Abstract
Escherichia coli K-12 provided with glucose and a mixture of amino acids depletes l-serine more quickly than any other amino acid even in the presence of ammonium sulfate. A mutant without three 4Fe4S l-serine deaminases (SdaA, SdaB, and TdcG) of E. coli K-12 is unable to do this. The high level of l-serine that accumulates when such a mutant is exposed to amino acid mixtures starves the cells for C1 units and interferes with cell wall synthesis. We suggest that at high concentrations, l-serine decreases synthesis of UDP-N-acetylmuramate-l-alanine by the murC-encoded ligase, weakening the cell wall and producing misshapen cells and lysis. The inhibition by high l-serine is overcome in several ways: by a large concentration of l-alanine, by overproducing MurC together with a low concentration of l-alanine, and by overproducing FtsW, thus promoting septal assembly and also by overexpression of the glycine cleavage operon. S-Adenosylmethionine reduces lysis and allows an extensive increase in biomass without improving cell division. This suggests that E. coli has a metabolic trigger for cell division. Without that reaction, if no other inhibition occurs, other metabolic functions can continue and cells can elongate and replicate their DNA, reaching at least 180 times their usual length, but cannot divide.
The Escherichia coli genome contains three genes, sdaA, sdaB, and tdcG, specifying three very similar 4Fe4S l-serine deaminases. These enzymes are very specific for l-serine for which they have unusually high Km values (3, 32). Expression of the three genes is regulated so that at least one of the gene products is synthesized under all common growth conditions (25). This suggests an important physiological role for the enzymes. However, why E. coli needs to deaminate l-serine has been a long-standing problem of E. coli physiology, the more so since it cannot use l-serine as the sole carbon source.
We showed recently that an E. coli strain devoid of all three l-serine deaminases (l-SDs) loses control over its size, shape, and cell division when faced with complex amino acid mixtures containing l-serine (32). We attributed this to starvation for single-carbon (C1) units and/or S-adenosylmethionine (SAM). C1 units are usually made from serine via serine hydroxymethyl transferase (GlyA) or via glycine cleavage (GCV). The l-SD-deficient triple mutant strain is starved for C1 in the presence of amino acids, because externally provided glycine inhibits GlyA and a very high internal l-serine concentration along with several other amino acids inhibits glycine cleavage. While the parent cell can defend itself by reducing the l-serine level by deamination, this crucial reaction is missing in the ΔsdaA ΔsdaB ΔtdcG triple mutant. We therefore consider these to be “defensive” serine deaminases.
The fact that an inability to deaminate l-serine leads to a high concentration of l-serine and inhibition of GlyA is not surprising. However, it is not obvious why a high level of l-serine inhibits cell division and causes swelling, lysis, and filamentation. Serine toxicity due to inhibition of biosynthesis of isoleucine (11) and aromatic amino acids (21) has been reported but is not relevant here, since these amino acids are provided in Casamino Acids.
We show here that at high internal concentrations, l-serine also causes problems with peptidoglycan synthesis, thus weakening the cell wall. Peptidoglycan is a polymer of long glycan chains made up of alternating N-acetylglucosamine and N-acetylmuramic acid residues, cross-linked by l-alanyl-γ-d-glutamyl-meso-diaminopimelyl-d-alanine tetrapeptides (1, 28). The glucosamine and muramate residues and the pentapeptide (from which the tetrapeptide is derived) are all synthesized in the cytoplasm and then are exported to be polymerized into extracellular peptidoglycan (2).
In this paper, we show that lysis is caused by l-serine interfering with the first step of synthesis of the cross-linking peptide, the addition of l-alanine to uridine diphosphate-N-acetylmuramate. This interference is probably due to a competition between serine and l-alanine for the ligase, MurC, which adds the first l-alanine to UDP-N-acetylmuramate (7, 10, 15). As described here, the weakening of the cell wall by l-serine can be overcome by a variety of methods that reduce the endogenous l-serine pool or counteract the effects of high levels of l-serine.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study, all derivatives of E. coli K-12, are described in Table 1. D. O. Wood kindly gave us the rickettsial SAM transporter gene (26) which we subcloned as described earlier (31).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or relevant characteristic(s) | Source and/or reference |
|---|---|---|
| Strains | ||
| Cu1008 | E. coli K-12 ilvA | L. S. Williams |
| MEW1 | Cu1008 Δlac | 24 |
| MEW999 | MEW1 ΔsdaA ΔsdaB ΔtdcG | 32 |
| JE7968 | Pmra::Plac; IPTG-dependent strain; Cmr | 16 |
| MEW999H | MEW999 derivative; Pmra::Plac; IPTG-dependent strain; Cmr | This study |
| Plasmids | ||
| JW0089 | pCA24N carrying murC | ASKA library (14) |
| JW0087 | pCA24N carrying ftsW | ASKA library (14) |
| JW0086 | pCA24N carrying murD | ASKA library (14) |
| JW0083 | pCA24N carrying murE | ASKA library (14) |
| JW0084 | pCA24N carrying murF | ASKA library (14) |
| JW0094 | pCA24N carrying lpxC | ASKA library (14) |
| JW0093 | pCA24N carrying ftsZ | ASKA library (14) |
| JW0081 | pCA24N carrying ftsL | ASKA library (14) |
| JW0091 | pCA24N carrying ftsQ | ASKA library (14) |
| JW0082 | pCA24N carrying ftsI | ASKA library (14) |
| JW0372 | pCA24N carrying ddlA | ASKA library (14) |
| JW0090 | pCA24N carrying ddlB | ASKA library (14) |
| psamT | pLtet plasmid carrying the rickettsial SAM transporter from pSMART | 26 |
| pGS146 | Expression plasmid carrying the gcvTHP operon | 23 |
Growth media, conditions, and genetic methods are as described in reference 32. Strain MEW999, the triple mutant, grew as wild-type E. coli in glucose minimal medium but had the various problems described in the text when grown in glucose minimal medium with 0.5% Casamino Acids (NGCAA). The cells were routinely grown overnight and diluted 104-fold in fresh medium to start the experiments unless otherwise noted. The cells were grown at 37°C unless otherwise stated.
Microscopy.
All photographs in this paper were taken with a Leica DMIRE2 microscope except for those in Fig. 7, which were taken with a National DC3-163. In all cases, the images in each figure come from a single experiment, which was repeated at least once more with similar results.
Fixation method.
To prepare the cells for microscopy, 500 μl of the culture was added with 20 μl of sodium phosphate buffer (pH 7.4) to 100 μl of fix solution (0.2 ml of 50% glutaraldehyde in 100 ml of 16% paraformaldehyde). This mixture was incubated at room temperature for 15 min and on ice for 15 min and centrifuged at 4,000 to 5,000 rpm for 5 min. The pellet was washed 2 or 3 times in 1 ml of phosphate-buffered saline (PBS) buffer and resuspended in PBS buffer at a concentration of 50 to 100 μl of PBS per 0.1 optical density at 600 nm (OD600) unit.
Staining of DNA with DAPI.
The fixed cells were incubated at room temperature in the dark with a mixture of 10 μl of 20-μg/ml DAPI (4′-6-diamidino-2-phenylindole) in 1 ml PBS buffer for 5 to 10 min, then washed 2 or 3 times with PBS buffer to reduce the background of DAPI, and finally resuspended in PBS buffer at a concentration of 50 to 100 μl PBS/0.1 OD600 unit.
Determination of amino acid concentrations in culture supernatants. (i) Growth and sample preparation.
Strain MEW1 and the triple mutant strain MEW999 grown in our potassium phosphate-based medium with glucose at 37°C overnight were chilled in ice water and diluted into 25-ml cultures of the same medium with 0.5% Casamino Acids (NGCAA). The NGCAA medium for all samples in an experiment was prepared in a single batch and then split into 25-ml aliquots for each sample. At 4, 6, 8, and 10 h, 0.5 ml was fixed and the cells were examined with a microscope as described previously, while 1.5 ml for amino acid analysis was immediately chilled and then centrifuged and the supernatant was filtered through a 0.2-μm filter. A portion (1.5 ml) of the initial NGCAA mixture was assayed and considered to represent the starting concentrations. The supernatants were stored at −86°C and taken to the Sick Children's Hospital in Toronto on dry ice for analysis.
