Abstract
Brochothrix belongs to the low-GC branch of Gram-positive bacteria (Firmicutes), closely related to Listeria, Staphylococcus, Clostridium, and Bacillus. Brochothrix thermosphacta is a nonproteolytic food spoilage organism, adapted to growth in vacuum-packaged meats. We report the first genome sequences and characterization of Brochothrix bacteriophages. Phage A9 is a myovirus with an 89-nm capsid diameter and a 171-nm contractile tail; it belongs to the Spounavirinae subfamily and shares significant homologies with Listeria phage A511, Staphylococcus phage Twort, and others. The A9 unit genome is 127 kb long with 11-kb terminal redundancy; it encodes 198 proteins and 6 tRNAs. Phages BL3 and NF5 are temperate siphoviruses with a head diameter of 56 to 59 nm. The BL3 tail is 270 nm long, whereas NF5 features a short tail of only 94 nm. The NF5 genome (36.95 kb) encodes 57 gene products, BL3 (41.52 kb) encodes 65 products, and both are arranged in life cycle-specific modules. Surprisingly, BL3 and NF5 show little relatedness to Listeria phages but rather demonstrate relatedness to lactococcal phages. Peptide mass fingerprinting of viral proteins indicate programmed −1 translational frameshifts in the NF5 capsid and the BL3 major tail protein. Both NF5 and BL3 feature circularly permuted, terminally redundant genomes, packaged by a headful mechanism, and integrases of the serine (BL3) and tyrosine (NF5) types. They utilize unique target sequences not previously described: BL3 inserts into the 3′ end of a RNA methyltransferase, whereas NF5 integrates into the 5′-terminal part of a putative histidinol-phosphatase. Interestingly, both genes are reconstituted by phage sequence.
Brochothrix thermosphacta is a Gram-positive, rod-shaped, nonmotile, non-spore-forming, facultative anaerobic, and psychrotrophic organism frequently involved in the spoilage of prepacked and vacuum-packaged meat or meat products (23). The conditions found in these foods selectively favor development of B. thermosphacta due to its ability to grow at 4°C under O2 depletion and in the presence of elevated CO2 concentrations (58). B. thermosphacta frequently constitutes the dominant portion of the spoilage microflora of aerobically and anaerobically stored meats (7), where it produces undesired volatile compounds such as acetoin, diacetyl (aerobic growth), or lactic acid and ethanol (anaerobic growth) (28, 58, 66). The taxonomic standing of the genus Brochothrix has been in dispute for a long time (65), until 16S rRNA sequences revealed its close phylogenetic relationship to the genus Listeria (16, 52), which led to the affiliation of Brochothrix, together with the genus Listeria into the Listeriaceae family. The two genera feature a similar GC content and other common characteristics, such as the same major fatty acids and menaquinones, meso-diamino pimelic acid in the peptidoglycan, production of catalase, and more (65).
The potential of bacteriophages and phage-encoded lytic enzymes for control of specific pathogens in foods has been intensively investigated in recent years (26, 31, 48). As an example, the application of phage to foods has turned out to be a useful approach in the control of Salmonella, Campylobacter, Listeria, and Escherichia coli (reviewed in reference 31). Another possibility is the direct application of purified bacteriophage endolysins to foods or raw products (reviewed in reference 48). These approaches might also be suitable for the biocontrol of B. thermosphacta to prevent food spoilage. Previous work has shown that Brochothrix phage A3 was able to limit off-odor formation and increase the storage life of pork adipose tissue (28).
Unfortunately, only very limited data and no genome sequences are available for bacteriophages infecting Brochothrix. Ackermann et al. classified 21 Brochothrix phages by electron microscopy and grouped them into three species based on morphology (1): species A19 contains 5 myoviruses with long tails that undergo extensive rearrangements upon contraction, species NF5 consists of 14 siphoviruses with rigid tails and complex baseplate structures with appendages, and species BL3 contains only 1 member of the siphovirus morphology. Due to the close phylogenetic relatedness of their hosts, phages of the genus Brochothrix were expected to share similarities with Listeria phages also on the molecular level.
We report here the complete genome sequences and molecular characterization of three bacteriophages infecting B. thermosphacta, namely, A9, BL3, and NF5. Their physical genome structures and attachment loci of the temperate bacteriophages were determined and revealed novel prophage insertion sites. Programmed translational frameshifts in structural genes and lytic activity of bacteriophage-encoded endolysin gene products were demonstrated. In addition, intra- and interspecies comparative genomics led to the placement of phage A9 into the recently proposed Spounavirinae subfamily, and indicated relatedness of phages BL3 and NF5 to the P335 quasispecies of Lactococcus phages.
MATERIALS AND METHODS
Bacterial strains and bacteriophages.
The bacterial strains used in the present study were B. thermosphacta HER1187 and HER1188 (Felix d'Hérelle Reference Center for Bacterial Viruses, Québec, Canada [www.phage.ulaval.ca]) and E. coli XL1-Blue MRF′ (Stratagene, Santa Clara, CA) and those listed in Table S1 in the supplemental material. Bacteria were grown at 24°C (Brochothrix), at 30°C (Listeria), or at 37°C (Escherichia, Bacillus, Enterococcus, and Staphylococcus) under constant agitation in half-strength brain heart infusion (BHI; Difco/BD, Basel, Switzerland) (for Listeria and Brochothrix) or Luria-Bertani (LB) broth (Oxoid, Hampshire, United Kingdom) (for E. coli). Bacteriophages A9, NF5, and BL3 were purchased from the Felix d'Hérelle Reference Center for Bacterial Viruses as lysates and stored at 4°C.
Phage growth and multiplication parameters.
Portions (10 ml) of a log-phase culture (optical density at 600 nm of 0.5, ∼107 CFU/ml) of B. thermosphacta HER1187 were harvested by centrifugation and resuspended in 2.5 ml of fresh BHI medium. Then, 5 × 104 PFU of phage A9 were added and allowed to adsorb for 5 min at 24°C. Free phage was removed by centrifugation, and the cell pellet was resuspended in 10 ml of fresh medium. Incubation was continued at 24°C, and samples were plated by soft-agar overlay (2) for free phage counts at various time points between 15 and 170 min postinfection. The log10 (PFU/ml) related to time (minutes) was plotted, and data points were fitted by using a sigmoidal five-parameter approximation (SigmaPlot version 11; Systat Software, Inc., Chicago, IL). The burst size was determined from triplicate experiments by phage numbers after the burst (t = 160 min) in relation to the initial titer at lag phase (t = 45 min).
Phage propagation and DNA isolation and manipulation.
Phages were purified and propagated by the standard soft-agar overlay method (2). SM buffer (100 mM NaCl, 8 mM MgSO4 and 50 mM Tris-HCl [pH 7.4]) was used to extract phages from plates. NF5 was propagated in B. thermosphacta HER1188 cells by using LB bottom and LC (LB agar supplemented with 10 mM CaCl) top agar. Phages BL3 and A9 were propagated in B. thermosphacta HER1187 cells using BHI-LC and LB-LC double layers, respectively. Phages were concentrated and purified by polyethylene glycol precipitation (PEG 8000, 7% in the presence of 1 M NaCl), stepped CsCl gradient ultracentrifugation (60), and dialysis against a 1,000-fold excess of SM buffer at 4°C. DNA was extracted with organic solvents as described previously (37).
Phage DNA (0.5-μg samples) was digested with restriction enzymes according to the manufacturer's instructions (Fermentas, St. Leon-Roth, Germany, or New England Biolabs, Ipswich, United Kingdom). The products were analyzed electrophoretically. Bal31 nuclease treatment was performed by digestion of 40 μg of phage DNA with 20 U of Bal31 (New England Biolabs) at 30°C. Samples were removed 0, 5, 10, 20, 40, 60, 90, and 120 min after enzyme addition and purified by organic extraction (60). An equal volume of each sample was digested with the indicated restriction enzyme, and products were electrophoresed in a GNA200 horizontal electrophoresis apparatus (GE Healthcare, Switzerland) at 2 V/cm in 1× Tris-acetate-EDTA buffer. Pulsed-field gel electrophoresis (PFGE) was performed in 0.5× Tris-borate-EDTA buffer with an initial switch time of 1 s and a final switch time of 6 s at 5 V/cm, at an angle of 120° and a 14°C buffer temperature (CHEF DR III apparatus; Bio-Rad, Rainach, Switzerland) for 20 h. Lambda Mix 19 (Fermentas) and MidRange I PFG and MidRange II PFG markers (New England Biolabs) were used as DNA size standards.
