Abstract
The degradosome is a multienzyme complex involved in mRNA degradation in Escherichia coli. The essential endoribonuclease RNase E contains a large noncatalytic region necessary for protein-protein interactions with other components of the RNA degradosome. Interacting proteins include the DEAD-box RNA helicase RhlB, the glycolytic enzyme enolase, and the exoribonuclease PNPase. Pseudoalteromonas haloplanktis, a psychrotolerant gammaproteobacterium distantly related to E. coli, encodes homologs of each component of the RNA degradosome. In P. haloplanktis, RNase E associates with RhlB and PNPase but not enolase. Plasmids expressing P. haloplanktis RNase E (Ph-RNase E) can complement E. coli strains lacking E. coli RNase E (Ec-RNase E). Ph-RNase E, however, does not confer a growth advantage to E. coli at low temperature. Ph-RNase E has a heterologous protein-protein interaction with Ec-RhlB but not with Ec-enolase or Ec-PNPase. The Ph-RNase E binding sites for RhlB and PNPase were mapped by deletion analysis. The PNPase binding site is located at the C-terminal end of Ph-RNase E at the same position as that in Ec-RNase E, but the sequence of the site is not conserved. The sequence of the RhlB binding site in Ph-RNase E is related to the sequence in Ec-RNase E. Together with the heterologous interaction between Ph-RNase E and Ec-RhlB, our results suggest that the underlying structural motif for the RNase E-RhlB interaction is conserved. Since the activity of Ec-RhlB requires its physical interaction with Ec-RNase E, conservation of the underlying structural motif over a large evolutionary distance could be due to constraints involved in the control of RhlB activity.
In the model Gram-negative bacterium Escherichia coli, RNase E is an essential RNase involved in RNA processing and mRNA degradation (6, 14, 28). RNase E is a large 1,061-residue protein. Residues 1 to 529 of RNase E correspond to the catalytic domain, which associates to form a tetrameric holoenzyme (3). In E. coli, RNase G is a paralog of RNase E, which is 497 residues in length (34, 52, 54). There is a high degree of sequence conservation between the catalytic domains of RNase E and RNase G, and these enzymes have similar catalytic properties. Both enzymes are 5′-end-dependent endoribonucleases with specificity for AU-rich single-strand cleavage sites. The 5′-end dependence involves a site in the catalytic domain that binds 5′ monophosphate ends (3, 36). Together, RNase E and RNase G are the founders of a family of bacterial ribonucleases conserved in many, but not all, bacteria (4, 13, 23, 30). A major difference between RNase E and RNase G is the large C-terminal extension of RNase E, which is equal in length to the region corresponding to the catalytic domain.
The noncatalytic region of RNase E, residues 530 to 1061, is mostly natively unstructured (2). However, it contains short regions involved in interactions with phospholipid membranes, RNA, or protein. In some cases, these regions have been shown to form secondary structures involved in the interaction, and the term “microdomain” has been used to describe these structures (37). “Segment A” of E. coli RNase E, corresponding to residues 568 to 582, has recently been shown to form a short amphipathic α-helix that binds to phospholipid membranes in vitro and that is necessary for the localization of RNase E to the inner cytoplasmic membrane in vivo (25). Residues 604 to 683, known as the arginine-rich RNA binding domain (AR-RBD), binds RNA in vitro (38, 51), and it is believed to enhance the activity of RNase E in vivo (35, 44). A second short arginine-rich segment (residues 796 to 819), known as AR2 (arginine-rich region 2), is also involved in RNA binding (32). Residues 701 to 1061 form a scaffold for protein-protein interactions with RNA helicase B (RhlB), enolase, and polynucleotide phosphorylase (PNPase) (23, 53). The multienzyme complex comprised of RNase E, RhlB, enolase, and PNPase is known as the RNA degradosome (7, 41, 47). RhlB is a DEAD-box RNA helicase involved in mRNA degradation (24, 47, 53). Enolase is a glycolytic enzyme whose role in RNA metabolism is unclear, although it has been suggested that it might link the mRNA degradation machinery to the control of central intermediary metabolism. PNPase is an exoribonuclease involved in RNA processing and mRNA degradation (12, 19). In the case of enolase and PNPase, the microdomains in the scaffold that are responsible for the protein-protein interactions have been characterized at atomic resolution by X-ray crystallography (8, 43). The microdomain involved in RhlB binding has been localized to a specific region in the scaffold by deletion and point mutation analysis, but its structure has not been elucidated (9, 27, 53).
In addition to the canonical RNA degradosome, a different complex containing RNase E, Hfq, and SgrS, a small regulatory RNA, is formed under conditions of phosphosugar stress (42, 55). The formation of the complex with Hfq and SgrS requires the same region of RNase E that is necessary for the formation of the RNA degradosome, and evidence suggests that the degradosome is remodeled because of the new interaction. RraA and RraB, protein factors that downregulate RNase E activity, are also believed to remodel the RNA degradosome, and there is evidence that RNase E can form a “cold shock” RNA degradosome in which CsdA, another DEAD-box RNA helicase, is recruited to the complex (18, 27, 31, 45, 48). The noncatalytic region of RNase E can therefore be viewed as an interaction hub involved in a variety of protein-protein interactions. It has been suggested that remodeling the canonical RNA degradosome in response to environmental changes facilitates the rapid adjustment of the program of gene expression by a posttranscriptional mechanism involving mRNA stability (37).
Authentic homologs of RNase E are found in many bacteria, and in the limited cases studied, these homologs have been found to interact with other enzymes to make RNA degradosome-like complexes (13, 37). The RNase E of the actinobacterium Streptomyces coelicolor has been shown to interact with PNPase (30). The RNase E of the photosynthetic alphaproteobacterium Rhodobacter capsulatus has been shown to interact with two DEAD-box RNA helicases and the transcription termination factor Rho (20, 21). The RNase E of gammaproteobacterium Pseudomonas syringae Lz4W has been shown to interact with a DEAD-box RNA helicase and the exoribonuclease RNase R (46). The composition of the RNase E-based complex thus varies with species, suggesting a specialized role for the RNA degradosome in molecular evolution (37). The variability in composition of the RNA degradosome likely involves changes in the sequences and structure of microdomains in the noncatalytic region of RNase E, although this idea awaits experimental validation.
Pseudoalteromonas haloplanktis TAC125 is a psychrotolerant gammaproteobacterium that was isolated from the Antarctic Ocean (1). Although described as a psychrophilic bacterium, we prefer the term psychrotolerant since it can grow at temperatures ranging from 4°C to 30°C. The genome of P. haloplanktis has been completely sequenced (39). Phylogenetic analysis shows that P. haloplanktis is a member of the Alteromonadales, an order that is separated from the Enterobacteriales (E. coli) by the Vibrionales, Aeromonadales, and Pasteurellales (17). Interest in P. haloplanktis is 2-fold, as follows: industrial applications involving psychrotolerant enzymes and environmental studies involving that adaptation of bacteria to a cold marine niche. Here we have identified and characterized the RNA degradosome of P. haloplanktis, showing that it is comprised of RNase E, RhlB, and PNPase but not enolase. The sites in RNase E for protein-protein interactions with RhlB and PNPase were mapped by deletion analysis. Together with a recently published two-hybrid analysis of protein-protein interactions in the RNA degradosome of Vibrio angustum S14 (16), this is the first work in which microdomains of RNase E involved in protein-protein interactions have been identified and characterized in a bacterium other than E. coli.
MATERIALS AND METHODS
Strains.
Strains used in this work are listed in Table 1. P. haloplanktis TAC125 was grown at 15°C in TYP broth (16 g/liter yeast extract, 16 g/liter Bacto tryptone, and 15 g/liter NaCl) supplemented with 50 μg/ml ampicillin when necessary.
TABLE 1.
