48 h of growth (Fig. 1). In the final 24 h of the experiment, little additional PHB was made in these strains, while polymer continued to accumulate in strains containing phaB1. Expression data show that phaB3 is expressed during growth on fructose but that expression decreases dramatically when nitrogen limitation is reached (Table 2). Without continued formation of PhaB3 protein, breakdown of PhaB3 in Re2112 would eventually lead to insufficient reductase activity for normal PHB synthesis. We also found that phaB3 was expressed in fructose cultures but not trioleate cultures, which explains the lack of PHB accumulation by Re2112 in Fig. 2. While there are many examples of genes whose expression is regulated by the presence of certain carbon sources (9), it is unclear why it might be advantageous for phaB3 to be regulated in this manner. Given that phaB1 is constitutively expressed at a high level, it is possible that there was little pressure driving the evolution of phaB3 regulation, so there may not be a satisfying explanation for the expression pattern of this gene.
We found that deletion of phaB2 did not lead to an observable phenotype under any of the conditions examined in this study. While the specific activity of purified PhaB2 protein was lower than the activities of PhaB1 and PhaB3, it is clear that phaB2 does encode an active acetoacetyl-CoA reductase. This observation, combined with the low levels of phaB2 expression measured in fructose and trioleate cultures (Table 2), suggests that this gene is not expressed under normal laboratory conditions. Another group similarly concluded that the phaC2 gene, which is immediately downstream of phaB2 in the R. eutropha genome, is unexpressed (27).
Complementation experiments showed that all three phaB genes could restore some level of PHB production. Despite the fact that native phaB2 expression was never observed, insertion of this gene at the phaB1 locus led to increased PHB storage in Re2115, although not to the level observed in the wild type. Insertion of fabG at the phaB1 locus had no significant impact on PHB accumulation. We found that while purified FabG was able to reduce acetoacetyl-CoA, the specific activity of FabG was significantly lower than that of the purified PhaB enzymes. While other groups have successfully used fabG genes from E. coli and pseudomonads to synthesize PHA precursors, plasmids were used for heterologous gene expression, supplying multiple gene copies per cell (24, 33, 42). It is therefore possible that higher levels of fabG expression in R. eutropha could increase PHB accumulation in Re2115. In addition, the previously studied FabGs showed preferences for substrates longer than C4 (24), so R. eutropha fabG may be useful for synthesis of MCL-PHA rather than PHB. The contribution of natively expressed fabG to PHB biosynthesis in Re2115 requires further study, but it is clear that FabG does not play a major role in PHB production in wild-type H16.
The data collected in this study allow us to determine which step limits PHB production during nitrogen-limited growth on fructose. Previous work in which PHA biosynthetic genes were overexpressed in R. eutropha indicated that increases in synthase activity do not affect the rate of PHB accumulation (15, 17, 23). These results imply that the β-ketothiolase and/or reductase reactions limit flux through the PHB pathway. The fact that reductase activity can be decreased from the wild-type level to the level for Re2111 and Re2112 with little change in PHB production suggests that the reduction of acetoacetyl-CoA is not the limiting step in polymer formation. Thiolase activities differed between H16 and Re2111/Re2112, which presents a possible complication in the analysis. We found, however, that Re2139 had thiolase activity similar to that of H16 and lower reductase activity while still making significant PHB, which confirms that the reductase step does not limit PHB production. Only when reductase activity drops below the level of Re2111/Re2112 does reduction of acetoacetyl-CoA become limiting in the pathway, as is observed with Re2115. The PHB-hyperproducing strain Re2143 had reductase activity above the limiting level and greater thiolase activity than H16, allowing for increased flux through the pathway and greater PHB accumulation than the wild-type organism.