Electron microscopy.
Purified phage particles recovered from density gradients were negatively stained with 2% sodium phosphotungstic acid (PTA), 2% uranyl acetate (UA), or 2% ammonium molybdate (AM) (67). Samples were observed in a Tecnai G2 Spirit instrument (at 120 kV) equipped with an Eagle charge-coupled-device camera (FEI Company, Hillsboro, OR).
Genome sequencing.
DNA shotgun-libraries were prepared after fragmentation by sonication of purified genomic DNA (Sonopuls; Bandelin Electronics, Berlin, Germany). Fragments of the desired lengths were recovered from agarose gels, DNA ends were repaired (EndIt kit; Epicentre Biotechnologies, Madison, WI), and ligated into EcoRV-digested pBluescript II SK(−) (Stratagene). Ligation products were transformed into E. coli XL1-Blue MRF′. Blue-white screening on ampicillin (100 μg/ml)-, IPTG (isopropyl-β-d-thiogalactopyranoside)-, and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing plates was used to identify plasmid-bearing clones. The presence of inserts was verified by PCR, and inserts were sequenced by using M13forward and M13reverse primers (see Table S2 in the supplemental material). Obtained nucleotide sequences were edited and aligned by using CLC Genomics Workbench (version 3.5; CLC Bio, Aarhus, Denmark) and SeqMan Pro (Lasergene SeqMan Pro version 8.1; DNASTAR, Madison, WI) software. Gaps between contigs were closed by a primer-walking strategy using purified genomic DNA as a template. Primers were derived from the contig sequences as they became available. Sequences with insufficient coverage or ambiguities were verified by primer walking.
Determination of the physical genome structure and packaging mode.
The presence of putative pac sites was evaluated by restriction profiling, which was also used to verify the assembled genome sequences. To exclude the presence of cohesive DNA ends, 0.5 μg of purified genomic DNA was digested with restriction enzymes, heated to 62°C for 10 min, and immediately separated by electrophoresis. A nonheated sample was used as a control. Digestion of genomic phage DNA with Bal31 exonuclease, prior to restriction digestion, was performed as described previously (see above and reference 37). Large terminase amino acid sequences of 86 phages with known DNA structure were compared as described previously (13); sequences were aligned by using the built-in alignment tool of CLC Genomics Workbench, and phylogenetic trees were generated with the neighbor-joining algorithm (1,000 bootstrap replicates).
Mass spectrometry (MS) and peptide mass fingerprinting.
Phage proteins were separated by horizontal sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on gradient gels (ExcelGel SDS, 8 to 18% gradient; GE Healthcare, Glattbrugg, Switzerland) as previously described (37, 74). Major protein bands were excised and, after tryptic in-gel digestion, analyzed by liquid chromatography-tandem MS and electrospray ionization-tandem MS to determine masses of the fragments (37, 74). The data were analyzed by using the Scaffold Proteome Software (version 2.0; Proteome Software, Inc., Portland, OR).
Identification of attB and attP.
In order to obtain stable lysogens, the respective Brochothrix host cells were infected with phage NF5 or BL3 for 1 h at a multiplicity of infection (MOI) of 1,000 in liquid culture at 24°C. Serial dilutions were plated, and surviving colonies were restreaked onto fresh agar plates. Selected lysogens were assayed for immunity against infection with the same phage (homoimmunity screening) and for the presence of phage-specific DNA sequences by PCR. B. thermosphacta HER1187::BL3 and HER1188::NF5 chromosomal DNA was purified (60). BL3 and NF5 integration sites (one transition position) were identified as previously described (54), using two divergent primers BL3_iPCR_fwd/BL3_iPCR_rev and NF5_iPCR_fwd/NF5_iPCR_rev, derived from the putative BL3 and NF5 phage integrase genes, respectively. Primers are listed in Table 2 in the supplemental material. Lysogen DNA was digested with PvuII (BL3) and HhaI (NF5) and self-ligated using T4 DNA ligase (New England Biolabs), and inverse PCR products were sequenced. Nonphage sequences were identified by alignments with the phage genomes. Second transition positions from phage to host were identified accordingly by using the primer pairs BL3_iPCR_2_rev/BL3_iPCR_2_fwd and NF5_iPCR_2_rev/NF5_iPCR_2_fwd, respectively. To confirm prophage locations, the transition regions between phage and host were PCR amplified and sequenced by using the primer pairs BL3_iPCR_to_int/BL3_iPCR_rev and BL3_iPCR_2_fwd/BL3_iPCR_to_ORF27_c2 for the BL3 prophage and NF5_iPCR_rev/NF5_iPCR_to_int and NF5_iPCR_2_fwd/NF5_iPCR_to_ORF24_v2 for the NF5 prophage. For final identification of attP, PCR using the primers BL3_iPCR_to_int/BL3_iPCR_to_ORF27_c2 and NF5_iPCR_to_int/NF5_iPCR_to_ORF24_v2 was performed, using genomic DNA of phage-free hosts as templates.
Bioinformatics and sequence submission.
Nucleotide and amino acid sequence manipulations were performed by using CLC Genomics Workbench (version 3.5; CLC Bio). Local sequence alignments were conducted by using the BLASTn, BLASTx, and BLASTp programs available at the National Center for Biotechnology Information website (4) or the CLC Genomics Workbench built-in blast engine. The alignment and pairwise comparison tools of CLC Genomics Workbench were used for multiple, global protein sequence alignments. The protein signatures of selected gene products were identified by InterProScan (http://www.ebi.ac.uk/Tools/InterProScan) (72) or by comparison to the Pfam database (Pfam 24.0) (21). tRNAs were predicted with tRNAScan SE (51), and GC content was calculated by using OligoCalculator (http://mbcf.dfci.harvard.edu/docs/oligocalc.html). Rho-independent bacterial transcription terminators were predicted by using Softberry FindTerm (http://www.softberry.ru/berry.phtml?topic=findterm&group=programs&subgroup=gfindb) using an energy threshold value of −11 (default setting). Hairpin stem-loop structures were predicted by using DNAsis Max 2.6 (Miraibio, San Francisco, CA), and RNA secondary structure was calculated by using CLC Genome Workbench. Nucleotide sequences of NF5 (HM144385), BL3 (HM144386), and A9 (HM242243), and partial sequences obtained from B. thermosphacta HER1187 (HM569776) and HER1188 (HM569777) have been deposited at GenBank under the accession numbers listed above.
RESULTS
Bacteriophages BL3, NF5, and A9 feature distinct morphologies.
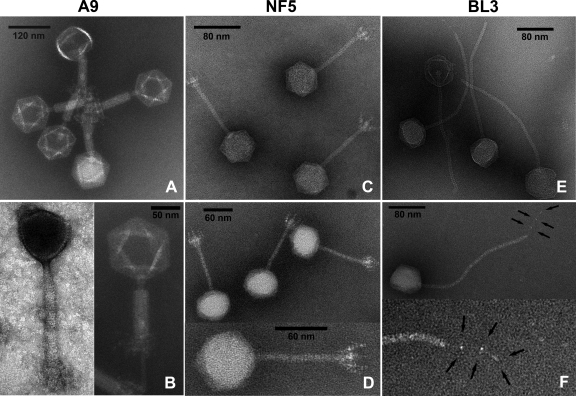
Transmission electron micrographs of phages A9, BL3, and NF5 were obtained from purified lysates (Fig. 1). A9 features an isometric head (89 nm) and a contractile, nonflexible tail (171 nm) with a complex baseplate structure (1). Upon contraction, the tail tube is exposed and a double baseplate structure becomes visible, as described earlier (1). Such extensive rearrangements of the distal tail structure have been described for a group of phages, such as A511, P100, K (37), LP65 (14), Bastille (35), φ812 (57), and others (38). Purified A9 particles all exhibited contracted tails; the phage seems sensitive to the force generated by ultracentrifugation. Electron micrographs of noncontracted virus particles were obtained from a filtered lysate (Fig. 1, left side of panel B). Phages BL3 and NF5 are members of the Siphoviridae family, featuring icosahedral heads and noncontractile tails. BL3 has a head diameter of 58.9 ± 2.4 nm and a long, flexible tail (length, 269.9 ± 4.8 nm; diameter, 10.4 ± 0.2 nm) with a single tail fiber (58.4 ± 2.0 nm) (Fig. 1F). The overall architecture is very basic, without recognizable fine structures. NF5 exhibits a smaller head diameter (55.7 ± 0.8 nm) with a short and rigid tail (length, 94.0 ± 2.0 nm; diameter, 8.7 ± 0.2 nm). In contrast to BL3, NF5 features a complex baseplate structure with appendages (Fig. 1C and D, Table 1).
