Abstract
Differential posttranslational modification of proliferating cell nuclear antigen (PCNA) by ubiquitin or SUMO plays an important role in coordinating the processes of DNA replication and DNA damage tolerance. Previously it was shown that the loss of RAD6-dependent error-free postreplication repair (PRR) results in DNA damage checkpoint-mediated G2 arrest in cells exposed to chronic low-dose UV radiation (CLUV), whereas wild-type and nucleotide excision repair-deficient cells are largely unaffected. In this study, we report that suppression of homologous recombination (HR) in PRR-deficient cells by Srs2 and PCNA sumoylation is required for checkpoint activation and checkpoint maintenance during CLUV irradiation. Cyclin-dependent kinase (CDK1)-dependent phosphorylation of Srs2 did not influence checkpoint-mediated G2 arrest or maintenance in PRR-deficient cells but was critical for HR-dependent checkpoint recovery following release from CLUV exposure. These results indicate that Srs2 plays an important role in checkpoint-mediated reversible G2 arrest in PRR-deficient cells via two separate HR-dependent mechanisms. The first (required to suppress HR during PRR) is regulated by PCNA sumoylation, whereas the second (required for HR-dependent recovery following CLUV exposure) is regulated by CDK1-dependent phosphorylation.
DNA damage occurs frequently in all organisms as a consequence of both endogenous metabolic processes and exogenous DNA-damaging agents. In nature, the steady-state level of DNA damage is usually very low. However, chronic low-level DNA damage can lead to age-related genome instability as a consequence of the accumulation of DNA damage (12, 27). Increasing evidence implicates DNA damage-related replication stress in genome instability (7, 21). Replication stress occurs when an active fork encounters DNA lesions or proteins tightly bound to DNA. These obstacles pose a threat to the integrity of the replication fork and are thus a potential source of genome instability, which can contribute to tumorigenesis and aging in humans (4, 11). Confronted with this risk, cells have developed fundamental DNA damage response mechanisms in order to faithfully complete DNA replication (8).
In budding yeast Saccharomyces cerevisiae, the Rad6-dependent postreplication repair (PRR) pathway is subdivided into three subpathways, which allow replication to resume by bypassing the lesion without repairing the damage (3, 22, 33). Translesion synthesis (TLS) pathways dependent on the DNA polymerases eta and zeta promote error-free or mutagenic bypass depending on the DNA lesion and are activated upon monoubiquitination of proliferating cell nuclear antigen (PCNA) at Lys164 (K164) (5, 16, 37). The Rad5 (E3) and Ubc13 (E2)/Mms2 (E2 variant)-dependent pathway promotes error-free bypass by template switching and is activated by polyubiquitination of PCNA via a Lys63-linked ubiquitin chain (16, 38, 41). It remains mechanistically unclear how polyubiquitinated PCNA promotes template switching at the molecular level. In addition to its ubiquitin E3 activity, Rad5 also has a helicase domain and was recently shown to unwind and reanneal fork structures in vitro (6). This led to the proposal that Rad5 helicase activity is required at replication forks to promote fork regression and subsequent template switching. It is possible that PCNA polyubiquitination acts to facilitate Rad5-dependent template switching by inhibiting monoubiquitination-dependent TLS activity and/or by recruiting alternative proteins to the fork.
In addition to modification by ubiquitin, PCNA can also be sumoylated on Lys164 by the SUMO E3 ligase Siz1 (16). A second sumoylation site, Lys127, is also targeted by an alternative SUMO E3 ligase, Siz2, albeit with lower efficiency (16, 30). PCNA SUMO modification results in recruitment of the Srs2 helicase and subsequent inhibition of Rad51-dependent recombination events (29, 32). The modification can therefore allow the replicative bypass of lesions by promoting the RAD6 pathway. Srs2 is known to act as an antirecombinase by eliminating recombination intermediates. This can occur independently of PCNA sumoylation, and when srs2Δ cells are UV irradiated or other antirecombinases, such as Sgs1, are concomitantly deleted, toxic recombination structures accumulate (1, 10). Such genetic data are consistent with the ability of Srs2 to disassemble the Rad51 nucleoprotein filaments formed on single-stranded DNA (ssDNA) in vitro (20, 40). In addition to directly inhibiting homologous recombination (HR), Srs2 is also involved in regulating HR outcomes to not produce crossover recombinants in the mitotic cell cycle (18, 34, 35).
The UV spectrum present in sunlight is a primary environmental cause of exogenous DNA damage. Sunlight is a potent and ubiquitous carcinogen responsible for much of the skin cancer in humans (17). In the natural environment, organisms are exposed to chronic low-dose UV light (CLUV), as opposed to the acute high doses commonly used in laboratory experiments. Hence, understanding the cellular response to CLUV exposure is an important approach complementary to the more traditional laboratory approaches for clarifying the biological significance of specific DNA damage response pathways. A recently developed experimental assay for the analysis of CLUV-induced DNA damage responses was used to show that the PCNA polyubiquitination-dependent error-free PRR pathway plays a critical role in tolerance of CLUV exposure by preventing the generation of excessive ssDNA when replication forks arrest, thus suppressing counterproductive checkpoint activation (13).
Mutants of SRS2 were first isolated by their ability to suppress the radiation sensitivity of rad6 and rad18 mutants (defective in PRR) by a mechanism that requires a functional HR pathway (23, 36). In this study, we analyzed the function of Srs2 in CLUV-exposed PRR-deficient cells. We established that Srs2 acts in conjunction with SUMO-modified PCNA to lower the threshold for checkpoint activation and maintenance by suppressing the function of HR in rad18Δ cells exposed to CLUV. We also showed that Srs2 is separately involved in an HR-dependent recovery process following cessation of CLUV exposure and that this second role for Srs2, unlike its primary role in checkpoint activation and maintenance, is regulated by CDK1-dependent phosphorylation. Thus, Srs2 is involved in both CLUV-induced checkpoint-mediated arrest and recovery from CLUV exposure in PRR-deficient cells, and these two functions, while both involving HR, are separable and thus independent.
MATERIALS AND METHODS
Strains and plasmids.
All yeast strains used in this study are listed in Table 1. Temperature-sensitive srs2-td strains were constructed by using a PCR-based method with W303-1A, modified for use with the degron system as described previously (19). A 3.9-kb BamHI genomic fragment containing SRS2 was cloned into pUC19. The srs2ΔC mutant was obtained from pUC19-Srs2 by site-directed PCR mutagenesis. The BamHI fragments of wild-type SRS2 and the srs2 mutant containing the promoter and open reading frame were cloned into pRS415 for expression. The SUMO-PCNAK127R/K164R expression plasmid was constructed as described previously (15). All other strains were constructed by standard genetic procedures (2). The DNA sequences of the PCR-amplified fragments were confirmed by sequencing the appropriate regions.
