Abstract
Epigenetic regulatory information must be retained during mammalian cell division to sustain phenotype-specific and physiologically responsive gene expression in the progeny cells. Histone modifications, DNA methylation, and RNA-mediated silencing are well-defined epigenetic mechanisms that control the cellular phenotype by regulating gene expression. Recent results suggest that the mitotic retention of nuclease hypersensitivity, selective histone marks, as well as the lineage-specific transcription factor occupancy of promoter elements contribute to the epigenetic control of sustained cellular identity in progeny cells. We propose that these mitotic epigenetic signatures collectively constitute architectural epigenetics, a novel and essential mechanism that conveys regulatory information to sustain the control of phenotype and proliferation in progeny cells by bookmarking genes for activation or suppression.
Each cell lineage exhibits distinct epigenetic instructions, which can be reversibly modified to regulate gene expression (30, 34, 76). Posttranslational modifications of nucleosomal histone proteins and the methylation of gene promoters are two extensively characterized epigenetic mechanisms that regulate gene expression and influence the cellular phenotype without altering the genotype (12, 63, 83, 88). Interrelationships between epigenetic mechanisms and cellular phenotype are well defined (30, 34, 76). For example, a series of specific posttranslational modifications that include phosphorylation, acetylation, and methylation on the amino-terminal tails of histones are associated with either physiologically responsive gene activation and suppression or irreversible silencing (83, 88). These patterns support the concept of a histone code in which specific histone modifications dictate the functional status of DNA regulatory regions (i.e., promoters) of genes (44). Similarly, the methylation of GC-rich gene promoters during embryonic development results in the permanent silencing of genes that are required only for early developmental events and are not expressed in adult organisms under physiological conditions (12, 34, 63). Noncoding RNA molecules are emerging as yet another mechanism of epigenetic control (27, 31, 64). In addition to developmental and regulatory roles, epigenetic control is required to sustain a transformed phenotype as well as to support tumor progression (26, 46). A subset is inheritable to maintain cellular identity (54, 68, 72, 75). Combined with recent observations that some transcription factors are retained on mitotic chromosomes (2, 98-100), the mitotically inheritable epigenetic mechanisms constitute a novel concept of “architectural epigenetics,” where the combinatorial outcome of mitotically retained epigenetic mechanisms dictates cellular potential for lineage commitment and growth in progeny cells (Fig. 1). We will briefly consider the principal components of epigenetic control in mammalian cells. For more extensive coverage, we refer to in-depth reviews discussing multiple dimensions of epigenetic control (12, 27, 30, 31, 34, 44, 63, 64, 76, 83, 88).
FIG. 1.
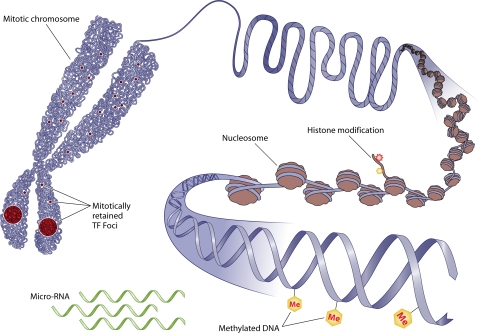
Mechanisms of inheritable epigenetics. Mammalian gene expression is tightly controlled by genetic as well as epigenetic mechanisms. Epigenetics modifies the phenotype without altering the genotype of a cell. Shown here are some well-defined epigenetic mechanisms that include histone modifications, DNA methylation, and the noncoding RNA-mediated modulation of gene expression. Some of these mechanisms are inheritable through successive cell divisions and contribute to the maintenance of cellular phenotype. Recent studies show that the association of components of transcriptional regulatory machinery with target genes on mitotic chromosomes is a novel epigenetic mechanism that poises genes involved in key cellular processes, such as growth, proliferation, and lineage commitment, for expression in progeny cells.
DNA METHYLATION
DNA methylation is a covalent modification that occurs at CpG dinucleotides and can be catalyzed by three different enzymes, the DNA methyl transferases (DNMTs) DNMT1, DMNT3a, and DNMT3b (7). DNA methylation plays a critical role in the long-term silencing of transcription and in heterochromatin formation either by direct interference with the binding of transcription factors to their target sites or by affecting histone modifications and nucleosome occupancy within the promoter regions of genes, thereby altering chromatin structure (12, 13, 63). Some methylation is reversible; cells can eliminate methyl moieties from CpG islands of certain genes enzymatically or through replication (7, 63). During the development of cancer, many CpG islands undergo hypermethylation, which leads to changes in chromatin structure and causes the silencing of tumor suppressor genes as well as the instability of the genome (46). CpG methylation is also an important constituent in X chromosome inactivation (35, 65). As an epigenetic modification, DNA methylation permits these silenced states to be inherited through multiple cellular divisions (54, 68, 72, 75).
HISTONE MODIFICATIONS
Nucleosomal histones are modified at more than 60 different residues (18, 83). These modifications are dynamic, and enzymes that add or remove the modifications are well characterized (19, 81). These modifications can lead either to the disruption of contacts between nucleosomes to increase chromatin accessibility or to the recruitment of nonhistone proteins. Thus, depending on the cohort of histone modifications, or histone code, the access of regulatory proteins to a DNA sequence motif can be either enhanced or reduced (9, 37, 74). Histone modifications as well as some histone variants have been implicated in a number of epigenetic phenomena (14, 54, 84). One example is H3K9 methylation in the transmission of heterochromatin. This modification, which in turn recruits the repression-associated factor HP1, brings in further H3K9-methylating activity that modifies nucleosomes on the daughter strand, thus ensuring the transmission of the H3K9me mark (40, 70). In addition, the incorporation of histone variants can dictate the transcriptional state of a gene, and some remain associated with bookmarked genes. It remains to be determined whether the histone modification signature inherited by the progeny chromatin is sufficient to impose the correct chromatin structure originating from the parent cell.
