Abstract
Activation of DNA damage checkpoints requires the rapid accumulation of numerous factors to sites of genomic lesions, and deciphering the mechanisms of this targeting is central to our understanding of DNA damage response. Histone modification has recently emerged as a critical element for the correct localization of damage response proteins, and one key player in this context is the fission yeast checkpoint mediator Crb2. Accumulation of Crb2 at ionizing irradiation-induced double-strand breaks (DSBs) requires two distinct histone marks, dimethylated H4 lysine 20 (H4K20me2) and phosphorylated H2AX (pH2AX). A tandem tudor motif in Crb2 directly binds H4K20me2, and this interaction is required for DSB targeting and checkpoint activation. Similarly, pH2AX is required for Crb2 localization to DSBs and checkpoint control. Crb2 can directly bind pH2AX through a pair of C-terminal BRCT repeats, but the functional significance of this binding has been unclear. Here we demonstrate that loss of its pH2AX-binding activity severely impairs the ability of Crb2 to accumulate at ionizing irradiation-induced DSBs, compromises checkpoint signaling, and disrupts checkpoint-mediated cell cycle arrest. These impairments are similar to that reported for abolition of pH2AX or mutation of the H4K20me2-binding tudor motif of Crb2. Intriguingly, a combined ablation of its two histone modification binding modules yields a strikingly additive reduction in Crb2 activity. These observations argue that binding of the Crb2 BRCT repeats to pH2AX is critical for checkpoint activity and provide new insight into the mechanisms of chromatin-mediated genome stability.
DNA damage response is an essential cellular guard that protects the genetic material from a constant barrage of genotoxic agents. To ensure their survival after genomic insult, cells orchestrate a signaling cascade that leads to checkpoint-mediated cell cycle arrest and the repair of damaged DNA (16, 35). A failure in this process can have catastrophic cellular consequences leading to the development of numerous disorders such as cancer (18, 30, 32). Because of its intimate connection with human health, deciphering the molecular mechanisms of DNA damage response is of high interest (16, 20).
Recently, histone posttranslational modification has emerged as one element that is critical for ensuring a faithful response to genomic challenge (7, 31). An octamer of the four core histones, H3, H4, H2A, and H2B, forms the core protein component of chromatin, and cells possess a considerable number of enzymes that target histones for posttranslation modification (21). These marks can impinge upon many aspects of DNA biology by acting to directly alter chromatin structure or by serving as a binding scaffold for the recruitment of regulatory factors (24).
In the context of DNA damage response, one factor that is intimately linked with histone modification is the fission yeast DNA damage checkpoint protein Crb2. After genomic insult, DNA damage checkpoints function to halt cell cycle progression, ensuring sufficient time for lesion repair (16, 35). In the fission yeast Schizosaccharomyces pombe, regulating the transition from G2 to mitosis (G2/M) represents the major DNA damage checkpoint and Crb2 is essential for this activity (4, 34). Crb2 is a member of a family of checkpoint regulators that have been termed mediators because they are thought to transmit the checkpoint signal from damage-sensing ATM/ATR-related kinases to effector kinases, such as Chk1, that trigger cell cycle arrest (11, 25). Crb2 is closely related to budding yeast Rad9 and mammalian p53 binding protein 53BP1, which all share two distinct domains, a tandem tudor motif and a pair of C-terminal BRCT repeats (Fig. 1A) (11, 25). Besides 53BP1, Crb2 also shares some functional similarities with other mammalian BRCT-containing checkpoint regulators, such as MDC1 and BRCA1 (11, 25). In response to ionizing irradiation (IR), the rapid accumulation of Crb2 and other checkpoint proteins can be readily visualized as nuclear foci that mark sites of double-strand breaks (DSBs) (9, 25). Understanding the mechanisms that govern this targeting has been an area of intense interest, and for Crb2 this accumulation requires two distinct histone marks: dimethylation of histone H4 lysine 20 (H4K20me2) and phosphorylated H2AX (pH2AX) (27, 36).
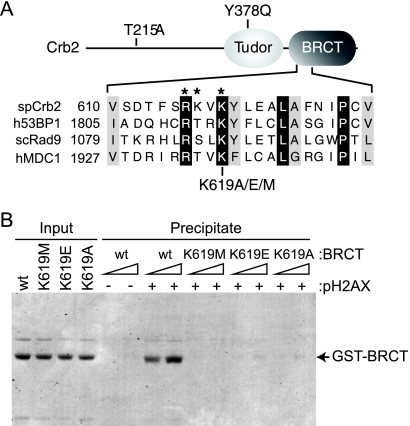
FIG. 1.
Crb2 pH2AX-binding mutations. (A) Top, schematic representation of Crb2 (not drawn to scale) with relevant mutations indicated. Bottom, protein sequence alignment of a portion of the BRCT phospho-binding motifs from Schizosaccharomyces pombe (sp) Crb2, human (h) 53BP1, human MDC1, and Saccharomyces cerevisiae (sc) Rad9. Identical residues are shaded black; similar residues are shaded gray. *, Crb2 phospho-binding residues. (B) The Crb2 BRCT domains specifically interact with pH2AX. Peptide pulldowns were performed as described in the text with C-terminal fission yeast H2A.1 peptides either unmodified or phosphorylated at Ser129 (see − or + pH2AX) and increasing amounts of the indicated recombinant Crb2 BRCT domain fragments (∼0.1 and 0.3 μM). After binding and washing, SDS-PAGE and Coomassie staining were used to visualize peptide-bound protein. A fraction of the total protein used for binding was also visualized (Input).
Mono-, di-, and trimethyl H4K20 are conserved chromatin marks that are readily detectable in fission yeast and mammalian cells (29, 36). In fission yeast, the Kmt5 methylase catalyzes all three H4K20 methyl modifications and its inactivation, or mutation of its H4K20 substrate, severely diminishes Crb2 accumulation at DSBs and compromises checkpoint activity (10, 36). Note that as outlined by the unified nomenclature for the naming of histone lysine methyltransferases (2), the fission yeast H4K20 methylase previously known as Set9 (36) is now termed Kmt5. The requirement for H4K20 methylation is mediated by the tandem tudor domains of Crb2 that preferentially bind H4 tail peptides dimethylated at lysine 20 (3, 14). Tudor motif mutations impair Crb2 DSB targeting and genome integrity in a manner analogous to loss of Kmt5 activity, and dimethylation of H4K20, but not trimethylation, is required for Crb2 activity (10, 14, 42). The tudor domain of 53BP1 can also directly bind H4K20me2, and this recognition event is required for its accumulation at IR-induced DSBs (3, 23, 45).