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| BY4741 | MATaleu2Δ0 met15Δ0 ura3Δ0 his3Δ1 | ATCC |
| TH601 | BY4741 rad18Δ::KanMX | 13 |
| TH602 | BY4741 srs2Δ::HIS3 | This study |
| TH603 | BY4741 srs2Δ::HIS3 rad18Δ::KanMX | This study |
| TH604 | BY4741 rad51Δ::URA3 | This study |
| TH605 | BY4741 rad18Δ:: KanMX rad51Δ::URA3 | This study |
| TH606 | BY4741 srs2Δ::HIS3 rad51Δ::URA3 | This study |
| TH607 | BY4741 srs2Δ::HIS3 rad18Δ::KanMX rad51Δ::URA3 | This study |
| TH608 | BY4741 rad5Δ::KanMX | 13 |
| TH609 | BY4741 srs2Δ::HIS3 rad5Δ::KanMX | This study |
| TH610 | BY4741 rad5Δ::KanMX rad51Δ::URA3 | This study |
| TH611 | BY4741 srs2Δ::HIS3 rad5Δ::KanMX rad51Δ::URA3 | This study |
| TH620 | BY4741 pol30K127R | This study |
| TH621 | BY4741 pol30K127R rad18Δ::KanMX | This study |
| TH623 | BY4741 pol30K164R | 13 |
| TH624 | BY4741 pol30K164R rad18Δ::KanMX | This study |
| TH625 | BY4741 pol30K164R/K127R | This study |
| TH626 | BY4741 pol30K164R/K127R rad18Δ::KanMX | This study |
| TH627 | BY4741 pol30K164R/K127R rad18Δ::KanMX srs2Δ::HIS3 | This study |
| TH612 | BY4741 siz1Δ::KanMX rad18Δ::KanMX | This study |
| TH631a | BY4741 rad18Δ::KanMX mec1::KanMX sml1Δ::LEU2 | 13 |
| TH632a | BY4741 rad18Δ::KanMX srs2Δ::HIS3 mec1::KanMX sml1Δ::LEU2 | This study |
| TH650 | BY4741 rad18Δ::KanMX sml1Δ::LEU2 | This study |
| TH655 | BY4741 rad18Δ::KanMX srs2Δ::HIS3 sml1Δ::LEU2 | This study |
| W3775-12C | MATaRFA-YFP | 25 |
| W3849-15C | MATaDDC2-YFP RAD52-CFP | 25 |
| TH896 | W3775-12C rad18Δ::KanMX | 13 |
| TH912 | W3849-15C rad18Δ::KanMX | 13 |
| TH937 | W3775-12C srs2Δ::HIS3 | This study |
| TH915 | W3775-15C srs2Δ::HIS3 | This study |
| TH900 | W3775-12C rad18Δ::KanMX srs2Δ::HIS3 | This study |
| TH913 | W3849-15C rad18Δ::KanMX srs2Δ::HIS3 | This study |
| TH948 | W3775-12C rad18Δ::KanMX srs2-7AV.HIS3 | This study |
| TH940 | W3775-12C rad51Δ::URA3 | This study |
| TH938 | W3775-12C rad51Δ::URA3 srs2Δ::HIS3 | This study |
| TH941 | W3775-12C rad51Δ::URA3 rad18Δ::KanMX | This study |
| TH935 | W3775-12C rad51Δ::URA3 rad18Δ::KanMX srs2Δ::HIS3 | This study |
| YKL200 | UBR1::GAL-HA-UBR1 (HIS3) | 19 |
| TH975 | YKL200 rad18Δ::KanMX | This study |
| TH991 | YKL200 rad18Δ::KanMX srs2Δ::HIS3 | This study |
| TH928 | YKL200 srs2-td (KanMX) | This study |
| TH925 | YKL200 srs2-td (KanMX) rad18Δ::TRP1 | This study |
| TH992 | YKL200 srs2-td (KanMX) rad18Δ::TRP1 RPA-YFP | This study |
| TH993 | YKL200 rad18Δ::TRP1 RPA-YFP | This study |
| W1588-4A | ade2-1 can1-100 his3-11,-15 leu2-3,-112 trp1-1 ura3-1 RAD5 | 28 |
| TH252 | W1588-4A rad18Δ::LEU2 | 14 |
| TH250 | W1588-4A srs2Δ::HIS3 | 14 |
| TH955 | W1588-4A srs2-7AV.HIS3 | This study |
| TH919 | W1588-4A srs2-7AV.HIS3 rad18Δ::LEU2 | This study |
| TH212 | W1588-4A rad52Δ::TRP1 | 14 |
| TH984 | W1588-4A rad52Δ::TRP1 rad18Δ::LEU2 | This study |
| TH988 | W1588-4A rad52Δ::TRP1 srs2-7AV.HIS3 | This study |
| TH971 | W1588-4A rad52Δ::TRP1 rad18Δ::LEU2 srs2-7AV.HIS3 | This study |
| TH903 | W1588-4A rad18Δ::LEU2 RAD52-YFP | This study |
| TH970 | W1588-4A rad18Δ::LEU2 srs2-7AV.HIS3 RAD52-YFP | This study |
A deletion of SML1 suppresses lethality without suppressing the checkpoint defect.
Media and growth conditions.
Cells were grown in yeast extract-peptone-dextrose (YPD) medium containing 0.01% adenine sulfate (YPAD) at 30°C. To synchronize cells in G1, 10 μg/ml alpha factor (Sigma) was added to cells in mid-log phase (∼5 × 106 cells/ml), followed by incubation for ∼3 h at 30°C. The cells were then washed with water and released into fresh medium containing 100 μg/ml pronase (Sigma). For depletion of Srs2, cells (∼1 × 107 cells/ml) grown at 25°C in yeast extract-peptone (YP)-2% raffinose (Raf) supplemented with 0.1 mM CuSO4 were collected, washed once in water, and resuspended in the same volume of fresh YP-2% galactose (Gal). Cells were further incubated at 37°C to trigger rapid degradation of the Srs2 protein.
CLUV irradiation.
CLUV exposure was carried out as described previously (13). Cells were incubated with horizontal shaking at 30°C under continuous exposure to UV irradiation (254 nm) at a dose of ≈0.12 J/m2 per min.
Preparation of yeast extracts and Western blotting.
Protein extracts were prepared from 2 × 107 logarithmically growing cells using the trichloroacetic acid (TCA) method, as described previously (31). Proteins were analyzed by SDS-PAGE, transferred to a polyvinylidene difluoride (PVDF) membrane, and probed with anti-Rad53 or anti-Srs2 monoclonal antibody (Santa Cruz).
