Abstract
The interferon consensus sequence binding protein (ICSBP) is an interferon regulatory transcription factor, also referred to as IRF8. ICSBP acts as a suppressor of myeloid leukemia, although few target genes explaining this effect have been identified. In the current studies, we identified the gene encoding growth arrest specific 2 (GAS2) as an ICSBP target gene relevant to leukemia suppression. We find that ICSBP, Tel, and histone deacetylase 3 (HDAC3) bind to a cis element in the GAS2 promoter and repress transcription in myeloid progenitor cells. Gas2 inhibits calpain protease activity, and β-catenin is a calpain substrate in these cells. Consistent with this, ICSBP decreases β-catenin protein and activity in a Gas2- and calpain-dependent manner. Conversely, decreased ICSBP expression increases β-catenin protein and activity by the same mechanism. This is of interest, because decreased ICSBP expression and increased β-catenin activity are associated with poor prognosis and blast crisis in chronic myeloid leukemia (CML). We find that the expression of Bcr/abl (the CML oncoprotein) increases Gas2 expression in an ICSBP-dependent manner. This results in decreased calpain activity and a consequent increase in β-catenin activity in Bcr/abl-positive (Bcr/abl+) cells. Therefore, these studies have identified a Gas2/calpain-dependent mechanism by which ICSBP influences β-catenin activity in myeloid leukemia.
The interferon consensus sequence binding protein (ICSBP) is an interferon regulatory transcription factor (IRF), also known as IRF8 (26). ICSBP is expressed in CD34+ progenitor cells, during myelopoiesis and lymphopoiesis, and in mature phagocytes and B cells (13, 32, 41). ICSBP interacts with a variety of DNA-binding consensus sequences and functions as a repressor or activator of transcription in a context-dependent manner (8, 9, 12, 22, 34). Although the level of ICSBP expression is relatively consistent during myelopoiesis, ICSBP becomes increasingly tyrosine phosphorylated as differentiation proceeds. Since ICSBP regulates various target genes in a tyrosine phosphorylation-dependent manner, cytokine-induced posttranslational modification contributes to the differentiation stage-specific activity of this transcription factor (9, 12, 13, 34).
The first ICSBP target genes identified encoded proteins involved in the innate immune response (8, 9). Consistent with this, mice with engineered disruption of the IRF8 gene exhibited defects in phagocyte and B-cell function (11, 19). However, ICSBP−/− mice also developed a myeloproliferative disorder (MPD) resembling human chronic myeloid leukemia (CML) (11, 19). The MPD in ICSBP−/− mice also progressed to myeloid blast crisis (BC) over time, similar to the course of human CML (11, 19). These studies suggested that ICSBP has myeloid leukemia suppressor functions.
A second murine model specifically implicated ICSBP in the pathogenesis of CML. In this model, mice that were transplanted with bone marrow transduced with a Bcr/abl expression vector developed an MPD which progressed to BC (29). The level of ICSBP expression was decreased in the bone marrow of these mice, and reexpression decreased MPD and delayed BC (10). Decreased ICSBP expression is also found in the bone marrow in human CML (35, 36). ICSBP expression increases during remission, but decreased expression is associated with drug resistance and progression to BC (36).
To identify target genes which contribute to the leukemia suppressor function of ICSBP, we used chromatin coimmunoprecipitation coupled with high-throughput screening. In previous studies, we validated the functional significance of several ICSBP target genes identified in these studies, including genes encoding neurofibromin (NF1), Fas-associated phosphatase 1 (Fap1; the PTPN13 gene), and Fanconi F (the FANCF gene) (12, 34, 41). ICSBP activated NF1 transcription in cytokine-treated myeloid progenitor cells (13, 41). Since Nf1 is a Ras-Gap, these studies identified a mechanism for cytokine hypersensitivity of ICSBP-deficient cells (13, 41). ICSBP repressed PTPN13 transcription in myeloid progenitor cells, and this activity increased during differentiation. Since Fap1 antagonizes Fas-induced apoptosis, this provided a mechanism for Fas resistance in CML (12, 25, 28). We also determined that ICSBP activated FANCF transcription in differentiating progenitors. Since FancF is a DNA repair protein, ICSBP deficiency increased sensitivity to DNA damage during the genotoxic stress of myelopoiesis (34).
The current studies investigate another potential ICSBP target gene identified by screening, the gene encoding growth arrest specific 2 (GAS2). Other investigators first isolated Gas2 while identifying genes expressed in serum-starved NIH 3T3 cells (37). Subsequent clinical studies found increased Gas2 expression in bone marrow samples from subjects with CML (15). Gas2 expression decreases with remission in CML and increases during progression to BC (15). No mutations in the GAS2 gene were found by these investigators, and they did not investigate the functional significance of Gas2 for CML pathogenesis (15). The expression profile of Gas2 in CML is the inverse of that of ICSBP, suggesting that Gas2 may have proleukemia activity. In contrast, other studies found decreased Gas2 in prostate cancer cells, suggesting a possible suppressor role in that disease (18).
Gas2 interacts directly with calpain and inhibits calpain protease activity (2). Previously described calpain substrates include β-catenin (3), suggesting that increased Gas2 expression in CML might increase the stability of the β-catenin protein. Consistent with this hypothesis, increased levels and activity of the β-catenin protein are associated with poor prognosis and BC in CML (14). Increased β-catenin activity in CML is hypothesized to expand the leukemia stem cell (LSC) compartment via transcription of target genes, such as MYC, CCND1, and BIRC5 (encoding c-myc, cyclin D1, and survivin, respectively) (1). However, the increased β-catenin activity in CML is not due to an increase in CTNNB1 transcription or Wnt expression (5, 40).
The hypothesis of the current studies is that decreased ICSBP expression in CML increases Gas2 expression, which inhibits calpain and increases the stability and, therefore, the activity of the β-catenin protein. This hypothesis mechanistically links isolated observations from clinical studies of CML and identifies a pathway of potential interest for the pathogenesis of this disease.
MATERIALS AND METHODS
Plasmids. (i) Protein expression vectors.
The ICSBP cDNA was obtained from Ben Zion-Levi (Technion, Haifa, Israel) (8). The retroviral plasmids MIGR1, MIG P210, and MIG P210 KI (K1176R; a kinase-inactive form) were kindly provided by Rhavi Bhatia (City of Hope National Medical Center, Duarte, CA) (33). The cDNA for Gas2 was obtained from Invitrogen; a dominant-negative (DN) form of Gas2 with a C-terminal truncation (Δ171-314 [DN-Gas2]) (3) and a form of Gas2 that represents the caspase cleavage product [Gas2(1-279)] were generated by PCR. These cDNAs were subcloned into the pcDNA mammalian expression vector.
(ii) shRNA expression vectors.
ICSBP- or β-catenin-specific small hairpin RNA (shRNA) and scrambled control sequences were designed using the Promega shRNA website (Promega Corporation, Madison, WI). Double-stranded oligonucleotides representing the complementary sequences separated by a hairpin loop were subcloned into the pLKO.1puro vector (a gift from Kathy Rundell, Northwestern University, Chicago, IL). Several sequences were tested, and the most efficient used. Plasmids with three different human or murine shRNAs specific to Gas2 (and control scrambled sequences) in the pRS retroviral vector were obtained from Origene (Origene USA, Rockville, MD). These sequences were validated by the manufacturer.
(iii) Reporter constructs.
Sequences from the 5′ flank of the GAS2 gene were obtained by PCR from U937 genomic DNA. Fragments were sequenced on both strands to verify identity with reported sequences. Constructs were generated with the pGL3-basic vector (Promega) using 1.0 kb, 250 bp, 130 bp, and 100 bp of the 5′ flank sequence. Other reporter constructs were generated with an artificial promoter/reporter vector with three copies of an ICSBP-binding cis element from the GAS2 promoter (bp −75 to −100) using the pGL3 promoter vector (Promega).
The TOPflash and FOPflash reporter constructs were purchased from Millipore (Billerica, MA). TOPflash contains three copies of a consensus binding site for Tcf-Lef linked to a minimal promoter and a luciferase reporter. FOPflash is a similar construct, but with a mutation which abolishes Tcf-Lef binding.
Oligonucleotides.
The oligonucleotide primers for PCR were custom synthesized by MWG Biotech (Piedmont, NC). The primers are as follows: human ICSBP (5′-CCAGGACTGATTTGGGAGAA-3′), murine ICSBP (5′-CAGTGGCTGATCGAACAGAT-3′ and 5′-CTCCTGATTGTAATCCTGCTT-3′), human β-catenin (5′-AAAATGGCAGTGCGTTTAG-3′ and 5′-TTTGAAGGCAGTCTGTCGTA-3′), murine β-catenin (5′-ACTGGCCTCTGATAAAGGCAACT-3′ and 5′-TAGTCGTGGAATAGCACCCTGTT-3′), human c-myc (5′-CTATGACCTCGACTACGACTCCGT-3′ and 5′-GCTCTGCTGTTGCTGGTGATAG-3′), murine c-myc (5′-AGAGCTCCTCGAGCTGTTTGAAGG-3′ and 5′-ACGGAGTCGTAGTCGAGGTCATAG-3′), human cyclin D1 (5′-TGGCCTCTAAGATGAAGGAGA-3′ and 5′-AGGAAGTGTTCGATGAAATCGT-3′), murine cyclin D1 (5′-TCACACAGGAGGCTTTTAAACACT-3′ and 5′-TGGTCATGGGCAGCCTTTC-3′), human survivin (5′-CCACCGCATCTCTACATTCA-3′ and 5′-CAAGTCTGGCTCGTTCTCAGT-3′), murine survivin (5′-CTGATTTGGCCCAGTGTTTT-3′ and 5′-CAGGGGAGTGCTTTCTATGC-3′), and p210 Bcr/abl (5′-CGTCCACTCAGCCACTGGAT-3′ and 5′-AGTTCCAACGAGCGGCTTC-3′).
Myeloid cell line culture.
The human leukemia cell line U937 (23) was obtained from Andrew Kraft (Hollings Cancer Center, Medical University of South Carolina, Charleston, SC). Cells were maintained as described previously (8, 9, 12). U937 cells were treated for 48 h with 500 U per ml human recombinant gamma interferon (IFN-γ) for differentiation (Roche, Indianapolis, IN).
Murine bone marrow culture.
Animal studies were performed according to a protocol approved by the Animal Care and Use Committees of Northwestern University and Jesse Brown VA Medical Center. C57/BL6 mice heterozygous for ICSBP knockout were obtained from Keiko Ozato (National Institutes of Health, Bethesda, MD). The mice have been previously described (11, 34).
Bone marrow mononuclear cells were obtained from the femurs of wild-type (WT) or ICSBP−/− C57/BL6 mice. Sca1+ cells were separated using the Miltenyi magnetic bead system (Miltenyi Biotechnology, Auburn, CA). Bipotential myeloid progenitor cells (granulocyte monocyte progenitors) were cultured (at 2 × 105 cells per ml) in Dulbecco's modified Eagle's (DME) medium supplemented with 10% fetal calf serum, 1% penicillin-streptomycin, 10 ng/ml murine granulocyte-macrophage colony-stimulating factor (GM-CSF; R&D Systems, Inc., Minneapolis, MN), 100 ng/ml stem cell factor (SCF; R&D Systems, Inc.), and 10 ng/ml murine recombinant interleukin-3 (IL-3; R&D Systems, Inc.). Cells were maintained in GM-CSF-SCF-IL-3 for 48 h or differentiated over 48 h in 10 ng/ml of G-CSF or M-CSF, as described previously (12, 13). Some WT or ICSBP−/− myeloid progenitors were transduced with a retroviral vector to express a β-catenin-specific shRNA or scrambled control shRNA, Gas2-specific shRNA or scrambled controls, p210 Bcr/abl or vector control, or dominant-negative Gas2 or vector control, as described previously (13). Some cells were treated with imatinib (IM; 1 μM) for 24 h.
Chromatin coimmunoprecipitation.
U937 cells were cultured with or without IFN-γ for 48 h (41). Cells were incubated briefly in medium supplemented with formaldehyde, and lysates were sonicated to generate chromatin fragments with an average size of 2.0 kb (27). Lysates were immunoprecipitated with ICSBP antiserum or preimmune serum (13). Antibody to ICSBP was a gift of Stephanie Vogel (University of Maryland, Baltimore, MD). Chromatin underwent two rounds of immunoprecipitation. Chromatin which coimmunoprecipitated with ICSBP was ligated into a plasmid vector and subjected to sequence analysis, as described previously (27, 41). Sequences representing potential ICSBP target genes were identified by GenBank analysis.
Specific coprecipitation of the GAS2 promoter was determined by PCR analysis of ICSBP-coprecipitating chromatin from an independent experiment. For these experiments, total input chromatin (not precipitated) was a positive control and chromatin precipitated by preimmune serum was a negative control. PCR products were analyzed by acrylamide gel electrophoresis.
In other studies, U937 cells were subjected to chromatin coimmunoprecipitation with an antibody to ICSBP (Santa Cruz Biotech, Santa Cruz CA), antibody to trimethylated K27 histone 3 (triMe-K27 H3; Cell Signaling Technology, Danvers MA), or irrelevant control antibody (anti-glutathione S-transferase [anti-GST] antibody; Santa Cruz). Immunoprecipitated chromatin was amplified by quantitative real-time PCR (using input chromatin to normalize for abundance). Primers were designed to flank the sequence from bp −75 to −150 in the GAS2 promoter.
Quantitative real-time PCR.
RNA was isolated using Trizol reagent (Gibco-BRL, Gaithersburg, MD) and tested for integrity by gel electrophoresis. Primers were designed with Applied Biosystems software, and real-time PCR was performed using SYBR green according to the “standard curve” method. The results were normalized to those for 18S RNA.
Myeloid cell line transfections. (i) Stable transfectant cell lines.
U937 cells were transfected by electroporation with equal amounts of an ICSBP expression vector, p210 Bcr/abl expression vector, or empty vector control (ICSBP/pcDNAamp, Bcr/abl/pcDNAamp, or pcDNAamp) plus a vector with a neomycin phosphotransferase cassette (pSRα) (30 μg each). Stable pools of cells were selected in G418 (0.5 mg/ml). Other cells were transfected by electroporation with a construct to express an ICSBP-specific shRNA or scrambled control shRNA using the pLKO.1puro vector. Stable pools of transfected cells were selected in puromycin (1.2 μg/ml). At least three independent pools of stable transfectants were selected and analyzed.
(ii) Transient transfections for GAS2 promoter activity.
U937 cells (32 × 106/ml) were transfected with various combinations of vectors to express ICSBP or control (50 μg), ICSBP-specific shRNA or scrambled control shRNA (50 μg), p210 Bcr/abl or control (50 μg), β-catenin-specific shRNA or scrambled control (50 μg), or Gas2, DN-Gas2, or shGas2 or the relevant controls (50 μg). Cells were cotransfected with a luciferase reporter construct with 1.0 kb, 250 bp, 130 bp, or 100 bp of the proximal GAS2 5′ flank (20 μg, using the pGL3 vector from Promega) and a cytomegalovirus (CMV)/β-gal reporter (10 μg, for transfection efficiency). The empty reporter vector was the control in these experiments. Luciferase assays were performed according to the manufacturer's instructions (Promega), and β-galactosidase (β-Gal) studies were performed as previously described (8, 9, 12, 13, 41). Some transfectants were treated with IFN-γ or trichostatin A as described previously (12).