NONCODING RNA MOLECULES
Recently, it has become evident that noncoding RNAs control multiple epigenetic phenomena (27, 31, 64). For example, dosage compensation mechanisms in Drosophila by rox RNA and in mammals mediated by XIST RNA, Piwi-interacting RNA (piRNA) in Drosophila, and small RNAs in Caenorhabditis elegans are established components of epigenetic control, as they can induce long-term inheritable gene silencing (21, 29, 47, 52, 58, 78). These noncoding RNAs often act in concert with components of chromatin and the DNA methylation machinery to establish and/or sustain silencing and may contribute to development, transformation, and tumor progression (26, 33). Additional species of small, noncoding RNA molecules are emerging as potentially significant contributors to the control of gene expression. These include microRNAs (miRs) that play a vital role in the regulation of key cellular processes, such as the cell cycle, cell growth, and differentiation. MicroRNAs modulate gene expression in an epigenetic manner and are candidates for novel components of regulatory networks in a broad spectrum of circumstances (27, 31, 64). To appreciate the mechanistic relationship of RNA-mediated silencing to epigenetic components of biological control, it is necessary to establish the extent to which RNA-mediated silencing is heritable, whether RNA control of gene expression is reversible or permanent, and the mechanisms that support RNA-directed epigenetic control.
ARCHITECTURAL EPIGENETICS: MITOTIC RETENTION OF TRANSCRIPTIONAL REGULATORY INFORMATION
Genome-wide global profiling of epigenetic parameters, including histone modifications, DNA methylation, and noncoding RNA expression, has made it possible to identify epigenetic signatures that are unique to specific biological conditions. Informational content provided by epigenetic signatures is equally important to sequence information encoded by nucleic acids and can be predictive of biological outcome or pathological conditions (10, 90). For example, epigenetic modifications influence structure and function; histone modifications reflect alterations in chromatin structure at a local level to permit the gene promoter access of transcription factors for activation and suppression (9, 19, 37, 63, 74, 81, 83). At a higher-order level, the clustering of epigenetic changes leads to specialized nuclear microenvironments (101). A similar epigenetic influence on gene structure and function can be attributed to DNA methylation (e.g., the presence of CpG islands in gene promoters) and RNA-mediated gene silencing (e.g., X chromosome inactivation) (31, 58, 78). Because there is a causal relationship between epigenetic signatures and biological outcome, it is critical to maintain some or all of these signatures through successive cell divisions. Recent studies have identified various mechanisms that may contribute to the mitotic retention of cellular phenotype (2, 98-100) (Fig. 2). We propose that these mitotically inheritable mechanisms constitute architectural epigenetics, a novel mechanism to “bookmark” genes for activation or suppression as cells exit mitosis and resume cell cycle progression. Here, we discuss various studies in support of this concept and present a case for its biological relevance and possible implications in sustaining the tumor phenotype.
FIG. 2.
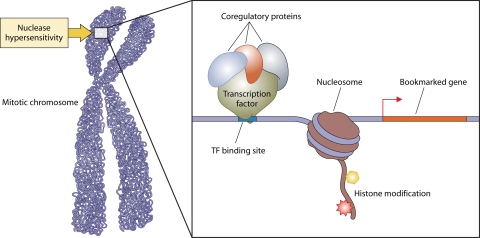
Gene bookmarking: the retention of transcriptional regulatory machinery with target genes on mitotic chromosomes. Mitotic chromosomes represent tightly packed, transcriptionally repressed chromatin and are characteristically resistant to nuclease accessibility. However, some regions of mitotic chromosomes remain sensitive to nuclease activity (depicted here by a white triangle), indicating that some genes remain in open chromatin conformation, thus giving rise to the concept of gene bookmarking. Advances in cell biological (e.g., in situ immunofluorescence) and biochemical (e.g., chromatin immunoprecipitation) assays have allowed a direct examination of protein complexes (shown here by green and gray ovals and a red star) that are retained on target gene promoters during mitosis. In addition, certain histone modifications and variants that are associated with open chromatin conformation also have been observed on mitotic chromosomes. These observations together indicate that certain genes are bookmarked for expression immediately after mitosis, a mechanism that may be pivotal to the maintenance of cellular memory for growth and proliferation potential as well as for lineage identity in progeny cells.
HALLMARKS OF GENES TO BE BOOKMARKED
It is an intriguing observation that only some genes are bookmarked for expression during mitosis while most remain in a repressed, closed chromatin conformation until after the cell has exited mitosis. Consistently with cellular requirements for the control of growth, proliferation, and phenotype, the retention of competency for the expression of genes transcribed by RNA polymerases I and II (Pol I and II) must be preserved during mitosis. Based on observations from our laboratory and others (2, 32, 45, 61, 85, 89, 92, 98-100), we have outlined below key characteristics of mammalian genes that are mitotically bookmarked for postmitotic regulation.
BOOKMARKED GENES ARE ESSENTIAL FOR MAINTENANCE OF KEY CELLULAR PROPERTIES
We and others have discovered that genes that are mitotically bookmarked and epigenetically preserved for expression often are components of essential cellular pathways, for example, cell growth, cell proliferation, and cell differentiation (2, 32, 45, 61, 85, 89, 92, 98-100). We will discuss in depth each of these examples and regulatory implications if gene bookmarking is perturbed and/or modified.
CELL GROWTH
rRNA gene transcription provides a striking link between specialized transcription and nuclear compartmentalization (8). In the mammalian genome, several hundred copies of the rRNA genes are organized as tandem arrays at multiple loci on several chromosomes and are transcribed by RNA polymerase I. During interphase, the ribosomal regulatory machinery in diploid cells is configured as two nucleoli, the sites of ribosome biogenesis. In proliferating cells, rRNA genes are actively transcribed to accommodate the demand for protein synthesis, an essential requirement for cell growth (8, 57, 95). The transcriptional output of rRNA genes fluctuates during the cell cycle; maximal activity occurs during G1, diminishes during G2, and ceases during mitosis (49). In mitotic cells, the tandem arrays of rRNA genes reside at nucleolar organizer regions (NORs) (91). Upstream binding factor (UBF), a regulatory protein that is required for rRNA transcription, remains associated with mitotic NORs (41, 82). Similarly, the SL1 complex that functions cooperatively with UBF interacts with components of RNA Pol I machinery and colocalizes with RNA Pol I to activate transcription (5). Consequently, the three components (RNA Pol I complex, UBF, and SL1) of the ribosomal DNA (rDNA) transcription machinery that are sufficient to promote rRNA gene transcription in vitro are associated with the NORs during mitosis (82). The mitotic silencing of the rRNA genes can be attributed to global chromatin condensation, the posttranslational modification of transcriptional activators, and/or the dissociation of key regulatory components from the rDNA repeats. While the extent to which these mechanisms contribute to the mitotic repression of rRNA transcription remains to be established, the retention of regulatory factors for rRNA gene expression during mitosis provides a novel component of epigenetic control to support cell growth when cell division is completed.
