Abstract
The transcription factor PU.1 is critical for multiple hematopoietic lineages, but different leukocyte types require strictly distinct patterns of PU.1 regulation. PU.1 is required early for T-cell lineage development but then must be repressed by a stage-specific mechanism correlated with commitment. Other lineages require steady, low expression or upregulation. Until now, only the promoter plus a distal upstream regulatory element (URE) could be invoked to explain nearly all Sfpi1 (PU.1) activation and repression, including bifunctional effects of Runx1. However, the URE is dispensable for most Sfpi1 downregulation in early T cells, and we show that it retains enhancer activity in immature T-lineage cells even where endogenous Sfpi1 is repressed. We now present evidence for another complex of conserved noncoding elements that mediate discrete, cell-type-specific regulatory features of Sfpi1, including a myeloid cell-specific activating element and a separate, pro-T-cell-specific silencer element. These elements yield opposite, cell-type-specific responses to Runx1. T-cell-specific repression requires Runx1 acting through multiple nonconsensus sites in the silencer core. These newly characterized sites recruit Runx1 binding in early T cells in vivo and define a functionally specific scaffold for dose-dependent, Runx-mediated repression.
The Ets family transcription factor PU.1 provides essential pleiotropic inputs regulating multiple cell fate decisions during differentiation of blood cells from hematopoietic stem cells (HSCs). Its roles all depend on tight regulation of PU.1 itself, with different levels and patterns of expression distinguishing various cell lineages and different developmental phases. PU.1 is essential for the development of myeloid and lymphoid lineages (22, 30), but inappropriately controlled expression can cause severe developmental defects and/or malignancy. The precise basis of PU.1 regulation is therefore important to resolve and could be a model for multifunctional transcription factor deployment in development from stem cells.
PU.1 is expressed specifically in HSCs and their derivatives. Upon differentiation of HSCs, PU.1 expression is silenced in erythroid cells but elevated in macrophages, continues at moderately high levels in neutrophils and most types of dendritic cells, and is fixed at lower levels in committed B cells (4, 24). A particularly dramatic shift of PU.1 expression occurs in the development of T cells. Although the earliest intrathymic precursors express PU.1 at HSC-like levels, PU.1 expression is silenced during the transition to the DN3 stage of T-cell development, as the cells undergo lineage commitment (3, 33, 35). This silencing is crucial, as forced expression of PU.1 beyond this stage causes a developmental block. PU.1 overexpression in DN3 thymocytes or a DN3-like immature T-cell line, Adh.2C2, can also cause the cells to gain myeloid characteristics (2, 10, 19), linking the silencing of PU.1 to exclusion of alternative fate choices during T-lineage commitment. The mechanism of this essential silencing event is not fully understood.
To date, most aspects of PU.1 regulation have been explained by invoking just two regulatory elements: the promoter and an upstream regulatory element (URE) at ∼14 kb upstream of the transcription start site of the Sfpi1 gene, which encodes PU.1. Both are suggested to contribute to cell type specificity (20). Thus, differential regulation would imply roles for different combinations of transcription factors working at these same elements. The Sfpi1 promoter contains octamer binding sites affecting B-cell expression (7), while PU.1 can bind its own promoter with Sp1 to regulate itself in myeloid cells (8). Sfpi1 promoter activity can also be directed in myeloid cells by C/EBPα and AP-1 (5). These regulatory inputs to Sfpi1 may be modulated by cell-type-specific DNA methylation as well (1). The promoter alone cannot drive reporter expression in a chromatin context, however, and the search for added regulatory function yielded the conserved URE (around kb −14), reported to be a myeloid-specific enhancer, enhancing promoter activity in a myeloid cell line but not in a mature T-cell line (20). In myeloid cells, the URE binds C/EBPα (6, 38) and PU.1 and may thus contribute to autoregulation as well (26, 31).
Data suggest that the URE could also play a role in silencing in T cells, and two mechanisms have been offered for this. First, a TCF/LEF site in the distal URE could promote repression as long as Wnt signals are absent (28). However, this mechanism does not explain continued PU.1 repression at stages of development when T cells are known to require canonical Wnt signaling (12, 37). Second, a Runx input into the URE was proposed to mediate silencing as well as activation (17). Initiation of PU.1 expression in HSCs depends on Runx1, which unfolds the chromatin structure of the Sfpi1 gene and primes it for expression (16, 25). The proximal URE enhancer has three conserved Runx1 sites able to bind Runx1. Mice with a deletion either of Runx1 itself or of these URE Runx sites showed a decrease in PU.1 expression in myeloid and B cells. In T-lineage cells, deletion of Runx1 produces a developmental block at the DN2 stage (13, 18), and the surviving cells have higher PU.1 expression, consistent with Runx1 repression of Sfpi1 (17). However, URE Runx sites are maintained in an open state of accessibility, with the Runx sites apparently occupied, in PU.1-expressing myeloid and B cells and PU.1-negative T-lineage cells alike (15). Thus, it remains unresolved how both the initial activation and the T-lineage-specific silencing of Sfpi1 can be mediated by the same factor binding to the same sites.
However, it is not proven that all regulation of Sfpi1, even by Runx1, goes through the URE. Deletion of the URE (UREΔ) neither fully blocked activation of PU.1 expression within hematopoietic cells nor fully blocked T-cell silencing (28, 29). Thus, while required for normal regulatory output, the URE was dispensable for turning on PU.1 (29). Also, the UREΔ mice still had T cells in which PU.1 expression was effectively silenced (28). Even though the Runx1 bound to the URE in T cells might be part of a repressive complex, lack of the URE had a milder effect on T-cell development than the severe DN2 developmental block observed with deletion of Runx1 itself. Thus, Runx factors might act in part through elements beyond the URE.
Here, we have identified a set of conserved, previously undefined cis-regulatory regions for Sfpi1. One is a novel enhancer that can amplify myeloid cell-specific expression of PU.1. Another new element is a bipartite silencer that is necessary and sufficient for full silencing within a chromatin context in an immature, DN3-like T-cell line. We show that in this immature T-cell line, unlike a mature T-cell line, the URE remains active as an enhancer, although endogenous PU.1 is fully repressed; the new repression element enables neutralization of URE enhancer activity. Mapping the sites required for the core silencer function revealed them to be novel Runx binding sites. These Sfpi1 silencer sites bind Runx1 in a dose-dependent, cell-type-specific way, and perturbations of Runx1 protein function blocked silencer activity mediated through these sites. Runx1 gain of function promotes repression through the silencer element in T cells but activation, through the myeloid-specific enhancer, in myeloid cells. The existence of these lineage-specific, functionally dedicated cis elements reveals a rich system of intermodule interactions to account for the cell-type-specific regulation of PU.1.
MATERIALS AND METHODS
Cell culture.
Adh.2C2 and EL4 cell lines were grown in RPMI medium supplemented with 10% fetal bovine serum, penicillin-streptomycin-glutamine, nonessential amino acids, sodium pyruvate, and 2-mercaptoethanol. RAW 264.7 and NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, penicillin-streptomycin-glutamine.
DNase I HS mapping and Southern blot analysis experiments.
DNase I hypersensitivity (HS) assays were carried out as described previously (23) with slight modifications. Briefly, about 100 million cells were harvested and washed three times with RS buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 5 mM MgCl2, and 2 mM CaCl2). Cells were resuspended in 0.5 ml RS buffer, mixed with 0.5 ml 1% NP-40, and incubated on ice for 10 min. The pelleted nuclei were washed twice with RS buffer and resuspended in 0.2 ml RS buffer. Twenty-five microliters of nuclei suspension was digested with 25 μl of DNase I (Roche) at concentrations ranging from 0 to 40 units/ml for 5 min at 20°C. Reactions were terminated by adding 450 μl of digestion buffer (100 mM NaCl, 20 mM EDTA [pH 8.0], 6 mM EDTA, 0.5% SDS) containing 100 μg of RNase A and incubated for 30 min at 56°C. Proteinase K was then added to 0.1 μg/μl, and the mixture was incubated at 56°C overnight. The genomic DNAs were then extracted by using phenol-chloroform, precipitated with ethanol, and resuspended in 50 μl of H2O. All obtained DNAs were digested with different restriction enzymes to completion according to the regions analyzed, and fragments were resolved by gel electrophoresis and transferred to a nylon membrane. Probes were amplified by PCR from the mouse genomic DNA using the primer sets in the selected regions. The PCR products were gel purified using the Qiagen gel extraction kit and radiolabeled by random priming. The filter was incubated in prehybridization solution (6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], 5× Denhardt's solution, 0.25% SDS, 100 μg/ml denatured salmon sperm DNA) in a heat-sealed plastic bag at 42°C for 4 h with agitation. The labeled DNA probe was denatured by heating at 100°C for 10 min, quickly chilled on ice, and added to the prehybridization solution in the plastic bag. After 16 to 24 h of incubation, the filter was washed first in wash buffer 1 (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.5% SDS) for 15 to 20 min at room temperature and then in wash buffer II (0.2× SSC, 0.5% SDS) twice at 65°C for 1 h. The filter was then wrapped in a plastic bag and exposed to film at −80°C for overnight.
Cloning of reporters and expression constructs.