The results of our study illustrate the challenges inherent in using genetics to study different genes in a cotranscribed operon. It has been established that a single nonsense mutation in phaC1 can dramatically alter PhaA and PhaB activities (23). In our study, deletion of phaB1 increased thiolase activity, likely by increasing expression of phaA. As phaA and phaC1 are cotranscribed (20), deletion of phaB1 could similarly increase PHA synthase expression. Strains in which phaB1 or phaB3 was inserted into the Re2115 genome had lower levels of reductase activity than strains in which the native phaB1 gene was present (Re2112). The gene insertion procedure leaves several additional base pairs at both ends of the open reading frame not normally present in the operon. It is known that modifications to the intergenic regions of an operon can influence gene expression by changing posttranscriptional processes (28), which may have been the case here. It is also possible that the new RBS used with the inserted genes could be less favorable than the native RBS of each gene. Inserting different reductase genes into the PHB operon will alter the secondary structure of the resulting polycistronic mRNA, potentially effecting the translation of all genes in the operon differently in each complemented strain (18), which could explain why different levels of thiolase activity were observed in the complemented strains.
Analysis of the PHB molecular weight data is complicated by the fact that the strains examined likely have different rates of HB-CoA synthesis and may also have different levels of PHA synthase activity. It has been shown with purified R. eutropha synthase in vitro and with recombinant E. coli in vivo that higher levels of synthase activity lead to shorter polymer chains (6, 37). The influence of substrate concentration on PHB molecular weight is less clear. A study with the R. eutropha synthase in vitro using HB-CoA as the substrate found that substrate concentration does not influence molecular weight (6). Later experiments using the PHA synthase from Allochromatium vinosum with (R)-3-hydroxybutyryl-N-acetylcysteamine as the substrate showed that low-molecular-weight polymer was made at the lowest substrate concentration tested (21). As the substrate was increased, however, PHB molecular weight reached a plateau and did not increase significantly with further increases in substrate concentration. We have shown that changes to the phaB1 locus alter thiolase activity, so it is probable that phaC1 expression is also changed by these mutations, thereby influencing PHB molecular weight. Despite this issue, valuable insights on the influence of in vivo HB-CoA availability on PHB molecular weight can still be gained from the data presented in this study. When the molecular weights of PHB from H16, Re2112, and Re2115 are compared, it is found that molecular weight decreases with the amount of PHB accumulated. This suggests that lower intracellular HB-CoA concentrations could lead to shorter polymer chains. Comparing Re2112 and Re2115 is especially valuable, as these strains have the same phaB1 deletion, which should have the same influence on synthase expression. Restoring PHB production in Re2115 by the addition of phaB3 increases PHB molecular weight, although not to the level observed in H16. All together, the molecular weight results gathered here suggest that diminished HB-CoA synthesis in vivo corresponds to lower PHB molecular weight. This presents a potential conflict with the previous in vitro results showing no effect of HB-CoA concentration on polymer chain length. Clearly, the in vitro system, which is a batch reaction, varies from the in vivo system, in which substrate is continuously synthesized. The substrate concentrations in the in vitro studies may not match the effective intracellular HB-CoA concentrations generated by the strains in this study and therefore miss the influence of substrate concentration on polymer length. Additionally, PHB made by R. eutropha in vivo is stored within granules, in which there are many proteins present on the granule surface (13). In vitro experiments are unlikely to capture the complexity of these granules and the effects that interacting proteins may have on the lengths of PHB chains.
We also observed that PHB molecular weight decreased as culture time increased from 48 to 72 h. Previous work has demonstrated turnover of PHB in R. eutropha, with a concurrent decrease in the molecular weight of the stored polymer (43). The reason for this decrease is not well understood, but it may be related to the finding that at least two PHA depolymerases are expressed in R. eutropha as PHB is being produced (20). The mechanisms by which PHB molecular weight is controlled in vivo are clearly an area for further investigation.
We have shown that in fructose minimal medium, the enzymes encoded by phaB1 and phaB3 provide the reductase activity for normal PHB biosynthesis in R. eutropha. In other media, only the product of phaB1 is important. Deletion of the phaB genes resulted in R. eutropha strains with significantly reduced PHB accumulation. In most prior work, decreases in PHB synthesis in R. eutropha were achieved by altering the PHA synthase (41). The ΔphaB1 ΔphaB2 ΔphaB3 mutant has diminished PHB accumulation, while still expressing the wild-type synthase. As previous efforts to study PhaC from the native host grown under PHB production conditions have been complicated by the presence of large quantities of stored PHB (7), our strains may represent useful experimental tools for future studies. The impact of decreased acetoacetyl-CoA reductase activity on metabolite pools in R. eutropha was not explored in this study but represents an intriguing opportunity for future work. Finally, the high level of HB-CoA synthesis in wild-type R. eutropha has led to difficulties in engineering this species to make PHA copolymers with substantial fractions of monomers other than HB. Efforts are under way in our laboratory to construct strains based on Re2115 that are able to produce useful PHA copolymers from a variety of carbon sources.
