Abstract
Telomeres are transcribed into telomeric repeat-containing RNA (TERRA), large, heterogeneous, noncoding transcripts which form part of the telomeric heterochromatin. Despite a large number of functions that have been ascribed to TERRA, little is known about its biogenesis. Here, we present the first comprehensive analysis of the molecular structure of TERRA. We identify biochemically distinct TERRA complexes, and we describe TERRA regulation during the cell cycle. Moreover, we demonstrate that TERRA 5′ ends contain 7-methylguanosine cap structures and that the poly(A) tail, present on a fraction of TERRA transcripts, contributes to their stability. Poly(A)− TERRA, but not poly(A)+ TERRA, is associated with chromatin, possibly reflecting distinct biological roles of TERRA ribonucleoprotein complexes. In support of this idea, poly(A)− and poly(A)+ TERRA molecules end with distinct sequence registers. We also determine that the bulk of 3′-terminal UUAGGG repeats have an average length of 200 bases, indicating that the length heterogeneity of TERRA likely stems from its subtelomeric regions. Finally, we find that TERRA is regulated during the cell cycle, being lowest in late S phase and peaking in early G1. Our analyses offer the basis for investigating multiple regulatory pathways that affect TERRA synthesis, processing, turnover, and function.
The ends of eukaryotic chromosomes, known as telomeres, play crucial roles as guardians of genome stability and tumor suppressors (1, 17). Human telomeres consist of tandem 5′-TTAGGG-3′/5′-CCCTAA-3′ repeats. The guanine-rich strand is directed in a 5′-to-3′ manner toward the chromosome end and extends beyond its complement strand to form a 3′ overhang. Telomeric DNA is associated with the six-polypeptide shelterin complex (18), and shelterin components have been demonstrated to protect the ends of chromosomes from being recognized as sites of DNA damage. Human telomeres vary in length between 5 and 15 kb, and they shorten during the course of life, due to the so-called DNA end replication problem and nucleolytic processing (19, 46, 47).
Telomere shortening is counteracted by telomerase, a ribonucleoprotein enzyme that can extend the 3′ ends of chromosomes. Telomerase works as a reverse transcriptase, using a small region of its tightly associated RNA moiety as a template for the synthesis of telomeric repeats at chromosome ends (22, 23, 29). The progressive erosion of telomeres and the activation of telomerase play key roles in chromosomal stability, cellular immortalization, and tumor progression (7, 26). Short telomeres elicit a DNA damage signal cascade that leads to growth arrest and replicative senescence (16). Reactivation of telomerase in human cancer cells allows overcoming of the senescence barrier and is a key requisite of cancer cells to attain unlimited proliferation potential.
Despite the fact that telomeres are heterochromatic structures, eukaryotic telomeres are transcribed into telomeric repeat-containing RNA (TERRA) (4, 31, 40). TERRA is an integral part of telomeric heterochromatin (4, 21, 40). TERRA molecules are transcribed from several if not all chromosome ends toward the telomere. They contain subtelomere-derived sequence and UUAGGG repeats originated from transcription of telomeric regions (4). Mammalian TERRA molecules range in size from 100 bases up to at least 9 kb and are detected exclusively in nuclear fractions. The nuclear localization of TERRA and its enrichment at telomeres indicate that TERRA is not translated but that it functions as noncoding RNA (ncRNA) in telomeric and subtelomeric heterochromatin structures. In addition, evidence exists that TERRA regulates telomerase. First, in human cells, TERRA is displaced or degraded at telomeres by effectors of nonsense-mediated RNA decay (NMD) (4). Among these factors, EST1A/SMG6 was identified through its sequence similarity with the Saccharomyces cerevisiae telomerase-associated protein Est1p. As yeast Est1p, human EST1A physically interacts with telomerase (35, 37, 42). Second, the TERRA-mimicking RNA oligonucleotides containing UUAGGG repeats inhibit telomerase activity in vitro, as determined in a telomeric repeat amplification protocol (TRAP) assay (40) as well as in direct telomerase assays (36). TERRA oligonucleotides base pair with the RNA template of telomerase RNA, and in addition, they are bound by the telomerase reverse transcriptase (TERT) polypeptide. Association of TERRA with endogenous telomerase can also be detected in cellular extracts (36). It has been proposed that telomerase may be regulated by TERRA in a telomere-length-dependent manner (40). Third, genetic experiments in S. cerevisiae provide evidence that TERRA regulates telomerase in vivo. In the rat1-1 mutant background, in which the function of the 5′-3′ exonuclease Rat1p is reduced, TERRA is upregulated and telomeres are shorter than in wild-type cells due to impairment of telomerase-mediated telomere elongation (31). Further support for the role of TERRA in inhibiting telomerase in vivo stems from an observation that forced telomere transcription (through the use of the strong Gal promoter) leads to telomere shortening of the transcribed telomere in cis (39). A recent study demonstrated that, in vitro, TERRA can form parallel G-quadruplex structures (49), but its relevance in vivo remains unclear. TERRA was also reported to establish association of ORC complexes with shelterin proteins at telomeres (21).
Despite the rather large number of functions that have been ascribed to TERRA, little is known about its biogenesis and its exact molecular nature. Here, we provide a detailed characterization of TERRA 5′ and 3′ ends, determine the UUAGGG tract length, and identify a role of the poly(A) tail in the promotion of TERRA stability and as a marker for non-chromatin-associated TERRA. We also report on the cell cycle regulation of TERRA and its low abundance in late S phase, which we suspect is important to prevent TERRA from interfering with replication of telomeric DNA. Overall, our analysis lays the basis for further investigation of the molecular pathways that affect TERRA homeostasis.
MATERIALS AND METHODS
Cell cultures.
The human cervical carcinoma cell line HeLa, the human primary lung fibroblast line HLF, and the human embryonic kidney cell line HEK293 were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin (Gibco). HeLa supertelomerase and HLF supertelomerase cell lines were obtained from their corresponding wild-type counterparts as described previously (15). Briefly, HeLa and HLF cell lines were first infected with pMND-Banshee-U1-hTR, selected with G418 and then with pBabe-Hygro-hTERT, and selected with hygromycin. After 10 to 15 days, when uninfected control cells were dead, the selection was considered complete. Both cell lines were checked for cooverexpression of human TERT (hTERT) and its RNA template (hTR) and then transferred to normal medium (DMEM containing 10% FCS and penicillin-streptomycin).
RNA preparation.