PU.1 DNA was obtained by PCR using the BAC RP23-20F9 construct (BACPAC Resource Center [bacpac.chori.org]). Reporter constructs were made by cloning PU.1 sequences into Promega's pGL3-Basic vector. Detailed maps of reporters and their construction are available upon request. For scanning mutagenesis analysis, CE4A core silencer fragments with M2, M14, or M5 plus M9 mutations were synthesized by GenScript and used to construct reporters. All other L98 plus 4A-5 mutants were made using overlap PCR to produce mutated core silencers for reporter construction. L9-3mRunx was made by excising the CE4 region and replacing it with a mutated sequence synthesized by GenScript. Runx1 dominant negative and full-length Runx1 cDNAs were kind gifts from Janice Telfer and were cloned into Invitrogen's pEF1/Myc-His B vector. The Ikaros dominant negative construct, Plastic, was synthesized by GenScript based on the published sequence (27) and cloned into pEF1. The following sequence with the M5a mutation and predicted Runx sites mutated was synthesized by GenScript and used to replace the wild-type sequence in L93 to construct L93-M5-mRunx: AGCTCTTAAGGGACTGAGGACTAAGCAAGATGCTGAGTTCTGGAGACGGGACTGTC TTCTTCCCCAGATTGAGATGCCAGGCATGTGTGTCTCACACAGACTCTGTGCCTACTACCTCAGTTAGCCTTGAGAAATCCCCACCTCCATTCC CAGAGGTATCTTCTATTATTGCTCCTATCTGGGGACAAAGAGCCTG AGGTCCCTAGAAGTGGGTTCCTGGCTCTCAGTTGTGAAGATAATTA GGTATAGGGAGTCACACTGCAGGTCACAGAAAGCACTGGCAGAAG CCAATGAAAGAGGCACATACTAAGTAGACTTTTAGTCTTGGAAACA AGGCTAGGAGGTGATTCTTGTTGATGTCTCTCTGTAGAGCTGAGCC TAAGTTCTGGAGAGGGGAAGGAACTCAGAAGGCTACATGGCCAAT CCATGGGGGTTGGGGGAGAACCCGTGGAGCTAGAGATGGGATGGTAGAGGGGGCGCCTTAGAGGAGGTAGGCCTGAGTGGGGAAGCAGC TCTTGTCCTTGGTGAGCAAGCTGGAGGTGTTCTGCTGCCCGTGGCG AGCAGACGACAGTTGCTGTTAGTTACGGTTAGTTTGATCTGCAGGA GACTGAGTGATGTTACCAGGAGGTGAGAGCTCCGCATCTGCAGGC CTGGTCAGCAGGAGACGGGGTTCAGTAAGATTCAGAGGAGTGTTA GCTGAACTGGAGATTTGTATCTCTCAGTCACCGGCCCTGGAACACA TGGGACCAGGAACCGGAATAGAACAGGAGGAGAAACTGAGGCAA GGCTTGGGAAAGACAGAGCAAACTGAAAAAAAAAAAAAAAGGGC ACTTAGAGGAGTGTCCAGTAGGGTGTTAAAGACAGTGAGAGCCTG TGTGAGCAAAGCCTGTTAGAGATTGAGAAAGAGCAGAGCTTCTGG ACATGTTGAGTCTTCTTACGCATCATGGGGGTAGGGCTAGCTGGAC TCCCAGTGTAGGAGGCTCCAGCACAGGCCTCCAAGGTATGGGCTCC AGCTCTGGACAGGTAAGAGCTGAGGAAGACTTCCAGGTAGGGAGA GACACAAGAAGCCAAGAGGTGAGACAGCTGAAGAAGGCCAGGCC CTAGG.
The M5a mutation in L93-M5-mRunx was corrected by PCR mutagenesis to generate L93mRunx. The M5 mutation was introduced into L93-M5 by overlap PCR of CE4, which was then used to replace the wild-type sequence. Sequences of primers used to construct reporters are available upon request. All reporter construct sequences were verified.
Transfections and luciferase assays.
Cells were transfected in some experiments with FuGENE 6 reagent, at a FuGENE:DNA ratio of 3:1. Alternatively, cells were transfected by using Nucleofection (Lonza/Amaxa). Solution-V kits were used when nucleofecting Adh.2C2 cells with program D-19 or RAW 264.7 cells with program D-32 (Lonza/Amaxa). EL4 cells were nucleofected with Solution-L kits and program C-09. Cells were harvested ∼48 h post-FuGENE treatment or ∼24 h post-nucleofection treatment. Cells were cotransfected with pRL-CMV, and lysates were analyzed using Promega's dual luciferase system. Three to 6 μg Sfpi1 reporter was used in transfection mixtures with 100 to 200 ng pRL-CMV control. For stable transfections, Sfpi1 reporters were linearized with NotI prior to transfection. The Renilla luciferase was cloned into Invitrogen's pTracer EF/blasticidin A and the construct was linearized for mixed transfection with the Sfpi1 reporters. After transfection, cells were aliquoted into six-well plates and then selected with 5 to 15 μg/ml blasticidin for their duration in culture, beginning 1 day posttransfection. The pTracer-Renilla control was linearized with FspI. Ten micrograms of Sfpi1 reporter was transfected with 1 to 2 μg of pTracer-Renilla. For some stable transfections, insert copy number was determined by quantitative PCR with primers specific to firefly luciferase and the pTracer-Renilla control vector's green fluorescent protein (GFP)-blasticidin sequence: Luc-F, ACGATTTTGTGCCAGAGTCC, and Luc-R, AGGAACCAGGGCGTATCTCT; GFP/Blast1-F, GTCAGTGGAGAGGGTGAAGG, and GFP/Blast1-R, ACGGGAAAAGCATTGAACAC. Expression in stable transfectants was approximately linear with copy number for all constructs tested, so this measurement was omitted in later experiments. All antisense morpholino transfections were performed by nucleofection with 2 nanomoles of morpholinos. The following morpholino antisense oligos were ordered from Gene Tools, Inc.: anti-Runx1, CAGGCAGGAGTACCTTGAAAGCGAT; anti-PU.1, GAGGACCAGGTACTCACCGCTATG; anti-CBFβ, CCTCCCCAACCGCCTCACCTCGCAC.
Transcription factor binding site predictions.
TRANSFAC analysis was used to predict potential transcription factor binding sites. The Biobase TRANSFAC suite's MATCH tool was used for the analysis. Matrix similarities of >0.925 are shown.
Gel shift assays.
Nuclear extracts were prepared by hypotonic swelling in buffer A, followed by NP-40 lysis, nuclei pelleting, and extraction with buffer C containing protease inhibitors (no. 11873580001; Roche). Buffer A contained 10 mM HEPES (pH 7.9), 60 mM KCl, 1 mM dithiothreitol (DTT), 0.1 mM EDTA, 0.1 mM EGTA, followed by addition of NP-40 to 0.625%. Buffer C contained 20 mM HEPES (pH 7.9), 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT. Protein was quantified by the Bradford method. Gel shifts were performed with ∼6 μg extract in a 30-μl volume containing 1 to 2 μg poly(dI-dC) and final concentrations of 15 mM HEPES (pH 7.9), 80 mM NaCl, 15 mM KCl, 0.02 mM EDTA, 1 mM DTT, and 3% glycerol. Five-picomole aliquots of probes were end labeled with T4 polynucleotide kinase followed by purification with G-50 columns (no. 100609; Roche). Complexes were allowed to form for 10 min with competitors prior to addition of radiolabeled probes. After probe addition, samples were incubated for an additional 30 min. All incubations were carried out on ice. Complexes shown below in Fig. 6D were resolved by 4% PAGE, run at a constant 350 V for 4 h. All other complexes were resolved by 6% PAGE run at a constant 350 V for 2.5 to 3.5 h. All gels were run at 4°C with 0.5× Tris-borate-EDTA (TBE) gels and 0.25× TBE running buffer. Gels (4%) were prerun for 30 min. Quantification was performed with a phosphorimager and ImageQuant 5.2 analysis. Runx1 N-terminal antibody was from Calbiochem (catalog no. PC284). Runx1 antibody against amino acids 231 to 245 was from Active Motif (catalog no. 39000). A pan-Runx antibody was a kind gift from Masanobu Satake (Tohoku University, Sendai, Japan). Ikaros (sc-13039), Myb (sc-516), and Ets2 (sc-351) antibodies used in gel shift assays were from Santa Cruz Biotechnology.
Western blot assays.
Nuclear extracts were mixed with 2× Laemmli sample buffer, boiled, and then run on an 8% SDS-PAGE gel. Gels were transferred to Immobilin (Millipore) by semidry transfer. Blots were blocked with 5% milk in TBS-T (Tris-buffered saline, 0.5% Tween 20) and then incubated overnight with primary antibody at a 1:3,000 dilution. After washing, blots were incubated for 90 min with secondary antibody at a 1:3,000 dilution, washed, then incubated with substrate (SuperSignal, catalog nos. 1859675 and 1859674; Pierce). Substrate was drained from blots and then blots were exposed to film. Primary Runx1 antibody PC284 was from Calbiochem. Sp1 antibody was from Santa Cruz Biotechnology (sc-59).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed as recommended by Upstate Biotechnology (Millipore). Briefly, 2 × 107 to 3 × 107 cells were fixed with 0.33 to 1% formaldehyde for 10 to 30 min and then lysed in 0.8 ml with protease inhibitors. Lysate was sonicated to produce an average fragment size of ∼ 250 bp. Lysate (130 μl) was diluted and used for each ChIP sample with 9 μg of antibody. Cross-linking was reversed by overnight incubation at 68°C. Proteinase K digests were for 30 min at 55°C. DNA was purified by ethanol precipitation and resuspended in 100 μl water. Analysis of recovered DNA was performed by SYBR green-based quantitative PCR (QPCR) with an AB 7900HT apparatus. One microliter of purified DNA was used per 10-μl PCR mixture, tested in triplicate. Whole thymi from Rag2−/− mice were excised, and thymocytes were recovered by cutting and scraping thymic lobes through steel mesh. Thymocytes were then immediately fixed and processed for ChIP assays. Approximately 4 × 106 thymocytes were used per ChiP assay. Runx ChIPs were performed with an equal mix of antibodies from Calbiochem and Active Motif (described above). Rabbit Ig (sc-2027) and GABPα (sc-22810) antibodies were from Santa Cruz Biotechnology. Primer pairs used for analysis of ChIP-enriched DNA by QPCR were the following: CE1-F, AGCTCAGCTGGATGTTACAGG, and CE1-R, AGATGGTCACACATCCCAAAG; −2kb-F, TTCTCACATCCCAGACCATTC, and −2kb-R, CGCCAGCAGTTGTAGTTCTTC; −2.8kb-F, GCAGCTCACTGCTCCAAGTT, and −2.8kb-R, GAGACGGGGAGTGGGTATGT; CE3-F, TGGAGCTCTGAGGGGCCTAA, and CE3-R, GGCTGGGAAAGCTGACCATAA; −8.4kb-F, AGAGGAGCTGACATTGGCATAC, and −8.4kb-R, TGAGCCTCTGAAGTGGCTTTAT; CE4B-F, AGCAAAGCCTGTGGGAGATT, and CE4B-R, ATACCTTGGAGGCCTGTGCT; CE4A-F, GGAAGCAGCTCTTGTCCTTGG, and CE4A-R, TCACCTCCTGGCCACATCACT; CE5-F, GCTCTGAAAAGCACCGTTTCC, and CE5B-R, CTGTGTTGGACCTGCAAGGAGT; −11.8kb-F, CTCTGCCCGCTCTTAACCTT, and −11.8kb-R, GATCTGACACGGGGATGAAA; CE76-F, CACACGGAGTCAGAGCGGGCAG, and CE76-R, AGGAAAGAGGAAGCCATGGGGAGA; CE8-F, AGGCAGAGCACACATGCTTC, and CE8-R, CTTCTGGGCAGGGTCAGAGT; CE9-F, CAGGAGAGGCAGGAGGAAGGA, and CE9-R, AGAGAGCAGAGCACTTCATGGCT; −17.8kb-F, CTGGACAAGTGGAAGGTGACA, and −17.8kb-R, TCAGAGGGCTTCAAAGTGGA; CD4-F, TGACGGAAGGGAGGATGTAG, and CD4-R, AGTGGGTGGGAGCTCTGTAA; MEF2C-F, AGCACACTCAGCCTGCTCTAC, and MEF2C-R, GGTGTAAAGGTGCTTCCTTCC; IL-7Rα-F, GTCTGAGCAAAAGGATTGCTG, and IL-7Rα-R, GGAGCTTCAGGGAATACCAAG.