CELL PROLIFERATION
Myc is a well-recognized mediator of growth control during development as well as tissue remodeling. The c-myc gene promoter is rapidly reactivated after mitosis (62). Several DNA binding proteins can modulate c-myc promoter activity. These include hnRNP K and FBP; both proteins are sequence-specific single-stranded DNA binding proteins (24, 60, 62). Another example is provided by the gene promoter of a crucial inhibitor of cyclin-dependent kinases, p21. The p21 promoter remains occupied by the osteogenic Runx2 transcription factor (100), which has an antiproliferative role, for postmitotic regulation in progeny cells. Although it remains to be seen how gene bookmarking contributes to postmitotic cellular potential for proliferation, these examples highlight the importance of bookmarking genes that are critical for postmitotic cell progression.
CELL DIFFERENTIATION
Runx proteins are master determinants of osteogenesis, hematopoeisis, neurogenesis, and gastrointestinal development (22, 25, 51). Runx transcription factors interact with cognate DNA elements through a specific DNA binding domain and with components of nuclear architecture through the nuclear matrix targeting sequence (NMTS) in microenvironments within the nucleus (51). The localization of Runx factors to subnuclear domains is required for biological activity; compromised intranuclear trafficking results in the mouse phenotype that occurs when the Runx genes are ablated (15, 23, 94). When the fidelity of Runx protein localization within the nucleus is perturbed, a transformed phenotype is observed in myeloid progenitors, and there is an inhibition of osteolytic disease caused by breast cancer cells that metastasize to bone (43, 94).
During mitosis, Runx proteins associate with RNA Pol I-transcribed rRNA genes as well as RNA Pol II-transcribed phenotypic genes that are involved in the control of the cell cycle and differentiation (99, 100). Runx regulatory proteins are equally partitioned to progeny cells at the completion of cell division (102). Runx association with ribosomal and cell cycle regulatory genes (e.g., p21) during mitosis marks these genes for repression during the early G1 phase of the cell cycle, which is consistent with the growth-inhibitory properties of these regulatory factors (71). The occupancy of differentiation-related genes by Runx proteins during mitosis provides a mechanistic basis for transcriptional memory in progeny cells. Only recently, it has become evident that some Runx coregulatory proteins, such as TLE1, are carried through mitosis (3). However, the extent to which the entire cohort of Runx coregulatory proteins remains in functional complexes that are associated with target genes on mitotic chromosomes, as well as the time line for the disassembly and reconstitution of other components of the Runx regulatory machinery, must be determined.
The basic helix-loop-helix myogenic regulatory factors MyoD, Myf5, myogenin, and MRF4 have critical roles in skeletal muscle development (6, 66). Together with the Mef2 proteins and E-box factors, these transcription factors are responsible for coordinating muscle-specific gene expression by negatively regulating proliferation and positively accelerating differentiation (6, 48). During interphase, muscle regulatory proteins are organized at punctate nuclear microenvironments. During the proliferative stage of uncommitted mesenchymal cells, MyoD is localized to mitotic chromosomes and associates with NORs where rRNA genes reside (2). The association of MyoD with the interphase nucleolus in early stages of myogenesis and its replacement by myogenin in later stages result in the suppression of rRNA genes concomitantly with the initiation of the skeletal muscle differentiation program (2, 6). These observations are consistent with a novel epigenetic mechanism in which phenotypic regulatory proteins associate with mitotic chromosomes to convey transcriptional machinery for cell growth and differentiation from one progeny to the next.
CCAAT/enhancer binding proteins β and δ (C/EBPβ/δ) are expressed early in the adipocyte differentiation program but are not immediately active (53). After a long lag, C/EBPβ/δ become competent to bind to the C/EBP regulatory element in the C/EBPα gene promoter, with C/EBPα being a transcriptional activator of numerous adipocyte genes. As C/EBPβ/δ acquire binding activity, they also become localized to centromeres as preadipocytes synchronously enter S phase at the onset of mitotic clonal expansion (92). Localization to centromeres occurs through C/EBP consensus binding sites in centromeric satellite DNA. C/EBPα, which is antiproliferative, becomes centromere associated much later in the differentiation program as mitotic clonal expansion ceases and the cells become terminally differentiated. The differential association of C/EBP transcription factors with mitotic chromosomes and their direct suppression of rRNA genes suggest that these phenotypic regulatory proteins mediate lineage commitment and maintenance through their association with metaphase chromosomes during the mitotic clonal expansion of preadipocytes (2, 92).
NUCLEASE ACCESSIBILITY DURING MITOSIS
Levens and colleagues have shown that several genes that are rapidly reactivated after mitosis share major regions of permanganate sensitivity in their promoter regions only during mitosis (61). Their observations point to the bookmarking of these promoters by regulatory protein(s) postmitotically, as the permanganate sensitivity of these regions is not detected in other cell cycle stages. These regulatory protein(s) may associate with the promoter regions in a cell cycle stage-specific manner to keep them in an open conformation and poise them for rapid transcriptional activation as cells exit mitosis. Here we discuss two examples.
The promoter regions of some genes, such as the stress-inducible hsp70i gene that encodes a heat shock protein, remain uncompacted during mitosis (98). It has been shown that the hsp70i bookmarking is mediated by the transcription factor HSF2, which binds this promoter in mitotic cells, recruits protein phosphatase 2A, and interacts with the CAP-G subunit of the condensin enzyme to promote the efficient dephosphorylation and inactivation of condensin complexes in the vicinity, thereby preventing compaction at this site. Blocking HSF2-mediated bookmarking by HSF2 RNA interference decreases hsp70i induction and the survival of stressed cells in the G1 phase, which demonstrates the biological importance of gene bookmarking and its pivotal role in postmitotic gene expression (85, 98).
