Abstract
The tolerance of Mycobacterium tuberculosis to antituberculosis drugs is a major reason for the lengthy therapy needed to treat a tuberculosis infection. Rifampin is a potent inhibitor of RNA polymerase (RNAP) in vivo but has been shown to be less effective against stationary-phase bacteria. Sigma factor F is associated with bacteria entering stationary phase and has been proposed to impact rifampin activity. Here we investigate whether RNAP containing SigF is more resistant to rifampin inhibition in vitro and whether overexpression of sigF renders M. tuberculosis more tolerant to rifampin. Real-time and radiometric in vitro transcription assays revealed that rifampin equally inhibits transcription by RNAP containing sigma factors SigA and SigF, therefore ruling out the hypothesis that SigF may be responsible for increased resistance of the enzyme to rifampin in vitro. In addition, overexpression or deletion of sigF did not alter rifampin susceptibility in axenic cultures of M. tuberculosis, indicating that SigF does not affect rifampin tolerance in vivo.
The treatment of drug-sensitive Mycobacterium tuberculosis infections is typically carried out using a cocktail of four drugs (isoniazid, rifampin, pyrazinamide, and ethambutol), for a 2-month period followed by 4 months of treatment with rifampin and isoniazid. Pharmacodynamic analysis of the sputum CFU following the start of this drug regimen typically shows a rapid decrease (1 to 2 logs) in bacterial burden in the first week of treatment. This early bactericidal stage is, however, followed by a far lower rate of bacterial elimination, thought to be due to the persistence of a bacterial subpopulation that is phenotypically less drug susceptible and often termed drug tolerant. Understanding the mechanisms underlying this drug tolerance is essential for efforts to shorten and improve tuberculosis drug therapy.
Numerous in vitro models have been proposed that potentially mimic conditions that give rise to the phenotypically drug-tolerant bacterial population found in vivo; these include nutrient starvation (3), growth in a hypoxic environment (28, 29), or nitric oxide challenge (12). These models all involve driving the bacteria into a nonreplicating state which consequently often decreases their drug susceptibility. This is especially true for those drugs that target the biogenesis of the bacterial cells wall (i.e., isoniazid and ethambutol), as this process is essential when the cells are actively growing. Intriguingly, inhibition of RNA polymerase (RNAP) by the currently most effective antitubercular drug, rifampin, is also subject to drug tolerance, with stationary-phase bacteria being less rifampin susceptible (11).
Transcription is an essential process, regardless of whether the bacteria are in log phase or in stationary phase. Interestingly, the addition of very high concentrations of rifampin to stationary-phase bacteria has revealed that transcription still occurs, though at a somewhat lower rate (11). M. tuberculosis must therefore have an alternative mechanism to circumvent rifampin inhibition and allow for continued transcription. Recent studies have revealed that some accessory proteins such as GroEL1 and RbpA can bind RNAP and prevent rifampin inhibition in vitro (8, 19). Here, we investigate whether altering the sigma factor usage can have a similar impact on the affinity of rifampin for RNAP, allowing transcription in its presence.
Rifampin binds to the β subunit of RNAP and forms a physical barrier that prevents de novo RNA from elongating out of the RNAP complex (18). It has been speculated that rifampin is also able to interact directly with domain 3.2 of the housekeeping sigma factor A (SigA) that inserts deeply into RNAP (1). In response to external stress factors, M. tuberculosis can utilize any of the 13 sigma factors (SigA to SigM) to alter its transcriptome. Interestingly, SigF is one of only three sigma factors with a domain 3.2 (24) (the others being SigA and B), and it has been implicated in the entry of mycobacteria into stationary phase (7). In addition, studies conducted in a different system revealed that changing sigma factor usage in Escherichia coli from Sig70 to an alternative sigma factor, Sig32, dramatically impacts rifampin susceptibility (30).
A role for SigF in rifampin tolerance was first suggested when it was shown that deletion of sigF from an M. tuberculosis strain (CDC1551) resulted in increased susceptibility to rifampin (5). That work, however, did not investigate the mechanism underlying this change in sensitivity to rifampin, a question that we seek to investigate in this report. The aim of this study is to determine if SigF can directly cause allosteric alterations in RNAP that will affect rifampin binding and therefore its activity. This is investigated by determining the rifampin inhibition of SigA- and SigF-specific in vitro transcription. Second, we sought to determine whether increased sigF expression led to increased bacterial tolerance to rifampin and to confirm that deletion of sigF affected rifampin's bactericidal activity.
MATERIALS AND METHODS
Reagents.
All PCRs were performed using Phusion DNA polymerase (Finnzymes) with primers from Microsynth (Balgach, Switzerland). Restriction enzymes were purchased from New England Biolabs, pUC19 and chemically competent Top10 E. coli from Invitrogen (Basel, Switzerland), and chemically competent C41 Escherichia coli from BioCat (Heidelberg, Germany). Middlebrook 7H9 and 7H11 medium, ADC (albumin-dextrose-catalase) and OADC (oleic acid-albumin-dextrose-catalase) were purchased from BD/Difco. All other chemicals and compounds used came from Sigma-Aldrich.
Purification of holo- and core RNAP from Mycobacterium smegmatis.
M. smegmatis mc2155 containing a histidine-tagged RNAP β′ subunit was kindly provided by AstraZeneca India, and the purification of RNAP from log-phase cultures (typically 6 liters) was performed as described previously (19). Purified RNAP contains both core and holoenzyme (here referred to as core and holo-RNAP), which were separated by cation-exchange chromatography over a Bio-Rex 70 resin (100 to 200 mesh; Bio-Rad). Briefly, Bio-Rex 70 resin columns were preequilibrated as described previously (4, 32) and equilibrated with 10 column volumes of TGEB (10 mM Tris-HCl, pH 7.9, 5% glycerol, 0.1 mM EDTA, 10 mM β-mercaptoethanol) with 75 mM KCl using an ÄKTApurifier (GE Healthcare). For the purification of holo-RNAP, contaminants were removed under an isocratic mobile-phase A (TGEB with 75 mM KCl, 10 column volumes) followed by elution of the RNAP with mobile-phase B (TGEB with 600 mM KCl). For the purification of core RNAP, holo- and core RNAP were separated on a very slow gradient (75 mM to 600 mM KCl in TGEB, 20 column volumes). The presence of native sigma factors was determined by running samples on a NuPAGE 4 to 12% Bis-Tris protein gel and staining by Simply Blue (both from Invitrogen). Holo-RNAP and core RNAP fractions were subsequently pooled, dialyzed against TGEB with 75 mM KCl, concentrated using size exclusion columns (Amicon Ultra, Ultracel 100k; Millipore), and stored in 50% (vol/vol) glycerol at −20°C.
Purification of recombinant SigA and SigF.