In other experiments, U937 cells were transfected with vectors to express various combinations of ICSBP and Tel or histone deacetylase 3 (HDAC3) or shTel or shHDAC3 (or the relevant control vectors). Cells were cotransfected with an artificial promoter vector with three copies of the bp −75 to −100 GAS2 promoter and a firefly luciferase reporter (GAS2-sv40-GL3) and a renilla luciferase promoter (2 μg, to control for transfection efficiency). The empty reporter vector was used as a control in these experiments. Reporter assays were performed using the dual luciferase reporter system according to the manufacturer's instructions (Promega).
(iii) Transient transfections for β-catenin activity.
U937 cells (32 × 106/ml) were transfected with various combinations of vectors to express ICSBP or control (50 μg), ICSBP-specific shRNA or scrambled control (50 μg), p210 Bcr/abl or control (50 μg), β-catenin-specific shRNA or scrambled control (50 μg), or Gas2, DN-Gas2, or shGas2 (or relevant controls) (50 μg). Cells were cotransfected with TOPflash reporter vector or FOPflash control (70 μg) and a CMV/β-Gal reporter (for transfection efficiency). Transfectants were assayed for luciferase expression using Promega's luciferase assay system according to the manufacturer's instructions (Promega, Madison, WI). Assays for β-galactosidase expression were performed as described previously (8, 9).
Western blot assays and immunoprecipitation.
U937 or murine bone marrow cells were lysed by boiling in 2× SDS sample buffer. Lysate proteins (50 μg) were separated by SDS-PAGE and transferred to nitrocellulose. Western blots were serially probed with combinations of antibodies to ICSBP (Santa Cruz Biotechnology, Santa Cruz, CA), Gas2 (Abcam, Cambridge, MA), calpain (small subunit), β-catenin, Bcr, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or tubulin (to control for loading). Each experiment was repeated several times with different batches of proteins, and representative blots are shown.
Cell lysate proteins were immunoprecipitated under nondenaturing conditions with an antibody to calpain (small subunit) or irrelevant control antibody (to glutathione S-transferase). Immunoprecipitates were collected with Staphylococcus protein A-agarose beads and separated by SDS-PAGE, and Western blots were serially probed with antibodies to Gas2, calpastatin, and calpain. The assays were repeated twice, and representative blots are shown.
In other studies, U937 cell lysates were immunoprecipitated with antibody to Gas2, calpastatin, or irrelevant control under denaturing conditions. Immunoprecipitates were collected with Staphylococcus protein A-agarose, and proteins renatured by serial washes in radioimmunoprecipitation (RIPA) buffer. The immunoprecipitates were analyzed by Western blot and calpain assays.
To verify overexpression of DN-Gas2, in vitro-transcribed RNA and [35S]methionine-labeled in vitro-translated protein were generated in reticulocyte lysate representing Gas2, DN-Gas2, and Gas2(1-279) using previously described techniques (8, 9). These proteins and lysates from U937 cells that were stably transfected with a DN-Gas2 expression vector or empty control vector were separated by SDS-PAGE on a 12% acrylamide gel. The lanes with in vitro-translated proteins were fixed, and proteins identified by autoradiography. The lanes with U937 cell lysates were analyzed by probing Western blots with a Gas2 antibody. Molecular weight markers were used to align the autoradiograph and the Western blot for comparison.
Calpain assays.
Calpain activity assays were performed using a commercially available kit (Biovision, Mountain View, CA). In most of the studies, cell lysates were assayed using the fluorescence-labeled substrate provided in the kit and calpain activity determined by the fluorometric change. For some of the studies, recombinant calpain (Biovision) was incubated with Gas2 or calpastatin which had been immunoprecipitated from U937 lysates and assayed as described above. In other assays, calpain was immunoprecipitated from cell lysates and incubated under calpain assay conditions with in vitro-translated, 35S-labeled β-catenin. β-Catenin degradation was determined by SDS-PAGE and autoradiography. In vitro transcription and translation were performed with rabbit reticulocyte lysate using commercially available kits (Promega, Madison, WI) (8, 9).
Immunofluorescence confocal microscopy.
Cultured cells were cytospun onto microscope slides (∼0.5 × 105 cells), fixed in 3.7% formaldehyde, permeabilized with methanol, and blocked with goat serum. Cells were treated with rabbit anti-β-catenin antibody and labeled with a fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG. Cells were mounted by adding ProLong Gold antifade reagent with 4′,6′-diamidino-2-phenylindole (DAPI). Stained cells were visualized using a Zeiss LSM 510 UV laser scanning confocal microscope (in the core imaging facility at Northwestern University).
Statistical analysis.
Statistical significance was determined by Student's t test and analysis of variance (ANOVA) using SigmaPlot and SigmaStat software.
RESULTS
Identification of GAS2 as an ICSBP target gene.
To identify ICSBP target genes, we used chromatin coimmunoprecipitation as reported in previous studies (12, 34, 41). ICSBP-coprecipitating chromatin fragments were subcloned into a plasmid vector and individually sequenced. We used the U937 myeloid leukemia cell line in these studies (23). These cells are arrested at the granulocyte-monocyte progenitor (GMP) stage but can be differentiated with various agents, including IFN-γ and retinoic acid (23). In studies with differentiated U937 cells, we identified genes encoding various inflammatory mediators (Nf1, Fap1, and FancF) as previously reported (12, 34, 41). We also identified potential target genes in experiments with undifferentiated U937 cells which we have not previously further characterized (12, 34). One of these genes was GAS2, the gene encoding growth specific arrest 2. This was of interest because increased Gas2 expression is found in CML.
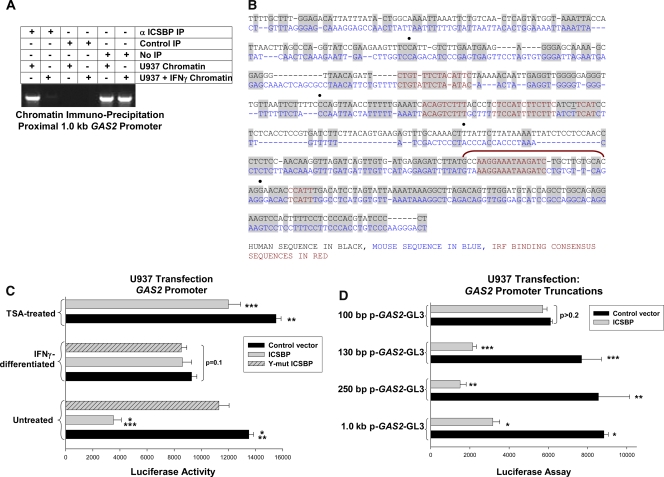
We performed independent chromatin coimmunoprecipitation studies with U937 cells to verify interaction between ICSBP and GAS2 and to determine the influence of differentiation on this interaction. ICSBP-coprecipitating chromatin was PCR amplified using GAS2-specific primers and analyzed by electrophoresis (with preimmune serum as a negative control). Based on the location of the ICSBP-coprecipitating fragment, primers flanking kb −2.0 to −1.0 and the kb −1.0 to bp +50 of the GAS2 gene were used. Primers to the first exon of GAS2 were a negative control and nonprecipitated chromatin a positive control for these studies. We found that only the proximal 1.0 kb of the GAS2 promoter coimmunoprecipitated with ICSBP from U937 cell lysates and that this interaction was decreased by IFN-γ-induced differentiation (Fig. 1A).
FIG. 1.
Identification of GAS2 as a potential ICSBP target gene. (A) ICSBP binds the GAS2 5′ flank in vivo. Chromatin that coimmunoprecipitated (IP) from U937 cells with an antibody to ICSBP was amplified with primers flanking the proximal 1.0 kb of the GAS2 5′ flank. Nonprecipitated input chromatin was a positive control, and chromatin that coprecipitated with preimmune serum was a negative control for these studies. (B) The GAS2 5′ flank includes DNA-binding consensus sequences for IRF protein binding. The proximal 500 bp of the GAS2 5′ flank was analyzed for ICSBP-binding consensus sequences. The human sequence is shown in black, and the murine sequence in blue. Conserved, IRF-binding consensus sequences are indicated in red. The ICSBP-binding cis element is identified by a red bracket. (C) ICSBP represses GAS2 transcription in undifferentiated myeloid cells. U937 cells were cotransfected with a GAS2 promoter/reporter vector (1.0 kb of 5′ flank) and a vector to overexpress ICSBP or the vector control. Because ICSBP tyrosine phosphorylation influences the activation or repression of other target genes, some cells were cotransfected with a vector to overexpress ICSBP with all tyrosine residues mutated to phenylalanine (Y-mut ICSBP). The transfectants were assayed for reporter activity with or without IFN-γ-induced differentiation or with or without trichostatin A (TSA) treatment. *, statistically significant decrease in reporter expression in ICSBP-overexpressing transfectants in comparison to the level of expression in the control (P < 0.001, n = 5); ** or ***, statistically significant increase in GAS2 promoter activity in TSA-treated transfectants without or with ICSBP overexpression, respectively (P < 0.001, n = 5). The lack of a statistically significant difference in reporter activity in IFN-γ-differentiated transfectants is indicated by the bracket (P = 0.2, n = 3). (D) ICSBP represses GAS2 transcription via a cis element between bp −110 and −130 in the GAS2 promoter. U937 cells were cotransfected with a series of reporter constructs with truncations of the GAS2 5′ flank, as indicated, and a vector to overexpress ICSBP or empty vector control. *, **, or ***, statistically significant decrease in activity of the 1.0-kb, 250-bp, or 130-bp reporter constructs, respectively, with ICSBP overexpression (P < 0.01, n = 4). The lack of a statistically significant difference in the activity of the 100-bp GAS2 promoter construct with versus without ICSBP overexpression is indicated by the bracket (P > 0.2, n = 4). Error bars indicate standard errors.
Sequences of the human and murine GAS2 5′ flanks were compared, and potential IRF-binding consensus sequences identified. ICSBP binds to a number of DNA sequences, including ets/IRF consensus elements (EICE), IRF/ets consensus elements (IECE), interferon-stimulated response elements (ISRE), and positive regulatory domain I (PRDI) sequences. We identified a number of potential ICSBP-binding sites within the proximal 350 bp of the GAS2 5′ flank which were conserved in the human and murine genes (Fig. 1B).
ICSBP represses GAS2 transcription in undifferentiated myeloid cells.
We investigated the influence of ICSBP on GAS2 promoter activity by cotransfecting U937 cells with a reporter construct containing 1.0 kb of the GAS2 5′ flank and a vector to overexpress either ICSBP or a form of ICSBP with all of the tyrosine residues mutated to phenylalanine (Y-mut ICSBP). The overexpression of WT ICSBP but not of Y-mut ICSBP significantly decreased GAS2 promoter activity in untreated U937 transfectants (Fig. 1C). In contrast, neither form of ICSBP influenced GAS2 promoter activity in IFN-γ-differentiated transfectants. ICSBP is somewhat tyrosine phosphorylated in undifferentiated U937 cells, and the amount of tyrosine phosphorylation increases with differentiation (19). Therefore, these results suggested that some tyrosine phosphorylation of ICSBP was necessary for GAS2 repression, but increasing tyrosine phosphorylation in differentiating cells did not augment ICSBP repression activity. In control experiments, Y-mut ICSBP and WT ICSBP were equivalently overexpressed in U937 cells (not shown) (19). In previous studies, both proteins repressed an artificial promoter construct containing the PRDI consensus sequence, indicating that Y-mut ICSBP is functionally competent (19).
ICSBP represses some target genes by recruiting histone deacetylase 3 (HDAC3), a transcriptional corepressor (22). To determine the role of HDAC activity in GAS2 repression, U937 transfectants were treated with the histone deacetylase inhibitor trichostatin A (TSA). TSA increased the GAS2 promoter activity in control U937 transfectants and abolished ICSBP-induced GAS2 repression (Fig. 1C). This was investigated further as described below.
We performed additional studies to identify the ICSBP-binding site in the GAS2 promoter. Based on the locations of IRF consensus sequences, reporter constructs were generated with 250 bp, 130 bp, or 100 bp of the GAS2 5′ flank. U937 cells were cotransfected with these GAS2 constructs and an ICSBP expression vector (or empty vector control). We found that ICSBP overexpression significantly decreased the activity of the 250-bp and 130-bp GAS2 promoter constructs but not that of the 100-bp construct (Fig. 1D). In these studies, the empty reporter vector had minimal activity which was not altered by ICSBP, IFN-γ, or TSA. This activity was subtracted as background.
ICSBP binds to a negative cis element in the GAS2 promoter.
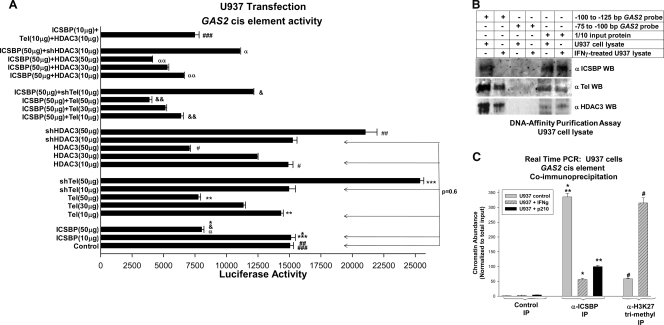
These studies identified a potential ICSBP-binding site between bp −100 and −130 in the GAS2 promoter. This region includes an EICE-like sequence. Therefore, we generated an artificial promoter construct with three copies of this 30-bp sequence linked to a minimal promoter and a reporter gene (GAS2-sv40-GL3). U937 cells were transfected with this construct (or control sv40-GL3) and an ICSBP expression vector (or vector control). We found that ICSBP repressed this EICE sequence in undifferentiated transfectants (Fig. 2 A).
FIG. 2.