Cells were trypsinized, and total RNA was prepared using an RNeasy minikit (Qiagen) according to the manufacturer instructions.
Immunoprecipitation of TMG-capped RNAs.
Immunoprecipitation (IP) of 2,2,7-trimethylguanosine (TMG)-capped RNAs was performed as described by Chanfreau et al. (11), with some modifications. Briefly, 50 μg of total RNA was extracted from HeLa and HLF cells and incubated with 5 μg of R1131 polyclonal anti-TMG antibody (generous gift from R. Lührmann) or with 5 μg of normal rabbit IgG (sc-2027; Santa Cruz) in 300 μl of IP buffer (50 mM Tris-HCl, pH 7.5, 5 mM EDTA, pH 8.0, 250 mM NaCl, 0.05% Nonidet P-40) overnight at 4°C, with continuous rotation. The RNA-antibody complexes were coupled to 50 μl of 50% slurry of protein A-Sepharose (GE Healthcare), prewashed three times with IP buffer for 2 h at 4°C. The supernatant (SN) and precipitate (beads) were separated by gentle centrifugation. RNAs in the supernatant were extracted using Trizol LS (Invitrogen). The beads were washed five times with IP buffer, and RNAs still bound to the beads were extracted with Trizol LS (Invitrogen). RNA was transferred to positively charged nylon membranes (Hybond N+; GE Healthcare), and blots were blocked in Church buffer (0.5 M sodium phosphate [pH 2.2], 1 mM EDTA, 7% sodium dodecyl sulfate [SDS], 1% bovine serum albumin [BSA]) for 1 h at 50°C and then incubated with a radiolabeled telomeric probe, obtained as described previously (4). Filters were then stripped and reprobed for β-actin and U1 snRNA.
Affinity purification of 5′-capped RNAs.
Affinity purification of 5′-capped RNAs was carried out as described previously (12). Briefly, purified glutathione S-transferase (GST)-eukaryotic translation initiation factor 4E (eIF4E) (1 mg) was nutated with glutathione agarose beads in 1× phosphate-buffered saline (PBS) for 1 h at 4°C. The beads were washed four times with 1× PBS. A portion (200 μg) of the immobilized GST-eIF4E fusion protein was washed three times with buffer A (10 mM KHPO4, pH 8, 100 mM KCl, 2 mM EDTA, 5% glycerol, 100 μg/ml yeast tRNA, 6 mM dithiothreitol [DTT], 1.3% polyvinyl alcohol, Triton X-100, 1 U/μl SuperaseIN [Ambion]) and then incubated with 50 μg of HeLa and HLF total RNA, preheated to 75°C for 5 min at room temperature. After 1 h, the supernatant was collected and the beads were washed five times with 1 ml of buffer B (same as buffer A but lacking yeast tRNA). RNA retained on the beads, as well as RNA present in the supernatant fraction, was extracted using an RNeasy minikit (Qiagen) according to the manufacturer instructions. As a control, the same amount of GST protein alone was used. RNA was then loaded onto 1.2% formaldehyde agarose gels and separated by electrophoresis. RNA was transferred to positively charged nylon membranes (Hybond N+; GE Healthcare), and blots were treated as described above. Filters were then stripped and reprobed for β-actin and 5.8S rRNA.
Isolation of poly(A)− and poly(A)+ RNA fractions.
Total RNA from HeLa cells was prepared, and poly(A)− and poly(A)+ fractions were separated using an oligo(dT) resin (Oligotex; Qiagen) according to the manufacturer instructions.
Subcellular fractionation.
HEK293 cells (107) were resuspended in 100 μl cytosolic extraction buffer (CEB) (10 mM HEPES-KOH, pH 7.9, 10 mM KCl, 0.34 M sucrose, 1.5 mM MgCl2, 1× Complete protease inhibitor cocktail [Roche], 0.5 mM DTT, 0.1 U/μl SuperaseIN [Ambion]). Next, 100 μl CEB containing in addition 0.2% Nonidet P-40 (AppliChem) was added, and the mixture was incubated for 5 min at 4°C on a rotating wheel. After centrifugation for 5 min at 1,290 × g, the supernatant (SN) was saved as cytosolic extract (cy). The pellet was washed once with 100 μl CEB. Nucleoplasmic extract was prepared by resuspending the 107-cell-equivalent pellet in 100 μl nucleoplasmic extract buffer (NPEB) (140 mM NaCl, 50 mM Tris-HCl, pH 7.5, 1.5 mM MgCl2, 0.5% Nonidet P-40, 1× Complete protease inhibitor cocktail, 0.5 mM DTT, 0.1 U/μl SuperaseIN), and nuclear membranes were disrupted by 20 strokes with a tight-fitting type B pestle using a Dounce homogenizer, followed by rotation for 20 min at 4°C. After centrifugation at 10,000 × g, the SN was saved as nuclear extract (nucl). The pellet was treated with 50 μl DNase digestion mix (74 kU/ml DNase I in 1× RDD buffer [Qiagen]) for 1 h at 37°C. After centrifugation at 10,000 × g and 4°C for 5 min, the chromatin was extracted with 50 μl chromatin extraction buffer (500 mM NaCl, 50 mM Tris-HCl, pH 7.5, 1.5 mM MgCl2, 0.5% Nonidet P-40, 1× Complete protease inhibitor cocktail, 0.5 mM DTT, 0.1 U/μl SuperaseIN) for 20 min at 4°C. Following centrifugation for 5 min at 10,000 × g and 4°C, the SN was kept as chromatin extract (chrom). Subcellular fractionation was evaluated by Western blot analysis. An equivalent of 5 × 105 cells was mixed with Laemmli SDS loading buffer (1.5% SDS, 62.5 mM Tris-HCl, pH 6.8, 10% sucrose, 0.04% bromophenol blue, 5% beta-mercaptoethanol), separated on a 4 to 20% gradient gel (Lonza), and transferred to a nitrocellulose membrane (GE Healthcare). After the mixture was blocked in blocking solution (5% milk, 0.2% Tween 20, 1× PBS) for 30 min at room temperature, the membrane was incubated with the corresponding primary antibodies in blocking solution overnight at 4°C. The following antibodies were used: anti-hnRNP A1, clone 4B10 (1:2,000; Santa Cruz Biotechnologies); anti-H3Ac, 06-599 (1:1,000; Upstate); anti-α-tubulin, clone B-5-1-2 (1:2,000; Sigma); anti-Dis3, clone T-20 (1:2,000; Santa Cruz Biotechnologies); and anticoilin, ab11822 (1:1,000; Abcam). After the mixture was washed three times for 5 min each in PBS-T (0.2% Tween 20, 1× PBS), the membrane was incubated with the corresponding anti-mouse horseradish peroxidase (HRP)-conjugated or anti-rabbit HRP-conjugated IgG in blocking solution for 1 h at room temperature (both 1:2,000; Promega). After three 5-min washes in PBS-T (0.2% Tween 20, 1× PBS), the signal was revealed using ChemiGlow ECL substrate (Alpha Innotech) and a FluorChem 8900 apparatus (Alpha Innotech).