Statistical methods.
In all graphs, error bars indicate standard deviations (not standard errors of the means). All P values reported were obtained using a two-tailed Student's t test for comparisons as indicated (unpaired, equal variance). For values obtained in scanning mutagenesis assays (see Fig. 5D), a Bonferroni correction for multiple comparisons was applied. Stable transfection results, which cover orders of magnitude in dynamic range and more closely conform to a log normal distribution, were log10 transformed before comparison by t test.
RESULTS
The PU.1 URE is a stage-specific T-cell enhancer.
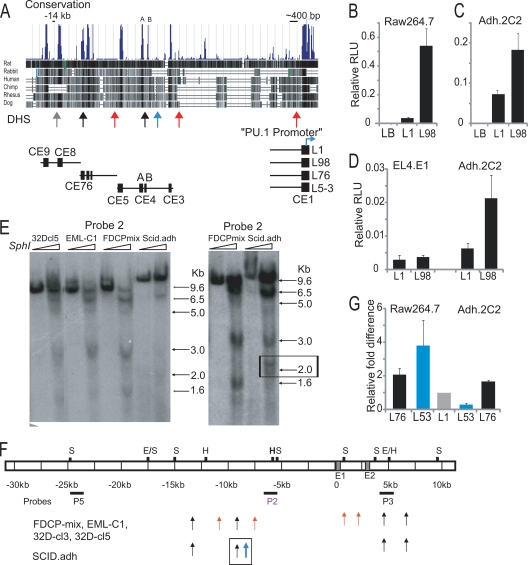
A 2.2-kb PU.1 promoter fragment could not drive reporter expression in myeloid cells in a chromatin context but was able to do so when joined to the 3.5-kb URE fragment. This URE failed to enhance promoter-driven expression in mature T cells (20). In order to elucidate the dynamic mechanism of PU.1 silencing during early T-cell development, we tested the regulatory function of the URE in a more immature T-cell line representing the DN3 stage, i.e., the developmental state in which endogenous PU.1 expression is actively being silenced. We made reporters with the PU.1 promoter, with or without the regulatory regions of the URE (Fig. 1A). These reporters were named L98 and L1, respectively, and were tested for activity by transient transfection into a mature myeloid cell line, RAW 264.7, and an immature DN3-like T-cell line, Adh.2C2. As expected, we observed L98 reporter activity enhanced by ∼14-fold relative to L1 in a myeloid cell line (Fig. 1B). We also transiently transfected a more mature T-cell line, EL4, and showed that in that case L98 failed to show enhanced activity relative to L1, as expected from previous reports (Fig. 1D). Unexpectedly, however, the L98 construct containing the URE reproducibly showed ∼3-fold-enhanced reporter expression in the immature T cells (Fig. 1C). These results suggest that the activity of the URE is not exclusively repressive in T cells, but rather is developmental stage dependent.
FIG. 1.
The Sfpi1 upstream region contains novel cis-regulatory elements. (A) Sfpi1 multigenome alignment from exon 1 to ∼15 kb upstream. Schematics of regions used in reporters are shown. (B and C) The CE9 to CE8 (URE) region is an enhancer in the RAW 264.7 myeloid cell line and in the immature DN3-like Adh.2C2 pro-T-cell line. The Renilla luciferase-expressing control vector pRL-CMV, was used as an internal standard. The empty pGL3-basic vector was used as a control (LB). The average relative light units (RLU) of triplicates from a representative experiment are shown with standard deviations. (D) The CE9 to CE8 region does not possess enhancer activity in a mature T-cell line, EL4. Data shown are the averages of three independent experiments performed in duplicate with standard deviations. (E) Novel DNase I HS sites were identified. Probe 2 Southern blotting results for SphI-digested DNA from nuclei of indicated cell lines, with or without DNase I, are shown. For sites at and downstream of promoter, see Fig. 2. Bands are defined by sites of DNase I sensitivity. The right panel, more extensively digested, shows a T-cell-specific doublet of HS sites at ∼2 kb from SphI site (box). (F) A schematic summarizing HS mapping with various probes. S, SphI; H, HindIII; E, EcoRV. (G) The L5-3 reporter showed lineage-specific activity. RLUs were normalized to L1. The average RLU of triplicates from a representative FuGENE transfection experiment are shown, with standard deviations.
Cell-type-specific patterns of DNase hypersensitivity associated with conserved regions outside the URE.
To look for other cis-regulatory elements that may contribute to PU.1 silencing in immature T cells, we used multigenome sequence alignments to find other conserved noncoding elements across the ∼50-kb PU.1 mouse locus (Fig. 1A). Besides the elements of the promoter (CE1) and the two previously identified within the URE, which we have termed CE9 and CE8, our alignments revealed other conserved regions, mapping from about kb −12.5 to kb −7.5 upstream of the Sfpi1 transcriptional start site, and these regions were designated CE7, CE6, CE5, CE4 (A+B), and CE3.
To assess whether any of these regions might show cell-type-specific differences in accessibility or transcription factor engagement, a range of hematopoietic cell lines, including myeloid (32D), multipotent progenitor (FDCP-mix and EML-c1), and pro-T-cell (Adh.2C2) lines were tested to map DNase HS sites across the 5′-flanking region and first two introns of Sfpi1 (Fig. 1E and F and 2). One DNase HS site related to the promoter was only formed in PU.1-expressing cells, as earlier reported (20). An HS site at kb −14 was previously identified (20) to mark the URE and was detected in all the hematopoietic cell lines tested (Fig. 1E, band at 9.6 kb). Also confirmed were two reported sites in the second intron, which were seen in both PU.1-expressing and -nonexpressing cell types (1) (Fig. 1F and 2). In addition, five new sites were found. Two novel DNase HS sites formed to varied extents in all the cell lines tested (Fig. 1A and E, black arrows). The first of these was found at kb −12.3, near CE7 and CE6. This site is just downstream of the boundary of the 3′-URE fragment. The second cell-type-nonspecific HS site was seen at kb −8.8, between CE4A and CE4B. Of more interest, two other upstream DNase HS sites were detected only in PU.1-expressing cell lines (Fig. 1A and F, red arrows). These two HS sites were found at kb −10.8 and kb −7.4 (5-kb and 1.6-kb bands in Fig. 1E), flanking CE5 and CE3, respectively. Notably, we also detected a doublet of DNase HS sites specific to immature T cells, at kb −8.5 (Fig. 1A and 1F, blue arrow, and E, right, band around 2 kb), and not seen in any of the PU.1-expressing cell types. We hypothesized that a regulatory feature associated with Sfpi1 silencing could be near this region.
FIG. 2.
DNase HS mapping detected HS sites in the first two Sfpi1 introns. (A) The pro-T-cell line scid.adh lacks HS sites downstream of exon 1 (E1). Probe 3 (P3) was used in a Southern blot assay of EcoRV-digested DNA from nuclei of the indicated cell lines, with or without DNase I. Bands correspond to sites of DNase I sensitivity. Size markers are shown. The schematic depicts HS mapping results with probe 3. Red arrows indicate HS sites detected in PU.1-expressing cells and correspond to the bands in the Southern blot. S, Sph I; H, Hind III; E, EcoRV. (B) Similar DNase HS sites were detected across intron 2 in PU.1-expressing cells and also weakly in a nonexpressing pro-T-cell line (scid.adh). Probe 3 was used in a Southern blot assay of Sph I-digested DNA, with or without DNase I, as described above.
Identification of a novel cell-type-specific PU.1 regulatory element with T-lineage-repressive activity.