Supplementary Material
Acknowledgments
This work was funded by the Malaysia MIT Biotechnology Partnership Programme (MMBPP), by NIH grant GM-049171 (A.J.S.), and by NIH grant T32GM081081 (J.L.).
We thank our MMBPP collaborators for useful discussions about this work. We also acknowledge Chris Brigham, Mimi Cho, and Dan MacEachran for their helpful suggestions concerning the manuscript.
Footnotes
Published ahead of print on 20 August 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akiyama, M., T. Tsuge, and Y. Doi. 2003. Environmental life cycle comparison of polyhydroxyalkanoates produced from renewable carbon resources by bacterial fermentation. Polym. Degradation Stab. 80:183-194. [Google Scholar]
- 2.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brigham, C. J., C. F. Budde, J. W. Holder, Q. Zeng, A. E. Mahan, C. Rha, and A. J. Sinskey. Elucidation of β-oxidation pathways in Ralstonia eutropha H16 by examination of global gene expression. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 4.Chen, G.-Q., and Q. Wu. 2005. Microbial production and applications of chiral hydroxyalkanoates. Appl. Microbiol. Biotechnol. 67:592-599. [DOI] [PubMed] [Google Scholar]
- 5.Fisher, M., J. T. M. Kroon, W. Martindale, A. R. Stuitje, A. R. Slabas, and J. B. Rafferty. 2000. The X-ray structure of Brassica napus β-keto acyl carrier protein reductase and its implications for substrate binding and catalysis. Structure 8:339-347. [DOI] [PubMed] [Google Scholar]
- 6.Gerngross, T. U., and D. P. Martin. 1995. Enzyme-catalyzed synthesis of poly[(R)-(-)-3-hydroxybutyrate]: formation of macroscopic granules in vitro. Proc. Natl. Acad. Sci. U. S. A. 92:6279-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerngross, T. U., K. D. Snell, O. P. Peoples, A. J. Sinskey, E. Csuhai, S. Masamune, and J. Stubbe. 1994. Overexpression and purification of the soluble polyhydroxyalkanoate synthase from Alcaligenes eutrophus: evidence for a required posttranslational modification for catalytic activity. Biochemistry 33:9311-9320. [DOI] [PubMed] [Google Scholar]
- 8.Goodrum, L. J., A. Patel, J. F. Leykam, and M. J. Kieliszewski. 2000. Gum arabic glycoprotein contains glycomodules of both extensin and arabinogalactan-glycoproteins. Phytochemistry 54:99-106. [DOI] [PubMed] [Google Scholar]
- 9.Gorke, B., and J. Stulke. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613-624. [DOI] [PubMed] [Google Scholar]
- 10.Haywood, G. W., A. J. Anderson, L. Chu, and E. A. Dawes. 1988. The role of NADH- and NADPH-linked acetoacetyl-CoA reductases in the poly-3-hydroxybutyrate synthesizing organism Alcaligenes eutrophus. FEMS Microbiol. Lett. 52:259-264. [Google Scholar]
- 11.Heath, R. J., and C. O. Rock. 1995. Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J. Biol. Chem. 270:26538-26542. [DOI] [PubMed] [Google Scholar]
- 12.Jendrossek, D. 2001. Microbial degradation of polyesters. Adv. Biochem. Eng. Biotechnol. 71:293-325. [DOI] [PubMed] [Google Scholar]
- 13.Jendrossek, D. 2009. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J. Bacteriol. 191:3195-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, K., Y. Jiang, R. Kleerebezem, G. Muyzer, and M. C. M. van Loosdrecht. 2009. Enrichment of a mixed bacterial culture with a high polyhydroxyalkanoate storage capacity. Biomacromolecules 10:670-676. [DOI] [PubMed] [Google Scholar]
- 15.Jung, Y.-M., J.-S. Park, and Y.-H. Lee. 2000. Metabolic engineering of Alcaligenes eutrophus through the transformation of cloned phbCAB genes for the investigation of the regulatory mechanism of polyhydroxyalkanoate biosynthesis. Enzyme Microb. Technol. 26:201-208. [DOI] [PubMed] [Google Scholar]