Strains and plasmids
| Strain/plasmid | Descriptiona | Reference/source |
|---|---|---|
| Strains | ||
| P. haloplanktis TAC125 | Wild type | 1 |
| KSL2000 | rne::cat recA::Tn10 | 31 |
| AC21 | MC1061 zce-726::Tn10 | 8 |
| AC27 | MC1061 zce-726::Tn10 rne131 | 34 |
| SVK438 | KSL2000 recA+ | V. Khemici |
| SVK446 | KSL2000 recA+ Δrhlb | V. Khemici |
| S17.1 | RP4 2-Tc::Mu-Km::Tn7 pro res mod1 | 52 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) dcm gal λ(DE3) | 53 |
| Plasmids | ||
| pAM238 | pGB2-derived plasmid | 12 |
| pAM-rne | Chromosomal PstI fragment containing the rne gene cloned into pAM238 (pSC101ori, spectinomycin resistance) | 56 |
| pVK200 | pAM238, Ec-RNase E, C-terminal FLAG | V. Khemici |
| pET15b-RneCTH | T7 expression of the RNase E CTH with an N-terminal His tag | 29 |
| pJB3 | Broad-host-range cloning vector, RK2 derivative plasmid | 2 |
| pIB3 (shuttle vector) | Fragment 1914 to 2786 of the resident plasmid pMtBL of P. haloplanktis containing the origin of replication was cloned into pJB3 | Ivona Bagdiul |
| pSAB1 | pET15b, Ph-rne CDS under the control of T7 expression signals | This work |
| pSAB11 | pAM238, lactose expression signals deleted | This work |
| pSAB12 | pSAB11, Ph-rne | This work |
| pSAB13 | pSAB11, E. coli expression signals fused to Ph-rne CDS | This work |
| pSAB15 | pSAB11, Ph-rne, C-terminal FLAG | This work |
| pSAB17 | pIB3, Ph-rne, C-terminal FLAG | This work |
| pSAB18 | pSAB15, E. coli expression signals fused to Ph-rne CDS | This work |
| pSAB29 | pSAB18, Ph-rne (deletion of HBS) | This work |
| pSAB20 | pSAB1, Ph-rne (deletion of residues 11-528) | This work |
| pSAB33 | pSAB20, Ph-rne CTH (deletion of residues 841-1071) | This work |
| pSAB34 | pSAB20, Ph-rne CTH (deletion of residues 996-1071) | This work |
| pSAB35 | pSAB20, Ph-rne CTH (deletion of residues 1035-1071) | This work |
CDS, coding sequence.
Plasmids.
Plasmids used in this work are listed in Table 1. pSAB11 was constructed by deleting the lactose promoter and operator of pAM238. The gene encoding FLAG-tagged P. haloplanktis RNase E (Ph-RNase E) under its own expression signals was constructed by PCR and inserted into pSAB11 to create pSAB15. In pSAB18, the coding sequence of Ph-RNase E was fused at AUG to the E. coli RNase E (Ec-RNase E) expression signals. pSAB29 was derived from pSAB18 by deleting the region corresponding to the putative RhlB binding site by inverse PCR.
Conjugation.
The shuttle vector pIB3 and E. coli strain S17.1 (kindly provided by M. Dreyfus) were used for transconjugation. pIB3 and pSAB17 were mobilized from E. coli S17.1 into P. haloplanktis TAC125 by conjugation. Aliquots (1 ml) of cultures in logarithmic growth phase were mixed and spotted onto a TYP plate. After 16 h at room temperature, the cells were suspended in 300 μl of TYP broth. Transconjugants were selected at 4°C by plating on TYP plates containing 100 μg/ml ampicillin.
Whole-cell extracts and immunopurification.
FLAG-tagged RNase E was immunopurified by a modification of a protocol described previously (42). Cells were grown in 200-ml cultures of LB (E. coli) or TYP (P. haloplanktis) to an optical density at 600 nm (OD600) of 0.6. The cultures were chilled on ice, and all subsequent steps were performed in the cold. The cells were harvested by centrifugation and washed in 10 ml of 50 mM Tris-HCl at pH 8.0 and 5 mM EDTA, except that EDTA was omitted for P. haloplanktis since the cells lysed prematurely in its presence. The cell pellets were suspended in 5 ml of IP buffer (20 mM Tris-HCl at pH 8.0, 0.1 M KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20, 1× Roche EDTA-free protease inhibitor cocktail, 150 μg/ml lysozyme). The cell suspension was subjected to three freeze-thaw steps, sonicated, and centrifuged at 10,000 × g for 1 h at 4°C. The supernatant (1 ml) was incubated with 50 μl of anti-FLAG M2-agarose suspension (Sigma) for 2 h at 4°C with gentle agitation. The mixture was filtered by using a mini-chromatography column (Bio-Rad). The bound proteins were eluted with 50 μl of TBS (50 mM Tris-HCl at pH 7.5 and 0.15 M NaCl) containing 5 μg of FLAG peptide (Sigma). The proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (7.5%). Western blotting was performed using peroxidase conjugated anti-FLAG antibody (Sigma) and an ECL kit (GE Healthcare).
Purification of RNase E CTH polypeptide and incubation with P. haloplanktis cell extracts.
The His-tagged RNase E C-terminal half (CTH) polypeptide was expressed using pET15b (Novagen). Briefly, pSAB1 was constructed by ligating the Ph-RNase E coding sequence into pET15b. pSAB20 was constructed by deleting the region corresponding to residues 11 to 528. Thus, the Ph-RNase E CTH polypeptide contains the His tag from pET15b and residues 1 to 10 of Ph-RNase E fused to residues 529 to 1071 of Ph-RNase E. pSAB33, pSAB34, and pSAB35 were derived from pSAB20 by deleting the region corresponding to the last 231, 76, and 37 residues of Ph-RNase E CTH, respectively.
BL21(DE3) was transformed with pSAB20, pSAB33, pSAB34, and pSAB35. Cultures were grown at 30°C and induced at an OD600 of 0.4 to 0.5 with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested 3 h after induction by centrifugation at 6,000 rpm at 4°C for 15 min and washed with ice-cold 50 mM Tris-HCl, pH 7.5. The following steps, which were done under denaturing conditions in 8 M urea, were performed at room temperature. A cleared lysate was prepared in buffer B according to the manufacturer's instructions (Qiagen). Approximately 100 μg of overexpressed protein was diluted in buffer B and incubated with 200 μl of Talon metal affinity resin (cobalt; Clontech). The beads were collected in a mini-chromatography column (Bio-Rad) and washed with 1 ml buffer C (Qiagen). The urea was removed by washing with buffer EQ (20 mM Tris-HCl at pH 7.5, 0.1 M KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20, 0.1 mg/ml bovine serum albumin [BSA], and 1× Roche EDTA-free protease inhibitor cocktail). The column was transferred to 4°C and washed again with cold buffer EQ. Subsequent steps were performed at 4°C. Whole-cell extract (1 ml) from P. haloplanktis was added and incubated for 2 h. The column was washed twice with buffer 1 (buffer EQ containing 20 mM imidazole without BSA). Elutions were performed with buffer 2 (buffer 1 containing 300 mM imidazole).
Mass spectrometry.
Proteins that copurified with FLAG-tagged Ph-RNase E were identified by mass spectrometry. Bands from SDS-PAGE were excised and sent to the proteomics service of the University of Bordeaux for analysis (Plateforme de Génomique Fonctionnelle Bordeaux, Université Victor Segalen). Coverage ranged from 47% for PNPase to 20% for RhlB.
PONDR and ClustalW.
The P. haloplanktis RNase E protein sequence was analyzed by the Predictor of Naturally Disordered Regions (PONDR) (Molecular Kinetics, Inc., Indianapolis, IN) (33). Protein sequence alignments were performed on the EMBL-EBI website (http://www.ebi.ac.uk/Tools/clustalw2/index.html) using CLUSTALW (10) and the Jalview editor (11).
RESULTS
Prediction of microdomains in the noncatalytic region of Ph-RNase E.