After DNA damage, serine 139 phosphorylation in the mammalian H2A variant H2AX, or a homologous site in canonical yeast H2A, specifically marks sites of genomic lesions (7, 12). The fission yeast genome encodes two H2A proteins, H2A.1 and H2A.2, which differ slightly in their primary amino acid sequence. Phosphorylation of S129 in H2A.1 and S128 in H2A.2 is collectively referred to as phosphorylated H2AX (pH2AX). The ATM/ATR family of PI3-like kinases that includes the fission yeast Rad3 and Tel1 enzymes catalyzes pH2AX (37). H2AX phosphorylation has a critical role in controlling both DNA repair and checkpoint activation in a variety of organisms from yeast to humans (7, 12). Central to its function is the ability of the pH2AX mark to coordinate the recruitment of a number of proteins to genomic lesions, and several factors can directly bind the modification (40). Serine-to-alanine substitutions at the H2AX phosphorylation site in fission yeast H2A (h2ax−) severely reduce Crb2 accumulation at IR-induced DSBs and compromise the ability of cells to maintain checkpoint cell cycle arrest in a manner very similar to loss of H4K20 methylation (10, 27).
The mechanism underlying the control of Crb2 DSB targeting and checkpoint activation by pH2AX is not understood. Because BRCT domains are known phospho-binding motifs (13), the initial demonstration that pH2AX is required for Crb2 function suggested that direct binding to the modification by Crb2 is critical for checkpoint activity (27). Supporting this idea, it has been demonstrated that the Crb2 BRCT repeats directly and specifically bind pH2AX peptides (22). Structural and biochemical studies have also identified a conserved pH2AX-binding motif in the BRCT repeats of Crb2, budding yeast Rad9, and human MDC1 and 53BP1 (Fig. 1A) (15, 22, 39). As would be expected, mutation of Crb2's critical phospho-binding motif impairs cell survival after DNA damage (22). Unexpectedly though, loss of its pH2AX-binding activity did not significantly affect the ability of Crb2 to localize to IR-induced DSBs (22). Rather, mutation of the Crb2 pH2AX-binding motif altered the kinetics of Rad22 accumulation at DSBs and triggered a prolonged checkpoint arrest after IR exposure (22). From these observations it was suggested that binding of the Crb2 BRCT repeats to pH2AX is critical for aspects of DNA repair but is not central to Crb2 targeting and checkpoint activity (22).
The apparent dispensability of its pH2AX-binding motif in controlling Crb2 localization to IR-induced DSBs (22) was a surprising observation because of the established requirement for the pH2AX modification (10, 27). The extended checkpoint delay seen in Crb2 pH2AX-binding mutants (22) was also unexpected because h2ax− cells cannot maintain checkpoint-mediated cell cycle arrest (10, 27). The prolonged checkpoint arrest was also surprising because a defect in IR-induced Chk1 phosphorylation was observed in the same Crb2 pH2AX-binding mutants (22). For these reasons we sought to reevaluate the requirement for the pH2AX-binding module of Crb2 in controlling DNA damage checkpoint activity. We demonstrate that the critical phospho-coordinating residue of Crb2 is required for binding to pH2AX peptides, Crb2 accumulation at IR-induced DSBs, cell survival after DNA damage, and maintenance of checkpoint-mediated cell cycle arrest. The observed impairments are similar to that reported for abolishment of pH2AX or mutation of the H4K20me2 binding tudor motif of Crb2. Strikingly, a combined ablation of the two modification binding modules of Crb2 produces an additive impairment in checkpoint dysfunction and genome integrity. These results argue that recognition of pH2AX by its BRCT repeats is critical for Crb2 accumulation at genomic lesions and its subsequent checkpoint activity. These observations also corroborate the independent findings of Sofueva et al. (38), who have observed a similar requirement for Crb2 binding to pH2AX in controlling DSB targeting and checkpoint activity.
MATERIALS AND METHODS
Strains and manipulations.
Relevant strains are listed in Table 1. Yeast cell growth, DNA damage phenotyping, DNA damage checkpoint assays, localization experiments, and generation of mutants were performed essentially as described previously (14, 36).
TABLE 1.
Fission yeast strains
| Strain | Genotype |
|---|---|
| YSLS702 | h−crb2Δkan leu1-32::leu1+pJK148- REP81-GFP-crb2+ |
| YSLS706 | h−crb2Δkan leu1-32::leu1+ pJK148- REP81-GFP-crb2K619M |
| YSLS707 | h−crb2Δkan leu1-32::leu1+pJK148- REP81-GFP-crb2K619A |
| YSLS738 | h−crb2Δkan leu1-32::leu1+pJK148- REP81-GFP-crb2K619E |
| YSLS710 | h− |
| YSLS711 | h−crb2T215A |
| YSLS712 | h−crb2Y378Q |
| YSLS714 | h−crb2K619M |
| YSLS715 | h−crb2K619A |
| YSLS716 | h−crb2Δkan |
| YSLS717 | h−crb2Y378Q-K619M |
| YSLS718 | h−kmt5Δkan |
| YSLS719 | h−kmt5Δkan crb2T215A |
| YSLS722 | h−kmt5Δkan crb2K619M |
| YSLS725 | h−crb2T215A-Y378Q |
| YSLS726 | h−crb2T215A-K619M |
| YSLS727 | h−crb2T215A-Y378Q-K619M |
| YSLS747 | h−crb2K619E |
| YARA36 | h−chk1-HA |
| YARA37 | h−chk1-HA crb2T215A |
| YARA38 | h−chk1-HA crb2Y378Q |
| YARA40 | h−chk1-HA crb2K619M |
| YARA41 | h−chk1-HA crb2Δkan |
| YARA42 | h−chk1-HA crb2Y378Q-K619M |
| YSLS762 | h−chk1-HA crb2K619A |
| YSLS764 | h−chk1-HA crb2K619E |
| YSLS778 | h−wee1-50 |
| YSLS779 | h−wee1-50 crb2Δkan |
| YSLS780 | h−wee1-50 crb2K619M |
| YSLS781 | h−wee1-50 crb2K619A |
| YSLS782 | h−wee1-50 crb2K619E |
Binding experiments.
N-terminally tagged glutathione S-transferase (GST) expression constructs for wild-type (WT) and mutant Crb2 BRCT domain proteins (amino acids 521 to 778) were generated by PCR cloning. Expression and purification of GST-tagged proteins from Escherichia coli was performed essentially as described previously (17). Fission yeast H2A.1 peptides (CRTGKPSQEL) encompassing amino acids 124 to 132 plus an N-terminal cysteine, with S129 either unmodified or phosphorylated, were cross-linked to SulfoLink resin (Pierce). Five μg of immobilized peptides was incubated with purified BRCT protein (∼0.1 and 0.3 μM final concentration) in binding buffer (25 mM Tris-HCl, pH 7.0, 1 mM dithiothreitol [DTT], 1 mM EDTA, 0.2 M NaCl, 0.5% NP-40) for 1 h at 4°C. After binding, the resin was washed with binding buffer and precipitated protein was subjected to SDS-PAGE and Coomassie staining.
