Abstract
Anabaena sp. strain PCC 7120 is a filamentous cyanobacterium that carries out N2 fixation in specialized cells called heterocysts, which exchange nutrients and regulators with the filament's vegetative cells that perform the photosynthetic fixation of CO2. The Anabaena genome carries two genes coding for alkaline/neutral invertases, invA and invB. As shown by Northern analysis, both genes were expressed monocistronically and induced under nitrogen deprivation, although induction was stronger for invB than for invA. Whereas expression of an InvA-N-GFP fusion (green fluorescent protein [GFP] fused to the N terminus of the InvA protein [InvA-N]) was homogeneous along the cyanobacterial filament, consistent with the lack of dependence on HetR, expression of an InvB-N-GFP fusion upon combined nitrogen deprivation took place mainly in differentiating and mature heterocysts. In an hetR genetic background, the InvB-N-GFP fusion was strongly expressed all along the filament. An insertional mutant of invA could grow diazotrophically but was impaired in nifHDK induction and exhibited an increased frequency of heterocysts, suggesting a regulatory role of the invertase-mediated carbon flux in vegetative cells. In contrast, an invB mutant was strongly impaired in diazotrophic growth, showing a crucial role of sucrose catabolism mediated by the InvB invertase in the heterocysts.
Many cyanobacteria, both unicellular and filamentous, are able to carry out the fixation of N2, and many can do it under oxic conditions. Because cyanobacteria perform oxygenic photosynthesis, the problem of protection of the N2 fixation machinery against oxygen is especially relevant in these organisms, which have to cope not only with ambient O2 but also with that produced inside the cells. A remarkable solution to this problem is the separation of the processes of photosynthetic CO2 fixation and N2 fixation in different cell types, as found in the filamentous, heterocyst-forming cyanobacteria (12, 19). In these organisms, under conditions of combined N limitation, certain vegetative cells, which occupy defined positions in the filament, differentiate into cells called heterocysts that are devoted to the fixation of N2. Heterocysts present multiple structural and functional differences with their parental vegetative cells. These differences result from a specific program of gene expression that involves activation of many genes and repression of others (37). The initiation of heterocyst differentiation requires the concourse of the N control transcription factor NtcA and the differentiation regulator HetR (15). Distinctive heterocyst features include the presence of additional glycolipid and polysaccharide layers in the cell envelope, as well as a narrowed septum of connection with neighboring vegetative cells to decrease the penetration of O2, the lack of activity of the water-splitting photosystem II and of key enzymes involved in the photosynthetic fixation of CO2, the expression of dedicated terminal oxidases to cope with residual O2, and the expression of the nitrogenase complex and accessory Nif proteins (35).
Thus, the diazotrophic filament of heterocyst-forming cyanobacteria represents a unique case of a truly multicellular bacterium with two different cell types, each specialized in a different metabolic function, which depend on each other and contribute to the operation of the filament as the organismic unit. Growth relies on the exchange of regulatory and nutritional compounds between the different cells of the filament, with the vegetative cells donating products of CO2 fixation to the heterocysts (33) and receiving in turn N2 fixation products from them (34). The nature of the exchanged products and transport conduits are the subject of intense debate and of past and present investigation (12, 13, 26, 35). Regarding the identification of fixed-N vehicles, glutamine is synthesized at high levels in the heterocysts and is commonly accepted to represent one such player. Less is known about the identity of the C compounds that are transferred from vegetative cells to heterocysts, which should support both the differentiation process, including the provision of building blocks for the heterocyst envelope, and the metabolism of the mature heterocyst, providing a source of electrons for the N2 reduction reaction and the C skeleton for the incorporation of the ammonium resulting from N2 fixation.
Few studies have directly addressed the issue of which compounds are responsible for the transport of reduced C from vegetative cells to heterocysts. Jüttner (16) proposed the migration of alanine from vegetative cells to heterocysts, where alanine could be metabolized through alanine dehydrogenase providing reducing equivalents (25). On the other hand, a movement of glucose together with the action of hexokinase was considered to explain the appearance of labeled glucose-6-phosphate (G6P) in heterocysts, even in short fixation times, and the fact that [14C]glucose was metabolized to G6P, fructose-6-P, and glutamate by isolated heterocysts (16).
Sucrose is a key sugar in eukaryotic photosynthetic organisms, and several enzymes for the synthesis and degradation of sucrose have also been studied in heterocyst-forming cyanobacteria. However, the relevance of sucrose metabolism in these phototrophic bacteria is not fully understood. Two sucrose-phosphate synthases (encoded by the genes spsA and spsB) and one sucrose-phosphate phosphatase (encoded by sppA) involved in sucrose biosynthesis have been identified in Anabaena spp. (5, 27). All these genes and their encoded enzymes are expressed at higher levels under diazotrophic conditions than in the presence of combined N, and using the green fluorescent protein (GFP) as a reporter, expression of spsA was observed only in vegetative cells, whereas expression of spsB and sppA was observed in both vegetative cells and heterocysts. Thus, during diazotrophic growth, sucrose could be synthesized in both cell types (6).