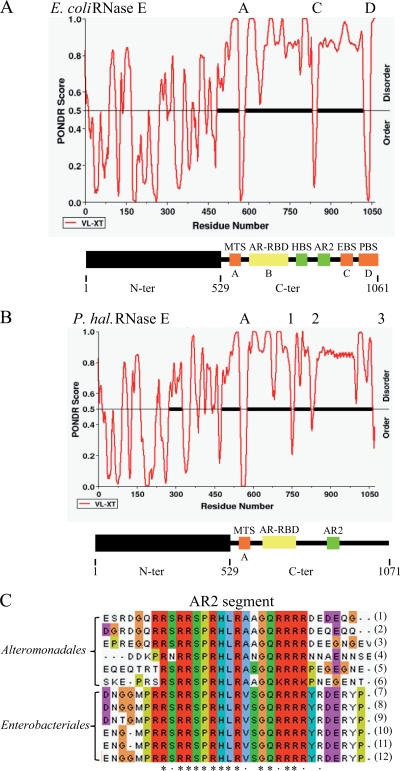
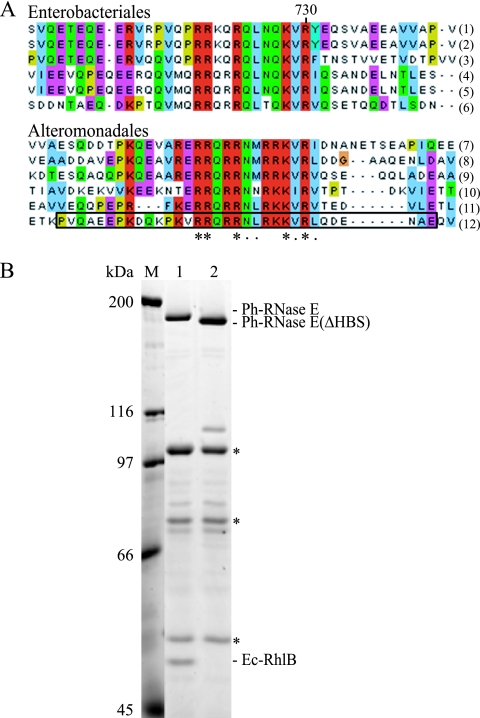
The genome of P. haloplanktis encodes orthologs of E. coli RNase E, RhlB, enolase, and PNPase that are conserved with overall sequence identities ranging from 52% for RNase E to 78% for enolase (data not shown). The score for the N-terminal catalytic domain (residues 1 to 529) of RNase E is 78% identity. No significant sequence conservation was detected between the C-terminal noncatalytic region of P. haloplanktis RNase E (Ph-RNase E) and E. coli RNase E (Ec-RNase E), a result consistent with previous work demonstrating a high degree of sequence variability in the noncatalytic region, even in closely related bacteria. Experimental work with E. coli RNase E has revealed six segments in the noncatalytic region that are involved in interactions with the phospholipid membrane, RNA, or protein (Fig. 1 A). The membrane-targeting sequence (MTS) has been shown to interact with phospholipid membranes (25). The arginine-rich RNA binding domain (AR-RBD) and a second arginine-rich region (AR2) have been shown to bind RNA. The helicase binding site (HBS), enolase-binding site (EBS), and PNPase binding site (PBS) have been shown to bind RhlB, enolase, and PNPase, respectively. Three of these segments overlap with or correspond to regions that have been predicted by PONDR to have the propensity to form secondary structures (Fig. 1A, segments A, C, and D), and experimental results have confirmed these predictions (2, 8, 25, 43). Segment B, which overlaps the AR-RBD, is predicted to have the propensity to form a coiled-coil structure, although there is no experimental evidence supporting this prediction.
FIG. 1.
Primary structure of P. haloplanktis RNase E. (A, B) PONDR analysis for prediction of RISPs (regions of increased structural propensity) and schematic representation of Ec-RNase E (A) and Ph-RNase E (B). RISPs correspond to regions with a score of less than 0.5. The analysis of Ec-RNase E, which has appeared previously (2), is presented to permit direct comparison with Ph-RNase E. In the schematic, the N-terminal catalytic domain is represented as a black box, and the noncatalytic C-terminal region is represented as a line. The noncatalytic region of Ec-RNase E contains six microdomains involved in membrane, RNA, and protein interactions. MTS, membrane targeting sequence; AR-RDB, arginine-rich RNA binding domain; HBS, helicase binding site; AR2, arginine-rich region 2; EBS, enolase binding site; PBS, PNPase binding site. Segments A, C, and D (red) correspond to regions predicted to form secondary structures by PONDR. Segment B (yellow), which overlaps with the AR-RBD, was predicted to form a coiled-coil structure (see reference 2). Two other microdomains (green) are known to make interactions but do not correspond to regions of predicted secondary structure. (C) Sequence alignment of the AR2 segments of RNase E homologs from the Alteromonadales and Enterobacteriales. The asterisks indicate strictly conserved residues; the dots indicate residues that are similar. The homologs used in this alignment correspond to Shewanella sp. ANA-3 (1), Shewanella amazonensis SB2B (2), Shewanella sp. PV-4 (3), Colwellia psychrerythraea 34H (4), Pseudoalteromonas haloplanktis TAC125 (5), Pseudoalteromonas tunicata D2 (6), Escherichia coli (7), Shigella boydii Sb227 (8), Salmonella enterica subsp. enterica (9), Yersinia pestis Antiqua (10), Yersinia pseudotuberculosis IP 31758 (11), and Erwinia carotovora subsp. atroseptica SCRI1043 (12).
We analyzed the protein sequence of Ph-RNase E by PONDR, and the profile shown in Fig. 1B is similar to that shown in results previously obtained with the RNase E of E. coli and Vibrio angustum S14 (2, 16). For comparison, the profile for Ec-RNase E is shown in Fig. 1A.The conserved N-terminal catalytic domain of Ph-RNase E is predicted to be mostly structured. Segment A in the noncatalytic region, which corresponds to the MTS, is clearly predicted to be structured, consistent with previous work showing that this microdomain is conserved throughout the gammaproteobacteria (25). Three other short segments of predicted structure were detected in the noncatalytic region of Ph-RNase E (Fig. 1B, regions 1 to 3), but by visual inspection and sequence alignment, there is no obvious correspondence to any of the microdomains identified in Ec-RNase E. The only other clearly discernible elements in the noncatalytic region of the Ph-RNase E are the AR-RBD and AR2, which were detected by visual inspection and alignment of the Ec-RNase E and Ph-RNase sequences. In summary, by structure prediction and sequence analysis, we detected segments in the noncatalytic region of Ph-RNase E that correspond to the MTS, AR-RBD, and AR2 of Ec-RNase E. For AR2, a sequence alignment of RNase E homologs from Enterobacteriales and Alteromonadales is shown in Fig. 1C, demonstrating a common core of 18 residues with 13 positions that are strictly conserved. By sequence alignment, we were unable to detect microdomains corresponding to the binding sites for RhlB, enolase, or PNPase. By structure prediction, the position of region 3 of Ph-RNase E corresponds to segment D of Ec-RNase E, which is the PNPase binding site.
RNA degradosome of P. haloplanktis.
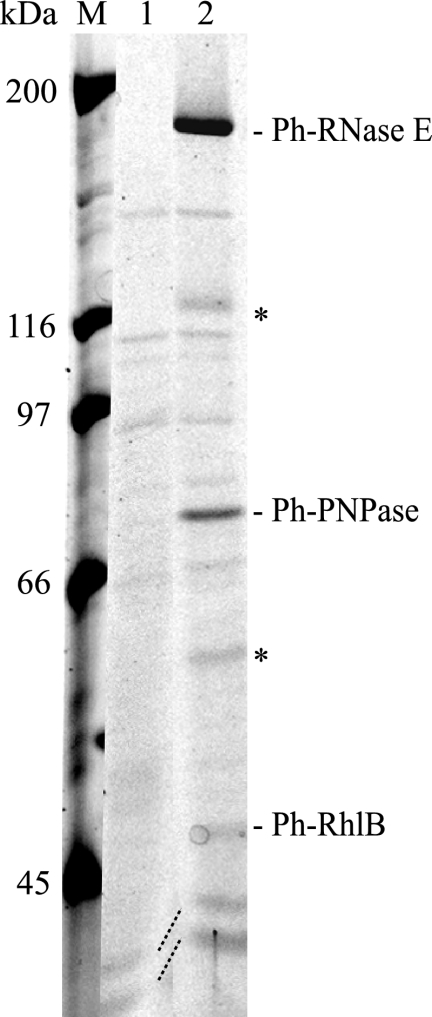
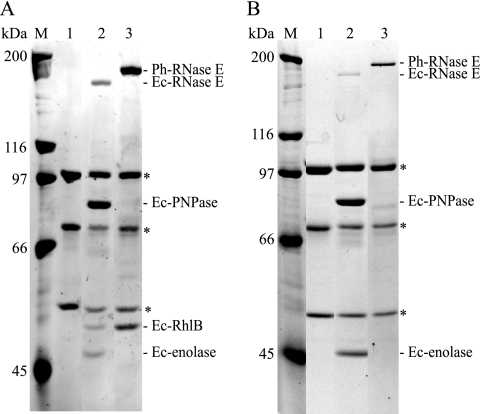
To test whether Ph-RNase E forms an RNA degradosome and to identify partner proteins, we employed pIB3, which is a shuttle vector permitting the transfer by conjugation of plasmids from E. coli to P. haloplanktis. A genomic fragment of Ph-rne, including flanking sequences necessary for expression, was cloned into the shuttle vector. The Ph-rne gene was modified to express Ph-RNase E with a C-terminal FLAG tag, creating the plasmid pSAB17 (Table 1). This plasmid was conjugated into P. haloplanktis. The expression of the FLAG-tagged Ph-RNase E was verified by Western blotting (data not shown). To detect proteins that interact with Ph-RNase E, whole-cell extracts were prepared, and the FLAG-tagged RNase E was affinity purified using anti-FLAG agarose beads. The FLAG-tagged Ph-RNase E was eluted with a competitor FLAG peptide and analyzed by SDS-PAGE (Fig. 2). Lane 1 of Fig. 2 shows the result of using a background control with an extract prepared from P. haloplanktis conjugated with the empty shuttle vector; lane 2 shows the result of using pSAB17. Five proteins, clearly visible in the eluate of the FLAG construct but absent in the background control, were identified by mass spectrometry. As indicated, the largest protein, migrating at about 180 kDa, is the full-length FLAG-tagged Ph-RNase E. Two smaller proteins (Fig. 2, asterisks) were identified as fragments of Ph-RNase E. Since these fragments were not detected by Western blotting when total protein was prepared by directly lysing the cells with SDS and heat, we believe that the fragments are due to proteolysis that occurs during the preparation of the whole-cell extract. Ec-RNase E is known to be exceptionally sensitive to proteolysis, and we observed previously that there is significant proteolysis of Ph-RNase E in native extracts prepared from P. haloplanktis, even in the presence of a cocktail of protease inhibitors (5).