The sigA and sigF genes were both cloned into the pHis9GW vector (20) by recombination cloning, using Invitrogen Gateway technology. Genes were first amplified with gene-specific primers containing part of attB1 and attB2 (Table 1, primers 1 and 2 for sigA and primers 3 and 4 for sigF), followed by a second PCR to add the remaining recombineering sequence (Table 1, primers 5 and 6 for both genes). For both genes the start codon was disrupted and a thrombin site was inserted between the gene and the AttB1 site. Linear PCR products were then recombined into pDONR207 vector by BP recombination cloning (recombination between attB-flanked DNA and attp-containing pDONR207), followed by sequence validation. Genes were transferred into pHIS9GW vector by LR recombination cloning (recombination between pDONR207 attL and pHIS9GW attR), giving pHis9SigA and pHis9SigF. Chemically competent TOP10 E. coli cells were used for plasmid production.
TABLE 1.
PCR primers used
| Role | Primer no. | Name | Sequence (5′-3′) |
|---|---|---|---|
| Recombinant sigma factor expression | 1 | SigA-GW-F | CTGGTTCCGCGTGGATCCAAAGCAAGCACGGCGAC |
| 2 | SigA-GW-R | CAAGAAAGCTGGGTCTCAGTCCAGGTAGTCGCGCAGG | |
| 3 | SigF-GW-F | CTGGTTCCGCGTGGATCCGGCGGTTCTGCATCGC | |
| 4 | SigF-GW-R | CAAGAAAGCTGGGTCCTACTCCAACTGATCCCGTAGCCGTG | |
| 5 | attB1-thrombin | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCTGGTTCCGCGTGGATC | |
| 6 | attB2 | GGGGACCACTTTGTACAAGAAAGCTGGGTC | |
| Real-time in vitro transcription template | 7 | F-rrnABt1(PlacZ) | TAGACTGAGCTCTCGAAGCTTATCGATGTCGACGTAG |
| 8 | R-rrnABt1 | AGTCAGTCGAGCTCAGAGATTTTGAGACACAACGTCGC | |
| 9 | F-PrsbW | CGGCGTCGGAGCTCTTGC | |
| 10 | R-PrsbW | CTAGTTAACTACGTCGACATCGATAAGCTTCGATCGACAGTACTCCCG | |
| 11 | F-rrnABt1(PrsbW) | CGGGAGTACTGTCGATCGAAGCTTATCGATGTCGACGTAGTTAACTAG | |
| 12 | BsiWI-polyA | GTACAAAAAAAAAAAAAAAAAAAAAAAAA | |
| 13 | BsiWI-polyT | GTACTTTTTTTTTTTTTTTTTTTTTTTTT | |
| Radiometric in vitro transcription template | 14 | IVT PCR F-Hsp60 | AATACCAGCCAGACGAGACGG |
| 15 | IVT PCR R-Hsp60 | AACCCCGCTACTTACGCTGG | |
| 16 | PrsbW IVT temp F | ATTGGCCTGGGTATCTGTTG | |
| 17 | PrsbW IVT temp R | TTGGTGGGTAAATCCGAGTC | |
| Pristinamycin-inducible SigF plasmid | 18 | F pri HindIII/Pptr | AGTCACAAGCTTACGAGTTCCGAGATCACCCCG |
| 19 | R pri Pptr/sigF | GGCAGCGCGCGCCGTCACTCAGGCTCCTTGTACGGTGTACGGG | |
| 20 | F Pri Pptr/sigF | CCCGTACACCGTACAAGGAGCCTGAGTGACGGCGCGCGCTG | |
| 21 | R Pri SigF/HindIII | AGTCACAAGCTTCTACTCCAACTGATCCCGTAGCCGT | |
| SigF knockout cassette | 22 | rsbwFKO(1)-F | CCCAGCGGATCCTGCTGG |
| 23 | rsbwKO(1)-R | CTGTGAGATACACTAGTGCGCAGCAGCGCC | |
| 24 | sigFKO(2)-F | GCTGCGCACTAGTGTATCTCACAGATGCACGTGTCG | |
| 25 | sigFKO(2)-R | CACCGAGGATCCCACCCC | |
| 26 | speI-HYG-F | GTACTAACTAGTCTGAGCTTGCATGCCTGC | |
| 27 | HYG-SpeI-R2 | TAACTAGTGACCACCCTGGAGGAGATGAT | |
| 28 | KO check F | GACCAGCTCGAAGACCAGAC | |
| 29 | KO check R | GCTCGCCGAGATCAAGTAAG |
For protein production, pHis9SigA or pHis9SigF was transformed into C41 chemically competent E. coli and grown in 6-liter LB broth cultures to an optical density at 600 nm (OD600) of 1 (30°C, 200 rpm). Sigma factors were induced with 150 μM IPTG (isopropyl-β-d-thiogalactopyranoside) overnight (18°C, 200 rpm). Bacteria were pelleted and resuspended in TGEBN (50 mM Tris-HCl, pH 7.9, 5% glycerol, 0.1 mM EDTA, 10 mM β-mercaptoethanol, 500 mM NaCl) with a complete mini-protease inhibitor cocktail tablet (Roche) and 50 mM imidazole. Following a freeze-thaw cycle, bacteria were sonicated (15 min, 30 s on, 30 s off, on ice) and the insoluble fraction was pelleted (30,000 × g, 30 min, 4°C). Histidine-tagged sigma factors were then isolated by affinity chromatography of the soluble fraction on Ni2+-NTA resin using an ÄKTApurifier (GE Healthcare), with a long wash in TGEBN-50 mM imidazole followed by a gradient to TGEBN 500 mM imidazole. Sigma factor-containing fractions were dialyzed against TGEBN, concentrated (Amicon Ultra, Ultracel 10k; Millipore), and subjected to size exclusion chromatography (Superdex 200 for SigA and Superdex 75 for SigF; GE Healthcare) with a TGEBN mobile phase. Sigma factor-containing fractions were again concentrated and stored in 50% (vol/vol) glycerol at −20°C.
Real-time in vitro transcription.
