Abstract
The biogenesis of the Tetrahymena telomerase ribonucleoprotein particle (RNP) is enhanced by p65, a La family protein. Single-molecule and biochemical studies have uncovered a hierarchical assembly of the RNP, wherein the binding of p65 to stems I and IV of telomerase RNA (TER) causes a conformational change that facilitates the subsequent binding of telomerase reverse transcriptase (TERT) to TER. We used purified p65 and variants of TERT and TER to investigate the conformational rearrangements that occur during RNP assembly. Nuclease protection assays and mutational analysis revealed that p65 interacts with and stimulates conformational changes in regions of TER beyond stem IV. Several TER mutants exhibited telomerase activity only in the presence of p65, revealing the importance of p65 in promoting the correct RNP assembly pathway. In addition, p65 rescued TERT assembly mutants but not TERT activity mutants. Taken together, these results suggest that p65 stimulates telomerase assembly and activity in two ways. First, by sequestering stems I and IV, p65 limits the ensemble of structural conformations of TER, thereby presenting TERT with the active conformation of TER. Second, p65 acts as a molecular buttress within the assembled RNP, mutually stabilizing TER and TERT in catalytically active conformations.
The catalytic core of the telomerase RNP is comprised of the protein reverse transcriptase TERT (for telomerase reverse transcriptase) and the RNA component TER (for telomerase RNA). TERT uses a region of TER that is complementary to the telomere DNA sequence to template the synthesis of telomeric repeats onto a single-stranded telomeric DNA primer (15). Like most reverse transcriptases, telomerase copies its template without dissociation, a phenomenon known as nucleotide addition processivity (NAP). In addition, telomerase is unique among reverse transcriptases in that it also undergoes repeat addition processivity (RAP), during which an already-extended primer is repositioned within its active site so the same intrinsic RNA sequence templates the addition of several telomeric repeats (14, 23).
Besides providing the template for telomere synthesis, TER primary, secondary, and tertiary structural elements play critical architectural and functional roles (34, 37, 39, 44, 45). In Tetrahymena thermophila TER (Fig. 1A), the base of stem II and part of the predicted single-stranded RNA on either side of stem II, called the template boundary element or the TERT-binding element (TBE), have been shown to be important for both TERT binding and delineating the 5′ boundary of the template sequence during synthesis (4, 18). The template recognition element (TRE), a single-stranded region of TER that is located 3′ of the template, has been shown to be involved in maintaining NAP and essential for RAP (4, 25). Just 3′ of the TRE is the pseudoknot, an interaction between stem-loop III and nucleotides preceding it, which contributes to NAP, RAP, RNA stability, and RNP assembly (12, 34, 39, 43, 44). The TER secondary structure continues 3′ of the pseudoknot to complete stem I through long-range base pairing with the nucleotides at the 5′ end of TER. Stem I is connected through a linker to stem-loop IV, which enhances NAP and RAP, potentially through its interaction with TERT (17). Since TER in ciliated protozoa is an RNA polymerase III transcript, there is an oligo(U) tail at the 3′ end of the molecule that makes a small contribution to RNP assembly (3).
FIG. 1.
p65 increases the amount of RNA associated with TERT and the enzymatic activity of telomerase. (A) A cartoon of Tetrahymena TER with secondary structural elements indicated. TBE, template boundary element or TERT-binding element; TRE, template recognition element. (B) An SDS polyacrylamide gel stained with Coomassie blue showing the purity of p65. (C) Direct activity assay performed on wild-type telomerase synthesized in the absence or presence of enough purified p65 to saturate TER. RC, 5′-32P-end-labeled DNA oligonucleotide added as a recovery control. Telomerase repeats added are indicated on the right side of the gel. Rel act, radiolabeled nucleotide incorporation relative to that in the absence of p65. (D) Duplicate SDS-PAGE analysis of TERT immunopurification of telomerase assembled in RRLs in the presence or absence of p65 (left) and the TERT-independent recovery of TER (right). The top panel shows the [35S]methionine incorporated into TERT and the coimmunoprecipitated 32P-labeled TER. The bottom panel is an exposure of the same gel with film inserted to block the detection of 35S, showing only the coimmunoprecipitation of 32P-labeled TER. Rel RNA, relative amount of TER pulldown normalized to the amount of TERT.
Telomerase holoenzymes contain other components besides TERT and TER, including both transiently and stably associated proteins (27, 31, 40). The initial purification of telomerase identified TER, TERT, and p43 in Euplotes aediculatus (22). p43 is a telomerase-specific protein containing a La motif and an RNA recognition motif (RRM); it binds primarily to stem-loop IV of TER, stimulates telomerase activity and RAP in vitro, and may contribute to the nuclear retention of TER (1-3).
The telomerase holoenzyme from T. thermophila contains a 65-kDa protein (p65) that is the ortholog of Euplotes p43 (47). p65, encoded by an essential gene, copurifies with TER, TERT, and telomerase activity, stimulates holoenzyme biogenesis, and stabilizes TER levels (33, 47). Homologs of p43/p65 have not been found outside the ciliates, but the complex of dyskerin, NHP2, NOP10, and GAR1 (11, 28, 32, 46) may be the functional analog in mammalian telomerase. These proteins are not telomerase specific but are components of small nucleolar RNPs (snoRNPs) (16).
Despite the presence of the La motif in p65 and an oligo(U) tail in TER, p65 appears to be recruited to TER mainly by the recognition of proximal stem IV and the GA bulge that delineates the proximal and distal regions of stem IV (33, 35). Biochemical data have uncovered a hierarchical assembly of the RNP whereby the binding of p65 to TER causes a conformational change in TER stem IV, increasing the angle between distal and proximal stem IV and facilitating the subsequent binding of the RNA binding domain (RBD) of TERT to TER (29). These sequential binding events have been further analyzed by single-molecule studies (42).
We set out to determine what other roles p65 plays in RNP biogenesis besides inducing the conformational change within stem IV. Specifically, we asked whether p65 interacts with TER beyond the established binding site of stems I and IV. Through a combination of electrophoretic mobility shift assays, nucleic acid structure probing experiments, and telomerase assays, we have determined that p65 stimulates holoenzyme formation by directly and indirectly promoting the proper folding of the entire TER structure, suggesting that p65 is a TER-specific RNA folding protein. We also have shown that the previously observed modest stimulation of telomerase activity by p65 becomes much more substantial in the context of TER and TERT mutations; several highly deleterious TER and TERT assembly mutants can be rescued to near-wild-type activity with p65, and in fact, p65 has emerged as a tool for sorting assembly and activity mutants of TER and TERT. Finally, these results suggest that p65 stabilizes the functional interactions between TERT and TER that are required for the telomerase reaction cycle.
MATERIALS AND METHODS
Protein purification.
The gene expressing p65 used in these studies was recloned from pET28-HisTEV-p65(1-542) into pET28, giving the full-length p65 protein an N-terminal His6 tag followed by a thrombin cleavage sequence. Escherichia coli BL21(DE3) cells transformed with HisThrombin-p65(1-542) were grown in LB supplemented with 30 μg/ml kanamycin at 37°C until an optical density at 600 nm of 0.4 to 0.6 was reached. The temperature then was decreased to 18°C for 30 min prior to induction with a final concentration of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 to 6 h. Cells were harvested and stored at −80°C.
Cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole, 10% glycerol) with Complete EDTA-free protease inhibitor tablets (Roche). Prior to lysis by sonication, cells were incubated with lysozyme at a final concentration of 5 mg/ml for 45 to 60 min. Lysate then was clarified and passed over a 5-ml HisTrap column (GE). Protein was eluted in two gradients of imidazole, one shallow (5 to 50 mM over 10 column volumes [cv]), to eliminate protein contaminated by nucleic acid, and one steep (50 to 500 mM over 10 cv), for collecting p65. The peak fractions were pooled and digested with thrombin in the presence of 0.1% β-mercaptoethanol (β-ME) at room temperature for 3 h. The protein solution then was dialyzed overnight to remove first the imidazole and then the β-ME. This sample then was passed over the nickel column again; the protein that had been cleaved by thrombin eluted in the shallow gradient. The p65 peak was pooled, diluted with 20 mM Tris-HCl, pH 8.0, 2 mM dithiothreitol (DTT) to reduce the NaCl concentration to 100 mM, loaded onto a HiTrap Q column (GE), and eluted with a 125-ml gradient from 100 mM to 1 M NaCl. The p65-containing fractions were pooled; concentrated; buffer exchanged into 20 mM HEPES, pH 7.5, 250 mM NaCl, 2 mM DTT; and loaded onto a gel filtration column (Sephadex 200; GE). The peak then was concentrated and stored in the presence of 5% glycerol at −80°C. The concentration of active p65 (typically 50 to 70%) was determined by the titration of 50 nM 32P-labeled TER RNA and analyzed by electrophoretic mobility shift assay (EMSA).
TERT-N (amino acids 1 to 516) was subcloned from mp190 (8) and inserted into the NcoI and BamHI sites of vector pET28a (Novagen), resulting in pET28-TERT516, which has a His6 tag followed by the first residue of TERT. The construct used for the RNA binding domain of TERT, RBD (amino acids 216 to 516), was a slightly smaller domain than that used in previous studies (18, 19, 29, 30) but had comparable TER and p65-TER binding affinity (C. O'Connor and K. Collins, unpublished results). This construct also was subcloned from mp190 (8) and inserted into the NdeI and BamHI sites of vector pET28a (Novagen), resulting in a His6 tag followed by a thrombin site and amino acids 216 to 516 of TERT.
An overnight culture of BL21(DE3) cells transformed with pET28-TERT516 or RBD was grown at 37°C and used to inoculate a 100-ml LB culture. This starter culture then was used to inoculate 1-liter cultures of LB (1:100) supplemented with 30 μg/ml kanamycin. These cultures then were grown until an optical density at 600 nm of 0.4 was reached. The temperature then was decreased to 18°C for 30 min prior to induction with a final concentration of 0.1 mM IPTG for 16 h. Cells were harvested and stored at −80°C.
Cells expressing TERT-N were resuspended in lysis buffer with Complete EDTA-free protease inhibitor cocktail tablets (Roche). The purification proceeded in the same manner as the purification of p65 through the first nickel column. The pooled fractions of TERT-N from the nickel column then were diluted with 50 mM MES, pH 6.5, 2 mM DTT to bring the concentration of NaCl to 250 mM. After pelleting any protein that precipitated, the sample was loaded onto tandem HiTrapQ/HiTrapS (GE) columns. The protein then was eluted off the HiTrapS column with a gradient from 250 mM to 1 M NaCl over 100 ml. Fractions containing TERT-N were concentrated; buffer exchanged into 20 mM HEPES, pH 7.5, 250 mM NaCl, 2 mM DTT; and loaded onto a gel filtration column (Sephadex 200; GE). The peak then was concentrated and stored in the presence of 5% glycerol at −80°C. The concentration of active TERT (typically 30%) was determined by a titration of 50 nM 5′-end-labeled TER RNA and analyzed by EMSA.
The protein purification protocol for RBD was similar to that for p65, except the peak fractions from the second nickel column were diluted with 50 mM MES, pH 6.5, 2 mM DTT to decrease the concentration of NaCl to 100 mM. This diluted sample was loaded onto a HiTrapS column and eluted with a gradient of 100 mM to 1 M NaCl. The sample then was concentrated and buffer exchanged into 20 mM HEPES, pH 7.0, 250 mM NaCl, 2 mM DTT prior to gel filtration (Sephadex 75; GE). The peak then was concentrated and stored in the presence of 5% glycerol at −80°C. The concentration of active RBD (typically 30%) was determined by the titration of 50 nM 5′-end-labeled TER RNA and analyzed by EMSA.
RNA preparation.
RNA was synthesized in vitro using T7 RNA polymerase for the runoff transcription of EarI-digested plasmids (ptetTELO) containing the template for TER (48). EarI digestion results in a DNA template encoding four U's at the TER 3′ end. All TER mutants were generated using the DpnI-based site-specific mutagenesis kit QuikChange (Stratagene) with ptetTELO as the template. The construct for the circularly permuted RNA (cpRNA) was generated by two rounds of PCR using ptetTELO as the template and then inserted into pUC19. After transcription at 37°C, samples were ethanol precipitated, and RNA was purified on a denaturing gel containing 6% polyacrylamide, 7 M urea, and 1× Tris-borate-EDTA and eluted and concentrated in 5 mM Na cacodylate, pH 6.5.
RNA was 5′ end labeled with [γ-32P]ATP using T4-polynucleotide kinase (NEB). After labeling, unincorporated nucleotide was removed with Illustra G-25 spin columns (GE) prior to 6% polyacrylamide-7 M urea-1× Tris-borate-EDTA gel purification. Bands were excised, eluted with 10 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, and 300 mM NaCl, and ethanol precipitated. Pellets were resuspended in 5 mM Na cacodylate, pH 6.5, and radioactivity was quantitated using a scintillation counter. When indicated, RNA was annealed in the presence of 10 mM Tris-HCl, pH 7.5, and 10 mM KCl by being heated to 80°C for 1 min and snap cooled on ice.
EMSAs.
EMSA was performed as previously described (33). Briefly, all 5× protein stocks were made in 1× binding buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT) with 1.25 μg/μl bovine serum albumin. All 10-μl binding reaction mixtures contained 2 μl 5× protein stock, 2 μl 5× RNA, 1× binding buffer, 2.5 μg yeast tRNA, and 0.25 μl RNasin Plus RNase inhibitor (Promega). Reactions were incubated at room temperature for 10 to 30 min and then loaded onto Novex 4 to 20% polyacrylamide gradient gels (Invitrogen) and run in 1× Tris-borate-EDTA at 120 V for 1.5 h. Gels then were dried and exposed to storage phosphor screens. Quantitation was performed with ImageQuant (GE).
Telomerase synthesis and assembly.