FIG. 1.
Electron micrographs of A9, NF5, and BL3. (A) A9 virions with contracted tails show double baseplate structures. Head symmetry is hexagonal; the darker heads are empty. Samples are negatively stained with phosphotungstic acid (PTA). (B) Close-up views of phage A9 with uncontracted (left panel, uranyl acetate [UA] stain) and contracted (right panel, PTA stain) tails; the thin whiskers are visible in the contracted state. Ammonium molybdate (AM) (C)- and PTA (D)-stained NF5 virions reveal a siphovirus morphology, with noncontractile tails and complex baseplates with appendages. (E) Phage BL3 (AM stained) is a siphovirus with a long flexible tail. (F) A single tail fiber with three characteristic nodes is visible in PTA-stained BL3 samples (indicated with arrows).
TABLE 1.
General characteristics of B. thermosphacta bacteriophages
| Name | Virus family | Viral species | Lifestyle | Capsid size (nm) | Tail length (nm) | G+C content (mol%) | Genome size (bp) | Genome structurea | No. of ORFs/no. of tRNAs |
|---|---|---|---|---|---|---|---|---|---|
| BL3 | Siphoviridae | BL3 | Temperate | 59 | 270 | 36 | 41,518 | tr, pp | 65/1 |
| NF5 | Siphoviridae | NF5 | Temperate | 56 | 94 | 37 | 36,953 | tr, pp | 57/0 |
| A9 | Myoviridae | A19 | Virulent | 89 | 171 | 41 | ∼138,000 | tr, np | 198/6 |
np, nonpermuted; pp, partially permuted; tr, terminally redundant.
Phage ecology and infection.
BL3 and NF5 were induced from lysogenic strains in France (1), whereas A9 was isolated from spoiled beef in Canada (27). A limited host range analysis was performed using the propagation strains HER1187 and HER1188, six B. thermosphacta strains isolated from pork loins (24), 13 strains isolated from beef loins (24), and 7 strains isolated from fish (see Table S1 in the supplemental material). As expected for temperate phages, BL3 and NF5 display a quite narrow host range. In contrast, phage A9 was able to lyse 12 of the 28 tested strains. Its broader host range is typical for virulent phages and has been reported earlier (27). None of the phages were able to infect any of the other tested Gram-positive species (see Table S1 in the supplemental material). A9 features a latency period of 85.3 ± 1.5 min, with a burst size of 81.6 ± 13.1.
Complete nucleotide sequences, genomic organization, and bioinformatics.
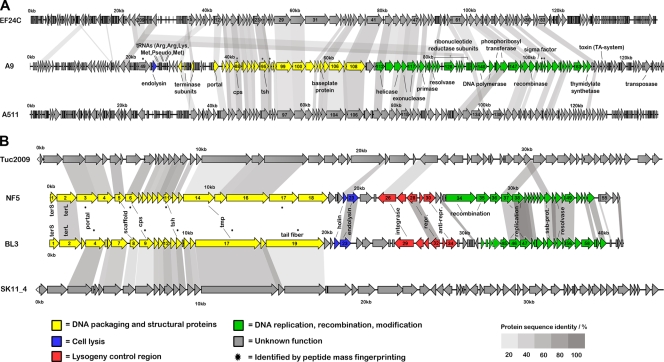
A graphical representation of the phage genomes is shown in Fig. 2, detailed annotations are provided as Tables S3 (BL3), S4 (NF5), and S5 (A9) in the supplemental material. Table 1 presents an overview.
FIG. 2.
Genome map alignments of Brochothrix phage A9, Listeria phage A511, and Enterococcus phage EF24C (A) and Brochothrix phages BL3 and NF5, Lactococcus phage Tuc2009, and prophage SK11_4 (B). Arrows indicate identified ORFs and are drawn to scale. Gene products with sequence identities ≥20% are linked by shaded areas. Color codes for functional clusters apply to Brochothrix bacteriophages only. Putative functions of selected gene products are indicated (see Tables S3 to S5 in the supplemental material). Abbreviations: terS and terL, small and large terminase subunits; scf, scaffold; cps, major capsid; tsh, tail sheath; tmp, tape measure; int, integrase; rep, repressor; ssb, single strand binding.
BL3 genome.
Shotgun sequencing and primer walking produced a genome length of 41,518 bp, which is in agreement with restriction enzyme digests. One tRNA gene and 65 open reading frames (ORFs) were identified, which cover 95.1% of the genome. Ribosomal binding sites (RBS) were identified for most ORFs, located 6 to 10 nucleotides (nt) upstream of the start codon and resembling those known for Listeria (AGGAGGTG [25, 61]). Significant BLAST hits were found for 46 gene products, and a putative function could be assigned to 22 (see Table S3 in the supplemental material). As expected for members of the Siphoviridae, their genome is organized into defined functional clusters, reflecting the phage life cycle. Starting from the small terminase subunit gene (gp1), these clusters comprise: (i) DNA packaging and structural protein cluster flanked by terminase and tail fiber (gp19) genes and (ii) cell lysis function consisting of a holin (gp22) with two predicted transmembrane domains and an endolysin (gp23) with N-terminal N-acetylmuramoyl-l-alanine amidase domain (InterPro ID IPR002502) and a C-terminal bacterial SH3 domain (IPR003646). The endolysin gene was cloned and purified (50), and the mureolytic activity of the product against B. thermosphacta HER1187 was demonstrated (data not shown). (iii) The lysogeny control module, encoded on the negative strand, includes integrase (gp29), repressor (gp32), and antirepressor (gp34), and (iv) the DNA replication, recombination, and modification cluster is located between gp41 (with a HTH motif, SM00530) and gp61.
NF5 genome.
The genome of phage NF5 is 36,953 bp in size, in agreement with restriction profiles, and 57 ORFs were found (see Table S4 in the supplemental material). It features a coding capacity of 98.4% and is also organized into functional modules. (i) DNA packaging and structural genes encompass all genes between the small terminase subunit (gp1) and gp18. Three large ORFs (orf16, orf17, and orf18) reflect the more complex baseplate and tail appendage architecture compared to phage BL3 (Fig. 1C and D). (ii) The lysis cassette (gp22-23) contains a holin protein (gp22) with a phage_holin_1 family domain (Pfam ID PF04531 [30]) and an endolysin (gp23) with an N-terminal N-acetylmuramoyl-l-alanine amidase domain, which is almost identical to the BL3 amidase, whereas the C-terminal cell wall binding domain is distinct. Again, the lytic activity of recombinant NF5 endolysin against host cells could be experimentally confirmed (data not shown). (iii) The lysogeny control region (gp26-30) is flanked by the integrase (gp26) and the antirepressor gene (gp30), and (iv) the replication, recombination, and modification cluster is defined by gp34, containing an N-terminal RecF/RecN/SMC_N domain (PF02463) and by gp53, a putative transcription regulator.
A9 genome.
Sequencing of phage A9 revealed an information genome of ∼127 kb, which is significantly shorter than that estimated by PFGE (Fig. 3 A). A total of 198 ORF and 6 tRNA genes were annotated, resulting in a coding capacity of 91.2%. Significant homologies were found for 88 gene products and putative functions were assigned to 34 (see Table S5 in the supplemental material). The overall A9 genome is structured in three blocks with opposite transcription directions: orf3-28, orf29-76, and orf77-196. The second block is interrupted by three short regions with inverted transcription direction (orf49-52, orf56-60, and orf66-71) (Fig. 2A). Two major functional clusters can be defined. (i) The DNA packaging and structural gene cluster is flanked by gp83 (portal protein) and gp108, a putative tail protein with homology to phage Lb338-1 gp134 (3). Surprisingly, the putative large terminase genes seems to be located away from the structural gene cluster and encoded on the opposite strand. Three gene products (gp63, gp65, and gp72) display homology to large terminase proteins of SPO1-related phages (Spounavirinae). (ii) The replication and DNA metabolism cluster is flanked by a putative helicase (gp112) and a thymidylate synthetase (gp165). gp171 displays homology to toxin components of toxin-antitoxin systems and contains an N-terminal PemK-like protein domain (PF02452) also found in other toxin components such as MazF (73). The endolysin gene product (gp52) with an N-terminal N-acetylmuramoyl-l-alanine amidase domain was identified, and the lytic activity of the recombinant protein was experimentally demonstrated (data not shown).