The developmentally regulated Globin gene loci contain several functionally related genes arranged in tandem (11). In hematopoietic progenitor cells, erythroid-specific DNase I-hypersensitive sites have been detected in the distal regulatory regions of the mouse globin gene cluster. Progeny cells can inherit these hypersensitive sites for at least 20 generations (59). Recent studies have provided mechanistic insight into the inheritance of globin gene transcription status through mitoses (97). The lineage-restricted expression of globin genes is controlled mainly by the transcription factors NF-E2 and GATA (55, 96). NF-E2 remains bound to mitotic chromosomes, while GATA1 is dissociated from the condensed chromatin during mitosis (97). This observation suggests that NF-E2 is an epigenetic marker that maintains the locally hypersensitive state of the globin gene clusters. NF-E2 also can recruit TAFII130 and CBP to the globin gene locus, further supporting its role in the rapid reactivation of the gene postmitotically. In addition, the distal regulatory regions of transcriptionally competent globin gene loci are marked by active histone modifications such as H3 acetylation, H3 K4 dimethylation, and K79 dimethylation (97).
RETENTION OF HISTONE MARKS
The remarkable stability of gene expression in somatic cells is exemplified by the retention of an active gene state when an endoderm cell nucleus is transplanted to an enucleated egg (38). Nuclear transplant experiments have revealed that transcriptional memory can persist through 24 cell divisions (67). The expression of the myogenic gene MyoD in non-muscle cell lineages of nuclear transplant embryos reflects this memory state. Transcriptional recollection is not explained by the methylation of promoter DNA. Rather, epigenetic memory correlates with the association of histone H3.3 with the MyoD promoter in embryos that display memory but not in those where memory has been lost. The association of a mutated histone H3.3 (H3.3 E4, which lacks the methylatable H3.3 lysine 4) with promoter DNA eliminates memory, further supporting a requirement of H3.3 K4 for memory. The overexpression of H3.3 can enhance memory in transplanted nuclei (67, 69). Thus, the association of histone H3.3 variant with the MyoD promoter makes a necessary contribution to transcriptional memory and suggests that epigenetic memory helps to stabilize gene expression in normal development; it also might partly account for the inefficient reprogramming in some transplanted nuclei.
WHY BOOKMARK A GENE?
Gene bookmarking may be important for lineage commitment at an early developmental stage. For example, in progenitor/stem cells, the genetic material is distributed in a symmetrically equivalent manner. However, to give rise to a lineage-committed progeny cell while maintaining a pool of stem cells, it is necessary to asymmetrically distribute epigenetic signatures that include histone marks, DNA methylation, and the retention of transcription factors. The resultant two classes of progeny cells are not equivalent, but both are necessary to support tissue-specific functions. One of the progeny will retain stemness for the maintenance of a perpetual, lineage-committed stem cell pool to support tissue renewal and the capacity for wound healing. The second progeny serves as an immediate source of specialized cells for organogenesis during development and for physiologically responsive tissue renewal, for example, during injury. Cancer stem cells represent yet another dimension to progenitor populations where active proliferation and/or long-term dormancy requires the perpetuation of the transformed and/or tumor phenotype.
The examples we have provided here offer mechanistic insights into postmitotic gene regulation and reflect experimental evidence that the mitotic retention of the gene regulatory machinery is a novel epigenetic mechanism for the coordination of lineage commitment, cell proliferation, and the differentiation of committed cells. We propose a model in which phenotypic transcription factors and components of the basal transcriptional machinery are retained on mitotic chromosomes to convey necessary information to initiate and sustain lineage commitment. However, it is unrealistic to suggest that all genes are epigenetically regulated by any one mechanism, and quite likely a series of epigenetic mechanisms is operative for the same genes. For example, a gene required for lineage maintenance in postmitotic cells may carry a unique histone modification and an accessible chromatin conformation and may be occupied by a sequence-specific transcription factor; taken together, these distinct yet functionally overlapping epigenetic mechanisms can provide a mitotic epigenetic signature for a gene that is bookmarked for postmitotic expression.
One of many regulatory implications of an epigenetic signature for gene bookmarking during mitosis may include poising genes involved in the progression of the G1 phase of the cell cycle for expression. The mitotic association of phenotypic regulatory proteins with genes in an allele-specific manner in omnipotent and pluripotent cells might facilitate the asymmetric distribution of transcription factors to cells destined to commit to a particular lineage. In committed cells, the equal partitioning of phenotypic transcription factors then can lead to the maintenance of the lineage. Further experimental exploration of our model will provide mechanistic insights into coordinate control of the cell cycle, proliferation, and differentiation.
ASYMMETRIC GENE BOOKMARKING
Asymmetric cell division that ensures the preservation of stem cells is characterized by the division of a pluripotent cell into genetically identical yet functionally distinct progeny cells. One of the progeny cells is destined for lineage commitment, while the other remains in an undifferentiated state. Recent studies of budding yeast, C. elegans, and Drosophila have provided some mechanistic insight into the process of asymmetric cell division (4, 20, 42, 93). For example, in budding yeast, the Ace2 transcription factor, a homologue of chromatin remodeling factor Swi5, accumulates only in the nucleus of the daughter cell. This asymmetry is achieved by the specific phosphorylation and dephosphorylation of Ace2. Ace2 protein is translated in G2 cells but is retained in the cytoplasm until late mitosis, when dephosphorylation leads to the unmasking of the nuclear localization signal and the nuclear translocation of the protein. Once in the daughter nucleus, Ace2 is phosphorylated by daughter cell-specific Cbk1 kinase, which results in the masking of the export signal and daughter cell-specific nuclear retention of Ace2 (17).
Asymmetric cell division also has been described during Drosophila neurogenesis, where it ensures diversity in the fate of cells originating from common progenitors (reviewed in reference 42). The neuroblast progenitor of the central nervous system undergoes several rounds of asymmetric division, each giving rise to a large cell that remains a neuroblast, and a smaller ganglion mother cell (GMC) that is the precursor to neurons and glia of the central nervous system. In parallel, the sensory organ precursor (SOP) undergoes several rounds of asymmetric cell division to give rise to five distinct cell types of the peripheral nervous system. In both GMC and SOP, Numb, an attenuator of Notch signaling, along with the Prospero transcription factor, segregates asymmetrically to only one of the daughter nuclei. The polarity in the localization of these proteins is accomplished by a protein complex that localizes to the apical cortex of dividing neuroblasts. The activity of the apical complex regulates mitotic spindle orientation along the apical basal axis, determines cell size asymmetry between the neuroblast and GMC, and targets Numb and associated proteins to the basal crescent of the cell during asymmetric cell division (36, 50, 77; also reviewed in reference 80).