ICSBP cooperates with Tel and HDAC3 to repress a GAS2 cis element found between bp −100 and −130 in the promoter. (A) ICSBP repression of the GAS2 cis element is increased by Tel or HDAC3. U937 cells were transfected with an artificial reporter construct with three copies of the GAS2 cis element linked to a minimal promoter and reporter (GAS2-sv40-GL3) or the empty vector control (sv40-GL3) and a vector to express ICSBP, a dose titration of HDAC3 or Tel, ICSBP with a dose titration of HDAC3 or Tel, an shRNA to HDAC3 or Tel, ICSBP with an shRNA to HDAC3 or Tel, or ICSBP with Tel and HDAC3, and reporter gene assays were performed. *, **, or #, statistically significant decrease in reporter activity in ICSBP-, Tel-, or HDAC3-overexpressing cells, respectively, in comparison to control cells (P < 0.01, n = 3); *** or ##, statistically significant difference in reporter expression in cells expressing shTel or shHDAC3, respectively (P < 0.01, n = 3); ###, statistically significant difference with ICSBP with Tel and HDAC3; & or &&, statistically significant difference in reporter expression in cells overexpressing ICSBP with shTel or Tel overexpression (P < 0.01, n = 3); α or αα, statistically significant difference in reporter expression in ICSBP-overexpressing cells with shHDAC3 or HDAC3 overexpression, respectively (P < 0.01, n = 3). (B) ICSBP binds to the bp −100 to −125 GAS2 promoter sequence in vitro. Nuclear proteins were isolated from U937 cells with or without IFN-γ-induced differentiation. Proteins were purified by affinity to a biotin-labeled, double-stranded oligonucleotide probe with the bp −102 to −127 or the bp −75 to −102 sequence from the GAS2 5′ flank. Affinity-purified proteins were analyzed by probing Western blots (WB) with an ICSBP antibody. Lanes with 1/10 the amount of affinity-purified protein were used as a loading control for ICSBP abundance. (C) In vivo ICSBP binding to the bp −100 to −130 GAS2 promoter sequence is decreased by differentiation or Bcr/abl expression. Chromatin coimmunoprecipitation (IP) was performed using U937 cells with or without IFN-γ (IFNg)-induced differentiation. Chromatin was coimmunoprecipitated with antibody to ICSBP, trimethylated lysine 27 histone 3 (H3 K27 tri-methyl), or irrelevant control antibody. Other U937 cells were stably transfected with a vector to express Bcr/abl, and chromatin was coimmunoprecipitated with anti-ICSBP or irrelevant control antibody. Precipitated chromatin was amplified by real-time PCR with primers flanking the bp −100 to −130 GAS2 cis element. The results were normalized to those for input (nonprecipitated) chromatin. * or **, statistically significant decrease in ICSBP binding to the GAS2 cis element in IFN-γ-differentiated or Bcr/abl+ cells, respectively (P < 0.001, n = 3); #, statistically significant increase in binding of H3 trimethyl K27 to the bp −100 to −130 GAS2 cis element in differentiated U937 cells (P < 0.001, n = 3). Error bars indicate standard errors.
ICSBP interacts with HDAC3 and Tel to repress an IECE in the 3′5′OAS gene (22). Because the GAS2 cis element was similar to this sequence, we investigated the involvement of Tel and HDAC3 in GAS2 repression. U937 cells were initially transfected with the GAS2-sv40-GL3 vector (or control) and a dose titration of vectors to express Tel or HDAC3 or with a Tel- or HDAC3-specific shRNA. The overexpression of either Tel or HDAC3 decreased the GAS2 cis element activity in U937 transfectants and knockdown increased this activity, in a dose-dependent manner (Fig. 2A). We next cotransfected U937 cells with the GAS2 cis element-containing reporter vector, a vector to overexpress ICSBP, and a vector to overexpress Tel or HDAC3. The overexpression of Tel or HDAC3 increased ICSBP-induced GAS2 repression in a dose-dependent manner (Fig. 2A). Conversely, the expression of a Tel or HDAC3 shRNA impaired the repression of the GAS2 cis element by ICSBP (Fig. 2A). We also cotransfected U937 cells with the GAS2-sv40-GL3 reporter and vectors to overexpress ICSBP, Tel, and HDAC3 at levels where none of them influenced GAS2 cis element activity independently (10 μg of each). We found that a combination of the three overexpressed proteins repressed the GAS2 cis element, although none did alone at this level, suggesting cooperation.
In control experiments, the Tel and HDAC3 shRNA vectors decreased Tel and HDAC3 expression, respectively, but did not influence the expression of each other or ICSBP (not shown). None of the proteins or shRNAs influenced reporter expression from the control sv40-GL3, which was subtracted as background.
We used an in vitro DNA-binding assay to further investigate interaction between ICSBP, Tel, HDAC3, and the GAS2 promoter. For these studies, nuclear proteins were isolated from either untreated or IFN-γ-differentiated U937 cells. Proteins were incubated with double-stranded, biotin-labeled, oligonucleotide probes representing bp −127 to −102 or bp −102 to −75 in the GAS2 promoter. Protein-DNA complexes were purified by affinity to streptavidin and separated by SDS-PAGE, and Western blots were sequentially probed with antibodies to ICSBP, Tel, and HDAC3. All three proteins bound the bp −127 to −102 GAS2 probe in experiments with nuclear proteins from U937 cells, but differentiation abolished ICSBP binding and greatly decreased Tel and HDAC3 binding (Fig. 2B). The input nuclear proteins were a loading control, and each study was repeated with two different batches of nuclear proteins (representative blots are shown).
We also investigated in vivo ICSBP binding to this GAS2 cis element by quantitative chromatin immunoprecipitation. Coprecipitating chromatin was amplified by real-time PCR using primers flanking the bp −100 to −130 GAS2 sequence. For these experiments, the sonication conditions were adjusted to generate chromatin fragments of <200 bp. Consistent with the results of the in vitro studies, in vivo interaction of ICSBP with this sequence decreased during differentiation (Fig. 2C). We similarly investigated in vivo ICSBP binding to the GAS2 cis element in U937 cells that were stably expressing Bcr/abl. Significantly less ICSBP bound the GAS2 promoter in Bcr/abl+ cells, consistent with decreased ICSBP expression in these cells (Fig. 2C). The influence of ICSBP on Gas2 expression in Bcr/abl+ cells is discussed below. Bcr/abl expression and activation (phosphorylation) in the transfectants were verified by Western blot analysis (not shown).
In studies with prostate cancer cells, decreased Gas2 expression correlated with the binding of trimethylated lysine 27 histone 3 (triMe-K27 H3) to the GAS2 promoter (18). Therefore, we used chromatin coimmunoprecipitation to investigate whether ICSBP binding to GAS2 is associated with a local increase in triMe-K27 H3 binding. We found little in vivo binding of triMe-K27 H3 to this sequence in undifferentiated U937 cells, but its binding increased significantly after IFN-γ-induced differentiation (Fig. 2C). These results were the reciprocal of the ICSBP binding and suggested that an ICSBP-independent mechanism resulted in trimethylation of local H3 binding to this region of the GAS2 promoter in differentiating cells.
ICSBP regulates Gas2 expression in myeloid progenitor cells.
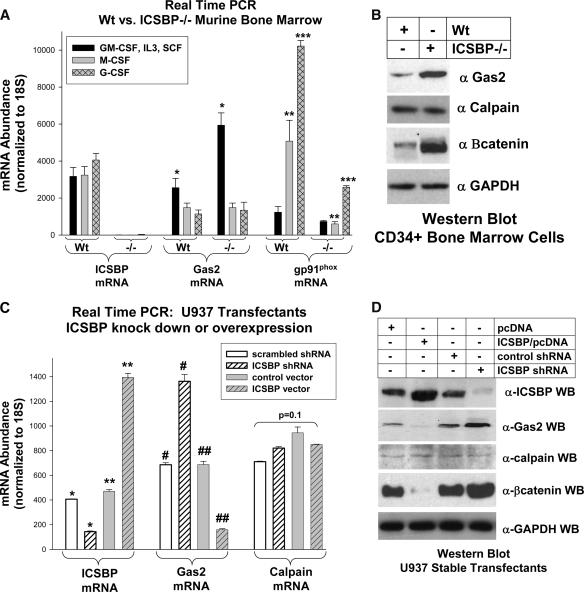
We next investigated whether the expression of endogenous Gas2 is ICSBP dependent in myeloid progenitor cells. In our initial studies, myeloid progenitor cells were isolated from the bone marrow of ICSBP−/− or WT mice and cultured in GM-CSF, IL-3, and SCF (see references 13 and 19). Both WT and ICSBP−/− cells cultured under these conditions represent granulocyte/monocyte progenitors (GMP) as determined by surface marker expression (Sca1− CD34+ Mac1low Gr1low) (19, 34). Some cells were differentiated to granulocytes with G-CSF or to monocytes with M-CSF (13, 19). Gas2 mRNA abundance in these cells was determined by real-time PCR. We found significantly more Gas2 mRNA in ICSBP−/− cells cultured in GM-CSF, IL-3, and SCF in than in WT cells (Fig. 3 A). However, Gas2 mRNA expression was not significantly different in WT than in ICSBP−/− cells after differentiation, suggesting that ICSBP only regulates Gas2 in myeloid progenitors. ICSBP mRNA was a negative control and gp91PHOX a positive control for ICSBP-dependent gene expression (8). The correlation between Gas2 mRNA and protein expression in WT and ICSBP−/− myeloid progenitors was determined by Western blot analysis (Fig. 3B).
FIG. 3.
ICSBP influences the expression of Gas2 and β-catenin in myeloid progenitor cells. (A) Gas2 mRNA expression is greater in myeloid progenitor cells from ICSBP−/− mice than in WT cells. Bone marrow cells were obtained from WT or ICSBP−/− mice, and myeloid progenitor cells cultured in GM-CSF, IL-3, and SCF. Some cells were differentiated ex vivo with M-CSF or G-CSF. The level of expression of Gas2 mRNA was determined by real-time PCR. ICSBP mRNA is a negative control and gp91PHOX mRNA a control for an ICSBP-dependent message. The results were normalized to those for 18S RNA. *, statistically significant increase in Gas2 mRNA in myeloid progenitors from ICSBP−/− versus WT cells (P < 0.001, n = 6); ** or ***, statistically significant difference in gp91PHOX mRNA expression in ex vivo-differentiated ICSBP−/− versus WT cells with M-CSF and G-CSF treatment, respectively (P < 0.005, n = 4). (B) The abundance of Gas2 and β-catenin protein is greater in primary myeloid progenitor cells from ICSBP−/− mice than in WT cells. Bone marrow was obtained from WT or ICSBP−/− mice and myeloid progenitor cells cultured in GM-CSF, IL-3, and SCF. Cell lysate proteins from CD34+ cells were analyzed by Western blot assay for Gas2 or GAPDH (to control for loading). Since Gas2 is a calpain inhibitor, the effect of ICSBP deficiency on calpain expression was determined (as a control for calpain activity studies). Because β-catenin is a calpain substrate, the effect of ICSBP knockout on β-catenin (Bcatenin) protein was also determined. (C) ICSBP overexpression decreases Gas2 mRNA and ICSBP knockdown increases Gas2 mRNA in U937 myeloid cells. U937 cells were stably transfected with a vector to overexpress ICSBP or with empty vector control or a vector to express an ICSBP-specific shRNA or a scrambled shRNA control. RNA was analyzed for Gas2 or ICSBP mRNA by real-time PCR. The results were normalized to those for 18S RNA. *, statistically significant decrease in ICSBP expression in cells expressing ICSBP-specific shRNA in comparison to its expression in the control (P < 0.001, n = 3); **, statistically significant increase in ICSBP expression in cells transfected with an ICSBP expression vector in comparison to its expression in the control (P < 0.0001, n = 3); #, statistically significant increase in Gas2 expression in cells with ICSBP knockdown (P = 0.003, n = 6); ##, decrease in Gas2 expression in ICSBP-overexpressing cells (P < 0.0001, n = 6). Since Gas2 is a calpain inhibitor, the effect of ICSBP overexpression or knockdown on calpain mRNA was determined (as a control for later experiments). The lack of a statistically significant change in calpain mRNA under these conditions is indicated by a bracket (P = 0.1, n = 3). (D) ICSBP overexpression decreases the abundance of Gas2 and β-catenin protein, and ICSBP knockdown increases Gas2 and β-catenin protein in U937 myeloid cells. These stable U937 transfectants were also studied by Western blot analysis (WB) for the expression of Gas2. Because Gas2 is a calpain inhibitor, the effect of ICSBP abundance on the amount of calpain protein was determined. Because β-catenin is a substrate for calpain, the level of expression of β-catenin was determined. GAPDH was a loading control in these studies. Error bars indicate standard errors.
The hypothesis of these studies is that ICSBP influences calpain activity by regulating Gas2. To exclude the possibility that ICSBP influences calpain expression directly, blots were also probed for the small subunit of calpain. Since the various large subunits of calpain all interact with the small subunit, this provided a measure of the total calpain. We found no difference in calpain expression (Fig. 3B) in WT and ICSBP−/− myeloid progenitors. The amount of β-catenin protein was increased in ICSBP−/− myeloid progenitor cells in comparison to its level in the WT, consistent with our hypothesis (Fig. 3B). CD34+ cells were isolated for these experiments to ensure that differences in Gas2 or β-catenin expression were not due to the stage of differentiation.
Since ICSBP is constitutively absent in ICSBP−/− mice, compensation for ICSBP deficiency may occur during development. To investigate the effect of direct manipulation of ICSBP on Gas2 expression, stable U937 transfectants were generated with a vector to express an ICSBP-specific shRNA (or scrambled control) or a vector to overexpress ICSBP (or control vector). ICSBP knockdown in U937 cells increased Gas2 mRNA and protein, increased β-catenin protein, and did not alter total calpain (Fig. 3C and D). Conversely, ICSBP overexpression decreased Gas2 mRNA and protein, decreased β-catenin protein, and did not alter total calpain (Fig. 3C and D). Functional connections between ICSBP, Gas2, calpain, and β-catenin are explored below.
Bcr/abl regulates Gas2 expression in myeloid progenitor cells in an ICSBP-dependent manner.
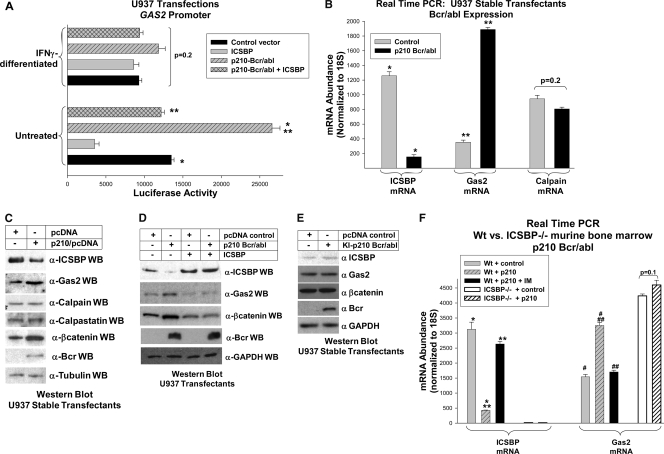
ICSBP is decreased and Gas2 and β-catenin are increased in CML (10, 35, 36). To determine if the Bcr/abl-induced decrease in ICSBP expression increases GAS2 promoter activity, additional transfection experiments were performed. U937 cells were cotransfected with the GAS2 promoter/reporter construct and a vector to express Bcr/abl, with or without an ICSBP expression vector. Bcr/abl significantly increased the activity of the GAS2 promoter in undifferentiated U937 cells but had no effect in differentiated transfectants (Fig. 4A). Consistent with our hypothesis, Bcr/abl-induced GAS2 promoter activity was reversed by reexpression of ICSBP (Fig. 4A).
FIG. 4.