Other methods.
Flow cytometry was performed as described previously (14). Yellow fluorescent protein signals were observed using a Zeiss Axioplan2 microscope equipped with a Hamamatsu C4742-95 charge-coupled-device (CCD) camera. Images were visualized using the Lumina Vision imaging software program (Mitani Corporation). More than 100 individual cells were scored for each strain. Data were the averages of three independent measurements.
RESULTS
Deletion of SRS2 suppresses CLUV sensitivity of rad18Δ cells.
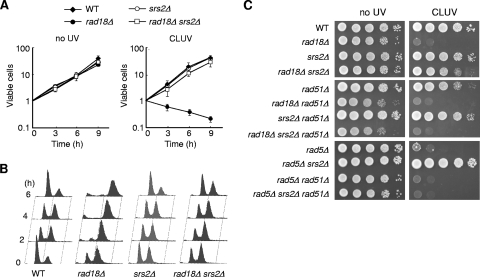
To test whether Srs2 is responsible for the CLUV sensitivity of PRR-deficient cells, we generated cells lacking SRS2. The deletion of SRS2 fully suppressed the CLUV sensitivity of rad18Δ cells, whereas the srs2Δ single mutant did not show any noticeable effects on growth (Fig. 1 A). The srs2 mutation also suppressed the CLUV-induced G2 arrest of rad18Δ cells, as determined by fluorescence-activated cell sorter (FACS) analysis (Fig. 1B). Moreover, suppression of the CLUV sensitivity of rad18Δ cells by concomitant srs2 mutation required RAD51, indicating an essential function of the HR pathway (Fig. 1C). Similar behavior was also observed in the rad5Δ strain (Fig. 1C).
FIG. 1.
Deletion of SRS2 suppresses the sensitivity of rad18Δ cells to CLUV in a recombination-dependent manner. (A) The plating efficiencies for WT, rad18Δ, srs2Δ, and rad18Δ srs2Δ strains during exposure to CLUV irradiation. Asynchronized log-phase cells were grown in YPAD under CLUV irradiation, and samples were taken every 3 h to determine plating efficiency. Cell viability is represented as relative CFU (= 1 at time zero). Data were obtained from at least three independent experiments. Error bars indicate standard deviation. (B) Flow cytometry of WT, rad18Δ, srs2Δ, and rad18Δ srs2Δ cells exposed to CLUV. Asynchronous cells were grown under CLUV, and samples were collected for FACS analysis for DNA content. (C) Spot assays were performed using 10-fold serial dilutions of exponential-phase cultures of the indicated strains. DNA damage was induced by CLUV exposure for 2 days.
PCNA sumoylation is required for CLUV-induced G2 arrest in rad18Δ cells.
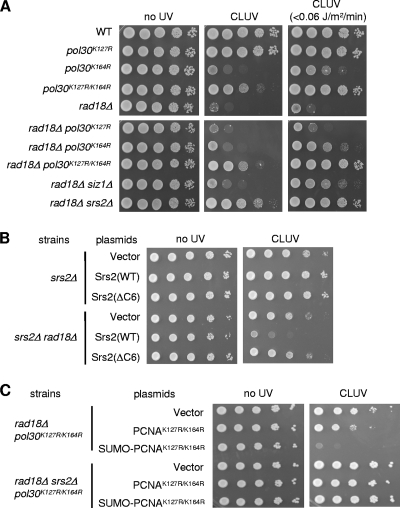
In response to DNA damage, PCNA is monoubiquitinated or polyubiquitinated on the highly conserved lysine residue K164 (16). Yeast PCNA is also modified by SUMO on K164 and to a lesser extent on a second residue, K127 (16). The hypersensitivity of rad18 mutants to acute UV exposure was recently shown to be related to Srs2 recruitment via the sumoylated form of PCNA (29, 32). Therefore, we performed experiments to determine the effect of PCNA sumoylation on the CLUV sensitivity of rad18Δ cells using mutants of the yeast POL30 gene, which encodes PCNA. A pol30K127R mutation had no effect on the CLUV sensitivity of wild-type or rad18Δ cells (Fig. 2 A). In contrast, the pol30K164R mutant was sensitive to CLUV exposure but to a lesser extent than rad18Δ cells (Fig. 2A). Furthermore, the pol30K164R mutation partially suppressed the CLUV sensitivity of rad18Δ cells; the double mutants showed a sensitivity equivalent to that of the pol30K164R mutant, the less sensitive of the two single mutants. This suppression effect was more pronounced when cells were exposed to a much lower dose of UV light (<0.06 J m−2 min−1; Fig. 2A). Consistent with a loss of the PCNA SUMO modification causing this effect, deletion of SIZ1, an E3 ligase required for PCNA-K164 sumoylation, also suppressed the CLUV sensitivity of the rad18Δ mutant cells (Fig. 2A). A PCNA mutant lacking both sumoylation sites (pol30K127R/K164R) suppressed the rad18Δ CLUV sensitivity to an even greater extent than pol30K164R (Fig. 2A). Thus, the CLUV hypersensitivity of rad18Δ cells is related to the sumoylation of PCNA at Lys164. In addition, PCNA-K127 sumoylation appears to play a cryptic role in CLUV tolerance, which can be unmasked by mutation of PCNA K164.
FIG. 2.
A deficiency of PCNA sumoylation suppresses the sensitivity of rad18Δ cells to CLUV. (A) Suppression of sensitivity of rad18Δ cells to CLUV by mutation of the SUMO accepter lysines K127 and/or K164. Cells were exposed to CLUV for 2 days. (B) Cells that carried the indicated SRS2 alleles on a plasmid were grown and spotted onto synthetic complete medium without leucine (SC-Leu) plates at 10-fold serial dilutions. The plates were incubated in the presence or absence of CLUV for 3days. (C) CLUV sensitivities of rad18Δ pol30K127R/K164R and rad18Δ pol30K127R/K164R srs2Δ cells expressing the indicated PCNA or SUMO-fused PCNA alleles on a plasmid. For quantitative assay, 10-fold serial dilutions of cells grown in SC-Leu were spotted on SC-Leu plates and incubated under CLUV conditions for 3 days.
A previous study showed that sumoylated PCNA interacts with Srs2 through a SUMO-interacting motif (SIM) in the C terminus of Srs2 (32). Indeed, an srs2 mutant (srs2ΔC6) lacking SIM in the six C-terminal amino acid residues also suppressed the CLUV sensitivity of rad18Δ cells with an efficiency similar to that for the srs2Δ mutant (Fig. 2B). Taken together, these results indicate that the CLUV hypersensitivity of PRR-deficient cells is due to Srs2 recruitment through PCNA sumoylation.