Telomerase was reconstituted in vitro in rabbit reticulocyte lysates (RRLs) as previously described (8), with minor changes. Briefly, 125-μl translation reactions were performed in TNT quick-coupled transcription/translation mix (Promega) containing plasmid encoding N-terminally T7-tagged TERT (8, 9), [35S]methionine, and in vitro-transcribed RNA that was prebound to purified p65 on ice for 10 min or preincubated with p65 buffer (20 mM Tris-HCl, pH 7.5, 250 mM NaCl, 2 mM DTT). The final concentrations of RNA and p65 in the translation reaction mixtures were 400 nM and 2 μM, respectively, unless otherwise stated. The translations were incubated at 30°C for 2 h and then for 2 h at 4°C with T7-tagged agarose beads (Novagen) at a ratio of 2.5 volumes of translation to 1 volume of 50% bead slurry. The beads then were pelleted and extensively washed with 1× tTB (50 mM Tris-HCl, pH 8.3, 1.25 mM MgCl2, 5 mM DTT) to remove all traces of blood. They then were snap-frozen in liquid nitrogen and stored at −80°C.
Telomerase translation reaction mixtures were resuspended by pipetting, and 5 or 10 μl of resuspended beads was added to the same volume of 2× Nu-PAGE LDS loading dye (Invitrogen) plus β-ME. The samples were heated to 80°C for 5 min and pelleted in a microcentrifuge, and then 5 μl of sample was loaded onto a prepoured 4 to 12% Bis-Tris Nu-PAGE gel (Invitrogen). Gels were run in 1× morpholinepropanesulfonic acid (MES) buffer (Invitrogen) at 180 V for 45 to 60 min, dried, and exposed overnight to a storage phosphor screen. These gels then were imaged and quantitated (ImageQuant TL; GE) for 35S-labeled protein and 32P-labeled RNA. The gels were reimaged overnight with a piece of X-ray film between the screen and the gel to image the 32P-labeled RNA only. The first exposure then was used to calculate the normalization factors for each sample with regard to TERT. Each translation then was diluted with 1× tTB plus 30% glycerol according to the normalization factors to equalize the amounts of TERT.
Telomerase assays.
Telomerase assays were performed as previously described (8). Briefly, reactions were carried out in 20 μl containing 0.33 μM [α-32P]dGTP, 0.1 mM dTTP, 9.67 μM dGTP, 1 mM oligonucleotide Tel3a [(dG2dT4)3], and 10 μl of diluted telomerase beads. Reaction mixtures were incubated at 30°C for 1 h and then stopped with stop buffer containing 3.6 M sodium acetate, pH 5.2, 1 μg glycogen, and 5,000 counts of a recovery control (a 5′-end-labeled and gel-purified 63-mer DNA oligonucleotide of random sequence). Samples then were ethanol precipitated, resuspended in 20 μl 1× formamide loading buffer, and heated to 95°C for 5 min. Eight microliters of each sample was loaded onto 10% polyacrylamide (40:1)-7 M urea-1× Tris-borate-EDTA sequencing gels. These gels were run at 90 W for 1.75 h, dried, and exposed to storage phosphor screens. Screens were scanned using a Typhoon Trio (GE) and analyzed using ImageQuant TL (GE).
The telomerase assays shown in Fig. 2D were performed with a modified protocol. In these assays, stem IV was added at 20 times the indicated final concentrations. In the samples that received p65-stem IV, 20 μM p65 was prebound to stem IV at a 20-fold level of the indicated final concentration.
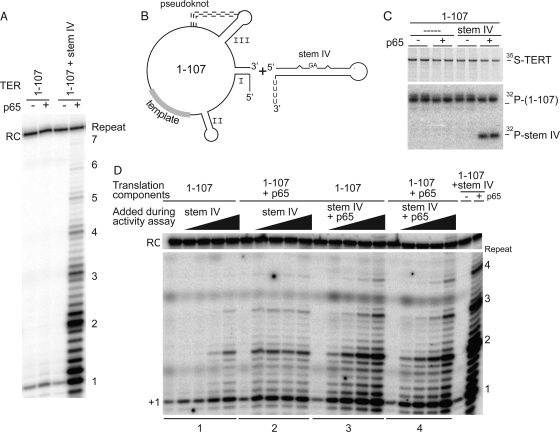
FIG. 2.
p65 restores robust activity to telomerase molecules containing a two-piece TER. (A) Telomerase activity assay of TERT reconstituted with various TER fragments in the presence and absence of saturating p65. (B) Diagram of the TER constructs added to the telomerase translation system. (C) SDS-PAGE analysis of TERT immunoprecipitates containing nucleotides 1 to 107 and stem IV shown in duplicate. (D) Telomerase activity assays on samples containing p65 added during the in vitro translation and adding stem IV or stem IV prebound to p65 in increasing amounts during the telomerase activity assay. Stem IV was added in the following final concentrations: 0, 20 nM, 200 nM, 2 μM, and 10 μM.
Structure-probing assays.
Nuclease digestion was performed in several buffer conditions (see below). While buffer conditions affected the intensities of the nuclease digestion patterns, the reported cleavage and protection patterns generally were consistent among multiple conditions.
RNase T1 assays.
RNase T1 (Ambion) digestions were performed on either trace amounts of 5′-end-labeled TER RNA or with 100 nM cold TER RNA spiked with a trace amount of 5′-end-labeled TER RNA. Digestions typically were performed in 10 mM Tris-HCl, pH 7.0, 100 mM KCl, 10 mM MgCl2, 100 ng/μl yeast tRNA, and either 1× binding buffer, 50 to 500 nM p65 (active concentration), 50 to 250 nM TERT-N (active concentration), or both proteins. Other buffer conditions tested include the buffer used by Bhattacharyya and Blackburn (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 20 mM MgCl2) (6), selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) buffer (100 mM HEPES, pH 8.0, 100 mM NaCl, 6.7 mM MgCl2), and high-salt buffer (25 mM HEPES, pH 7.0, 250 mM NaCl, 5 mM MgCl2, 5% glycerol). Proteins and RNA were incubated at room temperature for 5 to 30 min before the addition of T1 nuclease. T1 nuclease was added at 0.05, 0.025, or 0.013 U/μl and allowed to digest for 10 min at room temperature. Digests then were ethanol precipitated, resuspended, and counted in a scintillation counter. Equal counts for each were loaded onto a 12% polyacrylamide sequencing gel. T1 ladders were prepared by the T1 nuclease digestion of RNA in a denaturing buffer (Ambion).
RNase V1 assays.
RNase V1 (Ambion) digests were performed as for T1, except that 0.008, 0.004, or 0.002 U/μl was added to each reaction mixture and the reaction was allowed to proceed for 10 to 15 min at room temperature. RNase V1 assays were performed in 10 mM Tris-HCl, pH 7.0, 100 mM KCl, 10 mM MgCl2, 100 ng/ml yeast tRNA, or the buffer used by Bhattacharyya and Blackburn (6).
RNase ONE assays.
RNase ONE (Promega) digests were performed as for T1, except the buffer contained 10 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 200 mM sodium acetate, 15 mM magnesium acetate. Another buffer condition tested was 10 mM Tris-HCl, pH 7.5, 5 mM EDTA, 200 mM sodium acetate. Samples were not precipitated but stopped with 1% SDS and formamide gel-loading dye, and equal volumes were loaded directly onto the 12% sequencing gel.
RESULTS
p65 improves the activity of telomerase by facilitating functional interactions between TERT and TER.