Reporters were made as shown in Fig. 1A to test if any conserved elements from CE7 to CE3 might have regulatory activity independent of the URE in immature T cells. The L7-6 construct was made to combine the Sfpi1 promoter with a 2-kb region including CE7 and CE6, where an overlapping non-cell-type-specific HS was found in myeloid and T cells (Fig. 1A). L7-6 showed enhanced and nonspecific activity in both myeloid and immature T-cell lines, with ∼2-fold increased activity in transient-transfection assays, compared to L1(Fig. 1G, gray versus black bars). In contrast, a construct containing the promoter plus the new conserved elements from CE5 through CE3, L5-3, showed activity that was clearly cell type specific. L5-3 was strongly repressed in the Adh.2C2 cells compared to L1 (Fig. 1G, gray versus blue bars). Strikingly, the same L5-3 construct was found to have enhanced activity in RAW 264.7 cells. In summary, the conserved sequences CE9 to CE6 span a region of ∼4 kb and contain multiple regulatory elements that increase promoter activity in both myeloid and immature T cells. In contrast, the ∼3-kb region containing CE5 to CE3 was found to mediate cell-type-specific activating or repressive regulatory function.
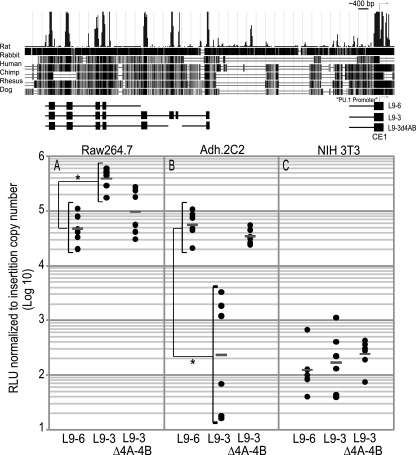
The regulatory elements in CE5 to -3 not only showed lineage-specific effects on promoter activity, but also strongly modulated the combined activity of the promoter and the URE. These effects were strongest when the reporters were stably integrated and expressed from a chromatin context in myeloid and immature T-cell lines. We stably transfected linearized reporters containing elements CE9 to CE6 (L9-6) or a longer sequence extending further to include CE9 to CE3 (L9-3). The L9-6 reporter efficiently expressed luciferase when stably integrated into chromatin, as expected, giving similar levels in myeloid cells (Fig. 3A) and in our DN3-like immature T-cell line (Fig. 3B). The CE9 to -6 enhancer activity was hematopoietic specific, as the L9-6 reporter generated a >100-fold increase in luciferase expression in both myeloid and immature T cells compared to nonhematopoietic NIH 3T3 fibroblasts (Fig. 3C).
FIG. 3.
The CE5 to CE3 region has cell-type-specific regulatory functions in stably transfected cells. A schematic of the reporters used is shown. Data are reported as the relative light units (RLU) per insert copy number. (A) Extension of L9-6 to include the CE5 to CE3 region increases activity in RAW 264.7 myeloid cells. (B) Inclusion of the CE5 to CE3 region silences activity in immature Adh.2C2 T cells. (C) Background activity of reporters in the nonhematopoietic NIH 3T3 fibroblast cell line. Dots represent independent wells containing multiple founders, and bars show the geometric mean for the wells. *, P < 0.001 in two-tailed t test comparisons of log10-transformed data.
The addition of the CE5 to CE3 region to L9-6 to make the L9-3 construct yielded sharply different results. This construct gave an ∼8-fold increase in reporter expression in myeloid cells compared to L9-6 (Fig. 3A). In the T cells, however, addition of CE5 to CE3 repressed reporter expression to a level comparable to the background level in NIH 3T3 fibroblasts (Fig. 3B and C). These data show that the CE5 to CE3 cis-regulatory region can contribute to a >500-fold difference in reporter expression between myeloid and immature T cells.
Mapping of a bipartite region necessary for silencing in a chromatin context and a core silencer sufficient for silencing in transient assays.
DNase HS mapping revealed a pan-hematopoietic HS site between CE4A and CE4B, as well as a T-lineage-specific HS site just downstream of CE4B (Fig. 1A, E, and F). This suggested that the CE4A-B region might be involved in the T-cell silencing effect. As shown in Fig. 3B, deletion of CE4A and CE4B did abolish the repressive function within the L9-3 construct. To map regions within CE5 to CE3 that contribute to the cell-type-specific regulatory function, we made more reporters by combining individual conserved regions together with the 2.2-kb PU.1 promoter and then tested their function in transient-transfection assays. While the dynamic range of these assays is far less than in the stable transfections, these rapid surveys yielded results that could be verified in the stable transfectants.
This functional mapping showed that the myeloid-enhancing activity and the pro-T-cell silencing activity are mediated by different cis-regulatory sequences. The CE5 region confers myeloid-specific enhancer activity. As shown in Fig. 4B, CE5 (in L5) was able to enhance promoter activity by ∼7-fold in myeloid cells, but by only 1.5-fold in the immature T cells (Fig. 4C). In contrast, all the repressive activity in immature T cells mapped to the CE4 region. Results with the L4A construct showed that the CE4A region alone could repress Sfpi1 promoter activity in the immature T cells (Fig. 4C), but with little or no effect in the myeloid cells (Fig. 4B). The CE4B region did not confer independent regulatory function in transient assays (Fig. 4B and C), although it contains one reported Stat3 site implicated in Sfpi1 induction by cytokines (14). However, the presence of both CE4A and CE4B was necessary for full silencing when integrated stably in a chromatin context (Fig. 4D), indicating that the CE4A-B region is a bipartite T-specific silencer.
FIG. 4.
T-cell- and myeloid cell-type-specific regulatory activities map to distinct regions within the CE5 to CE3 fragment. (A) Diagram of the CE5 to CE3 truncations used in transient-transfection experiments. (B and C) Myeloid enhancer activity mapped to the CE5 region. T-cell silencer activity mapped to the CE4A region. Representative experiments, with relative light units (RLU) normalized against L1 (with standard deviations) are shown. (D) Both CE4A and CE4B contribute to silencing in a chromatin context. The diagram shows reporters used for stable cell lines. Data points are as described for Fig. 3. Data are shown as RLUs. *, P < 0.007 in two-tailed t test comparisons of log10-transformed data.
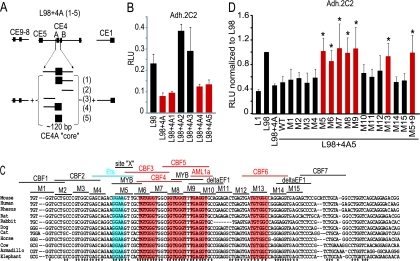
The CE4A region spans ∼450 nucleotides (nt) in which the central ∼120 nt are most conserved. To map the sequences within the CE4A region that are vital for silencing, we truncated this 450-nt region and made reporters with these truncations flanked by the UREs and promoter (Fig. 5A). These experiments showed that a minimal conserved core is necessary and sufficient for repression in the transient assays (Fig. 5B). Construct L98 plus 4A-5, containing only the minimal core silencer region of ∼120 nt from CE4A, was sufficient to repress the enhancer activity of the CE9 to CE8 region in immature T cells (Fig. 5B).
FIG. 5.
T-cell silencer activity mapped to a conserved core region. (A) Diagram of reporters with CE4A truncations flanked by the CE9-CE8 enhancer and the promoter. (B) The L98 plus 4A5 reporter contains the core silencer sufficient for silencing (red bar). Data shown are from a representative transient experiment performed in duplicate with Adh.2C2 T cells. (C) Multigenome alignment of the CE4A core silencer. Nucleotides conserved in all 11 organisms are marked by an asterisk. Six nucleotide blocks mutated in the scanning mutagenesis analysis are labeled (M1 to M15). Sequences with >0.925 similarity to TRANSFAC predicted binding sites are shown. The region affected by M5 is designated site X. (D) Scanning mutagenesis revealed multiple sites contributing to core silencer activity. Data shown are averages from four or more independent Adh.2C2 transfections ± standard deviations, with relative light units (RLU) normalized against L98 activity. Red bars mark reporters with mutations blocking silencer activity. *, P < 0.02 (Bonferroni corrected) compared to wild-type reporter data.
Scanning mutagenesis analysis of the core silencer.
The CE4A core silencer mapped to a peak of conservation identified by the multigenome alignment (Fig. 1A). The precise nucleotide alignment is shown in Fig. 5C, with asterisks marking nucleotides that are 100% conserved among 11 organisms and with predicted transcription factor binding sites shown (see Materials and Methods). To unmask the most influential repression sites in an unbiased way we carried out scanning mutagenesis and tested for loss of repression in transfection assays, mutating 6-nt blocks (M1 to M15) (Fig. 5C) across the core CE4A silencer region within the reporter construct L98 plus 4A-5. Mutants M1 to M4 did not affect the repressive function of the core silencer (Fig. 5D). However, mutants M5 to M9 caused a loss of core silencer function (Fig. 5D). These mutants span a region of 30 nt across the largest conserved block within the core silencer (Fig. 5C). Another mutant, M13, also blocked core silencer activity (Fig. 5D). Close examination of the sequences where mutations abolished core silencer function showed that 5/6 mutations, M6 to M9 and M13, overlap sequences with ∼90% similarity to the canonical Runx binding motif, (Py)G(Py)GGT (Fig. 4C, red boxes). Mutant M5 does not overlap a predicted Runx site, but it does cross a conserved site, “site X,” predicted to contain an Ets family target site (Fig. 5C, blue box).
Identification of T-cell-specific protein complexes.
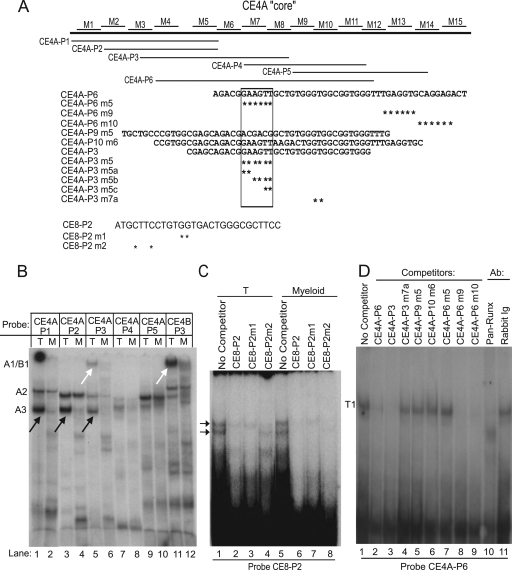
To identify transcription factors vital for the T-cell-specific repressive activity of the CE4A core silencer, probes spanning the CE4A core region were used in gel shift assays with nuclear extracts from Adh.2C2 and RAW 264.7 cells to determine the nature of cell-type-specific protein-DNA complexes. Complexes were identified by mobility and by cross-probe competition and antibody treatments. These assays showed that at least three regions of the CE4A and -B elements could nucleate cell-type-specific protein-DNA complexes that differed qualitatively when formed with T-cell or myeloid extracts (Fig. 6B). The cell type specificity of the complexes formed with these probes contrasted with those detected by the CE8 region of the URE, where similar patterns of complexes were formed with extracts from T-cells and myeloid cells alike (Fig. 6C).