(ii) Free amino acid analysis.
The free amino acid analysis was performed by Rey Interior of the Amino Acid Analysis Facility of the Sick Children's Hospital Advanced Protein Technology Center using a Waters Acquity UPLC system. An aliquot of each sample was transferred into a 6- by 55-mm glass culture tube and dried under vacuum using a centrifugal evaporator. The dried sample was treated with a redrying solution consisting of methanol-water-triethylamine (2:2:1), dried under vacuum for 15 min, and incubated for 20 min at room temperature with a derivatizing solution of methanol-water-triethylamine-phenylisothiocyanate (PITC) (7:1:1:1), after which the derivatizing solution was removed by 15-min incubation under vacuum. The derivatized sample was again washed with the redrying solution, vortex mixed, and dried under vacuum for 15 min.
The derivatized sample was dissolved in sample diluent (pH 7.40) and an aliquot was injected into the column, running on a modified Pico-Tag gradient. The column temperature was 48°C. The derivatized amino acids were detected at 254 nm.
The Waters Acquity UPLC system employed consists of a binary solvent manager, a sample manager, a tuneable UV (TUV) detector, and a Waters Acquity UPLC BEH C18 column (2.1 by 100 mm). Data were collected, stored, and processed using Waters Empower 2 chromatography software. Drying was done using a Tomy CC-181 centrifugal concentrator with a Sargent-Welch model 8821 vacuum pump.
(iii) Assessment of the validity of the results.
The entire determination of supernatant amino acid concentrations was carried out twice. In the first, the samples were derivatized and assayed once. In the second, which was entirely consistent with the first, each sample was derivatized twice and each derivatized sample was assayed. The results of these assays varied less than 3%, except for cysteine which occasionally varied much more. Table 2 presents the results of the second experiment as the average of the two determinations.
TABLE 2.
Amino acid composition of medium after incubationa
| Amino acid | Amt (μmol/liter) of amino acid in original medium | % amino acid remaining after incubation |
Amt (mol) of amino acid relative to 100 mol of glycine in: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MEW1 (parent strain) |
MEW999 (triple mutant) |
E. coli cell proteinb | Original medium | ||||||||
| 4 h | 6 h | 8 h | 10 h | 4 h | 6 h | 8 h | 10 h | ||||
| Asp | 1,170 | 105 | 94 | 6 | 6 | 103 | 90 | 59 | 32 | 79 | 245 |
| Glu | 1,890 | 113 | 103 | 64 | 2 | 112 | 102 | 84 | 51 | 86 | 396 |
| Ser | 770 | 83 | 3 | 1 | 0 | 94 | 76 | 35 | 1 | 35 | 161 |
| Gly | 480 | 111 | 110 | 68 | 0 | 103 | 103 | 89 | 80 | 100 | 100 |
| His | 140 | 111 | 107 | 104 | 101 | 98 | 102 | 112 | 103 | 15 | 30 |
| Arg | 270 | 107 | 91 | 11 | 3 | 105 | 87 | 57 | 15 | 48 | 56 |
| Thr | 410 | 118 | 94 | 24 | 1 | 119 | 107 | 96 | 64 | 41 | 85 |
| Ala | 680 | 112 | 107 | 83 | 0 | 111 | 105 | 100 | 85 | 84 | 142 |
| Pro | 1,220 | 115 | 102 | 68 | 2 | 114 | 106 | 106 | 94 | 36 | 255 |
| Tyr | 130 | 110 | 104 | 47 | 8 | 107 | 98 | 102 | 61 | 23 | 27 |
| Val | 4,640 | 108 | 104 | 88 | 100 | 105 | 98 | 104 | 87 | 69 | 971 |
| Met | 190 | 118 | 108 | 65 | 45 | 116 | 98 | 105 | 67 | 25 | 40 |
| Cys | 110 | 95 | 101 | 25 | 15 | 94 | 97 | 76 | 38 | 15 | 24 |
| Ile | 3,600 | 110 | 104 | 83 | 98 | 108 | 93 | 104 | 80 | 47 | 754 |
| Leu | 890 | 117 | 117 | 90 | 87 | 116 | 102 | 119 | 87 | 74 | 185 |
| Phe | 200 | 123 | 109 | 57 | 32 | 121 | 88 | 103 | 58 | 30 | 41 |
| Lys | 570 | 120 | 112 | 74 | 87 | 116 | 81 | 80 | 55 | 56 | 120 |
Cells of the parent strain (MEW1) and the triple mutant (MEW999) grown overnight in minimal medium containing glucose were subcultured into minimal medium containing glucose, ammonium sulfate, and Casamino Acids (NGCAA). Aliquots of the original medium and aliquots of medium in which cells were incubated for the times noted were chilled, centrifuged, and filtered. The supernatant was frozen, and the amino acids were detected as described in Materials and Methods. The amount of amino acid in the original medium is given in micromoles per liter in the second column from the left. This may be compared to the amount of each amino acid in average E. coli protein given in the second column from the right as micromoles relative to the amount of glycine and to the amount of each amino acid in the original medium expressed in the same way in the rightmost column. The amounts in the original medium and the percentages are averages of two determinations. The details of the method and assessment of its accuracy are described in Materials and Methods.
Data from reference 17.
RESULTS
Concentrations of amino acids in supernatants from cultures with glucose and Casamino Acids.
The triple mutant (strain MEW999) grows normally in minimal medium with glucose and ammonium sulfate, but its growth and morphology are badly distorted in the same medium with Casamino Acids (0.5%) added (NGCAA). Our explanation for this phenotype rests on the assumption that a cell deficient in l-serine deaminase would remove l-serine from an amino acid mixture more slowly than its parent strain (32). Therefore, if faced with a mixture of amino acids, including l-serine, the mutant will remain in the presence of inhibitory concentrations of serine for much longer. The following experiment demonstrates this clearly.
In this experiment, an overnight culture in glucose-containing minimal medium is diluted into the same medium with Casamino Acids (NGCAA). We expect that the parent E. coli K-12 (strain MEW1) should grow and take up amino acids at an increasing rate, relatively little in the first 4 h which starts at a low inoculum and faster as more cells accumulate. The concentration of free l-serine in the medium would drop as it is imported, degraded to pyruvate, assimilated as l-serine itself, and converted to several other biosynthetic intermediates. However, a strain which has no l-serine deaminase should deplete the l-serine in the medium more slowly and remain in the presence of inhibitory concentrations much longer.
To verify this, we compared the concentration of each of the amino acids in the medium during incubation of subcultures of the parent strain MEW1 and the triple mutant in NGCAA (see “Determination of amino acid concentrations in culture supernatants” in Materials and Methods). We also photographed the cells at every time point to verify that the parent maintained its usual shape and the mutant enlarged and lysed as described earlier (32). Because the strains behaved exactly as reported for other experiments, the pictorial data are not shown. The rates at which the amino acids decreased varied depending on the amino acid and strain (Table 2).
Because these cultures begin with a small inoculum, neither strain accumulated much biomass in the first 4 h. Nonetheless, the parent strain removed more than 10% of l-serine in the first 4 h and 97% by 6 h, a time when none of the other amino acids had decreased by more than 15%. Clearly, E. coli K-12 metabolism is organized to remove l-serine from its environment very quickly and efficiently, even when it is well supplied with glucose and ammonium sulfate. In contrast, at 6 and 8 h, the medium inoculated with the triple mutant retained 76% and 35% of its original serine, as opposed to 3% and 1% in the medium inoculated with the parent.