To assess the effect of p65 on telomerase RNP formation, we added 5′-end 32P-labeled TER either by itself or prebound to highly purified full-length p65 (Fig. 1A and B) to translations of TERT in rabbit reticulocyte lysates (RRLs). The use of radiolabeled TER facilitated the direct assessment of how much RNA was bound per molecule of TERT synthesized during the translation. The addition of p65 increased the activity of telomerase 3-fold (3.0 ± 0.3 [mean ± standard error of the mean {SEM}]) (Fig. 1C), which could be partially explained by more efficient complex formation (RNA pulldown was increased 1.4-fold ± 0.3-fold). In the absence of TERT, only trace amounts of TER were recovered from the antibody-coated beads used for the immunopurification of the complex (Fig. 1D). Consistent with an effect on the amount of complex, RAP was not affected by p65 (the R1/2 value, which describes the number of repeats at which half of the primers have dissociated from telomerase [20], was 0.40 ± 0.02 repeats for telomerase and 0.44 ± 0.02 repeats for telomerase assembled in the presence of p65).
It has been proposed that p65 bridges stems I and IV of TER (33). Telomerase activity was drastically reduced (Fig. 2A) when the RNA was broken between stems I and IV (nucleotides 1 to 107 and stem IV) (Fig. 2B). However, the addition of p65 to the translation system resulted in the recovery of robust telomerase activity (Fig. 2A). The p65-mediated restoration of activity was, at least in part, due to the retention in the RNP of stem IV (Fig. 2C), which has been shown to stimulate telomerase activity (5, 24, 41). The ability of p65 to bridge these 5′ and 3′ half molecules of TER, resulting in the recovery of stem IV during the pulldown of telomerase, was not seen in previously published experiments performed with similar RNA constructs (33).
To test the effect of stem IV on the stimulation of p65-assisted assembly and activity, we synthesized telomerase in the absence of stem IV. RNA containing nucleotides 1 to 107 was tested for telomerase activity in the presence or absence of p65 and with increasing amounts of stem IV. Without p65, the addition of a 500-fold excess of stem IV was required to obtain a modest recovery of activity and repeat addition processivity (Fig. 2D, lane set 1). However, when p65 was present in the translation, activity and RAP were recovered even at very low concentrations of stem IV. While robust activity and RAP were recovered in the presence of p65 at low concentrations of stem IV, an increase in telomerase activity was apparent at higher concentrations of stem IV when p65 was added during the activity assay (compare lane sets 2 and 3 in Fig. 2D), confirming the importance of the order of the addition of the components to the reaction (29). Furthermore, adding excess p65 during the telomerase assay to samples that contained p65 from the translation did not increase telomerase activity, suggesting that the system can be saturated by p65 (compare lane sets 3 and 4 in Fig. 2D).
Surprisingly, stem I was not required for telomerase activity in the presence of p65 (Fig. 3). A circularly permuted RNA (cpRNA31-25) (25) comprising the 5′ wheel of TER but missing stems I and IV bound to p65 with high affinity (Kd [apparent] of ∼38 nM; Kd stands for dissociation constant) but limited specificity (see Fig. S1 in the supplemental material). In a telomerase assay, RNPs containing this construct had minimal telomerase activity; the recovery of processive telomerase activity occurred only when stem IV and p65 were added in trans, presumably because p65 was necessary for binding stem IV in telomerase molecules containing cpRNA (Fig. 3B and C). These results suggest that p65 can bridge stem IV and the central wheel of TER. Alternatively, or in addition, p65 may interact with TERT to bring stem IV and the central wheel into the same complex.
FIG. 3.
p65 interacts with regions of TER outside stems I and IV. (A) Cartoon of the two-piece TER system consisting of stem IV and cpRNA(31-25), a circularly permuted region of TER that is missing stems I and IV. (B) Duplicate SDS-PAGE analysis of TERT immunoprecipitates following translations in RRLs in the presence or absence of p65 and the indicated RNA components. The top panel shows the [35S]methionine incorporated into TERT, while the bottom panel shows only the 32P-end-labeled TER components recovered by coimmunoprecipitation. (C) Telomerase activity assays using the indicated components. RC, recovery control.
We also found that the 5′ single-stranded RNA preceding stem I and the 3′ oligo(U) tail made modest contributions to p65 binding (see Fig. S2 in the supplemental material). These results are consistent with previously published experiments for Tetrahymena p65 (33) and Euplotes p43 (3).
p65 binding to TER causes conformational rearrangements in the telomerase RNP.
We then set out to define the specific p65-TER interactions and conformational rearrangements required for the functional reconstitution of the telomerase RNP. We used RNase T1 and RNase ONE to probe the single-stranded regions of TER and RNase V1 to probe its double-stranded and stacked regions (Fig. 4). In addition, several sites of nuclease-independent cleavage were observed. To understand the contribution of p65 to complex formation, we tested TER alone, the binary complexes of TER plus p65 and TER plus TERT*, and the ternary complex TER plus p65 plus TERT*. TERT* describes either the RBD of TERT (amino acids 216 to 516) or the N-terminal half of TERT containing the TEN and RBD domains, also known as TERT(1-516) (see Fig. S3 in the supplemental material); these regions account for most of the RNA-binding affinity of full-length TERT (9, 19). Both of these truncated proteins were tested as binary and ternary complexes in the nuclease protection assays and yielded equivalent results.
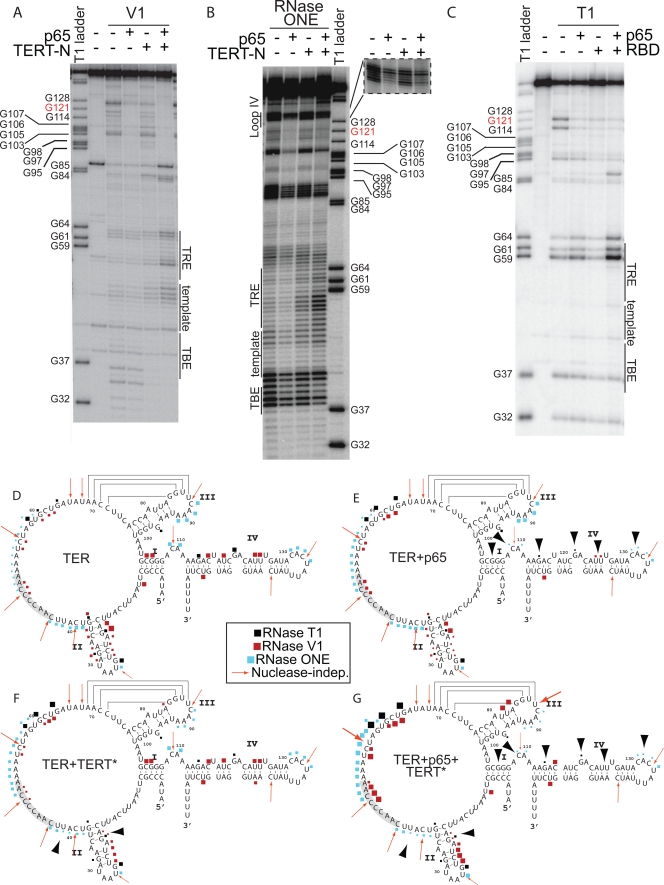
FIG. 4.