Real-time reverse transcription-PCR (RT-PCR) analysis.
Total RNA was reverse transcribed with β-actin- and telomere-specific primers [(CCCTAA)5] (listed in Table S1 in the supplemental material) at 55°C using SuperScript III reverse transcriptase (Invitrogen). For SYBR green reactions, four different TERRA transcripts, generated from four distinct chromosome ends (10q, 15q, XpYp, and XqYq), were analyzed using specific oligonucleotides listed in Table S1. TERRA levels were normalized against β-actin signals (primers listed in Table S2 in the supplemental material). Real-time PCR analysis was performed using Power SYBR green PCR master mix (Applied Biosystems). Relative quantification of TERRA and gene transcripts was performed using the comparative threshold cycle method.
In vitro RNA polyadenylation.
Total RNA (5 μg) was heated to 70°C for 5 min and then incubated with 360 U of yeast poly(A) polymerase (Pap1) for 10 min at 37°C (30).
Telomeric UUAGGG reverse transcription.
When indicated, 5 μg of total RNA was treated with yeast poly(A) polymerase (Pap1) as described above. Total or poly(A)+ RNA from different cell lines was reverse transcribed with SuperScript III (Invitrogen) in the presence of 5 μl of [α-32P]dCTP (3,000 Ci/mmol, 10 μCi/μl), 5 μM unlabeled dCTP, 500 μM dTTP, and 500 μM dATP. After 1 h, the reaction was chased with 500 μM unlabeled dCTP and 500 μM dGTP, and another 1 μl of SuperScript III (200 U/μl) was added. For the −dGTP reaction, only 500 μM unlabeled dCTP and 1 μl of SuperScript III were added during the chase. The reaction was carried out at 50°C using six different TERRA-specific RT primers (see Table S3 in the supplemental material), which contained oligo(dT)21 at their 5′ ends and which ended in -CCC-3′, -CCT-3′, -CTA-3′, -TAA-3′, -TTC-3′, or -TCC-3′. The length of the derived cDNA was measured on a 1% denaturing alkaline agarose gel.
In vitro transcription of UUAGGG repeat-containing RNAs with T7 RNA polymerase.
A DNA fragment containing 348 bp of pure telomeric TTAGGG repeats was cloned into HindIII/BamHI-cut pLitmus 28i (New England Biolabs), resulting in telo-350 pLitmus 28i. telo-350 pLitmus 28i was cleaved, and another DNA fragment, containing the chromosomal XpYp (Chr XpYp) subtelomere-derived sequence (100 bases) followed by a block of polymorphic TTAGGG-like repeats (250 bases), was inserted upstream of the TTAGGG repeats. The resulting plasmid (XpYp telo-350 pLitmus 28i) was digested with PvuII and BsmBI, gel purified, and used as a template (100 ng) for in vitro runoff transcription using T7 RNA polymerase (MEGAscript; Ambion) according to the manufacturer's instructions.
In order to generate a template for the transcription of an RNA containing 696 bases of pure UUAGGG repeats, telo-350 pLitmus 28i was cut with PvuII and BsmBI. The resulting fragment was gel purified and ligated in vitro with the pure telomeric DNA fragment obtained from the digestion of the telo-350 pLitmus 28i plasmid with BbsI and BamHI. The ligation was performed with T4 DNA ligase (New England Biolabs) for 5 min at 16°C. The ligation product was purified from the gel, digested with BsmBI, and then used as a template (100 ng) for T7-mediated in vitro transcription (MEGAscript; Ambion). Thus, a DNA template that ends with a TTAGGG repeat was generated. In vitro transcripts were purified on 4% polyacrylamide gels and used as substrates for polyadenylation reactions.
Terminal restriction fragment (TRF) analysis.
Genomic DNA was prepared using a Wizard genomic DNA kit (Promega). RsaI- and HinfI-digested DNA was fractionated on a 0.8% agarose 0.5× Tris-borate-EDTA (TBE) gel by using a pulsed-field gel electrophoresis (PFGE) apparatus (Rotaphor; Biometra) at 4.3 V/cm for 18 h at 10°C, followed by ethidium bromide staining. The gel was dried, denatured, and hybridized for 16 h at 50°C with a randomly labeled telomeric probe.
Cell cycle analysis.
HeLa cells were synchronized using a double thymidine block (early-S-phase block). Briefly, HeLa cells were seeded at 10% of confluence. After 24 h, they were incubated with 2 mM thymidine for 18 h. After the first thymidine block, the cells were released for 9 h by three washes in 1× PBS and by addition of fresh DMEM. Following the release, the cells were incubated again with 2 mM thymidine, for 17 h. After this second block, the thymidine was removed and cells were released as after the first block. Total RNA was extracted at the times indicated in Fig. 6 and analyzed by quantitative RT-PCR (qRT-PCR). Cell cycle synchronization was evaluated with propidium iodide (PI) staining and fluorescence-activated cell sorter (FACS) analysis. For PI staining, 5 × 105 cells were washed in 1× PBS and fixed in 1 ml of cold 70% ethanol. Cell pellets were resuspended in 0.5 ml of 1× PBS containing 0.2 μg/ml RNase A, incubated at 37°C for 15 min, and stained with 80 μg/ml PI (Sigma) for 15 to 20 min at 4°C. Cells were analyzed using a CyAn ADP analyzer.
RESULTS
7-Methylguanosine cap structure at TERRA 5′ ends.