To perform real-time in vitro transcription (IVT) as described previously (16, 17), it was decided to construct a sigma factor-specific template containing a specific promoter followed by a poly(A) and transcriptional termination region (rrnABT1 [25]) in a pUC19 backbone. To construct the control SigA-specific template (pUC19-PlacZpolyAT1), the native lacZ promoter of pUC19 was utilized, and a poly(A) stretch and an rrnABT1 transcriptional terminator sequence were inserted downstream. This was achieved by cloning a PCR-amplified rrnABT1 transcriptional terminator sequence (Table 1, primers 7 and 8) from pMV261 into the SacI restriction site located downstream of the lacZ promoter (PlacZ). A poly(A) stretch (formed by annealing primers 12 and 13) was subsequently cloned into a unique BsiWI site. A clone containing the poly(A) stretch in the sense strand was selected and confirmed by sequencing. The SigF reporter plasmid for real-time transcription (pUC19-PrsbWpolyAT1) was created by cloning a SigF-specific promoter (the −253 to +114 region of the transcription start point of the rsbW promoter, PrsbW [2]) into pUC19 followed by a poly(A) stretch and a T1 transcriptional termination site. Initially the rsbW promoter was attached to the terminator sequence by overlap PCR (Table 1, primers 9 and 10 for PrsbW and primers 11 and 8 for rrnABT1) and cloned into the SacI site. Clones with the insert in the direction opposite to that of the native lacZ promoter were selected, and a poly(A) stretch was cloned into the BsiWI site as described above. All constructs were confirmed by DNA sequencing. Chemically competent TOP10 E. coli cells were used for transformation and plasmid production.
To study the rate of RNA formation, real-time in vitro transcription using a molecular beacon targeting poly(A) RNA was performed (5′-FAM-cgcuuuuuuuuuuuugcg-DABCYL-3′, with a 2′-O-methylribonucleotide backbone) (17). Briefly, in a 40-μl reaction volume in a MicroAmp optical 96-well reaction plate (Applied Biosystems), each reaction mixture contained 40 mM Tris-HCl, pH 7.9, 75 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol (DTT), 0.1 mM EDTA, 20 units of RNasin (Promega), 500 nM molecular beacon, 2 μg of plasmid template, 4 pmol of core RNAP, and 20 pmol of recombinant sigma factor. To allow for the formation of holo-RNAP the reaction mixture was preincubated (30°C, 10 min) and returned on ice. Following the addition of a ribonucleotide triphosphate (rNTP) mix (150 μM each rNTP), the fluorescence (λex = 495 nm, λem = 515 nm) of the reaction mixture was determined (every 30 s for 2 h, 37°C) using an ABI Prism 7900HT sequence detection system (Applied Biosystems). For data analysis, the obtained fluorescence was background corrected (t = 0) and the rate of increase in the fluorescence was determined. Reactions were performed in the absence and presence of rifampin (3 nM to 10 μM) to determine the efficiency of inhibition.
Radiometric in vitro transcription.
Linear templates for in vitro transcription using [32P]CTP were generated by standard PCR to amplify the Mycobacterium bovis BCG hsp60 promoter (Phsp60) from pMV261 (SigA-recognized promoter [15, 22]; Table 1, primers 14 and 15) or the M. tuberculosis H37Rv rsbW promoter (PrsbW, SigF specific [2]; Table 1, primers 16 and 17).
Radiometric in vitro transcription was performed in a 100-μl reaction volume containing 40 mM Tris-HCl, pH 7.9, 75 mM KCl, 10 mM MgCl2, 10 mM DTT, 0.1 mM EDTA, 20 units of RNasin, 20 nM linear DNA template, 10 nM (1 pmol) of core RNAP, and 100 nM (10 pmol) of recombinant sigma factor. To allow for the formation of holo-RNAP, the reaction mixture was preincubated (37°C, 10 min) and returned on ice. An rNTP mix (final concentration of 150 μM each rATP, rUTP, rGTP and 30 μM rCTP with 1 μCi 32P-CTP) was added to initiate in vitro transcription (37°C, 30 min), which was terminated by the addition of 2× Novex TBE-urea sample buffer (Invitrogen), and the mixture was heated (80°C, 10 min). Samples were subsequently run (130 V, 80 min) on a Novex 6% TBE-urea PAGE gel (Invitrogen), before being dried, visualized, and quantified using Image Quant TL 2005/Typhoon software (GE Healthcare). Reactions were performed in the absence and presence of rifampin (1.25 nM to 320 nM) to determine the efficiency of inhibition of transcription.
Pristinamycin IA purification.
Pristinamycin IA was isolated and purified from pyostacin tablets by liquid chromatography. Briefly, 500 g pyostacin tablets were crushed in a mortar and dissolved in 100% methanol. The solution was centrifuged to remove insoluble materials and dried down under nitrogen. The resulting residue was dissolved in mobile phase A (55% acetonitrile, 45% water) and injected onto a VP 250/25 Nucleosil 100-5 C18 column (Macherey Nagel). To elute contaminants and pristinamycin IIA from the formulation, the column was washed in mobile phase A (5 ml/min, 200 ml) before being switched to mobile phase B (70% acetonitrile, 30% water) to elute a single peak of pristinamycin IA. Fractions of pristinamycin IA were subsequently pooled and dried down (speed vacuum centrifugation and freeze-drying), dissolved in dimethyl sulfoxide (DMSO) (50 mg/ml) and stored at −80°C. Pristinamycin IA purity was confirmed by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry.
Pristinamycin IA-inducible induction of SigF in M. tuberculosis.
For the inducible expression of SigF in M. tuberculosis H37Rv, advantage was taken of the pristinamycin IA inducible gene expression system developed previously and modified for use in M. tuberculosis (9). To construct this vector, the ptr promoter (Pptr) of Streptomyces pristinaespiralis was attached to sigF by overlap PCR (primers 18 to 22) and cloned into the unique HindIII site of pMY769 and confirmed by sequencing. The resultant plasmid (pMYSigF), as well as the control plasmid (pMY769), was transformed into M. tuberculosis strain H37Rv by electroporation, clones were selected on spectinomycin (100 μg/ml), and integration of plasmids was confirmed by PCR.
M. tuberculosis H37Rv containing the control vector (H37Rv::pMY769) or pMYSigF (H37Rv::pMYSigF) was grown and maintained in Middlebrook 7H9 complete medium (supplemented with 0.2% glycerol, 0.05% Tween 80, and 10% ADC). Log-phase cultures were diluted to an OD600 of 0.05 and induced with pristinamycin IA (0 to 2 μg/ml) for a 3-day period to allow for SigF induction and potential changes in downstream gene expression. Culture OD600 was determined daily, and SigF protein expression was confirmed on day 3 by Western blotting.
For Western blot analysis, bacteria were pelleted, washed in PBS-T (phosphate-buffered saline with 0.05% Tween 80) and resuspended in TEDPN (100 mM Tris-HCl, pH 7.9, 1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride [PMSF], 500 mM NaCl). Bacteria were disrupted in a sonic bath (15 min, 4°C) and centrifuged (12,000 × g, 10 min), and the supernatant was passed through a 0.2-μm filter. Following protein quantification using Bradford reagent, 10 μg of total protein was loaded and run on a NuPAGE 4 to 12% Bis-Tris gel (Invitrogen), transferred onto a nitrocellulose membrane, and blocked (2 h) in TNTM (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.2% Tween 20, and 5% nonfat milk). The membrane was then stained with a custom-generated primary rabbit anti-SigF polyclonal antibody (kindly generated by Ida Rosenkrands) (1:10,000 in TNTM, overnight, 4°C), washed and stained with a secondary goat anti-rabbit Hrp antibody (2 h, 1:100,000 in TNTM) (Sigma-Aldrich), and visualized using chemiluminescent peroxidase substrate.
