Abstract
The Mohawk homeobox (Mkx) gene encodes a new atypical homeodomain-containing protein with transcriptional repressor activity. Mkx mRNA exhibited dynamic expression patterns during development of the palate, somite, kidney, and testis, suggesting that it may be an important regulator of multiple developmental processes. To investigate the roles of Mkx in organogenesis, we generated mice carrying a null mutation in this gene. Mkx−/− mice survive postnatally and exhibit a unique wavy-tail phenotype. Close examination revealed that the mutant mice had smaller tendons than wild-type littermates and that the rapid postnatal growth of collagen fibrils in tendons was disrupted in Mkx−/− mice. Defects in tendon development were detected in the mutant mouse embryos as early as embryonic day 16.5 (E16.5). Although collagen fibril assembly initially appeared normal, the tendons of Mkx−/− embryos expressed significantly reduced amounts of collagen I, fibromodulin, and tenomodulin in comparison with control littermates. We found that Mkx mRNA was strongly expressed in differentiating tendon cells during embryogenesis and in the tendon sheath cells in postnatal stages. In addition to defects in tendon collagen fibrillogenesis, Mkx−/− mutant mice exhibited abnormal tendon sheaths. These results identify Mkx as an important regulator of tendon development.
The Mohawk homeobox (Mkx) gene, also known as Iroquois-related homeobox like 1 (Irxl1), was independently isolated in mice as a candidate gene for the spontaneous mutation Twirler, which causes inner ear defects in heterozygous mutants and cleft lip and/or cleft palate in homozygous mutants (21, 26, 27), and as a possible regulator of somite and testis differentiation (2). The Mkx gene encodes a protein of 353 amino acid residues with a putative homeodomain motif highly similar to those of the invertebrate Iroquois and vertebrate Irx subfamily members of the three-amino-acid loop extension (TALE) homeodomain family (2, 12, 26, 40). Orthologs of Mkx have been found throughout metazoans (2, 26). Biochemical studies have shown that, similar to the Irx proteins, Mkx has transcriptional repressor activity (3, 8, 30).
Mkx mRNA exhibits dynamic expression patterns during mouse embryogenesis (2, 26, 40). Mkx mRNA was strongly expressed in the frontonasal process and palatal mesenchyme during primary and secondary palate development (26). Strong Mkx mRNA expression was also detected in the dermomyotome of the differentiating somites (2, 26). In the developing limb bud, Mkx mRNA was detected in tendon progenitor cells. In addition, Mkx mRNA expression was detected in the otic vesicle, retinal ganglion cells, the ureteric bud in the developing metanephric kidney, and the testis cords of the male gonad (2, 26, 40). The dynamic expression patterns of Mkx mRNA suggested that Mkx might regulate multiple developmental processes.
Tendons connect muscles to bone and transmit the force generated during muscle contraction to the skeleton (7). Mature tendons are composed of tightly packed bundles of collagen fibers that connect to the muscle at the myotendinous junction and to the bone at the skeletal junction, the entheses (6, 7). The collagen fibers are composed of bundles of collagen fibrils whose organization represents a tightly regulated balance between collagen fibrillogenesis and cellular processes that package the collagen fibrils into higher-order structures. The tensile strength, the major functional feature of a tendon, is directly related to the diameter of the collagen fibrils (4, 43). Collagen fibril growth occurs in two very distinctive stages. In embryos, the collagen fibrils are small and homogeneous. The pace and mode of fibril growth change dramatically after birth, resulting in larger and highly heterogeneous fibrils. Tendon maturation in the postnatal stages is not only dependent on the assembly of monomeric units of collagen, but also mediated by fusion of collagen fibrils that enables the rapid pace of fibril growth (43). The mature tendon is further enclosed in ensheathing cell layers, the epitendon and paratendon, and in some positions in an outer tendon sheath that enables friction-free movement of the tendon (5, 6, 9, 22).