Regarding sucrose degradation in Anabaena spp., sucrose synthase activity has been reported at much higher levels in vegetative cells than in heterocysts (29). Sucrose synthase has been involved in sucrose degradation in vivo (7), and the susA gene, encoding sucrose synthase, is downregulated under diazotrophic conditions and in heterocysts (7, 8, 9). Whereas a strain carrying an inactivated susA gene had no significant growth defect, a strain overexpressing the gene was severely affected in diazotrophic growth (7). Because levels of insoluble polysaccharides correlate with sucrose synthase activity, a role of sucrose in the flux of reduced C in the filament, possibly in relation to glycogen biosynthesis, has been proposed (9). Finally, alkaline invertase activity (pH optimum of 7.5 to 7.8) has been detected to be associated almost exclusively with heterocysts (29), and more recently the genes invA and invB, encoding alkaline/neutral invertases, have been identified in Anabaena sp. strain PCC 7120 (31). Both InvA and InvB activities increase (3- and 6-fold, respectively) under diazotrophic conditions (31). Remarkably, alkaline/neutral invertases are restricted to cyanobacteria and plants. They are found in all the strains of cyanobacteria tested so far, including filamentous and unicellular strains, capable or not of N2 fixation. Although a role in the flux of C in the heterocyst (29) or as maintenance enzymes for the provision of sucrose degradation products when other sucrose hydrolytic pathways are low (31) has been considered, the physiological function(s) of cyanobacterial alkaline/neutral invertases are not established yet.
In this work, we have studied the spatiotemporal expression of the invA and invB genes in the filaments of Anabaena sp. strain PCC 7120 and have constructed strains carrying inactivated invA or invB genes. We have found that, whereas invA is expressed in all cells of the filament and is not required for diazotrophy, invB is mostly expressed in heterocysts and is essential for diazotrophic growth.
MATERIALS AND METHODS
Strains and growth conditions.
Anabaena sp. strain PCC 7120 (also known as Nostoc sp. strain PCC 7120) was grown in BG11 medium (containing NaNO3 and ferric citrate instead of ferric ammonium citrate [28]), BG110 medium (free of combined nitrogen), or BG110 medium plus ammonium (BG110 containing 4 to 6 mM NH4Cl and, respectively, 8 to 12 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid]-NaOH buffer [pH 7.5]) at 30°C in the light (25 μE m−2 s−1) in shaken (100 rpm) liquid cultures or in medium solidified with 1% Difco agar. Alternatively, cultures (referred to as bubbled cultures and named BG11C or BG110C) were supplemented with 10 mM NaHCO3 and bubbled with a mixture of air and 1% (vol/vol) CO2 in the light (75 μE m−2 s−1). In this case, the ammonium-containing medium was supplemented with 10 mM NH4Cl and 20 mM TES-NaOH buffer (pH 7.5). For the mutants described below, antibiotics were used at the following concentrations: 2 to 5 μg ml−1 (streptomycin [Sm]), 2 to 5 μg ml−1 (spectinomycin [Sp]), and 5 to 10 μg ml−1 (neomycin [Nm]) for liquid cultures and 5 μg ml−1 (Sm), 5 μg ml−1 (Sp), and 40 μg ml−1 (Nm) for solid cultures.
Escherichia coli DH5α was used for plasmid constructions. It and strains HB101 and ED8654, used for conjugations with Anabaena sp., were grown in LB medium, supplemented when appropriate with antibiotics at standard concentrations (1).
DNA and RNA isolation, manipulation, and analysis.
Isolation of genomic DNA (3) and of total RNA (1) from Anabaena sp. was done as described previously. For Northern blots, 10 to 20 μg of RNA was loaded per lane and electrophoresed in denaturing 1% agarose formaldehyde gels. DNA probes were generated by PCR using Anabaena DNA and oligodeoxynucleotide primer pairs, as follows: alr1521-7120-1/alr1521-7120-2 for invA, alr0819-7120-1/alr0819-7120-2 for invB, and NH1/NH4 for nifH (all oligodeoxynucleotide primers are listed in Table 1). Hybridizations were performed as previously described (24). As a control of RNA loading and transfer efficiency, the filters were hybridized with a probe of the RNase P RNA gene (rnpB) from strain PCC 7120 amplified by PCR with primers Universal and Reverse and plasmid pT7-7120 as the template (32). Probes were labeled with a DNA labeling kit (Ready to Go; GE Healthcare) and [α-32P]dCTP. Radioactive areas in Northern blot membranes were visualized with a Cyclone storage phosphor system (Packard).
TABLE 1.