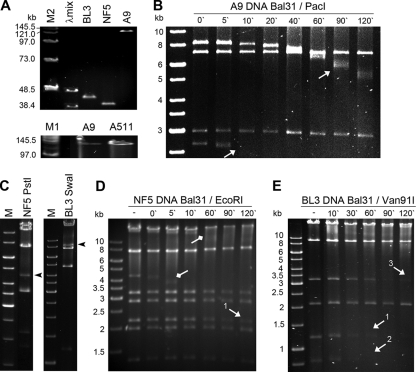
FIG. 3.
Structure analysis of the DNA molecules of phage A9, NF5, and BL3. (A) PFGE of full-length phage DNA. Size markers: λmix, lambda mix marker 19 (Fermentas); M1 and M2, PFG midrange markers I and II (NEB). BL3 and NF5 genomic DNAs run at slightly larger sizes than their unit genome lengths of 41.3 and 37 kb. A9 DNA exhibits approximately the same physical size as Listeria phage A511 DNA (137.62 kb) (37). (B) Gel electrophoresis of PacI-digested A9 DNA after Bal31 nuclease treatment for the indicated time intervals. Terminal fragments of the nonpermuted genomes disappear over time (indicated by arrows). (C) Restriction enzyme profiles of PstI-digested NF5 DNA and SwaI-digested BL3 DNA; the putative pac fragments are indicated by black arrowheads. (D and E) EcoRI (NF5)- and Van91I (BL3)-digested genomic DNA after Bal31 nuclease treatment for the indicated time intervals. Arrows with square heads indicate restriction fragments which were not predicted in silico from a circular molecule (pac fragment, 4.2 kb; smeary band, >10 kb). The “submolar” pac fragment disappears after 10 min of Bal31 treatment. Arrows with pointed heads indicate restriction fragments “downstream” from the putative pac site-containing fragment. Their consecutive appearance within the respective genome is indicated by numbering and correlates with the order of disappearance, indicating incomplete permutation of the DNA molecules based on the limited number of genomes in each replication concatemer.
The sequence between nucleotide positions 34900 and 36100 is highly ambiguous. It is characterized by the presence of large inverted repeats and long TAAA repetitive sequences located around nt 35420. Interestingly, when this fragment is PCR amplified and sequenced, it is ∼1 kb shorter than the corresponding restriction fragment and lacks most of the repeats. When sequencing directly on genomic DNA, said repeats are present, but the sequencing quality is poor, making it impossible to sequence the complete region from the flanking nonrepetitive sections. Therefore, the size of the A9 physical genome could only be estimated with ∼1 kb uncertainty.
BL3 and NF5 feature partially permuted genomes, while the A9 genome is nonpermuted and terminally redundant.
Elucidation of the DNA packaging mechanism was approached experimentally and in silico. Heating of restriction fragments did not alter the separation pattern, suggesting the absence of cohesive (cos) genome ends (data not shown). For phages BL3 and NF5, PFGE also indicated the presence of short terminal repeats (Fig. 3A). Restriction fragment patterns of NF5 were in agreement with in silico predictions from a circular molecule, except for some “submolar” bands observed, which indicate so-called pac site-containing fragments (Fig. 3C). These are likely generated from the first series initiation cleavage of the concatemeric phage DNA molecule during headful packaging, and the presence of such fragments is diagnostic for pac phages which feature partially permuted genomes (12). Similar evidence found in restriction patterns of phage BL3 DNA (Fig. 3C) also strongly suggests the usage of a headful packaging mechanism, although the predicted pac fragment was mostly overlaid by other restriction fragments or too small to be visible in gel electrophoresis (12). The pattern observed in a Van91I digest of Bal31 treated BL3 DNA (Fig. 3E) is characteristic for a partial permutation, which is to be expected, when the size of the terminal redundancy and the length of the concatemer is limited (12). An EcoRI digest of Bal31-treated NF5 DNA indicates similar findings for this phage (Fig. 3D). Putative pac fragments of NF5 and BL3 were extracted from restriction digests and sequencing confirmed the assumed location, at the small terminase subunit-encoding gene, as described for other phages (12; data not shown). The finding that both phages are related to Sfi11-type pac site phages of Streptococcus thermophilus (see below) and feature the characteristic presence of a scaffolding protein gene between portal and major head protein genes, as well as the absence of proteolytic processing of the major capsid protein (see below), yields further support for the proposed genome structure (9) (see Fig. S2 in the supplemental material).
The A9 DNA molecule has a structure and size similar to that of Listeria phage A511 (137.62 kb, Fig. 3B), strongly suggesting redundant termini. Exonuclease treatment, followed by digestion of genomic A9 DNA with XcmI, Van91I, or PacI, revealed that two fragments are consecutively shortened (Fig. 3), indicating a DNA molecule structure with invariable, fixed ends. The estimated 11-kb terminal redundancy is in perfect agreement with restriction patterns (37).
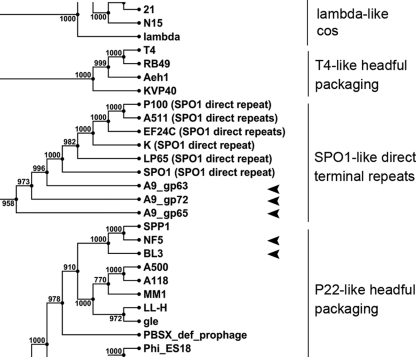
The genome structure and packaging strategy of bacteriophages largely depends on the function of the terminase holoenzyme. Large terminase amino acid sequences reliably cluster into groups with similar packaging strategies when aligned (13) (Fig. 4). Both BL3 and NF5 clustered with P22-like headful packaging phages, whereas all three putative gene products annotated as large terminase fragments in phage A9 clustered with terminases of SPO1-related phages, also confirming the experimentally determined genome structures for all three phages.
FIG. 4.
Phylogenetic relationships of the large terminase proteins of the three sequenced Brochothrix phages, including other phages with known packaging mechanisms (13). Groups with similar packaging strategies are indicated. The tree is based upon neighbor joining with 1,000 bootstrap replicates, calculated from an alignment with the following parameters: gap open cost, 10; gap extension cost, 1; and end gap cost, free (CLC Genomics Workbench).
Programmed translational frameshifts produce C-terminally modified structural proteins.
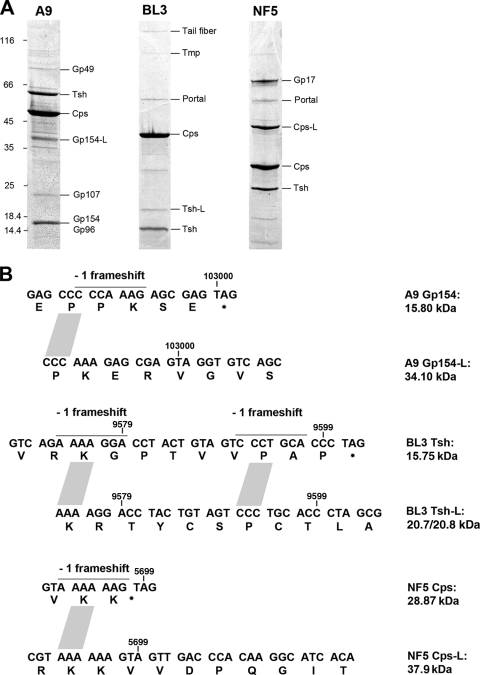
Structural proteins of the virus particles were analyzed by peptide mass fingerprinting (Fig. 5; see also Table S7 in the supplemental material). The most prominent bands generally represent major capsid (Cps) and major tail (Tsh) proteins. Bioinformatic analysis revealed potential −1 programmed translational frameshift sites at the 3′ end of Cps and Tsh-encoding genes, producing normal-length and larger (-L) protein variants of NF5 Cps (gp 6.1) and BL3 Tsh (gp14.1 and gp14.2). The in silico predicted masses of Cps (28.87 kDa), Cps-L (37.9 kDa), Tsh (15.75 kDa), and Tsh-L (20.7 or 20.8 kDa, two possible frameshift positions) correspond to the observed masses (Fig. 5). The Cps −1 frameshift in phage NF5 occurs at the 3′ end of the transcript when the ribosome reaches the AAA lysine codon of the consensus heptanucleotide slippery sequence (A.AAA.AAG → AAA.AAA.G; Fig. 5B) (56) and was experimentally confirmed by identification of tryptic fragments specifically generated from the longer protein variant Cps-L (see Table S6 in the supplemental material). Two potential −1 frameshift positions identified at the 3′ end of the tsh gene of phage BL3 do not follow the standard consensus heptanucleotide slippery sequence, and only a small amount of frameshift product was observed in the virion (Fig. 5). In phage A9, gp154 and gp155 could be identified as structural components, although located outside the structural gene cluster. A similar situation has been reported for other phages (20, 37). A perfect heptanucleotide slippery sequence (Fig. 5B) is present at the 3′ end of gp154, indicating a translational frameshift that results in a gp154/155 fusion protein (gp154-L). gp154 (15.8 kDa)- and gp154-L (34.10 kDa)-specific peptides were present in the corresponding protein bands, confirming the occurrence of this type recoding event.