Bcr/abl influences the expression of ICSBP, Gas2, and β-catenin in myeloid progenitor cells. (A) Bcr/abl increases GAS2 transcription in an ICSBP-dependent manner in undifferentiated myeloid cells. U937 cells were cotransfected with a GAS2 promoter/reporter vector (1.0 kb of the 5′ flank), a vector to express Bcr/abl or the vector control, and a vector to overexpress ICSBP or the vector control. Transfectants were assayed for reporter activity with or without IFN-γ-induced differentiation. *, statistically significant difference in reporter activity with Bcr/abl expression (P < 0.01, n = 4); **, statistically significant difference in reporter expression in Bcr/abl+ transfectants with versus without ICSBP (P < 0.01, n = 4). The lack of a statistically significant difference in reporter activity in IFN-γ-differentiated transfectants is indicated by the bracket. (B) Bcr/abl decreases ICSBP mRNA and increases Gas2 mRNA in U937 myeloid leukemia cells. Stable U937 transfectants were generated with a vector to express p210 Bcr/abl or with empty vector control. RNA was analyzed by real-time PCR for ICSBP and Gas2 expression. Since Gas2 is a calpain inhibitor, the level of expression of calpain was determined. The results were normalized to those for 18S RNA. *, statistically significant decrease in ICSBP expression in Bcr/abl+ cells; **, statistically significant increase in Gas2 mRNA (P < 0.0001, n = 3). The lack of a statistically significant change in calpain mRNA expression with or without Bcr/abl is indicated by the bracket (P = 0.2, n = 3). (C) Bcr/abl decreases ICSBP protein and increases Gas2 and β-catenin protein in U937 myeloid leukemia cells. Cell lysates from the U937 transfectants described above were analyzed by Western blot assay for the expression of ICSBP, Gas2, and tubulin (as a loading control). Since Gas2 inhibits calpain and β-catenin is a calpain substrate, the blots were also probed with antibodies to these proteins. Since calpastatin inhibits calpain in some cells, the blots were probed with an antibody to this protein. Anti-Bcr antibody was used as a control for Bcr/abl expression in the stable transfectants. (D) Increased Gas2 and β-catenin expression in Bcr/abl+ U937 cells is reversed by ICSBP overexpression. U937 stable transfectants were generated with a vector to express Bcr/abl or the empty vector control or with a vector to overexpress ICSBP or the empty vector control. Cell lysates were analyzed by Western blot assay for ICSBP, Gas2, β-catenin, or GAPDH (to control for loading). Blots were also probed with an antibody to Bcr as a control for Bcr/abl expression. (E) Kinase-inactive Bcr/abl does not alter the expression of ICSBP, Gas2, or β-catenin protein in U937 cells. Stable U937 transfectants were generated with a vector to express a kinase-inactive mutant of p210 Bcr/abl (KI-Bcr/abl) or the empty vector control. Cell lysate proteins were analyzed by Western blot assay for the expression of ICSBP, Gas2, β-catenin, Bcr, and GAPDH (loading control) as described for the p210 Bcr/abl-expressing cells. (F) Bcr/abl increases Gas2 mRNA in WT myeloid progenitors, but not in myeloid progenitors from ICSBP−/− mice. Myeloid progenitor cells were isolated from the bone marrow of WT or ICSBP−/− mice and cultured in GM-CSF, IL-3 and SCF. Cells were transduced with a retroviral vector to express p210 Bcr/abl or empty vector control. Some cells were treated with the imatinib (IM). RNA from the transduced cells was analyzed for ICSBP and Gas2 expression by real-time PCR. Results were normalized to those for 18S RNA. *, statistically significant decrease in ICSBP expression in Bcr/abl+ WT progenitor cells (P < 0.0001, n = 3); **, statistically significant increase in ICSBP expression in Bcr/abl+ WT progenitor cells treated with IM (P < 0.0001, n = 3); # or ##, statistically significant increase in the expression of Gas2 in Bcr/abl+ WT cells or decrease in the expression of Gas2 in Bcr/abl+ cells treated with IM, respectively (P < 0.001, n = 3). The lack of a statistically significant difference in Gas2 expression in ICSBP−/− progenitor cells with or without Bcr/abl is indicated by the bracket (P = 0.1, n = 3). Error bars indicate standard errors.
We also investigated the effect of ICSBP on Bcr/abl-induced Gas2 mRNA and protein. In the initial control experiments, the expression of Bcr/abl in U937 cells decreased ICSBP mRNA and protein, increased Gas2 mRNA and protein, and increased β-catenin protein, as anticipated (Fig. 4B and C). Bcr/abl did not influence the expression of either calpain or calpastatin (another calpain inhibitor, discussed below). To determine if decreased ICSBP was functionally relevant to Gas2 expression in Bcr/abl+ cells, stable U937 transfectants were generated with both Bcr/abl and ICSBP expression vectors. Reexpression of ICSBP decreased Gas2 and β-catenin protein in Bcr/abl+ U937 cells (Fig. 4D). In control experiments, stable U937 transfectants were generated that expressed a kinase-inactive (KI) form of Bcr/abl (33). The levels of expression of ICSBP, Gas2, and β-catenin were not altered in these cells (Fig. 4E).
U937 cells are transformed prior to the expression of Bcr/abl, which may influence the effect of Bcr/abl on various signaling pathways. Therefore, we also investigated the influence of ICSBP abundance on Gas2 expression in Bcr/abl+ primary murine bone marrow cells. In these studies, WT or ICSBP−/− murine myeloid progenitor cells were transduced with a vector to express Bcr/abl or control vector. Cells were analyzed for ICSBP and Gas2 mRNA by real-time PCR. As anticipated, ICSBP mRNA was decreased and Gas2 mRNA increased in Bcr/abl+ WT progenitor cells, and these changes were reversed by imatinib (IM) (Fig. 4F). In contrast, Bcr/abl did not increase the already elevated level of Gas2 expression in ICSBP−/− myeloid progenitor cells (Fig. 4F). Equivalent levels of Bcr/abl expression in WT and ICSBP−/− cells were verified by real-time PCR (not shown).
ICSBP and Bcr/abl regulate calpain activity in myeloid progenitor cells in a Gas2-dependent manner.
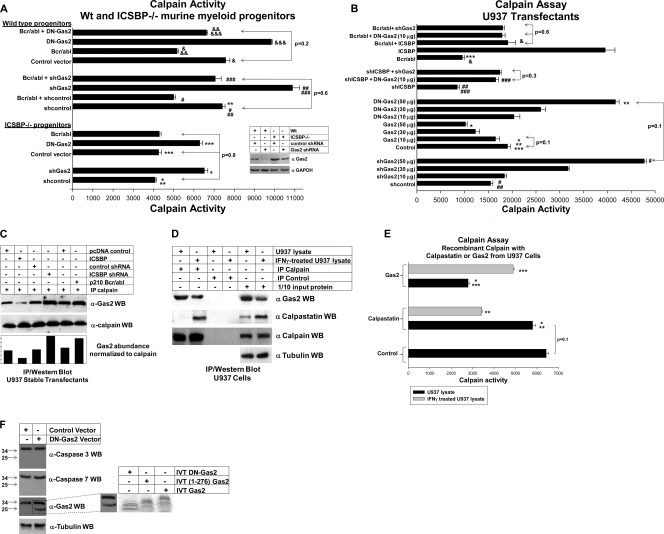
We next investigated whether ICSBP influences calpain activity in a Gas2-dependent manner. In the initial studies, we found that there was significantly less calpain activity in ICSBP−/− myeloid progenitor cells than in WT cells (Fig. 5A ). To determine if this was due to increased Gas2, ICSBP−/− and WT murine progenitors were transduced with a retroviral vector to express a Gas2-specific shRNA (or scrambled control) or a DN form of Gas2 (or empty vector). This DN-Gas2 protein binds to calpain but does not inhibit calpain activity (3). Gas2 inhibition by either method significantly increased calpain activity in ICSBP−/− cells. The levels of Gas2 expression and calpain activity in ICSBP−/− myeloid progenitors expressing shGas2 were equivalent to the levels of Gas2 expression and calpain activity in WT control progenitors (Fig. 5A). To determine if Bcr/abl also influenced calpain activity in a Gas2-dependent manner, WT murine progenitor cells were cotransduced with retroviral vectors to express Bcr/abl- and Gas2-specific shRNA or DN-Gas2 (or control vectors). The calpain activity in Bcr/abl+ myeloid progenitors was significantly less than that in control cells, and this effect was reversed by shGas2 or DN-Gas2 (Fig. 5A).
FIG. 5.
ICSBP expression influences calpain activity in undifferentiated myeloid cells. (A) There is less calpain activity in myeloid progenitor cells from ICSBP−/− mice than in WT cells. Myeloid progenitor cells were isolated from the bone marrow of WT or ICSBP−/− mice and cultured in GM-CSF, IL-3, and SCF. ICSBP−/− myeloid progenitors were transduced with retroviral vectors to express Gas2-specific shRNA (or scrambled control shRNA), a dominant-negative (DN) Gas2, or Bcr/abl (or empty vector control). WT progenitor cells were transduced with the Bcr/abl expression vector (or vector control) with or without a vector to express DN-Gas2 (or empty vector control) or a Gas2-specific shRNA (or scrambled control shRNA). Cell lysates were analyzed for calpain activity. *, statistically significant increase in calpain activity with the expression of Gas2-specific shRNA in ICSBP−/− myeloid progenitors (P < 0.001, n = 4); **, statistically significant increase in calpain activity in WT versus ICSBP−/− myeloid progenitors (P < 0.0001, n = 4); ***, statistically significant increase in calpain activity in ICSBP−/− myeloid progenitor cells expressing DN-Gas2 (P < 0.001, n = 4); # or &, statistically significant decrease in calpain activity in Bcr/abl+ WT myeloid progenitor cells (P < 0.001, n = 4). ##, statistically significant increase in calpain activity in WT cells expressing Gas2-specific shRNA (P < 0.001, n = 4); ###, statistically significant decrease in calpain activity in these cells also transduced with Bcr/abl expression vector (P < 0.001, n = 4); && or &&&, statistically significant increase in calpain activity in WT myeloid progenitors expressing DN-Gas2 or decrease in calpain activity in these cells, respectively, upon Bcr/abl expression (P < 0.001, n = 4). The relative expression levels of Gas2 in these cells are indicated by the results in the inset Western blot. (B) Calpain activity in U937 cells is increased by overexpression of ICSBP or Gas2 and decreased by ICSBP knockdown, Bcr/abl, or dominant-negative Gas2. U937 cells were transfected with a dose titration of vectors to overexpress Gas2, dominant-negative Gas2 (DN-Gas2), or a Gas2-specific shRNA (or relevant control vectors). An amount of Gas2, DN-Gas2, or shGas2 expression vector with minimal impact on calpain activity was identified. Based on these results, stable U937 transfectants expressing Bcr/abl (or vector control) or an ICSBP-specific shRNA (or scrambled shRNA) were cotransfected with DN-Gas2 expression vector (or vector control) or shGas2 expression vector (or control). Some Bcr/abl (or control) transfectants were cotransfected with an ICSBP expression vector. Cell lysates were analyzed for calpain activity. *, statistically significant decrease in calpain activity with Gas2 overexpression (P < 0.002, n = 4); ** or #, statistically significant increase in calpain activity with DN-Gas2 or shGas2 expression, respectively (P < 0.001, n = 4); *** or ##, statistically significant decrease in calpain activity with Bcr/abl or ICSBP-specific shRNA expression, respectively (P < 0.0001, n = 5); ###, statistically significant increase in calpain activity in cells with ICSBP-specific shRNA and DN-Gas2 or shGas2 (P < 0.01, n = 4); &, statistically significant increase in calpain activity in Bcr/abl-expressing cells with ICSBP overexpression (P = 0.001, n = 5). (C) Interaction between Gas2 and calpain is decreased by ICSBP overexpression and increased by ICSBP knockdown or Bcr/abl expression in U937 cells. Cell lysates from these U937 transfectants were analyzed for interaction between Gas2 and calpain by coimmunoprecipitation (IP). Cell lysates were immunoprecipitated with antibody to the small subunit of calpain or irrelevant control antibody. Immunoprecipitates were separated by SDS-PAGE, and Western blots (WB) probed with antibody to Gas2. Blots were reprobed with a calpain antibody as a loading control. The bottom panel shows the results for Gas2 abundance (as determined by pixel count) normalized to the abundance of calpain which immunoprecipitated in each experiment (in pixels). No proteins were detectable in control antibody immunoprecipitates, which are not shown. (D) Gas2 protein abundance decreases and calpastatin protein abundance increases during myeloid differentiation, and this influences the interaction of these proteins with calpain. Cell lysates from untreated or IFN-γ-differentiated U937 cells were immunoprecipitated with an antibody to the common small subunit of calpain (or irrelevant control antibody). Immunoprecipitated proteins were separated by SDS-PAGE, and coprecipitating proteins were identified by serially probing Western blots with antibodies to Gas2, calpastatin, and calpain. The input lysate protein (1/10 amount) was a loading control in these studies (blots were probed with antibody to tubulin). (E) The abundance of Gas2 and calpastatin proteins influences the relative inhibition of calpain by these proteins during myeloid differentiation. Lysate proteins from untreated or IFN-γ-differentiated U937 cells were immunoprecipitated with an antibody to Gas2 or calpastatin (or with irrelevant control antibody). Immunoprecipitates were incubated with recombinant calpain, and calpain assays were performed. *, statistically significant difference in calpain activity in experiments with Gas2 from undifferentiated U937 cells versus that in experiments with calpastatin or control studies (P < 0.001, n = 3); ** or ***, statistically significant difference in calpain activity in experiments with calpastatin or Gas2, respectively, from undifferentiated versus differentiated U937 cells (P < 0.001, n = 3). (F) The expression of various forms of Gas2 in U937 cells transfected with a DN-Gas2 expression vector is shown. Lysate proteins from stable U937 transfectants with a vector to express DN-Gas2 or the vector control were separated by SDS-PAGE, and Western blots were serially probed with antibodies to Gas2, caspase 3, caspase 7, and tubulin (as a loading control). In vitro-translated 35S-labeled proteins representing Gas2, Gas2(1-279), and DN-Gas2 were separated on the same SDS-PAGE gel. An autoradiograph of the in vitro-translated (IVT) protein lanes was compared to the Western blot probed for Gas2 to determine the forms of Gas2 present in these cells. Molecular sizes in kDa are shown to the left. Error bars indicate standard errors.
We used stable U937 transfectants to investigate the effect on calpain activity of directly altering ICSBP expression. ICSBP overexpression in U937 cells significantly increased the calpain activity (Fig. 5B). Decreased ICSBP expression due to either an ICSBP-specific shRNA or Bcr/abl significantly decreased calpain activity (Fig. 5B). To verify that the decreased calpain activity in Bcr/abl+ cells was ICSBP dependent, Bcr/abl+ cells were cotransfected with an ICSBP expression vector (Fig. 4E). Reexpression of ICSBP normalized the calpain activity in Bcr/abl+ cells (Fig. 5B).
We next investigated the effect of Gas2 expression on calpain activity in U937 cells. In these studies, U937 cells were transfected with dose titrations of vectors to express Gas2, DN-Gas2, or a Gas2-specific shRNA (or relevant vector controls). Gas2 overexpression induced a dose-dependent decrease in calpain activity, and Gas2 inhibition (by either method) a dose-dependent increase in calpain activity (Fig. 5B). The maximal increase in calpain activity by DN-Gas2 was not significantly different from that induced by shGas2.