Antibodies and Western blotting.
For quantitative analysis of Chk1 phosphorylation, cells were harvested from log-phase cultures grown at 30°C in rich media and IR irradiated at ∼10 Gy per minute, and cell pellets (4 × 107 cells) were frozen on dry ice. Frozen pellets were resuspended in 200 μl of SDS-PAGE load buffer, lysed with glass beads in a FastPrep FP120 (Q-Biogene), and immediately heated at 90°C for 5 min. After a brief centrifugation, samples were subjected to SDS-PAGE and transferred to a low-fluorescence nitrocellulose membrane (Li-Cor Bioscience). Membranes were blocked in TBST (Tris-buffered saline with 0.05% Tween 20) containing 2% ECL advance blocking reagent (GE), incubated with anti-HA (Santa Cruz Biotechnology, sc-7396) overnight at 4°C, briefly washed with TBST, incubated with Alexa Fluor 488 conjugated secondary antibody (Invitrogen) for 1 h at ambient temperature, washed extensively with TBST and briefly with TBS, and imaged on a Typhoon 8600 (GE). The ratio of the phosphorylated to unphosphorylated Chk1 bands was then calculated using Quantity One software (Bio-Rad).
To monitor Crb2 protein expression, extracts were prepared as detailed above and standard Western blotting was performed with polyclonal anti-Crb2 antibody (9) and anti-histone H3 antibody (Abcam ab1971).
RESULTS
Crb2 pH2AX-binding mutations.
To reinvestigate the requirement for pH2AX recognition in controlling Crb2 function, we first sought to generate several crb2 alleles deficient in phospho-binding activity. We focused our studies on Lys619 of Crb2 because this residue has a conserved role in coordinating pH2AX in the BRCT repeats of Crb2, Rad9, and MDC1 (Fig. 1A) (15, 22, 39). The phospho-binding capability of this lysine residue is also conserved in the BRCA1 BRCT repeats that bind phosphorylated BACH1 (6, 39). To allow a direct comparison with published observations, we generated the same K619E substitution used by Kilkenny et al. in their characterization of the Crb2 BRCT motif (22). A K619M mutation was also utilized because methionine substitutions were used to examine the phospho-binding lysine in the Rad9 and MDC1 BRCT repeats (15, 39). As a third allele, a K619A substitution was also generated.
We first investigated the pH2AX-binding activity of the three different Lys619 substitutions using a peptide-binding assay (Fig. 1B). Purified recombinant WT and mutant GST-tagged BRCT domain proteins were incubated with immobilized C-terminal H2A.1 peptides either unmodified or phosphorylated at Ser129 (see − or + pH2AX, Fig. 1B). After being bound and washed, peptide-bound protein was visualized by SDS-PAGE. As expected, WT BRCT protein efficiently bound the phosphorylated peptide but not the unmodified peptide (Fig. 1B). Importantly, this interaction was severely diminished by all three Lys619 substitutions (Fig. 1B). These results indicate that the Crb2 BRCT repeats can specifically bind pH2AX and that all three crb2K619 mutations significantly reduce this interaction.
The pH2AX-coordinating residue of Crb2 is required for DSB targeting.
After IR exposure, Crb2 rapidly accumulates at sites of DSBs in a pH2AX-dependent fashion (9, 27) and its BRCT repeats can directly bind the same modification (Fig. 1B) (22). To examine the requirement of this interaction in Crb2 DSB targeting, live cell imaging was used to measure the ability of the three Crb2K619 mutants to localize to IR-induced DSBs (Fig. 2A). In this assay a GFP-tagged crb2 allele, under the control of the moderately expressed nmt81 promoter, is integrated into the fission yeast genome as the sole source of the protein (36). Cells were grown in minimal media and processed for imaging either immediately before (Fig. 2A, 0 Gy) or immediately after (Fig. 2A, 36 Gy) IR exposure. To examine the stability of Crb2 foci, irradiated cells were allowed to recover for 1.5 h before being imaged (Fig. 2A, 1.5 h Post 36 Gy). Figure 2A demonstrates that WT GFP-Crb2 localized throughout the nucleus before irradiation, but distinct foci readily formed at sites representing DSBs after irradiation (9). In contrast, cells harboring any of the three Lys619 substitutions displayed a dramatic reduction in focal accumulation both immediately and 1.5 h after irradiation (Fig. 2A). These results indicate that the pH2AX-binding activity of Crb2 is required for its accumulation at IR-induced DSBs.
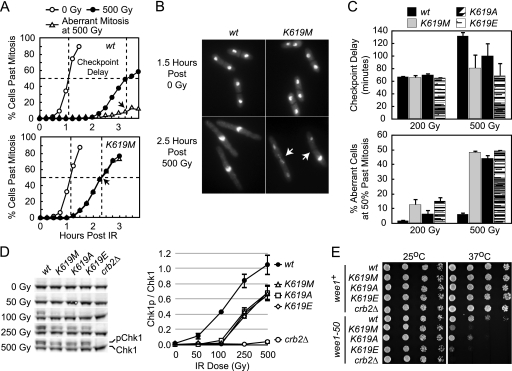
FIG. 2.
The pH2AX-binding motif of Crb2 is required for accumulation at IR-induced DSBs and cell survival after DNA damage. (A) Strains harboring a GFP-tagged crb2 allele were processed for live cell imaging either immediately before (0 Gy), immediately after (36 Gy), or 1.5 h after (1.5 h Post 36 Gy) IR exposure. Left shows a panel of representative images from cells either untreated (0 Gy) or immediately after irradiation (36 Gy). Right, quantification of nuclei with one or more Crb2 foci. Data are averaged from results of three independent experiments in which 200 to 300 cells were counted for each point. (B and C) Crb2 Lys619 mutations induce hypersensitivity to DNA damage. (B) Strains harboring the indicated mutation at the endogenous crb2 allele were grown in liquid culture, and fivefold serial dilutions were spotted onto either rich medium or rich medium containing the indicated amounts of CPT or HU. Cells spotted onto rich medium were then untreated (control) or irradiated with the indicated dose of IR (in grays [Gy]) or UV (in joules [J]/m2). All plates were then incubated at 30°C. For panel C, quantitative IR survival curves were performed. Log-phase cells grown in liquid culture at 30°C were treated with the indicated amount of IR (x axis), and 600 to 800 cells were plated in duplicate onto rich medium plates. Colony formation was measured after incubation at 30°C, and survival was plotted relative to the unirradiated 0 Gy sample (y axis). Data are averaged from results of three independent experiments with a standard error of 10% or less. (D) Expression of mutant Crb2 proteins. Total cell lysates were prepared from the top-labeled strains and processed for Western blotting using the indicated (left) polyclonal antibodies.