Little is known about the molecular regulation of tendon development, and only one transcriptional regulator of this process has been identified (11, 34, 41). The bHLH transcription factor Scleraxis (Scx) is a prototypic marker for tendon cells, with highly specific expression in tendon progenitor cells and persisting through tendon differentiation (11, 34). Scx is essential for early stages of tendon differentiation in force-transmitting tendons (25, 32, 36). In this report, we show, through generation and analysis of gene-targeted mice, that the Mkx transcription factor is required for tendon development during embryogenesis, as well as for tendon maturation after birth.
MATERIALS AND METHODS
Targeted disruption of the mouse Mkx gene.
A bacterial artificial chromosome (BAC) clone containing the Mkx genomic region was isolated from the RPCI-22 129/SvEvTac mouse BAC library (BACPAC Resources, Children's Hospital of Oakland, Oakland, CA). A 10.8-kb BamHI fragment containing exons 2 and 3 was subcloned for construction of the gene-targeting vector. The final targeting vector contained a 2.3-kb 5′ homology arm, a loxP-flanked PstI/HindIII fragment containing exon 3, a FLP recombination target (FRT)-flanked PGK-neo expression cassette, a 3.6-kb 3′ homology arm, and a PGK-DTA expression cassette (Fig. 1). The targeting vector was linearized and electroporated into CJ7 mouse embryonic stem (ES) cells as previously described (39). G418-resistant ES colonies were screened by Southern hybridization, with 6 out of 240 clones screened carrying the correctly targeted Mkx allele. Two independent targeted ES cell clones were injected into blastocysts from C57BL/6J mice to generate chimeric mice. The chimeric mice were crossed to 129/SvJ female mice to generate Mkxfneo/+ heterozygous mice, which were then crossed to the FLPeR mice (Jackson Laboratory, Bar Harbor, ME) to remove the PGK-neo cassette. The resultant Mkxflox/+ heterozygous mice were crossed to the EIIa-Cre transgenic mice (Jackson Laboratory, Bar Harbor, ME) to generate mice heterozygous for a deletion of exon 3 of the Mkx gene (Mkx+/−). The Mkx+/− mice were then intercrossed to generate Mkx−/− homozygous mice. The mice were genotyped by PCR using the following three primers: 5′-AGGAGAGGTCGAAGAAAGTGTC-3′, 5′-AACAGTGAGCAGAGCAGGATAC-3′, and 5′-GGGAGCTATTTCGATCCTAG-3′. The fragments amplified from wild-type and mutant alleles were 122 bp and 247 bp, respectively.
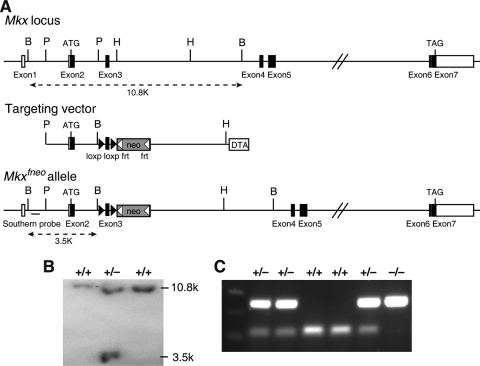
FIG. 1.
Targeted disruption of the mouse Mkx gene. (A) Targeting scheme. The top line shows the genomic organization of the Mkx gene. Exons are indicated by boxes, with the open reading frame shown in black. The positions corresponding to the translational initiation (ATG) and termination (TAG) codons are also marked. The restriction sites are as follows: B, BamHI; H, HindIII; P, PstI. The middle line represents the structure of the targeting vector, which was constructed with the 2.36-kb PstI fragment containing exon 2 as the 5′ arm and the 3.60-kb HindIII fragment from intron 3 as the 3′ arm. Between the 5′ and 3′ arms are the exon 3 fragment flanked by two loxP sequences and a FRT-flanked neo expression cassette. For selection against random integration, a DTA cassette was inserted downstream of the 3′ arm in the targeting vector. Shown at the bottom is the predicted structure of the targeted Mkxfneo locus. (B) Southern hybridization analyses of BamHI-digested genomic DNA samples from targeted ES clones using the probe 5′ to the 5′ arm. Wild-type and targeted alleles were detected as 10.8- and 3.5-kb fragments, respectively. (C) PCR genotyping of tail DNA samples from a litter of F2 progeny of an Mkx+/− heterozygous intercross. The sizes of the wild-type and mutant products were 122 and 247 bp, respectively.