Oligodeoxynucleotide primers used in this worka
| Primer name | Sequence (5′→3′) |
|---|---|
| alr0819-7120-1 | CATAGAAGAATCAGCATGGGAAG |
| alr0819-7120-2 (R1) | CGGTAGGCGGTTATCAATTGCTTC |
| alr0819-7120-3 | ATCGATGGTGTACAATATATGATCC |
| alr0819-7120-4 | GATATCCAGTCCGTTTAGCTTCTGC |
| alr0819-7120-5 | GTGTACAATATATGATCCTTGC |
| alr0819-7120-6 | ATCTACGATATCCAGTCCGTTTAGCTTCTGC |
| alr0819-7120-7 | GGACTGGATATCGTAGATGCTTCTATATCTCG |
| alr0819-7120-8 | CTTCAGCCAACCGGATATATTGCC |
| alr0819-7120-9 (F7) | GTATCATGGTTGTGGTGGGC |
| alr1521-7120-1 | GTACTGTAGCTGCTCAAGATCCAG |
| alr1521-7120-2 (R2) | GATGCGTTGAGATTCTTCCTCG |
| alr1521-7120-3 | ATCGATTGCTCAGAAGGCAGGAGTCAGCG |
| alr1521-7120-4 | GATATCATTAATTGGAGGGGTTTTC |
| alr1521-7120-5 | TGCTCAGAAGGCAGGAGTCAGCG |
| alr1521-7120-6 | GGGCCGGATATCATTAATTGGAGGGGT |
| alr1521-7120-7 | ATTAATGATATCCGGCCCTGGTTGTAGTC |
| alr1521-7120-8 | CTTATCAAGAGAGTATCCCACAAGC |
| alr1521-7120-9 (F8) | GGGACACGGGGTATCGGTATAG |
| CS.3-2 | GTTACCCGAGAGCTTGGC |
| CK.3-3′ | GGTGCCCTTAAACGCCTGGTG |
| GFP-4 | CAAGAATTGGGACAACTCC |
| NH1 | GTACTGCAAGGGGCGTGTGGC |
| NH4 | CCTATTGGTAGCTTCTGCGGG |
| M13-Reverse | GAGGAAACAGCTATGAC |
| M13-Universal | GTAAAACGACGGCCAGT |
Introduced restriction enzyme cutting sites are underlined.
Strain construction.
To produce a GFP fusion to the N terminus of the InvA protein, a 615-bp fragment from the alr1521 5′ and upstream regions was amplified by PCR using primers alr1521-7120-3 and alr1521-7120-4 (which contain a ClaI and an EcoRV restriction site, respectively) and DNA from strain PCC 7120 as the template. This fragment was cloned in vector pMBL-T (Dominion MBL) and transferred as a ClaI/EcoRV-ended fragment to ClaI/EcoRV-digested pCSEL21 (23), producing a fusion of the gfp-mut2 gene (4) to the 6th codon of alr1521. The resulting fusion was finally transferred as an EcoRI-ended fragment to EcoRI-digested pCSV3, producing pCSRL80 (Smr/Spr). (pCSV3 is a derivative of mobilizable vector pRL500 [10] in which the Apr gene has been excised with DraI and replaced by the DraI-ended gene cassette CS.3 encoding Smr and Spr in direct orientation [V. Rodríguez and A. Herrero, unpublished].) To produce a GFP fusion to the N terminus of the InvB protein (InvA-N-GFP), a 545-bp fragment from the alr0819 5′ and upstream regions was amplified by PCR using primers alr0819-7120-3 and alr0819-7120-4 (which contain a ClaI and an EcoRV restriction site, respectively) and DNA from strain PCC 7120 as the template. This fragment was cloned in vector pMBL-T (Dominion MBL) and transferred as a SacI/EcoRV-ended fragment to SacI/EcoRV-digested pCSAM135 (23), producing a fusion of the gfp-mut2 gene (4) to the 6th codon of alr0819. The resulting fusion was finally transferred as an EcoRI-ended fragment to EcoRI-digested pCSV3, producing pCSRL85 (Smr/Spr).
To inactivate invA, two DNA fragments, one encompassing 615 bp from the 5′ end of alr1521 and upstream sequences and the other including 568 bp from the 3′ end of alr1521 and downstream sequences, were amplified by standard PCR using DNA from strain PCC 7120 and primer pairs alr1521-7120-5/alr1521-7120-6 and alr1521-7120-7/alr1521-7120-8, respectively. The two DNA fragments were cloned in pMBL-T and, after digestion, ligated in direct orientation separated by gene cassette CS.3 encoding Smr and Spr (10). The insert of the resulting plasmid, excised with SpeI/NheI, was transferred to plasmid pRL278 carrying an Nmr determinant and the sacB gene for positive selection (2), producing plasmid pCSRL84. To inactivate invB, two DNA fragments, one encompassing 544 bp from the 5′ end of alr0819 and upstream sequences and the other including 536 bp from the 3′ end of alr0819 and downstream sequences, were amplified by standard PCR using DNA from strain PCC 7120 and primer pairs alr0819-7120-5/alr0819-7120-6 and alr0819-7120-7/alr0819-7120-8, respectively. The two DNA fragments were cloned in pMBL-T and, after digestion, ligated in direct orientation separated by gene cassette CK.3 encoding Nmr and Kmr (10). The insert of the resulting plasmid, excised with SpeI/NheI, was transferred to plasmid pRL277 (2) carrying an Smr/Spr determinant and the sacB gene for positive selection (2), producing plasmid pCSRL83.