Nuclease protection assays of TER in binary and ternary complexes reveal p65 and TERT* footprints and the induction of more single strandedness between stems II and III. (A) RNase V1 data. (B) RNase ONE data. (C) RNase T1 data. Regions of sequencing gels are labeled with corresponding RNA structural elements. T1 ladders are included on each gel for orientation. (D) Nuclease cleavage in the absence of proteins mapped onto the secondary structure of TER, taken from the experiments of panels A, B, and C and additional data not shown. The template is highlighted in gray. (E) Nuclease cleavage data for the complex of TER plus p65. (F) Nuclease cleavage data for the complex of TERT* plus TER. Two versions of TERT (TERT*), TERT-N and TERT RBD, gave equivalent results. (G) Nuclease cleavage data for the ternary complex of p65 plus TER plus TERT*. RNase T1 is in black, RNase ONE is in cyan, and RNase V1 is in red. The sizes of the geometric shapes in each map correlate with the relative strength of the cleavage. The red arrows represent nuclease-independent cleavages; these cleavages are enhanced in the presence of both proteins. The black arrowheads indicate key regions of the protection of the RNA upon protein binding.
The nuclease protection data are consistent with a direct footprint for p65 in stems I and IV of TER (Fig. 4A to E), as summarized by the large arrowheads in Fig. 4E. The single-stranded RNA probes revealed protections of the GA bulge (G121) and the linker between stems I and IV (Fig. 4D and E and data not shown). Also, proximal stem IV (G114) was susceptible to single-strand-specific RNase T1 in the absence of p65 but was protected from cleavage in the presence of p65 (Fig. 4C and E). Comparative sequence analysis indicates that this region is double stranded (37), suggesting that p65 is necessary to facilitate proper base pair formation within TER stems I and IV.
Notably, the 5′ half of hairpin loop IV became protected from the single-stranded RNA probe when p65 was bound (Fig. 4B, inset, and D and E). Cleavage by RNase ONE was reduced at nucleotides C132 to C134; selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) experiments also support this conclusion (data not shown). These protections by p65 might be direct or indirect, since the conformational change induced in stem IV by p65 brings loop IV into proximity with other regions of TER (35, 42).
The protection data reveal that TERT* binds at the TBE and the base of stem II. In the presence of TERT*, nucleotides at the base of stem II, such as A20 and G21, were protected from V1 digestion, while nucleotides closer to loop II, such as U23 to U25, had enhanced V1 reactivity. In addition, nucleotides U38 to U41 were protected from the single-strand-specific RNase ONE upon TERT* binding (Fig. 4B). TERT* binding to the TBE and stem II is consistent with published data (6, 19, 21, 48). An unanticipated finding was that TERT* binding causes structural rearrangements in positions within the central wheel of the RNA that are not directly bound by protein, such as nucleotides in the TRE.
The sequential binding of p65 to TER stems I and IV and TERT* to stem II induces conformational rearrangements that increase the single-stranded and stacked nature of the nucleotides in the region spanning the TBE, the template, and the TRE. For example, when both p65 and TERT* were bound to TER, G59, G61, and G64 had increased susceptibility to RNase T1, and TRE nucleotides A52 to U60 and template alignment nucleotides C49 to A51 exhibited enhanced cleavage by RNase ONE. Under similar conditions, U55, C56, and G61 to U63 had increased digestion by V1 nuclease (Fig. 4A and G), presumably indicative of base stacking (6). The observed conformational changes induced by protein binding may disrupt nonproductive base-pairing interactions within the central wheel of TER, which is consistent with in vivo DMS (dimethylsulfate) probing data (48). In addition, the binding of the proteins may stabilize the RNA into a more rigid structure that limits the sampling of various conformations by the nucleotides. Such rearrangements would facilitate template binding in the active-site cleft of the TERT RT domain.
p65 helps distinguish between assembly and activity mutants of TER.
The reorganization of the RNA between TER stems II and III upon p65 binding led us to ask how p65 affects the activity of telomerase containing different TER mutations. We generated a series of point, deletion, and sequence mutants that scan the secondary structural elements of TER (Fig. 5A). Conveniently, some of these mutants had been characterized previously in the absence of p65 (4, 5, 17, 18, 21). Our studies revealed three classes of TER mutants: (i) those that lose activity in the absence of p65 but are restored to near-wild-type activity levels or higher by p65, (ii) those that are rescued but not fully resuscitated, and (iii) those that are unresponsive to p65.
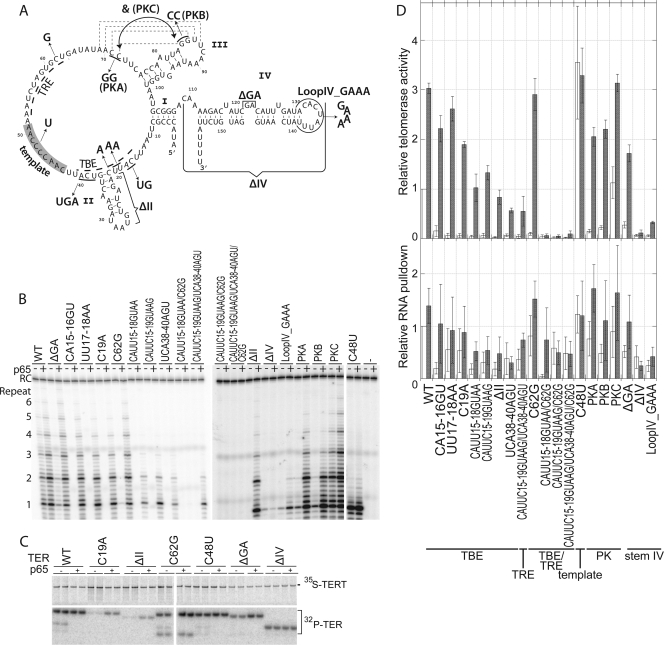
FIG. 5.
p65 rescue distinguishes TER assembly mutants from TER activity mutants. (A) Secondary-structure diagram of TER indicating the various mutations tested. (B) Telomerase assays for each of these mutants with equal amounts of 35S-labeled TERT. The activity for each mutant is shown in the absence and presence of p65. RC, recovery control. (C) SDS-PAGE analysis of a selection of telomerases containing the indicated TER mutations assembled in the absence and presence of p65. The samples are shown in duplicate, and the 35S-labeled TERT and coimmunoprecipitation of 32P-labeled mutant TER are indicated. (D) Quantitation of telomerase activity assays and RNA pulldowns. Mutants are listed at the bottom. The top panel shows relative telomerase activity for each mutant in the absence (white bars) and presence (gray bars) of p65. All activities were normalized to the activity of telomerase containing wild-type TER in the absence of p65. The error bars indicate SEM (n ≥ 3). The bottom panel shows the RNA recovered by the coimmunoprecipitation of TERT normalized to the amount of TERT and setting the amount of wild-type TER recovered in the absence of p65 to 1.0. White bars, without p65; gray bars, with p65. The error bars indicate the SEM from three independent measurements.