TERRA is transcribed from several telomeres by RNA polymerase II (Pol II) (4, 31, 40). mRNAs generated by Pol II have a 7-methylguanosine (m7G) cap structure (41), but snRNAs contain a peculiar 5′-end cap structure characterized by a double methylation of the nitrogen atom at position 2 of the m7G to form the 2,2,7-trimethylguanosine (TMG) cap (14). The presence of a TMG cap on snRNAs is believed to prevent leaking transport to the cytoplasm (14). Since TERRA is found exclusively in the nucleus, we first evaluated the presence of TMG cap structures on its 5′ end. Cap-containing RNAs were immunoprecipitated from HeLa and HLF total RNAs with the R1131 polyclonal antibody, which shows high specificity for the TMG cap structure (25). The TMG cap-containing U1 snRNA served as a positive control and β-actin mRNA as a negative control. Following selective immunoprecipitation, RNA was extracted and analyzed by Northern blotting using a strand-specific TERRA probe (4). This revealed that TERRA molecules do not contain a TMG cap structure (Fig. 1 A).
FIG. 1.
TERRA molecules contain an m7G cap structure but not a TMG cap structure at their 5′ ends. (A) Immunoprecipitation of total RNA from HeLa and HLF cells with R1131 anti-TMG antibody. Input and supernatant (SN) material were loaded at 25%. (B) Affinity purification of m7G cap-containing RNAs, followed by Northern blot analysis. Total RNA from HeLa and HLF cells was selectively enriched for RNAs containing the m7G cap by affinity purification using eIF4E (the high-affinity cap binding protein) fused to glutathione S-transferase (GST). Input and SN material were loaded at 100%.
To determine whether TERRA contains a canonical m7G cap, we isolated total RNA from HeLa and HLF cells and selected m7G cap-containing RNAs by using the high-affinity m7G cap-binding protein eIF4E fused to glutathione S-transferase (GST) that had been immobilized on a glutathione column. As a control, total RNA was incubated with GST protein alone. Northern blot analysis was carried out using a strand-specific TERRA probe. β-Actin and 5.8S rRNA were included as positive and negative controls, respectively. The analysis revealed that TERRA bound to the GST-eIF4E protein, indicating that the large majority of TERRA contains the m7G cap (Fig. 1B).
Poly(A) tail stabilizes TERRA transcripts.
Around 7% of human TERRA is polyadenylated (2). The mechanism of TERRA polyadenylation remains unclear, as a canonical 5′-AAUAAA-3′ poly(A) signal is predicted to be missing in primary TERRA transcripts. Canonical polyadenylation increases the transcript stability, whereas noncanonical poly(A) polymerases, which add short poly(A) tails to their substrate RNAs, usually trigger efficient decay of the RNAs by the recruitment of the nuclear exosome complex (28, 43, 44, 48). We blocked transcription in HeLa cells with 5 μg/ml actinomycin D and measured TERRA levels in poly(A)− and poly(A)+ RNA fractions in time course experiments by Northern blotting (Fig. 2). β-Actin, c-Myc, and U1 snRNA were included in the analysis for normalization (U1 snRNA and β-actin) and fractionation and to control for successful inhibition of Pol II (c-Myc) (half-life [t1/2] of 20 min [24, 27, 45]) (Fig. 2). Quantification of the hybridization signals on a PhosphorImager revealed that poly(A)− TERRA has a half-life of about 3 h and poly(A)+ TERRA has a very long half-life of more than 8 h (Fig. 2). Therefore, the poly(A) tail increases TERRA stability, suggesting that the canonical poly(A) polymerase is responsible for the polyadenylation of this TERRA population.
FIG. 2.
Poly(A)+ TERRA has a longer half-life than poly(A)− TERRA. HeLa cells were treated with 5 μg/ml actinomycin D for the indicated times. Total RNA was prepared from HeLa cells, and poly(A)+ RNA was separated from poly(A)− RNA by using an oligo(dT) resin. Both RNA fractions were subjected to Northern blot analysis using a telomeric DNA probe detecting UUAGGG repeats. Filters were then stripped and reprobed successively for c-Myc RNA and β-actin and U1 snRNA. The TERRA half-life in poly(A)− and poly(A)+ fractions was calculated using a scatter plot analysis after the TERRA signal was normalized to the U1 and β-actin signals, respectively. The best-fit exponential regression curve of the data points was determined for both fractions. R2 values are indicated.
Poly(A)− and poly(A)+ TERRA fractions have different subnuclear distributions.
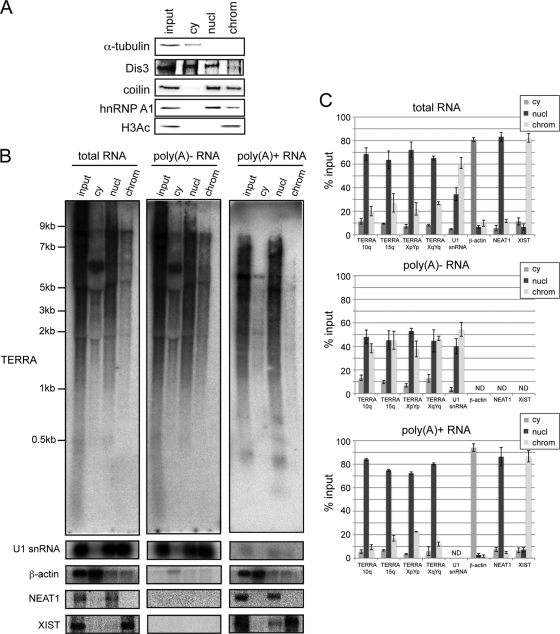
TERRA is nearly exclusively nuclear and partially colocalizes with telomeres, where it may elicit some of its biological functions. In order to further characterize poly(A)+ and poly(A)− TERRA, we developed a fractionating protocol to distinguish chromatin-associated, nucleoplasmic, and cytoplasmic TERRA populations (Fig. 3) (also see Materials and Methods). The protocol was applied to HEK293 cells, and the identity of subcellular fractions was confirmed by Western blot analysis with antibodies specific for α-tubulin (cytoplasm [cy]), Dis3 (cy and nucleoplasm [nucl]), coilin (nucl and chromatin associated [chrom]), hnRNP A1 (nucl and chrom), and H3Ac (chrom) (Fig. 3A). Poly(A)− and poly(A)+ RNA was isolated from each subcellular fraction, and TERRA levels were measured by Northern blotting and qRT-PCR (Fig. 3A and B). U1 snRNA, β-actin, NEAT1, and XIST were included in these analyses as RNA controls of the subcellular and poly(A)− and poly(A)+ fractionation. NEAT1 is a 4-kb polyadenylated ncRNA that is an integral component of nuclear paraspeckles (13). XIST ncRNA associates with and silences the inactive X chromosome (8, 9). As shown in Fig. 3B and C, ∼60% of poly(A)− TERRA was found in the nucleoplasmic extracts, and the remaining ∼40% was found associated with chromatin. On the contrary, the vast majority (80%) of poly(A)+ TERRA was present in the nucleoplasmic fraction, while it was barely detectable on chromatin (Fig. 3B and C). As expected, both TERRA populations were almost not present in the cytoplasmic extracts. The faint signals detected in this fraction (<10%) may be due to leakage out of the nucleus during cell fractionation (Fig. 3B and C). Overall, these findings indicated that chromatin-associated TERRA is not polyadenylated, whereas poly(A)+ TERRA accumulates in the nucleus, binding only weakly and/or transiently to telomeric or other chromatin.