FIG. 5.
Analysis of A9, BL3, and NF5 protein composition. (A) Identification of virus structural proteins by peptide fingerprinting. Protein molecular mass marker masses (in kilodaltons) and identified gene products are indicated. Abbreviations: Tsh, tail sheath protein; Cps, major capsid protein; Tmp, tape measure protein; gp, gene product; -L, longer protein variants produced by programmed translational frameshifting. (B) Proposed locations of frameshifts in structural protein genes of Brochothrix bacteriophages. The type of frameshift, location within the genome, and slippery sequences (lines) are indicated. The putative frameshifts in phages NF5 and A9 were experimentally verified by peptide mass fingerprinting (see Table S6 in the supplemental material).
Phages BL3 and NF5 utilize novel prophage insertion sites.
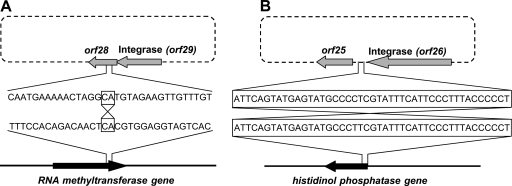
BL3 and NF5 integration sites were identified by an inverse PCR approach (54), since a Brochothrix genome sequence is not available. Analysis of the prophage flanking host sequences revealed that BL3 integrates into the 3′-end of a conserved RNA methyltransferase, partially reconstituting the remaining C-terminal amino acid residues (HVEVVTAFTLVD in the host versus HVEVVCALTLIE in the prophage). Similar methyltransferases are found in related Gram-positive organisms such as Bacillus mycoides (NZ_CM000744) or Listeria monocytogenes EGD-e (25), with similar C-terminal ends (HVECVAWLELV and HIEAVTVLHLN). The BL3 integrase recognizes a core region of only two identical nucleotides, flanked by inverted repeats both in the phage and in the host genome (Fig. 6 A). NF5 integrates at the 5′ end of a putative histidinol-phosphatase gene with homology (over the available 24 amino acids) to corresponding proteins from Lactobacillus and Listeria (Fig. 6B). Integration reconstitutes the gene including its ribosomal binding site. The site-specific recombinase of NF5 recognizes a region of identity extending over 45 nt between attP of NF5 and attB of B. thermosphacta HER1188. The intergenic sequence downstream of the NF5 integrase was not predicted to be able to form any potential secondary structure (stem-loop) that could function as a transcription terminator. This was a surprising observation, since other phages (BL3, A006, A500, A118, B025, ul36, and Tuc2009) generally feature transcription terminators located in between the 3′ end of the integrase gene and attP, with a free energy ΔG of −18.6 to −40.3 kcal/mol (details not shown).
FIG. 6.
Detail view of BL3 (A) and NF5 (B) integration site in the HER1187 and HER1188 genome, respectively. Regions of identity between phage and host sequence (core sequences) are boxed.
BL3 and NF5 share homologies and are related to the P335 quasispecies of Lactococcus phages.
Genomes of BL3 and NF5 were compared to each other and to a set of six Listeria phages (18, 49, 74), four Lactococcus phages (8, 40, 41), and four Lactococcus prophages (70) (see Fig. S1 and S2 in the supplemental material). Although NF5 and BL3 feature homologies primarily in the early gene region, no similarities to any of the Listeria phages could be identified, even though there is remarkable synteny in the genome organization. Instead, both Brochothrix phages reveal homology to distinct subgroups of the heterogeneous P335 quasispecies of Lactococcus phages (see Fig. S2 in the supplemental material). This also is restricted to the structural gene cluster and includes gene products of prophage SK11_4 and t712 for BL3, as well as phages Tuc2009, TP901-1, P335, ul36, and prophages SK11_2 and SK11_3 for NF5. With respect to their overall genome organization, all of these lactococcal phages and prophages were previously shown to be Sfi11-like Siphoviridae or demonstrated to be related to these phages (9, 40, 70). Therefore, we propose that BL3 and NF5 are also members of this group, which agrees well with the proposed headful packaging mechanism. Global protein sequence alignment confirmed the homology to P335 phage Tuc2009 (for NF5) and prophage SK11_4 (for BL3) (Fig. 2B). BL3 and NF5 share amino acid homologies mainly in early gene products and in TerL, endolysin (conserved amidase domain) and repressor. The homologies are not randomly distributed but rather focused in connected clusters, indicating a genome mosaicism, which is typical for many bacteriophage genomes, and likely originating from extensive horizontal gene transfer during evolution and adaptation (32).
A9 is a member of the large, virulent SPO1-related Myoviridae (Spounavirinae).
Assignment of phage A9 to the SPO1-related Myoviridae of the proposed new subfamily Spounavirinae (38, 44) is based on the following evidence: A9 (i) infects a low GC content, Gram-positive host (Firmicutes), (ii) is strictly virulent, (iii) displays a relatively broad host-range within the bacterial genus, and (iv) features a double-stranded DNA genome with invariable terminal repeats. Members of this subfamily share a common morphology, comparable genome sizes, marked synteny in their genome organization, and pronounced protein sequence relatedness (37, 38, 44). In A9, these homologies are largely confined to structural and few early gene products (see Table S5 in the supplemental material). An alignment of phage A9 with the Spounavirinae A511 (37) and EF24C (69) indicates striking colinearity in genome organization (Fig. 2A).
DISCUSSION
Genome sequencing and molecular characterization of representatives of each of the three known Brochothrix bacteriophage species provided basic information on morphology, genome organization, mode of DNA packaging, and integration into the host chromosome and revealed interesting details such as the occurrence of programmed translational frameshifts for synthesis of C-terminally modified structural proteins. The information genomes of the two small temperate siphoviruses BL3 and NF5 span 41,518 and 36,953 bp, respectively. The viral DNA is incorporated into the empty capsids via a headful packaging mechanism, producing a collection of circularly permuted, terminally redundant DNA molecules. The overall organization into defined functional modules is similar in both phages and resembles the well-known genome arrangements found in most temperate phages infecting the Firmicutes, mirroring the phage's life cycles (9, 18). The clearly different morphology of BL3 and NF5 (Fig. 1) is also reflected in a lack of amino acid sequence homology in the structural proteins (Fig. 2B). Sequence homologies dominate among the early genes, suggesting analogous host takeover and genome replication mechanisms. Such gene products are encoded by groups of adjacent genes, rather than being randomly dispersed over the genome (Fig. 2B). This pattern is indicative for a horizontal exchange of modules during evolution of these phages, which typically results in highly mosaic virus genomes (11, 33, 34). Both phages seem to lack genes coding for DNA polymerases and helicases and therefore depend on host enzymes for their replication. DNA and amino acid sequence alignments and the overall organization of structural genes indicate that both BL3 and NF5 are members of the Sfi11-like, pac site containing Siphoviridae (9, 59), which are structurally related to different subgroups of the heterogeneous P335 quasispecies of Lactococcus phages (40, 70). Brochothrix phages are strictly genus specific, and the cross-genus sequence homologies therefore suggest that any such genetic exchange probably occurred in the distant past or that the relationship is based on divergence from a common ancestor. Interestingly, only very limited amino acid sequence homologies to Listeria phages were observed, even though the host bacteria are closely related (16, 52) (see Tables S3 and S4 in the supplemental material). Assuming that horizontal gene transfer and exchange mechanisms are more important than vertical passage during the evolution of these phage genomes, it is tempting to speculate that phages sharing a common ecological niche are more likely to show relatedness than phages infecting phylogenetically related hosts. This might hold true for the relatedness of Brochothrix phages to phages infecting Lactococcus. However, other findings suggest that phage relatedness reflects the relatedness of their bacterial hosts (suggesting coevolution), at least for members of the Sfi21-like genus (9). Moreover, because these homologies are mostly restricted to structural proteins, relatedness may also reflect various degrees of a diverging (but still similar) morphology rather than being the result of constant selection based on functional requirements.