The pH2AX-binding activity of Crb2 is required for cell survival after DNA damage.
We next examined the requirement for the Crb2 pH2AX-binding module in controlling genome stability after DNA damage. The K619M, K619E, and K619A mutations were introduced into the endogenous crb2 allele, and DNA damage phenotyping was used to examine genome stability. Figures 2B and C demonstrate that cells harboring each of the crb2K619 mutations displayed a similar pattern of damage sensitivity. Compared to WT cells, these mutants are hypersensitive to IR, UV light (UV), and the topoisomerase I poison camptothecin (CPT). The levels of hypersensitivity of the crb2K619M and crb2K619E mutants were very similar, but the crb2K619A mutation yielded a moderately less severe phenotype (Fig. 2B and C). All three pH2AX-binding mutants only partially inactivated Crb2 function, as indicated by the more pronounced hypersensitivity seen with a complete loss of crb2+ (see Fig. 2B, crb2Δ). The crb2K619 mutants were also only very weakly sensitive to the replication inhibitor hydroxyurea (HU), in contrast to the significant sensitivity of crb2Δ cells (Fig. 2B). Importantly, immunoblotting with polyclonal Crb2 antibody revealed that all three mutants were expressed at levels similar to that of WT Crb2 protein (Fig. 2D). We conclude that the Crb2 pH2AX-binding activity is required for efficient cell survival after genotoxic challenge.
DNA damage checkpoint control requires Crb2 pH2AX-binding activity.
We next examined the activity of the G2/M DNA damage checkpoint in the substitution mutants (Fig. 3). IR-induced cell cycle arrest was first examined by synchronizing cells in G2 with size separation and then monitoring their rate of mitotic progression by DAPI staining after either no treatment or exposure to IR. A representative experiment comparing WT and crb2K619M cells after either 0 or 500 Gy of IR is shown in Fig. 3A. These data demonstrate that both WT and crb2K619M cells progressed through mitosis at a similar rate in the absence of irradiation. In contrast, after IR exposure crb2K619M cells prematurely released into mitosis ∼45 minutes prior to WT cells and the vast majority of these early-releasing cells displayed significant fragmentation and/or missegregation of nuclei (Fig. 3A and B). This catastrophic mitosis is indicative of cells prematurely entering the cell cycle with damaged DNA. A similar impairment in checkpoint maintenance was also observed in crb2K619A and crb2K619E cells (data not shown). To provide a quantitative assessment of checkpoint dysfunction in all three crb2K619 mutants, we determined the checkpoint delay after exposure to 200 and 500 Gy of IR by measuring the time difference required for 50% of the cells to pass mitosis with or without irradiation (Fig. 3A, dashed lines). We also determined the fraction of cells that underwent an aberrant mitosis at the time point at which 50% of the cells had passed mitosis after irradiation (see arrowheads in Fig. 3A). This analysis revealed that after 500 Gy of IR all three crb2K619 mutations displayed a reduction in checkpoint delay and a dramatic increase in aberrant mitosis relative to WT cells (Fig. 3C). Consistent with phenotypic observations (Fig. 2B and C), the observed defects were least pronounced in crb2K619A cells. After 200 Gy of IR, a reduction in checkpoint delay was not readily apparent in crb2K619 cells, but these mutants did display a marked increase in aberrant mitosis compared to that of WT cells (Fig. 3C). This less severe level of checkpoint dysfunction is consistent with the lower hypersensitivity of crb2K619 mutants at 200 Gy versus 500 Gy of IR (Fig. 2C).
FIG. 3.
The pH2AX-binding activity of Crb2 is required for a fully functional G2/M DNA damage checkpoint. (A) DNA damage checkpoint assay. Strains were grown in rich media at 30°C and G2 synchronized by lactose gradient sedimentation. G2 cells were then either untreated (0 Gy) or IR exposed (500 Gy) and allowed to recover at 30°C in liquid media. Cell aliquots were taken every 15 min (time indicated on x axis) and methanol fixed, and mitotic progression was assessed by DAPI staining (y axis). Shown are representative data from a single experiment with WT and crb2K619M cells in which 200 to 300 cells were counted for each point. (B) Representative images of DAPI-stained WT and crb2K619M cells after exposure to either 0 or 500 Gy of IR (time indicated at left). Arrowheads denote the septa of two crb2K619M cells that have undergone aberrant mitosis as indicated by unequal segregation of nuclei (right cell) or nuclear fragmentation (left cell). (C) Quantification of checkpoint delay (top) and aberrant mitosis (bottom) in crb2K619 mutants. Checkpoint delay was determined by measuring the difference in time required for 50% of the cells to pass mitosis with or without irradiation (see dashed lines in panel A). Aberrant mitosis was calculated by plotting the percentage of aberrant cells after IR exposure and then determining the intercept at the time point for which 50% of cells had passed mitosis (see arrowheads in panel A). Data are averaged from results of three independent experiments in which all four strains were processed at 0 and 200 Gy or 0 and 500 Gy simultaneously. (D) Lys619 of Crb2 is required for optimal IR-induced Chk1 phosphorylation. Strains harboring chk1-HA3 and the labeled crb2 mutations (top) were treated with the indicated dose of IR (left) and immediately processed for quantitative Western blotting using anti-HA and a fluorescent secondary antibody (see Materials and Methods). After fluorescent imaging, the ratio of phosphorylated (p) versus unphosphorylated Chk1 was determined. The left shows a representative image from a single experiment, and data from two independent experiments are averaged and plotted with standard deviation on the right. (E) Synthetic sick interaction between the crb2K619 and wee1-50 mutations. Serial dilutions of strains with the left-labeled mutations were spotted onto rich media and grown at the indicated temperature.
To investigate checkpoint activity at the molecular level we examined IR-induced phosphorylation of the Chk1 effector kinase because this is a well-established marker for G2/M checkpoint activation (4). Asynchronous WT, crb2K619 mutants, and crb2Δ cells were IR treated and then immediately processed by quantitative fluorescent Western blotting to monitor Chk1 phosphorylation (see Materials and Methods). Figure 3D demonstrates that all three Lys619 substitutions impaired Chk1 activation over a broad range of IR doses. Consistent with phenotypic observations (Fig. 2B and C), this decrement was only a partial inactivation and contrasts with crb2Δ cells that are essentially completely defective in Chk1 phosphorylation (Fig. 3D). Note that unlike the checkpoint delay observations in Fig. 3C, where crb2K619A cells displayed a lower level of dysfunction, all three crb2K619 mutations yielded a similar reduction in Chk1 phosphorylation (Fig. 3D). We reason it is likely that the Chk1 phosphorylation assay does not have sufficient sensitivity to resolve such differences because asynchronous cells are used in this experiment. Together, the results of Fig. 3A to D indicate that the pH2AX-binding activity of Crb2 is required for optimal checkpoint signaling and maintenance of checkpoint-mediated cell cycle arrest.