To further examine the relationship between Srs2 and PCNA sumoylation in CLUV-exposed PRR-deficient cells, we transformed rad18Δ pol30K127R/K164R and rad18Δ pol30K127R/K164R srs2Δ cells with a plasmid harboring a SUMO-PCNAK127R/K164R fusion construct. We have previously reported that the covalent attachment of SUMO to PCNA is capable of exerting an effect similar to that of PCNA sumoylated at K164, even though it occurs at a different position (15). Expression of the SUMO-PCNAK127R/K164R fusion protein inhibited the growth of rad18Δ pol30K127R/K164R cells under CLUV irradiation, whereas the expression of the PCNAK127R/K164R control did not (Fig. 2C). Importantly, the SUMO-PCNAK127R/K164R fusion protein failed to inhibit the growth of rad18Δ pol30K127R/K164R srs2Δ cells under CLUV conditions (Fig. 2C). Thus, the growth inhibition conferred by SUMO-PCNA conjugates depends on Srs2 function, supporting the idea that SUMO modification of PCNA recruits Srs2 to suppress HR events.
Srs2 is required for checkpoint activation in response to CLUV irradiation in rad18Δ cells.
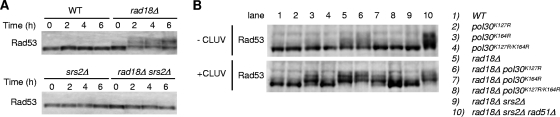
The deletion of RAD18 activates the Mec1/Rad9/Rad53-dependent DNA damage checkpoint in response to CLUV irradiation, resulting in cell cycle arrest in G2 (13). To clarify whether Srs2 is required for checkpoint activation in CLUV-exposed rad18Δ cells, we analyzed Rad53 phosphorylation by electrophoretic mobility shift. Two hours after initiating CLUV treatment of rad18Δ cells, hyperphosphorylated Rad53 was evident, showing a marked increase with exposure time, whereas Rad53 phosphorylation was strongly reduced in rad18Δ srs2Δ cells (Fig. 3 A). Moreover, Rad53 phosphorylation was still observed in CLUV-exposed pol30K164R cells but to a lesser extent than in rad18Δ cells (Fig. 3B). The pol30K164R rad18Δ double mutants exhibited Rad53 phosphorylation equivalent to that in pol30K164R cells. Mutation of both sumoylation sites (pol30K127R/K164R) suppressed Rad53 hyperphosphorylation in CLUV-exposed rad18Δ cells to an even greater extent than pol30K164R (Fig. 3B). In addition, substantial Rad53 phosphorylation was evident in rad18Δ srs2Δ rad51Δ cells in the absence of CLUV irradiation, and Rad53 hyperphosphorylation was significantly increased in CLUV-treated cells (Fig. 3B). These results are in good agreement with genetic data demonstrating that pol30K127R/K164R suppresses the CLUV sensitivity of rad18Δ cells to a greater extent than pol30K164R (Fig. 2A) and that suppression of CLUV sensitivity in rad18Δ cells by srs2 mutation does not occur with rad51Δ mutation (Fig. 1C).
FIG. 3.
Srs2 is required for checkpoint activation in CLUV-exposed rad18Δ cells. (A) CLUV-induced Rad53 phosphorylation. Protein extracts from cells exposed to CLUV for the indicated times were prepared and analyzed by 6% SDS-PAGE followed by Western blotting using an anti-Rad53 antibody. (B) Rad53 phosphorylation in CLUV-exposed PRR-deficient cells. Protein extracts from cells exposed to CLUV for 3 h in were prepared and analyzed by Western blotting with anti-Rad53 antibody.
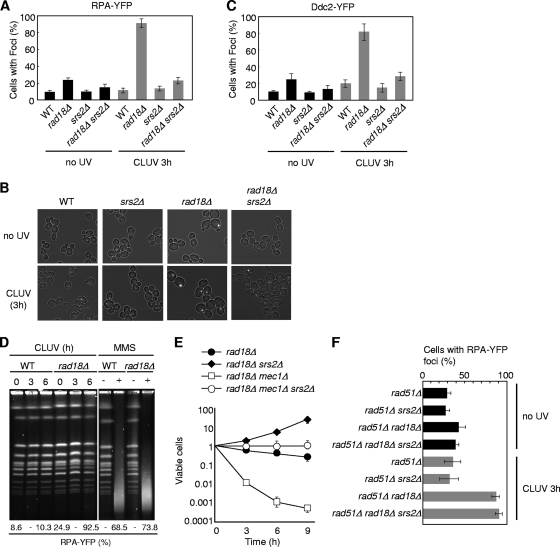
Ddc2 is a Mec1-binding partner that recognizes RPA-coated ssDNA (31). We therefore examined the formation of Ddc2-yellow fluorescent protein (YFP) and RPA-YFP foci in cells lacking Rad18 and Srs2, because these two YFP fusion proteins have been reported previously to form foci that correlate with DNA damage checkpoint activation (25). CLUV-induced Ddc2 and RPA foci were significantly reduced in rad18Δ srs2Δ cells compared to findings for rad18Δ cells (Fig. 4 A to C). Since RPA and Ddc2 foci are associated not only with ssDNA gaps but also with ssDNA regions processed from double-stranded breaks (DSBs), we examined whether DSBs occurred in CLUV-exposed rad18Δ cells by monitoring the appearance of subchromosomal fragments using pulsed-field gel electrophoresis (PFGE) (Fig. 4D). As a control, we also examined yeast chromosomes treated with methylmethane sulfonate (MMS), which generates DSBs. In the presence of MMS, all chromosomes were converted to a heterogeneous pool of subchromosomal fragments and RPA foci appeared in >70% of rad18Δ cells (Fig. 4D). Conversely, similar chromosome fragments were not observed in rad18Δ cells even after prolonged CLUV exposure, despite the fact that >90% of rad18Δ cells had RPA foci (Fig. 4D). These results suggest that the vast majority of RPA foci that accumulated in CLUV-exposed rad18Δ cells were associated with ssDNA gaps. Thus, Srs2 contributes to the accumulation of RPA foci and Ddc2 foci on ssDNA gaps in response to CLUV exposure in PRR-deficient cells, resulting in DNA damage checkpoint activation.