Rifampin susceptibility.
Resazurin microtiter assay (REMA) and CFU determination were used to determine if SigF impacts rifampin susceptibility. For both assays, H37Rv::pMY769 and H37Rv::pMYSigF (OD600 = 0.05) were grown in the absence and presence of pristinamycin IA (2 μg/ml, 3 days), followed by the dilution of the bacteria to an OD600 of 0.01. For viability analysis by CFU, the bacteria were then grown (4 days) in the absence and presence of rifampin (0.1, 1, and 10 μg/ml) before being plated onto Middlebrook 7H11 agar (supplemented with 0.05% glycerol and 10% OADC). Bacterial viability analysis by REMA was performed as described previously (21), using the bacterial dilution (OD600 of 0.001) grown in a 96-well plate in the presence and absence of rifampin (between 0.1 and 500 ng/ml, 7 days).
Construction of a sigF knockout mutant in M. tuberculosis H37Rv.
An M. tuberculosis H37Rv strain in which most of the rsbw/sigF operon is knocked out and replaced by a hygromycin cassette was constructed using a recombineering method described previously (26, 27). To achieve this, approximately 900 bp prior to rsbW and after sigF (including around 100 bp of the genes themselves) were PCR amplified using primers (Table 1, primers 22 to 25) that introduced a BamHI and a SpeI site into the amplicons. The two amplicons were then joined at the SpeI site, ligated into the BamHI site of pUC19 (named pUC19-rsbW/sigF), and confirmed by sequencing. A hygromycin cassette was PCR amplified from pYUB412 with primers introducing flanking SpeI sites (Table 1, primers 26 to 27) and subsequently introduced into the SpeI site of pUC19-rsbW/sigF, forming pUC19-rsbW/hygR/sigF. Clones were grown up, and plasmids were extracted by midiprep, BamHI digested to excise the linear allelic exchange substrate (AES), and gel extracted. Then, 100 ng of linear AES was transformed into M. tuberculosis H37Rv containing pJV53 (kindly provided by Graham F. Hatfull, University of Pittsburgh), and the strain was grown up and made electrocompetent as described previously (26, 27). Hygromycin-resistant clones were selected, and genomic DNA was checked for the correct homologous recombination and the knockout of the rsbW/sigF operon by PCR (Table 1, primers 28 to 29). Clones lacking the rsbW/sigF operon (H37RvΔrsbW/sigF) were grown up in parallel to M. tuberculosis H37Rv wild-type cells, and their rifampin susceptibility was determined by REMA (as described above) and by plating bacteria on 7H11 agar (supplemented with glycerol and OADC) containing rifampin (250 pg/ml to 250 ng/ml).
Neutral red staining.
To detect M. tuberculosis cell envelope-associated sulfolipids, neutral red staining was performed essentially as described previously (10). Briefly, M. tuberculosis strains H37Rv, H37Ra, H37RvΔrsbW/sigF, H37Rv::pMY769, and H37Rv::pMYSigF were grown in Middlebrook 7H9 with 0.2% glycerol (but without Tween 80). Three days prior to staining, bacteria were diluted (OD600 = 0.1) and pristinamycin (2 μg/ml) was added to H37Rv::pMY769 and H37Rv::pMYSigF. Bacteria were then fixed with 50% methanol (37°C for 60 min), pelleted, and resuspended in 5 ml of 5% NaCl in 0.5% Trizma base (pH 9.5), followed by the addition of 50 μl of 0.05% neutral red (60 min at 37°C).
RESULTS
Purification of core RNAP, rSigA, and rSigF.
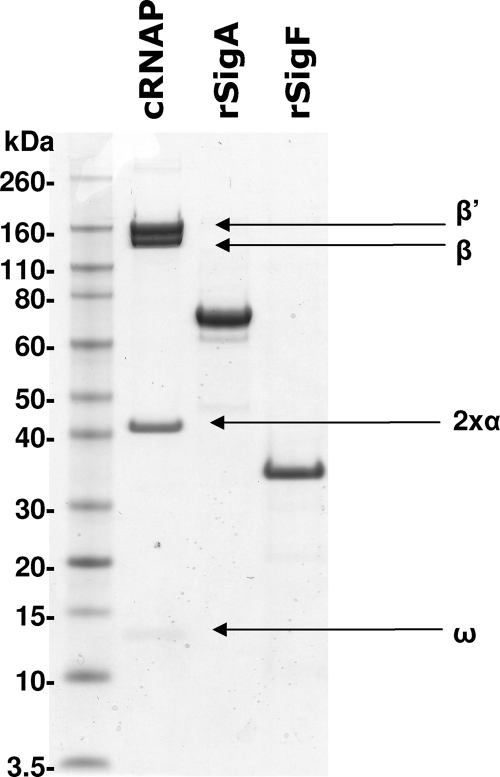
To determine if SigF is able to affect rifampin inhibition of RNAP, both real-time and radiometric in vitro transcription (IVT) assays were developed, both of which rely on purified core RNAP and recombinant sigma factors. The purification of M. smegmatis hexa-histidine-tagged RNAP was essentially as described previously (19), although an additional purification step using a Bio-Rex 70 column was found to improve the purity of the RNAP holoenzyme. Isolation of core RNAP from an RNAP mix using the Bio-Rex 70 was complicated by the fact that both holo- and core RNAP fractions have only slightly different affinities for the resin (270 mM and 340 mM KCl, respectively). Also, this method does not physically remove the sigma factor from the RNAP, and it relies on the purification and separation of the natural fraction of core RNAP present in the original sample. Here the best results were achieved when the M. smegmatis strain was grown only to early log phase (OD600 = 0.4), giving the greatest proportion of core RNAP (Fig. 1). Six liters of M. smegmatis culture processed by these means typically produced between 0.5 and 1 mg of core RNAP.
FIG. 1.
SDS-PAGE analysis of the core RNAP (cRNAP) from M. smegmatis and the recombinant sigma factors purified from C41 E. coli. Proteins were run on a 3 to 12% Bis-Tris SDS-PAGE gel and stained with Simply Blue (Invitrogen) for visualization. The core RNAP was separated into its five subunits and is clearly devoid of its native sigma factor. Both histidine-tagged recombinant SigA (rSigA) and SigF (rSigF) ran to their expected size, and their presence was confirmed by MALDI mass spectrometry.