FIG. 2.
RNA degradosome of P. haloplanktis. SDS-PAGE stained with Sypro orange. Whole-cell extracts were prepared from P. haloplanktis conjugated with the empty shuttle vector (pIB3) or the vector encoding FLAG-tagged Ph-RNase E (pSAB17). After absorption to the anti-FLAG agarose beads and being washed, Ph-RNase E was eluted with FLAG peptide. Lane 1, pIB3 (background control); lane 2, pSAB17 (FLAG-tagged Ph-RNase E). M, molecular mass markers. The proteins shown in lane 2 were identified by mass spectrometry. The asterisks indicate proteolytic products of Ph-RNase E.
Two other proteins detected and shown in Fig. 2 were identified as Ph-PNPase and Ph-RhlB. These proteins migrate at the expected sizes of 80 and 50 kDa, respectively. Enolase, which is expected to migrate at 48 kDa, was not detected by mass spectrometry nor was it detected by Western blotting using polyclonal rabbit serum raised against Ec-enolase (data not shown). Enolase is a highly conserved enzyme, and control experiments showed that our antiserum readily detects Ph-enolase in P. haloplanktis whole-cell extracts (data not shown). Taken together, these results show that the Ph-RNase E forms an RNA degradosome containing Ph-PNPase and Ph-RhlB and suggest that Ph-enolase is not tightly associated with Ph-RNase E (see below).
Complementation of E. coli strains lacking Ec-RNase E by Ph-RNase E.
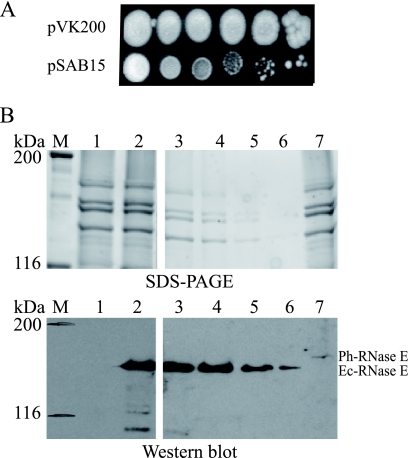
We wanted to test whether Ph-RNase E could replace the function of Ec-RNase E in E. coli and whether the substitution of a mesophilic RNase E by a psychrotolerant RNase E affects the growth of E. coli in the cold. We used the KSL2000 strain of E. coli, in which the gene encoding RNase E on the chromosome is disrupted (29). Since RNase E is essential, it is supplied in trans by pBAD-rne, which is a low-copy-number plasmid with the Ec-RNase E coding sequence under the control of the arabinose promoter. We transferred the Ph-rne gene with the FLAG tag from pSAB17 to pSAB11 to create pSAB15. Both pBAD-rne and pSAB15 have the same origin of DNA replication (pSC101) but different antibiotic resistance markers. By transforming the KSL2000 strain with pSAB15 and selecting for spectinomycin, it was possible to displace the pBAD-rne plasmid and complement the deletion of Ec-RNase E on the chromosome with Ph-rne on the plasmid. However, this strain formed only visible colonies after 6 days of growth at 25°C compared to the control strain complemented with Ec-rne (pVK200), which formed visible colonies after 2 days (Fig. 3A). A control experiment using Ph-RNase E without FLAG gave the same result (data not shown). Quantitative Western blotting (Fig. 3B), in which a cell extract from the Ec-rne control was serial diluted, showed that the amount of Ph-RNase E in the strain complemented with pSAB15 was at least 32-fold lower than the amount of Ec-RNase E in the strain complemented with pVK200 (Fig. 3B, compare lane 7, undiluted Ph-RNase E, to lane 6, 32-fold-diluted Ec-RNase E). Note that in Fig. 3B, the image corresponding to lanes 3 to 7 was the result of a 4-min exposure to permit detection of Ph-RNase E, whereas the panel corresponding to lanes 1 and 2 was the result of a 30-s exposure. We therefore hypothesized that the weak complementation of the KSL2000 strain by pSAB15 might be due to low levels of expression of Ph-RNase E.
FIG. 3.
Complementation of KSL2000 and expression of Ph-RNase E in E. coli. (A) Complementation of the KSL2000 strain (disruption of the chromosomal gene encoding Ec-RNase E) by plasmids expressing FLAG-tagged Ec-RNase E (pVK200) or FLAG-tagged Ph-RNase E (pSAB15). Serial dilutions (10-fold) were spotted on LB plates and grown at 25°C for 6 days. (B) SDS-PAGE stained with Sypro orange and Western blotting using anti-FLAG antibodies. Lane 1, pAM-rne (Ec-RNase E without FLAG); lane 2, pVK200; and lane 7, pSAB15. M, molecular mass markers. The protein loaded in lane 2 was diluted 4-fold (lane 3), 8-fold (lane 4), 16-fold (lane 5), and 32-fold (lane 6). In the panel showing lanes 1 and 2, the blot was exposed for 30 s, whereas in the panel showing lanes 3 to 7, the blot was exposed for 4 min to permit detection of Ph-RNase E.
Promoters of transcription in P. haloplanktis have essentially the same consensus sequence as those in E. coli and are therefore expected to act as promoters in E. coli (15). The genomic region containing the Ph-rne gene used in our experiments contains approximately 700 bp of DNA upstream of the rne coding sequence. This region, which includes a 400-bp intergenic spacer and part of an upstream coding sequence, contains signals for transcription and translation, as evidenced by expression of FLAG-tagged Ph-RNase E when the gene carried by pSAB17 was introduced into P. haloplanktis by conjugation (Fig. 2). The control of the expression of Ec-RNase E involves a complex system of posttranscriptional autoregulation in which a 361-nucleotide (nt)-long 5′ untranslated region is targeted by RNase E to destabilize the mRNA (22). Taken together, our results suggest that low-level expression is due to a dysfunction in either the transcriptional or posttranscriptional control of the Ph-rne gene in E. coli.
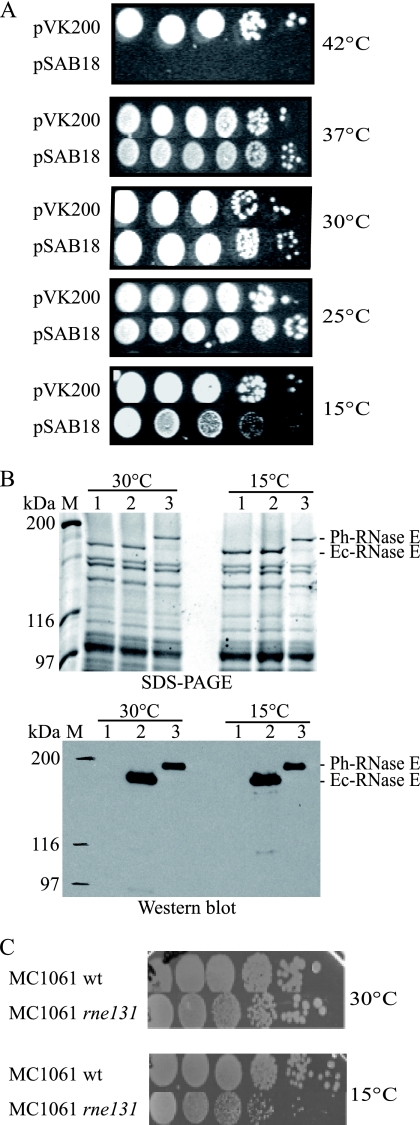
To express Ph-RNase E at levels comparable to those of Ec-RNase E in the KSL2000 strain, we constructed a hybrid rne gene in which the 5′ noncoding region of Ec-rne was fused to the coding sequence of Ph-RNase E. pSAB18, harboring the rne fusion, transformed into KSL2000 exhibited robust complementation at temperatures ranging from 25 to 37°C (Fig. 4A). Western blotting showed that the amount of Ph-RNase E in the strain complemented by pSAB18 is about 50% of the amount of Ec-RNase E in the strain complemented by pVK200 (Fig. 4B and quantification not shown). Taken together, these results suggest that the weak complementation by pSAB15 is due to low levels of expression of Ph-RNase E in E. coli when the coding sequence is under the control of the homologous P. haloplanktis expression signals. In Fig. 4A, Ph-RNase E failed to complement at 42°C, which is likely due to the thermolability of Ph-RNase E.