This clearly supports the hypothesis that the triple mutant is inhibited by continuing high levels of l-serine. However, Casamino Acids toxicity is caused by l-serine with five other amino acids (glycine, threonine, methionine, leucine, and lysine [32]). All of these are still present after 8 h in the mutant medium at 80% or more of the original concentration. Indeed, only threonine decreased to markedly lower levels even in the parent strain. We would then expect the medium in which the mutant grew to retain its inhibitory properties for the first 8 h of the experiment, though by 8 to 10 h, the inhibition should be over because of the depletion of l-serine. All the results reported here and in the earlier paper are consistent with this.
Effect of SAM on growth and lysis of the triple mutant in NGCAA.
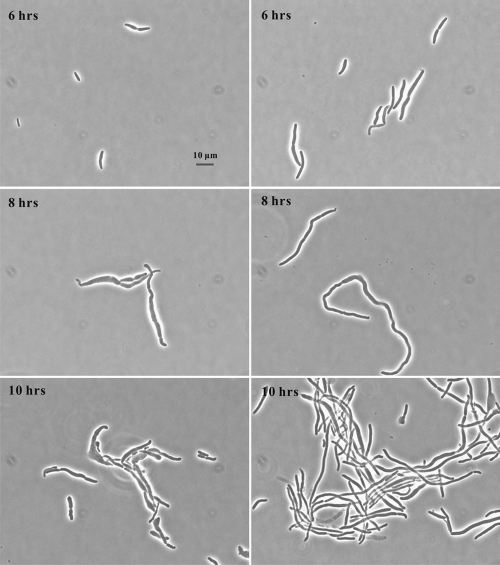
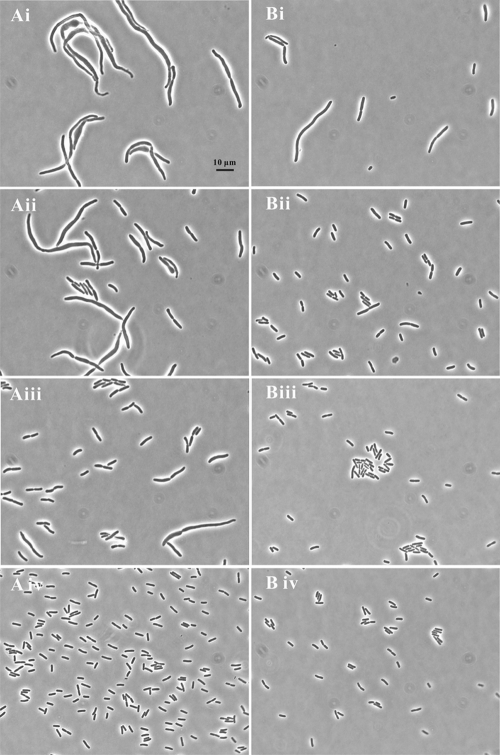
The metabolic problems caused by l-serine resulted in the triple mutant in NGCAA making deformed cells and frequently lysing, but this could be reversed by adding S-adenosylmethionine (SAM) (32). We examined here in more detail the effects of SAM on cell length and width of the triple mutant. To do this, we used a derivative of the triple mutant carrying the rickettsial SAM transporter on a plasmid, psamT, growing it in glucose minimal medium, and subculturing in NGCAA with 2 mM SAM and without SAM for 6, 8, and 10 h (Table 3 and Fig. 1). The addition of SAM resulted in much longer, narrower cells which grew quite rapidly but could not divide. Indeed, after 6 h of incubation, half of the cells subcultured with SAM were longer than the longest cell grown without SAM. This differential decreased at 8 and 10 h, but SAM-exposed cells were still much longer. The longest cell we measured was 179 μm in a 10-hour culture. Some degree of swelling could be seen at 6, 8, and 10 h in cells grown without SAM (Fig. 1), whereas cells grown with SAM were much more regular in width. Though ghosts could be seen in cultures with and without SAM, a survey of all photographs showed much less lysis in the presence of SAM (data not shown).
TABLE 3.
Effect of SAM on length of triple mutant cells carrying plasmid psamTa
| Expt | Time (h) | Addition of SAM (2 mM) | No. of cells measured | Length (μm) |
% of cells of the following length (μm) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Avg | Minimum | Maximum | <5 | 5 to 10 | 10 to 50 | >50 | ||||
| 1 | 6 | + | 54 | 23.6 | 7.3 | 58.5 | 0 | 6 | 87 | 7 |
| 2 | 8 | + | 53 | 46 | 3.6 | 143.5 | 4 | 10 | 43 | 43 |
| 3 | 10 | + | 132 | 34 | 4.6 | 179.8 | 2 | 17 | 60 | 22 |
| 4 | 6 | − | 23 | 10.0 | 4.4 | 17.2 | 13 | 39 | 48 | 0 |
| 5 | 8 | − | 30 | 25.4 | 6.6 | 57.5 | 0 | 7 | 80 | 13 |
| 6 | 10 | − | 95 | 28.6 | 4.6 | 80.6 | 1 | 14 | 68 | 18 |
Cells of the triple mutant carrying plasmid psamT were grown with 2 mM S-adenosylmethionine (SAM) (experiments 1 to 3) and without SAM (experiments 4 to 6) in NGCAA and photographed after 6, 8, or 10 h as noted. Photographed cells were measured, and average, maximum, and minimum lengths were determined.
FIG. 1.
Effect of S-adenosylmethionine (SAM) on growth of the triple mutant. The triple mutant carrying a SAM transporter, E. coli strain MEW999 psamT, was grown in glucose minimal medium, subcultured in the same medium with 0.5% Casamino Acids without SAM (left) and with 2 mM SAM (right) and photographed 6, 8, and 10 hours after subculture.
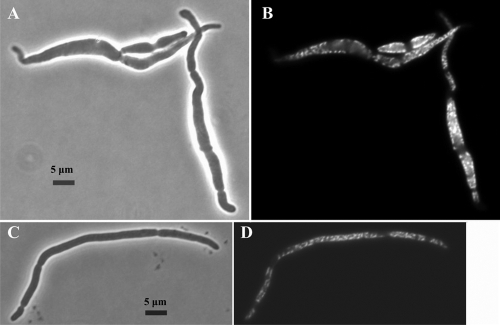
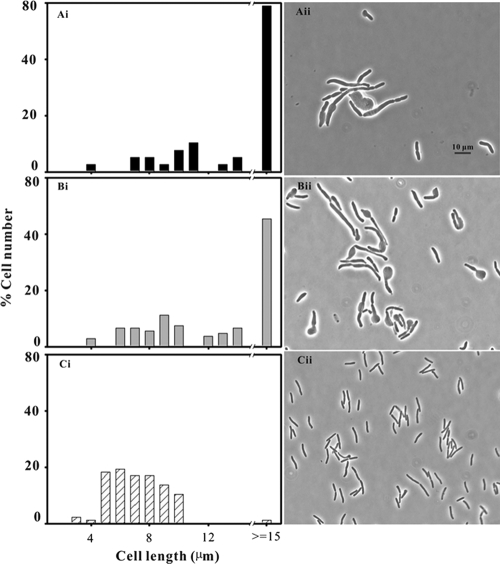
The difference between cells grown with and without SAM was much more obvious at a higher magnification after fixing and staining the cells with DAPI (Fig. 2). When examined with visible light, the cells grown without SAM were swollen, with many curves and kinks along their length, and showed some indication of lysis (Fig. 2A). The cells grown with SAM were longer and narrower and much more regular in contour, with fewer bends in the wall (Fig. 2C).
FIG. 2.
Effect of SAM on DNA distribution in the triple mutant. Strain MEW999 carrying plasmid psamT was grown in glucose-containing minimal medium and subcultured in the same medium with 0.5% CAA without SAM (A and B) and with 2 mM SAM (C and D) for 8 h, and then stained with DAPI and photographed with visible light (A and C) and fluorescent light (B and D).