Synthetic sick interaction between the crb2K619 and wee1-50 mutations.
To provide an independent assessment of checkpoint control in the crb2K619 mutants we next examined their genetic interaction with the Wee1 kinase. Wee1 acts as a negative regulator of cell cycle progression by inhibiting Cdc2 activity through Tyr15 phosphorylation (26). Although cells harboring a deletion of wee1+ are viable, the wee1Δ mutation is lethal when combined with checkpoint rad mutations (1, 33). In contrast, the wee1-50 temperature-sensitive allele is viable at the permissive temperature when coupled with checkpoint rad and chk1 mutations, but double mutants lose the ability to form colonies at the nonpermissive temperature (1, 33, 41). From these observations it has been argued that DNA damage checkpoint control becomes essential for cell viability in the absence of unrestrained Cdc2 activity. We reasoned that like checkpoint rad mutations, crb2 mutations would also display a synthetic sick interaction with wee1-50. To test this hypothesis the wee1-50 allele was combined with either the crb2Δ or the three different crb2K619 mutations and the abilities of these mutants to grow at the permissive (25°C) and nonpermissive (37°C) temperatures were assessed. Figure 3E demonstrates that all individual crb2 mutants grew similarly to WT cells at both the permissive and nonpermissive temperatures. As expected, wee1-50 cells grew similarly to WT cells at the permissive temperature but showed impaired growth at the nonpermissive temperature. Combining the wee1-50 and crb2 mutations produced a pronounced reduction in the ability of cells to form colonies at the nonpermissive temperature. Consistent with their partial inactivation of Crb2, crb2K619 mutations displayed a weaker interaction with wee1-50 than did crb2Δ (Fig. 3E). These observations indicate that combining the wee1-50 and crb2K619 mutations produces a synthetic sick interaction and provide independent genetic support for the defects in DNA damage checkpoint control observed in Fig. 3A to D.
A combined loss of H4K20me2 and pH2AX-binding activity additively impairs Crb2 function.
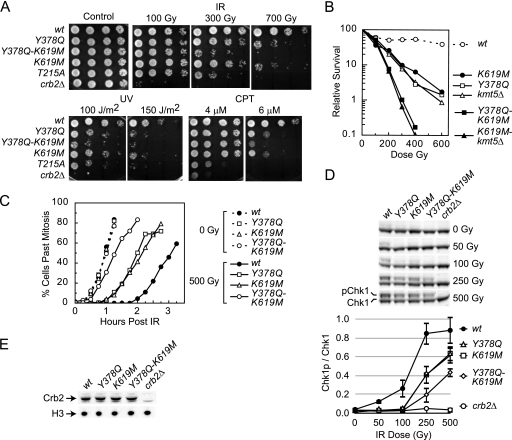
Because crb2K619 mutations only partially inactivate Crb2 (Fig. 2 and 3), we next explored whether a combined loss of H4K20me2 and pH2AX binding might further inactivate Crb2. To decipher the functional relationship between the two modification binding motifs, we compared cell survival after DNA damage and checkpoint activation in cells harboring the crb2Y378Q and crb2K619M mutations, either individually or in combination (Fig. 4). Tyr378 is one of five critical residues that make up the tudor methyl-lysine binding cage (3). We have previously demonstrated that the crb2Y378Q mutation disrupts H4K20me2 binding, diminishes Crb2 accumulation at IR-induced DSBs, and impairs checkpoint activity (14). Figures 4A and B show that inactivation of either the methyl-binding or phospho-binding motifs of Crb2 impairs cell survival after DNA damage to a similar degree (compare crb2Y378Q and crb2K619M). A similar impairment in both checkpoint-mediated cell cycle arrest and IR-induced Chk1 phosphorylation was also observed in each individual mutant (Fig. 4C and D). Interestingly, a combined loss of both binding activities (crb2Y378Q-K619M) produced a strikingly additive reduction in checkpoint activity and cell survival after DNA damage (Fig. 4A to D). The additive impairment in activity was not due to differences in Crb2 protein levels, because all mutants were expressed equally (Fig. 4E). Further, a similar additive effect was also observed when the crb2Y378Q and crb2K619A mutations were combined (data not shown). The severity of the defects in the double mutant were similar to that seen in crb2T215A cells that are defective in Cdc2-dependent Crb2 phosphorylation (references 5 and 28 and see below) but still less pronounced than that of crb2Δ cells. These observations indicate that a loss of either tudor or BRCT modification binding activity impairs Crb2 function similarly, but a combined ablation produces a marked increase in dysfunction.
FIG. 4.
A combined loss of H4K20me2 and pH2AX-binding activity impairs Crb2 function additively. (A and B) Phenotyping and IR survival curves were performed as described for Fig. 2B and C. IR data are averaged from results of three independent experiments with a standard error of 10% or less. (C and D) G2/M DNA damage checkpoint activity and Chk1 phosphorylation were monitored as detailed in Fig. 3A and D. The top of panel D shows a representative image from a single experiment, and data from two independent experiments are averaged and plotted with standard deviation on the bottom. (E) Crb2 protein levels in mutant strains. Strains with the top-labeled mutations were processed for Western blotting as described for Fig. 2D.
To probe the interaction between the two modification binding pathways further, we also combined the crb2K619M pH2AX-binding mutant with a loss of H4K20me2 by ablating the Kmt5 methylase (kmt5Δ). The crb2Y378Q and kmt5Δ mutations are genetically indistinguishable, whether introduced individually or in combination (14). From this it was expected that the crb2K619M and kmt5Δ mutants would display the same additive impairment in genome integrity as observed with the double crb2 mutant. This expectation was realized because the double crb2K619M-kmt5Δ mutant displayed IR hypersensitivity that was significantly more pronounced than that of either single mutant (Fig. 4B). Importantly, the severities and patterns of damage hypersensitivity for the double crb2K619M-kmt5Δ and crb2Y378Q-K619M mutants were essentially indistinguishable (Fig. 4B and data not shown). Predictably, a combined triple mutation was also equivalent to either double mutation (data not shown). These observations indicate that when combined, the crb2K619M and kmt5Δ mutations are additive in their inactivation of Crb2, similar to the crb2K619M and crb2Y378Q binding domain mutations.
Both histone modification binding modules are required for Crb2 activity in the absence of Thr215 phosphorylation.