FIG. 4.
Complementation of KSL2000, expression of Ph-RNase E at 30 and 15°C, and growth of the MC1061 rne131 strain at 15°C. (A) Complementation of the KSL2000 strain (the chromosomal gene encoding Ec-RNase E was disrupted) by a plasmid expressing FLAG-tagged Ph-RNase E (pSAB18). Serial dilutions (10-fold) were spotted on LB plates and grown at the indicated temperatures for times ranging from 1 day (37 and 42°C) to 10 days (15°C). (B) SDS-PAGE stained with Sypro orange and Western blotting using anti-FLAG antibodies. Lane 1, KSL2000 complemented with pAM-rne (Ec-RNase E without FLAG); lane 2, pVK200 (Ec-RNase E with FLAG); and lane 3, pSAB18 (Ph-RNase E with FLAG). M, molecular mass markers. (C) Growth of MC1061 (wild type [wt], AC21) and the rne131 derivative (AC27) at 30 and 15°C (10-fold serial dilutions).
It is notable that complementation by Ph-RNase E at 15°C is weak (Fig. 4A). The amount of Ph-RNase E does not change between 30 and 15°C, as shown by Western blotting (Fig. 4B). Therefore, the growth defect at 15°C is not due to small amounts of Ph-RNase E. Since the endoribonuclease activity of Ph-RNase E is expected to be robust at 15°C, relatively poor growth at this temperature suggests that Ph-RNase E fails to provide a noncatalytic function that is necessary for growth in the cold. Moreover, the E. coli MC1061 strain harboring the rne131 mutant lacking the last 477 residues of Ec-RNase E (32) grows slower than the wild-type strain at 15°C, whereas at 30°C, no growth defect was observed (Fig. 4C). Thus, the noncatalytic region of Ec-RNase E appears to have a function at low temperature that is not provided by Ph-RNase E.
Heterologous interaction between Ph-RNase E and Ec-RhlB.
Since we were able to complement the KSL2000 strain with FLAG-tagged Ph-RNase E, we decided to check for heterologous interactions with E. coli proteins. By following the same strategy described above for the characterization of the RNA degradosome in P. haloplanktis, FLAG-tagged Ph-RNase E was purified from E. coli whole-cell extracts using anti-FLAG agarose beads. In Fig. 5A, lane 1 contains a negative control showing KSL2000 complemented with pAM-rne (harboring Ec-RNase E without FLAG). The three proteins marked with the asterisks are subunits of pyruvate dehydrogenase, an E. coli enzyme that adventitiously reacts with the FLAG agarose beads (42). Lane 2 contains a control showing the results from the KSL2000 strain complemented with pVK200 (FLAG-tagged Ec-RNase E). As expected, Ec-PNPase, Ec-RhlB, and Ec-enolase copurify with Ec-RNase E. Lane 3 shows the results from the KSL2000 strain complemented with pSAB18 (FLAG-tagged Ph-RNase E). We detected neither Ec-PNPase nor Ec-enolase. This result was confirmed by Western blotting, which should have been sensitive enough to detect trace amounts of PNPase or enolase (data not shown). The failure to detect enolase in lane 3 of Fig. 5A corroborates the results in Fig. 2, adding support to the suggestion that Ph-RNase E does not tightly associate with enolase. There was, however, in Fig. 2, a protein associated with Ph-RNase E that migrated at 50 kDa with mobility identical to that of Ec-RhlB. This protein was identified as Ec-RhlB by Western blotting (data not shown) and by repeating the experiment using a strain of E. coli (SVK446) in which the gene encoding RhlB was disrupted (Fig. 5B). The absence of the band corresponding to Ec-RhlB in lane 2 (Ec-RNase E) and lane 3 (Ph-RNase E) confirms that the protein associated with Ph-RNase E is Ec-RhlB. Taken together, these results show that Ph-RNase E makes a heterologous interaction with Ec-RhlB but not Ec-PNPase or Ec-enolase.
FIG. 5.
Ph-RNase E expressed in E. coli interacts with Ec-RhlB. SDS-PAGE stained with Sypro orange. (A) Immunopurifications were performed as shown in Fig. 2. Lane 1, KSL2000 complemented with pAM-rne (Ec-RNase E without FLAG); lane 2, pVK200 (Ec-RNase E with FLAG); and lane 3, pSAB18 (Ph-RNase E with FLAG). M, molecular mass markers. The asterisks indicate subunits of pyruvate dehydrogenase, which interacts adventitiously with the FLAG antibody. (B) The experiment shown in panel A was repeated using a strain (SVK446) in which the gene encoding RhlB was disrupted.
Identification of the RhlB binding site in Ph-RNase E.
In order to identify the helicase binding site (HBS) in Ph-RNase E, we aligned the sequences of RNase E homologs from the Alteromonadales, the order of bacteria that includes P. haloplanktis. We identified a motif located in the same region as the HBS of RNase E homologs from the Enterobacteriales that is related by sequence (Fig. 6A). These motifs have in common a core of 13 residues, with 5 positions that are strictly conserved. To test if the motif identified in Ph-RNase E is necessary for RhlB binding, we deleted the corresponding region in pSAB18 to create pSAB29. The deletion corresponds to the boxed sequence in Fig. 6A. Figure 6B shows that the deletion results in the loss of the immunopurification of RhlB with Ph-RNase E (compare lanes 1 and 2). From this result, we conclude that the sequence motif identified in Fig. 5A corresponds to the microdomain necessary for the heterologous protein-protein interaction between Ec-RhlB and Ph-RNase E.
FIG. 6.
Identification of the RhlB binding site of Ph-RNase E. (A) Sequence alignment of the HBSs in homologs from the Enterobacteriales (top) and the corresponding regions in homologs from the Alteromonadales (bottom). The asterisks indicate residues that are conserved; the dots indicate residues that are similar. The homologs used in these alignments correspond to Escherichia coli (1), Shigella boydii Sb227 (2), Salmonella enterica subsp. enterica (3), Yersinia pseudotuberculosis IP 31758 (4), Erwinia carotovora subsp. atroseptica SCRI1043 (5), Yersinia pestis Antiqua (6), Shewanella sp. ANA-3 (7), Shewanella amazonensis SB2B (8), Shewanella sp. PV-4 (9), Colwellia psychrerythraea 34H (10), Pseudoalteromonas tunicata D2 (11), and Pseudoalteromonas haloplanktis TAC125 (12). The deletion of the region corresponding to the motif identified in Ph-RNase E in pSAB18 to create pSAB29 corresponds to the boxed sequence. (B) SDS-PAGE stained with Sypro orange. Immunopurifications were performed as shown in Fig. 2. Lane 1, pSAB18 (Ph-RNase E with FLAG); lane 2, pSAB29, which expresses a variant of Ph-RNase E deleted from residues 684 to 716 (boxed in panel A, last line). M, molecular mass markers. The asterisks correspond to the subunits of pyruvate dehydrogenase, which interacts adventitiously with the FLAG antibody.
Identification of the PNPase binding site in Ph-RNase E.