In situ hybridization and histological analyses.
Embryos at different stages were dissected and fixed in 4% paraformaldehyde (PFA) in PBS overnight at 4°C. Whole-mount in situ hybridization was performed as described previously (24). For section in situ hybridization, PFA-fixed embryos were dehydrated through graded alcohols, embedded in paraffin, and sectioned at 7-μm thickness, followed by hybridization with digoxigenin-labeled cRNA probes as described previously (44).
For histology, embryos were collected at predetermined stages, fixed in either Bouin's fixative or 4% PFA overnight, decalcified in EDTA, dehydrated through graded ethanol, embedded in paraffin wax, and sectioned at 7-μm thickness, followed by staining with hematoxylin and eosin.
Skeletal analysis.
Skeletal preparations of embryonic day 18.5 (E18.5) embryos and of postnatal day 14 (P14) mice were performed as described previously (29, 31).
Real-time reverse transcription (RT)-PCR.
For quantitative analysis of gene expression in tendons, limb and tail tendons were dissected from individual neonatal mice in cold diethyl pyrocarbonate (DEPC)-treated phosphate-buffered saline (PBS), collected in RNA Later (Ambion 7020), and stored for up to 1 week at 4°C. Following identification of the genotypes of the mice, the tendons were pooled by tissue type and genotype. RNA extraction was performed with the Qiagen RNeasy Mini Kit (Qiagen no. 74104) according to appendix C of the RNeasy manual. The tissue was disrupted with a mortar and pestle and homogenized using a QIAshredder (Qiagen no. 79654). Twenty microliters of collagenase type II (1% stock; Invitrogen no. 17101-015) was added to the proteinase K step of the protocol. DNase digestion was carried out using an on-column DNase set (Qiagen no. 79254). For analysis of Scx mRNA expression in the developing limb and tail tissues, total RNA was extracted from dissected tissues using Trizol reagents (Invitrogen).
Following total RNA extraction and quantification, first-strand cDNA was synthesized using a SuperScript First-Strand Synthesis System (Invitrogen). Quantitative PCR amplifications were performed in an iCycler real-time PCR machine (Bio-Rad) using the SYBR GreenER qPCR Supermix (Invitrogen). The reaction was run in a PCR program of 50°C for 2 min and 95°C for 8.5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s, with a melt curve generation cycle at the end. The PCR primers used for Col1a1, Col1a2, Col5a1, Col5a2, Dcn, Fmod, and Lum were as reported previously (28). The Scx gene-specific PCR primers were 5′-GCAAGCTCTCCAAGATTGAG-3′ and 5′-GCAGAAGGTGCAGATCTGTT-3′. The Hprt gene-specific primers were 5′-TGCTGGTGAAAAGGACCTCTCG-3′ and 5′-CTGGCAACATCAACAGGACTCC-3′. The Tnmd gene-specific primers were 5′-ACACTTCTGGCCCGAGGTAT-3′ and 5′-GACTTCCAATGTTTCATCAGTGC-3′. For each gene, the PCR was carried out in triplicate, and the relative levels of mRNAs were normalized to that of Hprt using the standard-curve method. Student's t test was used to analyze the significance of differences, and a P value of less than 0.05 was considered statistically significant.
Tendon measurements.
Tendon measurements were conducted on 10-μm cryosections of the forelimbs from five pairs of wild-type and Mkx−/− embryos at E16.5. Serial transverse sections were processed for in situ hybridization with a cRNA probe for the tubulin polymerization-promoting protein family member 3 (Tppp3) gene to highlight tendon circumference, followed by DAPI (4′,6-diamidino-2-phenylindole) staining of the cell nuclei. Tendon measurements were performed from sections at the digit, metacarpal, or wrist level, at which the particular tendon could be clearly identified and matched in the wild-type and control samples with great precision based on the patterns of tendon distribution, as recently described (42). Overlapping high-magnification images of the bright-field Tppp3 signal and fluorescent DAPI signal were captured using a Nikon E800 compound microscope. Area measurements were performed using Photoshop analytical tools. For cell counts, the DAPI and Tppp3 composite images of each tendon were aligned, and only cells within the tendon were counted. Student's t test was used to analyze the significance of differences.