FIG. 3.
Subcellular localization of poly(A)− and poly(A)+ TERRA fractions. (A) HEK293 cell cytoplasmic (cy), nucleoplasmic (nucl), and chromatin (chrom) soluble fractions were prepared and analyzed by Western blotting. Detection of α-tubulin (cy), Dis3 (cy and nucl), coilin (nucl and chrom), hnRNP A1 (nucl and chrom), and H3Ac (chrom) was used to confirm HEK293 cell fractionation. (B) Total, poly(A)−, and poly(A)+ RNA was prepared from each HEK293 subcellular fraction and monitored for TERRA transcriptional levels by Northern blot analysis. (C) TERRA fractionation was also evaluated by qRT-PCR after the TERRA signals were normalized to the enhanced green fluorescent protein (eGFP) signal. eGFP RNA was produced in a T7 in vitro reaction and added to each subcellular fraction before RNA extraction proceeded. β-Actin [poly(A)+] (cy), U1 snRNA [poly(A)−] (nucl and chrom), NEAT1 [poly(A)+] (nucl), and XIST [poly(A)+] (chrom) were used as controls for HEK293 cell fractionation. Error bars indicate standard deviations (SD).
The majority of TERRA UUAGGG tracts are shorter than 400 bases.
The length heterogeneity of TERRA may be due to heterogeneity of TERRA 5′ ends, variation of UUAGGG tract length at TERRA 3′ ends, or both. To address this issue, we developed an RT protocol to measure the UUAGGG tract length. Isolated RNA was reverse transcribed with a mixture of six oligonucleotides that were predicted to hybridize to the junction between the UUAGGG sequence in all six permutated registers and the poly(A) tail of poly(A)+ TERRA (Fig. 4 A). In order to also analyze the UUAGGG tract length in the poly(A)− TERRA population with the same RT primers, we polyadenylated TERRA in vitro with recombinant yeast Pap1 prior to the analysis (Fig. 4A). RT was first carried out in the presence of only three nucleotides ([32P]dCTP, dTTP, and dATP) in order to restrict reverse transcription to the pure UUAGGG tract. The length of the derived cDNA was measured on a 1% denaturing alkaline agarose gel (Fig. 4B). The 1-h pulse was followed by a chase reaction in which an excess of unlabeled dCTP and, for the +dGTP reaction, dGTP was added in order to allow RT beyond the cytosine-containing subtelomeric TERRA sequence. With dGTP, the majority of the shorter products were chased into longer cDNAs, indicating that the lack of dGTP, which is required to reverse transcribe beyond cytosine bases, was the major cause of the length restriction during the pulse.
FIG. 4.
The UUAGGG tract of TERRA molecules is not longer than 400 bases. (A) Schematic representation of UUAGGG tract-specific reverse transcription. (B) Total RNA from HeLa and HLF (wild type and supertelomerase [supertelo] [hTERT and hTR overexpressed]) cells was treated or not treated with yeast Pap1. A radioactive reverse transcription was performed with an oligo(dT)21NNN, where NNN corresponds to a sequence able to guide the annealing of the oligo(dT) itself at the junction between the poly(A) tail and the UUAGGG repeats. When the reaction is carried out in the absence of dGTP, only the UUAGGG portion of TERRA can be reverse transcribed.
In control experiments, we tested the efficiency of the RT on two different synthetic UUAGGG repeat-containing transcripts. The first transcript, referred to as XpYp telo-350, contains a subtelomere-derived sequence (100 bases, cloned from the terminal part of Chr XpYp) and a block of polymorphic UUAGGG-like repeats (250 bases) followed by pure telomeric UUAGGG repeats (348 bases). The second transcript, referred to as telo-700, contains only pure telomeric UUAGGG repeats, with a length of 696 bases. Under the employed experimental conditions, the reverse transcriptase efficiently reverse transcribed both UUAGGG repeat-containing transcripts (see Fig. S1A in the supplemental material). Also, as expected, the XpYp telo-350 RT product was efficiently chased into a longer RT product upon addition of dGTP. However, it is worth noting that the RT product from the telo-700 transcript was somewhat weaker and more smeary than the RT product of XpYp telo-350, indicating that an increased number of truncated cDNAs are produced when long UUAGGG repeats are reverse transcribed (see Fig. S1A). However, when we reverse transcribed the two in vitro transcripts together or in combination with total RNA from HeLa cells, we did observe the expected bands corresponding to XpYp telo-350 and telo-700, and no preferential RT of short-UUAGGG-tract-containing RNA was observed (see Fig. S1B).
In another control experiment, cDNAs derived from β-actin and GUSB mRNAs were undetectable when the RT was carried out in the absence of dGTP (see Fig. S2 in the supplemental material). On the contrary, after the chase, both of these mRNAs were reverse transcribed, as they could be detected by RT-PCR. The same was true for TERRA molecules derived from chromosome end XpYp, in which subtelomeric cDNA could be detected by PCR only after the chase (see Fig. S2).
The same protocol was applied to RNA from HeLa and HLF cells. As shown in Fig. 4B, this experiment suggests that the majority of both the total TERRA [93% of which is comprised of poly(A)− TERRA] and the poly(A)+ TERRA fraction has a pure UUAGGG tract length ranging from 100 bases to 400 bases, with a median length of 200 bases in both cell lines. Furthermore, to test whether changes in telomeric length may affect the size of the UUAGGG tract, we performed the same analysis in supertelomerase HeLa and HLF cells, which have overelongated telomeres (see Fig. S3 in the supplemental material) due to the cooverexpression of hTR and hTERT (15). However, even in the presence of overelongated telomeres, no significant change in the length of UUAGGG repeats was detected (Fig. 4B). This suggests that TERRA transcription does not proceed far into the telomeric tract, although posttranscriptional trimming cannot be excluded. Nonetheless, the results indicate that TERRA length heterogeneity is mainly due to size variability of its subtelomeric tract.