Integration of viral DNA into the host chromosome by site-specific recombination is essential for propagation of the prophage. Recombination is mediated by phage-encoded integrases (29, 64). The occurrence of highly conserved amino acids indicate that the BL3 integrase is a member of the serine recombinase family, which recognizes the only 2-bp core sequence shared in attB and attP but may also require additional recognition signals such as inverted repeats flanking the core sequence (64; Margaret Smith, unpublished data). In contrast, the NF5 integrase belongs to the tyrosine recombinases, which generally recognize longer core sequence, in this case 45 bp. Of particular interest is that both phages integrate into unique loci not previously known to be utilized for phage insertion. Phage NF5 inserts into the 5′ end of a putative histidinol-phosphatase gene, which first appeared to disrupt the gene. Upon closer inspection, however, it became evident that the 45-bp attachment site overlaps with the start codon, and phage integration does reconstitute the functional coding sequence with only one nucleotide exchange (silent) at the opposite end of the prophage. Moreover, since the NF5 integrase gene is apparently not followed by any transcription terminator, it appears that in the lysogenized host, expression of the histidinol-phosphatase is then driven by transcriptional coupling of the host gene to the phage integrase expression system.
Integration of the BL3 genome occurs close to the 3′ end of a putative RNA-methyltransferase gene but reconstitutes the interrupted reading frame to yield a product highly similar to the original polypeptide, probably resulting in a functional gene product. Such “reconstitutive” integration has been described for other temperate phages (10, 18, 53), including Listeria phage PSA, which inserts into the 3′ end of a tRNA coding sequence (74), and for B054, which integrates into the 3′ end of a transcription elongation factor gene (18). It is reasonable to assume that reconstitutive integration into specific and conserved sequences has evolved to secure insertion site sequences and, at the same time, maintains host fitness upon prophage insertion, which is beneficial for both participants.
Programmed translational frameshifting is not uncommon among bacteriophages, and alternative decoding is used to generate variant proteins utilized in phage morphogenesis. The mechanism has been reported for various phages, including A2, T3, and T7 (reviewed in reference 6), and a number of Listeria phages (18, 74). Recoding is quite frequent in siphoviruses and might have evolved to keep genomes small by increasing the coding capacity, which is surprisingly high in both phages. An elegant explanation for the functional role in capsid morphogenesis has been proposed for Listeria phage PSA. The longer frameshift variants are believed to produce pentameric ring structures of the capsomers, while the shorter variants produce the hexameric structures (74). We observed here a −1 translational frameshift site in the NF5 cps gene, featuring a classic X.XXY.YYZ heptanucleotide mRNA slippery sequence at which the ribosome can slip back on the mRNA (Fig. 5B) (71). However, the precise role of the longer polypeptide in head morphogenesis remains to be elucidated.
Programmed translational frameshifts in phage tail protein genes have also been frequently reported, and a role of the modified products in correct assembly of the tail was suggested (15, 18, 46, 71). We found a −1 frameshift site in tsh of BL3 (orf14; Fig. 5B), which does not follow the classic X.XXY.YYZ slippery sequence. However, not all phages with a verified −1 translational frameshift feature this consensus sequence. For instance, phage C31 features a slippery sequence of the type X.XXX.YYX (G.GGG.AAG) (63, 71), which is similar to the BL3 motif (A.AAA.GGA; Fig. 5B). Thus, it is likely that a frameshift in BL3 tsh occurs at the slippery sequence located between nt 9573 to 9579. The presence of a nonconsensus site might explain why only a small amount of frameshift product is detected in the mature virion (Fig. 5A). Again, the exact function of the frameshift products in the BL3 tail remains unclear, and yet an essential role for correct tail assembly may be assumed, similar to the situation in phages Lambda (46) and SPP1 (5).
Bacteriophage A9 is different from BL3 and NF5; it is a virulent (obligately lytic) virus and features an information genome of 127 kb with a large terminal redundancy of ∼11 kb. Genome organization, protein homologies, morphology, and lifestyle of the phage clearly place it within the recently proposed Spounavirinae subfamily of the Myoviridae. The terminally redundant, nonpermuted genome of A9 further strengthens the hypothesis that this genome structure is a characteristic hallmark of the Spounavirinae (37, 68). As previously shown for other phages of this subfamily (14, 37, 68, 69), the genome is organized into two clusters with discrete function, the structure-associated genes (gp83-gp108), and DNA synthesis- or replication-associated genes (gp112-gp165). When A9 is compared to other Spounavirinae, the gene synteny among the two clusters is clearly visible and yet not exhaustive; the loci of genes coding for tRNAs, endolysin, terminase, and ribonucleotide reductase appear to be particularly variable. The ribonucleotide reductase genes of A9 and A511 are located at similar sites within their genomes, which, however, are different from other family members such as phages phiEF24C (69) (Fig. 2A) or SPO1 (68).
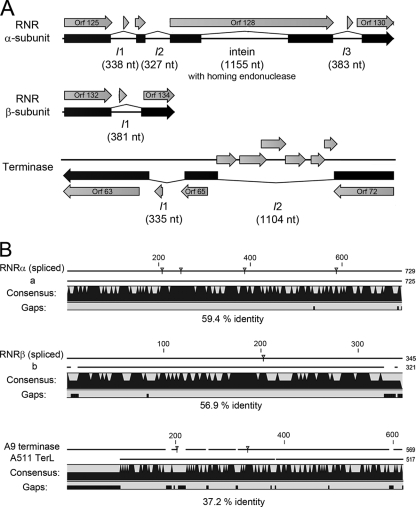
Another striking observation was the presence of large terminase subunits encoded in opposite transcription direction upstream of the portal protein in phage A9. Recent analyses in our lab have revealed such an unusual genome structure to be conserved among several SPO1-related Bacillus phages (unpublished data). Considering the strict genus specificity of A9, any cross-genus genetic exchange events probably occurred in the distant past, or represent the result of infrequent illegitimate recombination. Interestingly, genes exhibiting such variable positions within phage genomes are frequently interrupted by intervening sequences, often observed within genes involved in DNA metabolism (19). Several Spounavirinae have been shown to carry self-splicing group I introns or inteins in DNA-polymerase genes (phages G1, K, and SPO1), ribonucleotide-reductase genes (phage Twort), DNA helicase genes (phage Twort, intein), endolysin genes (phages G1 and K), and large terminase subunits (phage Twort) (39, 42, 43, 55). With respect to A9, sequence alignments indicated intervening sequences in both the large terminase and the ribonucleotide reductase (RNR) genes (Fig. 2A and 7A). Both are located at positions not previously described for SPO1-related phages. In fact, the putative A9 large terminase gene possibly contains two introns, which would explain the presence of three separate ORFs with partial terL homologies. A similar situation was reported for phage Twort (39), where the large terminase gene is also interrupted by two introns. The putative type I RNR of A9 is encoded on ORFs 125, 127, 128, 130, 132, and 134, with amino acid sequence homologies to the RNR alpha (nrdE-like) and beta (nrdF-like) subunits, respectively (Fig. 7; see Table S5 in the supplemental material). In addition, the phage encodes an nrdI-like (orf124) and an nrdH-like protein (orf135) (Table S5 in the supplemental material), all of which are typically found in type Ib RNR enzymes (62). The alpha subunit appears to be interrupted by three introns and one intein, located within orf128, and encoding a homing endonuclease. One putative intron could be identified in the RNR beta-subunit gene. Introns and inteins in RNR genes have previously been observed in phage (e.g., Twort and several Bacillus prophages [42, 45]) and in bacterial genomes (47). Whether the occurrence of introns and inteins has a regulatory role or whether these genes are particularly susceptible to the insertion of intervening sequences is not known (17).
FIG. 7.
Identification of intervening sequences (IVS) in the A9 genome. (A) Splicing variants producing the most significant database hits were annotated. The putative intein was identified by Blastp. RNR, ribonucleotide reductase. (B) Alignment of the spliced proteins with homologous sequences which do not contain IVS (a, RNRα subunit of Granulicatella elegans ATCC 700633; b, RNRβ subunit of Enterococcus casseliflavus EC30; A511 TerL, large terminase of Listeria phage A511). Amino acid sequence identities and gaps in the alignment are indicated. Small triangles mark the putative processing sites.