The examples described here underscore the existence of multiple mechanisms driving asymmetric cell division in lower eukaryotes. It is safe to assume that mammalian cells have equally diverse mechanisms to ensure the maintenance of stem cell pools while giving rise to distinct lineages from single progenitors. Recent studies indicate that the asymmetric distribution and localization of regulatory proteins is indeed a defining factor during the lineage commitment of a progenitor cell. Using gut epithelium, a continuously renewed tissue that contains a basal layer of progenitor cells from which the gut epithelium originates, as a paradigm, Quyn et al. (73) show that the spindle orientation of progenitor cells dictates which cells will become lineage committed and which will remain progenitor cells. The spindle is oriented perpendicularly to the apical surface in the stem cell compartment, and this orientation correlates with the asymmetric retention of labeled DNA in the basal cell. Importantly, this asymmetric cell division is lost in precancerous and cancerous tissues. While this study does not explore further the underlying mechanisms that control this asymmetric cell division, it can be assumed that components of transcriptional regulatory machinery dictate the specific orientation of the spindle and are lost in gastric cancer cells. Taken together, these studies point to a fundamental role for the asymmetric distribution of regulatory machinery in lineage commitment through mechanisms that include gene bookmarking.
MECHANISM(S) OF GENE BOOKMARKING
The studies discussed above show that there are at least three different mechanisms by which a gene may be bookmarked: (i) the retention of lineage-defining factors on target genes during mitosis, (ii) posttranslational modifications of regulatory factors, and (iii) the recruitment of Condensin dephosphorylation machinery. These mechanisms may not be mutually exclusive, as has been shown for the hsp70i gene.
OCCUPANCY BY LINEAGE-DEFINING FACTORS
The retention of lineage-defining factors on target genes during mitosis has multiple biological implications. A lineage-defining factor must orchestrate the concomitant activation of a differentiation program and an exit from the cell cycle. Because of these properties, most lineage-defining factors have cognate DNA binding elements in many genes involved in lineage commitment and cell growth and proliferation. As has been shown, and as discussed above, for osteogenic and hematopoietic Runx proteins, muscle regulatory factors, and adipogenic C/EBP proteins (2, 99, 100), the association of lineage determinants with target genes during mitosis appears to be a global mechanism of mitotic gene bookmarking to preserve cellular memory for lineage maintenance and growth.
POSTTRANSLATIONAL MODIFICATIONS OF REGULATORY FACTORS
In an interphase cell, posttranslational modifications play a central role in modulating the activity of a variety of regulatory proteins that include components of the basal transcription machinery, downstream effectors of many signaling pathways, and lineage-restricted transcription factors. It is possible that posttranslational alterations such as the phosphorylation and acetylation of regulatory proteins also regulate the retention or exclusion of these factors from mitotic chromatin. For example, it has been shown that the Oct1 transcription factor is displaced from mitotic chromosomes (28, 56), and this displacement is regulated by a mitosis-specific phosphorylation in the DNA binding domain of the protein (79, 87). In contrast, it is reported that components of the basal transcriptional machinery (e.g., TFIID) associate with condensed mitotic chromosomes, although their activities are inhibited during mitosis through phosphorylation (1, 16, 86). These findings suggest that posttranslational modifications, especially phosphorylation, play a critical role in determining the mitotic occupancy of target genes by transcriptional regulatory factors.
RECRUITMENT OF CONDENSIN DEPHOSPHORYLATION MACHINERY
Chromosomal condensation during mitosis is achieved by the activity of the condensin family of proteins (39). Condensin proteins associate with chromosomes prior to mitosis and induce the global condensation and packaging of nucleosomal DNA into higher-order chromatin. The phosphorylation of condensin proteins plays a pivotal role in their activity. Studies from the Sarge group have revealed that on the hsp70i promoter, HSF2 recruits PPA2, a phosphatase that dephosphorylates condensin proteins in the vicinity of the hsp70i promoter, thus preventing local chromosomal condensation (98). It is conceivable that similar mechanisms exist for other genes that are bookmarked during mitosis. Future studies identifying this as a universal mechanism should yield interesting insights into mitotic gene bookmarking.
PROSPECTS
The full implications for the mitotic association of regulatory machinery with gene loci in chromosomes remain to be explored experimentally. Compelling questions include the following. Do genes that do not retain cognate regulatory factors have mitotic bookmarks for activation or suppression in progeny cells? Are genes epigenetically regulated in a selective manner during asymmetric cell division? What are the proteins in the cohort of (co)regulatory proteins that are components of the transcriptional machinery occupying mitotic gene loci? Is the mitotic retention of epigenetic regulatory cues operative in cells that undergo endomitosis? Despite these unresolved questions, this epigenetic mechanism of postmitotic gene regulation may be clinically relevant. Current therapeutic strategies target major chromatin modification pathways that also are required for the physiological control of gene expression, thus resulting in detrimental off-target effects. The mitotic association of transcription factors offers novel options for targeted therapy with enhanced specificity and reduced off-target activity compared to that of the global inhibition of histone modifications or DNA methylation. For example, during mitosis, only minimal components of transcriptional complexes are present and may unmask epitopes to generate a drugable target. We have found that to be the case for Runx2, the osteogenic transcription factor that is upregulated in metastatic breast and prostate cancers. While Runx2 interacts with many coregulatory proteins, only TLE1 has been shown thus far to be carried to the progeny cells through its interaction with Runx2 (3). In this example, the therapeutic approach can be focused only on Runx2 and TLE1. Importantly, the mitotic association of regulatory proteins results in the focal concentration of factors that can favorably influence pharmacological kinetics, i.e., a minimal drug concentration will be required. In addition, the mitotic association of regulatory proteins, combined with the global, genome-wide assessment of histone modifications and DNA methylation, provides an epigenetic signature for diagnosis, prognosis, and cancer progression as well as for monitoring therapy.
Acknowledgments
We thank Patricia Jamieson for her assistance in the preparation of this article.
This work was supported by National Institutes of Health grants P01 AR048818, P01 CA082834, 5R37 DE012528, and 5 P30 DK32520.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print on 9 August 2010.