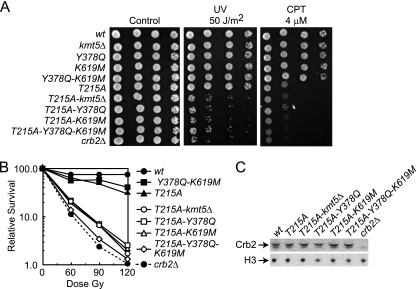
After DNA damage, Thr215 of Crb2 is hyperphosphorylated in a Rad3- and Cdc2-dependent fashion (5, 28). Ablation of this phosphorylation site with the crb2T215A mutation does not impair accumulation of Crb2 at IR-induced DSBs but does abolish targeting to a persistent HO-induced site-specific DSB and partially inactivates checkpoint activity (10, 28). Others and we have previously demonstrated that recognition of H4K20me2 becomes essential for Crb2 function in the absence of Thr215 phosphorylation (10, 14, 28, 36). When Thr215 phosphorylation is abolished, pH2AX is also indispensable for Crb2 activity, as crb2T215A-h2ax− cells display phenotypic defects that are similar to a complete deletion of crb2+ (28). We next asked if the pH2AX-binding motif of Crb2 is also essential for its activity in the absence of Thr215 phosphorylation. To address this question, a double crb2T215A-K619M mutant was generated and its sensitivity to DNA damage was compared to that of each individual mutant. Figure 5A and B demonstrate that the double mutation yields a near-complete loss of Crb2 function because crb2T215A-K619M cells are almost as hypersensitive to DNA damage as crb2Δ cells. This additive impairment in Crb2 function is identical to that observed when the crb2T215A mutation is combined with either the kmt5Δ or crb2Y378Q mutations. Again, these additive interactions are not due to differing levels of Crb2 protein because all mutants are expressed equally well (Fig. 5C). Interestingly, a mutant with a combined triple ablation of Thr215 phosphorylation and both modification binding modules (crb2T215A-Y378Q-K619M) was modestly more sensitive to IR than either double mutant (Fig. 5B). This is consistent with the additive nature of the crb2K619M and crb2Y378Q mutations (Fig. 4). The results in Fig. 5 indicate that the pH2AX-binding activity of Crb2 is essential for its function in the absence of Thr215 phosphorylation.
FIG. 5.
Both histone modification binding modules are required for Crb2 activity in the absence of Thr215 phosphorylation. (A and B) Phenotyping and IR survival curves were performed with strains harboring the indicated mutations as described for Fig. 2B and C. IR data are averaged from results of three independent experiments with a standard error of 10% or less. (C) Crb2 protein levels in mutant strains. Strains with the top-labeled mutations were processed for Western blotting as described for Fig. 2D.
DISCUSSION
H2AX phosphorylation plays a central role in coordinating fission yeast checkpoint activation and genome stability. A key aspect of this role is the ability of pH2AX to control the targeting and activity of the checkpoint mediator Crb2, but the mechanism underlying this control has been unclear. Initial studies suggested that a direct interaction between its BRCT repeats and pH2AX may control Crb2 activity (27). The recent demonstration that the Crb2 BRCT repeats can directly bind the modification through a conserved pH2AX-binding motif has added support to this idea (22). We have extended this work by demonstrating that amino acid substitutions in its critical pH2AX-coordinating residue severely diminish the ability of Crb2 to accumulate at IR-induced DSBs (Fig. 2). This Crb2 targeting defect is similar to that shown for h2ax− cells that are deficient in pH2AX (10, 27). The requirement for its pH2AX-binding motif in DSB targeting is also consistent with the demonstration that a heterologous dimerization motif can substitute for the majority of BRCT function but cannot mediate Crb2 accumulation at IR-induced DSBs (8). The Crb2 pH2AX-binding mutants are also hypersensitive to genotoxic challenge, display an inability to maintain cell cycle arrest mediated by the G2/M DNA damage checkpoint, and impair checkpoint signaling (Fig. 2 and 3). These defects are less severe than a complete deletion of crb2+ and indicate that loss of pH2AX binding only partially inactivates Crb2. This partial inactivation is also comparable to that observed after ablation of pH2AX, because h2ax− cells display similar defects in genome stability and checkpoint activity (10, 27). Consistent with the observed defects in DNA damage checkpoint control, we have also demonstrated a synthetic sick interaction between the crb2K619 and wee1-50 mutations (Fig. 3E). Further, as with the pH2AX modification, we have also shown that the pH2AX-binding activity of Crb2 is essential for its function in the absence of Thr215 phosphorylation (Fig. 5). This underscores the genetic similarities between the pH2AX-binding BRCT motif of Crb2 and the pH2AX chromatin mark. Together these observations argue that direct recognition of pH2AX by its BRCT repeats is a key mechanism for controlling Crb2 targeting and checkpoint activity. Our findings are also corroborated by the independent work of Sofueva et al. (38), who have observed a similar requirement for the pH2AX-binding activity of Crb2 in DSB targeting and checkpoint control.
As discussed in the introduction, these studies were prompted by a previous report suggesting that the pH2AX-binding activity of Crb2 is not required for DSB targeting or maintenance of checkpoint-mediated cell cycle arrest (22). Using a K619E substitution, Kilkenny et al. reported that a loss of pH2AX-binding activity induced a prolonged checkpoint arrest and did not alter Crb2 accumulation at IR-induced DSBs (22). These results contrast with our demonstration that crb2K619 mutants cannot efficiently localize to IR-induced DSBs or maintain checkpoint-mediated cell cycle arrest (Fig. 2 and 3). Because we have used the same K619E substitution as Kilkenny et al., as well as K619M and K619A substitutions, the reason for this discrepancy is not completely clear. In the case of Crb2 targeting, the difference may be reflective of the different protocols used to monitor accumulation at IR-induced DSBs. We have used an established live cell imaging protocol (9, 36), whereas a cell fixation method was used by Kilkenny et al. (22). The reason for the observed differences in checkpoint defects is less clear because we have used protocols similar to those of Kilkenny et al. (1). Interestingly, although Kilkenny et al. observed a prolonged checkpoint arrest in crb2K619E cells, IR-induced Chk1 phosphorylation and cell survival after DNA damage was impaired to a degree similar to what we have observed. Kilkenny et al. also reported that the kinetics of Rad22 accumulation at IR-induced DSBs was altered in crb2K619E cells (22). We have not attempted to examine DNA repair in our crb2K619 mutants.
Interestingly, the level of IR and UV sensitivity we have observed in the crb2K619 mutants is similar to that reported for h2ax− cells, but h2ax− cells display a much more pronounced hypersensitivity to CPT and HU (10, 27). This suggests that after exposure to either CPT or HU, pH2AX may have roles in promoting genome stability that occur independently of its interaction with the Crb2 BRCT repeats. This idea is supported by the recent characterization of fission yeast Brc1 as a second pH2AX-binding protein that functions independently of Crb2 (44). How pH2AX may discriminate between its different effector molecules is unknown and will be of high interest in the future.