Recombinant M. tuberculosis sigma factors are usually generated by refolding sigma factors from denatured inclusion bodies following protein induction in E. coli (13, 14). Here we found it possible to generate sufficient amounts of recombinant SigA (rSigA) and SigF (rSigF) by inducing at a lower IPTG concentration and at a lower temperature, albeit requiring a larger culture volume (6 liters). Protein purification by Ni2+ affinity and size exclusion chromatography resulted in purified recombinant sigma factor without the need to refold the proteins (Fig. 1). Interestingly, it was noted that induction of rSigF in E. coli led to growth arrest and that Ni2+ affinity chromatography resulted in the copurification of some E. coli RNAP subunits (which were removed by subsequent size exclusion chromatography). The purified components of RNAP were used for IVT in the assays described below.
Rifampin inhibition of sigma factor-specific in vitro transcription (IVT).
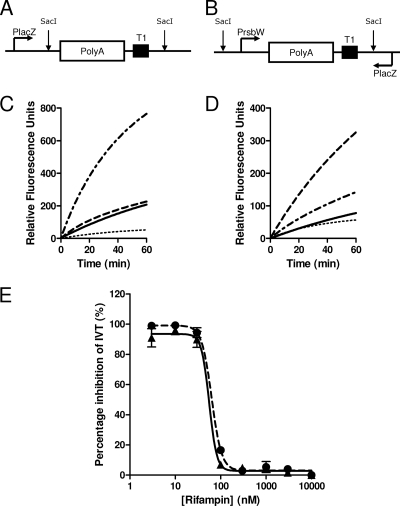
Real-time IVT was first introduced by Liu and colleagues (16) and improved by the utilization of molecular beacons that possess a 2′-O-methylribonucleotide backbone (17). Here we took advantage of this technology by using the same published molecular beacon (17) to recognize the poly(A) stretch introduced into the plasmid. The natural lacZ promoter of pUC19 (PlacZ) was found to be recognized by SigA holo-RNAP. To make the SigF-specific template, it was therefore important to insert the IVT promoter in the direction opposite to that of PlacZ, minimizing any potential SigA-mediated transcription mediated through residual holo-RNAP contamination (Fig. 2 A and B). Control experiments illustrate that for both templates, in the absence of sigma factors, the core RNAP has a basal level of transcription, most likely due to nonspecific transcription of the plasmid template (Fig. 2C and D). Addition of rSigA greatly increases the transcription from PlacZ and only slightly increases transcription on the pUC19-PrsbWpolyAT1 template (Fig. 2C). Addition of SigF to the core RNAP greatly increases transcription from PrsbW but not from PlacZ (Fig. 2D). In both cases, addition of high concentrations of rifampin (10 μM) results in the total inhibition of transcription, resulting in a very low background increase in fluorescence (which is equal in the two assays and is possibly due to some beacon instability). When performing a rifampin titration no significant difference was observed between the inhibition of SigA- and SigF-mediated IVT (Fig. 2E). In both cases rifampin concentrations above 100 nM (4 pmol rifampin in 40 μl) resulted in the total inhibition of IVT, a concentration that equates to approximately a 1:1 molar ratio of rifampin to RNAP.
FIG. 2.
Real-time in vitro transcription. A graphical representation of part of pUC19-PlacZpolyAT1 (A) and pUC19-PrsbWpolyAT1 (B) indicating the SigA- and SigF-specific templates, respectively. The SigA template utilizes the native lacZ promoter, while the SigF template contains an inserted rsbW promoter (placed in the antisense direction to the native lacZ promoter). Each promoter is followed by an inserted poly(A) stretch and a T1 transcriptional terminator. IVT specificity of SigA and SigF for pUC19-PlacZpolyAT1 (C) and pUC19-PrsbWpolyAT1 (D) was demonstrated using only core RNAP ( ), core RNAP with rSigA (
), core RNAP with rSigA ( ), and core RNAP with rSigF (
), and core RNAP with rSigF ( ). Also, IVT was shown to be abolished to background in the presence of 10 μM rifampin (
). Also, IVT was shown to be abolished to background in the presence of 10 μM rifampin ( ). Dose-dependent rifampin inhibition of the rate of real-time IVT (E) was determined for SigA-mediated IVT (100 nM core RNAP with SigA) on pUC19-PlacZpolyAT1 (solid circles, dashed line) and SigF-mediated IVT (100 nM core RNAP with SigF) on pUC19-PrsbWpolyAT1 (solid triangles, solid line) (mean and standard deviation of results of three independent experiments).
). Dose-dependent rifampin inhibition of the rate of real-time IVT (E) was determined for SigA-mediated IVT (100 nM core RNAP with SigA) on pUC19-PlacZpolyAT1 (solid circles, dashed line) and SigF-mediated IVT (100 nM core RNAP with SigF) on pUC19-PrsbWpolyAT1 (solid triangles, solid line) (mean and standard deviation of results of three independent experiments).
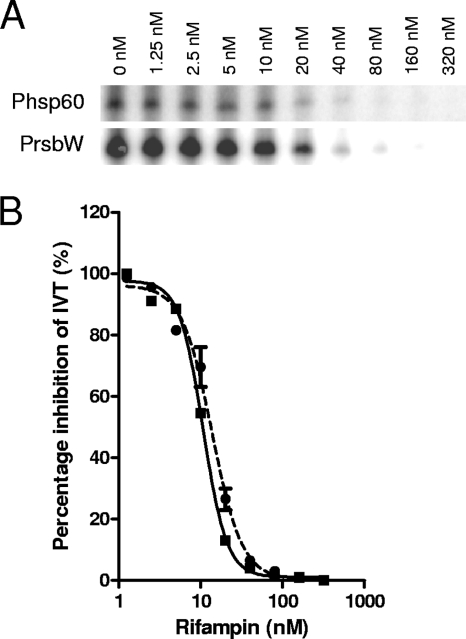
Radiometric IVT showed no SigF-mediated transcription from the SigA-recognized promoter, Phsp60 (15, 22), and very little SigA-mediated transcription from the SigF-specific promoter, PrsbW (2) (data not shown). As with the real-time IVT assay, the inhibition of SigA- and SigF-mediated transcription by rifampin occurred at the same concentration (Fig. 3). In both cases, IVT transcription by the 10 nM RNAP was fully inhibited at concentrations above 40 nM, with 50% inhibition at approximately 10 nM.
FIG. 3.
Radiometric in vitro transcription. (A) A phosphorimage of [32P]CTP-labeled SigA and SigF transcripts formed in the absence and presence of rifampin (1.25 to 320 nM). In vitro transcription was performed using 10 nM core RNAP combined with 100 nM rSigA or rSigF on a PCR-amplified Phsp60 (20 nM) or PrsbW (20 nM) template, respectively. (B) Rifampin-mediated decrease in transcript intensity as quantified by densitometry of the phosphorimage.