RESULTS
Generation of Mkx−/− mice.
Since Mkx mRNA is expressed in multiple tissues during embryogenesis, we designed the gene-targeting construct to generate mice carrying a conditional allele containing two direct repeats of loxP sequences flanking the third exon of the Mkx gene (see Materials and Methods) (Fig. 1A). Deletion of exon 3 results in a frameshift immediately following the exon 2 sequence and premature stop codons in all possible alternatively spliced Mkx transcripts, which is expected to trigger nonsense-mediated decay of the transcripts and to completely disrupt Mkx gene function. Gene-targeted ES colonies were screened and identified by Southern blot hybridization (Fig. 1B). Following generation of chimeric mice using the correctly targeted ES clones, founder Mkxfneo mice were bred to FLPeR mice, which ubiquitously express the FLP recombinase (20), to remove the neo expression cassette in the targeted locus to generate mice carrying the Mkxflox conditional allele.
To investigate the developmental roles of Mkx, we crossed the Mkxflox mice to the EIIa-Cre transgenic mice (23) to generate Mkx+/− mice, which were then intercrossed to generate Mkx−/− mice (Fig. 1C). Mkx−/− homozygous mutant mice were born at the expected Mendelian ratio and exhibited normal fertility and life span. Despite the dynamic expression patterns of Mkx mRNA during craniofacial, kidney, and testis development (2, 26), Mkx−/− mutant mice did not have obviously detectable defects in these tissues (data not shown). Interestingly, Mkx−/− mice exhibited a wavy-tail phenotype by 2 weeks of age, and the tail abnormality was more obvious during mouse movement (Fig. 2).
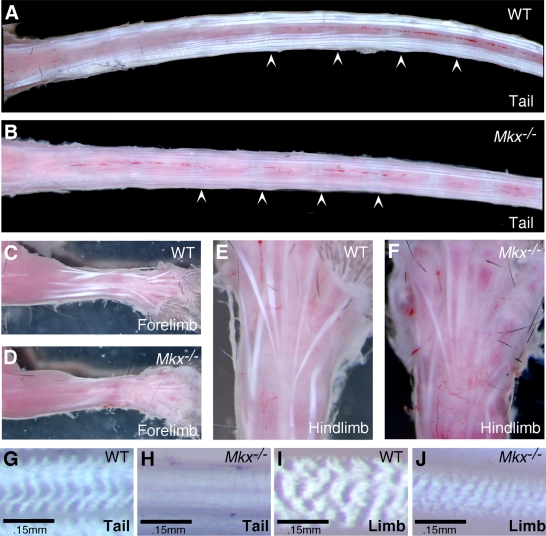
FIG. 2.
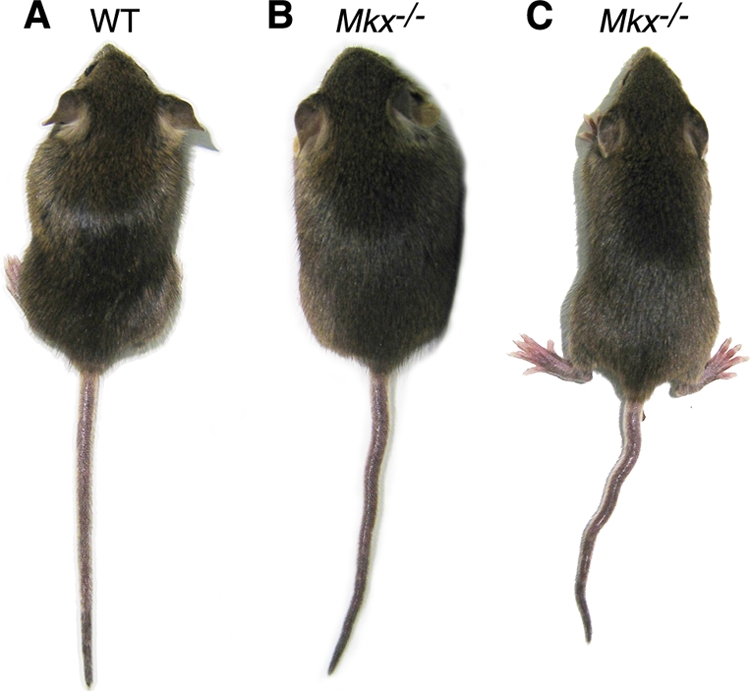
Mkx−/− homozygous mice exhibited a wavy-tail phenotype. In comparison with wild-type mice (A), Mkx−/− mutant mice exhibited wavy tails (B), which were more obvious when the mice were running (C).