Poly(A)− and poly(A)+ TERRA differ in their termination sites.
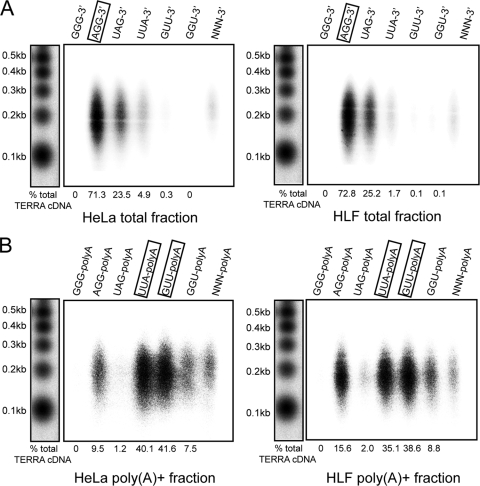
TERRA 3′ ends can potentially terminate at six different positions within the UUAGGG repeat sequence. To determine TERRA 3′-end sequences, we performed UUAGGG-specific reverse transcription reactions as described above but using in separate reactions the six different TERRA-specific primers that contained oligo(dT) at their 5′ ends and that annealed with their three most-3′-terminal bases to the TERRA 3′ end, in case of base complementarity. Therefore, these primers ended in -CCC-3′, -CCT-3′, -CTA-3′, -TAA-3′, -TTC-3′, or -TCC-3′. As described above, in order to also characterize termination sites in the poly(A)− TERRA population, we polyadenylated total RNA with recombinant yeast Pap1 prior to reverse transcription. In a control experiment with synthetic RNA oligonucleotides that ended in all possible telomeric registers, we found that Pap1 elongates all 3′ ends with similar efficiencies (see Fig. S4 in the supplemental material). As seen with total RNA extracted from HeLa and HLF cells (Fig. 5 A), we obtained the most efficient TERRA reverse transcription with the 5′-(dT)21CCT-3′ primer, indicating that poly(A)− TERRA preferentially terminates with UUAGG-3′.
FIG. 5.
Poly(A)− and poly(A)+ TERRA fractions have different termination sequences. (A) UUAGGG tract reverse transcription was performed on total RNA from HeLa and HLF cells treated with yeast Pap1. Six different primers were used in six different reactions. Each primer consists of an oligo(dT)21NNN, where the last three positions are a single combination of dCTP, dTTP, and dATP. The use of six different oligonucleotides allows discrimination between all of the possible termination sites. (B) The same approach was used to identify a preferential register between the UUAGGG repeats and the poly(A) tail in the TERRA poly(A)+ fraction. Numbers at the bottom of each panel indicate the percentage of TERRA cDNA produced by each specific primer. The total TERRA cDNA value was calculated as the sum of the single signals.
With the poly(A)+ TERRA fraction, however, we found that two different primers, 5′-(dT)21-TAA-3′ and 5′-(dT)21-AAC-3′, produced the strongest signals. Both of these primers hybridize to and recognize the same frame, indicating that poly(A) addition occurs most frequently after the second uracil or the adenosine within the UUAGGG repeats (Fig. 5B). The different termination sites of poly(A)+ and poly(A)− TERRA transcripts support the idea of unrelated 3′ processing events for these distinct RNA populations.
TERRA is regulated in a cell cycle-dependent manner.
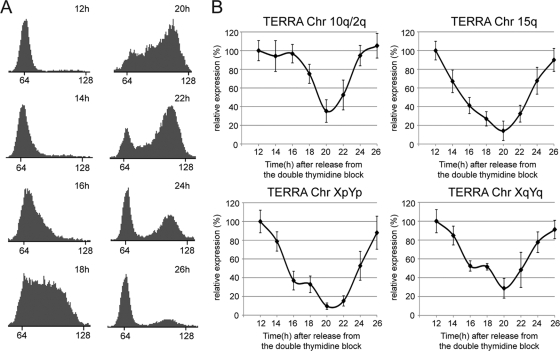
To assess whether TERRA is regulated during the cell cycle, we synchronized HeLa cells near the G1/S boundary by using a double thymidine block. In order to exclude possible effects of the cell cycle arrest and checkpoint activation on TERRA abundance, we analyzed TERRA levels only after the second G1 phase from the release in a time course experiment. Total RNA from HeLa cells was extracted from 12 h (second G1 phase) to 26 h (third G1 phase) after the release from the block, at intervals of 2 h. Cell cycle progression was monitored by FACS analysis (Fig. 6 A). TERRA molecules derived from four different chromosome ends (10q, 15q, XpYp, and XqYq) were assessed by qRT-PCR and normalized to β-actin mRNA. For all TERRA molecules, we observed the highest levels in early G1. TERRA levels decreased slightly in late G1 and plunged in S phase, reaching the lowest expression levels as cells progressed from late S to G2 phase. After cells had passed through G2 and M phases and reentered the next G1 phase, TERRA expression increased, again reaching the initial expression values (Fig. 6B). Notably, TERRA levels in G1 were 3 to 10 times higher than in late S/G2. Low levels of TERRA in late S phase may prevent deleterious interference with DNA replication of telomeric tracts, which is seen under circumstances in which TERRA is upregulated at telomeres (4, 50).
FIG. 6.
TERRA levels are regulated during the cell cycle. (A) FACS analysis of DNA content (x axis, arbitrary units) in HeLa cells synchronized at the G1/S boundary with a double thymidine block. The cells were analyzed 12 h to 26 h after the release from the thymidine block. (B) TERRA expression was assessed during the cell cycle and evaluated by qRT-PCR, after normalization to β-actin mRNA. Error bars indicate SD.
DISCUSSION
The notion that the ends of eukaryotic chromosomes are transcribed into RNA and that telomeric chromatin contains RNA molecules adds a new fascinating level of complexity to telomere metabolism and provides another example of a structural RNA molecule that may prove to be of relevance. A detailed understanding of TERRA structure and biogenesis is of fundamental importance in order to elucidate its enigmatic functions. Here we provide a first in-depth analysis of the primary structures of TERRA molecules.