Our observations argue that the checkpoint activity of pH2AX in fission yeast cells is mediated through its interaction with the Crb2 BRCT repeats. This is similar to the situation for budding yeast, where the interaction between pH2AX and Rad9 is key for checkpoint control (15). Interestingly, the BRCT repeats of the Crb2-related mammalian 53BP1 protein are also capable of binding pH2AX (22), but the 53BP1 BRCT motif is not required for its accumulation at DSBs (19, 43). Rather, in mammalian cells recognition of pH2AX by the BRCT repeats of the MDC1 mediator is thought to be a primary mechanism by which the modification exerts its coordinating activity (39). This indicates that although some divergence in recognition factors has occurred, the mechanisms of pH2AX-mediated genome integrity have been conserved through evolution.
A very intriguing finding from this work is the strikingly additive reduction in Crb2 activity that is seen by a combined abolishment of H4K20me2 and pH2AX binding (Fig. 4). The crb2Y378Q and crb2K619M mutations display very similar defects, but a combined ablation results in a marked decrease in both cell survival after DNA damage and checkpoint activity. Despite this striking effect, the double binding domain mutant still only partially inactivates Crb2. We presume that this indicates that even in the absence of histone modification binding, some fraction of Crb2 must still localize to lesion-adjacent chromatin to mediate checkpoint signaling. This recruitment is not readily observable by microscopy because the crb2Y378Q and crb2K619M mutations severely compromise IR-induced focal accumulation (Fig. 2) (14). Histone modification-independent targeting appears to be mediated by Thr215 phosphorylation because both the tudor and BRCT binding motifs are essential for Crb2 function in crb2T215A cells (Fig. 5). We speculate that mutation of its two histone modification binding modules ablates the ability of Crb2 to directly bind nucleosomes. In the absence of modification binding, Crb2 targeting to lesion-adjacent chromatin occurs indirectly through its interaction with other checkpoint regulators, such as Cut5, and is controlled by Thr215 phosphorylation (10). This is in agreement with the idea that binding to histone modifications and Thr215 phosphorylation represent two independent pathways to control Crb2 recruitment (10).
The additive nature of the crb2Y378Q and crb2K619M mutations has additional mechanistic implications for Crb2 function. Previous work argues that the H4K20me2 and pH2AX modifications function within the same genetic pathway because the kmt5Δ and h2ax− mutations display an epistatic, nonadditive impairment in genome integrity (10). This is in contrast to the significant additive decrease in Crb2 activity seen after combining the crb2Y378Q and crb2K619M mutations (Fig. 4). These observations indicate that double mutation of its two modification binding domains additively inactivates Crb2 but double mutation of the two binding targets of these domains does not. One possible explanation for this genetic difference is that the tudor or BRCT domain may have unknown interaction partners important for Crb2 activity. The Crb2 tudor motif and the Kmt5 methylase display a very intimate functional link (10, 14), suggesting that unknown tudor binding substrates are unlikely. This suggests that pH2AX may not be the sole target of the phospho-binding BRCT motif. Such postulated interactions are likely to occur in a phospho-dependent manner and be regulated by DNA damage.
An alternative, but not mutually exclusive, explanation for the observed genetic interactions is that Crb2 can independently bind H4K20me2 and pH2AX, but dual engagement increases the stability of nucleosome recognition to produce maximal Crb2 activity. As isolated domains, the tudor and BRCT motifs of Crb2 can directly bind their respective modified targets on peptide substrates (3, 14, 22) (Fig. 1B), but how this recognition occurs on native chromatin is not understood. It is also unknown whether some interplay between the tudor and BRCT motifs might modulate modification binding in the context of full-length Crb2. This is a very intriguing idea because BRCT-mediated dimerization is essential for Crb2 activity but the molecular mechanism that underlies this requirement is unknown (8, 22). One possibility is that binding to two pH2AX and two H4K20me2 modifications by a dimeric Crb2 protein is required for nucleosome recognition at lesion-adjacent chromatin. Structural modeling supports this idea and suggests that Crb2 can simultaneously bind four modified residues on a single nucleosome (22), but direct biochemical evidence is lacking. It is also possible that multimerization could promote Crb2 spreading from lesion sites into flanking chromatin regions by mediating internucleosome recognition of H4K20me2 and/or pH2AX. Whatever the role of dimerization, future experiments to examine the interplay between the dimerization and modification binding modules of Crb2 offer a unique opportunity to decipher the mechanisms of chromatin-controlled genome integrity.
Acknowledgments
We give a special “thank you” to Edward Stavnezer for critical reading of the manuscript and to Paul Russell for gifts of reagents and communication of results prior to publication. We are also grateful to all of our colleagues in the Department of Biochemistry at Case Western Reserve University for their helpful advice and encouragement during the course of this study.
This work was supported by Career Development Award 0017/2006-C from the International Human Frontier Science Program Organization and funding from the Case Comprehensive Cancer Center and Radiation Resource Core Facility, National Institutes of Health grant P30 CA43703.
Footnotes
Published ahead of print on 2 August 2010.