FIG. 6.
The CE4 region nucleates T-cell-specific protein complexes in vitro. (A) Schematic showing alignment of probes relative to the core silencer. Wild-type probe sequences are also shown with asterisks to mark bases mutated in competitors. Sequences of the CE8 probes are also shown. (B) Multiple CE4A probes form T-cell-specific complexes in vitro. Probes were incubated with nuclear extracts from Adh.2C2 (T) or RAW 264.7 cells (M). T-cell-specific complexes A1/B1 and A3 are indicated by arrows. (C) Unlike CE4A-P3, an Ets-Runx CE8 probe forms similar complexes with both pro-T-cell (Adh.6D4) and myeloid (P388D1) extracts. (D) Mapping of sites contributing to “T1” complex formation on probe CE4A-P6 by using Adh.2C2 nuclear extracts.
Figure 6D shows that a large probe spanning the whole critical repression region (CE4A-P6) could nucleate a single complex from Adh.2C2 extracts. This large, slow-migrating complex (T1) depended on binding to two distinct regions, as defined by competition with mutant and wild-type competitors. One critical site was element X (lanes 2 versus 7), and the other was the region of predicted Runx site CBF3 (laned 3 versus 4). To define the distinct component complexes that might contribute to the large T1 complex and to identify those that might be cell type specific, an overlapping series of smaller CE4A probes were used. Two distinct T-cell-specific complexes were formed on these smaller probes: one named A3 and a larger, slow-migrating complex named A1 (Fig. 6B). Using probe CE4A-P3, which formed both complexes, Fig. 7A shows that the sequence requirements for these two complexes can be dissociated. Complex A3 mapped to site X, which is mutated in CE4A-P3m5a, while complex A1 depended on the region of the CBF3 site that is mutated in CE4A-P3m7a.
FIG. 7.
Cross-reactive, pro-T-cell-specific complexes formed at the CE4A and CE4B regions. CE4B can nucleate a pro-T-cell-specific complex in vitro and can cross-compete with the T-lineage-specific A1 complex formed by the CE4A region probe. (A) The sequence requirements for formation of different T-lineage complexes on probe CE4A-P3 can be separated. A homologous probe from the CE4A region cannot compete for complex A1 if the Runx sites are mutated (compare lanes 2 and 3), indicating that a Runx site is needed for A1 formation. Complexes A2 and A3 are competed by the homologous CE4A-P3 probe, but not when the GGAA core of the Ets site is mutated (lane 4). A probe from the CE4B region competes for complex A1 but not for complexes A2 or A3 (lane 5), and the ability to compete for A1 depends on a Runx site here as well (lanes 6 and 7). Complex A1 may also contain Ikaros, as it can be shifted by an anti-Ikaros antibody (lane 9). (B) The CE4B-P3 probe nucleates a pro-T-cell-specific complex, B1 (lane 1; “B”) and a myeloid cell-specific complex, B3. Complex B1 does not depend on the STAT site previously shown to contribute to PU.1 expression (lanes 3 and 4). In contrast, the myeloid complex B3 and the weaker T-lineage complex B2 do require the STAT site for formation of complexes (lanes 3 and 4 and lanes 10 and 11). (C) Complex B1 depends on the sequences spanning nonconsensus Runx and Ikaros sites (lanes 2 and 3). Probe CE4A-P3 can be used in competition to eliminate complex B1 formation, and the ability to compete requires the Runx site (lanes 4 and 5). (D) Like A1, complex B1 can be supershifted by an anti-Ikaros antibody (lane 3). (E) Interaction of Runx factors with complexes at multiple sites in CE4A and CE4B. A pan-Runx antibody inhibits complex formation on multiple probes by Adh.2C2 pro-T-cell nuclear extracts (red arrows, lanes 2, 5, and 15). Anti-Ets2 antibody had no effect on any complex. (F) The sequence of probe CE4B-P3 is shown, with asterisks indicating mutated nucleotides. TRANSFAC predicted transcription factor binding sites are shown with their matrix similarity scores in parentheses.
Because the full silencing activity in a chromatin context depends on region CE4B as well as CE4A, we carried out a screen for potential T-cell-specific binding complexes to CE4B probes too. Of seven CE4B probes tested, only two formed complexes and only probe CE4B-P3 formed cell-type-specific complexes (Fig. 7B). This probe was also found to form a very-slow-moving complex, designated B1, which was T-cell specific (Fig. 6B, lane 11, and 7B). The CE4B B1 complex was similar to the large A1 complex formed with CE4A, based on cross-competition and mutational analysis (Fig. 7A and C), and both depended on the integrity of predicted Runx sites. Both large complexes, A1 and B1, were also supershifted by antibodies against Ikaros, although not by anti-Myb (Fig. 7A and D,), suggesting a higher-order complex structure.
Together, the analyses suggest that the T-cell-specific bands A1 and B1 represent T-cell-specific complexes dependent on predicted Runx sites, while A3 is a T-cell-specific complex that depends on site X. These may interact to form a higher-order T1 complex. The possibility that the A1 and A3 complexes on CE4A interact at the protein level was supported by the ability of a pan-Runx antibody to interfere not only with T1 and A1 but also with A3 (Fig. 7E, lane 5, and 6D, lane 10). The composition and precise role of the complexes depending on the X site are still unresolved (data not shown). However, further experiments strongly support direct involvement of the Runx1 protein in the T-cell-specific repression activity of the bipartite silencer.
The CE4A silencer contains multiple nonconsensus Runx binding sites that contribute to T-cell-specific complexes.
Although similar to Runx sites, the core silencer defined by mutations does not contain precise matches to the Runx consensus sequence (Py)G(Py)GGT, and more evidence was needed to determine whether Runx proteins could be part of complexes A1, B1, and T1. The pan-Runx antibodies shifted or inhibited these complexes and Runx1-specific antibodies seemed to cross-link them, but the slow initial mobility of these complexes made the results difficult to interpret (data not shown). Therefore, the binding of Runx proteins to CE4A was confirmed by competition in gel shifts against a validated Runx-dependent silencer sequence, the Cd4 silencer (32) (Fig. 8).
FIG. 8.
Multiple Runx sites are present across the CE4A core silencer region. (A) The strong complex nucleated by the Cd4 reference probe is dependent on Runx sites (lane 3) and can be supershifted by anti-Runx1 antibodies (lanes 13 and 14). The CE4A probes P1, P3, P4, and P5 (Fig. 6) and CE4B-P3 (Fig. 7) compete with the Cd4 probe for Runx protein binding. Competition by the CE4A-P3 and CE4B-P3 probes depends on the integrity of their Runx sites (lanes 7 and 11). (B) The CE4A-P3 probe's site X region is needed to compete with the Cd4 probe for Runx binding, but the GGAA motif (Fig. 5C) is dispensable (lane 4 versus lanes 5 and 7). (C) All Runx sites across the extended CE4A-P8 probe must be mutated to abolish competition for Runx complex formation on the Cd4 probe. Compare CE4A-P8 M-all (lane 15) with lanes 7, 9, 11, and 13. (D) Diagram showing where the CE5, CE4A-P3, and CE4B-P3 probes are located. (E) Diagram of probes. Asterisks indicate mutated nucleotides in competitors.
The Cd4 silencer probe contains two canonical Runx motifs (Fig. 9B) and formed a strong band with nuclear extract prepared from Adh.2C2 cells (Fig. 8, black arrows), which was specifically supershifted by Runx1-specific antibodies (Fig. 8A, white arrows). The complex is competed by cold Cd4 probe, and this competition depends on the two Runx motifs (Fig. 8A, lane 3). Competition for this complex thus affords an assay for Runx1 binding to other sequences. Probe CE4A-P1, although outside the core region required for silencing, does contain a canonical Runx motif, CBF1, and competed with the Cd4 probe for Runx binding (Fig. 8A). CE4A-P2 lacks the canonical Runx motif and did not compete for Runx binding. Competitor probe CE4A-P3 competed for Runx binding (Fig. 8A), but when the potential Runx site CBF3 was mutated (Fig. 6A, CE4A-P3m7a), it could no longer compete for the Cd4 silencer Runx complex (Fig. 8A). However, this is not the only site in the CE4A region where Runx could bind. CE4A-P4 contains predicted Runx sites CBF4 and -5 that also competed with the Cd4 probe for Runx binding (Fig. 8A). Note that the Runx sites in all these probes, except for CE4A-P1, map to the locations of mutations that damaged repression in Fig. 5, i.e., mutants M6 to M9. The predicted CBF6 sequence in probe CE4A-P5, which is disrupted by mutant M13, also competed with the Cd4 probe for Runx binding (Fig. 8A). When a longer probe that spanned the M5 to M13 regions of CE4A was used as a competitor, mutations in at least four distinct sites were needed to abolish all Runx competition activity (Fig. 8C, M-all). The CE4B-P3 probe also competed against Cd4, again indicating that this region has a Runx binding site (Fig. 8A). These results suggest not one but multiple potential Runx binding sites in the functionally vital regions of the bipartite silencer.
FIG. 9.