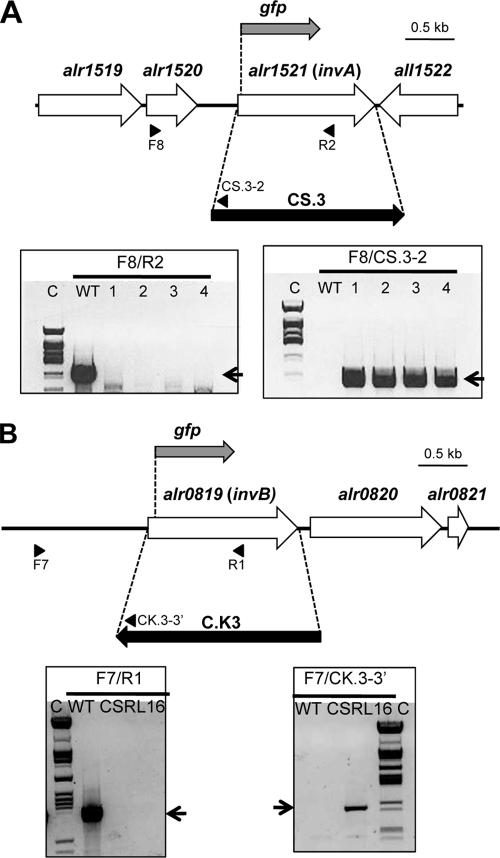
Conjugation of Anabaena sp. strain PCC 7120 or strain 216 (hetR) with E. coli HB101 carrying pCSRL80, pCSRL85, pCSRL83, or pCSRL84 with helper and methylation plasmid pRL623 was effected by the conjugative plasmid pRL443, carried in E. coli ED8654 and performed as described previously (11), with selection for resistance to Sm/Sp or Nm. The genetic structure of the selected clones was studied by PCR with DNA from those clones and the primer pairs indicated in Fig. 1 for the insertion mutants (strains CSRL16 and CSRL17) and gfp-4/alr1521-7120-9 for the InvA-N-GFP (CSRL19 and CSRL20) and gfp-4/alr0819-7120-9 for the InvB-N-GFP (CSRL18 and CSRL21) fusion strains.
FIG. 1.
Schematic of gfp reporter fusions and insertional inactivation of the invA (A) and invB (B) genes. Strategies of insertion of gene cassettes to produce strains CSRL17 (invA) and CSRL16 (invB) and sites of insertion of the gfp gene to produce strains CSRL19 (invA-gfp) and CSRL18 (invB-gfp) are shown. The approximate positions of the primers used to check segregation of the mutant chromosomes by PCR are indicated in each case. PCR analysis of the genetic structure in the invA (A) and invB (B) regions in strains PCC 7120, CSRL17, and CSRL16 is also shown, with the indication of the primer pairs used for amplification. Electrophoresis gel lanes: C, λ DNA digested with ClaI; WT, reactions with DNA from strain PCC 7120; 1 to 4 in panel A, reactions with DNA from 4 different exconjugants; CSRL16 in panel B, reaction with DNA from strain CSRL16. The expected amplification products are as follows: for F8/R2, 1,630 bp (WT) and none in the mutant clones (note that the R2 primer is within the deleted DNA sequences); for F8/CS.3-2, none in the WT (the CS3-2 primer is within the CS.3 gene cassette) and 805 bp in the mutants; for F7/R1, 1,577 bp (WT) and none in CSRL16 (the R1 primer is within the deleted DNA sequences); for F7/CK.3-3′, none in the WT (the CK3-3′ primer is within the CK.3 gene cassette) and 995 bp in CSRL16.
Confocal microscopy.
Samples from cultures of Anabaena sp. set atop solidified medium (BG11 or BG110) were visualized using a Leica HCX PLAN-APO 63× 1.4-numerical aperture (NA) oil immersion objective lens attached to a Leica TCS SP2 confocal laser-scanning microscope. GFP was excited using 488-nm irradiation from an argon ion laser. Fluorescent emission was monitored by collection across windows of 500 to 538 nm (GFP imaging) and 630 to 700 nm (cyanobacterial autofluorescence).
Growth rates.
The growth rate constant (μ = ln 2/td, where td is the doubling time) was calculated from the increase of protein content, determined in 0.2-ml samples, of shaken liquid cultures (22). Protein concentration was determined by a modified Lowry procedure (21). The chlorophyll a (Chl) content of cultures was determined by the method of Mackinney (20).
Nitrogenase activity.
Nitrogenase activity was determined by the acetylene reduction technique in filaments incubated in bubbled BG110C medium as described previously (22). For assays under anoxic conditions, 10 μM DCMU [3-(3,4-dichlorophenyl)-1,1-dimethylurea] was added to the cell suspension, and the flask containing the cells was sealed with a rubber stopper, bubbled with argon for 3 min, and further incubated under culture conditions for 1 h before the assay was started by the addition of acetylene.
RESULTS
Expression of invertase-encoding genes.
In the genome of Anabaena sp. strain PCC 7120, open reading frames (ORFs) alr1521 and alr0819 correspond to the genes invA and invB, encoding alkaline and neutral invertases, respectively (17, 31). ORF alr1521 is flanked by the upstream ORF alr1520, which is separated from invA by 316 nucleotides (nt), and by the downstream ORF all1522, which is oriented opposite to invA (Fig. 1A). ORF alr0819 is followed by the downstream ORFs alr0820, separated from invB by 125 nucleotides, and alr0821 (Fig. 1B).
The expression of the Anabaena invA and invB genes was studied by Northern analysis using RNA isolated from filaments grown with ammonium and incubated in the absence of combined nitrogen. For invA, hybridization signals detected were smeared, which suggests the presence of unstable transcripts, as has also been described for other Anabaena genes (e.g., 14), showing an upper limit that correspond to RNAs of ca. 2 kb (alr1521 is 1,407 nt long) (Fig. 2). Expression of this gene increased slightly upon combined nitrogen deprivation, and no clear effect of the mutation of either ntcA (in strain CSE2) or hetR (in strain 216) could be observed (Fig. 2). For invB, a transcript of ca. 1.9 kb (alr0819 is 1,452 nt long) could be detected, whose abundance increased until ca. 9 to 12 h of combined nitrogen deprivation, decreasing thereafter (Fig. 2). This increase was impaired in the ntcA mutant strain (Fig. 2). In the hetR mutant, expression of invB upon combined N deprivation was higher than that in the wild type, especially at the later times tested (Fig. 2). The two genes appear, therefore, to be transcribed monocistronically.