DAPI staining showed clusters of nucleoids separated spatially at irregular intervals along the periphery of the cells at all time points both with and without SAM (Fig. 2D and B, respectively). This was more obvious in the swollen cells that formed without SAM (Fig. 2B). In both cases, partial septation is indicated on the phase-contrast images by indentations along the length of the cells, and these correspond to areas with no DNA on the DAPI images. Whether the cells can complete these septa cannot be seen from these pictures.
We conclude that triple mutant cells exposed to Casamino Acids and SAM are essentially healthy, metabolically competent cells capable of elongating to astonishing lengths but with the single but major problem that they cannot divide. Many cases of much less filamentation after cellular damage have been reported, as for instance after DNA damage (sfi response [13]). In these cases, the cells stop dividing until some damage is repaired and elongate somewhat until the inhibition of cell division is lifted. The problem of the triple mutant seems to be specific to cell division and perhaps nucleoid segregation.
Cells grown without SAM are also essentially healthy cells, capable of most metabolic functions, including DNA synthesis and nucleoid segregation. However, superimposed on their division problems is an inability to maintain their integrity. They therefore swell and lyse long before they can attain the length of the cells grown with SAM, which can maintain their integrity even though they cannot divide.
Spheroplast formation precedes lysis on exposure to NGCAA.
The triple mutant lyses extensively for the first several hours in NGCAA. Apparently the cells subcultured in NGCAA make a membrane of sufficient strength to maintain the integrity of the cell, but they make a weakened cell wall. Lysis occurs whenever and wherever the wall is weakened so much that the membrane cannot keep the cell intact.
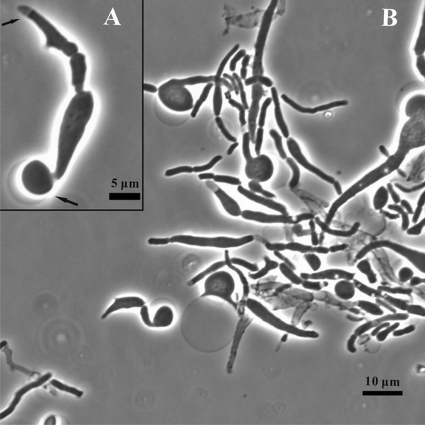
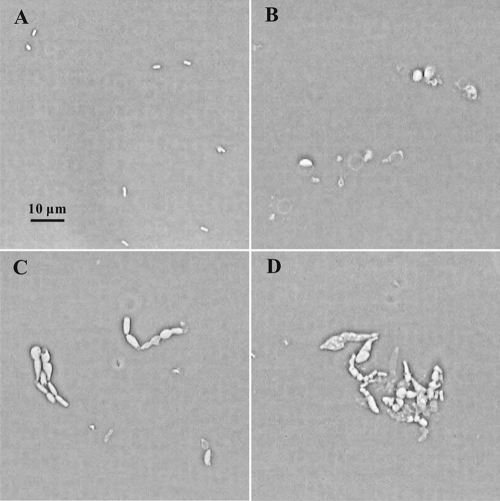
This implies that lysing cells pass through a spheroplast stage, which is usually too transient to be noticed. We photographed such spheroplasts on agar slides where they last slightly longer than in liquid medium. For this, cells subcultured in NGCAA as usual for 7 h were plated on agar slides of the same medium for 1 h more and photographed. While many cells had already lysed by 7 h, others produced spheroplasts in the next hour. In Fig. 3, one can see ghosts, swelling cells and emerging spheroplasts, as well as a few possibly normal cells. The enlarged view in Fig. 3B shows a single unit, including a spheroplast forming at the bottom end, with its connection to the cell clearly visible. At the other end, one can see a clear area inside the cell wall where the cytoplasm has been pulled away from the wall with its membrane as the cell swells and cell material is extruded elsewhere. The spheroplast did not always emerge from a particular point on the cell but could apparently arise anywhere the cell wall was sufficiently weak.
FIG. 3.
Spheroplasts formed from the triple mutant. The triple mutant grown in glucose minimal medium overnight was subcultured into glucose minimal medium with 0.5% CAA and incubated for 7 h. An aliquot was streaked on a slide culture of the same medium (NGCAA) and photographed after 1 h at 37°C. (Inset) Magnified portrait from the same culture.
We conclude that when exposed to Casamino Acids, the triple mutant can continue to make a strong membrane, but its cell wall is impaired, leading to the swelling and lysis reported here and previously (32). This lysis is much decreased in the presence of SAM so that the cells have time to grow much longer.
Effect of increased expression of peptidoglycan synthesis gene murC and increased l-alanine supply.
The weakening of the cell wall and extensive lysis suggest that incubation in NGCAA causes problems in peptidoglycan synthesis. This might happen if one or more of the peptidoglycan-synthesizing enzymes were either underexpressed or inhibited or both in the triple mutant. We therefore studied the effect of overexpression of a number of genes from the 2-min dcw (division cell wall) cluster, each carried on a plasmid from the ASKA collection under the control of the lac promoter (14). As a preliminary screen, we transformed the triple mutant with these plasmids one by one, subcultured the transformants from overnight cultures in glucose minimal medium to NGCAA with and without isopropyl-β-d-thiogalactopyranoside (IPTG), and photographed the cells after 10 h.
The most striking effect was seen with a clone of the murC gene. Inducing murC expression with IPTG considerably increased the likelihood that the triple mutant would divide rather than filament, resulting in shorter cells. Clones carrying the genes for the next 4 steps, namely, murD, murE, ddlA, and ddlB, did not restore cell division, suggesting that the inhibition in the triple mutant was indeed at the first step.
The murC gene product, UDP-N-acetylmuramate l-alanine ligase, adds the first amino acid of the 5-amino acid chain used to connect peptidoglycan strands. While this ligase is designed to use l-alanine as the substrate, both l-serine and glycine serve inefficiently as substrates (7, 10, 15). It seems likely that the high concentration of intracellular l-serine that accumulates in triple mutant cells results in serine incorporation or inhibits l-alanine addition, preventing cross-linking in either case. This failure of cross-linking would weaken the cell wall wherever it occurred, leading eventually to lysis. An increase in the amount of ligase would improve growth by making more of the l-alanyl derivative even if it also produced more of the other derivatives. However, provision of high l-alanine concentrations to compete with the internal l-serine might be even more effective.
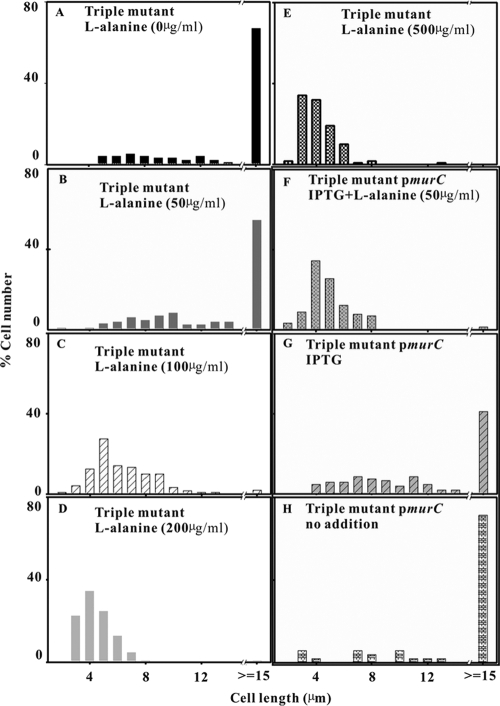
We therefore studied the effect of increased murC expression, provision of high concentrations of l-alanine, and a combination of the two (Fig. 4). After a 6-h incubation in NGCAA with l-alanine, but not any of the other physiological amino acids, the average length of the triple mutant cells was decreased markedly by 100, 200, and 500 μg/ml l-alanine, but not by 50 μg/ml (Fig. 4A to E). By 8 h, when l-serine in the medium had started to decline, even 50 μg/ml had a marked effect (data not shown). After 6-h incubation with 75% of l-serine remaining, it took 100 μg/ml l-alanine to rescue the triple mutant (Fig. 4). By 8 h with only 35% l-serine remaining, rescue needed only 50 μg/ml.