The nonconsensus CE4A-P3 Runx site has weaker binding affinity than the consensus sites of CE8. (A) Western blot of Runx1 and Sp1 protein. The same blot was probed for Runx1 and then Sp1. (B) Sequences of Cd4 and CE8 oligos are shown. Asterisks mark bases mutated in competitors. (C and D) CE4A-P3 binds Runx more weakly than CE8 probes. The CD4 silencer probe was used to assay Runx1 binding potential. Competitors were used at a 100-fold excess except where indicated. Band intensity was quantified by phosphorimaging, with the background subtracted, and data are plotted as relative intensities normalized against lane 1 (one experiment, representative of three).
The one mutation that affected repression in transient assays that seemed not to alter a Runx site was the M5 mutant, a 6-bp mutation which spans the Ets-like site we call X (Fig. 5C). Consistent with the specificity of the assay, smaller mutations crossing the X site did not prevent the CE4A-P3 oligo from competing for Runx binding to the Cd4 silencer probe (Fig. 8B, lanes 5 to 7). Thus, the GGAA core of the X site is not required for the binding of Runx to this probe. However, the full 6-nucleotide M5 mutation also destroys the ability to compete with the Cd4 Runx complex (Fig. 8B, lane 4), implying that the sequence between the X site and CBF3 is important to stabilize Runx binding. Thus, all of the mutations that abolished core silencer function in the scanning mutagenesis assay are involved in formation of Runx complexes in vitro.
Quantitative impact of Runx levels on binding to the CE4A silencer.
Runx1 was already clearly known to play activating and repressive roles in PU.1 expression (17) but was assumed to work only through the CE8 element, where a doublet of canonical Runx sites are separated by ∼50 nt from a Runx/Ets site pair. Runx1 is expressed in myeloid, B, and T cells alike, raising the question of how it mediates such diverse effects in different cells. One potential mechanism could depend on dose. Western blot analysis of Adh.2C2 and RAW 264 nuclear extracts showed that the T-lineage cells had ∼4 times more Runx1 protein than the myeloid cells (Fig. 9A). We therefore asked whether the ability of Runx1 to bind CE4 is quantitatively distinct from its ability to bind CE8.
We noted that the CE4 Runx sites in functionally important regions affected by mutants M6 to M9 and M13 all deviate from the consensus, (Py)G(Py)GGT, unlike CE8 sites (Fig. 9B) Thus, we hypothesized that occupancy of the repression-linked Runx sites in CE4A-4B might require higher Runx expression levels than the sites in CE8.
We examined the binding affinities of the CE8 and CE4A-P3 Runx sites by titrated competitions against the Cd4 Runx probe, with self-competition by cold Cd4 probe as a standard (Fig. 9C). The regions of CE8 containing Runx sites, in probes CE8-P1 and CE8-P3, competed equally strongly, and mutation of the Runx sites proved that this was site specific (CE8-P1m1 and CE8-P3m1) (Fig. 9D). In contrast, the CE4A-P3 probe was a notably weaker competitor, although it included both the highest-scoring Runx site across the functionally important silencer region and the adjacent X site which seemed to enhance Runx binding. Taken together, these data suggest that even the most dominant Runx site in CE4A indeed has a weaker binding affinity than the Runx sites in the URE and might require higher Runx1 levels for occupancy.
Runx protein perturbations and Runx binding site mutations abolish silencer function.
To establish whether Runx1 itself was functionally important for the silencing mediated by CE4A in the T-cell context, we carried out Runx1 gain-of-function experiments in T and myeloid cells (Fig. 10A and B). We also blocked Runx1 function two ways. First, we used cotransfections of a Runx1 dominant negative expression construct (d190) (34) together with the reporters to compete against endogenous Runx1 (Fig. 10A). This Runx1 DN construct includes the full Runx1 DNA binding domain but lacks the C-terminal effector domains. Second, we transfected morpholino antisense oligonucleotides to knock down Runx1 protein levels and morpholinos against the Runx complex partner, CBFβ (Fig. 10C). In both assays, we assessed the effects of the perturbation on CE4-dependent silencing activity by comparing effects on expression of L98 with or without the silencer (the full CE5 to CE3 region or CE4A alone). For controls, cells were also transfected with a morpholino against PU.1 itself. Finally, to assess whether Ikaros, which also appeared to be associated with the T-cell-specific complexes (Fig. 7A and D), might promote silencing, we further transfected the cells with a “dominant negative” mutant form of Ikaros called Plastic (27).
FIG. 10.
Runx perturbations abolish silencing activity in immature T cells (Adh.2C2). (A) Cells were nucleofected with PU.1 reporters, plus 1 to 2 μg of the indicated plasmid. Only cotransfection with the Runx dominant negative expression vector relieved silencing (red bars). (B) Runx1 overexpression in RAW 264.7 cells enhances CE5 to CE3 activity and does not lead to repression. *, P < 0.03. Data are from three experiments performed in duplicate and are reported relative to L1 activity. (C) Western blot of whole-cell lysates from Adh.2C2 cells transfected with or without Runx1 morpholino, showing Runx1 loss. (D) Antisense morpholino knockdown of endogenous Runx1 blocks silencing (red bar). Data from two independent experiments are shown as the L98+4/L98 reporter activity ratio. *, P < 0.02 compared to the no-morpholino data set. (E) Runx sites are essential for silencing in a chromatin context in stably transfected Adh.2C2 cells. With L93 mRunx, all predicted Runx sites in the CE4A-B region mutated. L93 M5, L93 with the M5 mutation; L93 mRunx M5, mutations combined. Data are plotted as for Fig. 4D with data from two independent experiments, each with four to six independent mixed pools of stable cells. *, P < 0.001 compared to L96; #, P < 0.001 compared to L9-3.
Figure 10A shows that added full-length Runx1 sustained or intensified repression mediated by the CE4 silencer in T-lineage cells, whereas competition with Runx1 DN dramatically blocked repression. Figure 10A shows the clear enhancer activity of the CE9 to CE8 URE in the T cells, which was eliminated by inclusion of the CE5 to CE3 regions (Fig. 10A, lanes 11 and 12 versus lanes 3 and 4). Cotransfection with the dominant negative Plastic mutant of Ikaros or exogenous full-length Runx1 had no effect. However, cotransfection of the Runx1 DN relieved silencer activity in a dose-dependent way (Fig. 10A, red bars). Some repression-alleviating effect of the Runx1 DN was also detectable when assayed with the L98 reporter, consistent with evidence that endogenous Runx1 can be repressive at CE8 as well (17); however, this effect was much weaker than when the CE5 to CE3 region is present.
The effect of Runx1 and the general potency of the CE4 element were both cell type specific. We also tested Runx1 overexpression on reporter activity in myeloid cells and found that added Runx1 exerts only a weak repression effect on constructs containing CE4. This effect is dwarfed by its augmentation of the enhancer function of CE5 to CE3 (Fig. 10B), reflecting possible action through stimulatory Runx sites present in the CE5 element.
Endogenous Runx1 protein in the Adh.2C2 cells could be knocked down by transfection with an antisense morpholino (Fig. 10C). This treatment also blocked silencer activity of CE4A, as measured by the ratio of expression driven by L98 plus the CE4 element to expression driven by L98 alone (Fig. 10D). As Runx1 binds DNA in a heterodimer with CBFβ, a morpholino was made to block CBFβ expression, and this also relieved some repression (Fig. 10D). All these effects were cell type specific. In contrast, a morpholino against PU.1 had no effect, nor did a control morpholino against the inverse of the Runx1 sequence.
Finally, the role of the Runx sites in silencing was confirmed in stable transfections by testing the L9-3mRunx reporter in which all predicted Runx sites across CE4A-B were mutated. The impact of the M5 mutation in L9-3 was also evaluated, alone and in combination with mRunx (L9-3 M5 mRunx), as the X site stabilized Runx1 binding in vitro. As shown in stably transfected cells, the L9-3mRunx reporter, with or without the M5 mutation, could no longer be silenced in Adh.2C2 cells (Fig. 10E). In contrast, the L9-3 M5 construct, which retains all Runx sites but has the M5 mutation, was still moderately silenced.
Runx1 binds to the CE4A core silencer in vivo specifically in immature T cells.
These results raised the question why Runx1 could exert repression via CE4 in T cells but not in myeloid cells. To determine whether Runx1 recruitment to CE4 was lineage specific, we carried out ChIP assays against Runx1 on chromatin from RAW 264.7 and Adh.2C2 cells. The results confirmed that Runx1 binding on CE4A is cell type specific in vivo (Fig. 11A and B). No strong Runx1 binding was detected across any part of the PU.1 upstream region in the myeloid cells (Fig. 11A, right), consistent with evidence that it may act there in a hit-and-run style (16). In contrast, there was a strong peak of Runx1 binding at CE4A in the immature T-cell line, stronger than its binding to the URE (Fig. 11B, right). The disparity between strong Runx1 binding at CE4A and weaker signals at CE8 in the T cells was unexpected in view of the established activity of Runx at CE8 and the open chromatin at CE8 in T and B cells alike (15), but this was highly reproducible. Both T and myeloid cells showed similar binding of the Ets family factor GABPα to CE8 (Fig. 11A and B, left panels), confirming that protein-DNA complexes could be detected at CE8 at least as efficiently in the T cells as in myeloid cells, if they were there. Thus, in immature T cells but not in myeloid cells Runx1 is selectively recruited to CE4A, even more than to the URE, consistent with a role in T-cell-specific silencing activity of this element in vivo.
FIG. 11.
Runx1 binds the CE4A silencer region in vivo in T cells. A schematic of the PU.1 upstream region with conserved elements and their approximate location is shown. (A and B) ChIP assays in RAW 264.7 cells (A; red) and Adh.2C2 (B; green). The left panel shows ChIP analysis with antibody against the Ets factor GABPα. The right panel shows the Runx1 ChIP. Results shown are from three or more independent experiments. (C) ChIP assays with Ig failed to enrich any region in Rag2−/− thymocytes (left panel). ChIP assays demonstrated Runx1 binding to the CE4A core silencer in primary thymocytes (right panel). Runx binding to the CD4 silencer is shown for comparison (green bars). Results shown are from two independent experiments. Standard deviations are shown (error bars.).