Taken together, these results clearly indicate that rifampin inhibition of holo-RNAP is not different when SigA or SigF is incorporated into the core RNAP in vitro.
Pristinamycin IA-mediated induction of SigF in M. tuberculosis H37Rv.
To evaluate if sigF expression in M. tuberculosis can lead to rifampin tolerance in a whole bacterium, a pristinamycin IA-inducible system was developed to overexpress sigF in M. tuberculosis H37Rv (Fig. 4 A and B). As has been shown in previous work using this pristinamycin IA-inducible system (9), induction of SigF in H37Rv::pMYSigF was found to be dose dependent, with induction observed at pristinamycin IA concentrations above 50 ng/ml to maximal induction at 2 μg/ml (data not shown). No induction of SigF was seen by Western blot analysis in the absence of pristinamycin IA or by pristinamycin IA in H37Rv::pMY769 (Fig. 4C). This system is therefore a good tool to investigate the impact of SigF induction in M. tuberculosis H37Rv.
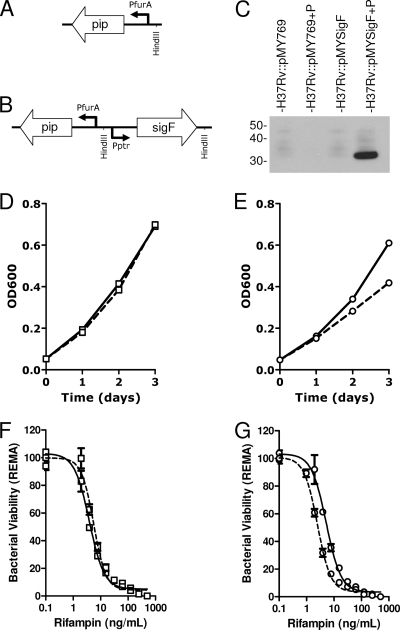
FIG. 4.
Bactericidal activity of rifampin following sigF induction. A representation of part of the M. tuberculosis H37Rv-integrated pristinamycin IA-inducible plasmid pMY769 (A) and pMYsigF (B) where Pptr-sigF is inserted into the HindIII site of pMY769. SigF protein expression following growth (3 days) in the absence and presence or absence of 2 μg/ml pristinamycin IA (P) is shown by Western blot (C) using antibodies specific for SigF. Growth curves of M. tuberculosis strains H37Rv::pMY769 (D) and H37Rv::pMYsigF (E) in the absence (solid line) and presence (dashed line) of 2 μg/ml pristinamycin IA. REMA-determined rifampin-mediated antibacterial activity for H37Rv::pMY769 (F) and H37Rv::pMYsigF (G) following 3 days culture in the absence (solid line) or presence (dashed line) of 2 μg/ml pristinamycin IA.
Analysis of the growth rate of H37Rv::pMYSigF in the presence of 2 μg/ml pristinamycin IA revealed that SigF induction significantly decreased the growth rate of M. tuberculosis H37Rv 24 h after induction (Fig. 4E). This change in growth rate was not observed when pristinamycin IA was added to the H37Rv::pMY769 (Fig. 4D). Also it was observed that this decrease in growth rate was pristinamycin IA dose dependent, with intermediate changes in growth rate observed when concentrations of pristinamycin IA were decreased to 500 and 200 ng/ml (data not shown).
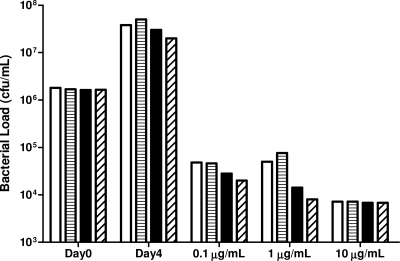
Analysis of the bactericidal activity of rifampin on H37Rv::pMYSigF following 3 days of SigF induction was determined by both REMA and CFU evaluation. REMA revealed that the induction of SigF in H37Rv::pMYSigF did not lead to increased bacterial tolerance to rifampin compared to that in the vector control or uninduced samples (Fig. 4F and G). Indeed, these bacteria appear more susceptible to rifampin than under the control conditions. Similarly, CFU determination showed that SigF induction did not affect the number of viable bacteria following 4 days exposure to 100 ng/ml, 1 μg/ml, or 10 μg/ml rifampin (Fig. 5). These data show that sigF overexpression in M. tuberculosis H37Rv does not increase resistance or tolerance to rifampin.
FIG. 5.
Bacterial CFU determination of H37Rv::pMY769 and H37Rv::pMYsigF following 4 days of rifampin exposure (0.1, 2, and 10 μg/ml). M. tuberculosis H37Rv clones were grown for 3 days in the absence and presence of 2 μg/ml pristinamycin IA prior to rifampin exposure. White bars, H37Rv::pMY769; horizontally lined bars, H37Rv::pMY769 with pristinamycin IA; black bars, H37Rv::pMYsigF; diagonally lined bars, H37Rv::pMYsigF and pristinamycin IA.
Drug susceptibility of M. tuberculosis H37Rv lacking sigF.
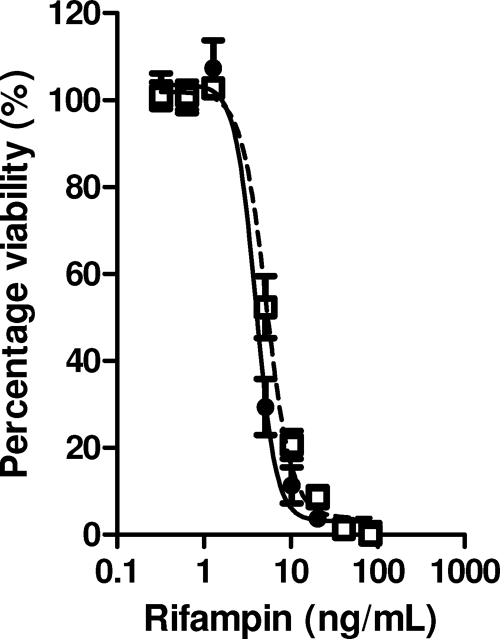
Chen and colleagues have previously shown that deletion of sigF in CDC1551 resulted in greater sensitivity of the bacterium to rifampin (5). Here we decided to construct an M. tuberculosis H37Rv knockout mutant that lacked the full anti-SigF/SigF operon (rsbW/sigF) and compare its rifampin susceptibility. A mycobacterial recombineering system was used to successfully delete the majority of the rsbW/sigF operon (leaving around 100 bp of each split by a hygromycin cassette). Growth curves showed no significant difference between wild-type M. tuberculosis H37Rv and H37RvΔrsbW/sigF. The rifampin susceptibilities of both these strains growing in log phase were determined by REMA and were found to be identical (Fig. 6). Similarly, plating bacteria from both cultures on solid media containing rifampin revealed an identical MIC of 25 ng/ml (data not shown). These data clearly showed that, under the experimental conditions tested, deletion of rsbW/sigF has no influence on M. tuberculosis H37Rv survival in the presence of rifampin and is therefore unlikely to cause tolerance to the antibiotic.