- 16.Karr, D. B., J. K. Waters, and D. W. Emerich. 1983. Analysis of poly-β-hydroxybutyrate in Rhizobium japonicum bacteroids by ion-exclusion high-pressure liquid chromatography and UV detection. Appl. Environ. Microbiol. 46:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kichise, T., T. Fukui, Y. Yoshida, and Y. Doi. 1999. Biosynthesis of polyhydroxyalkanoates (PHA) by recombinant Ralstonia eutropha and effects of PHA synthase activity on in vivo PHA biosynthesis. Int. J. Biol. Macromol. 25:69-77. [DOI] [PubMed] [Google Scholar]
- 18.Kozak, M. 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361:13-37. [DOI] [PubMed] [Google Scholar]
- 19.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 20.Lawrence, A., J. Schoenheit, A. He, J. Tian, P. Liu, J. Stubbe, and A. Sinskey. 2005. Transcriptional analysis of Ralstonia eutropha genes related to poly-(R)-3-hydroxybutyrate homeostasis during batch fermentation. Appl. Microbiol. Biotechnol. 68:663-672. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence, A. G., J. Choi, C. Rha, J. Stubbe, and A. J. Sinskey. 2005. In vitro analysis of the chain termination reaction in the synthesis of poly-(R)-β-hydroxybutyrate by the class III synthase from Allochromatium vinosum. Biomacromolecules 6:2113-2119. [DOI] [PubMed] [Google Scholar]
- 22.Martin, D. P., and S. F. Williams. 2003. Medical applications of poly-4-hydroxybutyrate: a strong flexible absorbable biomaterial. Biochem. Eng. J. 16:97-105. [Google Scholar]
- 23.Mifune, J., S. Nakamura, and T. Fukui. 2008. Targeted engineering of Cupriavidus necator chromosome for biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from vegetable oil. Can. J. Chem. 86:621-627. [Google Scholar]
- 24.Nomura, C. T., K. Taguchi, Z. Gan, K. Kuwabara, T. Tanaka, K. Takase, and Y. Doi. 2005. Expression of 3-ketoacyl-acyl carrier protein reductase (fabG) genes enhances production of polyhydroxyalkanoate copolymer from glucose in recombinant Escherichia coli JM109. Appl. Environ. Microbiol. 71:4297-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peoples, O. P., and A. J. Sinskey. 1989. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J. Biol. Chem. 264:15298-15303. [PubMed] [Google Scholar]
- 26.Peoples, O. P., and A. J. Sinskey. 1989. Poly-β-hydroxybutyrate biosynthesis in Alcaligenes eutrophus H16. Characterization of the genes encoding β-ketothiolase and acetoacetyl-CoA reductase. J. Biol. Chem. 264:15293-15297. [PubMed] [Google Scholar]
- 27.Peplinski, K., A. Ehrenreich, C. Doring, M. Bomeke, F. Reinecke, C. Hutmacher, and A. Steinbuchel. 2010. Genome-wide transcriptome analyses of the ‘Knallgas’ bacterium Ralstonia eutropha H16 with regard to polyhydroxyalkanoate metabolism. Microbiology 156:2136-2152. [DOI] [PubMed] [Google Scholar]
- 28.Pfleger, B. F., D. J. Pitera, C. D. Smolke, and J. D. Keasling. 2006. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat. Biotechnol. 24:1027-1032. [DOI] [PubMed] [Google Scholar]
- 29.Pohlmann, A., W. F. Fricke, F. Reinecke, B. Kusian, H. Liesegang, R. Cramm, T. Eitinger, C. Ewering, M. Potter, E. Schwartz, A. Strittmatter, I. Vosz, G. Gottschalk, A. Steinbuchel, B. Friedrich, and B. Bowien. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24:1257-1262. [DOI] [PubMed] [Google Scholar]
- 30.Price, A. C., Y.-M. Zhang, C. O. Rock, and S. W. White. 2001. Structure of β-ketoacyl-[acyl carrier protein] reductase from Escherichia coli: negative cooperativity and its structural basis. Biochemistry 40:12772-12781. [DOI] [PubMed] [Google Scholar]