The addition of p65 to several extremely debilitated mutants restored telomerase activity to within 50% of wild-type levels. Comparing the telomerase activity of these class 1 mutants in the absence of p65 (Fig. 5D, top panel, white bars) to their activity in the presence of p65 (gray bars) indicates a recovery of approximately 30-fold (i.e., 3000%), but the accuracy of these estimates is limited by the weak signals for telomerase activity in the absence of p65. Included in this class are several mutations in the sites of direct TERT binding, such as CA15-16GU, UU17-18AA, C19A, and the TRE mutant C62G, which display recovery to near-wild-type activity with p65. Resuscitation in activity was much greater than can be explained by the increase in RNA association between TERT and the mutant TER in the presence of p65 (Fig. 5C and D, bottom). The pseudoknot mutants PKA and PKB also had minimal activity in the absence of p65 but near-wild-type levels of activity in the presence of p65. The rescue of the activity of these mutants cannot be correlated with an increase in RNA association (Fig. 5D, bottom). The compensatory mutations in the pseudoknot (PKC) were included as a control and demonstrate wild-type activity and RNA association (Fig. 5D).
In our system, p65 rescued the TER mutant containing a deletion of the GA bulge in stem IV to ∼50% of the level of wild-type activity (1.7-fold ± 0.2-fold for ΔGA and 3.0-fold ± 0.3-fold for wild-type TER) by p65. The presence of p65 resulted in a modest increase (2-fold) in RNA association with TERT and an ∼5-fold increase in telomerase activity (Fig. 5) for this mutant. In contrast, other studies have indicated that the deletion of the GA bulge prevents p65 from affecting telomerase RNP assembly (1, 33, 42).
In class 2, p65 increased the activity of several TER assembly mutants and stem IV mutants to a more modest extent. For example, CAUU15-18GUAA, CAUUC15-19GUAAG, UCA38-40AGU, CAUUC15-19GUAAG/UCA38-40AGU, and the stem II deletion mutant (ΔII) were recovered to ∼15 to 40% of wild-type levels in the presence of p65 (Fig. 5D). While the presence of p65 resulted in an increase in RNA pulldown in some of these cases, the levels of RNA recovery were too little to explain the activity recovery (Fig. 5D). The mutation of loop IV to a GAAA tetraloop resulted in the incomplete recovery of activity in the presence of p65 despite comparable levels of RNA association with TERT; although stem IV of this construct is a binding substrate for p65, these results confirm that the sequence of loop IV is important for telomerase function (36).
In class 3, several TER mutants were not suppressed by the presence of p65. Although the mutants combining altered TRE and the TBE regions (such as CAUU15-18GUAA/C62G) had an increase in RNA pulldown in the presence of p65 (Fig. 5D), activity was not recovered. p65 also was unable to affect TER that lacked stem IV (ΔIV); since stem IV is a major site of p65 interaction (3, 29, 33), the presence of p65 in the translation and assembly reactions was not able to rescue RNA recovery or activity. Only one round of replication occurred with ΔIV (Fig. 5B), which is consistent with the role loop IV has been shown to play in RAP (17). Finally, another subgroup of class 3 included the template mutation C48U that was unaffected by p65. This was expected, as the template is not involved in assembly. This mutant gave a single round of addition because the extended primer cannot base pair properly with the mutated template.
p65 rescues TERT assembly mutants but not activity mutants.
We then asked if the assembly and activity of telomerases containing TERT mutants could be rescued by p65. The panel of mutants that we tested spanned the CP and T motifs of the RBD, which are involved in RNP assembly, and motifs 1, A, B, and E of the RT domain (Fig. 6) (9). We predicted that the RT motif mutants previously characterized as activity mutants would not be rescued by p65, while some of the RBD mutants would be aided by p65 because they are defective in TER binding and, therefore, RNP assembly.
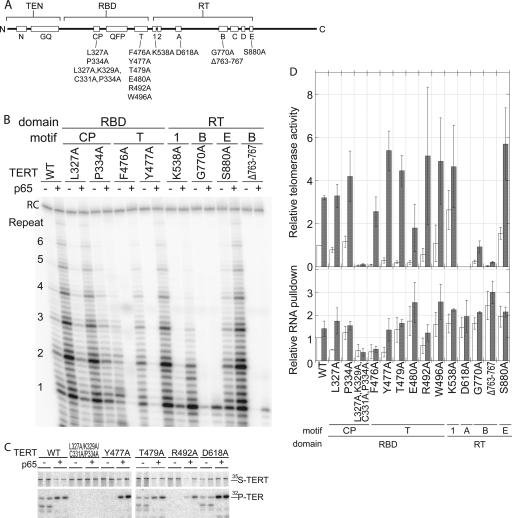
FIG. 6.
p65 rescues TERT assembly mutants but not TERT activity mutants. (A) Linear diagram outlining the domains and motifs of TERT. The mutations studied are indicated below the motif in which they occur. (B) Telomerase assays for several of the mutants tested with wild-type TER in the absence and presence of p65. (C) SDS-PAGE analysis of a selection of telomerases containing the indicated TERT mutations assembled in the absence and presence of p65. The samples are shown in duplicate, and the 35S-labeled TERT and coimmunoprecipitation of 32P-labeled TER are indicated. (D) Quantitation of the telomerase activity and RNA pulldown of each mutant in the absence (white bars) and presence (gray bars) of p65. The top panel shows the relative telomerase activity for each mutant; error bars indicate SEM (n ≥ 3). The bottom panel shows relative RNA pulldown in the absence and presence of p65 for each mutant; error bars indicate SEM (n ≥ 3). Samples in both panels were normalized to wild-type TERT without p65.
The point mutants in the T motif of the RBD showed notable p65 stimulation of telomerase activity. While the recovery can be explained partially by an increase in RNA binding (typically a <2-fold increase), this does not completely account for the magnitude of activity stimulation observed for these mutants, which is typically greater than 10-fold (Fig. 6D). As before, the magnitude of recovery for some mutant pairs is difficult to measure accurately because of the extremely weak activity in the absence of p65 (Fig. 6B). CP point mutants (L327A and P334A) were increased by p65 by a factor similar to that for wild-type TERT, correlating with similar moderate increases in RNA binding.
Several point mutants of amino acids in the RT domain showed near-wild-type telomerase activity and RNA binding (K538A and S880A). The catalytic carboxylate mutant (D618A) and the 5-amino-acid deletion in the B motif (Δ763-767) were not activated by p65, which is consistent with severe disruptions of the enzyme active site (9), despite RNA binding that was comparable to that of wild-type TERT (Fig. 6C and D). Taken together, these data demonstrate that p65 can compensate for misfolded TERT or TERT that is compromised in its RNA-binding ability, but it cannot stimulate TERT mutants that are deficient in the ability to perform catalysis.
DISCUSSION
Previous work established p65 as a telomerase-specific protein that enhances telomerase RNP assembly by binding to the TER RNA and inducing a structural change in the RNA, thereby enhancing the binding of TERT; furthermore, the site of p65 action was localized to the proximal half of stem IV, a conserved GA bulge, and stem I of TER (3, 29, 33, 42, 47). In agreement, the most-straightforward interpretation of our nuclease protection data is that stem I, proximal stem IV, and the GA bulge in stem IV constitute the binding site of p65 (Fig. 4E). Moreover, our binding data show p65 binding beyond stems I and IV, perhaps most convincingly demonstrated by Fig. 3C. In addition, we find that p65 reorganizes the RNA structure outside the regions of its binding, which helps explain why p65 enhances telomerase activity above and beyond its increasing the amount of complex formed. Finally, we show that the normally modest effect of p65 on telomerase activity is greatly amplified in the case of TER and TERT assembly mutants to the extent that many severe mutants are fully resuscitated. Thus, p65 is a tool for distinguishing between assembly and activity mutants of Tetrahymena telomerase.
p65-assembled telomerase RNPs are of higher quality.