To determine if decreased ICSBP expression decreases calpain activity in a Gas2-dependent manner, U937 cells were cotransfected with a vector to express an ICSBP-specific shRNA or Bcr/abl and a vector to express Gas2-specific shRNA or DN-Gas2. For these studies, shGas2 and DN-Gas2 expression vectors were used at levels which did not alone influence calpain activity. Therefore, these studies investigated the effect of Gas2 inhibition on calpain activity specifically related to decreased ICSBP. Gas2 inhibition by either approach significantly increased calpain activity in cells expressing either ICSBP-shRNA or Bcr/abl (Fig. 5B).
We also investigated the effect of ICSBP expression on the interaction between Gas2 and calpain. For these studies, lysates from U937 transfectants were immunoprecipitated with an antibody to the small subunit of calpain and the immunoprecipitates analyzed by Western blotting. We found that ICSBP overexpression decreased the interaction of Gas2 with calpain, consistent with decreased Gas2 expression (Fig. 5C). Conversely, decreased ICSBP expression, due to either ICSBP-specific shRNA or Bcr/abl, increased the amount of Gas2 that coprecipitated with calpain. Neither Gas2 nor calpain immunoprecipitated with control, irrelevant antibody (not shown).
Calpain activity is also inhibited by calpastatin in various cell types (2, 3). The DN-Gas2 used in these studies may antagonize calpain inhibition by both Gas2 and calpastatin. Therefore, we determined the relative contributions of Gas2 and calpastatin to calpain activity in myeloid cells. We found that calpastatin was expressed at low levels in U937 cells and its expression increased during 48 h of IFN-γ-induced differentiation (Fig. 5D). Consistent with this, the amount of calpastatin that coimmunoprecipitated with calpain was low in untreated U937 cells but increased during differentiation (Fig. 5D). Conversely, coimmunoprecipitation of Gas2 with calpain was decreased by differentiation of U937 cells.
To correlate Gas2 and calpastatin expression with calpain inhibition, calpain assays were performed using recombinant calpain and Gas2 or calpastatin that had been isolated from U937 cells. Gas2 immunoprecipitates from untreated U937 cells had a significant inhibitory effect on recombinant calpain, which was not observed with Gas2 immunoprecipitates from differentiated cells (Fig. 5E). This was consistent with the greater abundance of Gas2 in undifferentiated cells in comparison to its level in differentiated cells. Conversely, calpastatin immunoprecipitates from untreated U937 cells did not alter calpain activity. However, calpastatin immunoprecipitates from IFN-γ-differentiated cells significantly inhibited calpain activity (Fig. 5E). This was consistent with the greater abundance of calpastatin in differentiated cells in comparison to its level in undifferentiated cells. Therefore, the calpain-inhibitory activity of calpastatin or Gas2 in U937 cells increased or decreased according to the abundance of the protein. These results were also consistent with the results of the calpain activity assays, shown in Fig. 5A and B. If both calpastatin and Gas2 were significant calpain inhibitors in myeloid progenitor cells, the maximum effect of DN-Gas2 on calpain activity would be greater than the effect of complete knockdown of Gas2. In control studies, calpastatin and Gas2 did not coprecipitate (not shown).
The DN-Gas2 used in these studies is a C-terminal truncation (Δ171-314) (3) which generates a 26-kDa protein that is recognized by the Gas2 antibody used in our studies. Therefore, we anticipated the expression of Gas2-immunoreactive proteins of 26 and 35 kDa (representing endogenous Gas2) in U937 cells transfected with a DN-Gas2 expression vector. However, Gas2 may be cleaved by caspase 3 or 7 in apoptotic cells (at amino acid 279), resulting in a protein of ∼31 kDa. If DN-Gas2 induces caspase activation in U937 cells, this 31-kDa cleavage product might also be detected. To test this, in vitro-translated proteins representing Gas2, Gas2(1-279), and DN-Gas2 were analyzed on the same SDS-PAGE gel as lysates from U937 cells which were stably transfected with a DN-Gas2 expression vector or control vector. The in vitro-translated proteins were used as size markers for Gas2 species in the transfectants. We identified two Gas2-immunoreactive proteins in stable DN-Gas2 transfectants that were comparable in size to in vitro-translated Gas2 and DN-Gas2 (Fig. 5F). No activation of caspase 3 or 7 was found when the blots were reprobed with antibodies which recognize both the inactive and activated forms of these proteins (Fig. 5F). In addition, we found no increase in annexin V staining in flow cytometry studies of DN-Gas2-expressing U937 cells in comparison to its level in control cells (<10% apoptotic cells for both).
ICSBP regulates β-catenin activity in myeloid progenitor cells.
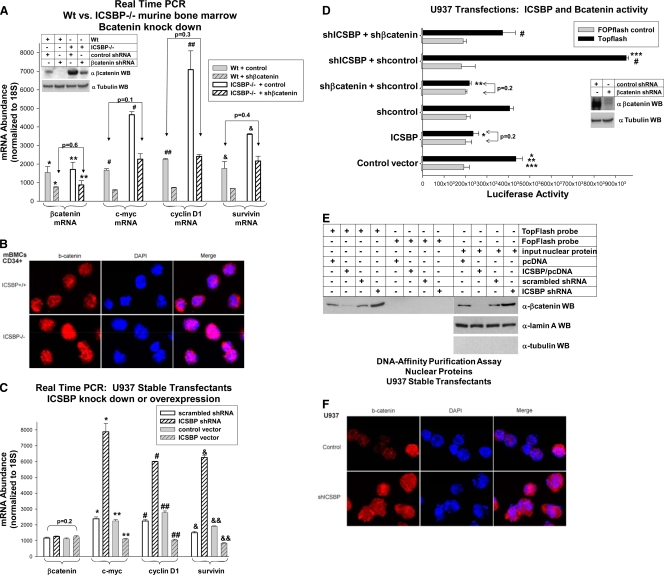
In the studies described above, the ICSBP expression level influenced β-catenin protein abundance (Fig. 3B and D). We next investigated whether β-catenin activity was regulated by ICSBP. Since the expression of β-catenin target genes (c-myc, cyclin D1, and survivin) is an indicator of β-catenin activity, the levels of expression of these genes in WT or ICSBP−/− myeloid progenitors were determined. We found increased expression of c-myc, cyclin D1, and survivin mRNA in ICSBP−/− murine myeloid progenitors compared to their levels in the WT (Fig. 6A). There was no difference in β-catenin mRNA in these cells, consistent with our hypothesis that ICSBP alters the stability of β-catenin protein via Gas2 and calpain.
FIG. 6.
β-Catenin activity is decreased by ICSBP overexpression and increased by ICSBP deficiency in myeloid progenitor cells. (A) The expression of β-catenin (Bcatenin) target gene mRNA is increased in ICSBP−/− murine myeloid progenitors in comparison to its expression in WT cells, but that of β-catenin mRNA is not altered. Myeloid progenitor cells were isolated from WT or ICSBP−/− mice and cultured in GM-CSF, IL-3, and SCF. Cells were transduced with a retroviral vector to express a β-catenin-specific shRNA or scrambled shRNA control. The levels of expression of mRNA for β-catenin and β-catenin target genes (c-myc, cyclin D1, and survivin) were determined by real-time PCR. The results were normalized to those for 18S RNA. * or **, statistically significant decrease in β-catenin mRNA in WT or ICSBP−/− cells expressing β-catenin-specific shRNA (versus control), respectively; #, ##, or &, statistically significant increase in the expression of c-myc, cyclin D1, or survivin, respectively, in ICSBP−/− control cells in comparison to their expression in WT cells (P < 0.01, n = 3). The lack of a significant difference in c-myc, cyclin D1, or survivin mRNA in WT control cells versus ICSBP−/− cells with β-catenin-specific shRNA is indicated by the arrows (P > 0.1, n = 3). The effect of β-catenin-specific shRNA expression on β-catenin protein expression in these cells is indicated by the results in the inset Western blot. (B) Increased β-catenin (b-catenin) protein is present in the nucleus of ICSBP−/− myeloid progenitors in comparison to the amount in WT cells. Bone marrow was harvested from WT and ICSBP−/− mice, and myeloid progenitor cells were selected and stained with an antibody to β-catenin. β-Catenin staining (red) was compared to staining of the nuclei with DAPI (blue) by confocal microscopy. Nuclear β-catenin is purple/pink in the images. (C) The levels of expression of β-catenin target gene mRNA are decreased by ICSBP overexpression and increased by ICSBP knockdown in U937 myeloid leukemia cells, but that of β-catenin mRNA is not altered. U937 cells were stably transfected with a vector to overexpress ICSBP or the empty control vector or with a vector to express an ICSBP-specific shRNA or scrambled control shRNA. The expression of mRNA for β-catenin or β-catenin target genes (c-myc, cyclin D1, or survivin) was determined by real-time PCR. The results were normalized to those for 18S RNA. *, #, or &, statistically significant increase in c-myc, cyclin D1, or survivin mRNA, respectively, with ICSBP knockdown (P < 0.001, n = 6); **, ##, or &&, statistically significant decrease in expression of c-myc, cyclin D1, or survivin, respectively, due to ICSBP overexpression (P < 0.0002, n = 6). The lack of a difference in β-catenin mRNA expression under all of these conditions is indicated by the bracket (P = 0.2, n = 6). (D) The activity of a β-catenin-binding reporter construct is increased by ICSBP knockdown and decreased by ICSBP overexpression in U937 myeloid cells. U937 cells were cotransfected with a β-catenin activated reporter vector (TOPflash) or its mutant control (FOPflash) and a vector to overexpress ICSBP or the empty vector control. Other U937 cells were cotransfected with the TOPflash or FOPflash reporter vectors, a vector to express an ICSBP-specific shRNA or scrambled shRNA control, or a vector to express a β-catenin-specific shRNA or scrambled shRNA control. * or **, statistically significant decrease in TOPflash reporter activity with ICSBP overexpression or β-catenin-specific shRNA expression, respectively (P < 0.01, n = 4); ***, statistically significant increase in TOPflash activity with expression of ICSBP-specific shRNA (P < 0.002, n = 4); #, statistically significant decrease in TOPflash activity in cells with ICSBP knockdown with expression of β-catenin-specific shRNA versus the control (P < 0.005, n = 4). Complete knockdown of β-catenin by the specific shRNA under these conditions is indicated by the results in the inset Western blot (WB). (E) ICSBP overexpression decreases and ICSBP knockdown increases β-catenin DNA-binding activity in the nucleus of U937 myeloid leukemia cells. Nuclear proteins were isolated from the stable U937 transfectants with ICSBP knockdown or overexpression. Proteins were purified by affinity to biotin-labeled oligonucleotide probes with either a β-catenin DNA-binding site or a mutant, nonbinding sequence. Purified proteins were analyzed by probing Western blots with antibody to β-catenin. To control for protein content in these nuclear fractions, Western blots of these proteins (1/10 the amount used for affinity purification) were probed with antibodies to β-catenin, lamin A (as a nuclear protein loading control), and tubulin (as a cytoplasmic protein loading control). (F) Increased β-catenin staining is present in the nucleus of U937 cells with ICSBP knockdown compared to the staining in control cells. Stable U937 transfectants with an ICSBP-specific shRNA or scrambled control shRNA were stained with an antibody to β-catenin. β-Catenin staining (red) was compared to staining of the nuclei with DAPI (blue) by confocal microscopy. Nuclear β-catenin stains are purple/pink in these images. Error bars indicate standard errors.
We determined whether the increase in c-myc, cyclin D1, and survivin in ICSBP−/− cells was β-catenin dependent by transducing these cells and WT myeloid progenitor cells with a retroviral vector to express a β-catenin-specific shRNA (or scrambled control). In ICSBP−/− cells, β-catenin shRNA decreased β-catenin protein to a level that was equivalent to the amount of β-catenin protein in WT control cells (Fig. 6A). Consistent with this, β-catenin target gene expression in ICSBP−/− cells with β-catenin knockdown was equivalent to the expression of these genes in control WT cells.
To investigate the effect of ICSBP expression on the abundance of nuclear β-catenin, myeloid progenitor cells from WT or ICSBP−/− murine bone marrow were analyzed by immunofluorescence confocal microscopy (Fig. 6B). Staining with a β-catenin antibody was compared to nuclear staining with 4′,6-diamidino-2-phenylindole (DAPI). We found that the increase in total β-catenin in ICSBP−/− cells in comparison to its level in WT cells was associated with increased nuclear β-catenin.
Since directly manipulating ICSBP expression in U937 cells alters β-catenin protein abundance (Fig. 3D), we also investigated the effect on β-catenin activity. The levels of expression of c-myc, cyclin D1, and survivin mRNA were significantly increased by ICSBP knockdown and decreased by ICSBP overexpression in U937 cells (Fig. 6C). As anticipated, the β-catenin mRNA abundance was not influenced by the ICSBP expression level.
β-Catenin forms a complex with Lef/Tcf transcription factors and activates gene transcription (20). Therefore, β-catenin activity can also be studied using an artificial promoter/reporter construct with multiple copies of an Lef/Tcf binding site (referred to as TOPflash). As another approach to determining the influence of ICSBP on β-catenin activity, U937 cells were cotransfected with TOPflash reporter vector (or the mutant FOPflash control) and a vector to overexpress ICSBP (or control) or to express an ICSBP-specific shRNA (or scrambled control). ICSBP overexpression decreased TOPflash activity to a level that was equivalent to the level in the FOPflash control (Fig. 6D). Conversely, ICSBP knockdown significantly increased TOPflash activity. To verify that ICSBP knockdown produced this effect by increasing β-catenin, U937 cells were cotransfected with TOPflash or FOPflash vectors, a vector to express ICSBP-specific shRNA, and another vector to express a β-catenin-specific shRNA (or relevant controls). The expression of β-catenin-specific shRNA abrogated the increase in TOPflash activity in U937 cells with ICSBP knockdown (Fig. 6D). The expression of β-catenin-specific shRNA at this level abolished β-catenin protein expression and decreased TOPflash activity so that it was not significantly different than that of the FOPflash control (Fig. 6D).
We next determined whether these ICSBP-dependent changes in TOPflash activity correlated with targeting of nuclear β-catenin to the Lef/Tcf DNA-binding site. Nuclear protein fractions from U937 transfectants with ICSBP knockdown or overexpression were incubated with a biotin-labeled, double-stranded oligonucleotide probe with the TOPflash consensus sequence or the nonbinding FOPflash mutant. Interacting proteins were affinity purified and separated by SDS-PAGE, and the presence of β-catenin determined by Western blotting. We found that ICSBP overexpression decreased and knockdown increased the amount of β-catenin protein in the affinity-purified complex (Fig. 6E). The expression of lamin A but not that of tubulin validated the nuclear origin of the proteins. Western blots of total cell lysates were positive for both proteins (not shown).
We also used immunofluorescence confocal microscopy to determine if ICSBP knockdown increased nuclear β-catenin in U937 cells. We found increased total and nuclear β-catenin staining in these cells in comparison to the staining in the control (Fig. 6F).
Bcr/abl regulates β-catenin activity in myeloid progenitor cells in an ICSBP-dependent manner.