A −1 translational frameshift is programmed into the coding sequence and mRNA for gp154, a protein identified as a component of the A9 virion. This alternative recoding event is especially interesting, since the coding sequence is not located within the structural gene cluster, and its expression therefore is probably not driven by a late promoter. In addition, gp154-L contains a bacterial Ig-like domain (PF02368 [36]), raising the question of whether it might be involved in nonspecific binding of host surface structures involved in phage adsorption. The role of virion-associated Ig-like domains in other tailed phages was recently reconsidered, and an accessory role during infection was proposed (22). Apparently, several of these domains are produced by translational frameshifting (22).
In conclusion, the characterization and genome sequencing of three different phages infecting the genus Brochothrix revealed a diversity of morphotypes, genetic make-up, and relationship to phages of other Firmicutes. Our data support the classification of Brochothrix phages into at least three families, as proposed by Ackermann et al. (1). Although phage A9 is a member of the large but poorly characterized group of SPO1-related large myoviruses (Spounavirinae), NF5 and BL3 are small temperate siphoviruses, primarily related to lactococcal phages. All phages feature a high degree of flexibility and mosaicism among their genomes. The availability of more genome sequences will facilitate the development of a reliable classification scheme and enable substantiated evolutionary comparisons between bacteriophages infecting Firmicutes. The presented data on phage A9, NF5, and BL3 highlight the phylogenetic relationships between bacteriophages across genus boundaries from a new angle.
Supplementary Material
Acknowledgments
We are grateful to George Nychas (University of Athens, Athens, Greece) and to Bryan Dilts (Agriculture and Agri-Food Canada, Lacombe, Canada) for the gift of B. thermosphacta strains. We thank Hans-Wolfgang Ackermann (University of Laval, Quebec City, Quebec, Canada) for fruitful discussions; Margaret Smith (University of Aberdeen, Aberdeen, United Kingdom) for helpful discussions on phage integrases; and Rudi Lurz (Max Planck Institute for Molecular Genetics, Berlin, Germany) for electron microscopic images of A9, BL3, and NF5. We are grateful to Vanessa Bernbach for assistance with some experiments.
Footnotes
Published ahead of print on 3 August 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ackermann, H.-W., G. G. Greer, and J. Rocourt. 1988. Morphology of Brochothrix thermosphacta phages. Microbios 56:19-26. [PubMed] [Google Scholar]
- 2.Adams, M. H. 1959. Methods of study of bacterial viruses, p. 443-457. In Bacteriophages. Interscience Publishers, Inc., New York, NY.
- 3.Alemayehu, D., R. P. Ross, O. O'Sullivan, A. Coffey, C. Stanton, G. F. Fitzgerald, and O. McAuliffe. 2009. Genome of a virulent bacteriophage Lb338-1 that lyses the probiotic Lactobacillus paracasei cheese strain. Gene 448:29-39. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auzat, I., A. Droge, F. Weise, R. Lurz, and P. Tavares. 2008. Origin and function of the two major tail proteins of bacteriophage SPP1. Mol. Microbiol. 70:557-569. [DOI] [PubMed] [Google Scholar]
- 6.Baranov, P. V., O. Fayet, R. W. Hendrix, and J. F. Atkins. 2006. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 22:174-181. [DOI] [PubMed] [Google Scholar]
- 7.Borch, E., M. L. Kant-Muermans, and Y. Blixt. 1996. Bacterial spoilage of meat and cured meat products. Int. J. Food Microbiol. 33:103-120. [DOI] [PubMed] [Google Scholar]
- 8.Brondsted, L., S. Ostergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 9.Brüssow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 10.Bruttin, A., S. Foley, and H. Brussow. 1997. The site-specific integration system of the temperate Streptococcus thermophilus bacteriophage phiSfi21. Virology 237:148-158. [DOI] [PubMed] [Google Scholar]
- 11.Casjens, S. R. 2005. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8:451-458. [DOI] [PubMed] [Google Scholar]
- 12.Casjens, S. R., and E. B. Gilcrease. 2009. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions, p. 91-111. In M. R. J. Clokie and A. M. Kropinski (ed.), Bacteriophages: methods and protocols, vol. 2. Molecular and applied aspects. Humana Press, Inc., New York, NY. [DOI] [PMC free article] [PubMed]
- 13.Casjens, S. R., E. B. Gilcrease, D. A. Winn-Stapley, P. Schicklmaier, H. Schmieger, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2005. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 187:1091-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chibani-Chennoufi, S., M. L. Dillmann, L. Marvin-Guy, S. Rami-Shojaei, and H. Brussow. 2004. Lactobacillus plantarum bacteriophage LP65: a new member of the SPO1-like genus of the family Myoviridae. J. Bacteriol. 186:7069-7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie, G. E., L. M. Temple, B. A. Bartlett, and T. S. Goodwin. 2002. Programmed translational frameshift in the bacteriophage P2 FETUD tail gene operon. J. Bacteriol. 184:6522-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins, M. D., S. Wallbanks, D. J. Lane, J. Shah, R. Nietupski, J. Smida, M. Dorsch, and E. Stackebrandt. 1991. Phylogenetic analysis of the genus Listeria based on reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:240-246. [DOI] [PubMed] [Google Scholar]
- 17.Derbyshire, V., and M. Belfort. 1998. Lightning strikes twice: intron-intein coincidence. Proc. Natl. Acad. Sci. U. S. A. 95:1356-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorscht, J., J. Klumpp, R. Bielmann, M. Schmelcher, Y. Born, M. Zimmer, R. Calendar, and M. J. Loessner. 2009. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J. Bacteriol. 191:7206-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgell, D. R., M. Belfort, and D. A. Shub. 2000. Barriers to intron promiscuity in bacteria. J. Bacteriol. 182:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eyer, L., R. Pantucek, Z. Zdrahal, H. Konecna, P. Kasparek, V. Ruzickova, L. Hernychova, J. Preisler, and J. Doskar. 2007. Structural protein analysis of the polyvalent staphylococcal bacteriophage 812. Proteomics 7:64-72. [DOI] [PubMed] [Google Scholar]
- 21.Finn, R. D., J. Mistry, J. Tate, P. Coggill, A. Heger, J. E. Pollington, O. L. Gavin, P. Gunasekaran, G. Ceric, K. Forslund, L. Holm, E. L. Sonnhammer, S. R. Eddy, and A. Bateman. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211-D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser, J. S., Z. Yu, K. L. Maxwell, and A. R. Davidson. 2006. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J. Mol. Biol. 359:496-507. [DOI] [PubMed] [Google Scholar]
- 23.Gardner, G. A. 1981. Brochothrix thermosphacta (Microbacterium thermosphactum) in the spoilage of meats: a review, p. 139-173. In T. A. Roberts, G. A. Hobbs, J. H. B. Christian, and N. Skovgaard (ed.), Psychrotrophic microorganisms in spoilage and pathogenicity. Academic Press, London, England.