The addition of p65 to in vitro translation and assembly systems of T. thermophila telomerase results in telomerase RNPs with higher telomerase activity. This can be explained partially by the enrichment of TER-bound TERT molecules that occurs with p65. However, the 1.4-fold ± 0.3-fold increase in RNA association with TERT does not completely account for the 3.0-fold ± 0.3-fold (means ± SEM) increase in telomerase activity. As described below, some mutant telomerases show much more dramatic effects of p65 on activity above and beyond assembly; that is, they have greater activity per RNP molecule. Thus, p65 contributes not only to the quantity of RNP formed but also to the quality. This conclusion is consistent with published FRET data indicating that the presence of p65 increases the homogeneity of telomerase RNPs (42), thereby improving the quality of the complexes overall.
Nuclease probing experiments identified RNA structural changes that may explain the improvement of activity of telomerase assembled in the presence of p65. Nucleotides 3′ of the template become more single stranded and yet more organized in nature, as evidenced by RNase ONE and RNase V1 cleavage, respectively. These changes are expected to facilitate template accessibility and primer binding. Perhaps these nucleotides participate in nonproductive base-pairing interactions that are disrupted when p65 binds to their illegitimate pairing partners.
In addition, several loop IV nucleotides are protected from nuclease digestion by p65 and remain protected in the p65-TERT-TER ternary complex. In combination with the telomerase activity assay for the loop IV_GAAA TER mutant (Fig. 5D), these results suggest that loop IV is poised for proper holoenzyme formation upon the complexation of p65 and TER, with the sequence of loop IV contributing to the assembly of TERT, as suggested by Robart et al. (36). It also agrees with the single-molecule studies that suggest loop IV is brought into the core of the telomerase RNP during assembly (42).
p65 is thought to serve as a bridge between stems I and IV (33); in agreement, we find greater activity of p65-assembled telomerase with nucleotides 1 to 107 (which contains stem I) plus stem IV than with cpRNA (no stem I) plus stem IV (Fig. 2A and 3C). In addition, the cpRNA plus stem IV TER system revealed that p65 may have direct interactions with other regions of TER. The binding experiments of Fig. S1 in the supplemental material indicate a direct interaction between p65 and the central wheel of TER that is relatively high affinity and low specificity. The presence of TERT may enhance these interactions. Such interactions may contribute to the quality of the RNP; if p65 promotes proper secondary and tertiary structure formation for all of TER, TERT would bind TER molecules that were folded in their active conformation more often.
p65 is a tool for distinguishing between assembly and activity mutants.
Since p65 participates in telomerase RNP assembly, we predicted that p65 would be able to rescue mutations in regions of TER that have been shown to be responsible for the TER-TERT interaction. Indeed, altering nucleotides in the TBE, stem II, and/or the TRE abrogated assembly and therefore activity in the absence of p65, which is consistent with previously published results (4, 21), but many of these mutants were rescued by p65.
The pseudoknot mutants were rescued by p65, but this increase in telomerase activity was not due to an increase in RNA association (Fig. 7). In this case, it is likely that the quality rather than the quantity of the complex is enhanced, and in fact for yeast and human telomerases it has been proposed that the pseudoknot triple-helix region of the RNA is involved in the alignment of the primer-template relative to the active site (34). Previously, Autexier and Greider reported that pseudoknot mutations were not deleterious to telomerase activity in vitro (5). Their starting preparations of telomerase were assembled in vivo, so they can be assumed to include the p65 subunit; thus, it is not surprising that their activity data agree with our data for PKA and PKB in the presence of p65.
FIG. 7.
Summary of the effect of p65 on mutants of TER and TERT.
Finally, the mutation in the template region, a segment of TER that is not involved in assembly but in the activity of the holoenzyme, was not rescued by p65. Thus, the RNA mutants studied can be sorted into three classes: those aided by p65 because they are defective in RNP assembly, mutants helped by p65 not in assembly per se but because of the quality of the assembly, and mutants unaffected by p65 because they have a major defect in RNP activity (Fig. 7).
All of the TERT mutants studied followed the trends in RNA association and telomerase activity reported by Bryan et al. (9). In general, the mutation of amino acids responsible for binding TER (in the RBD) decreased telomerase activity because these mutations destroyed the TERT-TER association, while the mutation of several conserved amino acids in the active site of the RT domain decreased or eliminated telomerase activity without decreasing RNA binding. In the presence of p65, mutations in the RBD, especially those in the T motif, were suppressed; telomerase activity was significantly stimulated (4- to 30-fold), and this was partially due to stimulation in the TERT-TER association (less than or equal to 3-fold) (Fig. 6D). Thus, our data suggest that the T motif has a function beyond simple RNA binding, a conclusion that is consistent with the recent protein crystal structure showing that the T motif may structurally support part of the RT domain; the β-hairpin of the T motif extends toward the active site of the RT domain and may form functionally relevant interactions with the fingers and the presumed thumb subdomains, which typically stabilize the incoming nucleotide and the primer-template duplex, respectively (13, 38). Additionally, the T motif may form a hinge between the RBD and the RT domain, possibly playing a role in coordinating the movements of these subdomains during polymerization and translocation. The TERT mutants in several conserved amino acids of the RT domain were not rescued by p65, as expected. Thus, p65 rescued mutants that were deficient in assembly (7, 19, 30) but could not rescue mutants deficient in polymerization activity (Fig. 7).
Model for the mechanism of p65-induced stimulation of telomerase activity.
The binding of p65 to stems I and IV [and the 3′-oligo(U) tail] sequesters these nucleotides from the rest of the TER molecule. This may help the template and surrounding regions of TER assume the correct conformations required for telomerase activity. Indeed, the structural effects of p65 seen far from its primary binding site in stem IV are consistent with the idea that improving the single-stranded and stacked nature of the template and the surrounding regions of TER are important for increasing the efficiency of complex formation. In short, p65 may limit the ensemble of structural possibilities to those that are catalytically active.
Beyond interacting with TER, several observations suggest that p65 also participates in interactions with TERT. The TRE and template become more accessible only in the presence of both proteins, while stem II assumes its predicted secondary structure only in the presence of both proteins. Even more compelling, the TER stem II deletion mutant data (ΔII) suggest that p65 directly contacts TERT. In this example, the main site in TER of TERT binding (18) is missing, yet p65 stimulates a robust recovery of telomerase activity; here, the simplest explanation is a direct contact between the two proteins. Previous studies have tested for the direct interaction between p65 and TERT, and none was found in the absence of TER (33). However, it is possible that the binding of p65 to TER induces a conformational change in p65 that allows it to interact with TERT.
Several of the TERT mutants also hint at a direct interaction between the two proteins. For example, point mutations in the conserved T motif demonstrate a substantial rescue in the presence of p65, yet point mutations in another RNA-binding element, the CP motif (10, 26), do not. These data support the hypothesis that p65 directly interacts with the T motif of the RBD. Alternatively, by helping to organize the RNA structures necessary for productive RBD binding, p65 may indirectly suppress T motif mutants.