REFERENCES
- 1.Akoulitchev, S., and D. Reinberg. 1998. The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev. 12:3541-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, S. A., S. K. Zaidi, C. S. Dacwag, N. Salma, D. W. Young, A. R. Shakoori, M. A. Montecino, J. B. Lian, A. J. van Wijnen, A. N. Imbalzano, G. S. Stein, and J. L. Stein. 2008. Phenotypic transcription factors epigenetically mediate cell growth control. Proc. Natl. Acad. Sci. U. S. A. 105:6632-6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali, S. A., S. K. Zaidi, J. R. Dobson, A. R. Shakoori, J. B. Lian, J. L. Stein, A. J. van Wijnen, and G. S. Stein. 2010. Transcriptional corepressor TLE1 functions with Runx2 in epigenetic repression of ribosomal RNA genes. Proc. Natl. Acad. Sci. U. S. A. 107:4165-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armakolas, A., M. Koutsilieris, and A. J. Klar. 2010. Discovery of the mitotic selective chromatid segregation phenomenon and its implications for vertebrate development. Curr. Opin. Cell Biol. 22:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell, S. P., R. M. Learned, H. M. Jantzen, and R. Tjian. 1988. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241:1192-1197. [DOI] [PubMed] [Google Scholar]
- 6.Berkes, C. A., and S. J. Tapscott. 2005. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 16:585-595. [DOI] [PubMed] [Google Scholar]
- 7.Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9:2395-2402. [DOI] [PubMed] [Google Scholar]
- 8.Boisvert, F. M., K. S. van, J. Navascues, and A. I. Lamond. 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell. Biol. 8:574-585. [DOI] [PubMed] [Google Scholar]
- 9.Cairns, B. R. 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461:193-198. [DOI] [PubMed] [Google Scholar]
- 10.Calin, G. A., and C. M. Croce. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6:857-866. [DOI] [PubMed] [Google Scholar]
- 11.Cao, A., and P. Moi. 2002. Regulation of the globin genes. Pediatr. Res. 51:415-421. [DOI] [PubMed] [Google Scholar]
- 12.Cedar, H., and Y. Bergman. 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10:295-304. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, X., and R. M. Blumenthal. 2010. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry 49:2999-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung, P., and P. Lau. 2005. Epigenetic regulation by histone methylation and histone variants. Mol. Endocrinol. 19:563-573. [DOI] [PubMed] [Google Scholar]
- 15.Choi, J.-Y., J. Pratap, A. Javed, S. K. Zaidi, L. Xing, E. Balint, S. Dalamangas, B. Boyce, A. J. van Wijnen, J. B. Lian, J. L. Stein, S. N. Jones, and G. S. Stein. 2001. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc. Natl. Acad. Sci. U. S. A. 98:8650-8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christova, R., and T. Oelgeschlager. 2002. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 4:79-82. [DOI] [PubMed] [Google Scholar]
- 17.Colman-Lerner, A., T. E. Chin, and R. Brent. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107:739-750. [DOI] [PubMed] [Google Scholar]
- 18.Cosgrove, M. S. 2007. Histone proteomics and the epigenetic regulation of nucleosome mobility. Expert Rev. Proteomics 4:465-478. [DOI] [PubMed] [Google Scholar]
- 19.Couture, J. F., and R. C. Trievel. 2006. Histone-modifying enzymes: encrypting an enigmatic epigenetic code. Curr. Opin. Struct. Biol. 16:753-760. [DOI] [PubMed] [Google Scholar]
- 20.Cowan, C. R., and A. A. Hyman. 2004. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu. Rev. Cell Dev. Biol. 20:427-453. [DOI] [PubMed] [Google Scholar]
- 21.Cox, D. N., A. Chao, J. Baker, L. Chang, D. Qiao, and H. Lin. 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12:3715-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Bruijn, M. F., and N. A. Speck. 2004. Core-binding factors in hematopoiesis and immune function. Oncogene 23:4238-4248. [DOI] [PubMed] [Google Scholar]
- 23.Dowdy, C. R., R. Xie, D. Frederick, S. Hussain, S. K. Zaidi, D. Vradii, A. Javed, X. Li, S. N. Jones, J. B. Lian, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 2010. Definitive hematopoiesis requires Runx1 C-terminal-mediated subnuclear targeting and transactivation. Hum. Mol. Genet. 19:1048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duncan, R., I. Collins, T. Tomonaga, T. Zhang, and D. Levens. 1996. A unique transactivation sequence motif is found in the carboxyl-terminal domain of the single-strand-binding protein FBP. Mol. Cell. Biol. 16:2274-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durst, K. L., and S. W. Hiebert. 2004. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene 23:4220-4224. [DOI] [PubMed] [Google Scholar]
- 26.Esteller, M. 2008. Epigenetics in cancer. N. Engl. J. Med. 358:1148-1159. [DOI] [PubMed] [Google Scholar]
- 27.Filipowicz, W., S. N. Bhattacharyya, and N. Sonenberg. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9:102-114. [DOI] [PubMed] [Google Scholar]
- 28.Fletcher, C., N. Heintz, and R. G. Roeder. 1987. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell 51:773-781. [DOI] [PubMed] [Google Scholar]
- 29.Franke, A., and B. S. Baker. 1999. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol. Cell 4:117-122. [DOI] [PubMed] [Google Scholar]
- 30.Goldberg, A. D., C. D. Allis, and E. Bernstein. 2007. Epigenetics: a landscape takes shape. Cell 128:635-638. [DOI] [PubMed] [Google Scholar]
- 31.Goodrich, J. A., and J. F. Kugel. 2006. Non-coding-RNA regulators of RNA polymerase II transcription. Nat. Rev. Mol. Cell. Biol. 7:612-616. [DOI] [PubMed] [Google Scholar]
- 32.Groudine, M., and H. Weintraub. 1982. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell 30:131-139. [DOI] [PubMed] [Google Scholar]
- 33.Guil, S., and M. Esteller. 2009. DNA methylomes, histone codes and miRNAs: tying it all together. Int. J. Biochem. Cell Biol. 41:87-95. [DOI] [PubMed] [Google Scholar]
- 34.Hanna, J., B. W. Carey, and R. Jaenisch. 2008. Reprogramming of somatic cell identity. Cold Spring Harb. Symp. Quant. Biol. 73:147-155. [DOI] [PubMed] [Google Scholar]