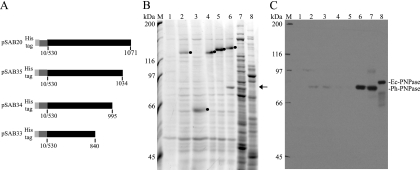
Using the same approach that led to the identification of the HBS, we failed to identify a sequence motif in the RNase E homologs of the Alteromonadales that was related to the PNPase binding site (PBS) of Ec-RNase E. Since the PBS is located in the C-terminal region of Ec-RNase E and since this region in Ph-RNase E is predicted to have the propensity to form secondary structure (Fig. 1B, region 3), we decided to construct a series of nested deletions extending from the C-terminal end into the protein. The protein interaction was tested in vitro using an N-terminal His-tagged derivative corresponding to the noncatalytic C-terminal half (CTH) of Ph-RNase E (Fig. 7A). We showed previously that the Ec-CTH interacts with Ec-PNPase in vitro to form a complex that can be purified using a His tag (27). In the experiment performed here, the Ph-CTH was overexpressed in E. coli and purified by affinity chromatography. The resin with the bound Ph-CTH was then incubated with a whole-cell extract prepared from P. haloplanktis, washed, and eluted with imidazole. Figure 7B shows SDS-PAGE of the eluates. Lane 1 shows a negative control with a mock reaction that did not contain the CTH, lane 2 shows the result using the Ec-CTH, and lane 6 shows the result using the Ph-CTH. In lane 6, a protein of about 80 kDa copurified with Ph-CTH (indicated by the arrow). Western blotting identified the protein in lane 6 of Fig. 7B as Ph-PNPase (Fig. 7C). In this Western blot, trace amounts of Ph-PNPase were detected in lanes 2 and 3, but this result, which is not reproducible, is likely due to incomplete removal during the wash step. Taken together, these results suggest that the interaction of PNPase and RNase E is species specific. This conclusion is supported by the in vivo results shown in Fig. 5, in which we failed to detect an interaction between Ec-PNPase and Ph-RNase E. Each of the deletions (Fig. 7B, lanes 3 to 5) failed to interact with Ph-PNPase, demonstrating that residues 1035 to 1071 of Ph-RNase E are necessary for the interaction with Ph-PNPase. In subsequent work, we overexpressed Ph-PNPase at 25°C in an E. coli strain in which the gene encoding Ec-PNPase was disrupted, and we prepared a highly enriched fraction of Ph-PNPase by high-speed centrifugation and ammonium sulfate precipitation. For unknown reasons, we have not been able to demonstrate specific binding to the Ph-CTH using purified Ph-PNPase. Our results therefore need to be interpreted with caution, since we lack proof that Ph-PNPase by itself is sufficient for the interaction with Ph-RNase E.
FIG. 7.
Interaction between Ph-RNase E and Ph-PNPase. (A) Schematic diagram of His-tagged polypeptides corresponding to the CTH of Ph-RNase E (pSAB20) and a nested set of deletions extending from the C-terminal end (pSAB35, pSAB34, and pSAB33). (B) SDS-PAGE stained with Sypro orange of proteins that copurify with the His-tagged CTH peptides. Lane 1, negative control without His-tagged polypeptide; lane 2, control using the His-tagged CTH of Ec-RNase E; lanes 3 to 6, pSAB33, pSAB34, pSAB35, and pSAB20, respectively; lanes 7 and 8, controls showing P. haloplanktis and E. coli whole-cell extracts, respectively. M, molecular mass markers. The black dots in lanes 2 to 6 indicate the positions of the His-tagged polypeptides. The arrow to the right of the panel indicates the position of a protein in lane 6 that copurifies with the CTH of Ph-RNase E and migrates at a position corresponding to the molecular mass of Ph-PNPase. (C) Western blot. A gel comparable to the gel shown in panel B was blotted and probed with antibodies against Ec-PNPase. The major signal detected in lane 8 corresponds to Ec-PNPase, which runs as a slightly larger protein than Ph-PNPase (lanes 6 and 7).
DISCUSSION
We report here the identification of the RNA degradosome of P. haloplanktis and the characterization of microdomains in Ph-RNase E involved in the protein-protein interactions. We have also shown that Ph-RNase E can restore the viability of E. coli strains lacking Ec-RNase E. The introduction of the Ph-rne gene on a low-copy-number plasmid (pSAB15) into E. coli in which the chromosomal Ec-rne gene was disrupted resulted in weak complementation, with detectable colony formation only after 6 days at 25°C. Analysis of Ph-RNase E in this strain showed that the amount was at least 32-fold lower than the normal amount of Ec-RNase E. Thus, the Ph-rne gene under endogenous expression signals is poorly expressed in E. coli. To improve expression of Ph-RNase E in E. coli, we constructed a hybrid gene in which the 5′ expression signals of the Ec-rne gene were fused to the coding sequence of Ph-rne. The resulting fusion on a low-copy-number plasmid (pSAB18) complemented E. coli at temperatures ranging from 15 to 37°C. The amount of Ph-RNase E in this strain is about 50% of the normal amount of Ec-RNase E. Taken together, these results show that the poor complementation with the Ph-rne gene is due to the low level of Ph-RNase E expression in E. coli.
Since Ph-RNase E is obtained from a psychrotolerant bacterium, we were surprised to observe that complementation in E. coli at 15°C is weak compared to that of the Ec-RNase E control. We have presented experimental evidence that the amount of Ph-RNase E detected at 15°C is comparable to the amount detected at 30°C. Since the endoribonuclease activity of Ph-RNase E is expected to be comparable to or higher than the activity of Ec-RNase E at 15°C, this result suggests that Ph-RNase E fails to supply a noncatalytic function necessary for growth in the cold. In addition, we have shown that the E. coli MC1061 strain harboring the rne131 allele, expressing Ec-RNase E lacking the noncatalytic region (32), grows slower than the parental MC1061 strain at 15°C. These results suggest that the noncatalytic region of Ec-RNase E provides a function that facilitates growth at low temperature. This function could, for example, involve the interaction between Ec-RNase E and a protein component of the RNA degradosome.
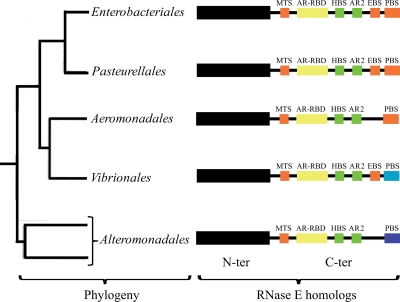
The noncatalytic region of Ec-RNase E contains two well-characterized microdomains that are involved in protein-protein interactions with enolase and PNPase. The structures of these microdomains complexed with their partner proteins have been elucidated by X-ray crystallography. The microdomain involved in enolase binding forms an α-helix that makes an extensive network of hydrogen-bonding interactions at the enolase dimer interface (8). The sequence motif corresponding to the microdomain involved in enolase binding is conserved throughout the Enterobacteriales. A recent analysis of Vibrio angustum S14, a member of the Vibrionales, involving sequence comparisons and two hybrid tests has demonstrated the conservation of the EBS in RNase E homologs of these bacteria (16). Taken together with our results, we can predict that the EBS is conserved in RNase E homologs of the Enterobacteriales, Pasteurellales, and Vibrionales but absent in the Aeromonadales, Alteromonadales, and more distantly related Gammaproteobacteria (Fig. 8). These observations suggest that the interaction with enolase was acquired during the evolution of the Gammaproteobacteria. The association of enolase with RNase E in the Enterobacteriales could link the mRNA degradation machinery to the control of central intermediary metabolism. P. haloplanktis inhabits a marine environment that is poor in sources of carbon, nitrogen, and phosphorus. It notably lacks the cyclic AMP-catabolite gene activator protein (cAMP-CAP) system responsible for catabolite repression and the phosphotransferase system (PTS) required for the transport of glucose (39). The association of enolase with RNase E in E. coli might therefore be an adaptation to a niche such as the animal gut, where nutrient levels vary dramatically and rapid adjustments in the control of nutrient uptake and utilization give a competitive advantage.
FIG. 8.
Primary structure of RNase E homologs in the Enterobacteriales, Pasteurellales, Aeromonadales, Vibrionales, and Alteromonadales. The tree on the left shows in outline form the relationship of five orders of the Gammaproteobacteria (17). The schematic diagrams on the right summarize our understanding of the primary structures of the RNase E homologs of each of these orders. The N-terminal catalytic domain is represented as a black box; the C-terminal noncatalytic region is represented as a line. Microdomains are represented as colored boxes following the scheme described in the legend to Fig. 1. The RNase E homologs of the Aeromonadales have a PNPase binding site similar to the homologs from the Enterobacteriales and Pasteurellales but lack identifiable enolase binding sites based on sequence comparisons. For the RNase E homologs of the Vibrionales and Alteromonadales, the light and dark blue boxes, respectively, represent PNPase binding sites that are unrelated by sequence to each other or to the corresponding sites in homologs from the Enterobacteriales and Pasteurellales.