Runx1 binding to CE4 also occurs in normal T-cell precursors at the stage when they first turn off PU.1 expression, as shown by ChIP assays on chromatin of primary thymocytes from Rag2−/− mice. These are populations in which >90% of cells are blocked in vivo at the pro-T-cell DN3 stage and have newly silenced Sfpi1. Runx1-specific antibodies were strongly enriched for CE4A in Rag2−/− thymocytes, at least as strongly as CE8 (Fig. 11C). The magnitude of enrichment was comparable to the enrichment of the Cd4 silencer in these cells, an internal positive control, because Runx1 is known to repress Cd4 at this stage of development (33) (Fig. 11C). Thus, Runx1 is recruited to CE4A preferentially and lineage specifically under physiological conditions within primary T-cell precursors.
DISCUSSION
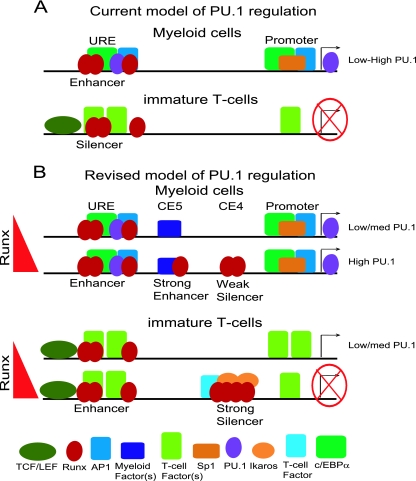
A challenging mechanistic question has been how all the intricate lineage-specific regulation of PU.1 could be achieved through the only two fairly simple cis-regulatory elements known for the Sfpi1 gene, the promoter and the URE. The problem has been intensified by accumulating evidence that has shown the same factor, Runx1, to be both a major positive and negative regulator for the Sfpi1 gene, yet apparently both are through the fixed context of the URE sites (Fig. 12A). Additional mechanisms recently shown to modulate PU.1 expression, such as antisense transcription (11) and miRNA suppression (36), have explained only modest differences in the level, inadequate to account for the 2 to 3 orders of magnitude of repression that Sfpi1 undergoes during T-lineage commitment. Here, we have shown that the true regulatory system for this gene is more complex, involving deployment of multiple, functionally dedicated, conserved elements of Sfpi1 that can mediate developmentally lineage-specific transcriptional regulation. We propose that it is the modulation of URE function by dominant enhancing or repressive inputs at these elements that yields the richness of developmental regulatory patterns for this crucial gene (Fig. 12B).
FIG. 12.
Schematic showing proposed cell-type-specific interactions between the URE (CE8/9) and novel elements CE5 and CE4. (A) Current model of Runx1 dosage-independent bifunctional URE-regulated PU.1 expression. (B) Revised model of Runx1 dosage-sensitive PU.1 regulation. For full details of factors known to bind the promoter and URE, see reference 15.
The nature of these new elements contrasts with the roles proposed for the promoter and the URE, which are essentially bifunctional. The new elements seem specific not only for context-dependent activity but also for valency of function, i.e., activation versus repression. They correspond to conserved sequences and are thus likely to have been evolutionarily selected for these functions. Although the factors that bind these new modules include ones like Runx1, which also binds to three conserved sites in the URE, our results show that the new element CE4A can provide these factors with an alternative site organization and an alternative selection of interaction partners. Thus, at CE4A and CE4B, Runx factors participate in cell-type-specific complexes that they do not generate at CE8 (Fig. 12B). These alternative complexes, organized by binding to the distinct cis-regulatory DNA “scaffolds,” are likely to explain why the impact of Runx1 binding at sites like CE4A can be focused on mediating a repressive transcriptional response.
Identification of novel regulatory elements was needed to explain the early repression of Sfpi1 during T-cell development, because the Sfpi1 UREs CE9 and CE8 retain their enhancer function in an immature DN3-like T-cell line even though they lose enhancer function in more mature T cells. The residual expression of PU.1 observed in UREΔ hematopoietic cells (29) already implied the existence of additional positive regulatory sites outside the URE. Our results in fact identify at least two novel, conserved cis-elements: one that can act as an enhancer in myeloid cells at about kb −10, CE5, as well as the bipartite CE4A plus -4B element that mediates profound silencing in immature T cells, ∼9 kb upstream of the Sfpi1 transcriptional start site. At least CE4A appears to be fully T-cell specific in its function and a selective mediator of negative regulation.
The discovery of CE4 and the ability of this element to exert dominant silencer activity over continued URE enhancer function in immature T cells together offer an elegant explanation for how the dual functionality of the URE is temporally switched in T-cell development. We show that Runx1 in these immature T cells is dose limiting for the effect of this silencer, that Runx1 binds to the CE4A and CE4B regions, and that its sites in the CE4A conserved element are crucial for repressive function of the silencer. In vivo, Runx1 rises to its highest level in immature T cells at the DN3 stage, just as PU.1 expression is first shut off (21, 32). T cells then continue to express one or more of the Runx family members throughout their continued development and mature function, potentially preserving the silence of PU.1 by active repression. Even if this were not the case, the Runx-dependent silencing of the Cd4 gene provides a precedent for a hit-and-run silencing mechanism that Runx proteins can use for lasting effects (33).
Nevertheless, Runx1 is clearly not T-cell specific, and so its effectiveness at the CE4 silencer must be subject to other conditions. Its selective recruitment to the CE4 repression module in T cells likely depends both on increase of Runx1 beyond a dose-dependent threshold, from our evidence that the silencer Runx1 sites are relatively weak, and also on interaction with a T-cell-specific repression cofactor. The evidence for the latter is indirect, based on two findings. First, Runx1 is recruited to the CE4 element in T cells more strongly than to the CE8 element, even though individual Runx binding sites in CE8 are “stronger.” This could be explained if Runx1 interacted with a partner that binds at CE4 but not at CE8. Our results suggest that a T-cell-specific partner might bind at the site X region of CE4A, a region that has an effect on Runx binding affinity as well as on repression in the context of the whole silencer. Clearly, the factor or complex that binds here is of great interest. One motif within site X suggests that a T-cell-specific Ets family factor could be involved, but to date this has been impossible to confirm with a large range of antibodies tested in both gel shift and ChIP assays (M. Zarnegar, unpublished data). However, note that a critical mass of Runx1 binding is still required in order to mediate repression and that in the most rigorous stable transfection assays, the multiplex Runx sites can mediate silencing in the absence of the X element.
Formally, the T-cell-specific recruitment of Runx factors to CE4 could depend on the removal of an antisilencing factor. Against this possibility is the evidence that the CE4 region shows more “open” histone marks in T cells than in myeloid or B cells (M. Zarnegar, unpublished results). The CE4 region is marked by H3K4m1 and H3K4me2 even in mature CD4+ human T cells, as defined in ChIP-seq studies (5) (Fig. 13). Nevertheless, it is interesting that the CE4B region includes a Stat3 binding site shown to mediate Sfpi1 activation (13). The Stat3 site does not overlap but is adjacent to the sites needed for complex B1 formation, and it is possible that Stat3 mobilization by growth factor receptors such as Flt3 or gp130 could prevent assembly of the silencer complex until a key stage of T-cell development and until inflammatory cytokines are absent.
FIG. 13.
CD4+ T-cell ChIP-seq experiments suggested open chromatin across CE4 in normal, mature human T cells. H3K4me2 ChIP-seq data are borrowed from Barski et al. (5) (http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hgtcell.aspx). CE4 is marked by H3K4me2 to a similar extent as the UREs.
In a larger sense, the characterization of the Runx-dependent CE4 silencer provides a prototype for other functionally dedicated, developmentally specific cis-regulatory elements that may collaborate with the URE and the promoter to shape the complex expression pattern of PU.1. For example, we have identified a new myeloid enhancer at kb −10.3, CE5. There is an additional conserved cluster, CE6 and CE7, located from kb −12.5 to −12 that can mediate enhancer activity in a non-cell-type-specific way in transient assays, but it may have more restricted functions in a chromatin context (Zarnegar, unpublished). Additional lineage-specific elements may refine PU.1 expression in other cell types. Recent evidence from others shows that in erythroid cells GATA factors may modulate PU.1 gene expression through binding to another conserved site at kb −17.8, upstream of the URE (9). Although T-cell GATA-3 should also recognize these sites, in vivo GATA-3 does not appear to bind to the −17.8 sites or promoter-associated GATA sites in immature T cells as they silence PU.1 expression (M. Zarnegar and J. Zhang, unpublished results), so these sites may be specific for regulation in erythroid and megakaryocytic lineages. Thus, the complex lineage-specific regulation of Sfpi1 is not played out simply through transcription factor interactions at the URE and the promoter, but also through lineage-specific intermodular interactions between the URE and a variable set of other conserved regulatory elements.
Acknowledgments
We are grateful to Masanobu Satake (Tohoku University, Sendai, Japan) for donating pan-Runx antibody and to Janice Telfer (University of Massachusetts, Amherst, MA) for the Runx1 DN construct. We also thank Jingli Zhang and Georgi Marinov for sharing data before publication and Rochelle Diamond for help in isolating primary mouse tissues and excellent lab management.
This work was supported by grants from the U.S. PHS, R21 DK073658 and R01 CA90233, and by funds from the Louis A. Garfinkle Memorial Laboratory Fund, the Al Sherman Foundation, and the Albert Billings Ruddock Professorship to E.V.R.
Footnotes
Published ahead of print on 9 August 2010.