FIG. 4.
srs2Δ mutation suppresses ssDNA production in CLUV-exposed rad18Δ cells. (A and C) Asynchronous cultures of the indicated strains were treated with CLUV for 3 h and examined by fluorescence microscopy. The percentage of cells with RPA-YFP (A) or Ddc2-YFP (C) foci is shown. The results represent the means of three independent measurements. The error bars indicate the standard deviations. (B) Representative RPA-YFP foci are shown. (D) Asynchronous cultures were treated with CLUV for the indicated times or with 0.1% MMS for 30 min. Chromosomal DNA was separated by PFGE and detected by staining with SYBR gold. The percentage of cells with RPA-YFP foci is indicated at the bottom. (E) Cell viability was measured as described for Fig. 1A. The error bars indicate the standard deviations. All strains contain a deletion of SML1, which suppresses mec1Δ lethality without suppressing the checkpoint defect. (F) Deletion of SRS2 does not affect CLUV-induced RPA foci in rad18Δ cells when HR function is impaired. Cells grown to logarithmic phase were treated with CLUV and examined by fluorescence microscopy as with panel A. The results represent the means of at least three independent measurements. The error bars indicate the standard deviations.
Cells deficient in both PRR and DNA damage checkpoint pathways do not arrest in G2 during CLUV exposure and fail to form viable colonies (13). We therefore tested whether srs2 mutation affects cell viability. We found that rad18Δ mec1Δ srs2Δ cells did not arrest in G2, and the cell number as judged by the optical density at 600 nm (OD600) and microscopic analysis increased with CLUV exposure time (data not shown). However, rad18Δ mec1Δ srs2Δ cells, unlike rad18Δ mec1Δ cells, maintained a high degree of viability (Fig. 4E). It should be noted that rad18Δ mec1Δ srs2Δ cells were more sensitive to CLUV than rad18Δ srs2Δ cells (Fig. 4E). Consistent with this, an increased number of RPA-YFP foci were detected in rad18Δ mec1Δ srs2Δ cells (data not shown). These results suggest that a Mec1-dependent function(s) is required for full viability of rad18Δ srs2Δ cells.
Srs2 is not directly involved in ssDNA generation in CLUV-exposed rad18Δ cells.
The data presented above indicate that Srs2 facilitates the recruitment of RPA and checkpoint proteins to ssDNA by inhibiting Rad51 loading. However, it remains unclear whether Srs2 helicase activity is also required to generate ssDNA. If Srs2 is directly involved in the generation of ssDNA, RPA focus formation should be suppressed in rad18Δ srs2Δ rad51Δ cells but not in rad18Δ rad51Δ cells; therefore, we next examined RPA focus formation after CLUV irradiation in the rad51Δ genetic backgrounds. Conversely, the results showed a noticeable increase in RPA foci after CLUV exposure in both rad18Δ rad51Δ and rad18Δ rad51Δ srs2Δ cells (Fig. 4F), implying that Srs2 helicase itself is not directly involved in CLUV-induced ssDNA formation in PRR-deficient cells. Thus, we conclude that Srs2 is involved in the establishment of checkpoint activation by preventing Rad51-dependent HR repair.
Srs2 is required for maintenance of the checkpoint activation state during CLUV exposure in rad18Δ cells.
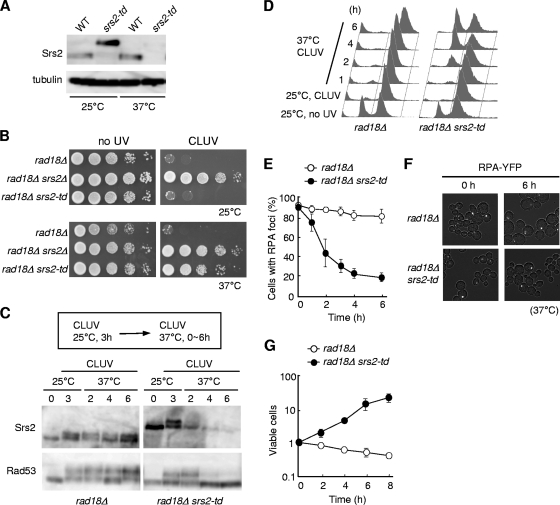
The CLUV-induced G2 arrest of rad18Δ cells persists throughout CLUV exposure (13). Therefore, we assessed whether the Srs2 protein is required for this prolonged G2 arrest. To deplete the Srs2 protein specifically after Srs2-dependent G2 arrest is established, we replaced the endogenous SRS2 gene with a temperature-dependent degron allele (srs2-td), which is expressed by a CUP1 promoter (19). This mutant strain allows the degradation of the degron-fused Srs2 protein when cells are shifted to the restrictive temperature of 37°C. The UBR1 gene, encoding the degron recognition factor, is induced by the presence of galactose. We confirmed with an immunoblot that the Srs2-td protein is degraded upon temperature shift (Fig. 5 A). Consistent with this, the CLUV sensitivity of rad18Δ cells was suppressed at 37°C but not at 25°C (Fig. 5B). Thus, Srs2-td is functional in vivo at 25°C and is efficiently depleted and nonfunctional at 37°C.
FIG. 5.
Srs2 is required to maintain the DNA damage checkpoint in CLUV-exposed rad18Δ cells. (A) Degron-mediated proteolysis of Srs2. Wild-type or srs2-td strains were grown in YP (Raf) at 25°C. The cells were transferred to YP (Gal) to induce UBR1 expression and then cultured at 25 or 37°C for 3 h. Srs2 was detected by Western blotting using an anti-Srs2 antibody. α-Tubulin was used as a loading control. (B) The indicated strains were cultured in YP (Raf) at 25°C. Tenfold serial dilutions of cells were spotted onto YP (Gal) plates and incubated at 25 or 37°C under CLUV conditions. (C) rad18Δ and rad18Δ srs2-td cells were grown at 25°C for 3 h under CLUV conditions to arrest the cell cycle at G2 phase and then transferred to the restrictive conditions to degrade Srs2-td. Samples were collected at the indicated times for immunoblotting with anti-Rad53 and anti-Srs2. (D) Cells were treated with CLUV for the indicated times as for panel C and analyzed by flow cytometry. (E, F, and G) Cells arrested in G2 under CLUV exposure as for panel C and were transferred to restrictive conditions (time = 0). At the indicated time points, cells were analyzed to determine the percentage of cells with RPA foci (E) or for plating efficiency (G). The results represent the averages of at least three independent measurements. The error bars indicate the standard deviations. Representative RPA-YFP foci observed after transfer to the restrictive conditions are shown (F).