Since TERRA is very heterogeneous in length, it seemed conceivable that TERRA 5′ ends are trimmed posttranscriptionally and therefore are uncapped. However, our data demonstrate that the vast majority of TERRA 5′ ends harbor a canonical m7G cap structure. This therefore indicates that TERRA 5′ ends are generated cotranscriptionally and are not due to posttranscriptional processing. The 5′-end cap modification should protect TERRA from the action of 5′-to-3′ exonucleases, in analogy to other capped RNAs (33). Although transcribed by Pol II, the majority (∼90%) of TERRA molecules are not polyadenylated, resembling in terms of 5′- and 3′-end structures some other Pol II transcripts (histone mRNAs) that contain a 5′ cap but not a poly(A) tail (34).
We identify remarkable differences in biochemical behavior between the poly(A)− and poly(A)+ TERRA fractions. Poly(A)+ TERRA is more stable, making it unlikely that a noncanonical polyadenylation pathway, which channels RNAs toward exosome-mediated RNA degradation, is responsible for polyadenylation (43). How the canonical poly(A) polymerase may polyadenylate TERRA remains to be explored, as the classical cleavage and polyadenylation signals appear to be missing in the TERRA sequence. Moreover, poly(A)− and poly(A)+ TERRA fractions differ in termination sequence. Finally, poly(A)+ TERRA behaves differently during nuclear fractionation, being mostly in a non-chromatin-associated pool whereas a considerable fraction of poly(A)− TERRA is chromatin associated. Together, these findings suggest distinct processing pathways and biological functions for poly(A)− and poly(A)+ TERRA populations. Why poly(A)+ but not poly(A)− TERRA is released from the chromatin remains unclear. It will also be interesting to determine whether or not telomerase associates specifically with the nucleoplasmic or chromatin-associated TERRA pool. If TERRA regulates telomerase in a telomere-specific manner, one would expect specific association of telomerase with chromatin-associated poly(A)− TERRA molecules. The nucleoplasmic TERRA pool might, on the other hand, have effects on overall telomere length and/or might be involved in biological functions not yet revealed.
Another fundamental question concerning the biology and the homeostasis of TERRA regards its length heterogeneity. Human TERRA ranges from ∼100 bases up to more than ∼9 kb (4). Here, we develop a new reverse transcription strategy to measure the pure telomeric UUAGGG tract length. Recently, it has been shown that TERRA transcription start sites at 19 different chromosome ends contain CpG islands, which, according to database entries, are located not more than 1 kb upstream of the predicted telomeric regions (32). This finding led us to consider that the heterogeneity of TERRA might stem from its telomeric region. However, the reference sequences from the NCBI database do not take into account the blocks of polymorphic (TTAGGG)n-like repeats (38), which are interspersed with canonical (TTAGGG)n repeats and extend for at least 1 kb (5). Our results now suggest that the majority of both poly(A)− and poly(A)+ fractions have UUAGGG tract sizes of approximately 200 bases, with little length heterogeneity. Although we cannot rule out the existence of a subset of TERRA molecules with long UUAGGG repeats, our data suggest that, mostly, TERRA transcription does not proceed far into the telomeric tract. In support of this conclusion, a recent study demonstrates partial transcriptional blockage occurring during transcription of guanine-rich DNA in vitro (6). In summary, we conclude that TERRA length heterogeneity is mostly due to length differences of the sequences that are derived from polymorphic (TTAGGG)n-like repeats and other sequences of the subtelomeric region.
Finally, we also identify that TERRA levels change in a cell cycle-dependent manner. TERRA accumulates in early G1, continuously decreases in S phase, and reaches the lowest expression levels at the transition between late S and G2. TERRA inhibits telomerase both in vitro (36, 40) and in vivo (31, 36). Therefore, downregulation of TERRA in S phase might unleash telomerase, allowing the extension of the telomeric G-rich strand in a cell cycle-dependent manner. Downregulation of TERRA in S phase may also be critical to allow efficient replication of chromosome ends by the canonical DNA replication machinery. Indeed, depletion of RNA surveillance factors, which were shown to interact with the replication machinery (3, 10), leads to accumulation of TERRA at chromosome ends and coincides with stochastic loss of entire telomere tracts (4). A similar phenotype is seen in cells derived from patients suffering from ICF (immunodeficiency, centromere instability, facial anomalies) syndrome, which contain defective DNA methyltransferase DNMT3b. Cell lines derived from ICF patients show high TERRA levels, and they suffer from frequent telomere loss (20, 32, 50).
In conclusion, our study investigates fundamental aspects of TERRA homeostasis, providing evidence for the existence of multiple regulatory pathways for TERRA biogenesis, turnover, and subnuclear localization. This lays the basis for further investigations on TERRA regulatory pathways, which promise to provide insight into the functions of this fascinating RNA.
Supplementary Material
Acknowledgments
We thank Verena Pfeiffer and Alix Christen for technical advice, Brian Luke for comments on the manuscript, and George Martin and Walter Keller (University of Basel) for recombinant Pap1.
This work was supported by an EMBO long-term fellowship to A.P. and grants to J.L. from the Swiss National Science Foundation, the European Community's Seventh Framework Programme FP7/2007-2011 (grant agreement number 200950), and a European Research Council advanced investigator grant (grant agreement number 232812).