The microdomain of Ec-RNase E involved in the protein-protein interaction with Ec-PNPase extends a preexisting β-sheet on the surface of PNPase (43). β-sheet extension has been suggested as a potentially common element in protein-protein interactions (49). The RNase E-PNPase interaction appears to be widespread in the Proteobacteria. Our results, however, suggest that the interaction is species specific. That is, Ec-RNase E binds Ec-PNPase but not Ph-PNPase, and Ph-RNase E binds Ph-PNPase but not Ec-PNPase. Our results, together with the recent two-hybrid analysis of the RNase E-PNPase interaction between Vibrio angustum S14 homologs (16), shows that this interaction does not correspond to a sequence motif conserved among the Alteromonadales or the Vibrionales. Since the interaction is species specific, one possibility is that the underlying structural motif for the interaction is different in different bacteria, although this seems unlikely. Another possibility is that the secondary structure involved in β-sheet extension is conserved but that the constraint on protein sequence of the RNase E microdomains involved in β-sheet extension is low. These considerations suggest that the species-specific RNase E-PNPase interaction could involve a conserved β-sheet extension that imposes minimal constrains on the protein sequence.
PNPase is a phosphate-dependent exoribonuclease. Although PNPase has been found to be associated with RNase E in several different bacteria, this is not always the case. Notably, RNase E of the psychrophilic gammaproteobacterium Pseudomonas syringae Lz4W has been shown to be associated with RNase R, which is a hydrolytic exoribonuclease (46). Exoribonucleases related to PNPase and RNase R are widely distributed in bacteria, plants, and animals (13, 40). These enzymes represent two distinct families of exoribonucleases. Like P. haloplanktis, Pseudomonas syringae Lz4W is a cold-adapted marine gammaproteobacterium (50). Since both of these bacteria inhabit similar environmental niches, the formation of an RNA degradosome with a phosphorolytic rather than hydrolytic exoribonuclease does not appear to correlate with the conditions of growth in a cold marine environment.
Our results show that the Ph-RhlB interacts with Ph-RNase E as a component of the RNA degradosome of P. haloplanktis. In Ec-RNase E, the minimal binding site for RhlB has been mapped to the region comprising residues 698 to 762 (9), and a sequence motif within the region is well conserved in the Enterobacteriales (Fig. 6A). The substitution of a highly conserved arginine residue in this motif for alanine (R730A) abolishes RhlB binding (27). Ph-RNase E contains a related sequence motif in the same region, which includes a conserved arginine residue corresponding to R730. We have shown that deletion of the region corresponding to the motif abolishes the interaction between Ph-RNase E and Ec-RhlB. In Fig. 1C, region 1, corresponding to residues 747 to 758 of Ph-RNase E, is predicted to have the capacity to form secondary structure. We mapped the Ph-HBS to residues 683 to 716. Thus, the region predicted to have the capacity to form secondary structure and the region necessary for binding to RhlB are distinct. Like other DEAD-box RNA helicases, the catalytic core of RhlB is comprised of two structurally related domains. The second domain of RhlB has been shown to be necessary and sufficient for the interaction with RNase E (9). The surface of RhlB involved in this interaction and the structure of the microdomain corresponding to the binding site in RNase E have not yet been elucidated (see reference 56). The heterologous interaction of Ec-RhlB with Ph-RNase E suggests that the underlying structural motif for the protein-protein interaction is conserved over a large evolutionary distance. Experimental evidence has shown that RhlB needs to interact with RNase E as part of the RNA degradosome to have biological activity (24, 26). The interaction with RNase E activates the ATPase and RNA unwinding activities of RhlB (9, 27, 53, 56). These considerations suggest that the conservation of the RhlB binding site over a large evolutionary distance could be due to structural constraints involved in the control of RhlB activity.
Figure 8 shows a simplified phylogenetic tree of the Enterobacteriales, Pasteurellales, Aeromonadales, Vibrionales, and Alteromonadales (17) and summarizes our understanding of the primary structure of the RNase E homologs in these orders of the Gammaproteobacteria. The identification of the RNA degradosome of P. haloplanktis has revealed two widely conserved interactions with RNase E homologs of the Gammaproteobacteria. Although the interaction is conserved, the PNPase binding site (PBS) does not correspond to a conserved sequence motif. The light and dark blue boxes for the RNase E homologs of the Vibrionales and the Alteromonadales, respectively, indicate that the protein sequences of the PBSs are distinct from the sequences in the homologs of the Enterobacteriales, Pasteurellales, and Aeromonadales. The binding site for RhlB (HBS) corresponds to a conserved sequence motif. Despite the lack of overall sequence conservation of the CTH of the RNase E homologs, our result suggests that there is nevertheless a conserved “module” composed of the MTS, AR-RBD, HBS, and AR2. Based on experimental results, we suggested previously that the AR-RBD and AR2 cooperate in the RNA unwinding activity of RhlB (9). The conservation of the MTS/AR-RBD/HBS/AR2 module over a relatively large evolutionary distance suggests an important conserved function, which for example, could be the control of the activity or specificity of RhlB within the RNA degradosome.
Acknowledgments
We thank I. Bagdiul, M. Dreyfus, and V. Khemici for providing the plasmids and strains; members of the Carpousis group, M. Dreyfus and B. Luisi, for helpful discussions; and B. Clouet-d'Orval and P. Genevaux for critical reading of the manuscript.
S.A.-B. was supported by a predoctoral fellowship from the MESR (Ministère de l'Enseignement Supérieur et de la Recherche). Research in our group is supported by the CNRS (Centre National de la Recherche Scientifique) and the ANR (Agence Nationale de la Recherche, projects CARMa NT05_1-44659 and mRNases BLAN08-1_329396).
Footnotes
Published ahead of print on 20 August 2010.
REFERENCES
- 1.Birolo, L., M. L. Tutino, B. Fontanella, C. Gerday, K. Mainolfi, S. Pascarella, G. Sannia, F. Vinci, and G. Marino. 2000. Aspartate aminotransferase from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC 125. Cloning, expression, properties, and molecular modelling. Eur. J. Biochem. 267:2790-2802. [DOI] [PubMed] [Google Scholar]
- 2.Callaghan, A. J., J. P. Aurikko, L. L. Ilag, J. Gunter Grossmann, V. Chandran, K. Kuhnel, L. Poljak, A. J. Carpousis, C. V. Robinson, M. F. Symmons, and B. F. Luisi. 2004. Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J. Mol. Biol. 340:965-979. [DOI] [PubMed] [Google Scholar]
- 3.Callaghan, A. J., M. J. Marcaida, J. A. Stead, K. J. McDowall, W. G. Scott, and B. F. Luisi. 2005. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437:1187-1191. [DOI] [PubMed] [Google Scholar]
- 4.Carpousis, A. J. 2002. The Escherichia coli RNA degradosome: structure, function and relationship to other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 30:150-155. [PubMed] [Google Scholar]
- 5.Carpousis, A. J., V. Khemici, S. Ait-Bara, and L. Poljak. 2008. Co-immunopurification of multiprotein complexes containing RNA-degrading enzymes. Methods Enzymol. 447:65-82. [DOI] [PubMed] [Google Scholar]
- 6.Carpousis, A. J., B. F. Luisi, and K. J. McDowall. 2009. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog. Mol. Biol. Transl. Sci. 85:91-135. [DOI] [PubMed] [Google Scholar]
- 7.Carpousis, A. J., G. Van Houwe, C. Ehretsmann, and H. M. Krisch. 1994. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76:889-900. [DOI] [PubMed] [Google Scholar]
- 8.Chandran, V., and B. F. Luisi. 2006. Recognition of enolase in the Escherichia coli RNA degradosome. J. Mol. Biol. 358:8-15. [DOI] [PubMed] [Google Scholar]
- 9.Chandran, V., L. Poljak, N. F. Vanzo, A. Leroy, R. N. Miguel, J. Fernandez-Recio, J. Parkinson, C. Burns, A. J. Carpousis, and B. F. Luisi. 2007. Recognition and cooperation between the ATP-dependent RNA helicase RhlB and ribonuclease RNase E. J. Mol. Biol. 367:113-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 12.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 62:55-108. [DOI] [PubMed] [Google Scholar]