To establish whether Srs2 function is required for the maintenance of the prolonged checkpoint arrest, rad18Δ and rad18Δ srs2-td cells were cultured at 25°C for 3 h under CLUV conditions. In both strains, G2 arrest was induced, Rad53 was hyperphosphorylated, and RPA-YFP foci were evident (Fig. 5C to F). Cells were subsequently shifted to the restrictive conditions to promote degradation of the Srs2 protein. In the control strain (rad18Δ), cells remained arrested in G2 phase under CLUV conditions, Rad53 was extensively hyperphosphorylated, and RPA-YFP foci remained (Fig. 5C to F). After the degradation of Srs2 in rad18Δ srs2-td cells, G2 arrest was lost and cells continued to grow during CLUV exposure (Fig. 5D and G). The levels of Rad53 hyperphosphorylation and RPA-YFP foci were significantly reduced (Fig. 5C, E, and F). Taken together, these results demonstrated that in addition to promoting the activation of the DNA damage checkpoint in CLUV-exposed rad18Δ cells, Srs2 is also required for maintenance of the checkpoint activation state.
Srs2 phosphorylation is required for reversible G2 arrest in CLUV-exposed rad18Δ cells.
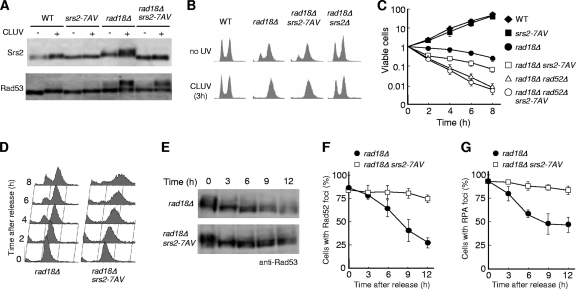
Srs2 became phosphorylated in rad18Δ cells under CLUV conditions (see Fig. 5C). Previous studies have shown that Srs2 phosphorylation is specifically induced by DNA damage and requires a functional Cdk1 kinase (24). Consistent with this, the srs2-7AV mutant, in which the seven putative Cdk1 phosphorylation sites have all been replaced with Ala or Val residues, shows no mobility shift in response to DNA damage (9). As expected, we detected CLUV-induced Srs2 phosphorylation in rad18Δ cells but not in rad18Δ srs2-7AV cells (Fig. 6 A). This suggests that the Srs2-7AV protein mimics the unphosphorylated form of Srs2. To examine the potential role of Cdk1-mediated phosphorylation, we investigated whether the srs2-7AV mutation affects the CLUV-induced checkpoint activation of rad18Δ cells. rad18Δ srs2-7AV cells remained competent for CLUV-induced Rad53 hyperphosphorylation and G2 arrest (Fig. 6A and B). Thus, the phosphorylation of Srs2 on these seven residues is not required for checkpoint activation and maintenance. However, although the srs2-7AV mutation had no effect on the growth of wild-type cells under CLUV conditions, it led to a loss of viability in CLUV-exposed rad18Δ cells (Fig. 6C). Therefore, we next investigated whether these seven residues are important for the resumption of the cell cycle after the cessation of CLUV treatment, a phenotype reflecting recovery from prolonged checkpoint arrest. Cells were released from CLUV exposure after 3 h of treatment, and samples were taken at 2-h intervals for FACS to measure the DNA content. When released from CLUV exposure, the majority of rad18Δ G2-arrested cells resumed the cell cycle after a 4-h or greater delay, whereas rad18Δ srs2-7AV cells showed a broadened peak of DNA content (Fig. 6D). These results suggest that rad18Δ srs2-7AV mutants are not checkpoint defective but rather are unable to resume the cell cycle after release from CLUV-induced G2 arrest.
FIG. 6.
Phosphorylation of Srs2 is required for HR-mediated recovery after release from CLUV exposure in rad18Δ cells. (A) The indicated strains were grown in the presence or absence of CLUV for 3 h, and the phosphorylation status of Srs2 and Rad53 was analyzed by immunoblotting. (B) Same experiment as in panel A, except that samples were subjected to FACS analysis. (C) Cell survival after CLUV irradiation was determined as for Fig. 1A. (D and E) Cells were treated with CLUV to activate the checkpoint and then removed from CLUV exposure to allow recovery. Aliquots were taken at the indicated times and processed by flow cytometry (D) or immunoblotting with anti-Rad53 (E). (F and G) rad18Δ and rad18Δ srs2-7AV mutants were analyzed for the presence of Rad52-YFP (F) or RPA-YFP (G) foci at the indicated time points after release from CLUV exposure. The error bars indicate the standard deviations.
One explanation for the failure of rad18Δ srs2-7AV mutants to recover is that the DNA damage checkpoint is persistently activated even after the cells are no longer exposed to CLUV irradiation. Therefore, Rad53 status during the recovery process was examined. Cells were treated with CLUV to activate the checkpoint and then removed from CLUV exposure to allow recovery. In the rad18Δ mutant, Rad53 was dephosphorylated during recovery (Fig. 6E). We observed that Rad53 remains somewhat phosphorylated after the cell cycle has resumed. This seems to be due to the asynchronous resumption of the cell cycle during the recovery process, because a heterogeneous population of cells with small, large, or no buds is evident in rad18Δ cells at 4 h after release from CLUV (data not shown). Interestingly, the extent of Rad53 phosphorylation during the recovery of rad18Δ srs2-7AV cells was comparable to that of rad18Δ cells (Fig. 6E). These results suggest that the recovery defect in rad18Δ srs2-7AV cells is not due to constitutive checkpoint activation.
Srs2 phosphorylation is involved in HR function following release from CLUV-induced G2 arrest.
HR is required for the viability of rad18Δ cells after their release from CLUV exposure (13). This led us to postulate that the phosphorylation of Srs2 may be required for this HR-dependent recovery process. Therefore, we examined the genetic relationship between the rad52Δ, srs2-7AV, and rad18Δ mutations. We observed that rad18Δ rad52Δ and rad18Δ srs2-7AV double mutants are much more sensitive to CLUV than the rad18Δ single mutant (Fig. 6C), but the rad18Δ rad52Δ srs2-7AV triple mutant was not more sensitive than the rad18Δ rad52Δ double mutant (Fig. 6C). Moreover, when released from CLUV exposure, the number of RPA-YFP and Rad52-YFP foci progressively decreased in rad18Δ cells after the termination of CLUV exposure compared to findings for rad18Δ srs2-7AV cells (Fig. 6F and G). Microscopic examination of rad18Δ srs2-7AV cells revealed a significant increase in the percentage of abnormal cells with protruding and multiple buds after release from CLUV irradiation (data not shown), suggesting that these cells arrest in G2/M phase due to a failure to complete HR prior to cell division. These data are consistent with the idea that the CDK1-dependent phosphorylation of Srs2 is important for the promotion of the HR repair of ssDNA gaps following release from CLUV irradiation.