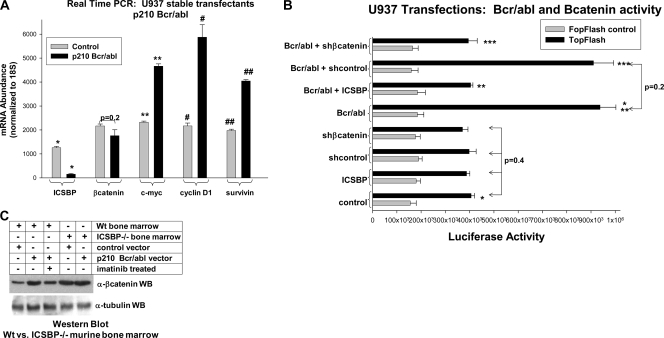
Since ICSBP reexpression reversed the increase in β-catenin protein in Bcr/abl+ U937 cells (Fig. 4D), we investigated the ICSBP dependence of increased β-catenin activity in Bcr/abl+ cells. In the initial experiments, the effect of Bcr/abl on β-catenin activity was investigated by determining the expression of β-catenin target genes. We found that the amounts of c-myc, cyclin D1, and survivin mRNA were significantly greater in Bcr/abl+ U937 cells than in control cells (Fig. 7A). ICSBP expression was decreased in Bcr/abl+ cells, and the level of β-catenin mRNA was not altered.
FIG. 7.
Bcr/abl increases β-catenin activity in myeloid progenitor cells. (A) Bcr/abl increases the expression of β-catenin target gene mRNA but not of β-catenin mRNA in U937 myeloid cells. U937 cells were stably transfected with a vector to express Bcr/abl or empty vector control. The levels of expression of mRNA for ICSBP, β-catenin, and β-catenin target genes (c-myc, cyclin D1, or survivin) were determined by real-time PCR. The results were normalized to those for 18S RNA. *, statistically significant decrease in ICSBP mRNA in Bcr/abl+ cells (P < 0.0001, n = 6); **, #, or ##, statistically significant increase in mRNA for c-myc, cyclin D1, or survivin, respectively, in Bcr/abl-expressing cells (P < 0.002, n = 3). The lack of a statistically significant difference in β-catenin mRNA expression with versus without Bcr/abl is indicated by the bracket (P = 0.2, n = 3). (B) The activity of a β-catenin-binding (Bcatenin) reporter construct is increased by Bcr/abl in U937 myeloid cells. U937 cells were cotransfected with a β-catenin-activated reporter vector (TOPflash) or its mutant control (FOPflash) and a vector to express Bcr/abl or the empty vector control. Some cells were also cotransfected with a vector to overexpress ICSBP or the control vector or a vector for β-catenin-specific shRNA or scrambled shRNA control. The amounts of the ICSBP expression vector and the β-catenin-specific shRNA expression vector were titrated to have a minimal impact on TOPflash reporter activity alone (P = 0.2, n = 6, for comparison between ICSBP vector, control vector, β-catenin-specific shRNA vector, or scrambled shRNA control vector). *, statistically significant increase in TOPflash activity in Bcr/abl-expressing cells (P = 0.005, n = 6); ** or ***, statistically significant decrease in TOPflash activity in Bcr/abl-expressing cells cotransfected with a vector to overexpress ICSBP or β-catenin-specific shRNA, respectively (P < 0.005, n = 6). (C) Bcr/abl increases β-catenin protein expression in WT myeloid progenitor cells but does not further increase β-catenin in progenitors from ICSBP−/− mice. Bone marrow myeloid progenitor cells were isolated from WT and ICSBP−/− mice, cultured in GM-CSF, IL-3, and SCF, and transduced with a retroviral vector to express p210 Bcr/abl or the empty vector control. Some cells were treated with imatinib (IM). The cell lysates were analyzed by probing Western blots (WB) with antibodies to β-catenin or tubulin (as a loading control). Error bars indicate standard errors.
As another approach to investigate the influence of ICSBP on β-catenin activity in Bcr/abl+ cells, U937 cells were cotransfected with TOPflash or FOPflash reporter vectors and vectors to express Bcr/abl and ICSBP (or relevant control vectors). Bcr/abl significantly increased TOPflash activity in these cells, but this increase was blocked by reexpression of ICSBP (Fig. 7B). For these studies, we used an amount of ICSBP expression vector which did not alone influence TOPflash expression (based on dose titration studies, not shown). Therefore, these studies assessed the effect of ICSBP reexpression on TOPflash activity that was specifically due to Bcr/abl. To verify that the increased TOPflash activity in Bcr/abl+ U937 cells was β-catenin dependent, the experiment was repeated with a vector to express a β-catenin-specific shRNA. The β-catenin shRNA vector was titrated to have a minor influence on TOPflash activity alone to assess the specific effect of Bcr/abl (not shown). As anticipated, β-catenin knockdown prevented the Bcr/abl-induced increase in TOPflash activity (Fig. 7B).
Because U937 cells are transformed prior to Bcr/abl expression, we also determined the ICSBP dependence of increased β-catenin in Bcr/abl+ cells using primary myeloid progenitor cells. For these studies, WT or ICSBP−/− murine myeloid progenitors were transduced with a Bcr/abl expression vector or control. As anticipated, Bcr/abl expression increased the amount of β-catenin protein in WT myeloid progenitors and this was reversed by IM (Fig. 7C). However, the already elevated level of β-catenin in ICSBP−/− cells was not further increased by Bcr/abl (Fig. 7C). The results of real-time PCR demonstrated equivalent levels of Bcr/abl expression in WT and ICSBP−/− cells (not shown).
ICSBP regulates β-catenin activity in a Gas2-dependent manner.
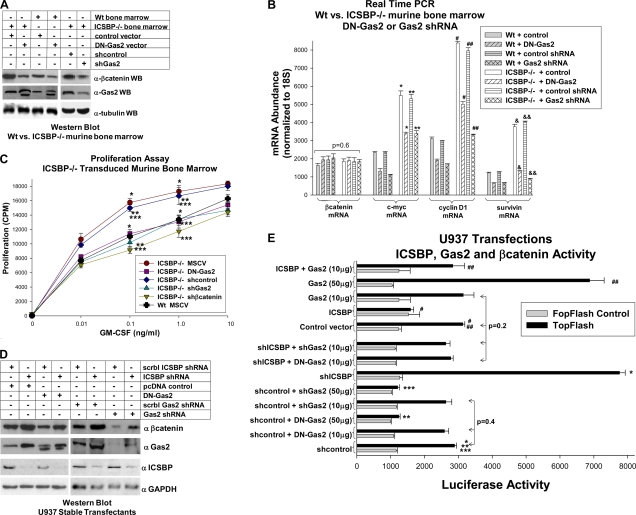
We next determined if the increase in β-catenin protein in ICSBP−/− cells was Gas2 dependent. For these studies, bone marrow myeloid progenitor cells from WT or ICSBP−/− mice were transduced with retroviral vectors to express DN-Gas2 (or its control) or a Gas2-specific shRNA (or the scrambled control shRNA). DN-Gas2 and shGas2 induced equivalent decreases in β-catenin protein in ICSBP−/− cells (Fig. 8A). Since Gas2 was more abundant in ICSBP−/− cells than in WT cells, β-catenin was also more abundant in ICSBP−/− than in WT cells, with or without Gas2 inhibition. We also used these cells to investigate the role of Gas2 in ICSBP-dependent β-catenin activity. Either DN-Gas2 or shGas2 decreased the expression of c-myc, cyclin D1, and survivin in both WT and ICSBP−/− myeloid progenitor cells (Fig. 8B). Consistent with the greater abundance of β-catenin in ICSBP−/− myeloid progenitors, the expression of β-catenin target genes was greater in ICSBP−/− cells than in WT control cells, even with Gas2 inhibition (Fig. 8B).
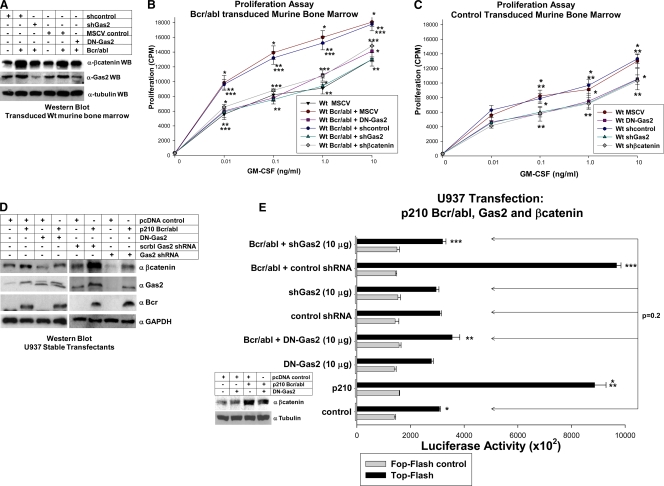
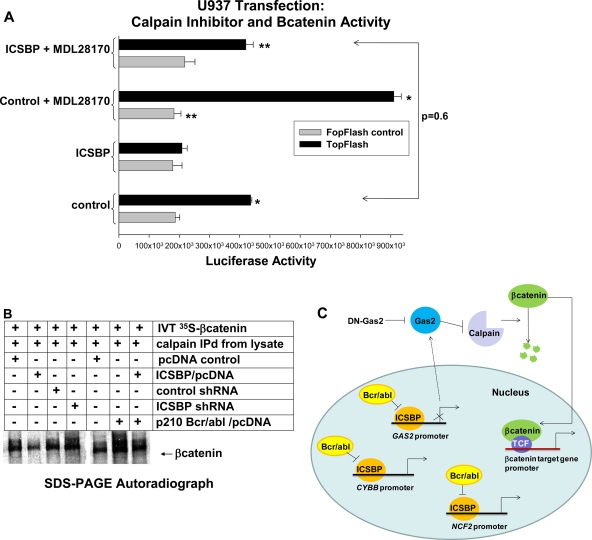
FIG. 8.
The increased levels of β-catenin protein and activity in myeloid progenitor cells with ICSBP knockdown are Gas2 dependent. (A) The expression of DN-Gas2 or a Gas2-specific shRNA decreases β-catenin protein expression in ICSBP−/− myeloid progenitor cells. Bone marrow myeloid progenitors were isolated from WT and ICSBP−/− mice and cultured in GM-CSF, IL-3, and SCF. Cells were transduced with a vector to express dominant-negative Gas2 (DN-Gas2) or empty vector control. Some ICSBP−/− myeloid progenitors were transduced with a vector to express a Gas2-specific shRNA or scrambled control. The lysate proteins from the transduced cells were analyzed by Western blot assay for β-catenin and Gas2 protein expression. The blots were also probed with an antibody to tubulin as a loading control. (B) The expression of DN-Gas2 in WT or ICSBP−/− myeloid progenitor cells decreases the expression of β-catenin target genes. WT or ICSBP−/− myeloid progenitor cells were transduced with a vector to express DN-Gas2 or a Gas2-specific shRNA (or relevant controls). Total cellular RNA was isolated from the transduced myeloid progenitor cells and analyzed by real-time PCR for the expression of β-catenin or β-catenin target genes. The results were normalized to those for 18S RNA. *, #, or & ***, statistically significant decrease in the expression of c-myc, cyclin D1, or survivin, respectively, in ICSBP−/− cells with DN-Gas2 in comparison to their expression with the control vector (P < 0.001, n = 3); **, ##, or &&, statistically significant decrease in mRNA expression of c-myc, cyclin D1, or survivin, respectively, in cells transduced with shGas2 vector. (C) Gas2 knockdown or the expression of DN-Gas2 reverses GM-CSF hypersensitivity in ICSBP−/− myeloid progenitor cells. Murine myeloid progenitor cells from WT or ICSBP−/− mice were cultured in GM-CSF, IL-3, and SCF. Cells were transduced with a vector to express a Gas2-specific shRNA, a β-catenin-specific shRNA, dominant-negative Gas2 (DN-Gas2), or the relevant control vectors. WT cells were also transduced with empty murine stem cell virus (MSCV) vector. The cells were cytokine deprived and treated with a dose titration of GM-CSF as indicated. Proliferation was determined by the incorporation of 3H-thymidine. *,**, or ***, statistically significant decrease in proliferation in ICSBP−/− cells transduced with vector to express DN-Gas2 versus control MSCV vector or Gas2 shRNA or β-catenin shRNA versus control, respectively. CPM, counts per minute. (D) The expression of dominant-negative Gas2 reverses the increase in β-catenin protein in U937 cells with ICSBP knockdown. Stable U937 transfectants were generated with a vector to express an ICSBP-specific shRNA or scrambled control shRNA and a vector to express dominant-negative Gas2 (DN-Gas2) or a Gas2-specific shRNA or the relevant control vectors. The cell lysates were analyzed by Western blot assay for the expression of β-catenin, Gas2, ICSBP, or GAPDH (as a loading control). (E) The increased activity of a β-catenin-binding reporter construct in U937 cells with ICSBP knockdown is reversed by dominant-negative Gas2, and its decreased activity in cells with ICSBP-overexpression is reversed by the overexpression of Gas2. U937 cells were cotransfected with the TOPflash reporter vector or FOPflash control, a vector to express an ICSBP-specific shRNA or scrambled shRNA control vector, and a vector to express dominant-negative Gas2 (DN-Gas2) or a Gas2-specific shRNA or the relevant control vectors. Other cells were cotransfected with the reporter vectors and vectors to overexpress ICSBP or the empty vector control and a vector to express Gas2 or the empty vector control. A dose titration of DN-Gas2, shGas2, and Gas2 expression vectors was performed. *, statistically significant increase in reporter activity in cells with an ICSBP-specific shRNA; ** or ***, statistically significant decrease in TOPflash reporter activity in transfectants with DN-Gas2 or Gas2-specific shRNA, respectively (P < 0.0001, n = 4); # and ##, statistically significant decrease in TOPflash reporter activity in transfectants with ICSBP and increase in transfectants with ICSBP plus Gas2, respectively, in comparison to ICSBP alone (P < 0.01, n = 4). The reporter activity in transfectants with shICSBP and either DN-Gas2 or shGas2 is not significantly different than that in the control, as indicated. Error bars indicate standard errors.
We were also interested in the influence of Gas2 on functional activities mediated by β-catenin target gene products. c-myc and cyclin D1 are implicated in the proliferative response of myeloid progenitor cells to GM-CSF (4, 16), and ICSBP−/− cells are hypersensitive to GM-CSF (increased proliferation at low doses). Therefore, we investigated the role of Gas2 in GM-CSF hypersensitivity in ICSBP−/− cells. Transduced ICSBP−/− murine myeloid progenitor cells were treated with a dose titration of GM-CSF, and proliferation was determined by 3H-thymidine incorporation. Gas2 inhibition by either approach decreased GM-CSF hypersensitivity in ICSBP−/− cells (Fig. 8C). The role of β-catenin in GM-CSF hypersensitivity was also investigated. For these studies, ICSBP−/− murine myeloid progenitors were transduced with a retroviral vector to express a β-catenin-specific shRNA (or scrambled control). The results of proliferation assays demonstrated a reversal of GM-CSF hypersensitivity in ICSBP−/− cells with β-catenin knockdown (Fig. 8C).