- 31.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 32.Rehm, B. H. A. 2003. Polyester synthases: natural catalysts for plastics. Biochem. J. 376:15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren, Q., N. Sierro, B. Witholt, and B. Kessler. 2000. FabG, an NADPH-dependent 3-ketoacyl reductase of Pseudomonas aeruginosa, provides precursors for medium-chain-length poly-3-hydroxyalkanoate biosynthesis in Escherichia coli. J. Bacteriol. 182:2978-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saegusa, H., M. Shiraki, C. Kanai, and T. Saito. 2001. Cloning of an intracellular poly[D(-)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Schubert, P., N. Kruger, and A. Steinbuchel. 1991. Molecular analysis of the Alcaligenes eutrophus poly(3-hydroxybutyrate) biosynthetic operon: identification of the N terminus of poly(3-hydroxybutyrate) synthase and identification of the promoter. J. Bacteriol. 173:168-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sim, S. J., K. D. Snell, S. A. Hogan, J. Stubbe, C. Rha, and A. J. Sinskey. 1997. PHA synthase activity controls the molecular weight and polydispersity of polyhydroxybutyrate in vivo. Nat. Biotechnol. 15:63-67. [DOI] [PubMed] [Google Scholar]
- 38.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology (NY) 1:784-791. [Google Scholar]
- 39.Slater, S., K. L. Houmiel, M. Tran, T. A. Mitsky, N. B. Taylor, S. R. Padgette, and K. J. Gruys. 1998. Multiple β-ketothiolases mediate poly(β-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J. Bacteriol. 180:1979-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivasan, S., G. C. Barnard, and T. U. Gerngross. 2003. Production of recombinant proteins using multiple-copy gene integration in high-cell-density fermentations of Ralstonia eutropha. Biotechnol. Bioeng. 84:114-120. [DOI] [PubMed] [Google Scholar]
- 41.Stubbe, J., and J. Tian. 2003. Polyhydroxyalkanoate (PHA) homeostasis: the role of the PHA synthase. Nat. Prod. Rep. 20:445-457. [DOI] [PubMed] [Google Scholar]
- 42.Taguchi, K., Y. Aoyagi, H. Matsusaki, T. Fukui, and Y. Doi. 1999. Co-expression of 3-ketoacyl-ACP reductase and polyhydroxyalkanoate synthase genes induces PHA production in Escherichia coli HB101 strain. FEMS Microbiol. Lett. 176:183-190. [DOI] [PubMed] [Google Scholar]
- 43.Taidi, B., D. A. Mansfield, and A. J. Anderson. 1995. Turnover of poly(3-hydroxybutyrate) (PHB) and its influence on the molecular mass of the polymer accumulated by Alcaligenes eutrophus during batch culture. FEMS Microbiol. Lett. 129:201-205. [Google Scholar]
- 44.Tian, J., A. J. Sinskey, and J. Stubbe. 2005. Kinetic Studies of polyhydroxybutyrate granule formation in Wautersia eutropha H16 by transmission electron microscopy. J. Bacteriol. 187:3814-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uchino, K., T. Saito, B. Gebauer, and D. Jendrossek. 2007. Isolated poly(3-hydroxybutyrate) (PHB) granules are complex bacterial organelles catalyzing formation of PHB from acetyl coenzyme A (CoA) and degradation of PHB to acetyl-CoA. J. Bacteriol. 189:8250-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.York, G. M., B. H. Junker, J. Stubbe, and A. J. Sinskey. 2001. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J. Bacteriol. 183:4217-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.York, G. M., J. Lupberger, J. M. Tian, A. G. Lawrence, J. Stubbe, and A. J. Sinskey. 2003. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[D-(-)-3-hydroxybutyrate] depolymerase genes. J. Bacteriol. 185:3788-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zor, T., and Z. Seliger. 1996. Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal. Biochem. 236:302-308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.