Footnotes
Published ahead of print on 16 August 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Artandi, S. E., and R. A. DePinho. 2010. Telomeres and telomerase in cancer. Carcinogenesis 31:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azzalin, C. M., and J. Lingner. 2008. Telomeres: the silence is broken. Cell Cycle 7:1161-1165. [DOI] [PubMed] [Google Scholar]
- 3.Azzalin, C. M., and J. Lingner. 2006. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 16:433-439. [DOI] [PubMed] [Google Scholar]
- 4.Azzalin, C. M., P. Reichenbach, L. Khoriauli, E. Giulotto, and J. Lingner. 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318:798-801. [DOI] [PubMed] [Google Scholar]
- 5.Baird, D. M., A. J. Jeffreys, and N. J. Royle. 1995. Mechanisms underlying telomere repeat turnover, revealed by hypervariable variant repeat distribution patterns in the human Xp/Yp telomere. EMBO J. 14:5433-5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belotserkovskii, B. P., R. Liu, S. Tornaletti, M. M. Krasilnikova, S. M. Mirkin, and P. C. Hanawalt. 2010. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl. Acad. Sci. U. S. A. 107:12816-12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 8.Brockdorff, N., and S. M. Duthie. 1998. X chromosome inactivation and the Xist gene. Cell. Mol. Life Sci. 54:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, S. D. 1991. XIST and the mapping of the X chromosome inactivation centre. Bioessays 13:607-612. [DOI] [PubMed] [Google Scholar]
- 10.Carastro, L. M., C. K. Tan, M. Selg, H. M. Jack, A. G. So, and K. M. Downey. 2002. Identification of delta helicase as the bovine homolog of HUPF1: demonstration of an interaction with the third subunit of DNA polymerase delta. Nucleic Acids Res. 30:2232-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chanfreau, G., G. Rotondo, P. Legrain, and A. Jacquier. 1998. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17:3726-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi, Y. H., and C. H. Hagedorn. 2003. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc. Natl. Acad. Sci. U. S. A. 100:7033-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clemson, C. M., J. N. Hutchinson, S. A. Sara, A. W. Ensminger, A. H. Fox, A. Chess, and J. B. Lawrence. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33:717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cougot, N., E. van Dijk, S. Babajko, and B. Seraphin. 2004. ‘Cap-tabolism.’ Trends Biochem. Sci. 29:436-444. [DOI] [PubMed] [Google Scholar]
- 15.Cristofari, G., and J. Lingner. 2006. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 25:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.d'Adda di Fagagna, F., P. M. Reaper, L. Clay-Farrace, H. Fiegler, P. Carr, T. Von Zglinicki, G. Saretzki, N. P. Carter, and S. P. Jackson. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426:194-198. [DOI] [PubMed] [Google Scholar]
- 17.de Lange, T. 2009. How telomeres solve the end-protection problem. Science 326:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lange, T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19:2100-2110. [DOI] [PubMed] [Google Scholar]
- 19.de Lange, T., L. Shiue, R. M. Myers, D. R. Cox, S. L. Naylor, A. M. Killery, and H. E. Varmus. 1990. Structure and variability of human chromosome ends. Mol. Cell. Biol. 10:518-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng, Z., A. E. Campbell, and P. M. Lieberman. 2010. TERRA, CpG methylation and telomere heterochromatin: lessons from ICF syndrome cells. Cell Cycle 9:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng, Z., J. Norseen, A. Wiedmer, H. Riethman, and P. M. Lieberman. 2009. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 35:403-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 23.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 24.Hann, S. R., and R. N. Eisenman. 1984. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol. Cell. Biol. 4:2486-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia, D., L. Cai, H. He, G. Skogerbo, T. Li, M. N. Aftab, and R. Chen. 2007. Systematic identification of non-coding RNA 2,2,7-trimethylguanosine cap structures in Caenorhabditis elegans. BMC Mol. Biol. 8:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, S. H., P. Kaminker, and J. Campisi. 2002. Telomeres, aging and cancer: in search of a happy ending. Oncogene 21:503-511. [DOI] [PubMed] [Google Scholar]
- 27.King, M. W., J. M. Roberts, and R. N. Eisenman. 1986. Expression of the c-myc proto-oncogene during development of Xenopus laevis. Mol. Cell. Biol. 6:4499-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121:713-724. [DOI] [PubMed] [Google Scholar]
- 29.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 30.Lingner, J., and W. Keller. 1993. 3′-end labeling of RNA with recombinant yeast poly(A) polymerase. Nucleic Acids Res. 21:2917-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luke, B., A. Panza, S. Redon, N. Iglesias, Z. Li, and J. Lingner. 2008. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 32:465-477. [DOI] [PubMed] [Google Scholar]
- 32.Nergadze, S. G., B. O. Farnung, H. Wischnewski, L. Khoriauli, V. Vitelli, R. Chawla, E. Giulotto, and C. M. Azzalin. 2009. CpG-island promoters drive transcription of human telomeres. RNA 15:2186-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neugebauer, K. M. 2002. On the importance of being co-transcriptional. J. Cell Sci. 115:3865-3871. [DOI] [PubMed] [Google Scholar]
- 34.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 35.Redon, S., P. Reichenbach, and J. Lingner. 2007. Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res. 35:7011-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redon, S., P. Reichenbach, and J. Lingner. 11 May 2010. The noncoding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 37.Reichenbach, P., M. Hoss, C. M. Azzalin, M. Nabholz, P. Bucher, and J. Lingner. 2003. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 13:568-574. [DOI] [PubMed] [Google Scholar]
- 38.Riethman, H. 2008. Human telomere structure and biology. Annu. Rev. Genomics Hum. Genet. 9:1-19. [DOI] [PubMed] [Google Scholar]
- 39.Sandell, L. L., D. E. Gottschling, and V. A. Zakian. 1994. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc. Natl. Acad. Sci. U. S. A. 91:12061-12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoeftner, S., and M. A. Blasco. 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10:228-236. [DOI] [PubMed] [Google Scholar]
- 41.Shatkin, A. J. 1976. Capping of eucaryotic mRNAs. Cell 9:645-653. [DOI] [PubMed] [Google Scholar]
- 42.Snow, B. E., N. Erdmann, J. Cruickshank, H. Goldman, R. M. Gill, M. O. Robinson, and L. Harrington. 2003. Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 13:698-704. [DOI] [PubMed] [Google Scholar]
- 43.Vanacova, S., and R. Stefl. 2007. The exosome and RNA quality control in the nucleus. EMBO Rep. 8:651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanacova, S., J. Wolf, G. Martin, D. Blank, S. Dettwiler, A. Friedlein, H. Langen, G. Keith, and W. Keller. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waters, C. M., T. D. Littlewood, D. C. Hancock, J. P. Moore, and G. I. Evan. 1991. c-myc protein expression in untransformed fibroblasts. Oncogene 6:797-805. [PubMed] [Google Scholar]
- 46.Wellinger, R. J., K. Ethier, P. Labrecque, and V. A. Zakian. 1996. Evidence for a new step in telomere maintenance. Cell 85:423-433. [DOI] [PubMed] [Google Scholar]
- 47.Wellinger, R. J., and D. Sen. 1997. The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer 33:735-749. [DOI] [PubMed] [Google Scholar]
- 48.Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121:725-737. [DOI] [PubMed] [Google Scholar]
- 49.Xu, Y., K. Kaminaga, and M. Komiyama. 2008. G-quadruplex formation by human telomeric repeats-containing RNA in Na+ solution. J. Am. Chem. Soc. 130:11179-11184. [DOI] [PubMed] [Google Scholar]
- 50.Yehezkel, S., Y. Segev, E. Viegas-Pequignot, K. Skorecki, and S. Selig. 2008. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 17:2776-2789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.