FIG. 2.
Northern blot analysis of the expression of alr1521 (invA) and alr0819 (invB). RNA isolated from bubbled cultures of Anabaena sp. strain PCC 7120, strain CSE2 (ntcA), and strain 216 (hetR) grown with ammonium (0) and incubated in the absence of combined nitrogen for the times indicated in hours was electrophoresed and hybridized with the probes indicated at the left, which were generated by PCR (see Materials and Methods). The positions of some size markers are indicated at the right. Hybridization with an rnpB gene probe was used as a loading and transfer control.
The spatial expression of invA and invB along the filament was studied using GFP as a reporter. The gfp-mut2 gene was inserted, maintaining the translation frame, after the first six codons of either invA or invB both in the wild-type and hetR genetic backgrounds (Fig. 1). The selected clones, strains CSRL19 (invA-gfp in PCC 7120), CSRL18 (invB-gfp in PCC 7120), CSRL20 (invA-gfp in 216), and CSRL21 (invB-gfp in 216), had incorporated the construct by a single crossover. As a result, the fusion gene was expressed in each strain from the whole promoter and sequences upstream from the corresponding inv gene, whereas an intact copy of the inv gene was preceded by the cloned promoter fragment. Fluorescence from GFP was analyzed in ammonium-grown filaments subjected to different periods of combined nitrogen deprivation (Fig. 3). For InvA-N-GFP in the wild-type background (strain CSRL19), some fluorescence increase was observed upon combined-nitrogen deprivation, consistent with Northern analysis results. Maximum fluorescence was observed after 9 h of incubation, and at all time points, fluorescence was distributed homogenously along the filament. Similar fluorescence emission and distribution were observed for InvA-N-GFP in the hetR background (strain CSRL20). Fluorescence from InvB-N-GFP in the wild-type background (strain CSRL18) was observed to increase to an extent higher than that of InvA-GFP. Moreover, for InvB-N-GFP, the fluorescence increase was stronger in certain cells of the filament, which after 9 h could be identified as proheterocysts or heterocysts, and was particularly strong in mature heterocysts after 21 h of incubation (Fig. 3). In contrast, in the hetR background (strain CSRL21), fluorescence was higher and evenly distributed along the filament.
FIG. 3.
Time course of expression of InvA-N-GFP and InvB-N-GFP in the wild-type and hetR genetic backgrounds after N stepdown. Bright field (left) and GFP fluorescence (right) micrographs of filaments of strains CSRL19 (InvA-N-GFP in PCC 7120), CSRL18 (InvB-N-GFP in PCC 7120), CSRL20 (InvA-N-GFP in 216), and CSRL21 (InvB-N-GFP in 216) grown with ammonium and incubated without a source of combined nitrogen for the time periods indicated (in hours). Settings to capture GFP fluorescence were identical for all the samples. Size marker, 10 μm.
Inactivation of invA and invB.
To study the roles of invA and invB in Anabaena sp. strain PCC 7120, mutant strains bearing inactivated versions of each of these genes were constructed. To generate an invA mutant, plasmid pCSRL84 bearing invA upstream and downstream regions joined by gene cassette CS.3, encoding resistance to Sm/Sp, was transferred to strain PCC 7120 by conjugation. Clones exhibiting Smr/Spr, resistance to sucrose, and sensitivity to Nm (sensitivity to sucrose and resistance to Nm are encoded in the vector portion of pCSRL84), which were expected to have substituted the invA gene by the gene cassette through double crossover events, were selected. PCR analysis of several such clones was consistent with the expected genomic structure and complete segregation of the mutants (Fig. 1A). One of these clones was selected for further study and named strain CSRL17. To generate an invB mutant, plasmid pCSRL83 bearing invB upstream and downstream regions joined by gene cassette CK.3, encoding resistance to Nm, was transferred to strain PCC 7120 by conjugation. Clones exhibiting Nmr, resistance to sucrose, and sensitivity to Sm/Sp (sensitivity to sucrose and resistance to Sm/Sp are encoded in the vector portion of pCSRL83), which were expected to have substituted the invB gene by the gene cassette through double crossover events, were selected. The genomic structure in one selected clone was analyzed by PCR (Fig. 1B). The results obtained indicated the correct insertion of the C.K3 gene cassette and segregation of the mutation. The selected clone was named strain CSRL16. Lack of a negative effect of the insertion of the C.K3 cassette on the expression of the downstream gene alr0820 was confirmed by Northern analysis of the expression of this gene (not shown).