- 13.Condon, C., and H. Putzer. 2002. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 30:5339-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deutscher, M. P. 2006. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 34:659-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duilio, A., S. Madonna, M. L. Tutino, M. Pirozzi, G. Sannia, and G. Marino. 2004. Promoters from a cold-adapted bacterium: definition of a consensus motif and molecular characterization of UP regulative elements. Extremophiles 8:125-132. [DOI] [PubMed] [Google Scholar]
- 16.Erce, M. A., J. K. Low, P. E. March, M. R. Wilkins, and K. M. Takayama. 2009. Identification and functional analysis of RNase E of Vibrio angustum S14 and two-hybrid analysis of its interaction partners. Biochim. Biophys. Acta 1794:1107-1114. [DOI] [PubMed] [Google Scholar]
- 17.Gao, B., R. Mohan, and R. S. Gupta. 2009. Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 59:234-247. [DOI] [PubMed] [Google Scholar]
- 18.Gao, J., K. Lee, M. Zhao, J. Qiu, X. Zhan, A. Saxena, C. J. Moore, S. N. Cohen, and G. Georgiou. 2006. Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Mol. Microbiol. 61:394-406. [DOI] [PubMed] [Google Scholar]
- 19.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 20.Jager, S., O. Fuhrmann, C. Heck, M. Hebermehl, E. Schiltz, R. Rauhut, and G. Klug. 2001. An mRNA degrading complex in Rhodobacter capsulatus. Nucleic Acids Res. 29:4581-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jager, S., M. Hebermehl, E. Schiltz, and G. Klug. 2004. Composition and activity of the Rhodobacter capsulatus degradosome vary under different oxygen concentrations. J. Mol. Microbiol. Biotechnol. 7:148-154. [DOI] [PubMed] [Google Scholar]
- 22.Jain, C., and J. G. Belasco. 1995. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: unusual sensitivity of the rne transcript to RNase E activity. Genes Dev. 9:84-96. [DOI] [PubMed] [Google Scholar]
- 23.Kaberdin, V. R., A. Miczak, J. S. Jakobsen, S. Lin-Chao, K. J. McDowall, and A. von Gabain. 1998. The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl. Acad. Sci. U. S. A. 95:11637-11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khemici, V., and A. J. Carpousis. 2004. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 51:777-790. [DOI] [PubMed] [Google Scholar]
- 25.Khemici, V., L. Poljak, B. F. Luisi, and A. J. Carpousis. 2008. The RNase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol. 70:799-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khemici, V., L. Poljak, I. Toesca, and A. J. Carpousis. 2005. Evidence in vivo that the DEAD-box RNA helicase RhlB facilitates the degradation of ribosome-free mRNA by RNase E. Proc. Natl. Acad. Sci. U. S. A. 102:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khemici, V., I. Toesca, L. Poljak, N. F. Vanzo, and A. J. Carpousis. 2004. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol. Microbiol. 54:1422-1430. [DOI] [PubMed] [Google Scholar]
- 28.Kushner, S. R. 2002. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 184:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, K., J. A. Bernstein, and S. N. Cohen. 2002. RNase G complementation of rne null mutation identifies functional interrelationships with RNase E in Escherichia coli. Mol. Microbiol. 43:1445-1456. [DOI] [PubMed] [Google Scholar]
- 30.Lee, K., and S. N. Cohen. 2003. A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol. Microbiol. 48:349-360. [DOI] [PubMed] [Google Scholar]
- 31.Lee, K., X. Zhan, J. Gao, J. Qiu, Y. Feng, R. Meganathan, S. N. Cohen, and G. Georgiou. 2003. RraA: a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell 114:623-634. [PubMed] [Google Scholar]
- 32.Leroy, A., N. F. Vanzo, S. Sousa, M. Dreyfus, and A. J. Carpousis. 2002. Function in Escherichia coli of the non-catalytic part of RNase E: role in the degradation of ribosome-free mRNA. Mol. Microbiol. 45:1231-1243. [DOI] [PubMed] [Google Scholar]
- 33.Li, X., P. Romero, M. Rani, A. K. Dunker, and Z. Obradovic. 1999. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 10:30-40. [PubMed] [Google Scholar]
- 34.Li, Z., S. Pandit, and M. P. Deutscher. 1999. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 18:2878-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez, P. J., I. Marchand, S. A. Joyce, and M. Dreyfus. 1999. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 33:188-199. [DOI] [PubMed] [Google Scholar]
- 36.Mackie, G. A. 1998. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395:720-723. [DOI] [PubMed] [Google Scholar]
- 37.Marcaida, M. J., M. A. DePristo, V. Chandran, A. J. Carpousis, and B. F. Luisi. 2006. The RNA degradosome: life in the fast lane of adaptive molecular evolution. Trends Biochem. Sci. 31:359-365. [DOI] [PubMed] [Google Scholar]
- 38.McDowall, K. J., and S. N. Cohen. 1996. The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding site. J. Mol. Biol. 255:349-355. [DOI] [PubMed] [Google Scholar]
- 39.Medigue, C., E. Krin, G. Pascal, V. Barbe, A. Bernsel, P. N. Bertin, F. Cheung, S. Cruveiller, S. D'Amico, A. Duilio, G. Fang, G. Feller, C. Ho, S. Mangenot, G. Marino, J. Nilsson, E. Parrilli, E. P. Rocha, Z. Rouy, A. Sekowska, M. L. Tutino, D. Vallenet, G. von Heijne, and A. Danchin. 2005. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 15:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mian, I. S. 1997. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 25:3187-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miczak, A., V. R. Kaberdin, C. L. Wei, and S. Lin-Chao. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. U. S. A. 93:3865-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morita, T., K. Maki, and H. Aiba. 2005. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 19:2176-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nurmohamed, S., B. Vaidialingam, A. J. Callaghan, and B. F. Luisi. 2009. Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly. J. Mol. Biol. 389:17-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ow, M. C., Q. Liu, and S. R. Kushner. 2000. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol. Microbiol. 38:854-866. [DOI] [PubMed] [Google Scholar]
- 45.Prud'homme-Genereux, A., R. K. Beran, I. Iost, C. S. Ramey, G. A. Mackie, and R. W. Simons. 2004. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome.’ Mol. Microbiol. 54:1409-1421. [DOI] [PubMed] [Google Scholar]
- 46.Purusharth, R. I., F. Klein, S. Sulthana, S. Jager, M. V. Jagannadham, E. Evguenieva-Hackenberg, M. K. Ray, and G. Klug. 2005. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J. Biol. Chem. 280:14572-14578. [DOI] [PubMed] [Google Scholar]
- 47.Py, B., C. F. Higgins, H. M. Krisch, and A. J. Carpousis. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169-172. [DOI] [PubMed] [Google Scholar]
- 48.Regonesi, M. E., M. Del Favero, F. Basilico, F. Briani, L. Benazzi, P. Tortora, P. Mauri, and G. Deho. 2006. Analysis of the Escherichia coli RNA degradosome composition by a proteomic approach. Biochimie 88:151-161. [DOI] [PubMed] [Google Scholar]
- 49.Remaut, H., and G. Waksman. 2006. Protein-protein interaction through beta-strand addition. Trends Biochem. Sci. 31:436-444. [DOI] [PubMed] [Google Scholar]
- 50.Shivaji, S., N. S. Rao, L. Saisree, V. Sheth, G. S. Reddy, and P. M. Bhargava. 1989. Isolation and identification of Pseudomonas spp. from Schirmacher Oasis, Antarctica. Appl. Environ. Microbiol. 55:767-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taraseviciene, L., G. R. Bjork, and B. E. Uhlin. 1995. Evidence for an RNA binding region in the Escherichia coli processing endoribonuclease RNase E. J. Biol. Chem. 270:26391-26398. [DOI] [PubMed] [Google Scholar]
- 52.Tock, M. R., A. P. Walsh, G. Carroll, and K. J. McDowall. 2000. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem. 275:8726-8732. [DOI] [PubMed] [Google Scholar]
- 53.Vanzo, N. F., Y. S. Li, B. Py, E. Blum, C. F. Higgins, L. C. Raynal, H. M. Krisch, and A. J. Carpousis. 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12:2770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wachi, M., G. Umitsuki, M. Shimizu, A. Takada, and K. Nagai. 1999. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem. Biophys. Res. Commun. 259:483-488. [DOI] [PubMed] [Google Scholar]
- 55.Worrall, J. A., M. Gorna, N. T. Crump, L. G. Phillips, A. C. Tuck, A. J. Price, V. N. Bavro, and B. F. Luisi. 2008. Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J. Mol. Biol. 382:870-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Worrall, J. A., F. S. Howe, A. R. McKay, C. V. Robinson, and B. F. Luisi. 2008. Allosteric activation of the ATPase activity of the Escherichia coli RhlB RNA helicase. J. Biol. Chem. 283:5567-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]