REFERENCES
- 1.Amaravadi, L., and M. J. Klemsz. 1999. DNA methylation and chromatin structure regulate PU.1 expression. DNA Cell Biol. 18:875-884. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M., A. Weiss, G. Hernandez-Hoyos, C. Dionne, and E. Rothenberg. 2002. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity 16:285-296. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, M. K., G. Hernandez-Hoyos, R. A. Diamond, and E. V. Rothenberg. 1999. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development 126:3131-3148. [DOI] [PubMed] [Google Scholar]
- 4.Arinobu, Y., S.-I. Mizuno, Y. Chong, H. Shigematsu, T. Iino, H. Iwasaki, T. Graf, R. Mayfield, S. Chan, P. Kastner, and K. Akashi. 2007. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell 1:416-427. [DOI] [PubMed] [Google Scholar]
- 5.Barski, A., S. Cuddapah, K. Cui, T.-Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. [DOI] [PubMed] [Google Scholar]
- 6.Cai, D. H., D. Wang, J. Keefer, C. Yeamans, K. Hensley, and A. D. Friedman. 2008. C/EBPα:AP-1 leucine zipper heterodimers bind novel DNA elements, activate the PU.1 promoter and direct monocyte lineage commitment more potently than C/EBPα homodimers or AP-1. Oncogene 27:2772-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., D. Ray-Gallet, P. Zhang, C. J. Hetherington, D. A. Gonzalez, D. E. Zhang, F. Moreau-Gachelin, and D. G. Tenen. 1995. PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene 11:1549-1560. [PubMed] [Google Scholar]
- 8.Chen, H., P. Zhang, H. S. Radomska, C. J. Hetherington, D. E. Zhang, and D. G. Tenen. 1996. Octamer binding factors and their coactivator can activate the murine PU.1 (spi-1) promoter. J. Biol. Chem. 271:15743-15752. [DOI] [PubMed] [Google Scholar]
- 9.Chou, S., E. Khandros, L. C. Bailey, K. Nichols, C. Vakoc, Y. Yao, Z. Huang, J. Crispino, R. Hardison, G. Blobel, and M. Weiss. 2009. Graded repression of PU.1/Sfpi1 gene transcription by GATA factors regulates hematopoietic cell fate. Blood 114:983-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dionne, C., K. Tse, A. Weiss, C. Franco, D. Wiest, M. Anderson, and E. Rothenberg. 2005. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev. Biol. 280:448-466. [DOI] [PubMed] [Google Scholar]
- 11.Ebralidze, A. K., F. C. Guibal, U. Steidl, P. Zhang, S. Lee, B. Bartholdy, M. A. Jorda, V. Petkova, F. Rosenbauer, G. Huang, T. Dayaram, J. Klupp, K. B. O'Brien, B. Will, M. Hoogenkamp, K. L. B. Borden, C. Bonifer, and D. G. Tenen. 2008. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev. 22:2085-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goux, D., J. Coudert, D. Maurice, L. Scarpellino, G. Jeannet, S. Piccolo, K. Weston, J. Huelsken, and W. Held. 2005. Cooperating pre-T-cell receptor and TCF-1-dependent signals ensure thymocyte survival. Blood 106:1726-1733. [DOI] [PubMed] [Google Scholar]
- 13.Growney, J., H. Shigematsu, Z. Li, B. Lee, J. Adelsperger, R. Rowan, D. Curley, J. Kutok, K. Akashi, I. Williams, N. Speck, and D. G. Gilliland. 2005. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106:494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegde, S., S. Ni, S. He, D. Yoon, G. S. Feng, S. S. Watowich, R. F. Paulson, and P. A. Hankey. 2009. Stat3 promotes the development of erythroleukemia by inducing Pu.1 expression and inhibiting erythroid differentiation. Oncogene 28:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoogenkamp, M., H. Krysinska, R. Ingram, G. Huang, R. Barlow, D. Clarke, A. Ebralidze, P. Zhang, H. Tagoh, P. Cockerill, D. Tenen, and C. Bonifer. 2007. The Pu.1 locus is differentially regulated at the level of chromatin structure and noncoding transcription by alternate mechanisms at distinct developmental stages of hematopoiesis. Mol. Cell. Biol. 27:7425-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoogenkamp, M., M. Lichtinger, H. Krysinska, C. Lancrin, D. Clarke, A. Williamson, L. Mazzarella, R. Ingram, H. Jorgensen, A. Fisher, D. Tenen, V. Kouskoff, G. Lacaud, and C. Bonifer. 2009. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 114:299-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, G., P. Zhang, H. Hirai, S. Elf, X. Yan, Z. Chen, S. Koschmieder, Y. Okuno, T. Dayaram, J. Growney, R. Shivdasani, D. G. Gilliland, N. Speck, S. Nimer, and D. Tenen. 2008. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat. Genet. 40:51-60. [DOI] [PubMed] [Google Scholar]
- 18.Kawazu, M., G. Yamamoto, M. Yoshimi, K. Yamamoto, T. Asai, M. Ichikawa, S. Seo, M. Nakagawa, S. Chiba, M. Kurokawa, and S. Ogawa. 2007. Expression profiling of immature thymocytes revealed a novel homeobox gene that regulates double-negative thymocyte development. J. Immunol. 179:5335-5345. [DOI] [PubMed] [Google Scholar]
- 19.Laiosa, C., M. Stadtfeld, H. Xie, L. de Andres-Aguayo, and T. Graf. 2006. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity 25:731-744. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y., Y. Okuno, P. Zhang, H. S. Radomska, H. Chen, H. Iwasaki, K. Akashi, M. J. Klemsz, S. R. McKercher, R. A. Maki, and D. G. Tenen. 2001. Regulation of the PU.1 gene by distal elements. Blood 98:2958-2965. [DOI] [PubMed] [Google Scholar]
- 21.Lorsbach, R., J. Moore, S. Ang, W. Sun, N. Lenny, and J. Downing. 2004. Role of RUNX1 in adult hematopoiesis: analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood 103:2522-2529. [DOI] [PubMed] [Google Scholar]
- 22.McKercher, S. R., B. E. Torbett, K. L. Anderson, G. W. Henkel, D. J. Vestal, H. Baribault, M. Klemsz, A. J. Feeney, G. E. Wu, C. J. Paige, and R. A. Maki. 1996. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15:5647-5658. [PMC free article] [PubMed] [Google Scholar]
- 23.Ninomiya, Y., N. Kotomura, and O. Niwa. 1996. Analysis of DNase I hypersensitive site of the ELP gene. Biochem. Biophys. Res. Commun. 222:632-638. [DOI] [PubMed] [Google Scholar]
- 24.Nutt, S., D. Metcalf, A. D'Amico, M. Polli, and L. Wu. 2005. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J. Exp. Med. 201:221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okada, H., T. Watanabe, M. Niki, H. Takano, N. Chiba, N. Yanai, K. Tani, H. Hibino, S. Asano, M. L. Mucenski, Y. Ito, T. Noda, and M. Satake. 1998. AML1(-/-) embryos do not express certain hematopoiesis-related gene transcripts including those of the PU.1 gene. Oncogene 17:2287-2293. [DOI] [PubMed] [Google Scholar]
- 26.Okuno, Y., G. Huang, F. Rosenbauer, E. Evans, H. Radomska, H. Iwasaki, K. Akashi, F. Moreau-Gachelin, Y. Li, P. Zhang, B. Gttgens, and D. Tenen. 2005. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol. Cell. Biol. 25:2832-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papathanasiou, P., A. Perkins, B. Cobb, R. Ferrini, R. Sridharan, G. Hoyne, K. Nelms, S. Smale, and C. Goodnow. 2003. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity 19:131-144. [DOI] [PubMed] [Google Scholar]
- 28.Rosenbauer, F., B. Owens, L. Yu, J. Tumang, U. Steidl, J. Kutok, L. Clayton, K. Wagner, M. Scheller, H. Iwasaki, C. Liu, B. Hackanson, K. Akashi, A. Leutz, T. Rothstein, C. Plass, and D. Tenen. 2006. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat. Genet. 38:27-37. [DOI] [PubMed] [Google Scholar]
- 29.Rosenbauer, F., K. Wagner, J. Kutok, H. Iwasaki, M. Le Beau, Y. Okuno, K. Akashi, S. Fiering, and D. Tenen. 2004. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 36:624-630. [DOI] [PubMed] [Google Scholar]
- 30.Scott, E. W., M. C. Simon, J. Anastasi, and H. Singh. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265:1573-1577. [DOI] [PubMed] [Google Scholar]
- 31.Spooner, C., J. Cheng, E. Pujadas, P. Laslo, and H. Singh. 2009. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity 31:576-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taghon, T., M. Yui, R. Pant, R. Diamond, and E. Rothenberg. 2006. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity 24:53-64. [DOI] [PubMed] [Google Scholar]
- 33.Taniuchi, I., M. Osato, T. Egawa, M. Sunshine, S. Bae, T. Komori, Y. Ito, and D. Littman. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111:621-633. [DOI] [PubMed] [Google Scholar]
- 34.Telfer, J., E. Hedblom, M. Anderson, M. Laurent, and E. Rothenberg. 2004. Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J. Immunol. 172:4359-4370. [DOI] [PubMed] [Google Scholar]
- 35.Tydell, C. C., E.-S. David-Fung, J. Moore, L. Rowen, T. Taghon, and E. Rothenberg. 2007. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J. Immunol. 179:421-438. [DOI] [PubMed] [Google Scholar]
- 36.Vigorito, E., K. L. Perks, C. Abreu-Goodger, S. Bunting, Z. Xiang, S. Kohlhaas, P. P. Das, E. A. Miska, A. Rodriguez, A. Bradley, K. G. C. Smith, C. Rada, A. J. Enright, K.-M. Toellner, I. C. M. MacLennan, and M. Turner. 2007. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 27:847-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, Y., D. Banerjee, J. Huelsken, W. Birchmeier, and J. Sen. 2003. Deletion of beta-catenin impairs T cell development. Nat. Immunol. 4:1177-1182. [DOI] [PubMed] [Google Scholar]
- 38.Yeamans, C., D. Wang, I. Paz-Priel, B. Torbett, D. Tenen, and A. Friedman. 2007. C/EBPα binds and activates the PU.1 distal enhancer to induce monocyte lineage commitment. Blood 110:3136-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]