Taken together, these data suggest that p65 acts as a molecular buttress within the telomerase RNP. In the context of a wild-type system, p65 enhances the number of TERT molecules that are stably associated with TER, thereby increasing the activity of the bulk telomerase population. When TER is broken into two pieces, p65 is required for the association of stem IV with TERT, and activity is recovered only in the presence of p65. In the context of many TERT and TER assembly mutants, p65 is not simply a useful accessory factor but is essentially required for stable RNA association. Thus, we conclude not only that p65 is a telomerase-specific assembly protein but also that its presence in the telomerase RNP maintains active TER-TERT conformations.
Supplementary Material
Acknowledgments
We are grateful to the members of the Cech laboratory for helpful discussions. We also thank Kathleen Collins (University of California, Berkeley) for the plasmid encoding p65 and for helpful discussions in the initial phases of the project. We also thank Catherine O'Connor and Kathleen Collins for sharing unpublished studies in which they designed RBD 216-516 and determined that it gave robust soluble expression and retained full interaction with TER and TER-p65.
A.J.B. is a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research. This investigation has been aided by a grant from the Jane Coffin Childs Memorial Fund for Medical Research.
Footnotes
Published ahead of print on 16 August 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aigner, S., and T. R. Cech. 2004. The Euplotes telomerase subunit p43 stimulates enzymatic activity and processivity in vitro. RNA 10:1108-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aigner, S., J. Lingner, K. J. Goodrich, C. A. Grosshans, A. Shevchenko, M. Mann, and T. R. Cech. 2000. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 19:6230-6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aigner, S., J. Postberg, H. J. Lipps, and T. R. Cech. 2003. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry 42:5736-5747. [DOI] [PubMed] [Google Scholar]
- 4.Autexier, C., and C. W. Greider. 1995. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 9:2227-2239. [DOI] [PubMed] [Google Scholar]
- 5.Autexier, C., and C. W. Greider. 1998. Mutational analysis of the Tetrahymena telomerase RNA: identification of residues affecting telomerase activity in vitro. Nucleic Acids Res. 26:787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharyya, A., and E. H. Blackburn. 1994. Architecture of telomerase RNA. EMBO J. 13:5721-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosoy, D., Y. Peng, I. S. Mian, and N. F. Lue. 2003. Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem. 278:3882-3890. [DOI] [PubMed] [Google Scholar]
- 8.Bryan, T. M., K. J. Goodrich, and T. R. Cech. 2000. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem. 275:24199-24207. [DOI] [PubMed] [Google Scholar]
- 9.Bryan, T. M., K. J. Goodrich, and T. R. Cech. 2000. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell 6:493-499. [DOI] [PubMed] [Google Scholar]
- 10.Bryan, T. M., J. M. Sperger, K. B. Chapman, and T. R. Cech. 1998. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl. Acad. Sci. U. S. A. 95:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan, E. D., and K. Collins. 2010. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol. Cell. Biol. 30:2775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilley, D., and E. H. Blackburn. 1999. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc. Natl. Acad. Sci. U. S. A. 96:6621-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillis, A. J., A. P. Schuller, and E. Skordalakes. 2008. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455:633-637. [DOI] [PubMed] [Google Scholar]
- 14.Greider, C. W. 1991. Telomerase is processive. Mol. Cell. Biol. 11:4572-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 16.Kiss, T., E. Fayet, B. E. Jady, P. Richard, and M. Weber. 2006. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 71:407-417. [DOI] [PubMed] [Google Scholar]
- 17.Lai, C. K., M. C. Miller, and K. Collins. 2003. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell 11:1673-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, C. K., M. C. Miller, and K. Collins. 2002. Template boundary definition in Tetrahymena telomerase. Genes Dev. 16:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, C. K., J. R. Mitchell, and K. Collins. 2001. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 21:990-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latrick, C. M., and T. R. Cech. 2010. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 29:924-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Licht, J. D., and K. Collins. 1999. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes Dev. 13:1116-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lingner, J., and T. R. Cech. 1996. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc. Natl. Acad. Sci. U. S. A. 93:10712-10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lue, N. F. 2004. Adding to the ends: what makes telomerase processive and how important is it? Bioessays 26:955-962. [DOI] [PubMed] [Google Scholar]
- 24.Mason, D. X., E. Goneska, and C. W. Greider. 2003. Stem-loop IV of Tetrahymena telomerase RNA stimulates processivity in trans. Mol. Cell. Biol. 23:5606-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, M. C., and K. Collins. 2002. Telomerase recognizes its template by using an adjacent RNA motif. Proc. Natl. Acad. Sci. U. S. A. 99:6585-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, M. C., J. K. Liu, and K. Collins. 2000. Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J. 19:4412-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min, B., and K. Collins. 2010. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J. Biol. Chem. 285:16434-16443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell, J. R., E. Wood, and K. Collins. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402:551-555. [DOI] [PubMed] [Google Scholar]
- 29.O'Connor, C. M., and K. Collins. 2006. A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol. Cell. Biol. 26:2029-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Connor, C. M., C. K. Lai, and K. Collins. 2005. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 280:17533-17539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osterhage, J. L., and K. L. Friedman. 2009. Chromosome end maintenance by telomerase. J. Biol. Chem. 284:16061-16065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pogaciæ, V., F. Dragon, and W. Filipowicz. 2000. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 20:9028-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prathapam, R., K. L. Witkin, C. M. O'Connor, and K. Collins. 2005. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 12:252-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiao, F., and T. R. Cech. 2008. Triple-helix structure in telomerase RNA contributes to catalysis. Nat. Struct. Mol. Biol. 15:634-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards, R. J., H. Wu, L. Trantirek, C. M. O'Connor, K. Collins, and J. Feigon. 2006. Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. RNA 12:1475-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robart, A. R., C. M. O'Connor, and K. Collins. 2010. Ciliate telomerase RNA loop IV nucleotides promote hierarchical RNP assembly and holoenzyme stability. RNA 16:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romero, D. P., and E. H. Blackburn. 1991. A conserved secondary structure for telomerase RNA. Cell 67:343-353. [DOI] [PubMed] [Google Scholar]
- 38.Rouda, S., and E. Skordalakes. 2007. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure 15:1403-1412. [DOI] [PubMed] [Google Scholar]
- 39.Shefer, K., Y. Brown, V. Gorkovoy, T. Nussbaum, N. B. Ulyanov, and Y. Tzfati. 2007. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol. Cell. Biol. 27:2130-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 41.Sperger, J. M., and T. R. Cech. 2001. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry 40:7005-7016. [DOI] [PubMed] [Google Scholar]
- 42.Stone, M. D., M. Mihalusova, M. O'Connor, C. R. Prathapam, K. Collins, and X. Zhuang. 2007. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 446:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ten Dam, E., A. van Belkum, and K. Pleij. 1991. A conserved pseudoknot in telomerase RNA. Nucleic Acids Res. 19:6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theimer, C. A., C. A. Blois, and J. Feigon. 2005. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell 17:671-682. [DOI] [PubMed] [Google Scholar]
- 45.Tzfati, Y., T. B. Fulton, J. Roy, and E. H. Blackburn. 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288:863-867. [DOI] [PubMed] [Google Scholar]
- 46.Wang, C., and U. T. Meier. 2004. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 23:1857-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witkin, K. L., and K. Collins. 2004. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 18:1107-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaug, A. J., and T. R. Cech. 1995. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA 1:363-374. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.