The ability to grow using different nitrogen sources was studied in solid (Fig. 4 A) and liquid (Fig. 4B) culture media in strains CSRL17 (invA) and CSRL16 (invB) in comparison to the wild type. Using nitrate or ammonium, both mutants were able to grow to levels similar to those of strain PCC 7120. However, at the expense of N2, growth of strain CSRL16, but not of CSRL17, was severely impaired (Fig. 4A). In liquid medium, strain CSRLl7 showed growth rates similar to those of the wild type independently of the N source (Fig. 4B). In contrast, whereas the growth rate of strain CSRL16 using nitrate was similar to that of the wild type, its growth rate fixing N2 was less than 10% that of the wild type (Fig. 4B). Thus, inactivation of invB, but not of invA, has a strong negative effect specifically on diazotrophic growth.
FIG. 4.
Growth of strains CSRL16 and CSRL17 with different N sources. (A) Cell suspensions of Anabaena sp. strains PCC 7120, CSRL16, and CSRL17 grown with ammonium (with antibiotics in the case of the mutants) and washed with combined-nitrogen-free medium were used to inoculate (10 ng Chl per spot) plates of BG11 medium (NO3−) or of BG110 medium supplemented (NH4+) or not (N2) with ammonium that were incubated for 14 days under culture conditions (see Materials and Methods for details). (B) Cell suspensions of Anabaena sp. strains PCC 7120, CSRL16, and CSRL17 grown with ammonium (and antibiotics in the case of the mutants) and washed with combined-nitrogen-free medium were used to inoculate liquid cultures with the indicated N source, which were incubated under culture conditions. The numbers are specific growth rate constants (day−1) and are the averages from two independent experiments with similar results.
Microscopic observation of diazotrophic cultures of strains CSRL16 and CSRL17 showed that the frequency of heterocysts was slightly (7.0% of total cells in the filament) and considerably (9.9% of total cells in the filament) higher in the invB and invA mutants, respectively, than in the wild type (5.9%). Regarding heterocyst distribution, the number of cells between heterocysts was lower in the invA mutant than in either the wild type or the invB mutant (Fig. 5). Whereas the frequency of heterocysts clustered in doublets amounted to 3.7% of total heterocysts in the wild type and to 10% in the invB mutant, it was 26.3% in the invA mutant, which moreover exhibited 3.6% of heterocysts in triplets and one observed quartet in the sample analyzed (12,572 cells). No heterocyst triplet or quartet was observed in either the wild-type or invB strains. Thus, in strain CSRL16, and especially CSRL17, heterocyst frequency is higher than that in the wild type. Of note, in strain CSRL17 the pattern of heterocyst distribution significantly differs from that found in strain PCC 7120 and is characterized by shorter intervals of vegetative cells between heterocysts than those in the wild type.
FIG. 5.
Length of vegetative cell intervals between heterocysts in Anabaena sp. strains PCC 7120, CSRL16, and CSRL17. Filaments grown with ammonium (and antibiotics in the case of the mutants) were incubated in the absence of combined nitrogen in bubbled cultures for 48 hours and visualized by standard light microscopy.
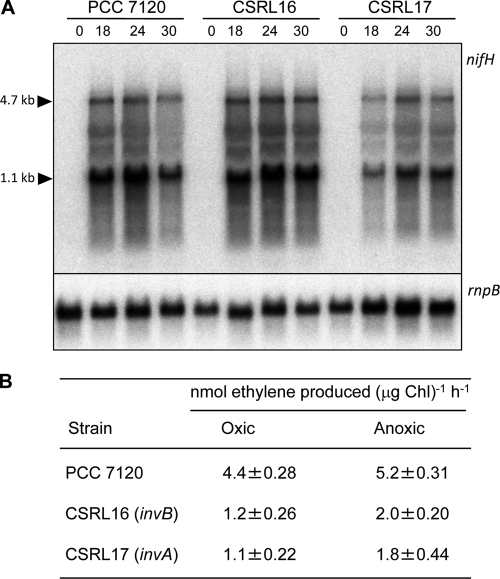
Expression of the nifHDK genes, encoding nitrogenase, and nitrogenase activity under oxic and anoxic conditions were studied in strains CSRL16 and CSRL17 in comparison to the wild-type strain. Whereas activation of nifHDK expression upon ammonium deprivation was somewhat stronger in strain CSRL16 (invB) than in the wild type, activation was slower in strain CSRL17 (invA), although after 30 h levels similar to the wild-type levels were observed (Fig. 6 A). In both mutants, nitrogenase activity was significantly lower than that in the wild type both under oxic (27 and 25% of the wild-type levels for CSRL16 and CSRL17, respectively) and anoxic (39 and 25%, respectively) conditions (Fig. 6B).
FIG. 6.
Expression of nifHDK and nitrogenase activity in strains CSRL16 and CSRL17. (A) RNA was isolated from cultures of the indicated strains grown with ammonium (0) and incubated in the absence of combined nitrogen for the times indicated in hours. Northern analysis was carried out with a probe of nifH, which was generated by PCR (see Materials and Methods). Sizes of previously identified nifH and nifHDK transcripts are indicated on the left. Hybridization with an rnpB gene probe was used as a loading and transfer control. (B) Ammonium-grown filaments were incubated in the absence of combined nitrogen in bubbled cultures for 24 h and then used for the determination of nitrogenase activity under oxic and anoxic conditions as described in Materials and Methods. Data are the means and standard deviations from 3 to 6 independent determinations.