- 35.Heard, E. 2004. Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 16:247-255. [DOI] [PubMed] [Google Scholar]
- 36.Hirata, J., H. Nakagoshi, Y. Nabeshima, and F. Matsuzaki. 1995. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature 377:627-630. [DOI] [PubMed] [Google Scholar]
- 37.Ho, L., and G. R. Crabtree. 2010. Chromatin remodelling during development. Nature 463:474-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hochedlinger, K., and R. Jaenisch. 2006. Nuclear reprogramming and pluripotency. Nature 441:1061-1067. [DOI] [PubMed] [Google Scholar]
- 39.Hudson, D. F., K. M. Marshall, and W. C. Earnshaw. 2009. Condensin: architect of mitotic chromosomes. Chromosome Res. 17:131-144. [DOI] [PubMed] [Google Scholar]
- 40.Iida, T., J. Nakayama, and D. Moazed. 2008. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol. Cell 31:178-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jantzen, H. M., A. Admon, S. P. Bell, and R. Tjian. 1990. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature 344:830-836. [DOI] [PubMed] [Google Scholar]
- 42.Januschke, J., and C. Gonzalez. 2008. Drosophila asymmetric division, polarity and cancer. Oncogene 27:6994-7002. [DOI] [PubMed] [Google Scholar]
- 43.Javed, A., G. L. Barnes, J. Pratap, T. Antkowiak, L. C. Gerstenfeld, A. J. van Wijnen, J. L. Stein, J. B. Lian, and G. S. Stein. 2005. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc. Natl. Acad. Sci. U. S. A. 102:1454-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 45.John, S., and J. L. Workman. 1998. Bookmarking genes for activation in condensed mitotic chromosomes. BioEssays 20:275-279. [DOI] [PubMed] [Google Scholar]
- 46.Jones, P. A., and S. B. Baylin. 2007. The epigenomics of cancer. Cell 128:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelley, R. L., V. H. Meller, P. R. Gordadze, G. Roman, R. L. Davis, and M. I. Kuroda. 1999. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98:513-522. [DOI] [PubMed] [Google Scholar]
- 48.Kitzmann, M., and A. Fernandez. 2001. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell Mol. Life Sci. 58:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein, J., and I. Grummt. 1999. Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. U. S. A. 96:6096-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knoblich, J. A., L. Y. Jan, and Y. N. Jan. 1995. Asymmetric segregation of Numb and Prospero during cell division. Nature 377:624-627. [DOI] [PubMed] [Google Scholar]
- 51.Lian, J. B., A. Javed, S. K. Zaidi, C. Lengner, M. Montecino, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 2004. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit. Rev. Eukaryot. Gene Expr. 14:1-41. [PubMed] [Google Scholar]
- 52.Lin, H., and A. C. Spradling. 1997. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124:2463-2476. [DOI] [PubMed] [Google Scholar]
- 53.MacDougald, O. A., and M. D. Lane. 1995. Transcriptional regulation of gene expression during adipocyte differentiation. Annu. Rev. Biochem. 64:345-373. [DOI] [PubMed] [Google Scholar]
- 54.Martin, C., and Y. Zhang. 2007. Mechanisms of epigenetic inheritance. Curr. Opin. Cell Biol. 19:266-272. [DOI] [PubMed] [Google Scholar]
- 55.Martin, D. I., and S. H. Orkin. 1990. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 4:1886-1898. [DOI] [PubMed] [Google Scholar]
- 56.Martínez-Balbás, M. A., A. Dey, S. K. Rabindran, K. Ozato, and C. Wu. 1995. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83:29-38. [DOI] [PubMed] [Google Scholar]
- 57.Mayer, C., and I. Grummt. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25:6384-6391. [DOI] [PubMed] [Google Scholar]
- 58.McCarrey, J. R., and D. D. Dilworth. 1992. Expression of Xist in mouse germ cells correlates with X-chromosome inactivation. Nat. Genet. 2:200-203. [DOI] [PubMed] [Google Scholar]
- 59.McGhee, J. D., W. I. Wood, M. Dolan, J. D. Engel, and G. Felsenfeld. 1981. A 200 base pair region at the 5′ end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell 27:45-55. [DOI] [PubMed] [Google Scholar]
- 60.Michelotti, E. F., G. A. Michelotti, A. I. Aronsohn, and D. Levens. 1996. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 16:2350-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michelotti, E. F., S. Sanford, and D. Levens. 1997. Marking of active genes on mitotic chromosomes. Nature 388:895-899. [DOI] [PubMed] [Google Scholar]
- 62.Michelotti, G. A., E. F. Michelotti, A. Pullner, R. C. Duncan, D. Eick, and D. Levens. 1996. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 16:2656-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miranda, T. B., and P. A. Jones. 2007. DNA methylation: the nuts and bolts of repression. J. Cell. Physiol. 213:384-390. [DOI] [PubMed] [Google Scholar]
- 64.Moazed, D. 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457:413-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohandas, T., R. S. Sparkes, and L. J. Shapiro. 1981. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science 211:393-396. [DOI] [PubMed] [Google Scholar]
- 66.Mohun, T. 1992. Muscle differentiation. Curr. Opin. Cell Biol. 4:923-928. [DOI] [PubMed] [Google Scholar]
- 67.Ng, R. K., and J. B. Gurdon. 2005. Epigenetic memory of active gene transcription is inherited through somatic cell nuclear transfer. Proc. Natl. Acad. Sci. U. S. A. 102:1957-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ng, R. K., and J. B. Gurdon. 2008. Epigenetic inheritance of cell differentiation status. Cell Cycle 7:1173-1177. [DOI] [PubMed] [Google Scholar]
- 69.Ng, R. K., and J. B. Gurdon. 2008. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 10:102-109. [DOI] [PubMed] [Google Scholar]
- 70.Peng, J. C., and G. H. Karpen. 2009. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 5:e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pratap, J., M. Galindo, S. K. Zaidi, D. Vradii, B. M. Bhat, J. A. Robinson, J.-Y. Choi, T. Komori, J. L. Stein, J. B. Lian, G. S. Stein, and A. J. van Wijnen. 2003. Cell growth regulatory role of Runx2 during proliferative expansion of pre-osteoblasts. Cancer Res. 63:5357-5362. [PubMed] [Google Scholar]