REFERENCES
- 1.al-Khodairy, F., and A. M. Carr. 1992. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 11:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allis, C. D., S. L. Berger, J. Cote, S. Dent, T. Jenuwien, T. Kouzarides, L. Pillus, D. Reinberg, Y. Shi, R. Shiekhattar, A. Shilatifard, J. Workman, and Y. Zhang. 2007. New nomenclature for chromatin-modifying enzymes. Cell 131(4):633-636. [DOI] [PubMed] [Google Scholar]
- 3.Botuyan, M. V., J. Lee, I. M. Ward, J. E. Kim, J. R. Thompson, J. Chen, and G. Mer. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127:1361-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carr, A. M. 2002. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair (Amst.). 1:983-994. [DOI] [PubMed] [Google Scholar]
- 5.Caspari, T., J. M. Murray, and A. M. Carr. 2002. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clapperton, J. A., I. A. Manke, D. M. Lowery, T. Ho, L. F. Haire, M. B. Yaffe, and S. J. Smerdon. 2004. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat. Struct. Mol. Biol. 11:512-518. [DOI] [PubMed] [Google Scholar]
- 7.Downs, J. A., M. C. Nussenzweig, and A. Nussenzweig. 2007. Chromatin dynamics and the preservation of genetic information. Nature 447:951-958. [DOI] [PubMed] [Google Scholar]
- 8.Du, L. L., B. A. Moser, and P. Russell. 2004. Homo-oligomerization is the essential function of the tandem BRCT domains in the checkpoint protein Crb2. J. Biol. Chem. 279:38409-38414. [DOI] [PubMed] [Google Scholar]
- 9.Du, L. L., T. M. Nakamura, B. A. Moser, and P. Russell. 2003. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 23:6150-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du, L. L., T. M. Nakamura, and P. Russell. 2006. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 20:1583-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.FitzGerald, J. E., M. Grenon, and N. F. Lowndes. 2009. 53BP1: function and mechanisms of focal recruitment. Biochem. Soc. Trans. 37:897-904. [DOI] [PubMed] [Google Scholar]
- 12.Foster, E. R., and J. A. Downs. 2005. Histone H2A phosphorylation in DNA double-strand break repair. FEBS J. 272:3231-3240. [DOI] [PubMed] [Google Scholar]
- 13.Glover, J. N., R. S. Williams, and M. S. Lee. 2004. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 29:579-585. [DOI] [PubMed] [Google Scholar]
- 14.Greeson, N. T., R. Sengupta, A. R. Arida, T. Jenuwein, and S. L. Sanders. 2008. Di-methyl H4 lysine 20 targets the checkpoint protein CRB2 to sites of DNA damage. J. Biol. Chem. 283:33168-33174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammet, A., C. Magill, J. Heierhorst, and S. P. Jackson. 2007. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep. 8:851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harper, J. W., and S. J. Elledge. 2007. The DNA damage response: ten years after. Mol. Cell 28:739-745. [DOI] [PubMed] [Google Scholar]
- 17.Hinks, J. A., M. Roe, J. C. Ho, F. Z. Watts, J. Phelan, M. McAllister, and L. H. Pearl. 2003. Expression, purification and preliminary X-ray analysis of the BRCT domain from Rhp9/Crb2. Acta Crystallogr. D Biol. Crystallogr. 59:1230-1233. [DOI] [PubMed] [Google Scholar]
- 18.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 19.Iwabuchi, K., B. P. Basu, B. Kysela, T. Kurihara, M. Shibata, D. Guan, Y. Cao, T. Hamada, K. Imamura, P. A. Jeggo, T. Date, and A. J. Doherty. 2003. Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J. Biol. Chem. 278:36487-36495. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, S. P., and J. Bartek. 2009. The DNA-damage response in human biology and disease. Nature 461:1071-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khorasanizadeh, S. 2004. The nucleosome: from genomic organization to genomic regulation. Cell 116:259-272. [DOI] [PubMed] [Google Scholar]
- 22.Kilkenny, M. L., A. S. Dore, S. M. Roe, K. Nestoras, J. C. Ho, F. Z. Watts, and L. H. Pearl. 2008. Structural and functional analysis of the Crb2-BRCT2 domain reveals distinct roles in checkpoint signaling and DNA damage repair. Genes Dev. 22:2034-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J., J. Daniel, A. Espejo, A. Lake, M. Krishna, L. Xia, Y. Zhang, and M. T. Bedford. 2006. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 7:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128:693-705. [DOI] [PubMed] [Google Scholar]
- 25.Mochan, T. A., M. Venere, R. A. J. DiTullio, and T. D. Halazonetis. 2004. 53BP1, an activator of ATM in response to DNA damage. DNA Repair (Amst.) 3:945-952. [DOI] [PubMed] [Google Scholar]
- 26.Murakami, H., and P. Nurse. 2000. DNA replication and damage checkpoints and meiotic cell cycle controls in the fission and budding yeasts. Biochem. J. 349:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura, T. M., L. L. Du, C. Redon, and P. Russell. 2004. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 24:6215-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura, T. M., B. A. Moser, L. L. Du, and P. Russell. 2005. Cooperative control of Crb2 by ATM family and Cdc2 kinases is essential for the DNA damage checkpoint in fission yeast. Mol. Cell. Biol. 25:10721-10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishioka, K., J. C. Rice, K. Sarma, H. Erdjument-Bromage, J. Werner, Y. Wang, S. Chuikov, P. Valenzuela, P. Tempst, R. Steward, J. T. Lis, C. D. Allis, and D. Reinberg. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9:1201-1213. [DOI] [PubMed] [Google Scholar]
- 30.O'Driscoll, M., and P. A. Jeggo. 2006. The role of double-strand break repair—insights from human genetics. Nat. Rev. Genet. 7:45-54. [DOI] [PubMed] [Google Scholar]
- 31.Peterson, C. L., and J. Cote. 2004. Cellular machineries for chromosomal DNA repair. Genes Dev. 18:602-616. [DOI] [PubMed] [Google Scholar]
- 32.Rass, U., I. Ahel, and S. C. West. 2007. Defective DNA repair and neurodegenerative disease. Cell 130:991-1004. [DOI] [PubMed] [Google Scholar]
- 33.Rowley, R., S. Subramani, and P. G. Young. 1992. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 11:1335-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saka, Y., F. Esashi, T. Matsusaka, S. Mochida, and M. Yanagida. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11:3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39-85. [DOI] [PubMed] [Google Scholar]
- 36.Sanders, S. L., M. Portoso, J. Mata, J. Bahler, R. C. Allshire, and T. Kouzarides. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119:603-614. [DOI] [PubMed] [Google Scholar]
- 37.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 38.Sofueva, S., L.-L. Du, O. Limbo, J. S. Williams, and P. Russell. 2010. BRCT domain interactions with phospho-histone H2A target Crb2 to chromatin at double-strand breaks and maintain the DNA damage checkpoint. Mol. Cell. Biol. 30:4722-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stucki, M., J. A. Clapperton, D. Mohammad, M. B. Yaffe, S. J. Smerdon, and S. P. Jackson. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123:1213-1226. [DOI] [PubMed] [Google Scholar]
- 40.Stucki, M., and S. P. Jackson. 2006. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst.). 5:534-543. [DOI] [PubMed] [Google Scholar]
- 41.Walworth, N., S. Davey, and D. Beach. 1993. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature 363:368-371. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Y., B. Reddy, J. Thompson, H. Wang, K. Noma, J. R. Yates III, and S. Jia. 2009. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol. Cell 33:428-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward, I. M., K. Minn, K. G. Jorda, and J. Chen. 2003. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 278:19579-19582. [DOI] [PubMed] [Google Scholar]
- 44.Williams, J. S., R. S. Williams, C. L. Dovey, G. Guenther, J. A. Tainer, and P. Russell. 2010. gammaH2A binds Brc1 to maintain genome integrity during S-phase. EMBO J. 29:1136-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, H., J. J. Pesavento, T. W. Starnes, D. E. Cryderman, L. L. Wallrath, N. L. Kelleher, and C. A. Mizzen. 2008. Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J. Biol. Chem. 283:12085-12092. [DOI] [PMC free article] [PubMed] [Google Scholar]