DISCUSSION
In the genomic sequence of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120, two alkaline/neutral invertase-encoding genes are found (31). The invA gene is expressed in a monocistronic transcript. Only small changes in the expression levels of this gene were observed upon combined N deprivation, consistent with an apparent lack of effect of the inactivation of the ntcA gene, encoding the general N control transcription factor. Homogeneous expression of invA is found along the filament, consistent with an apparent lack of effect of inactivation of the hetR gene, encoding a regulator required for heterocyst differentiation.
Because Anabaena sp. strain PCC 7120 is a strict photoautotroph, intracellular sucrose should be formed from intermediates of the CO2 assimilation pathways or from degradation of internal polymers (glycogen). Because in Anabaena sp. strain PCC 7120 the activity of sucrose synthase, the other enzyme involved in sucrose degradation, is downregulated under diazotrophic conditions (7), it can be assumed that under these conditions InvA has an important role in sucrose degradation in vegetative cells and, thus, in the flux of C compounds inside these cells. Inactivation of invA has a clear effect on heterocyst differentiation, affecting both the frequency of heterocysts and the pattern of heterocyst distribution along the filament. The mutant shows a higher tendency to differentiation than the wild type, resulting in more heterocysts separated by shorter vegetative-cell intervals. These effects could be the result of the above-discussed alterations in the distribution of C compounds provoked by the mutation, which could also affect the availability of substrates for the assimilation of C and/or N. All these changes would impact the C-to-N balance of the filament, a key parameter in the determination of heterocyst differentiation (15). However, a more specific role of sucrose, or a product of its degradation, in the regulation of heterocyst differentiation cannot be excluded. Finally, in strain CSRL17, the decrease in nitrogenase activity could result from the observed decreased expression levels of nifHDK (Fig. 6). Nonetheless, the nitrogenase levels of strain CSRL17 allow diazotrophic growth rates similar to those found in the wild-type strain, at least under the conditions here used.
The invB gene is also expressed in a monocistronic transcript. At the whole-filament level (Fig. 2), invB expression increases transiently upon combined N deprivation, dependent on NtcA. In the wild-type strain, a small increase of expression is observed in vegetative cells, as shown by a GFP translational fusion, but activation of invB takes place mainly in the cells that are undergoing differentiation and in mature heterocysts (Fig. 3). Remarkably, in the hetR mutant, in which heterocyst differentiation does not take place, invB expression is high in all the cells of the filament and higher than in vegetative cells of the wild type. At advanced times after ammonium withdrawal, this effect could result from activation of the invB gene under conditions of severe N stress. However, high-level expression of InvB-N-GFP in the hetR background is also observed earlier after combined nitrogen deprivation, at least at 14 h, a time at which N2 fixation has not started in the wild type. Thus, it is possible that a specific signal of heterocyst differentiation (or activity) has a role in the downregulation of invB expression in vegetative cells. Alternatively, the HetR protein might have a repressor role in vegetative cells. A negative role of HetR on gene expression has been previously noticed (18).
Inactivation of invB leads to severe impairment in the ability to grow specifically under diazotrophic conditions. This result, together with the specificity of invB expression in the heterocysts, supports the notion that sucrose catabolism in the heterocysts mediated by InvB plays an important role in the activity of these differentiated cells. Nitrogenase activity, measured as acetylene reduction, in the mutant is ca. one third of that in the wild type. However, the differences in the in vivo reduction of N2 to NH4+ could be higher, because a limitation of reductant or ATP is expected to affect the electron flow during the catalytic cycle of the nitrogenase enzyme complex, which would result in a preferential reduction of H+ versus N2 (see reference 30). Also, the lack of sucrose degradation could affect the availability of ATP for the incorporation of the ammonium resulting from N2 fixation into glutamine mediated by glutamine synthetase, which takes place in the heterocysts (36). Because degradation of sucrose by overexpressed sucrose synthase in vegetative cells (7) or lack of degradation by invertase in heterocysts (this work) impairs diazotrophic growth, sucrose appears to represent a vehicle for the transfer of reduced carbon from vegetative cells to heterocysts.
Acknowledgments
This work was supported by grant BFU2007, cofinanced by FEDER, from the Ministerio de Ciencia e Innovación (Spain).
Footnotes
Published ahead of print on 20 August 2010.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2010. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience, New York, NY.