- 24.Gardner, G. A. 1966. A selective medium for the enumeration of Microbacterium thermosphactum in meat and meat products. J. Appl. Bacteriol. 29:455-460. [DOI] [PubMed] [Google Scholar]
- 25.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 26.Greer, G. G. 2005. Bacteriophage control of food-borne bacteria. J. Food Prot. 68:1102-1111. [DOI] [PubMed] [Google Scholar]
- 27.Greer, G. G. 1983. Psychrotrophic Brochothrix thermosphacta bacteriophages isolated from beef. Appl. Environ. Microbiol. 46:245-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greer, G. G., and B. D. Dilts. 2002. Control of Brochothrix thermosphacta spoilage of pork adipose tissue using bacteriophages. J. Food Prot. 65:861-863. [DOI] [PubMed] [Google Scholar]
- 29.Groth, A. C., and M. P. Calos. 2004. Phage integrases: biology and applications. J. Mol. Biol. 335:667-678. [DOI] [PubMed] [Google Scholar]
- 30.Grundling, A., M. D. Manson, and R. Young. 2001. Holins kill without warning. Proc. Natl. Acad. Sci. U. S. A. 98:9348-9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagens, S., and M. J. Loessner. 2007. Application of bacteriophages for detection and control of food-borne pathogens. Appl. Microbiol. Biotechnol. 76:513-519. [DOI] [PubMed] [Google Scholar]
- 32.Hatfull, G. F. 2008. Bacteriophage genomics. Curr. Opin. Microbiol. 11:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendrix, R. W. 2003. Bacteriophage genomics. Curr. Opin. Microbiol. 6:506-511. [DOI] [PubMed] [Google Scholar]
- 34.Hendrix, R. W., G. F. Hatfull, and M. C. Smith. 2003. Bacteriophages with tails: chasing their origins and evolution. Res. Microbiol. 154:253-257. [DOI] [PubMed] [Google Scholar]
- 35.Jarvis, A. W., L. J. Collins, and H.-W. Ackermann. 1993. A study of five bacteriophages of the Myoviridae family which replicate on different gram-positive bacteria. Arch. Virol. 133:75-84. [DOI] [PubMed] [Google Scholar]
- 36.Kelly, G., S. Prasannan, S. Daniell, K. Fleming, G. Frankel, G. Dougan, I. Connerton, and S. Matthews. 1999. Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli. Nat. Struct. Biol. 6:313-318. [DOI] [PubMed] [Google Scholar]
- 37.Klumpp, J., J. Dorscht, R. Lurz, R. Bielmann, M. Wieland, M. Zimmer, R. Calendar, and M. J. Loessner. 2008. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: a model for the SPO1-like myoviruses of gram-positive bacteria. J. Bacteriol. 190:5753-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klumpp, J., R. Lavigne, M. J. Loessner, and H.-W. Ackermann. 2010. The SPO1-related bacteriophages. Arch. Virol. 155:1547-1561. [DOI] [PubMed] [Google Scholar]
- 39.Kwan, T., J. Liu, M. DuBow, P. Gros, and J. Pelletier. 2005. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl. Acad. Sci. U. S. A. 102:5174-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labrie, S. J., and S. Moineau. 2002. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within the P335 quasi-species of lactococcal phages. Virology 296:308-320. [DOI] [PubMed] [Google Scholar]
- 41.Labrie, S. J., J. Josephsen, H. Neve, F. K. Vogensen, and S. Moineau. 2008. Morphology, genome sequence, and structural proteome of type phage P335 from Lactococcus lactis. Appl. Environ. Microbiol. 74:4636-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landthaler, M., U. Begley, N. C. Lau, and D. A. Shub. 2002. Two self-splicing group I introns in the ribonucleotide reductase large subunit gene of Staphylococcus aureus phage Twort. Nucleic Acids Res. 30:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landthaler, M., and D. A. Shub. 1999. Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes. Proc. Natl. Acad. Sci. U. S. A. 96:7005-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavigne, R., P. Darius, E. J. Summer, D. Seto, P. Mahadevan, A. S. Nilsson, H. W. Ackermann, and A. M. Kropinski. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lazarevic, V. 2001. Ribonucleotide reductase genes of Bacillus prophages: a refuge to introns and intein coding sequences. Nucleic Acids Res. 29:3212-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levin, M. E., R. W. Hendrix, and S. R. Casjens. 1993. A programmed translational frameshift is required for the synthesis of a bacteriophage lambda tail assembly protein. J. Mol. Biol. 234:124-139. [DOI] [PubMed] [Google Scholar]
- 47.Liu, X. Q., J. Yang, and Q. Meng. 2003. Four inteins and three group II introns encoded in a bacterial ribonucleotide reductase gene. J. Biol. Chem. 278:46826-46831. [DOI] [PubMed] [Google Scholar]
- 48.Loessner, M. J. 2005. Bacteriophage endolysins-current state of research and applications. Curr. Opin. Microbiol. 8:480-487. [DOI] [PubMed] [Google Scholar]
- 49.Loessner, M. J., R. B. Inman, P. Lauer, and R. Calendar. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35:324-340. [DOI] [PubMed] [Google Scholar]
- 50.Loessner, M. J., A. Schneider, and S. Scherer. 1996. Modified Listeria bacteriophage lysin genes (ply) allow efficient overexpression and one-step purification of biochemically active fusion proteins. Appl. Environ. Microbiol. 62:3057-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ludwig, W., K.-H. Schleifer, and E. Stackebrandt. 1984. 16S rRNA analysis of Listeria monocytogenes and Brochothrix thermosphacta. FEMS Microbiol. Lett. 25:6. [Google Scholar]
- 53.McShan, W. M., Y. F. Tang, and J. J. Ferretti. 1997. Bacteriophage T12 of Streptococcus pyogenes integrates into the gene encoding a serine tRNA. Mol. Microbiol. 23:719-728. [DOI] [PubMed] [Google Scholar]
- 54.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Flaherty, S., A. Coffey, R. Edwards, W. Meaney, G. F. Fitzgerald, and R. P. Ross. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 186:2862-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olia, A. S., and G. Cingolani. 2008. A shifty stop for a hairy tail. Mol. Microbiol. 70:549-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pantucek, R., A. Rosypalova, J. Doskar, J. Kailerova, V. Ruzickova, P. Borecka, S. Snopkova, R. Horvath, F. Gotz, and S. Rosypal. 1998. The polyvalent staphylococcal phage phi 812: its host-range mutants and related phages. Virology 246:241-252. [DOI] [PubMed] [Google Scholar]
- 58.Pin, C., G. D. Garcia de Fernando, and J. A. Ordonez. 2002. Effect of modified atmosphere composition on the metabolism of glucose by Brochothrix thermosphacta. Appl. Environ. Microbiol. 68:4441-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Proux, C., D. van Sinderen, J. Suarez, P. Garcia, V. Ladero, G. F. Fitzgerald, F. Desiere, and H. Brussow. 2002. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J. Bacteriol. 184:6026-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sambrook, J., and D. W. Russell. 2001. Molecular cloning, vol. 1 to 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 61.Schurr, T., E. Nadir, and H. Margalit. 1993. Identification and characterization of Escherichia coli ribosomal binding sites by free energy computation. Nucleic Acids Res. 21:4019-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sjöberg, B.-M. 1997. Ribonucleotide reductases: a group of enzymes with different metallosites and a similar reaction mechanism, p. 139-173. In P. J. Sadler and A. J. Thomson (ed.), Structure and bonding: metal sites in proteins and models. Springer, Berlin, Germany.
- 63.Smith, M. C., R. N. Burns, S. E. Wilson, and M. A. Gregory. 1999. The complete genome sequence of the Streptomyces temperate phage straight φC31: evolutionary relationships to other viruses. Nucleic Acids Res. 27:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith, M. C., and H. M. Thorpe. 2002. Diversity in the serine recombinases. Mol. Microbiol. 44:299-307. [DOI] [PubMed] [Google Scholar]
- 65.Stackebrandt, E., and D. Jones. 2006. The genus Brochothrix, p. 477-491. In S. Falkow, E. Rosenberg, K.-H. Schleifer, E. Stackebrandt, and M. Dworkin (ed.), The prokaryotes. Springer, New York, NY.
- 66.Stanley, G., K. J. Shaw, and A. F. Egan. 1981. Volatile compounds associated with spoilage of vacuum-packaged sliced luncheon meat by Brochothrix thermosphacta. Appl. Environ. Microbiol. 41:816-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steven, A. C., B. L. Trus, J. V. Maizel, M. Unser, D. A. Parry, J. S. Wall, J. F. Hainfeld, and F. W. Studier. 1988. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200:351-365. [DOI] [PubMed] [Google Scholar]
- 68.Stewart, C. R., S. R. Casjens, S. G. Cresawn, J. M. Houtz, A. L. Smith, M. E. Ford, C. L. Peebles, G. F. Hatfull, R. W. Hendrix, W. M. Huang, and M. L. Pedulla. 2009. The genome of Bacillus subtilis bacteriophage SPO1. J. Mol. Biol. 388:48-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uchiyama, J., M. Rashel, I. Takemura, H. Wakiguchi, and S. Matsuzaki. 2008. In silico and in vivo evaluation of bacteriophage φEF24C, a candidate for treatment of Enterococcus faecalis infections. Appl. Environ. Microbiol. 74:4149-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ventura, M., A. Zomer, C. Canchaya, M. O'Connell-Motherway, O. Kuipers, F. Turroni, A. Ribbera, E. Foroni, G. Buist, U. Wegmann, C. Shearman, M. J. Gasson, G. F. Fitzgerald, J. Kok, and D. van Sinderen. 2007. Comparative analyses of prophage-like elements present in two Lactococcus lactis strains. Appl. Environ. Microbiol. 73:7771-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu, J., R. W. Hendrix, and R. L. Duda. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11-21. [DOI] [PubMed] [Google Scholar]
- 72.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, J., Y. Zhang, L. Zhu, M. Suzuki, and M. Inouye. 2004. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 279:20678-20684. [DOI] [PubMed] [Google Scholar]
- 74.Zimmer, M., E. Sattelberger, R. B. Inman, R. Calendar, and M. J. Loessner. 2003. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed +1 translational frameshifting in structural protein synthesis. Mol. Microbiol. 50:303-317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.