DISCUSSION
The loss of error-free PRR generates ssDNA gaps spanning the fork-blocking lesion during exposure to CLUV, which activates the DNA damage checkpoint (13); these gaps are subsequently repaired by the HR repair pathway following release from CLUV exposure. ssDNA is an important and universal component for recruiting both checkpoint and HR complexes. However, how the DNA damage checkpoint and the HR pathways are coordinated on ssDNA at sites of DNA damage has remained unclear.
In this study, we have provided evidence that Srs2 is required for DNA damage checkpoint activation in response to CLUV exposure in PRR-deficient cells and that it acts through its recruitment to sumoylated PCNA. The CLUV sensitivity of rad18Δ cells was suppressed by deleting SRS2, by inhibiting PCNA sumoylation, or by preventing Srs2 recruitment to sumoylated PCNA. Furthermore, the suppression of PRR mutant sensitivity to CLUV by the loss of this Srs2 function required a functional HR repair pathway, indicating that suppressing recombination in the absence of PRR prevents efficient DNA repair. In accordance with this, Srs2 has been shown to block Rad52/Rad51-mediated HR by actively disrupting the Rad51 nucleoprotein filament in vitro (20, 40). Taken together, these results support a model whereby Srs2 facilitates the recruitment of checkpoint proteins such as the Mec1/Ddc2 complex onto the ssDNA by inhibiting Rad51 loading, at the expense of limiting the efficiency of repair.
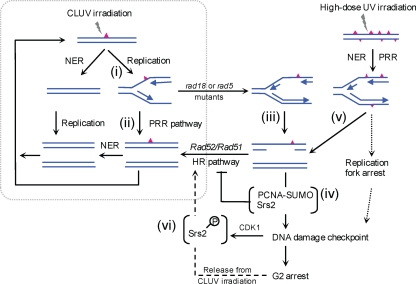
Based on these findings, we developed the model presented in Fig. 7. In response to CLUV exposure, most UV lesions are quickly repaired by nucleotide excision repair (NER). However, lesions remaining at the onset of S phase block replication fork progression [Fig. 7 (i)]. The error-free PRR pathway promotes replication across the damaged template, likely by using the newly synthesized sister chromatid as a template. This enables cells to complete replication without ssDNA gap accumulation [Fig. 7 (ii)]. In PRR-deficient cells, DNA replication of the damaged template is completed by other replication bypass mechanisms, possibly involving repriming downstream of the damaged region [Fig. 7 (iii)]. When ssDNA regions are created by replicating damaged templates, PCNA-SUMO-Srs2 facilitates the recruitment of checkpoint proteins, such as the Mec1-Ddc2 complex, to the ssDNA by inhibiting Rad51 loading, which establishes the DNA damage checkpoint [Fig. 7 (iv)]).
FIG. 7.
A model summarizing CLUV damage tolerance in yeast. See Discussion for details. Red triangles represent UV lesions.
The prevention of recombination and the activation of the DNA damage checkpoint by Srs2 via PCNA-SUMO are evident only in PRR-deficient cells. Thus, extrapolating these activities to wild-type cells remains questionable. One likely explanation is that in wild-type cells, Srs2 facilitates the PRR pathway by preventing complementary recombination events (32). In addition to this role, we propose that Srs2 acts in conjunction with SUMO-modified PCNA to lower the threshold for checkpoint activation by suppressing the functional HR in response to ssDNA generated when the PRR pathway does not, or cannot, efficiently bypass damaged templates. This may be particularly important when DNA lesions overwhelm cells: these lesions likely result in discontinuities of daughter strands in S phase because of the limited ability of NER and PRR to repair and bypass the lesions, respectively [Fig. 7 (v)]. Under such conditions, Srs2 might constitute a first-line mechanism to ensure an appropriate checkpoint response to ssDNA. This model is consistent with findings of previous studies showing that acute high-dose UV treatments result in the accumulation of ssDNA gaps when cells initiate replication (26), and it is also supported by reports that srs2Δ mutants fail to properly activate the DNA damage checkpoint when exposed to genotoxic agents (24).
The srs2-7AV mutant, which was not phosphorylated by CDK1 in response to damage, still functions in the same manner as wild-type Srs2 in CLUV-dependent checkpoint activation and maintenance in a PRR-deficient strain. On the other hand, srs2-7AV rad18Δ cells are deficient in recombination-dependent viability and cell cycle reentry following release from CLUV irradiation, two phenotypes that reflect recovery from prolonged checkpoint arrest. In contrast, the results obtained by using Srs2-td indicate that depletion of the Srs2 protein in PRR-deficient cells allows cell growth even under CLUV conditions. Therefore, these results suggest that an unphosphorylated Srs2 protein disturbs the recovery from CLUV irradiation. How phosphorylated Srs2 functions after release from CLUV exposure is unclear. One possibility is that Srs2 phosphorylation directly interferes with an interaction between Srs2 and Rad51, which might terminate the inhibition of HR by Srs2 to allow repair. Alternatively, Srs2 phosphorylation might be required for HR-dependent ssDNA gap repair by processing the recombination intermediates and/or recruiting the additional proteins to sites of DNA damage. The latter possibility would be consistent with observations that the phosphorylated form of Srs2 is involved in a complex containing Sgs1 and Mre11 that is formed in response to DNA damage (9) and that Srs2 phosphorylation promotes recombination repair through a synthesis-dependent strand annealing (SDSA) mechanism (35). These results suggest that Srs2 phosphorylation plays an important role in controlling the HR-dependent recovery process, which would otherwise impede HR function. Thus, we propose that the phosphorylation of Srs2 in response to CLUV exposure is actively involved in HR-dependent ssDNA gap repair [Fig. 7 (vi)], although unphosphorylated Srs2, together with sumoylated PCNA, impedes HR repair [Fig. 7 (iv)]. It would be of great interest to determine how the two distinct HR functions of Srs2 are regulated during CLUV exposure.
Vaze et al. (39) reported that srs2Δ mutants are able to arrest in response to a single DSB introduced by an endonuclease but fail to resume growth following repair of the DSB. Their findings demonstrated that the DSB can be repaired only through the single-strand annealing (SSA) pathway, which is dependent on Rad52 but not on Rad51. They also showed that the recovery defect of srs2Δ was suppressed by loss of the checkpoint or Rad51 function, suggesting that the lethality is due to persistent checkpoint-mediated arrest and/or toxic HR intermediates. In the study presented here, however, the recovery defect of rad18Δ mutant cells was seen only in srs2-7AV cells and not in srs2Δ cells, indicating that the unphosphorylated form of the Srs2 protein specifically disrupts recovery from CLUV irradiation. Furthermore, Rad51-dependent HR is required for CLUV tolerance in rad18Δ srs2Δ cells. It is important to note that CLUV-induced checkpoint activation in PRR-deficient cells is associated with DNA replication and is dependent on sumoylated PCNA and Srs2, whereas endonuclease-induced DSBs occur independently of DNA replication. Thus, the apparent differences between their results and ours may be due to the two different experimental systems; one is for repair of single DSB damage, and the other is a model of CLUV tolerance.