FIG. 6.
Rifampin susceptibility of M. tuberculosis H37Rv (closed circles, solid line) and H37RvΔrsbW/sigF (open squares, dashed line) as determined by REMA (mean and standard deviation of results of three independent experiments).
Neutral red staining of M. tuberculosis strains.
It has previously been reported that M. tuberculosis CDC1551 lacking sigF was unable to retain neutral red staining compared to the wild-type strain (10). Here, following neutral red staining of M. tuberculosis strains grown in liquid media in the absence of Tween 80, H37Rv stained positive (pink/red), while the negative control H37Ra stained negative. H37RvΔrsbW/sigF and the pristinamycin-induced M. tuberculosis H37Rv::pMY769 and H37Rv::pMYSigF showed similar neutral red staining to M. tuberculosis H37Rv (data not shown).
DISCUSSION
RNAP is a well-validated target for the treatment of M. tuberculosis, targeted by what is currently the most important antituberculosis drug, rifampin. Nonetheless, there are bacterial niches that are phenotypically tolerant to rifampin (3, 12, 28, 29), posing a giant hurdle to the rate of bacterial elimination from the human host. Under particular stress conditions, bacterial sigma factor usage of RNAP is often changed, allowing the bacteria to respond to different environments. Recent studies suggest that allosteric interactions between the housekeeping sigma factor A and rifampin bound to the RNAP are possible and that changing the sigma factor may impact rifampin binding (1). Considering that SigF is associated with bacterial entry into stationary phase (7) and that a sigF knockout mutant was reported to be more rifampin susceptible (5), we decided to investigate if RNAP containing SigF is more tolerant to rifampin inhibition.
In vitro transcription results show clearly that rifampin is able to inhibit both SigA- and SigF-mediated transcription at the same efficiency. Differences in the sensitivity of the two assays mean that more RNAP is needed to perform the real-time IVT, and therefore more rifampin is needed to inhibit this transcription. Nonetheless, by both methods a near-equimolar amount of rifampin is sufficient to inhibit transcription. These data suggest that even though domain 3.2 of SigA may interact with RNAP-bound rifampin (1), natural changes in this domain between SigA and SigF have no impact on rifampin binding. Of the remaining 11 alternative sigma factors (24), only SigB contains domain 3.2, but it is very similar in sequence arrangement to that of SigA and unlikely to affect rifampin binding. It cannot be excluded, however, that any of the other alternative sigma factors can allosterically change the conformation of RNAP and affect rifampin binding.
In addition to its not having a direct influence on rifampin inhibition of RNAP, we found here that the artificial induction of sigF in M. tuberculosis H37Rv did not render the bacterium more tolerant to rifampin. Indeed, data from the REMA experiments even suggest that the bacteria are somewhat more sensitive to rifampin following sigF induction; however, this difference could also be attributed to the decreased bacterial growth rate following sigF induction. It was initially hypothesized that increased sigF expression may result in altered membrane composition, as observed in M. smegmatis (23), or potentially an increased expression of drug efflux proteins or RNAP-binding proteins, all of which can affect rifampin accumulation or activity. A 3-day induction of sigF should be sufficient to allow for such potential downstream alterations to the bacterium (and indeed a change in growth rate was observed); however, it did not increase tolerance to rifampin. These data are in contradiction to those previously published, where the deletion of sigF from M. tuberculosis strain CDC1551 leads to an 8-fold increase in rifampin susceptibility (5). Here we were unable to reproduce these data and have shown that wild-type M. tuberculosis H37Rv and H37RvΔrsbW/sigF had identical MIC values for rifampin as determined by both REMA and plating on solid media. Furthermore, the same M. tuberculosis CDC1551 strain lacking sigF was shown to stain negative with neutral red (10), while here H37Rv, H37RvΔrsbW/sigF, and pristinamycin-induced H37Rv::pMYsigF all stained positive. This observed discrepancy in rifampin susceptibility, growth rate, and neutral red staining may in part be explained by differences in the M. tuberculosis strain used (CDC1551 versus H37Rv) and possibly due to the added removal of the anti-sigma factor, RsbW in H37RvΔrsbW/sigF, used here. Furthermore, differences in liquid culture medium components (0.2% glycerol here versus 5% glycerol used by Chen and colleagues [5]) and in the media used for neutral red staining (liquid culture versus Lowenstein-Jensen medium by Geiman and colleagues [10]) may also explain discrepancies.
It was observed that induction of sigF led to a consistent decrease in the growth rate of M. tuberculosis H37Rv (interestingly, this observation was also made when the recombinant protein was expressed in E. coli). Previous data from experiments in which sigF was artificially induced using an acetamide-inducible promoter in M. tuberculosis CDC1551 showed no difference in the bacterial growth rate (14, 31); however, it was found that overexpression of sigF from a strong promoter in BCG caused growth arrest (6). The effect on growth rate observed here could be caused by the competitive displacement of SigA by SigF from the RNAP, leading to decreased expression of housekeeping genes and thereby decreased growth rate. Alternatively, a downstream effector protein induced by SigF itself may be responsible for the change in growth rate, a possibility supported by the observed 24-h time lag between the start of induction and the observed decline in growth rate.
In conclusion, the data obtained in this study contradict the proposal that SigF is able to directly or indirectly alter bacterial rifampin susceptibility of M. tuberculosis H37Rv. Though study of the involvement of sigma factors in rifampin tolerance is not exhausted, recent promising research on the origin of rifampin tolerance in M. tuberculosis is being focused on RNAP accessory proteins. Both GroEL1 (19) and RbpA (8) have been suggested to stabilize RNAP and affect the accessibility of rifampin to its binding site, though it remains to be determined if these proteins cause rifampin tolerance at physiological concentrations in an experimental model of infection.
Acknowledgments
R. C. Hartkoorn was the recipient of a postdoctoral fellowship from the Heiser Program for Research in Leprosy and Tuberculosis of the New York Community Trust. This work was supported in part by the European Commission (contract no. LSHP-CT-2005-018923).
We thank Suresh Solapure and Sharma Umender (AZ India) for providing the M. smegmatis strain with the histidine-tagged RNA polymerase, Daniela Ghisotti, Francesca Forti (Università degli Studi di Milano, Milan, Italy), and Graham Hatfull (University of Pittsburgh) for providing plasmids, and Ida Rosenkrands (Statens Serum Institut, Copenhagen, Denmark) for anti-SigF antibodies.