- 72.Probst, A. V., E. Dunleavy, and G. Almouzni. 2009. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell. Biol. 10:192-206. [DOI] [PubMed] [Google Scholar]
- 73.Quyn, A. J., P. L. Appleton, F. A. Carey, R. J. Steele, N. Barker, H. Clevers, R. A. Ridgway, O. J. Sansom, and I. S. Nathke. 2010. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell 6:175-181. [DOI] [PubMed] [Google Scholar]
- 74.Racki, L. R., and G. J. Narlikar. 2008. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr. Opin. Genet. Dev. 18:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rando, O. J., and K. J. Verstrepen. 2007. Timescales of genetic and epigenetic inheritance. Cell 128:655-668. [DOI] [PubMed] [Google Scholar]
- 76.Reik, W. 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447:425-432. [DOI] [PubMed] [Google Scholar]
- 77.Rhyu, M. S., L. Y. Jan, and Y. N. Jan. 1994. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76:477-491. [DOI] [PubMed] [Google Scholar]
- 78.Richler, C., H. Soreq, and J. Wahrman. 1992. X inactivation in mammalian testis is correlated with inactive X-specific transcription. Nat. Genet. 2:192-195. [DOI] [PubMed] [Google Scholar]
- 79.Roberts, S. B., N. Segil, and N. Heintz. 1991. Differential phosphorylation of the transcription factor Oct1 during the cell cycle. Science 253:1022-1026. [DOI] [PubMed] [Google Scholar]
- 80.Roegiers, F., and Y. N. Jan. 2004. Asymmetric cell division. Curr. Opin. Cell Biol. 16:195-205. [DOI] [PubMed] [Google Scholar]
- 81.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 82.Roussel, P., C. Andre, C. Masson, G. Geraud, and D. Hernandez-Verdun. 1993. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J. Cell Sci. 104:327-337. [DOI] [PubMed] [Google Scholar]
- 83.Ruthenburg, A. J., H. Li, D. J. Patel, and C. D. Allis. 2007. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8:983-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Santoro, R., and L. F. De. 2005. Many players, one goal: how chromatin states are inherited during cell division. Biochem. Cell Biol. 83:332-343. [DOI] [PubMed] [Google Scholar]
- 85.Sarge, K. D., and O. K. Park-Sarge. 2005. Gene bookmarking: keeping the pages open. Trends Biochem. Sci. 30:605-610. [DOI] [PubMed] [Google Scholar]
- 86.Segil, N., M. Guermah, A. Hoffmann, R. G. Roeder, and N. Heintz. 1996. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 10:2389-2400. [DOI] [PubMed] [Google Scholar]
- 87.Segil, N., S. B. Roberts, and N. Heintz. 1991. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science 254:1814-1816. [DOI] [PubMed] [Google Scholar]
- 88.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75:243-269. [DOI] [PubMed] [Google Scholar]
- 89.Smith, C. M., Z. W. Haimberger, C. O. Johnson, A. J. Wolf, P. R. Gafken, Z. Zhang, M. R. Parthun, and D. E. Gottschling. 2002. Heritable chromatin structure: mapping “memory” in histones H3 and H4. Proc. Natl. Acad. Sci. U. S. A. 99(Suppl. 4):16454-16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith, L. T., G. A. Otterson, and C. Plass. 2007. Unraveling the epigenetic code of cancer for therapy. Trends Genet. 23:449-456. [DOI] [PubMed] [Google Scholar]
- 91.Spence, M. A., and F. W. Luthardt. 1975. Mitotic association patterns of nucleolar organizing chromosomes in the mouse. Cytogenet. Cell Genet. 15:276-280. [DOI] [PubMed] [Google Scholar]
- 92.Tang, Q. Q., T. C. Otto, and M. D. Lane. 2003. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 100:850-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thorpe, P. H., J. Bruno, and R. Rothstein. 2008. Modeling stem cell asymmetry in yeast. Cold Spring Harb. Symp. Quant. Biol. 73:81-88. [DOI] [PubMed] [Google Scholar]
- 94.Vradii, D., S. K. Zaidi, J. B. Lian, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 2005. Point mutation in AML1 disrupts subnuclear targeting, prevents myeloid differentiation, and effects a transformation-like phenotype. Proc. Natl. Acad. Sci. U. S. A. 102:7174-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.White, R. J. 2005. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 6:69-78. [DOI] [PubMed] [Google Scholar]
- 96.Whitelaw, E., S. F. Tsai, P. Hogben, and S. H. Orkin. 1990. Regulated expression of globin chains and the erythroid transcription factor GATA-1 during erythropoiesis in the developing mouse. Mol. Cell. Biol. 10:6596-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xin, L., G. L. Zhou, W. Song, X. S. Wu, G. H. Wei, D. L. Hao, X. Lv, D. P. Liu, and C. C. Liang. 2007. Exploring cellular memory molecules marking competent and active transcriptions. BMC Mol. Biol. 8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xing, H., D. C. Wilkerson, C. N. Mayhew, E. J. Lubert, H. S. Skaggs, M. L. Goodson, Y. Hong, O. K. Park-Sarge, and K. D. Sarge. 2005. Mechanism of hsp70i gene bookmarking. Science 307:421-423. [DOI] [PubMed] [Google Scholar]
- 99.Young, D. W., M. Q. Hassan, J. Pratap, M. Galindo, S. K. Zaidi, S. H. Lee, X. Yang, R. Xie, A. Javed, J. M. Underwood, P. Furcinitti, A. N. Imbalzano, S. Penman, J. A. Nickerson, M. A. Montecino, J. B. Lian, J. L. Stein, A. J. van Wijnen, and G. S. Stein. 2007. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature 445:442-446. [DOI] [PubMed] [Google Scholar]
- 100.Young, D. W., M. Q. Hassan, X.-Q. Yang, M. Galindo, A. Javed, S. K. Zaidi, P. Furcinitti, D. Lapointe, M. Montecino, J. B. Lian, J. L. Stein, A. J. van Wijnen, and G. S. Stein. 2007. Mitotic retention of gene expression patterns by the cell fate determining transcription factor Runx2. Proc. Natl. Acad. Sci. U. S. A. 104:3189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zaidi, S. K., D. W. Young, A. Javed, J. Pratap, M. Montecino, A. van Wijnen, J. B. Lian, J. L. Stein, and G. S. Stein. 2007. Nuclear microenvironments in biological control and cancer. Nat. Rev. Cancer 7:454-463. [DOI] [PubMed] [Google Scholar]
- 102.Zaidi, S. K., D. W. Young, S. H. Pockwinse, A. Javed, J. B. Lian, J. L. Stein, A. J. van Wijnen, and G. S. Stein. 2003. Mitotic partitioning and selective reorganization of tissue specific transcription factors in progeny cells. Proc. Natl. Acad. Sci. U. S. A. 100:14852-14857. [DOI] [PMC free article] [PubMed] [Google Scholar]