In conclusion, we provide evidence that Srs2 plays an important role in checkpoint-mediated reversible G2 arrest in PRR-deficient cells via involvement in at least two distinct HR functions in CLUV-exposed PRR-deficient cells. One depends on PCNA sumoylation to suppress functional HR and is required for both checkpoint activation and checkpoint maintenance. The other promotes HR, is regulated by CDK1 phosphorylation, and is required for recovery after release from CLUV-induced G2 arrest.
Acknowledgments
We thank R. Rothstein, K. Labib, M. Kanemaki, and G. Liberi for strains and plasmids.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
Footnotes
Published ahead of print on 16 August 2010.
REFERENCES
- 1.Aboussekhra, A., R. Chanet, A. Adjiri, and F. Fabre. 1992. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 12:3224-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberg, D. C., D. J. Burke, and J. N. Strathern. 2005. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Andersen, P. L., F. Xu, and W. Xiao. 2008. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 18:162-173. [DOI] [PubMed] [Google Scholar]
- 4.Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864-870. [DOI] [PubMed] [Google Scholar]
- 5.Bergink, S., and S. Jentsch. 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458:461-467. [DOI] [PubMed] [Google Scholar]
- 6.Blastyak, A., L. Pinter, I. Unk, L. Prakash, S. Prakash, and L. Haracska. 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 28:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branzei, D., and M. Foiani. 2007. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amsterdam) 6:994-1003. [DOI] [PubMed] [Google Scholar]
- 8.Branzei, D., and M. Foiani. 2008. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 9:297-308. [DOI] [PubMed] [Google Scholar]
- 9.Chiolo, I., W. Carotenuto, G. Maffioletti, J. H. Petrini, M. Foiani, and G. Liberi. 2005. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell. Biol. 25:5738-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 11.Gorgoulis, V. G., L. V. Vassiliou, P. Karakaidos, P. Zacharatos, A. Kotsinas, T. Liloglou, M. Venere, R. A. Ditullio, Jr., N. G. Kastrinakis, B. Levy, D. Kletsas, A. Yoneta, M. Herlyn, C. Kittas, and T. D. Halazonetis. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907-913. [DOI] [PubMed] [Google Scholar]
- 12.Hasty, P., J. Campisi, J. Hoeijmakers, H. van Steeg, and J. Vijg. 2003. Aging and genome maintenance: lessons from the mouse? Science 299:1355-1359. [DOI] [PubMed] [Google Scholar]
- 13.Hishida, T., Y. Kubota, A. M. Carr, and H. Iwasaki. 2009. RAD6-RAD18-RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature 457:612-615. [DOI] [PubMed] [Google Scholar]
- 14.Hishida, T., T. Ohno, H. Iwasaki, and H. Shinagawa. 2002. Saccharomyces cerevisiae MGS1 is essential in strains deficient in the RAD6-dependent DNA damage tolerance pathway. EMBO J. 21:2019-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hishida, T., T. Ohya, Y. Kubota, Y. Kamada, and H. Shinagawa. 2006. Functional and physical interaction of yeast Mgs1 with PCNA: impact on RAD6-dependent DNA damage tolerance. Mol. Cell. Biol. 26:5509-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 17.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 18.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanemaki, M., A. Sanchez-Diaz, A. Gambus, and K. Labib. 2003. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423:720-724. [DOI] [PubMed] [Google Scholar]
- 20.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 21.Lambert, S., B. Froget, and A. M. Carr. 2007. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Repair (Amst.) 6:1042-1061. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence, C. W. 2007. Following the RAD6 pathway. DNA Repair (Amst.) 6:676-686. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, C. W., and R. B. Christensen. 1979. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol. 139:866-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberi, G., I. Chiolo, A. Pellicioli, M. Lopes, P. Plevani, M. Muzi-Falconi, and M. Foiani. 2000. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 19:5027-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisby, M., J. H. Barlow, R. C. Burgess, and R. Rothstein. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699-713. [DOI] [PubMed] [Google Scholar]
- 26.Lopes, M., M. Foiani, and J. M. Sogo. 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21:15-27. [DOI] [PubMed] [Google Scholar]
- 27.Maslov, A. Y., and J. Vijg. 2009. Genome instability, cancer and aging. Biochim. Biophys. Acta 1790:963-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald, J. P., A. S. Levine, and R. Woodgate. 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147:1557-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papouli, E., S. Chen, A. A. Davies, D. Huttner, L. Krejci, P. Sung, and H. D. Ulrich. 2005. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19:123-133. [DOI] [PubMed] [Google Scholar]
- 30.Parker, J. L., A. Bucceri, A. A. Davies, K. Heidrich, H. Windecker, and H. D. Ulrich. 2008. SUMO modification of PCNA is controlled by DNA. EMBO J. 27:2422-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellicioli, A., C. Lucca, G. Liberi, F. Marini, M. Lopes, P. Plevani, A. Romano, P. P. Di Fiore, and M. Foiani. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18:6561-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfander, B., G. L. Moldovan, M. Sacher, C. Hoege, and S. Jentsch. 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436:428-433. [DOI] [PubMed] [Google Scholar]
- 33.Prakash, S., R. E. Johnson, and L. Prakash. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317-353. [DOI] [PubMed] [Google Scholar]
- 34.Robert, T., D. Dervins, F. Fabre, and S. Gangloff. 2006. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 25:2837-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saponaro, M., D. Callahan, X. Zheng, L. Krejci, J. E. Haber, H. L. Klein, and G. Liberi. 2010. Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet. 6:e1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiestl, R. H., S. Prakash, and L. Prakash. 1990. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124:817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulrich, H. D. 2009. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst.) 8:461-469. [DOI] [PubMed] [Google Scholar]
- 38.Ulrich, H. D., and S. Jentsch. 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO. J. 19:3388-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaze, M. B., A. Pellicioli, S. E. Lee, G. Ira, G. Liberi, A. Arbel-Eden, M. Foiani, and J. E. Haber. 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10:373-385. [DOI] [PubMed] [Google Scholar]
- 40.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309-312. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, H., and C. W. Lawrence. 2005. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl. Acad. Sci. U. S. A. 102:15954-15959. [DOI] [PMC free article] [PubMed] [Google Scholar]