FIG. 4.
Effects of MurC and l-alanine on cell length. Cells of the triple mutant (A to E) were grown overnight in glucose minimal medium and subcultured into NGCAA for 6 h with no l-alanine (A) or with l-alanine added at 50 (B), 100 (C), 200 (D), and 500 (E) μg/ml. The triple mutant carrying plasmid JW0089 (pmurC) (F to H) grown in the same way was subcultured into NGCAA for 6 h with 100 μM IPTG and l-alanine (50 μg/ml) (F), with 100 μM IPTG alone (G), and with no further additions (H).
Expression of murC combined with even the lowest concentration of l-alanine was more effective than increased murC expression alone at 6 h (Fig. 4F versus B). Increased MurC by itself also improved growth as seen by comparing Fig. 4G with Fig. 4H. It is clear that l-alanine works in concert with MurC. At low l-alanine concentrations, increasing murC expression increased the cells' ability to divide. At higher l-alanine concentrations, induction of murC was not necessary (Fig. 4F and G).
The appearance of these cells was also much improved (Fig. 5). With 50 or 100 μg/ml l-alanine, the cells were thinner and much less swollen (Fig. 5Aii) and shorter (Fig. 5Aiii). At 500 μg/ml, the population consisted entirely of single and dividing rods, still somewhat larger than is normal for the parent strain or the triple mutant in glucose minimal medium (Fig. 5 Aiv). With murC expression induced and alanine added, almost all cells were short rods (Fig. 5B panels).
FIG. 5.
Effects of MurC and l-alanine on cell appearance. Cells of the triple mutant (A) and the triple mutant carrying plasmid JW0089 (pmurC) (B) were grown overnight in glucose minimal medium and subcultured into NGCAA without l-alanine (i) or with l-alanine added at 50 (ii), 100 (iii), and 500 (iv) μg/ml for 6 h. The images are taken from one experiment and representative of two.
Effect of overexpression of ftsW on the triple mutant.
Overexpression of only one other 2-min cluster gene, ftsW, had a strong effect on the size and shape of the triple mutant. This gene codes for a transmembrane protein which localizes to the septum and recruits FtsI (penicillin binding protein 3 [PBP3]) to follow it. Overproduction of FtsW at 100 μM IPTG skewed the distribution of cell lengths to shorter lengths when viewed at 8 h (Fig. 6Ci). Without IPTG, the cells show a variety of lengths, with swelling and lysis Fig. 6Aii). At 100 μM IPTG, the cells were of normal width and uniform length but 2 to 3 times longer than the parent strain growing in the same medium (Fig. 6Cii). The genes tested that did not have a marked effect on the appearance of the triple mutant cells were ftsZ, ftsA, ftsQ, ftsL, ftsI, ddlA, ddlB, lpxC, murD, murE, and murF.
FIG. 6.
Effect of FtsW overexpression on cell length and appearance. Cells of the triple mutant carrying plasmid JW0087 (pftsW) were grown overnight in glucose minimal medium and subcultured into NGCAA without IPTG (A) or with IPTG at concentrations of 50 (B) and 100 (C) μM. The cells were photographed after subculture for 8 h.
Effect of a bypass of the 2-min promoter on amino acid toxicity.
Both the ftsW and murC genes are transcribed from the 2-min promoter, Pmra. Since overexpression of either gene overcomes the problems caused by Casamino Acids, we thought that expression of the mra promoter might be set too low in the triple mutant, so that neither gene was expressed sufficiently but either could compensate. An increase in Pmra transcription for whatever reason might then allow normal growth. H. Hara and J. A. Ayala kindly provided us with a strain in which the expression of the 2-min cluster could be increased (12, 16). This strain, JE7968, carries an insert of the lac promoter and chloramphenicol resistance gene interrupting the promoter Pmra of the 2-min cluster and putting the cluster genes under the control of the lac promoter (12). This makes the strain dependent on IPTG for expression of the 2-min cluster and for growth, as shown by the fact that without IPTG, it lyses within 2 h.
We transduced the Hara promoter from strain JE7968 into the triple mutant, selecting chloramphenicol resistance and screening for IPTG dependence. The resultant transductant, strain MEW999H, depended on IPTG for growth as expected. If the effect of CAA involved inhibition of Pmra as the sole site of inhibition, we would expect the strain to grow in NGCAA and IPTG without problem. However, cells in a slide culture of NGCAA with IPTG were deformed and lysed just as the triple mutant did (Fig. 7). We conclude that inhibition of growth in the triple mutant does not result exclusively from an effect on transcription of the 2-min promoter Pmra, though growth in this medium may decrease transcription of both Pmra and some other promoter(s) essential for cell division.
FIG. 7.
Effect of control of the Pmra promoter by IPTG. Cells of the triple mutant with the Hara promoter (strain MEW999H) were grown overnight in glucose minimal medium and subcultured into NGCAA with 1 mM IPTG (C and D) or without IPTG (B). The pictures were taken at 0 (A), 3 (B and C), and 9 (D) hours.
Effect of increased gcv expression on growth of the triple mutant.
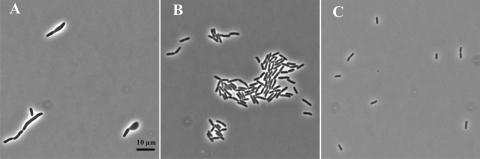
We suggested previously (32) that serine-dependent amino acid toxicity resulted in a starvation for C1 units due to inhibition of expression of the gcv operon, causing an increase in glycine concentration which together with high l-serine and other amino acids would inhibit serine hydroxymethyltransferase (SHMT). This simultaneous decrease in serine and glycine cleavage would starve the cells for C1-tetrahydrofolic acid (C1-THF) derivatives, and the lack of C1-THF and SAM would hinder septum formation and cell division through an as yet unknown mechanism (32).
If the growth problems were in fact due to a lack of C1-THF, overexpression of the gcv operon should overcome some or all of the growth problems. Plasmid pGS146, a kind gift of George Stauffer, carries the entire coding sequence of the gcvTHP operon as well as the PurR binding site. The triple mutant transformed with pGS146 grew in NGCAA as the usual E. coli small rods throughout the growth cycle (data shown for 5 h only [Fig. 8B]).
FIG. 8.
Effect of overexpression of the glycine cleavage operon on growth of the triple mutant. After growth overnight in glucose minimal medium, the strains were subcultured into the same medium with 0.5% Casamino Acids and photographed after 5 h. The triple mutant (A), the triple mutant strain carrying the glycine cleavage operon on plasmid pGS146 (B), and the parent strain (C) are shown.
It is clear that all the growth problems of the triple mutant can be overcome by increased expression of the gcv operon. Increased glycine cleavage would have two effects: it would of course increase the availability of C1 units, but it would also decrease the concentration of glycine, which would in turn relieve the inhibition of SHMT and decrease the serine concentration. Since as we showed earlier (32), removal of glycine from the inhibitory mixture of amino acids renders the mixture nontoxic, either decreased serine and glycine or increased C1 supply, or a combination of these factors could account for the effect of the gcv plasmid.
To see whether a higher glycine concentration would actually be toxic, we grew strain MEW999 carrying pGS146 in NGCAA with various concentrations of glycine from 0 to 700 μg/ml added. If the beneficial effect of pGS146 was due solely to a lowered glycine concentration, one would expect higher glycine concentrations to counteract it. However, the cells were of normal size and appearance at all concentrations tested (data not shown). We conclude that the triple mutant problems in Casamino Acids are caused by decreased gcv expression and not by excess glycine.
Effects of purines on the growth of the triple mutant.
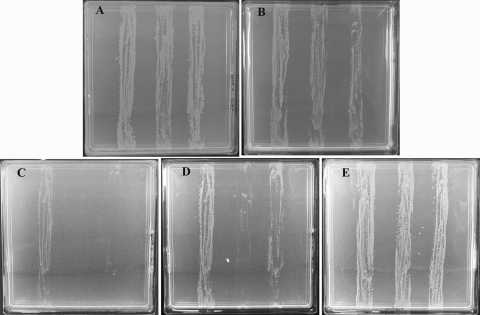
The triple mutant probably has a higher than usual serine pool and underexpresses the gcv operon even when grown in glucose minimal medium without CAA. In that case, one might expect the triple mutant to be particularly sensitive to the addition of purines to its growth medium, since expression of the gcv operon is inhibited by purines (22). In fact, the triple mutant is indeed more sensitive to purine, but only when l-serine is also present. A gradient of 0 to 40 μg/ml hypoxanthine had little effect on the growth of either the triple mutant in glucose minimal medium, with or without plasmid pGS146, or its parent strain MEW1 (Fig. 9A). All three strains also grew well in the presence of l-serine (300 μg/ml) (Fig. 9B). However, a combination of serine and hypoxanthine very much delayed growth of all three strains, and this delay could be circumvented by plasmid pGS146. With the hypoxanthine gradient in a medium with a uniform l-serine concentration of 300 μg/ml, by 16 h, only the parent strain grew well, the other two showing slight growth at the low hypoxanthine end of the gradient (Fig. 9C). By 20 h, the triple mutant grew at low hypoxanthine without the gcv plasmid, but with the plasmid, it grew throughout the gradient (though less well than the parent strain) (Fig. 9D). By 48 h, all three strains grew (Fig. 9E).
FIG. 9.
Inhibition by hypoxanthine and l-serine of growth of the triple mutant. Strains MEW1 (parent) (left streaks), MEW999 (triple mutant) (center streaks), and MEW999 carrying pGS146 (right streaks) were streaked on the plates containing minimal medium and glucose and the following additions and gradients (if any): hypoxanthine gradient (0 to 40 μg/ml) (A), l-serine (300 μg/ml) throughout the plate (B), and hypoxanthine gradient (0 to 40 μg/ml) with l-serine (300 μg/ml) throughout the plate (C, D, and E). The gradients are shown as follows: low concentration at the top of the plate to high concentration at the bottom of the plate. The plates were incubated at 37°C, examined at regular intervals, and photographed after 48 h (A, B, and E), 16 h (C), and 20 h (D). No difference was seen between the strains in panels A, B, and E at any interval prior to the photograph.
We conclude that even in the absence of Casamino Acids, the triple mutant has problems maintaining its C1 pool. Inhibition of gcv expression by purines at an increased l-serine concentration suffices to inhibit its growth. However, these problems are circumvented by increased gcv expression, which restores normal growth.
DISCUSSION
In this work, we show that l-serine is so toxic to E. coli that it has evolved three very specific 4Fe4S defensive l-serine deaminases to detoxify it. These enzymes with high Km values rapidly remove high concentrations of l-serine from the medium but do not act upon l-serine at physiological concentration. Rapid degradation of l-serine and other amino acids by cells using them as energy and nitrogen sources is well-known (20). We show here that this is the only amino acid that is rapidly degraded even in the presence of other carbon, energy, and nitrogen sources. Indeed, E. coli K-12 removed 75% of the l-serine in an amino acid mix before it significantly reduced any of the others (Table 2).
That the enzymes play a critical role is clear from the fact that their elimination from the cell leads to extensive disturbance of growth and cell division (32). In this work, we show that this is attributable to a very high internal concentration of l-serine that accumulates when l-SD-deficient cells are grown in the presence of a mixture of amino acids. This establishes for the first time the metabolic role of the three l-serine deaminases as the maintenance of tolerable levels of l-serine. Their high Km values (3) ensure that the l-serine is not depleted to levels that would starve the cell for l-serine. Because there is no danger of excessive l-serine degradation, the enzyme can be made, as it is, even in glucose minimal medium.
The l-serine pool was much more stable in triple mutant cultures which still contained 76% of their original l-serine after 6 h of incubation, when the parent strain retained only 0.3%. This clearly demonstrates that the l-serine concentration remains high enough to account for inhibitory effects on the cells and begins to drop at the time that the population starts to recover. This delayed decrease in l-serine occurred without any of the l-SDs, probably in part by assimilation of l-serine and derivatives of l-serine into new cells. However, other degrading enzymes (e.g., hydroxyacid deaminases) might also be involved.
l-Serine toxicity due to interference with biosynthesis of isoleucine and aromatic amino acids is well-known (11, 21) but not relevant here, since these amino acids are provided in Casamino Acids. The high internal l-serine concentration that accumulates in the triple mutant creates at least two other difficulties. First, it weakens the cell wall, and second, together with several other amino acids, it results in inhibition of division. We attribute the weakening of the cell wall to l-serine itself, directly or indirectly decreasing activity and/or synthesis of an enzyme involved in peptidoglycan biosynthesis, the murC-encoded N-acetylmuramate l-alanine ligase. The inhibition of septation and cell division is related in an unknown manner to problems in synthesis of single-carbon units (32) and overcome, as shown here, by overproduction of the glycine cleavage enzymes.
The growth problems seen in NGCAA were also entirely or partially circumvented by overproduction of FtsW or MurC or by supplying a large excess of l-alanine. These counteracted the weakening of the cell wall which apparently allowed the division machinery to function. Surprisingly, provision of S-adenosylmethionine (SAM) also largely avoided the problems of cell wall synthesis and lysis without restoring cell division and permitted instead extensive increases in cell mass and length.
Involvement of the murC gene product in the lysis of the triple mutant.
When the triple mutant was incubated with amino acids, it became deformed due to defects in the cell wall, though it could still make an intact membrane as seen in the transient formation of spheroplasts (Fig. 3). This lysis was counteracted by supplying l-alanine or overproducing MurC ligase, the enzyme catalyzing the first step in the formation of the pentapeptide that cross-links the peptidoglycan chains.
Since overproduction of the other cross-linking enzymes does not restore growth, we ascribe the fragility of the wall to a competition between l-serine and l-alanine for the active site of the MurC ligase. High internal l-serine would decrease availability of the MurC product, UDP-N-acetylmuramate-alanine. Providing a high concentration of l-alanine would counteract this because MurC has a much lower Km for alanine than for serine. Increased murC expression would help, but it would not counteract l-serine inhibition unless some l-alanine was also provided—exactly as described here. Competition between glycine and l-alanine was suggested as a cause of glycine-induced lysis in pyridoxine-starved E. coli B (4). This is also reminiscent of the inhibition of a relA mutant by an amino acid mixture containing serine, glycine, and other amino acids (27). Whether this was also accompanied by elongation or lysis was not reported.
While this alanine/serine competition probably explains lysis, the actual mechanism through which serine inhibition of the MurC ligase weakens the peptidoglycan cannot be specified even with the aid of the fine set of reviews on peptidoglycan synthesis and degradation in FEMS Microbiology Reviews (1, 2, 5, 28, 29). The problem is that the peptidoglycan precursors are made in the cytoplasm, but peptidoglycan synthesis is extracellular. MurC catalyzes the first of several cytoplasmic steps, attaching l-alanine to uridinediphospho-N-acetylmuramate. l-Serine might decrease the formation of UDP-N-acetylmuramate-alanine or even cause the formation of some UDP-N-acetylmuramate-serine (Fig. 10). How either eventuality (and other possibilities) would be translated into an extracellular weakened peptidoglycan or even how they would be transported across the membrane is not obvious. Alternatively, even unincorporated, any of the unusual intermediates that might accumulate in the presence of high l-serine, like UDP-N-acetylmuramate-serine or free N-acetylmuramate, might be toxic.
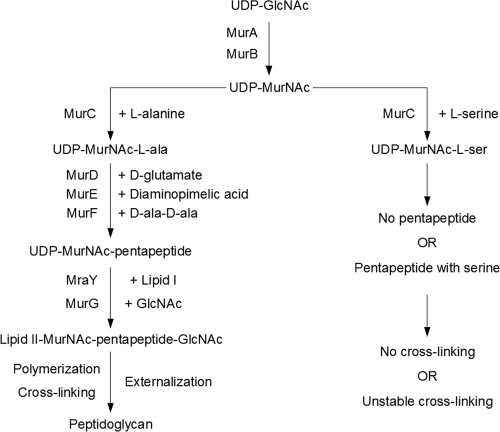
FIG. 10.
Role of MurC in peptidoglycan synthesis. Uridine-diphospho-N-acetylglucosamine (UDP-GlcNAc) is converted via MurA and MurB to UDP-N-acetylmuramic acid (UDP-MurNAc), to which alanine is added by MurC. The pentapeptide is completed by the addition of d-glutamate, diaminopimelic acid, and d-alanine-d-alanine via MurD, MurE, and MurF, respectively. These are transferred to lipid I by MraY, followed by the addition of GlcNAc and transfer to lipid II by MurG. All these are cytoplasmic reactions. Lipid II with its peptidoglycan precursor is flipped to the periplasmic side of the inner membrane, followed by polymerization of one chain and cross-linking to another, all by unknown mechanisms. Serine in sufficiently high concentrations can act as a substrate for MurC and might either inhibit pentapeptide synthesis or become incorporated into a pentapeptide analog, which may interfere with further steps of peptidoglycan synthesis.
A further complicating factor is that the cell replaces 40 to 50% of its peptidoglycan in every generation, and the cell has a large number of hydrolases at its disposal to do this (29). The lysis we see could be explained if l-serine simply decreased the rate of synthesis of UDP-N-acetylmuramate-alanine by competition for the ligase, and the rate of peptidoglycan hydrolysis was not decreased. That is, new peptidoglycan synthesis might not be able to keep up with peptidoglycan hydrolysis.
Switch from elongation to cell division.
Peptidoglycan is synthesized by two different enzyme complexes (6). The first one called an elongase extends the cell length and is presumably active during filamentation. For cell division to occur, a second enzyme complex, the divisome, must be activated and must synthesize peptidoglycan perpendicular to the cell wall, coordinated with intercellular septum formation. The longer the delay before the divisome is activated, the longer elongation continues, and the longer the filaments formed.
In this work, we show a clear competition between elongation and septation (5) in that the length of the cell could vary with the concentration of l-alanine provided. When no l-alanine was added to NGCAA, long filaments were made, and the divisome activation was delayed. At 500 μg/ml, l-alanine allows septation in NGCAA as soon as E. coli reached its usual length. With lower concentrations, e.g., 100 and 200 μg/ml, the cells were longer—but not as long as when no alanine was provided, and the average cell length was roughly inversely proportional to the alanine concentration. This suggests that growth in NGCAA delays divisome activation, and the higher the concentration of l-alanine, the greater the chance of divisome activity and the shorter the cell formed. The length of the cell at septation thus depended on the metabolic state of the cell. This would be consistent with NGCAA starving the cells for C1-THF and methylation being needed for divisome activation.
Formation of the septum is usually considered a linear process beginning with formation of the FtsZ ring, after which a number of proteins assemble at midcell to form the septum: ZipA and FtsA, followed by others, including FtsW. However, in a series of elegant experiments, Goehring et al. (8, 9) targeted septal proteins to the FtsZ ring in such a way that some arrived earlier than expected. They found that proteins could arrive in preformed groups and that those proteins that arrived could draw other proteins with them, unconstrained by their “usual” position in the hierarchy, later proteins even drawing in earlier ones (8, 9). Septation thus occurred whenever the required proteins reached the correct site.
Similarly, we showed here that the delayed septation of the triple mutant in NGCAA could be overcome by high overexpression of the ftsW gene. When ftsW carried on a moderately high-copy-number plasmid was induced with 100 μM IPTG (Fig. 6), the average cell length was decreased. This required a large amount of FtsW: half as much (50 μM) had much less effect. As in the experiments of Goehring et al., this may indicate that excess FtsW can accumulate at the FtsZ ring and pull the other septation proteins into position, allowing peptidoglycan to be formed by the divisome and cell division to be completed, and thus avoiding prolonged elongation.
If swelling and lysis require an accumulation of several errors to weaken the wall sufficiently, the high FtsW concentration may avoid lysis by allowing the cell to switch from elongation to septation before enough damage has accumulated. The smaller the individual cell formed, the fewer the errors in the peptidoglycan, and perhaps the more the likelihood of repairing them as the peptidoglycan is recycled.
The two genes ftsW and murC are expressed from the same promoter Pmra so that overexpression of Pmra might lead to overexpression of both genes and overcome CAA toxicity. However, overexpression of the Pmra operon using the interesting strain kindly provided by H. Hara and J. A. Ayala did not counteract CAA toxicity. The organization of the 2-min cluster is so complex, with so many major and minor promoters, that overexpression of the entire operon might not mimic overexpression of just ftsW or murC. In any case, exposure to CAA cannot inhibit solely at Pmra and may not inhibit Pmra at all.
Rescue of the triple mutant by overproduction of the glycine cleavage enzymes.
Overexpression of the glycine cleavage operon completely restored growth of the triple mutant in NGCAA, avoiding both lysis and filamentation. This is consistent with our previous suggestion that the high l-serine concentration accumulated by the triple mutant in NGCAA, together with other amino acids in NGCAA, results in a deficiency of C1 units, and that this is responsible for the growth defects observed (32). Increased glycine cleavage would restore the supply of C1-THF and decrease the glycine concentration. The decreased glycine would reduce inhibition of serine hydroxymethyltransferase (GlyA), allowing that enzyme to decrease the serine concentration and increase C1-THF production even more.
The effects of the gcv clone can thus be understood in terms of an increase in C1-THF acting directly on the unidentified growth problem, and/or a lowered internal serine concentration decreasing competition of l-serine with l-alanine. The triple mutant clearly has problems with C1 metabolism, as seen by its sensitivity to purines (e.g., hypoxanthine) even during growth in glucose minimal medium without Casamino Acids. Purine biosynthesis is one of the major consumers of C1 units (19). Since the cell therefore needs less C1-THF when purines are added to the medium, it is not surprising that hypoxanthine is a major inhibitor of gcv expression (22). We suggest that even in minimal medium, the triple mutant has such a low expression of gcv that hypoxanthine pushes the pool below the level where the cell can function. The triple mutant can tolerate either hypoxanthine or a moderate exogenous concentration of l-serine, but its growth is greatly delayed when both are present (Fig. 9). It is much more sensitive to this mixture than the parent strain, which is nonetheless inhibited to a considerable extent. A similar inhibition, also explained by C1 starvation, occurred when glycine cleavage was inhibited by prior growth with serine and a purine (19).
The fact that decreased C1 availability has such drastic effects suggests that reactions involving C1 units, e.g., methylation, may have a regulatory role in E. coli growth and division. Because starvation for S-adenosylmethionine led to a failure in septum formation (30), we suggested that methylation is needed in the course of septum formation. The addition of the major methyl donor, SAM, to NGCAA indeed had enormous structural effects, avoiding lysis and allowing extensive elongation. These cells were not limited in growth as the metK84 filaments we described earlier were (18), and both had a regular distribution of nucleoids for the entire cell length. The triple mutant with SAM grew into the longest filaments we have seen, with some over 150 μm long. These cells seem to be specifically inhibited in division but are otherwise metabolically competent and structurally organized, and like the metK84 filaments, rarely swell or lyse. This suggests that E. coli can function well even without dividing, as long as no part of it is far removed from a surface, and that complete horizontal integration along the cell is not needed.
Though SAM restores so much of the cell's competence, it is striking that it cannot restore cell division. This suggests that C1 starvation results in a requirement for SAM and for some other C1 derivative as well. This might involve methyl-THF as a specific methyl donor in the modification of rRNA, or some other as yet unknown reaction. We suggest that E. coli normally keeps its supply of C1 at a low level which allows a role for C1 derivatives in regulation of metabolism but makes E. coli very vulnerable to any growth conditions in which the supply of C1 derivatives would be even lower.