Footnotes
Published ahead of print on 20 August 2010.
REFERENCES
- 1.Artsimovitch, I., M. N. Vassylyeva, D. Svetlov, V. Svetlov, A. Perederina, N. Igarashi, N. Matsugaki, S. Wakatsuki, T. H. Tahirov, and D. G. Vassylyev. 2005. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell 122:351-363. [DOI] [PubMed] [Google Scholar]
- 2.Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brzezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-sigma factor antagonists control sigmaF activity by distinct mechanisms. Mol. Microbiol. 45:1527-1540. [DOI] [PubMed] [Google Scholar]
- 3.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 4.Burgess, R. R., and J. J. Jendrisak. 1975. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry 14:4634-4638. [DOI] [PubMed] [Google Scholar]
- 5.Chen, P., R. E. Ruiz, Q. Li, R. F. Silver, and W. R. Bishai. 2000. Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect. Immun. 68:5575-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMaio, J., Y. Zhang, C. Ko, and W. R. Bishai. 1997. Mycobacterium tuberculosis sigF is part of a gene cluster with similarities to the Bacillus subtilis sigF and sigB operons. Tuber. Lung Dis. 78:3-12. [DOI] [PubMed] [Google Scholar]
- 7.DeMaio, J., Y. Zhang, C. Ko, D. B. Young, and W. R. Bishai. 1996. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 93:2790-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dey, A., A. K. Verma, and D. Chatterji. 2010. Role of an RNA polymerase interacting protein, MsRbpA, from Mycobacterium smegmatis in phenotypic tolerance to rifampicin. Microbiology 156:873-883. [DOI] [PubMed] [Google Scholar]
- 9.Forti, F., A. Crosta, and D. Ghisotti. 2009. Pristinamycin-inducible gene regulation in mycobacteria. J. Biotechnol. 140:270-277. [DOI] [PubMed] [Google Scholar]
- 10.Geiman, D. E., D. Kaushal, C. Ko, S. Tyagi, Y. C. Manabe, B. G. Schroeder, R. D. Fleischmann, N. E. Morrison, P. J. Converse, P. Chen, and W. R. Bishai. 2004. Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis. Infect. Immun. 72:1733-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu, Y., J. A. Mangan, J. Dhillon, K. M. Sole, D. A. Mitchison, P. D. Butcher, and A. R. Coates. 2000. Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J. Bacteriol. 182:6358-6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussain, S., M. Malik, L. Shi, M. L. Gennaro, and K. Drlica. 2009. In vitro model of mycobacterial growth arrest using nitric oxide with limited air. Antimicrob. Agents Chemother. 53:157-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacques, J. F., S. Rodrigue, R. Brzezinski, and L. Gaudreau. 2006. A recombinant Mycobacterium tuberculosis in vitro transcription system. FEMS Microbiol. Lett. 255:140-147. [DOI] [PubMed] [Google Scholar]
- 14.Lee, J. H., P. C. Karakousis, and W. R. Bishai. 2008. Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network. J. Bacteriol. 190:699-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin, M. E., and G. F. Hatfull. 1993. Mycobacterium smegmatis RNA polymerase: DNA supercoiling, action of rifampicin and mechanism of rifampicin resistance. Mol. Microbiol. 8:277-285. [DOI] [PubMed] [Google Scholar]
- 16.Liu, J., P. A. Feldman, J. S. Lippy, E. Bobkova, M. G. Kurilla, and T. D. Chung. 2001. A scintillation proximity assay for RNA detection. Anal. Biochem. 289:239-245. [DOI] [PubMed] [Google Scholar]
- 17.Marras, S. A., B. Gold, F. R. Kramer, I. Smith, and S. Tyagi. 2004. Real-time measurement of in vitro transcription. Nucleic Acids Res. 32:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, L. P., J. T. Crawford, and T. M. Shinnick. 1994. The rpoB gene of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 38:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee, R., and D. Chatterji. 2008. Stationary phase induced alterations in mycobacterial RNA polymerase assembly: a cue to its phenotypic resistance towards rifampicin. Biochem. Biophys. Res. Commun. 369:899-904. [DOI] [PubMed] [Google Scholar]
- 20.O'Maille, P. E., M. D. Tsai, B. T. Greenhagen, J. Chappell, and J. P. Noel. 2004. Gene library synthesis by structure-based combinatorial protein engineering. Methods Enzymol. 388:75-91. [DOI] [PubMed] [Google Scholar]
- 21.Palomino, J. C., A. Martin, M. Camacho, H. Guerra, J. Swings, and F. Portaels. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 46:2720-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Predich, M., L. Doukhan, G. Nair, and I. Smith. 1995. Characterization of RNA polymerase and two sigma-factor genes from Mycobacterium smegmatis. Mol. Microbiol. 15:355-366. [DOI] [PubMed] [Google Scholar]
- 23.Provvedi, R., D. Kocincova, V. Dona, D. Euphrasie, M. Daffe, G. Etienne, R. Manganelli, and J. M. Reyrat. 2008. SigF controls carotenoid pigment production and affects transformation efficiency and hydrogen peroxide sensitivity in Mycobacterium smegmatis. J. Bacteriol. 190:7859-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodrigue, S., R. Provvedi, P. E. Jacques, L. Gaudreau, and R. Manganelli. 2006. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30:926-941. [DOI] [PubMed] [Google Scholar]
- 25.Sarmientos, P., J. E. Sylvester, S. Contente, and M. Cashel. 1983. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell 32:1337-1346. [DOI] [PubMed] [Google Scholar]
- 26.van Kessel, J. C., and G. F. Hatfull. 2008. Mycobacterial recombineering. Methods Mol. Biol. 435:203-215. [DOI] [PubMed] [Google Scholar]
- 27.van Kessel, J. C., and G. F. Hatfull. 2007. Recombineering in Mycobacterium tuberculosis. Nat. Methods 4:147-152. [DOI] [PubMed] [Google Scholar]
- 28.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 30.Wegrzyn, A., A. Szalewska-Palasz, A. Blaszczak, K. Liberek, and G. Wegrzyn. 1998. Differential inhibition of transcription from sigma70- and sigma32-dependent promoters by rifampicin. FEBS Lett. 440:172-174. [DOI] [PubMed] [Google Scholar]
- 31.Williams, E. P., J. H. Lee, W. R. Bishai, C. Colantuoni, and P. C. Karakousis. 2007. Mycobacterium tuberculosis SigF regulates genes encoding cell wall-associated proteins and directly regulates the transcriptional regulatory gene phoY1. J. Bacteriol. 189:4234-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu, Q. L., D. Kong, K. Lam, and R. N. Husson. 1997. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 179:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]