- 2.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 3.Cai, Y. P., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 5.Cumino, A., L. Curatti, L. Giarrocco, and G. L. Salerno. 2002. Sucrose metabolism: Anabaena sucrose-phosphate synthase and sucrose-phosphate phosphatase define minimal functional domains shuffled during evolution. FEBS Lett. 517:19-23. [DOI] [PubMed] [Google Scholar]
- 6.Cumino, A. C., C. Marcozzi, R. Barreiro, and G. L. Salerno. 2007. Carbon cycling in Anabaena sp. PCC 7120. Sucrose synthesis in the heterocysts and possible role in nitrogen fixation. Plant Physiol. 143:1385-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curatti, L., E. Flores, and G. Salerno. 2002. Sucrose is involved in the diazotrophic metabolism of the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 513:175-178. [DOI] [PubMed] [Google Scholar]
- 8.Curatti, L., L. Giarrocco, and G. L. Salerno. 2006. Sucrose synthase and RuBisCo expression is similarly regulated by the nitrogen source in the nitrogen-fixing cyanobacterium Anabaena sp. Planta 223:891-900. [DOI] [PubMed] [Google Scholar]
- 9.Curatti, L., L. E. Giarrocco, A. C. Cumino, and G. L. Salerno. 2008. Sucrose synthase is involved in the conversion of sucrose to polysaccharides in filamentous nitrogen-fixing cyanobacteria. Planta 228:617-625. [DOI] [PubMed] [Google Scholar]
- 10.Elhai, J., and C. P. Wolk. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119-138. [DOI] [PubMed] [Google Scholar]
- 11.Elhai, J., A. Vepritskiy, A. M. Muro-Pastor, E. Flores, and C. P. Wolk. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179:1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores, E., and A. Herrero. 2010. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 8:39-50. [DOI] [PubMed] [Google Scholar]
- 13.Flores, E., A. Herrero, C. P. Wolk, and I. Maldener. 2006. Is the periplasm continuous in filamentous multicellular cyanobacteria? Trends Microbiol. 14:439-443. [DOI] [PubMed] [Google Scholar]
- 14.Frías, J. E., E. Flores, and A. Herrero. 1997. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 179:477-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrero, A., A. M. Muro-Pastor, A. Valladares, and E. Flores. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28:469-487. [DOI] [PubMed] [Google Scholar]
- 16.Jüttner, F. 1983. 14C-labeled metabolites in heterocysts and vegetative cells of Anabaena cylindrica filaments and their presumptive function as transport vehicles of organic carbon and nitrogen. J. Bacteriol. 155:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 18.Khudyakov, I., and C. P. Wolk. 1997. hetC, a gene coding for a protein similar to bacterial ABC protein exporters, is involved in early regulation of heterocyst differentiation in Anabaena sp. strain PCC 7120. J. Bacteriol. 179:6971-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar, K., R. A. Mella-Herrera, and J. W. Golden. 2010. Cyanobacterial heterocysts. Cold Spring Harb. Perspect. Biol. 2:a000315. doi: 10.1101/cshperspect.a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackinney, G. 1941. Absorption of light by chlorophyll solutions. J. Biol. Chem. 140:109-112. [Google Scholar]
- 21.Markwell, M. A. K., S. M. Hass, L. L. Bieber, and N. E. Tolbert. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210. [DOI] [PubMed] [Google Scholar]
- 22.Montesinos, M. L., A. Herrero, and E. Flores. 1995. Amino acid transport systems required for diazotrophic growth in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 177:3150-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olmedo-Verd, E., A. M. Muro-Pastor, E. Flores, and A. Herrero. 2006. Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 188:6694-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paz-Yepes, J., A. Herrero, and E. Flores. 2007. The NtcA-regulated amtB gene is necessary for full methylammonium uptake activity in the cyanobacterium Synechococcus elongatus. J. Bacteriol. 189:7791-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pernil, R., A. Herrero, and E. Flores. 2010. Catabolic function of compartmentalized alanine dehydrogenase in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 192:5165-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picossi, S., M. L. Montesinos, R. Pernil, C. Lichtlé, A. Herrero, and E. Flores. 2005. ABC-type neutral amino acid permease N-I is required for optimal diazotrophic growth and is repressed in the heterocysts of Anabaena sp. strain PCC 7120. Mol. Microbiol. 57:1582-1592. [DOI] [PubMed] [Google Scholar]
- 27.Porchia, A. C., and G. L. Salerno. 1996. Sucrose biosynthesis in a prokaryotic organism: presence of two sucrose-phosphate synthases in Anabaena with remarkable differences compared with the plant enzymes. Proc. Natl. Acad. Sci. U. S. A. 93:13600-13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 29.Schilling, N., and K. Ehrnsperger. 1985. Cellular differentiation of sucrose metabolism in Anabaena variabilis. Z. Naturforsch. 40c:776-779. [Google Scholar]
- 30.Seefeldt, L. C., B. M. Hoffman, and D. R. Dean. 2009. Mechanism of Mo-dependent nitrogenase. Annu. Rev. Biochem. 78:701-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vargas, W., A. Cumino, and G. L. Salerno. 2003. Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta 216:951-960. [DOI] [PubMed] [Google Scholar]
- 32.Vioque, A. 1997. The RNase P RNA from cyanobacteria: short tandemly repeated repetitive (STRR) sequences are present within the RNase P RNA gene in heterocyst-forming cyanobacteria. Nucleic Acids Res. 25:3471-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolk, C. P. 1968. Movement of carbon from vegetative cells to heterocysts in Anabaena cylindrica. J. Bacteriol. 96:2138-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolk, C. P., S. M. Austin, J. Bortins, and A. Galonsky. 1974. Autoradiographic localization of N after fixation of N-labeled nitrogen gas by a heterocyst-forming blue-green alga. J. Cell Biol. 61:440-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 36.Wolk, C. P., J. Thomas, P. W. Shaffer, S. M. Austin, and A. Galonsky. 1976. Pathway of nitrogen metabolism after fixation of 13N-labeled nitrogen gas by the cyanobacterium, Anabaena cylindrica. J. Biol. Chem. 251:5027-5034. [PubMed] [Google Scholar]
- 37.Xu, X., J. Elhai, and C. P. Wolk. 2008. Transcriptional and developmental responses by Anabaena to deprivation of fixed nitrogen, p. 383-422. In A. Herrero and E. Flores (ed.), The cyanobacteria. Molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom.