Abstract
AmrZ is a putative ribbon-helix-helix (RHH) transcriptional regulator. RHH proteins utilize residues within the β-sheet for DNA binding, while the α-helices promote oligomerization. AmrZ is of interest due to its dual roles as a transcriptional activator and as a repressor, regulating genes encoding virulence factors associated with both chronic and acute Pseudomonas aeruginosa infection. In this study, cross-linking revealed that AmrZ forms oligomers in solution but that the amino terminus, containing an unordered region and a β-sheet, were not required for oligomerization. The first 12 unordered residues (extended amino terminus) contributed minimally to DNA binding. Mutagenesis of the AmrZ β-sheet demonstrated that residues 18, 20, and 22 were essential for DNA binding at both activation and repressor sites, suggesting that AmrZ utilizes a similar mechanism for binding to these sites. Mice infected with amrZ mutants exhibited reduced bacterial burden, morbidity, and mortality. Direct in vivo competition assays showed a 5-fold competitive advantage for the wild type over an isogenic amrZ mutant. Finally, the reduced infection phenotype of the amrZ-null strain was similar to that of a strain expressing a DNA-binding-deficient AmrZ variant, indicating that DNA binding and transcriptional regulation by AmrZ is responsible for the in vivo virulence defect. These recent infection data, along with previously identified AmrZ-regulated virulence factors, suggest the necessity of AmrZ transcriptional regulation for optimal virulence during acute infection.
The Pseudomonas aeruginosa transcriptional regulator AmrZ is a proposed member of the ribbon-helix-helix (RHH) family of DNA-binding proteins, sharing structural similarity with the Arc and Mnt repressors of Salmonella enterica serovar Typhimurium bacteriophage P22. This family is grouped by structural similarity and includes several transcriptional regulators found in prokaryotes, archaea and their viruses, and other bacteriophages (2, 7, 12, 20, 24, 26, 34, 46). Based on amino acid identity as well as secondary-structure prediction models, AmrZ likely possesses a ribbon-helix-helix motif (i.e., one β-strand and two α-helices) (Fig. 1 B) responsible for DNA-binding activity in this family of proteins (44). RHH proteins function through the oligomerization of the α-helices, which allows the two β-strands to form an antiparallel β-sheet that recognizes and binds in the major groove of the operator site (31, 38). Arc exists as a dimer in solution, while Mnt utilizes an extra carboxy-terminal α-helical domain to maintain a tetramer configuration (43). When binding DNA, these oligomers are maintained, and the inhibition of oligomerization negatively impacts DNA-binding activity (44). To facilitate higher-order oligomers at the RHH binding site, operator sites often contain sequences in either a direct repeat or palindromic orientation (34, 46). Because there are specific contacts between residues of the DNA-binding β-sheet and bases in the operator site, mutations of critical bases within the operator sites typically abolish DNA-binding activity by these proteins.
FIG. 1.
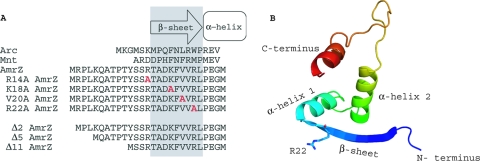
Alignment and predicted secondary structure of the putative ribbon-helix-helix transcriptional regulator AmrZ. (A) An amino acid alignment of the Arc-like DNA-binding domains of Arc (residues 1 to 18), Mnt (residues 1 to 15), and AmrZ (residues 1 to 27) reveals conserved residues in the DNA-binding β-sheet as well as the presence of the extended amino terminus. Residues in gray indicate the DNA-binding β-sheet. Residues to the left are part of the extended amino acid. Residues within the DNA-binding β-sheet that were targeted for site-specific mutagenesis are shown in red. (B) Predicted three-dimensional (3D) structure of AmrZ residues 13 to 80 provided by the secondary-structure prediction program pyMol. The major structural components (N and C termini, β-sheet, and α-helices) are indicated. The location of arginine-22 (R22) is also indicated, given the frequent references to the R22A mutant.
Of the 108 AmrZ amino acids, residues 1 to 66 share 34% amino acid identity with Arc and suggest the presence of an Arc-like DNA-binding domain (2). Figure 1A presents an amino acid alignment of the amino termini of Arc, Mnt, and AmrZ to illustrate conserved residues, particularly in the proposed DNA-binding β-sheet region. The first 12 AmrZ residues (extended amino terminus) are not present in Arc or Mnt, although the two proposed AmrZ orthologs in Pseudomonas putida and Pseudomonas syringae each have an extended amino terminus with a highly conserved sequence (47). An extended amino terminus has been observed for several other RHH proteins, and for those regions that have been studied, it typically serves a unique role (5, 11, 22, 23, 28, 35, 38). For example, this region functions in protein-protein interactions (plasmid-partitioning protein ParG), metal cofactor specificity (Helicobacter pylori NikR), and ATP hydrolysis or oligomerization (plasmid pSK41-encoded ArtA) (5, 8, 23). Residues 13 to 25 of AmrZ share amino acid identity with β-sheet residues in Arc and Mnt, and protein prediction models suggest that AmrZ residues 15 to 23 form a β-sheet (Fig. 1B) (32). A majority of the amino acid identity with Arc and Mnt falls between AmrZ residues 26 and 66, and this region is predicted to form a coiled-coil motif that mimics the dimerization domain found in Arc used to stabilize protein-DNA interactions (Fig. 1B). The carboxyl terminus of AmrZ may function similarly to the tetramerization domain of Mnt and contribute to oligomerization and high-affinity DNA binding, since residues 67 to 108 of AmrZ are proposed to form a second coiled-coil domain.
AmrZ is highly expressed in alginate-overproducing mucoid variants of P. aeruginosa, such as those isolated in chronic infections of cystic fibrosis (CF) patients. Previous work from our laboratory demonstrated that AmrZ is required for the activation of algD transcription, the first gene in the alginate biosynthetic operon (4). AmrZ also acts as a transcriptional repressor by binding to two sites upstream of its own promoter (30). In addition, AmrZ represses fleQ, a gene encoding an activator of flagellum expression (41). Finally, AmrZ is required for the control of genes involved in type IV pilus localization and twitching motility, although its specific gene target(s) is still being elucidated (3). Due to its regulatory role in both alginate production and motility, the protein formerly called AlgZ has been designated AmrZ (alginate and motility regulator Z) (3).
AmrZ regulates the transcription of algD and amrZ using protein-nucleotide contacts (2, 30). Although the binding sites at both promoters have some nucleotide sequences in common, it is unclear whether AmrZ utilizes similar amino acid residues contained in the Arc-like DNA-binding domain to modulate its opposing activities of activation and repression. In this study, glutaraldehyde cross-linking data showed for the first time that AmrZ forms oligomers naturally in solution. The truncation of the extended amino terminus minimally reduced DNA-binding activity in vitro, although this truncation did not affect the oligomerization capability. Mutations of Lys18, Val20, and Arg22 also caused an elimination of algD transcription and alginate production in vivo, but this was not always correlated with a loss of in vitro DNA-binding activity. Additionally, Lys18, Val20, and Arg22 were required for the repression of amrZ transcription in vivo, indicating that AmrZ utilizes a conserved binding mechanism to control activated and repressed target genes. Finally, to assess the biological impact of AmrZ transcriptional regulation on virulence during an infection, a murine model of acute infection was used to compare the levels of virulence of the wild type, an amrZ-null mutant, and a mutant expressing a DNA-binding-deficient AmrZ variant. The defect in bacterial colonization and morbidity shown by strains without transcriptionally active AmrZ established the contribution of AmrZ DNA-binding activity and transcriptional regulation to virulence. Taken together, these data identify key residues in the oligomeric protein AmrZ that contribute to DNA binding and the transcriptional regulation of virulence factors in a biologically relevant infection model.
MATERIALS AND METHODS
Bacterial strains, plasmids, chemicals, oligonucleotides, and growth conditions.
All strains, plasmids, and oligonucleotides used in this study are listed in Table S1 in the supplemental material. For all manipulations, Escherichia coli and P. aeruginosa were cultured as previously described (49). E. coli strains were grown in LB (10 g liter−1 tryptone, 5 g liter−1 yeast extract, 5 g liter−1 sodium chloride) or on LA (LB with 15 g liter−1 agar). When necessary for plasmid maintenance, broth and plates were supplemented with 100 μg/ml ampicillin. All P. aeruginosa strains were cultured in LBNS (10 g liter−1 tryptone and 5 g liter−1 yeast extract) or on LANS (LBNS with 15 g liter−1 agar). Incubations were carried out at 37°C. All manipulations with E. coli were performed with either strain JM109 or C41(DE3) (Promega). All oligonucleotides used in this study were synthesized either by the DNA Synthesis Core Laboratory at Wake Forest University School of Medicine or by MWG and are listed in Table S1. Chemicals and molecular biology reagents were purchased from Sigma or Promega unless stated otherwise.
All strains generated from P. aeruginosa strain PAO1 were described previously (3). All P. aeruginosa strains used in Fig. 5 and Tables 2 and 3 were derived from FRD1 (mucA22), a mucoid CF isolate (25). The specifics regarding strain construction (15) can be found in Table S1 in the supplemental material. For complementation studies, wild-type amrZ from pDJW586 was substituted in place of the tetracycline cassette in FRD1224 (mucA22 ΔamrZ::Ωtet) to generate FRD2514. PCR amplification and DNA sequencing of the corresponding locus verified all gene replacement constructs.
Transcriptional reporter assays.
Transcriptional reporter assays were essentially performed as described previously (30). The absorbances of the samples at 420 nm, 550 nm, and 660 nm were read, and the β-galactosidase activity (amount of o-nitrophenyl-β-d-galactopyranoside [ONPG] hydrolyzed per minute as a function of cell density) was calculated. Statistical analysis of data was performed by using GraphPad InStat software.
Construction and purification of AmrZ mutant proteins.
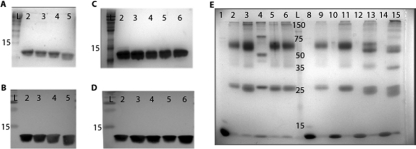
For protein assays, pET19b (Novagen, Madison, WI) was utilized due to its cleavable polyhistidine tag. The generation-and-purification protocol was based on protocols detailed previously (17, 30). Briefly, the amrZ allele of interest was cloned into the multiple-cloning site of the pET19b vector in frame with a DNA sequence encoding an 8-amino-acid (aa) rhinovirus 3C protease recognition sequence and a polyhistidine affinity tag. The resulting construct has an amino-terminal histidine affinity tag that can be cleaved with PreScission protease (Amersham Pharmacia) after purification of the AmrZ variant. To generate the alanine mutants of AmrZ, the mutant alleles were cloned into pET19b using primers amrZ9 and amrZ85 and template pJLV1 (AmrZ R14A), pPJ148 (AmrZ K18A), pPJ149 (AmrZ V20A), or pPJ150 (AmrZ R22A). To generate the extended amino terminus truncation proteins, the mutant alleles were cloned into pET19b by using template pDJW585 and the following primers: amrZ90 and amrZ85 for Δ2AmrZ, amrZ91 and amrZ85 for Δ5AmrZ, and amrZ89 and amrZ85 for Δ11AmrZ. The initial purification protocol followed a methodology similar to that described previously (17). Plasmid-expressing E. coli cells were grown at 37°C to mid-log phase, induced with 1 mM IPTG (isopropyl-β-d-1-thiogalactopyranoside) overnight at 16°C to increase protein yield, and then pelleted, resuspended in lysis buffer (100 mM KH2PO4 [pH 7.5], 500 mM NaCl, 10% glycerol, 4 M urea), sonicated, and applied over a nickel-chelating column (Qiagen). The purified protein samples were eluted with imidazole. Eluted, purified AmrZ was dialyzed overnight into dialysis buffer (100 mM Bis-Tris [pH 5.5], 100 mM NaCl, 5% glycerol, 2 mM dithiothreitol [DTT], and 0.5 mM EDTA). The polyhistidine tag was cleaved by the addition of excess PreScission protease. The final step of purification separated the pure cleaved protein from the PreScission protease and any residual uncleaved portion via a MonoS cation exchange column (GE Healthcare). The fractions containing cleaved AmrZ were pooled and dialyzed into storage buffer (100 mM Bis-Tris [pH 6.5], 100 mM NaCl, and 5% glycerol). Samples were stored at −80°C. Proteins were verified by Western blotting (Fig. 2 B and D) and by electrospray mass spectroscopy (see Table S2 in the supplemental material).
FIG. 2.
Purified AmrZ forms oligomers in solution, and oligomerization does not require the first 26 residues. (A and B) Purification and verification of the wild type and the extended amino truncation proteins Δ2AmrZ, Δ5AmrZ, and Δ11AmrZ by SDS-PAGE and visualization by GelCode staining (A) and Western blotting (B). Lane 2, wild-type AmrZ; lane 3, Δ2AmrZ; lane 4, Δ5AmrZ; lane 5, Δ11AmrZ. (C and D) Purification and verification in parallel of the wild type and the R14A AmrZ (lane 3), K18A AmrZ (lane 4), V20A AmrZ (lane 5), and R22A AmrZ (lane 6) β-sheet mutants by SDS-PAGE and visualization by GelCode staining (C) and Western blotting (D). (E) Samples were cross-linked by incubation with glutaraldehyde to form a stable complex that would withstand separation by SDS-PAGE. Lanes 1 to 6 are the β-sheet mutants. AmrZ in lane 1 was not incubated with glutaraldehyde to indicate the size of a wild-type AmrZ monomer. Lanes 2 to 6 are cross-linked and contain wild-type AmrZ (lane 2), R14A AmrZ (lane 3), K18A AmrZ (lane 4), V20A AmrZ (lane 5), and R22A AmrZ (lane 6). Lanes 8 to 15 represent the truncation proteins with alternating untreated monomeric and glutaraldehyde-treated samples. Lanes 8 and 9, wild-type AmrZ; lanes 10 and 11, Δ2AmrZ; lanes 12 and 13, Δ5AmrZ; lanes 14 and 15, Δ11AmrZ. All lanes contain 40 μmol of protein and were separated by SDS-PAGE on a 12% polyacrylamide gel, and complexes were visualized by GelCode staining.
Isolation and labeling of DNA.
Previously used protocols for the generation of target DNA for the electrophoretic mobility shift assays (EMSAs) utilized radioactive DNA (4, 30); however, this was modified for the use of fluorescently labeled target DNA. In brief, fragments containing portions of algD or amrZ were amplified by PCR using Taq polymerase and the oligonucleotides listed below. Template DNA was contributed by pDJW585 (amrZ) (49) or pDJW221 (algD) (48). A 5′-6-carboxyfluorescein (6-FAM)-labeled DNA primer amplified the desired target region in a similar PCR, generating a fluorescein-labeled PCR product at either the algD or amrZ promoter site. algD promoter sequences were amplified from pDJW221 with primers algD5 and algD7, while amrZ promoter sequences were amplified from pDJW585 with primers amrZ33 and amrZ37. For fluorescence anisotropy, each 22-bp segment was ordered individually, with a 5′ FAM label on one strand. The labeled and unlabeled strands were mixed for a stock concentration of 50 nM total.
DNA binding studies and Western blotting.
DNA binding assays and conditions were similar to those previously reported, although a fluorescent label was used in place of radioactivity to detect the promoter fragment (4). In general, wild-type or mutant AmrZ was incubated with 5′-end-labeled DNA fragments, 0.75 μg of poly(dI-dC), and binding buffer (4 mM Tris-HCl [pH 8.0], 40 mM NaCl, 4 mM MgCl2, 4% glycerol). All DNA binding assay mixtures were incubated at 25°C for 20 min in a reaction volume of 10 μl, unless noted otherwise. All EMSAs utilized a Typhoon scanner in fluorescence mode and were visualized with ImageQuant software.
Fluorescence anisotropy was chosen as a method of DNA binding analysis in equilibrium (14). Previously described protocols were used for analyses of in vitro activity (16, 27). Briefly, increasing protein titrations were incubated with a solution containing 1 nM 6-FAM-labeled DNA, 100 nM competitive nonspecific DNA, 100 mg/liter bovine serum albumin (BSA), 100 mM Bis-Tris (pH 6.5), 150 mM NaCl, and 5% glycerol and added to a constant volume of 25 μl. Protein samples were assayed by using a Safire2 microplate reader with a fluorescence polarization module (Tecan Group, Ltd.) using excitation and emission wavelengths of 490 nm and 520 nm, respectively. Anisotropy data were computed, and the background was subtracted. It was then normalized to the maximum value and plotted as a function of the AmrZ concentration by using SigmaPlot software. The Kd (dissociation constant) was calculated based on the equation generated by the best-fit curve using a single-ligand binding model. Each AmrZ variant was analyzed at each site by three individual replicates.
In all Western blots, AmrZ-specific antiserum was used as previously described (30, 49). For the Western blot shown in Fig. 5B, P. aeruginosa strains were grown at 37°C with shaking to an optical density (OD) (600 nm) of 1.0. Cells were pelleted, boiled for 5 min at 95°C, and pelleted again. The supernatants were collected, resolved on a 12% SDS-PAGE gel, and probed with an AmrZ-specific antibody.
Glutaraldehyde cross-linking.
Glutaraldehyde cross-linking was performed based on a previously described protocol (6). Briefly, AmrZ (40 μmol) was incubated with 2 μmol glutaraldehyde for 5 min before the reaction mixture was reduced on ice with 2 μmol of sodium borohydride for 20 min. The reaction mixture was neutralized by the addition of Tris-HCl (pH 8.0) to a final concentration of 167 mM. Cross-linked species were resolved on a 12% SDS-PAGE gel and visualized by GelCode blue staining.
Animal studies.
Wild-type nonmucoid P. aeruginosa laboratory strain PAO1, strain WFPA205 (an isogenic ΔamrZ::Ωtet strain), and strain WFPA513 (the R22A AmrZ-expressing strain) were previously described (3). All strains were grown in LBNS with shaking at 37°C to mid-log phase before centrifugation and resuspension in sterile phosphate-buffered saline (PBS).
The intranasal infection protocol was based on a previously described protocol (37) and modified for P. aeruginosa. Five- to six-week-old female C57BL/6 mice (Jackson Laboratory) were lightly sedated with isoflurane (Butler) and intranasally inoculated with 30 μl of sterile PBS either alone or with 1 × 108 CFU of P. aeruginosa. At designated times postinoculation, mice were euthanized, and the blood and lungs were aseptically harvested. Blood was directly plated to detect bacteria. Lungs were homogenized in PBS, and dilutions were plated onto LANS. Colonies were enumerated after overnight growth at 37°C.
For the coinfection experiment, a modified protocol was utilized wherein PAO1 and WFPA205 were mixed at a 1:1 ratio for a total of 1 × 108 CFU in 30 μl. Following infection, lung homogenates and blood were serially diluted and plated in duplicate onto both LANS and LANS supplemented with 100 μg/ml tetracycline to provide total (PAO1 and WFPA205) bacteria and WFPA205 counts, respectively. PAO1 counts were determined by the subtraction of the tetracycline-resistant (WFPA205) values from the total. The competitive index was calculated by dividing the ratio of the amount of WFPA205 recovered to the amount of PAO1 recovered by the ratio of the amount of WFPA205 inoculated to the amount of PAO1 inoculated. All animal experiments were carried out in accordance with Wake Forest University Health Sciences guidelines under an IACUC-approved protocol and were repeated at least twice. Statistical analysis was carried out by using an unpaired two-tailed Student's t test.
RESULTS
AmrZ forms oligomers in solution, and residues in the amino terminus are not required for oligomerization.
Members of the RHH family of transcriptional regulators use the DNA-binding β-sheet to recognize and bind specific sequences. In order to identify the mechanism of AmrZ-mediated gene regulation, we focused our attention on the amino terminus of AmrZ. This region contains two predicted elements, an extended amino terminus which is absent in Arc or Mnt and the putative DNA-binding β-sheet. To further clarify the residues necessary for DNA-binding activity, amino-terminal truncation mutants and site-specific mutations in the proposed β-sheet were used to evaluate the contribution of these regions to the DNA-binding activity.
To determine if the extended amino terminus contributes to either AmrZ-specific DNA binding or oligomerization, three AmrZ truncation proteins (Δ2AmrZ, Δ5AmrZ, and Δ11AmrZ) were generated (Fig. 1A). The proteins were purified, examined by SDS-PAGE and Western blotting, and verified by electrospray mass spectrometry to express the anticipated deletion (Fig. 2A and B and see Table S2 in the supplemental material). In order to evaluate the residues within the putative AmrZ β-sheet, we identified conserved residues to target for mutagenesis. The mutation of critical residues within the DNA-binding β-sheet of RHH proteins typically abrogates DNA-binding activity (20, 36). Arg14, Lys18, and Arg22 were chosen based on identity or charge similarity with required amino acids in Arc and Mnt (31, 36). Val20 was chosen because it was different from the conserved Asn and may contribute to an AmrZ-specific sequence interaction. Our previous studies showed that the substitution of alanines in place of residues Lys18 and Arg22 resulted in a loss of AmrZ-mediated DNA-binding activity at algD and a loss of twitching motility, suggesting that these residues supplied critical protein-nucleotide contacts required for DNA binding and type IV-mediated motility (3). However, these AmrZ variants were not purified or rigorously tested for either in vitro or in vivo activity. In addition, we hoped to utilize P. aeruginosa mutants expressing these variants for effects on in vivo gene expression (see below). To resolve the role of specific residues in the putative AmrZ β-sheet, several point mutations that contained alanine substitutions in AmrZ at Arg14, Lys18, Val20, or Arg22 were generated. These AmrZ β-sheet mutants were purified and examined by SDS-PAGE and Western blotting; mass spectrometry was used to verify the amino acid identity of the anticipated substitution (Fig. 2C and D and Table S2).
Most of the characterized RHH proteins exist naturally as dimers or tetramers in solution (34, 43). While oligomerization is essential for Mnt and Arc activity (44), no such studies have been performed with AmrZ. Therefore, to determine if AmrZ oligomerizes and if amino acids within the amino terminus were required for oligomerization, the wild type and the above-mentioned variant proteins were analyzed by glutaraldehyde cross-linking (Fig. 2E). Untreated AmrZ migrated at ∼12.5 kDa, consistent with the monomeric form (Fig. 2E, lane 1). However, cross-linked AmrZ showed two distinct migration bands, and taking into account the additional mass of glutaraldehyde, the bands were consistent with a dimer (∼27 kDa) and a tetramer (∼60 kDa) (Fig. 2E, lane 2). Similar species were also observed when each of the β-sheet mutants was evaluated (Fig. 2E, lanes 3 to 6). K18A AmrZ (lane 4) migrated to a position consistent with a dimer, but higher-molecular-weight oligomers had a slightly different migration compared with those of wild-type AmrZ and the other mutants. This is likely due to the fact that glutaraldehyde preferentially links the ɛ-amino group of lysine (13). Residue 18 is one of two lysine residues in AmrZ, and upon dimerization, the antiparallel β-strands would place two Lys18 residues in close proximity to each other within the β-sheet. The mutation of these lysine residues likely removed a target for the glutaraldehyde to cross-link, resulting in an incomplete cross-linking of K18A AmrZ oligomers, consistent with the altered migration of oligomers for K18A AmrZ.
Recent work indicates that the extended amino terminus of ArtA may contribute to dimer-dimer interactions at some target sites (23). This does not appear to be the case for AmrZ, since each of the AmrZ amino-terminal truncation mutants showed an oligomerization pattern similar to that of wild-type AmrZ (Fig. 2E, lanes 8 to 15), suggesting that the function of the extended amino terminus is different for AmrZ. Taken together, these data indicate that purified AmrZ does oligomerize in solution, a key feature of RHH transcriptional regulators, and that residues within the amino terminus do not contribute to this function. To further establish the role of these residues in the DNA-binding activity, we utilized the truncation and β-sheet mutants to rigorously test their contribution to DNA-binding activity in vitro and in vivo.
The extended amino terminus of AmrZ is not absolutely required for DNA-binding activity.
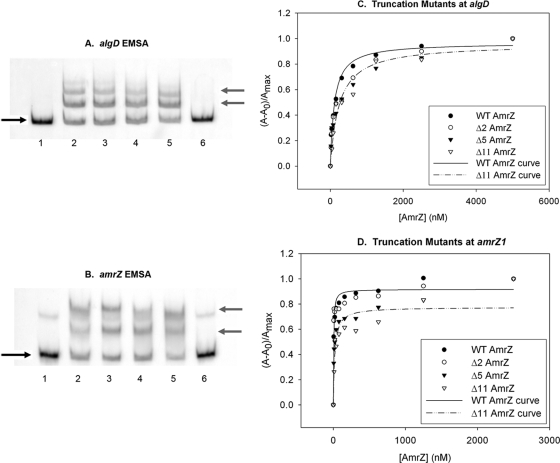
To evaluate the role of the extended amino terminus in DNA-binding activity, each truncation variant was assayed for DNA-binding activity via electrophoretic mobility shift assays (EMSAs) and fluorescence anisotropy at both an activator site (algD) and a repressor site (amrZ). The purified wild-type and AmrZ variant proteins were analyzed by EMSA at nonsaturating concentrations to directly compare DNA-binding activities. Δ2AmrZ, Δ5AmrZ, and Δ11AmrZ each retained DNA-binding activity at both templates (Fig. 3 A and B, lanes 3, 4, and 5, respectively).
FIG. 3.
The AmrZ extended amino terminus is not required for DNA-binding activity. (A and B) Protein activity was analyzed with 5′-FAM-labeled PCR-amplified targets of the algD (A) or amrZ (B) promoter region. Lanes 1 and 6 have no protein. lane 2, wild-type (WT) AmrZ; lane 3, Δ2AmrZ; lane 4, Δ5AmrZ; lane 5, Δ11AmrZ (at 125 nM each). The black arrows on the left indicate free unbound DNA, while the gray arrows indicate the migration of DNA-protein complexes. The fluorescence anisotropy data (see Materials and Methods) were assembled into a table, separated for each target site (Table 1). (C and D) A representative experiment is illustrated for the activator site algD (C) and the high-affinity repressor site amrZ1 (D).
Analysis by fluorescence anisotropy (Table 1 and Fig. 3C and D) provided a quantitative assessment of Δ2AmrZ, Δ5AmrZ, and Δ11AmrZ DNA binding at equilibrium. The ability of the protein to bind the labeled DNA was measured over increasing protein concentrations, and the Kd was calculated. Previous work has defined two distinct binding sites in the amrZ promoter sequence; based on footprinting data, there is a higher-affinity site (amrZ1) and a lower-affinity site (amrZ2), and both binding sites were individually analyzed (30). The anisotropy data revealed that the AmrZ truncation mutants had moderately reduced DNA-binding affinity at all three sites. This finding was unexpected given that there was no visible loss of DNA-binding activity by EMSA (compare data in Fig. 3C and Table 1 with Fig. 3A and B). This was most notable with Δ11AmrZ, which had a 4-fold reduction in affinity at amrZ1 compared to that of wild-type AmrZ (Table 1). The difference between the EMSA data and the anisotropy data may be due to the limitations of each assay. Anisotropy is performed at equilibrium with a small segment of DNA (22 bp in size), while EMSA is performed with a much larger piece of DNA (∼300 bp) and in the context of a cage formation by the gel matrix. Regardless of the subtle difference, both assays showed a binding activity of the truncation mutants and suggested that the extended amino terminus may contribute to, but is not absolutely required for, in vitro DNA-binding activity.
TABLE 1.
Fluorescence anisotropy data
| AmrZ protein |
algD |
amrZ1 |
amrZ2 |
|||
|---|---|---|---|---|---|---|
| Avg Kd (nM) ± SDa | Fold over WTb | Avg Kd (nM) ± SDa | Fold over WTb | Avg Kd (nM) ± SDa | Fold over WTb | |
| WT | 107 ± 17 | 1.0 | 4.34 ± 0.74 | 1.0 | 59 ± 9.7 | 1.0 |
| D2 | 143 ± 17 | 1.33 | 3.87 ± 0.76 | 0.89 | 89 ± 15 | 1.52 |
| Δ5 | 140 ± 21 | 1.30 | 9.77 ± 1.92 | 2.25 | 134 ± 13 | 2.28 |
| Δ11 | 170 ± 25 | 1.58 | 17.9 ± 5.70 | 4.12 | 106 ± 25 | 1.81 |
| R14A | 613 ± 86 | 5.72 | 3.96 ± 0.73 | 0.91 | 306 ± 28 | 5.21 |
| K18A | 2,300 ± 472 | 21.46 | 1,190 ± 210 | 274.2 | 306 ± 78 | 5.22 |
| V20A | 474 ± 97 | 4.42 | 45.4 ± 10.6 | 10.46 | 988 ± 98 | 16.83 |
| R22A | 1,347 ± 133 | 12.57 | 192 ± 26 | 44.24 | 875 ± 89 | 14.91 |
The Kd was calculated based on the averages of data from three independent experiments. The data were graphed with SigmaPlot (a representative experiment is graphed in Fig. 3 and 4C and D) and analyzed based on a single-ligand-binding model with saturation.
Fold over the wild type (WT) is defined as (calculated Kd of sample)/(calculated Kd of the wild type) for each binding site.
Mutation of critical residues within the AmrZ DNA-binding β-sheet abrogates DNA-binding activity.
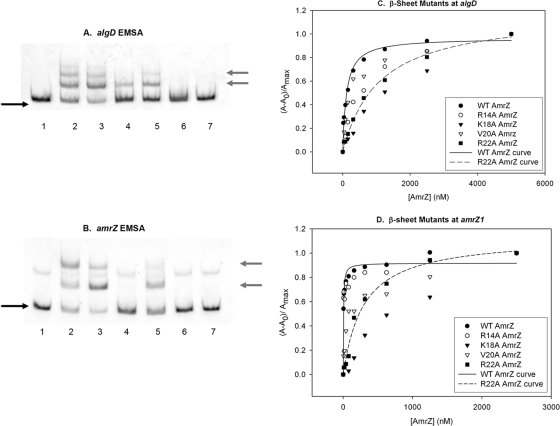
The AmrZ β-sheet variants were also rigorously assayed for DNA-binding activity at algD and amrZ by EMSA and fluorescence anisotropy (Table 1 and Fig. 4). As documented previously (2, 30), AmrZ forms several protein-DNA complexes (Fig. 4A and B, lanes 2), which increase in number as the concentration of AmrZ increases. Our prior footprinting studies showed that these species likely represent AmrZ oligomers binding to a single site rather than AmrZ occupying new sites as the protein concentration increases (30). The R14A AmrZ mutant showed a DNA-binding activity similar to that of AmrZ at both algD and amrZ (Fig. 4A and B, lanes 3). Interestingly, K18A AmrZ exhibited reduced DNA-binding activity at algD yet had no visible activity at amrZ (compare lanes 4 in Fig. 4A and B). This finding initially suggested that the K18A AmrZ mutant is able to differentiate between the activator and repressor sites. V20A AmrZ showed reduced binding activity at both sites (Fig. 4A and B, lanes 5). As seen previously with hexahistidine-tagged versions (3), R22A AmrZ showed no DNA-binding activity at either site (Fig. 4A and B, lane 6). The reduced binding affinity of V20A AmrZ and K18A AmrZ (at algD only) was compensated for by the increased protein concentration (data not shown); however, this was not the case for R22A AmrZ.
FIG. 4.
Mutation of specific residues causes a loss of AmrZ-mediated DNA-binding activity. (A and B) Protein activity was analyzed with 5′-FAM-labeled PCR-amplified targets of the algD activator (A) or amrZ (B) promoter region. Lanes 1 and 7 have no protein. Lane 2, wild-type AmrZ; lane 3, R14A AmrZ; lane 4, K18A AmrZ; lane 5, V20A AmrZ; lane 6, R22A AmrZ (at 125 nM each). The black arrows on the left indicate free unbound DNA, while the gray arrows indicate the migration of DNA-protein complexes. The fluorescence anisotropy data (see Materials and Methods) were assembled into a table, separated for each target site (Table 1). (C and D) A representative experiment is illustrated for the activator site algD (C) and the high-affinity repressor site amrZ1 (D).
Analysis of the DNA-binding activities of these proteins by fluorescence anisotropy revealed that each protein had reduced binding compared to that of wild-type AmrZ (Fig. 4C and Table 1). R14A AmrZ and V20A AmrZ showed only a modest increase in the Kd at algD, which correlates with the minor reduction of activity seen by EMSA. The R22A mutation exhibited measurable activity by this assay, although it was greatly reduced at all three sites compared to that of wild-type AmrZ. Interestingly, fluorescence anisotropy studies showed that the K18A mutation had a lower affinity than the R22A mutation at algD (Fig. 4C), which conflicts with the moderate activity seen by EMSA (Fig. 4A). One potential explanation may be that the K18A mutation recognizes an altered DNA-binding sequence that exists within the EMSA target fragment and is not the same target site on the anisotropy DNA fragment; the disparity between experiments may be due to the size and sequence of the target DNA used in these two assays. We will later pursue this topic as it relates to in vivo protein activity (Fig. 5 A and Tables 2 and 3).
FIG. 5.
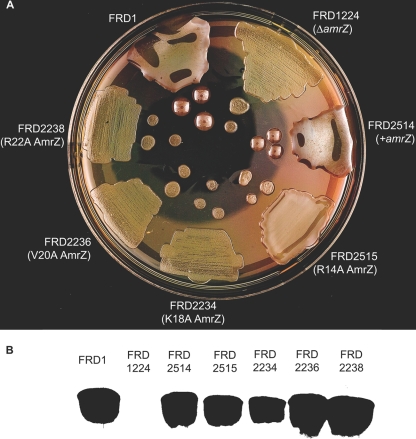
Residues Lys18, Val20, and Arg22 are required for alginate production. (A) The wild-type amrZ strain (FRD1), an amrZ-null strain (FRD1224), and FRD1224 complemented with wild-type amrZ (FRD2514), amrZ28 (AmrZ R14A) (FRD2515), amrZ17 (AmrZ K18A) (FRD2234), amrZ18 (AmrZ V20A) (FRD2236), or amrZ19 (AmrZ R22A) (FRD2238) was plated onto LANS and incubated at 37°C for 24 h to visualize the alginate phenotype. (B) Western blot of strains expressing the wild type or the null or alanine substitution mutants of AmrZ. Strain names are identical to those listed in A.
TABLE 2.
AmrZ residues Lys18, Val20, and Arg22 are absolutely required for activation of algD transcriptiona
| Strain | Genotype | AmrZ expressed | Avg β-galactosidase activity (Miller units) ± SD | % of wild-type activity |
|---|---|---|---|---|
| FRD2606 | mucA22 attB::algD-lacZ | Wild type | 10,011 ± 44 | 100 |
| FRD2526 | mucA22 amrZ::Ωtet attB::algD-lacZ | 5 ± 0.9 | 0.05 | |
| FRD2534 | mucA22 amrZ37 attB::algD-lacZ | AmrZ R14A | 285 ± 5 | 3 |
| FRD2517 | mucA22 amrZ17 attB::algD-lacZ | AmrZ K18A | 0 ± 0.9 | 0 |
| FRD2519 | mucA22 amrZ18 attB::algD-lacZ | AmrZ V20A | 65 ± 2 | 0.6 |
| FRD2521 | mucA22 amrZ19 attB::algD-lacZ | AmrZ R22A | 0 ± 0.6 | 0 |
Single-copy algD-lacZ transcriptional fusions were placed at the neutral attB site in isogenic amrZ+ (FRD2606), AmrZ R14A (FRD2531), AmrZ K18A (FRD2517), AmrZ V20A (FRD2519), and AmrZ R22A (FRD2521) strains of P. aeruginosa. All strains were engineered in an FRD1 (mucA22) background. β-Galactosidase activity was recorded in Miller units, and results are average data from three separate experiments. Percentages of wild-type values are also listed for comparison purposes.
TABLE 3.
AmrZ residues Lys18, Val20, and Arg22 are absolutely required for repression of amrZ transcriptiona
| Strain | Genotype | AmrZ expressed | Avg β-galactosidase activity (Miller units) ± SD | % of wild-type activity |
|---|---|---|---|---|
| FRD1310 | mucA22 attB::amrZ-lacZ | Wild type | 2,054 ± 41 | 100 |
| FRD1312 | mucA22 amrZ::xylE aacC1 attB::amrZ-lacZ | 3,157 ± 214 | 154 | |
| FRD2528 | mucA22 amrZ37 attB::amrZ-lacZ | AmrZ R14A | 1,588 ± 38 | 77 |
| FRD2530 | mucA22 amrZ17 attB::amrZ-lacZ | AmrZ K18A | 3,913 ± 100 | 191 |
| FRD2532 | mucA22 amrZ18 attB::amrZ-lacZ | AmrZ V20A | 4,377 ± 109 | 213 |
| FRD2503 | mucA22 amrZ19 attB::amrZ-lacZ | AmrZ R22A | 3,946 ± 104 | 192 |
Single-copy amrZ-lacZ transcriptional fusions were placed at the neutral attB site in isogenic amrZ+ (FRD1310), AmrZ R14A (FRD2528), AmrZ K18A (FRD2530), AmrZ V20A (FRD2532), and AmrZ R22A (FRD2503) strains of P. aeruginosa. All strains were engineered in an FRD1 (mucA22) background. β-Galactosidase activity was recorded in Miller units, and results are average data from three separate experiments. Percentages of wild-type values are also listed for comparison purposes.
At the higher-affinity amrZ1 site (Fig. 4D), the anisotropy data correlated with the EMSA data; the R14A AmrZ mutant showed activity similar to that of the wild type, with the V20A mutant having moderately reduced activity and the K18A and R22A AmrZ mutants showing little detectable binding. At the lower-affinity amrZ2 site, all of the mutants had a moderate-to-high reduction in activity. Val20 appears to contribute to binding activity but is not absolutely required, as shown by the EMSA data (Fig. 4A and B). Taken together with previous data (3), these results indicate that Lys18 and Arg22 are required for proper DNA-binding activity in vitro.
Residues Lys18, Val20, and Arg22 are required for alginate production in vivo.
AmrZ is a DNA-binding protein that functions as a transcriptional regulator, and DNA-binding activity is likely required, but not necessarily sufficient, for transcriptional activity. Since several of the AmrZ variants had reduced or no detectable DNA-binding activity in vitro, we wanted to assess the effect of these mutant proteins on AmrZ transcriptional activity in vivo. To do so, we examined alginate production by strains harboring the amrZ mutant alleles as well as an amrZ deletion strain and its chromosomally complemented amrZ+ strain (Fig. 5). All strains displayed similar growth kinetics in rich medium (data not shown). Protein extracts from each strain were analyzed by SDS-PAGE and probed with an AmrZ-specific antibody in an immunoblot assay to verify AmrZ expression (Fig. 5B).
The parental wild-type strain (FRD1) displayed a characteristic mucoid phenotype due to the overproduction of alginate (Fig. 5A). As expected, the deletion of amrZ resulted in a nonmucoid phenotype; alginate production was restored upon the chromosomal complementation of wild-type amrZ. Interestingly, strains expressing K18A AmrZ, V20A AmrZ, and R22A AmrZ all displayed a nonmucoid phenotype, indicating that AmrZ residues Lys18, Val20, and Arg22 are required for alginate production. Despite R14A AmrZ, K18A AmrZ, and V20A AmrZ all exhibiting some DNA-binding activity at algD in vitro, only P. aeruginosa strains expressing the R14A AmrZ mutant displayed visible levels of alginate production (Fig. 5A). This finding indicated that despite the apparent DNA-binding activity by EMSA (Fig. 4A), the 5-fold decline in activity determined by fluorescence anisotropy (Table 1) is a better reflection of the impact of the R14A mutation in vivo.
Lys18, Val20, and Arg22 are required for transcriptional activation and repression in vivo.
We next evaluated the effect of β-sheet mutations on the AmrZ-mediated activation of algD and autorepression at amrZ using single-copy algD-lacZ or amrZ-lacZ transcription fusions. Consistent with the alginate production assay, P. aeruginosa strains expressing K18A AmrZ, V20A AmrZ, or R22A AmrZ (Table 2) showed no detectable algD-lacZ transcription. Also, FRD2515, which expresses the R14A AmrZ mutant and displayed a semimucoid phenotype (Fig. 5), had significantly reduced levels of algD transcription (3% compared to the wild type) (Table 2). Despite sharing identity with a critical residue in the Mnt β-sheet, Arg14 is not absolutely essential for the activation of algD transcription. Collectively, data in Table 2 and Fig. 5 demonstrated that proposed β-sheet residues Lys18, Val20, and Arg22 of AmrZ are required for algD transcription and subsequent alginate production.
In a parallel manner, we assayed the effect of the β-sheet mutations on autorepression at amrZ. A lack of repression by an AmrZ variant would be seen as an increase in β-galactosidase activity. Like the amrZ-null strain, an increase in amrZ-lacZ transcription was seen for the strains expressing K18A AmrZ, V20A AmrZ, and R22A AmrZ compared to wild-type levels (Table 3), indicating a loss of AmrZ-mediated repression by these variants. Likely, insufficient DNA binding to the amrZ promoter by K18A, V20A, and R22A (Fig. 4B and D) lifts the repression of amrZ transcription (Table 3). R14A AmrZ levels of amrZ-lacZ transcription were similar to those of the wild type, illustrating that the DNA-binding activity of the AmrZ R14A mutant was sufficient to mediate the repression of amrZ transcription (Fig. 4B and Tables 1 and 3). Collectively, these data show that AmrZ residues Lys18, Val20, and Arg22 are required for the repression of amrZ transcription and suggest that identical residues within a single DNA-binding domain are required for AmrZ-mediated gene regulation.
AmrZ DNA-binding activity and transcriptional regulation contribute to P. aeruginosa virulence in an acute model.
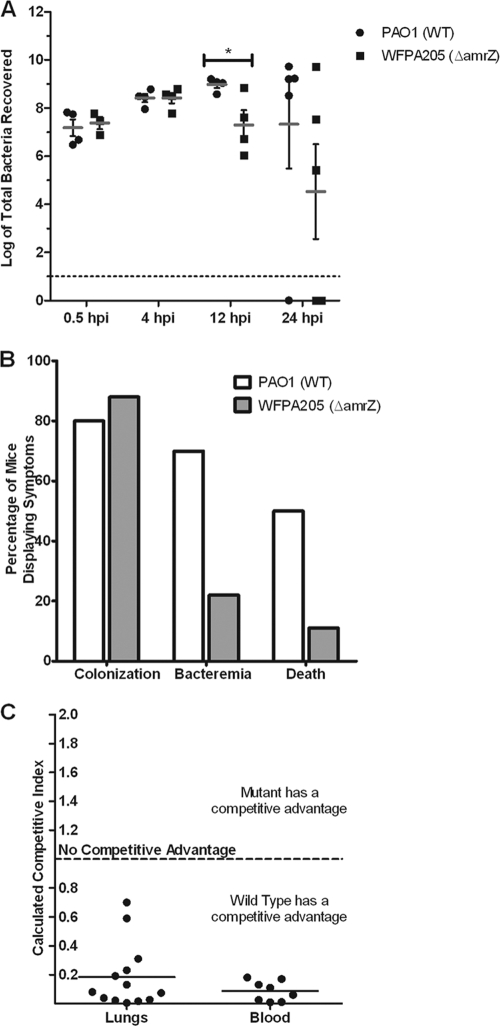
The role of AmrZ transcriptional regulation during the course of infection has never been tested. As a regulator of known P. aeruginosa virulence determinants, its contribution was analyzed by comparisons of the relative levels of virulence of strains expressing wild-type AmrZ, amrZ-null mutants, and strains expressing AmrZ R22A in an acute pulmonary model of infection. First, wild-type PAO1 or the isogenic amrZ-null mutant strain WFPA205 was intranasally inoculated into mice. At the indicated time points, mice were euthanized, the lungs were aseptically harvested, and bacterial counts were determined. At early time points (0.5 and 4 h postinfection [hpi]), there were similar bacterial counts recovered from mice infected with both strains (Fig. 6 A), suggesting no defect in growth or the ability to recover bacteria from both strains and that similar bacterial numbers were inoculated into mice. In contrast, at 12 hpi, there was a significant reduction in the number of WFPA205 bacteria recovered compared with that of the wild type (P = 0.0360) (Fig. 6A), indicating that the amrZ mutant had a reduction in bacterial colonization of the lungs. By 24 hpi, the recovered bacterial counts had a larger distribution, and no statistical observation could be made. While the incidence of bacteria present in the lung did not differ between strains, levels of bacteremia and mortality associated with PAO1 infection were higher than those of mice infected with WFPA205 (Fig. 6B). This finding suggests that the loss of AmrZ results in a less virulent strain and that AmrZ does regulate virulence factors that contribute to the establishment of an acute infection.
FIG. 6.
amrZ mutants exhibit an in vivo virulence defect. (A) Wild-type strain PAO1 and strain WFPA205 (amrZ::tet) were used to intranasally inoculate 6-week-old C57BL/6 mice with 1 × 108 bacteria. At the indicated time points (hpi), lungs were aseptically harvested, and the bacteria were enumerated. Each symbol indicates an individual mouse (n = 5 for each group per time point). Gray bars indicate averages along with standard deviations (*, P = 0.036). (B) Mice (n = 15) were assessed for bacterial colonization (>104 bacteria recovered from the lungs), bacteremia (bacteria recovered from the blood), or death (requiring euthanasia due to severe disease symptoms) at 12 hpi. (C) Strains PAO1 and WFPA205 were coinoculated at a 1:1 ratio (total of 108 bacteria). At 24 hpi, lungs and blood were harvested, and bacterial counts were determined. The competitive index from the lungs is plotted for each mouse (n = 13) and for those that showed evidence of bacteremia (n = 8).
To reduce natural variation between mice and more directly test relative fitness during an infection, a coinfection model used the same two strains to determine if wild-type P. aeruginosa had an in vivo fitness advantage compared with WFPA205. In vitro coculturing showed no growth advantage for either strain (data not shown). After 12 hpi, compared with WFPA205, PAO1 showed a ∼5-fold competitive advantage in the lungs and a 12-fold advantage in the bloodstream (Fig. 6C). This finding indicates that the transcriptional regulation of virulence factors by AmrZ during an infection provides protection from the immune system specifically for the wild-type bacteria within the host.
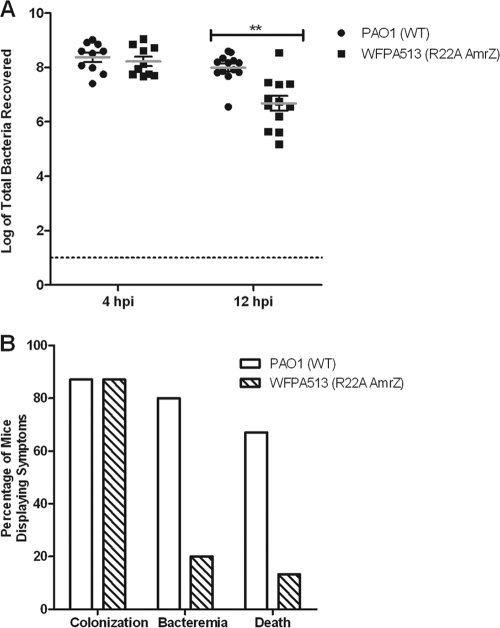
The data shown in Fig. 6 suggest that the presence of amrZ provides P. aeruginosa a competitive advantage, hypothesized to be via AmrZ's transcriptional regulation of virulence factors. To directly test the role of AmrZ DNA binding and transcriptional regulation in vivo, WFPA513, a strain expressing the DNA-binding-defective R22A AmrZ, was used. Based on EMSA data and anisotropy data (Fig. 4 and Table 1) as well as in vivo data (Fig. 5 and Tables 2 and 3) (3), the R22A mutation has no DNA-binding activity or any transcriptional regulatory capacity. Similar to the effect shown in Fig. 6A, WFPA513 also exhibited a decrease in the lung bacterial burden at 12 hpi compared with that of wild-type PAO1 (P = 0.003) (Fig. 7 A). Additionally, WFPA513 had reductions in rates of morbidity and mortality (Fig. 7B) similar to those seen for amrZ-null mutant strain WFPA205 (Fig. 6B). Taken together, this indicates that the loss of virulence correlates with the loss of DNA binding and transcriptional regulation of virulence factors mediated by AmrZ.
FIG. 7.
AmrZ DNA-binding activity is required for optimum virulence. (A) A total of 108 PAO1 or WFPA513 (R22A AmrZ) bacteria were used to intranasally inoculate 6-week-old C57BL/6 mice. At 4 and 12 hpi, lungs were aseptically harvested, and the bacteria were enumerated. Each symbol indicates an individual mouse (n = 15 for each group per time point). Gray bars indicate averages along with standard deviations (**, P = 0.003). (B) Mice (n = 15) were assessed for bacterial colonization (>104 bacteria recovered from the lungs), bacteremia (bacteria recovered from the blood), or death (requiring euthanasia due to severe disease symptoms) at 12 hpi.
DISCUSSION
In this study, we found that AmrZ residues Lys18, Val20, and Arg22 within the proposed β-sheet are essential for the activation of algD expression and alginate production. AmrZ residues Lys18, Val20, and Arg22 are also required for autorepression, signifying that these residues are important for modulating both the repression and activation of these two target genes. Another discovery is that AmrZ has the capability of oligomerizing into species that are consistent with dimers and tetramers in solution. Previous studies revealed that oligomerization is essential for Mnt repressor activity (44), and most studied RHH proteins exist as dimers or tetramers (34, 43). Evidence for AmrZ oligomerization contributes to its inclusion as an RHH protein. In addition, each of the AmrZ variants exhibited oligomerization properties similar to that of native AmrZ, indicating that our observed in vitro and in vivo effects were not due to the inability of these proteins to oligomerize. Cross-linking of the truncation proteins also indicated that the extended amino terminus is not required for the formation of higher-order oligomers in solution.
AmrZ functions as a putative RHH transcriptional regulator, wherein residues in the DNA-binding β-sheet are responsible for creating sequence-specific contacts with the target DNA. In vivo analysis revealed that Lys18, Val20, and Arg22 are absolutely required for transcriptional regulation by AmrZ (Tables 2 and 3) and downstream alginate production (Fig. 5). AmrZ residues 15 to 23 are predicted to form a β-sheet (Fig. 1), and previous studies of the Arc and Mnt repressors have shown that β-sheet residues determine binding specificity (19, 20). The specific mutation of conserved residues from the Arc and Mnt amino acid template (Fig. 1A) supports the hypothesis that similar amino acids in the AmrZ putative β-sheet (Fig. 1B) are required for DNA-binding activity. This not only supports the classification of AmrZ as an RHH protein but also supports the hypothesis that AmrZ uses similar residues within the β-sheet to recognize both transcriptionally activated and repressed targets.
Over the course of this investigation, we analyzed V20A AmrZ, which had reduced binding activity; K18A AmrZ, which had differential activity; and R22A AmrZ, which had no DNA binding in vitro (Fig. 4). However, strains expressing all three mutant proteins had similar defects in transcriptional regulation (Tables 2 and 3) and in alginate production (Fig. 5). With regard to the data for V20A AmrZ, the disparity between in vitro and in vivo binding activities may be due to a more transient interaction with the DNA, which is not conducive to transcriptional regulation. In EMSAs, K18A AmrZ appears to be able to differentiate between algD activator and amrZ repressor sites. However, K18A AmrZ shows no in vivo activity, which is consistent with the in vitro anisotropy data.
AmrZ residues in the extended amino terminus contribute minimally to binding at algD and amrZ (Table 1). Since these 12 residues are absent in Arc or Mnt but have functions in other RHH proteins, the contribution of these residues to AmrZ transcriptional regulation is an interesting area for future study. Notably, studies of other RHH proteins with an extended amino terminus indicate that this portion of the protein often contributes to a protein-specific activity, including DNA-binding activity. The flexible 30-amino-acid amino-terminal tail of ParG is not required for dimerization but is required for repression and high-affinity protein-DNA interactions (8). More-recent work has identified this ParG sequence as an arginine finger-like motif that has two vital functions in vivo, stimulating ATP hydrolysis and ParF-ATP filamentation (1). Like AmrZ, H. pylori NikR has several DNA targets (nixA, ureA, fur, nikR, exbB, fecA3, and frpB4), where it also acts as a transcriptional activator and repressor. The 9-amino-acid amino-terminal tail of H. pylori NikR prevents low-affinity binding. In fact, within this region, two specific residues have been linked to high-affinity binding, while two others are linked to nickel-specific DNA-binding activity in vitro (5). Recent work indicated that ArtA has an amino terminus that is necessary for target recognition and binding (23). However, it appears to be necessary for only one of the six targets studied. Further experiments may show that this is also true for an AmrZ target other than algD or amrZ. Future studies will also examine the role of the extended amino terminus in AmrZ structural stability and its modest contribution to AmrZ binding.
AmrZ utilizes a single DNA-binding domain to regulate the expression of at least two genes. The DNA-binding activity of the amino-terminal portion of Mnt is distinct from the oligomerization properties of the C-terminal domain (44), and truncated Mnt repressors lacking the C-terminal oligomerization domain form dimers in solution and retain DNA-binding activity (21). The AmrZ C-terminal truncation that lacks the proposed oligomerization domain (AmrZ residues 1 to 66) binds DNA in a manner similar to that of the wild type, suggesting that separate regions of AmrZ may mediate binding and oligomerization (data not shown). Future studies will address the potential contribution of C-terminal residues to higher-order oligomerization and high-affinity DNA binding.
Alginate production and algD expression in P. aeruginosa are controlled by a variety of transcriptional regulators, including the proposed RHH DNA-binding protein AmrZ. A majority of soil-dwelling pseudomonads do not produce alginate in the absence of specific environmental cues, although they have the genetic capacity to do so (42). Homologs of genes encoding regulators of alginate biosynthesis are found in the genome of P. putida WCS358. Some of the alginate biosynthesis and regulatory genes in P. syringae, a plant pathogen that utilizes environmental signals such as copper to trigger alginate production, have been studied (10, 18). However, in P. syringae, the expression of the alginate transcriptional regulator AlgT, rather than alginate production alone, is required for virulence (33). Amino acid comparisons of AmrZ with its proposed orthologs PP4470 (P. putida) and PSPTO1847 (P. syringae pv. tomato) showed that greater than 80% of AmrZ residues are conserved. Although these loci appear to be AmrZ orthologs, no function has been ascribed to these open reading frames (ORFs) in either Pseudomonas species. Their extensive amino acid identity with AmrZ suggests that these proteins have the potential to serve as regulators of alginate genes and possibly others. Furthermore, similar virulence factors are necessary for both human and plant colonization by P. aeruginosa (29, 45), suggesting conserved transcriptional regulation in host pathogenesis. It is reasonable to hypothesize that the mechanisms of regulation may be similar in the pseudomonads and increase the likelihood of the AmrZ orthologs contributing to the regulation of virulence factors in a manner similar to that displayed by P. aeruginosa.
The impact of AmrZ in the context of an acute-infection model has not previously been assessed. In this study it is clear, both by bacterial enumeration at 12 hpi as well as by indicators of morbidity and mortality, that AmrZ-mediated transcriptional regulation is necessary for optimal virulence. In the PAO1 background, the loss of functional AmrZ (both in an amrZ-null mutant and in R22A AmrZ-expressing strains) results in the improper expression of type IV pili and a loss of twitching motility (3). The loss of these factors in other P. aeruginosa strains and in other models of infection (neonatal mouse and corneal) also reduced virulence (9, 39, 40, 50). However, AmrZ likely regulates other virulence factors, and future work aims to elucidate the targets of AmrZ transcriptional regulation that contribute to virulence in models of both chronic and acute infection.
Supplementary Material
Acknowledgments
We acknowledge Jessica L. VonCannon for technical support as well as Cameron Dennis and William Willner from Creative Communications at Wake Forest University School of Medicine for photography assistance.
This work was supported by American Heart Association predoctoral fellowships 0815249E (E.A.W.) and 0215191U (D.M.R.) and Public Health Service grants AI061396 and HL58334 (D.J.W).
Footnotes
Published ahead of print on 13 August 2010.
Present address: Institute for Cellular Therapeutics, University of Louisville, Donald Baxter I Bldg., Ste. 404, 570 S. Preston Street, Louisville, KY 40202.
REFERENCES
- 1.Barillà, D., E. Carmelo, and F. Hayes. 2007. The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. Proc. Natl. Acad. Sci. U. S. A. 104:1811-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baynham, P. J., A. L. Brown, L. L. Hall, and D. J. Wozniak. 1999. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 33:1069-1080. [DOI] [PubMed] [Google Scholar]
- 3.Baynham, P. J., D. M. Ramsey, B. V. Gvozdyev, E. M. Cordonnier, and D. J. Wozniak. 2006. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 188:132-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baynham, P. J., and D. J. Wozniak. 1996. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 22:97-108. [DOI] [PubMed] [Google Scholar]
- 5.Benanti, E. L., and P. T. Chivers. 2007. The N-terminal arm of the Helicobacter pylori Ni2+-dependent transcription factor NikR is required for specific DNA binding. J. Biol. Chem. 282:20365-20375. [DOI] [PubMed] [Google Scholar]
- 6.Biswas, E. E., P.-H. Chen, and S. B. Biswas. 1995. Overexpression and rapid purification of biologically active yeast proliferating cell nuclear antigen. Protein Expr. Purif. 6:763-770. [DOI] [PubMed] [Google Scholar]
- 7.Burgering, M. J. M., R. Boelens, D. E. Gilbert, J. N. Breg, K. L. Knight, R. T. Sauer, and R. Kaptein. 2002. Solution structure of dimeric Mnt repressor (1-76). Biochemistry 33:15036-15045. [DOI] [PubMed] [Google Scholar]
- 8.Carmelo, E., D. Barillà, A. P. Golovanov, L.-Y. Lian, A. Derome, and F. Hayes. 2005. The unstructured N-terminal tail of ParG modulates assembly of a quaternary nucleoprotein complex in transcription repression. J. Biol. Chem. 280:28683-28691. [DOI] [PubMed] [Google Scholar]
- 9.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fakhr, M. K., A. Penaloza-Vazquez, A. M. Chakrabarty, and C. L. Bender. 1999. Regulation of alginate biosynthesis in Pseudomonas syringae pv. syringae. J. Bacteriol. 181:3478-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golovanov, A. P., D. Barillà, M. Golovanova, F. Hayes, and L.-Y. Lian. 2003. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol. Microbiol. 50:1141-1153. [DOI] [PubMed] [Google Scholar]
- 12.Guillière, F., N. Peixeiro, A. Kessler, B. Raynal, N. Desnoues, J. Keller, M. Delepierre, D. Prangishvili, G. Sezonov, and J. Guijarro. 2009. Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1. J. Biol. Chem. 284:22222-22237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayat, M. A. 2000. Principles and techniques of electron microscopy: biological applications, 4th ed. Cambridge University Press, Cambridge, United Kingdom.
- 14.Heyduk, T., and J. Lee. 1990. Application of fluorescence energy transfer and polarization to monitor Escherichia coli cAMP receptor protein and lac promoter interaction. Proc. Natl. Acad. Sci. U. S. A. 87:1744-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 16.Holland, P. J., and T. Hollis. 2010. Structural and mutational analysis of Escherichia coli AlkB provides insight into substrate specificity and DNA damage searching. PLoS One 5:e8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollis, T., J. M. Stattel, D. S. Walther, C. C. Richardson, and T. Ellenberger. 2001. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc. Natl. Acad. Sci. U. S. A. 98:9557-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keith, R. C., L. M. W. Keith, G. Hernandez-Guzman, S. R. Uppalapati, and C. L. Bender. 2003. Alginate gene expression by Pseudomonas syringae pv. tomato DC3000 in host and non-host plants. Microbiology 149:1127-1138. [DOI] [PubMed] [Google Scholar]
- 19.Knight, K. L., and R. T. Sauer. 1992. Biochemical and genetic analysis of operator contacts made by residues within the beta-sheet DNA binding motif of Mnt repressor. EMBO J. 11:215-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knight, K. L., and R. T. Sauer. 1989. DNA binding specificity of the Arc and Mnt repressors is determined by a short region of N-terminal residues. Proc. Natl. Acad. Sci. U. S. A. 86:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight, K. L., and R. T. Sauer. 1988. The Mnt repressor of bacteriophage P22: role of C-terminal residues in operator binding and tetramer formation. Biochemistry 27:2088-2094. [DOI] [PubMed] [Google Scholar]
- 22.Murayama, K., P. Orth, A. B. de la Hoz, J. C. Alonso, and W. Saenger. 2001. Crystal structure of [omega] transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 Å resolution. J. Mol. Biol. 314:789-796. [DOI] [PubMed] [Google Scholar]
- 23.Ni, L., S. O. Jensen, N. K. Tonthat, T. Berg, S. M. Kwong, F. H. X. Guan, M. H. Brown, R. A. Skurray, N. Firth, and M. A. Schumacher. 2009. The Staphylococcus aureus pSK41 plasmid-encoded ArtA protein is a master regulator of plasmid transmission genes and contains a RHH motif used in alternate DNA-binding modes. Nucleic Acids Res. 37:6970-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberer, M., K. Zangger, S. Prytulla, and W. Keller. 2002. The anti-toxin ParD of plasmid RK2 consists of two structurally distinct moieties and belongs to the ribbon-helix-helix family of DNA-binding proteins. Biochem. J. 361:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohman, D. E., and A. M. Chakrabarty. 1981. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect. Immun. 33:142-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavlov, N. A., D. I. Cherny, I. V. Nazimov, A. I. Slesarev, and V. Subramaniam. 2002. Identification, cloning and characterization of a new DNA-binding protein from the hyperthermophilic methanogen Methanopyrus kandleri. Nucleic Acids Res. 30:685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perrino, F. W., U. de Silva, S. Harvey, E. E. Pryor, D. W. Cole, and T. Hollis. 2008. Cooperative DNA binding and communication across the dimer interface in the TREX 3′-5′-exonuclease. J. Biol. Chem. 283:21441-21452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popescu, A., A. Karpay, D. A. Israel, R. M. Peek, Jr., and A. M. Krezel. 2005. Helicobacter pylori protein HP0222 belongs to Arc/MetJ family of transcriptional regulators. Proteins 59:303-311. [DOI] [PubMed] [Google Scholar]
- 29.Rahme, L., E. Stevens, S. Wolfort, J. Shao, R. Tompkins, and F. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 30.Ramsey, D. M., P. J. Baynham, and D. J. Wozniak. 2005. Binding of Pseudomonas aeruginosa AlgZ to sites upstream of the algZ promoter leads to repression of transcription. J. Bacteriol. 187:4430-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raumann, B. E., M. A. Rould, C. O. Pabo, and R. T. Sauer. 1994. DNA recognition by β-sheets in the Arc repressor-operator crystal structure. Nature 367:754-757. [DOI] [PubMed] [Google Scholar]
- 32.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 33.Schenk, A., H. Weingart, and M. S. Ullrich. 2008. The alternative sigma factor AlgT, but not alginate synthesis, promotes in planta multiplication of Pseudomonas syringae pv. glycinea. Microbiology 154:413-421. [DOI] [PubMed] [Google Scholar]
- 34.Schreiter, E. R., and C. L. Drennan. 2007. Ribbon-helix-helix transcription factors: variations on a theme. Nat. Rev. Microbiol. 5:710-720. [DOI] [PubMed] [Google Scholar]
- 35.Schreiter, E. R., M. D. Sintchak, Y. Guo, P. T. Chivers, R. T. Sauer, and C. L. Drennan. 2003. Crystal structure of the nickel-responsive transcription factor NikR. Nat. Struct. Biol. 10:794-799. [DOI] [PubMed] [Google Scholar]
- 36.Silbaq, F. S., S. E. Ruttenberg, and G. D. Stormo. 2002. Specificity of Mnt ‘master residue’ obtained from in vivo and in vitro selections. Nucleic Acids Res. 30:5539-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sloan, G. P., C. F. Love, N. Sukumar, M. Mishra, and R. Deora. 2007. The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J. Bacteriol. 189:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somers, W. S., and S. E. Phillips. 1992. Crystal structure of the met repressor-operator complex at 2.8 angstrom resolution reveals DNA recognition by β-strands. Nature 359:387-393. [DOI] [PubMed] [Google Scholar]
- 39.Tang, H., M. Kays, and A. Prince. 1995. Role of Pseudomonas aeruginosa pili in acute pulmonary infection. Infect. Immun. 63:1278-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang, H. B., E. DiMango, R. Bryan, M. Gambello, B. H. Iglewski, J. B. Goldberg, and A. Prince. 1996. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect. Immun. 64:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tart, A. H., M. J. Blanks, and D. J. Wozniak. 2006. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol. 188:6483-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venturi, V., M. Otten, V. Korse, B. Brouwer, J. Leong, and P. Weisbeek. 1995. Alginate regulatory and biosynthetic gene homologs in Pseudomonas putida WCS358: correlation with the siderophore regulatory gene pfrA. Gene 155:83-88. [DOI] [PubMed] [Google Scholar]
- 43.Vershon, A. K., P. Youderian, M. M. Susskind, and R. T. Sauer. 1985. The bacteriophage P22 Arc and Mnt repressors. Overproduction, purification, and properties. J. Biol. Chem. 260:12124-12129. [PubMed] [Google Scholar]
- 44.Waldburger, C. D., and R. T. Sauer. 1995. Domains of Mnt repressor: roles in tetramer formation, protein stability, and operator DNA binding. Biochemistry 34:13109-13116. [DOI] [PubMed] [Google Scholar]
- 45.Walker, T. S., H. P. Bais, E. Deziel, H. P. Schweizer, L. G. Rahme, R. Fall, and J. M. Vivanco. 2004. Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 134:320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weihofen, W. A., A. Cicek, F. Pratto, J. C. Alonso, and W. Saenger. 2006. Structures of ω repressors bound to direct and inverted DNA repeats explain modulation of transcription. Nucleic Acids Res. 34:1450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winsor, G. L., T. Van Rossum, R. Lo, B. Khaira, M. D. Whiteside, R. E. W. Hancock, and F. S. L. Brinkman. 2009. Pseudomonas Genome Database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 37:D483-D488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wozniak, D. J. 1994. Integration host factor and sequences downstream of the Pseudomonas aeruginosa algD transcription start site are required for expression. J. Bacteriol. 176:5068-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wozniak, D. J., A. B. Sprinkle, and P. J. Baynham. 2003. Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J. Bacteriol. 185:7297-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zolfaghar, I., D. J. Evans, and S. M. J. Fleiszig. 2003. Twitching motility contributes to the role of pili in corneal infection caused by Pseudomonas aeruginosa. Infect. Immun. 71:5389-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.