Acknowledgments
This work has been supported by grant A6050 from the National Science and Engineering Research Council of Canada for which we continue to be extremely grateful in its 39th year.
We are also grateful to the Japanese E. coli program which made available the deletion mutants and plasmids used in this work and in several other projects in our laboratory. We thank George Stauffer (University of Iowa) for plasmid pGS146. We also thank R. Kilgour, chairman of the Department of Exercise Science, for the use of the Leica microscope. We also thank Marc Champagne, Hongsheng Su, J. A. Kornblatt, R. D'Ari, C. Woldringh, and Mark Paci for their continuing advice and encouragement.
Footnotes
Published ahead of print on 20 August 2010.
REFERENCES
- 1.Barreteau, H., A. Kovac, A. Boniface, M. Sova, S. Gobec, and D. Blanot. 2008. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:168-207. [DOI] [PubMed] [Google Scholar]
- 2.Bouhss, A., A. E. Trunkfield, T. D. H. Bugg, and D. Mengin-Lecreulx. 2008. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 32:208-233. [DOI] [PubMed] [Google Scholar]
- 3.Cicchillo, R. M., M. A. Baker, E. J. Schnitzer, E. B. Newman, C. Krebs, and S. J. Booker. 2004. Escherichia coli l-serine deaminase requires a [4Fe-4S] cluster in catalysis. J. Biol. Chem. 279:32418-32425. [DOI] [PubMed] [Google Scholar]
- 4.Dempsey, W. B. 1973. Lysis of Escherichia coli by glycine is potentiated by pyridoxine starvation. J. Bacteriol. 169:373-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.den Blaauwen, T., M. A. de Pedro, M. Nguyen-Disteche, and J. A. Ayala. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32:321-344. [DOI] [PubMed] [Google Scholar]
- 6.Derouaux, A., B. Wolf, C. Fraipont, E. Breukink, M. Nguyen-Disteche, and M. Terrak. 2008. The monofunctional glycosyltransferase of Escherichia coli localizes to the cell division site and interacts with penicillin-binding protein 3, FtsW, and FtsN. J. Bacteriol. 190:1831-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emanuele, J., Jr., H. Jin, B. L. Jacobson, C. Y. Chang, H. M. Einspahr, and J. J. Villafranca. 1996. Kinetic and crystallographic studies of Escherichia coli UDP-N-acetylmuramate:L-alanine ligase. Protein Sci. 5:2566-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goehring, N. W., M. D. Gonzalez, and J. Beckwith. 2006. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol. Microbiol. 61:33-45. [DOI] [PubMed] [Google Scholar]
- 9.Goehring, N. W., I. Petrovska, D. Boyd, and J. Beckwith. 2007. Mutants, suppressors, and wrinkled colonies: mutant alleles of the cell division gene ftsQ point to functional domains in FtsQ and a role for domain 1C of FtsA in divisome assembly. J. Bacteriol. 189:633-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubler, M., Y. Appoldt, and W. Keck. 1996. Overexpression, purification, and characterization of UDP-N-acetylmuramyl:l-alanine ligase from Escherichia coli. J. Bacteriol. 178:906-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hama, H., T. Kayahara, M. Tsuda, and T. Tsuchiya. 1991. Inhibition of homoserine dehydrogenase 1 by l-serine in Escherichia coli. J. Biochem. 109:604-608. [DOI] [PubMed] [Google Scholar]
- 12.Hara, H., S. Yasuda, K. Horiuchi, and J. T. Park. 1997. A promoter for the first nine genes of the Escherichia coli mra cluster of cell division and cell envelope biosynthesis genes, including ftsI and ftsW. J. Bacteriol. 179:5802-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huisman, O., R. D'Ari, and J. George. 1980. Inducible Sfi-dependent division inhibition in Escherichia coli. Mol. Gen. Genet. 177:619-636. [DOI] [PubMed] [Google Scholar]
- 14.Kitagawa, M., T. Ara, M. Arifuzzaman, T. Ioka-Nakamichi, E. Inamoto, M. Toyonaga, and H. Mori. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291-299. [DOI] [PubMed] [Google Scholar]
- 15.Liger, D., A. Masson, D. Blanot, J. van Heijenoort, and C. Parquet. 1995. Overproduction, purification and properties of the uridine-diphosphate-N-acetylmuramate:l-alanine ligase from Escherichia coli. Eur. J. Biochem. 230:80-87. [DOI] [PubMed] [Google Scholar]
- 16.Mengin-Lecreulx, D., J. Ayala, A. Bouhss, J. Van Heijenoort, C. Parquet, and H. Hara. 1998. Contribution of the Pmra promoter to expression of genes in the Escherichia coli mra cluster of cell envelope biosynthesis and cell division genes. J. Bacteriol. 180:4406-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach, p. 96. Sinauer Associates, Inc., Sunderland, MA.
- 18.Newman, E. B., L. I. Budman, E. C. Chan, R. C. Greene, R. T. Lin, C. L. Woldringh, and R. D'Ari. 1998. Lack of S-adenosylmethionine results in a cell division defect in Escherichia coli. J. Bacteriol. 180:3614-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman, E. B., and B. Magasanik. 1963. The relation of serine-glycine metabolism to the formation of single-carbon units. Biochim. Biophys. Acta 78:437-448. [DOI] [PubMed] [Google Scholar]
- 20.Prüss, B. M., J. M. Nelms, C. Park, and A. J. Wolfe. 1994. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 176:2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito, K., S. Ishida, and K. Yamada. 2003. Inhibition of growth by l-serine in E. coli. Bull. Mukagawa Women's University Nat. Sci. 51:51-53. [Google Scholar]
- 22.Stauffer, L. T., and G. V. Stauffer. 1994. Characterization of the gcv control region from Escherichia coli. J. Bacteriol. 176:6159-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stauffer, L. T., A. Ghrist, and G. V. Stauffer. 1993. The Escherichia coli gcvT gene encoding the T-protein of the glycine cleavage enzyme system. DNA Seq. 3:339-346. [DOI] [PubMed] [Google Scholar]
- 24.Su, H. S., and E. B. Newman. 1991. A novel l-serine deaminase activity in Escherichia coli K-12. J. Bacteriol. 173:2473-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su, H. S., B. F. Lang, and E. B. Newman. 1989. l-Serine degradation in Escherichia coli K-12: cloning and sequencing of the sdaA gene. J. Bacteriol. 171:5095-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tucker, A. M., H. H. Winkler, L. O. Driskell, and D. O. Wood. 2003. S-Adenosylmethionine transport in Rickettsia prowazekii. J. Bacteriol. 185:3031-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uzan, M., and A. Danchin. 1976. A rapid test for the relA mutation in E. coli. Biochem. Biophys. Res. Commun. 69:751-758. [DOI] [PubMed] [Google Scholar]
- 28.Vollmer, W., D. Blanot, and M. A. de Pedro. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149-167. [DOI] [PubMed] [Google Scholar]
- 29.Vollmer, W., B. Joris, P. Charlier, and S. Foster. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32:259-286. [DOI] [PubMed] [Google Scholar]
- 30.Wang, S., S. J. Arends, D. S. Weiss, and E. B. Newman. 2005. A deficiency in S-adenosylmethionine synthetase interrupts assembly of the septal ring in Escherichia coli K-12. Mol. Microbiol. 58:791-799. [DOI] [PubMed] [Google Scholar]
- 31.Wei, Y., and E. B. Newman. 2002. Studies on the role of the metK gene product of Escherichia coli K-12. Mol. Microbiol. 43:1651-1656. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, X., and E. B. Newman. 2008. A deficiency in L-serine deaminase results in abnormal cell division and growth of E. coli K-12. Mol. Microbiol. 69:870-881. [DOI] [PubMed] [Google Scholar]