We also investigated the role of Gas2 in the increase in β-catenin protein in U937 cells with ICSBP knockdown. For these studies, U937 cells were cotransfected with vectors to express an ICSBP-specific shRNA (or empty vector) and DN-Gas2 or a Gas2-specific shRNA (or control vectors). Inhibition of Gas2 by either approach proportionately decreased the amount of β-catenin protein in ICSBP knockdown and control cells (Fig. 8D).
We next investigated the role of Gas2 in the ICSBP-dependent regulation of β-catenin activity. In preliminary studies, U937 cells were cotransfected with TOPflash or FOPflash reporter vector and a dose titration of vector expressing DN-Gas2, shGas2, or Gas2 (or control vectors). Either method of Gas2 inhibition resulted in dose-dependent repression of TOPflash activity (Fig. 8E). Conversely, Gas2 overexpression increased TOPflash activity. Therefore, U937 cells were cotransfected with the reporter vectors, a vector to express ICSBP-specific shRNA, and a vector to express DN-Gas2 or shGas2 (or relevant controls). An amount of DN-Gas2 or shGas2 vector was used that did not alone influence TOPflash activity. Both DN-Gas2 and shGas2 normalized TOPflash reporter activity in transfectants with ICSBP knockdown (Fig. 8E). U937 cells were also cotransfected with TOPflash or FOPflash, a vector to overexpress ICSBP, and a vector to overexpress Gas2 (or control vectors). Gas2 was overexpressed at a level which did not alone influence TOPflash activity. Consistent with our hypothesis, Gas2 reexpression reversed the ICSBP-induced inhibition of TOPflash activity (Fig. 8E).
Bcr/abl regulates β-catenin activity in an ICSBP and Gas2-dependent manner.
We next investigated the role of Gas2 in Bcr/abl-induced β-catenin expression and activity. For these studies, myeloid progenitor cells were isolated from WT murine bone marrow and transduced with retroviral vectors to express Bcr/abl and DN-Gas2 or a Gas2-specific shRNA (or control vectors). The inhibition of Gas2 by either approach decreased the amount of β-catenin protein in Bcr/abl+ myeloid progenitors (Fig. 9A). In these studies, the β-catenin protein abundance in Bcr/abl+ cells with Gas2 inhibition was slightly greater than that in control vector-transduced cells.
FIG. 9.
The increased levels of β-catenin protein and activity in Bcr/abl+ myeloid progenitor cells are Gas2 dependent. (A) The increase in β-catenin protein in Bcr/abl+ myeloid progenitor cells is reversed by Gas2 knockdown or the expression of DN-Gas2. WT murine myeloid progenitor cells were transduced with retroviral vectors expressing Bcr/abl (or the murine stem cell virus [MSCV] vector control) and either a Gas2-specific shRNA (or scrambled shRNA control) or DN-Gas2 (or the vector control). The lysate proteins from the transduced cells were analyzed by Western blot (WB) assay for β-catenin, Gas2, and tubulin (as a loading control). (B) Gas2 knockdown or the expression of DN-Gas2 reverses GM-CSF hypersensitivity in Bcr/abl+ murine myeloid progenitor cells. Murine myeloid progenitor cells from WT mice were cultured in GM-CSF, IL-3, and SCF and transduced with retroviral vectors as described above. Other cells were cotransduced with retroviral vectors to express Bcr/abl and a β-catenin-specific shRNA (or scrambled control). The cells were deprived of cytokines and treated with a dose titration of GM-CSF as indicated. Proliferation was determined by the incorporation of 3H-thymidine. *, **, or ***, statistically significant decrease in proliferation in Bcr/abl+ cells transduced with a DN-Gas2 vector versus the control MSCV vector, with a Gas2 shRNA vector versus the control, or with a β-catenin shRNA vector versus the control, respectively. CPM, counts per minute. (C) Gas2 knockdown or the expression of DN-Gas2 slightly decreases GM-CSF-induced proliferation in WT murine myeloid progenitor cells. Murine myeloid progenitor cells were transduced with retroviral vectors to express DN-Gas2 (or the empty vector control) or a Gas2-specific shRNA or a β-catenin-specific shRNA (or the scrambled shRNA control vectors). Proliferation in response to a dose titration of GM-CSF was determined as described above. *, **, or ***, statistically significant decrease in proliferation under the respective conditions. (D) The increase in β-catenin protein in Bcr/abl+ U937 cells is reversed by the overexpression of Gas2. Stable U937 transfectants were generated with a vector to express Bcr/abl or the empty vector control and a vector to express dominant-negative Gas2 (DN-Gas2) or a Gas2-specific shRNA (or the relevant control vectors). The cell lysates were analyzed by Western blot assay for the expression of β-catenin, ICSBP, Gas2, Bcr, or GAPDH (as a loading control). (E) The increased activity of a β-catenin-activated reporter construct in Bcr/abl+ U937 cells is reversed by Gas2 inhibition. U937 cells were cotransfected with the TOPflash reporter vector or the FOPflash control, a vector to express Bcr/abl or the empty vector, and a vector to express DN-Gas2 or a Gas2-specific shRNA (or the relevant control vectors). *, statistically significant increase in TOPflash reporter activity in cells expressing Bcr/abl (P < 0.0001, n = 4); ** or ***, statistically significant decrease in TOPflash reporter activity in transfectants with Bcr/abl and DN-Gas2 or Bcr/abl and shGas2, respectively, versus Bcr/abl alone (P < 0.0001, n = 4). The effect of these vectors on β-catenin protein expression is indicated by the results in the inset Western blot. Error bars indicate standard errors.
We also investigated the effect of Gas2 on activities downstream from β-catenin by determining the proliferative response to a dose titration of GM-CSF in these cells. We found that Bcr/abl+ myeloid progenitors were GM-CSF hypersensitive but that this was reversed by Gas2 inhibition by either mechanism (Fig. 9B). To determine the role of β-catenin in Bcr/abl-induced GM-CSF hypersensitivity, some cells were cotransduced with a vector to express a β-catenin-specific shRNA. β-Catenin knockdown decreased GM-CSF hypersensitivity in Bcr/abl+ cells (Fig. 9B). Gas2 inhibition and sh-β-catenin expression also decreased GM-CSF-induced proliferation of control myeloid progenitors but to a lesser extent (Fig. 9C).
The role of Gas2 in the increased β-catenin expression in Bcr/abl+ cells was further investigated using U937 transfectants. U937 cells were stably transfected with vectors to express Bcr/abl and DN-Gas2 or shGas2 (or relevant control vectors). DN-Gas2 decreased β-catenin protein abundance in both Bcr/abl+ and control cells (Fig. 9D). Consistent with a higher level of Gas2 expression in Bcr/abl+ cells, the amount of β-catenin protein in these cells was greater than the amount in control cells, with or without DN-Gas2. The expression of shGas2 also decreased the amount of β-catenin protein in Bcr/abl+ control cells (Fig. 9D). The expression of Gas2 in Bcr/abl+ cells with shGas2 was approximately equivalent to the Gas2 expression in control cells. Consistent with this, the amounts of β-catenin protein were equivalent in control cells and Bcr/abl+ cells with Gas2 knockdown.
To investigate the effect of Gas2 on β-catenin activity, U937 cells were cotransfected with TOPflash or FOPflash reporter, a vector to express Bcr/abl, and a vector to express DN-Gas2 or shGas2 (or control vectors). As in previous studies, DN-Gas2 and shGas2 were expressed at levels that did not alone influence β-catenin expression or activity (Fig. 9E and inset). Gas2 inhibition by either method reversed the Bcr/abl-induced increase in TOPflash activity. The level of TOPflash activity in Bcr/abl+ cells with DN-Gas2 or shGas2 was not significantly different than in cells transfected with control vectors.
ICSBP regulates β-catenin activity in a calpain-dependent manner.
We next investigated whether the ICSBP-induced regulation of β-catenin was dependent on calpain activity. U937 cells were cotransfected with TOPflash or FOPflash vectors and a vector to overexpress ICSBP (or control). Reporter assays were performed with or without the calpain inhibitor MDL28170. We found that MDL28170 rescues TOPflash reporter activity in ICSBP-overexpressing transfectants (Fig. 10A). In control experiments, this treatment did not alter ICSBP expression (not shown).
FIG. 10.
The increased levels of β-catenin protein and activity in Bcr/abl+ or ICSBP-deficient U937 cells are calpain dependent. (A) The decreased activity of a β-catenin (Bcatenin)-activated reporter construct in U937 cells overexpressing ICSBP is reversed by a calpain inhibitor. U937 cells were cotransfected with TOPflash reporter vector or the FOPflash control and a vector to overexpress ICSBP or the empty vector control. Reporter gene assays were performed with or without treatment with the calpain inhibitor MDL28170. * or **, statistically significant increase in TOPflash reporter activity in control transfectants or ICSBP overexpression transfectants, respectively, with versus without a calpain inhibitor (P < 0.01, n = 3). (B) Calpain isolated from U937 cells degrades β-catenin in an ICSBP- and Bcr/abl-dependent manner. Stable U937 transfectants were generated with a vector to express ICSBP, p210 Bcr/abl with or without ICSBP overexpression, or ICSBP-specific shRNA or scrambled control shRNA. Cell lysates were immunoprecipitated (IPd) with an antibody to the small subunit of calpain, and the immunoprecipitates were incubated with in vitro-translated (IVT) β-catenin (35S labeled). After the incubation, β-catenin protein was analyzed by SDS-PAGE, followed by autoradiography. (C) Schematic representation of the regulation of β-catenin by Bcr/abl and ICSBP. Functional interactions between Bcr/abl, ICSBP, Gas2, calpain, and β-catenin indicated by these studies. Error bars indicate standard errors.
We also performed a β-catenin degradation assay using calpain that was isolated from U937 cells. In these studies, calpain was immunoprecipitated from lysates of stable U937 transfectants either overexpressing ICSBP or with decreased ICSBP expression due to shICSBP or Bcr/abl (or with relevant control vectors). Immunoprecipitated proteins were incubated with in vitro-translated, 35S-labeled β-catenin, and the stability of β-catenin protein determined by autoradiography of SDS-PAGE gels. The degradation of recombinant β-catenin was increased by incubation with calpain from ICSBP-overexpressing cells and decreased after incubation with calpain from cells with decreased ICSBP (Fig. 10B).
DISCUSSION
In CML, a poor prognosis and progression to BC are characterized by decreased expression of ICSBP, increased expression of Gas2, and increased β-catenin activity. These isolated clinical observations are mechanistically linked in the current study. We found that ICSBP cooperates with Tel and HDAC3 to repress GAS2 transcription in myeloid progenitor cells, an effect not observed in differentiating phagocytes. Since Gas2 is a calpain inhibitor, ICSBP-induced repression of GAS2 results in increased calpain protease activity and the consequent degradation of β-catenin protein (Fig. 10C). Conversely, a Gas2- and calpain-dependent increase in the stability and activity of β-catenin was found in myeloid progenitor cells with decreased ICSBP expression. And, Bcr/abl expression resulted in an increase in β-catenin activity which was dependent on ICSBP, Gas2, and calpain. Therefore, these studies identified modulation of the stability of β-catenin protein as a novel mechanism for ICSBP-induced leukemia suppression.
We found that ICSBP interacts with a negative cis element in the GAS2 promoter in myeloid progenitors but not in differentiating phagocytes. The tyrosine phosphorylation state influences the interaction of ICSBP with some previously described target genes by regulating protein-DNA and protein-protein interactions (9, 13, 38). Since ICSBP tyrosine phosphorylation increases gradually during myelopoiesis, this provides a mechanism for regulation specific to the differentiation stage of these genes. We found that tyrosine phosphorylation was necessary for the repression of GAS2 transcription by ICSBP in myeloid cells, suggesting that ICSBP decreases β-catenin activity as hematopoietic stem cells (HSC) become granulocyte/monocyte progenitors (GMP). This was consistent with our hypothesis that decreased ICSBP increases β-catenin activity in CML-LSC, which resemble GMP more closely than HSC.
Since ICSBP tyrosine phosphorylation is required for GAS2 repression, one might anticipate increasing ICSBP-induced repression as differentiation proceeds. However, ICSBP was irrelevant to GAS2 transcription in differentiating phagocytes. Instead, trimethyl-K27 histone H3 (triMe-K27 H3) binding to GAS2 increased in these cells. This silencer mark is induced by Ezh2, which is part of a highly conserved protein complex involved in histone methylation (reviewed in reference 24). Since the repression of GAS2 by ICSBP involves histone deacetylation, this suggests different mechanisms for GAS2 repression in myeloid progenitors than in differentiating phagocytes. It is possible that ICSBP-induced repression in myeloid progenitors is reversible and that the triMe-K27 H3 mark represents permanent repression in mature cells. However, the results of recent studies suggest that this mark may be reversible by Jumonji proteins during phagocyte activation (6). It is possible that the binding of an Ezh2-containing complex to the GAS2 promoter inhibits ICSBP binding as differentiation proceeds. This will be explored in subsequent studies.
Gas2 inhibits calpain by a direct interaction. The DN-Gas2 used in these studies interacts with calpain but does not inhibit calpain protease activity (2, 3). Calpastatin is another endogenous calpain inhibitor, and both Gas2 and calpastatin inhibit calpain in epithelial cells and fibroblasts (2, 3). We found that Gas2 was abundantly expressed in myeloid progenitors but that its expression decreased during myelopoiesis. In contrast, calpastatin expression was low in myeloid progenitors but increased during differentiation. In myeloid progenitors, calpain was equivalently inhibited by either DN-Gas2 or a Gas2 shRNA, supporting the conclusion that Gas2 is the major calpain inhibitor in these cells. Since DN-Gas2 impairs calpain inhibition by both Gas2 and calpastatin, one would anticipate that DN-Gas2 would have a greater effect on calpain activity than Gas2 knockdown if calpastatin and Gas2 both contribute to calpain inhibition.
In previous studies, we identified two calpain genes (CAPN2 and CAPN12) as potential ICSBP targets in IFN-γ-differentiated U937 cells but not in untreated cells (12). Consistent with this, we found that neither ICSBP knockout nor Bcr/abl expression influenced the expression of calpain 2 or 12 in myeloid progenitor cells (not shown). Calpain regulates several aspects of the phagocyte response, including neutrophil migration, degranulation, and chemotaxis, and specific calpains are expressed during macrophage activation (17, 21, 30, 31, 39). The regulation of calpain gene expression by ICSBP in mature and activated phagocytes will be the topic of future studies.
Bcr/abl expression in myeloid progenitor cells decreases ICSBP mRNA, suggesting that IRF8 transcription is inhibited in CML. Studies investigating IRF8 promoter function in CML are ongoing in the laboratory. Since ICSBP is tyrosine phosphorylated, we considered the possibility that ICSBP is a Bcr/abl substrate. However, we found no increase in ICSBP phosphorylation that was attributable to Bcr/abl tyrosine kinase activity in myeloid cells (not shown).
Consistent with our results, the results of previous studies suggested that Bcr/abl renders β-catenin resistant to degradation (5). However, Bcr/abl is likely to influence β-catenin by mechanisms in addition to the Gas2-calpain pathway identified in our studies. Perhaps consistent with this, Gas2 inhibition in Bcr/abl+ murine myeloid progenitors did not completely return the β-catenin protein abundance to normal. In contrast, Gas2 inhibition in Bcr/abl+ U937 cells brought the amount of β-catenin protein back to the level in control cells. U937 cells are transformed prior to Bcr/abl expression and exhibit preexisting abnormality in β-catenin expression and activity. Our results suggest that aberrant pathways in U937 cells (which are part of the transformed phenotype of these cells) are redundant with unidentified pathways in Bcr/abl+ cells which increase the level of β-catenin protein. The identification of non-Gas2-calpain pathways which increase β-catenin activity in Bcr/abl+ cells will be the focus of future studies.
The goal of our investigations is to identify target genes relevant to the leukemia suppressor function of ICSBP. We have identified target genes involved in the phagocyte phenotype (CYBB and NCF2), DNA repair (FANCF), apoptosis sensitivity (PTPN13), and proliferation (NF1 and GAS2) (12, 34, 41). Therefore, ICSBP regulates multiple events during myelopoiesis, including phenotypic differentiation, proliferation arrest, and increased sensitivity to apoptosis. These results suggest that proliferation and differentiation are inversely regulated by a common mechanism, ICSBP activity (Fig. 10C). In other words, our studies suggest that ICSBP activity is integral to the selection between proliferation (via regulation of target genes such as NF1 and GAS2) and differentiation (via regulation of target genes such as CYBB, NCF2, TLR4, and others). These target genes also suggest mechanisms for leukemia suppression by ICSBP. In previous studies, we reported additional putative ICSBP target genes of potential significance to the pathogenesis of leukemia, including APC, PDCD10, and IRF2 (34). These genes will be the subject of future studies.
Although the development of specific kinase inhibitors has revolutionized the treatment of CML, drug resistance and BC remain significant clinical problems (7). Increased levels of β-catenin protein and activity in CML-LSC correlate with these adverse events. This increases the potential significance of the ICSBP-Gas2-calpain pathway for the regulation of β-catenin in CML. Dysregulation of this pathway may provide a molecular marker of disease progression in CML, suggesting the need for aggressive approaches like stem cell transplantation. Or, this pathway may identify targets for molecular therapeutic approaches for poor-risk patients.
Acknowledgments
This work was supported by National Institutes of Health grants number R01 HL088747 (to E.A.E.) and CA77816 (to L.C.P.), a Leukemia and Lymphoma Society of America Translational Research award (to E.A.E.), and Veteran's Administration Merit Review awards (to E.A.E. and L.C.P.).
Footnotes
Published ahead of print on 2 August 2010.
REFERENCES
- 1.Baba, Y., T. Yokota, H. Spits, K. P. Garrett, S. Hayashi, and P. W. Kincade. 2006. Constitutively active beta-catenin promotes expansion of multipotent hematopoietic progenitors in culture. J. Immunol. 177:2294-2303. [DOI] [PubMed] [Google Scholar]
- 2.Benetti, R., G. Del Sal, M. Monte, G. Paroni, C. Brancolini, and C. Schneider. 2001. The death substrate Gas2 binds m-calpain and increases susceptibility to p53-dependent apoptosis. EMBO J. 20:2702-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benetti, R., T. Copetti, S. Dell'Orso, E. Melloni, C. Brancolini, M. Monte, and C. Schneider. 2005. The calpain system is involved in the constitutive regulation of β-catenin signaling functions. J. Biol. Chem. 280:22070-22080. [DOI] [PubMed] [Google Scholar]
- 4.Chen, A. R., and L. R. Rohrschneider. 1993. Mechanism of differential inhibition of factor-dependent cell proliferation by transforming growth factor beta 1; selective uncoupling of FMS from MYC. Blood 81:2539-2546. [PubMed] [Google Scholar]
- 5.Coluccia, A. M., A. Vacca, M. Duñach, L. Mologni, S. Redaelli, V. H. Bustos, D. Benati, L. A. Pinna, and C. Gambacorti-Passerini. 2007. Bcr-Abl stabilizes beta-catenin in chronic myeloid leukemia through its tyrosine phosphorylation. EMBO J. 26:1456-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Santa, F., M. G. Totaro, E. Prosperini, S. Notarbartolo, G. Testa, and G. Natoli. 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130:1083-1094. [DOI] [PubMed] [Google Scholar]
- 7.Druker, B. J., F. Guilhot, O'Brien, S. G., I. Gathmann, H. Kantarjian, N. Gattermann, M. W. Deininger, R. T. Silver, J. M. Goldman, R. M. Stone, F. Cervantes, A. Hochhaus, B. L. Powell, J. L. Gabrilove, P. Rousselot, J. Reiffers, J. J. Cornelissen, T. Hughes, H. Agis, T. Fischer, G. Verhoef, J. Shepherd, G. Saglio, A. Gratwohl, J. L. Nielsen, J. P. Radich, B. Simonsson, K. Taylor, M. Baccarani, C. So, L. Letvak, and R. A. Larson. 2006. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 355:2408-2417. [DOI] [PubMed] [Google Scholar]
- 8.Eklund, E. A., A. Jalava, and R. Kakar. 1998. The transcription factors PU.1, IRF-1 and ICSBP cooperate to increase gp91-phox expression. J. Biol. Chem. 273:13957-13965. [DOI] [PubMed] [Google Scholar]
- 9.Eklund, E. A., and R. Kakar. 1999. Recruitment of CREB-binding protein by PU.1, IFN-regulatory factor 1, and the IFN consensus sequence-binding protein is necessary for IFNγ induced p67phox and gp91phox expression. J. Immunol. 163:6095-6105. [PubMed] [Google Scholar]
- 10.Hao, S. X., and R. Ren. 2000. Expression of interferon consensus sequence binding protein (ICSBP) is down regulated in Bcr-Abl-induced murine chronic myelogenous leukemia-like disease, and forced coexpression of ICSBP inhibits Bcr-Abl-induced myeloproliferative disorder. Mol. Cell. Biol. 20:1149-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holtschke, T., J. Löhler, Y. Kanno, T. Fehr, N. Giese, F. Rosenbauer, J. Lou, K. P. Knobeloch, L. Gabriele, J. F. Waring, M. F. Bachmann, R. M. Zinkernagel, H. C. Morse III, K. Ozato, and I. Horak. 1996. Immuno-deficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87:307-317. [DOI] [PubMed] [Google Scholar]
- 12.Huang, W., C. Zhu, H. Wang, E. Horvath, and E. A. Eklund. 2008. The interferon consensus sequence-binding protein (ICSBP/IRF8) represses PTPN13 gene transcription in differentiating myeloid cells. J. Biol. Chem. 283:7921-7935. [DOI] [PubMed] [Google Scholar]
- 13.Huang, W., C. Zhu, G. Saberwal, E. Horvath, S. Lindsey, and E. A. Eklund. 2006. Leukemia associated, constitutively active mutants of SHP2 protein tyrosine phosphatase inhibit NF1-transcriptional activation by the interferon consensus sequence binding protein. Mol. Cell. Biol. 26:6311-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamieson, C. H., L. E. Ailles, S. J. Dylla, M. Muijtjens, C. Jones, J. L. Zehnder, J. Gotlib, K. Li, M. G. Manz, A. Keating, C. L. Sawyers, and I. L. Weissman. 2004. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351:657-667. [DOI] [PubMed] [Google Scholar]
- 15.Janssen, J. J., S. M. Klaver, Q. Waisfisz, G. Pasterkamp, D. P. de Kleijn, G. J. Schuurhuis, and G. J. Ossenkoppele. 2005. Identification of genes potentially involved in disease transformation in CML. Leukemia 19:998-1004. [DOI] [PubMed] [Google Scholar]
- 16.Kamijo, T., K. Koike, Y. Nakazawa, K. Takeuchi, E. Ishii, and A. Komiyama. 2002. Synergism between SCF and GM-CSF on cell proliferation by induction of cyclins. Cytokine 19:267-275. [DOI] [PubMed] [Google Scholar]
- 17.Knepper-Nicolai, B., J. Savill, and S. B. Brown. 1998. Constitutive apoptosis in human neutrophils requires synergy between calpains and the proteasome downstream of caspases. J. Biol. Chem. 273:30530-30536. [DOI] [PubMed] [Google Scholar]
- 18.Kondo, Y., L. Shen, A. S. Cheng, S. Ahmed, Y. Boumber, C. Charo, T. Yamochi, T. Urano, K. Furukawa, B. Kwabi-Addo, D. L. Gold, Y. Sekido, T. H. Huang, and J. P. Issa. 2008. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat. Genet. 40:741-750. [DOI] [PubMed] [Google Scholar]
- 19.Konieczna, I., E. Horvath, H. Wang, S. Lindsey, G. Saberwal, L. Bei, L. Platanias, and E. A. Eklund. 2008. Constitutive activation of SHP2 in mice cooperates with ICSBP deficiency to accelerate progression to acute myeloid leukemia. J. Clin. Invest. 118:853-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 21.Kunimatsu, M., S. Higashiyama, K. Sato, I. Ohkubo, and M. Sasaki. 1989. Calcium dependent cysteine proteinase is a precursor of a chemotactic factor for neutrophils. Biochem. Biophys. Res. Commun. 164:875-882. [DOI] [PubMed] [Google Scholar]
- 22.Kuwata, T., C. Gongora, Y. Kanno, K. Sakaguchi, T. Tamura, T. Kanno, V. Basrur, R. Martinez, E. Appella, T. Golub, and K. Ozato. 2002. Gamma interferon triggers interaction between ICSBP (IRF8) and TEL, recruiting the histone deacetylase HDAC3 to the interferon responsive element. Mol. Cell. Biol. 22:7439-7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larrick, J. W., D. G. Fischer, S. J. Anderson, and H. S. Koren. 1980. Characterization of a human macrophage-like cell line stimulated to differentiate in vitro: a model of macrophage functions. J. Immunol. 125:6-14. [PubMed] [Google Scholar]
- 24.Lund, A. H., and M. van Lohuizen. 2004. Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 16:239-246. [DOI] [PubMed] [Google Scholar]
- 25.Michor, F., T. P. Hughes, Y. Iwasa, S. Branford, N. P. Shah, C. L. Sawyers, and M. A. Nowak. 2005. Dynamics of chronic myeloid leukaemia. Nature 435:1267-1270. [DOI] [PubMed] [Google Scholar]
- 26.Nelson, N., M. S. Marks, P. H. Driggers, and K. Ozato. 1993. Interferon consensus sequence binding protein, a member of the interferon regulatory factor family, represses interferon-induced gene transcription. Mol. Cell. Biol. 13:588-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberley, M. J., and P. J. Farnham. 2003. Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 371:577-596. [DOI] [PubMed] [Google Scholar]
- 28.Otten, H. G., W. G. van Ginkel, A. Hagenbeek, and E. J. Petersen. 2004. Prevalence and clinical significance of resistance to perforin- and FAS-mediated cell death in leukemia. Leukemia 18:1401-1405. [DOI] [PubMed] [Google Scholar]
- 29.Pear, W. S., J. P. Miller, L. Xu, J. C. Pui, B. Soffer, R. C. Quackenbush, A. M. Pendergast, R. Bronson, J. C. Aster, M. L. Scott, and D. Baltimore. 1998. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 92:3780-3792. [PubMed] [Google Scholar]
- 30.Pontremoli, S., and E. Melioni. 1988. The role of calpain and protein kinase C in activation of human neutrophils. Prog. Clin. Biol. Res. 282:195-208. [PubMed] [Google Scholar]
- 31.Pontremoli, S., E. Melloni, M. Michetti, O. Sacco, F. Salamino, B. Sparatore, and B. L. Horecker. 1986. Biochemical responses in activated human neutrophils mediated by PKC and a Ca2+-requiring proteinase. J. Biol. Chem. 261:8309-8313. [PubMed] [Google Scholar]
- 32.Qian, Z., A. A. Fernald, L. A. Godley, R. A. Larson, and M. M. Le Beau. 2002. Expression profiling of CD34+ hematopoietic stem/progenitor cells reveals distinct subtypes of therapy-related acute myeloid leukemia. Proc. Natl. Acad. Sci. U. S. A. 99:14925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramaraj, P., H. Singh, N. Niu, S. Chu, M. Holtz, J. K. Yee, and R. Bhatia. 2004. Effect of mutational inactivation of tyrosine kinase activity on Bcr/abl-induced abnormalities in cell growth and adhesion in human hematopoietic progenitors. Cancer Res. 64:5322-5331. [DOI] [PubMed] [Google Scholar]
- 34.Saberwal, G., E. Horvath, L. Hu, C. L. Zhu, E. Hjort, and E. A. Eklund. 2009. The interferon consensus sequence binding protein (ICSBP/IRF8) activates transcription of the FANCF gene during myeloid differentiation. J. Biol. Chem. 284:33242-33254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt, M., S. Nagel, J. Proba, C. Thiede, M. Ritter, J. F. Waring, F. Rosenbauer, D. Huhn, B. Wittig, I. Horak, and A. Neubauer. 1998. Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemias. Blood 91:22-29. [PubMed] [Google Scholar]
- 36.Schmidt, M., M. Schmidt, A. Hochhaus, A. Nitsche, R. Hehlmann, and A. Neubauer. 2001. Expression of nuclear transcription factor interferon consensus sequence binding protein in chronic myeloid leukemia correlates with pretreatment risk features and cytogenetic response to interferon-alpha. Blood 97:3648-3650. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, C., R. King, and L. Philipson. 1988. Genes specifically expressed at growth arrest of mammalian cells. Cell 54:787-793. [DOI] [PubMed] [Google Scholar]
- 38.Sharf, R., D. Meraro, A. Azriel, A. M. Thornton, K. Ozato, E. F. Petricoin, A. C. Larner, F. Schaper, H. Hauser, and B. Z. Levi. 1997. Phosphorylation events modulate the ability of interferon consensus sequence binding protein to interact with interferon regulatory factors and bind DNA. J. Biol. Chem. 272:9785-9792. [DOI] [PubMed] [Google Scholar]
- 39.Trost, M., L. English, S. Lemieux, M. Courcelles, M. Desjardins, and P. Thibault. 2009. The phagosomal proteome in interferon γ activated macrophages. Immunity 30:143-154. [DOI] [PubMed] [Google Scholar]
- 40.Ysebaert, L., G. Chicanne, C. Demur, F. De Toni, N. Prade-Houdellier, J. B. Ruidavets, V. Mansat-De Mas, F. Rigal-Huguet, G. Laurent, B. Payrastre, S. Manenti, and C. Racaud-Sultan. 2006. Expression of beta-catenin by acute myeloid leukemia cells predicts enhanced clonogenic capacities and poor prognosis. Leukemia 20:1211-1216. [DOI] [PubMed] [Google Scholar]
- 41.Zhu, C., G. Saberwal, Y. F. Lu, L. C. Platanias, and E. A. Eklund. 2004. The interferon consensus sequence binding protein (ICSBP) activates transcription of the gene encoding Neurofibromin 1 (NF1). J. Biol. Chem. 279:50874-50885. [DOI] [PubMed] [Google Scholar]