Mkx−/− mutant mice have tendon defects.
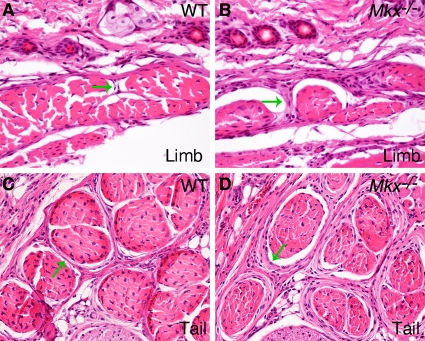
Mkx mRNA is strongly expressed in the dermomyotome of the differentiating somites and in the tendon progenitor cells during embryogenesis (2, 26). We therefore examined the musculoskeletal tissues for an underlying cause of the abnormal tail phenotype in Mkx−/− mutant mice. Skeletal preparations of E18.5 embryos and of 2-week-old mice were carefully examined, but no skeletal defect was detected in Mkx−/− mutants (data not shown). Histological analyses carried out to examine the muscles at the same developmental stages showed no differences between wild-type and Mkx−/− mutant mice (data not shown). To examine whether Mkx−/− mice had tendon defects, the skin was removed from the tails of newly euthanized mice, and the tails were visualized under a stereomicroscope. In the mouse tail, there are four large groups of tendons that originate in muscles at the sacrum and extend through the length of the tail to insert at the rostral or caudal end of each of the tail vertebrae (33, 35) (Fig. 3A). Individual tendons are composed of bundles of collagen fibrils that weave in a highly stereotypic fashion, generating perfectly aligned surfaces that reflect incident light directly to create a white and vibrant appearance (7) (Fig. 3A). The tail tendons of 2-week-old Mkx−/− mice were much smaller than those of wild-type counterparts and appeared less vibrant, likely due to a disruption of fibril packaging in these tendons (Fig. 3A and B). Moreover, tendon crimp, the irregular undulations of the collagen fibers in tendons that results in a banding pattern in wild-type tendons when they are examined with top illumination (17, 18), was missing in Mkx−/− mutant tail tendons (Fig. 3G and H), again indicating disruption of the tertiary organization of the collagen fibers in these tendons. Since tail movement is controlled by a coordinated use of tail tendons, the wavy-tail phenotype in Mkx−/− mice likely resulted from stochastic differences in the force transmitted by the tail tendons inserted at each tail vertebra. Tendon insertions were easy to detect in the wild-type tail (Fig. 3A). Although fainter and less conspicuous in the Mkx−/− tail, most tendon insertions could be identified in the mutant tails upon close examination (Fig. 3B).
FIG. 3.
Tendon defects in Mkx−/− mutant mice. (A and B) Wild-type (A) and mutant (B) tail samples. The arrowheads point to representative tendon insertion sites in the tail vertebrae. (C and D) Wild-type (C) and mutant (D) forelimbs. (E and F) Wild-type (E) and mutant (F) hind limbs. (G to J) High-magnification views of crimp patterns of the tendons. The wild-type tail (G) and limb (I) tendons showed the typical crimp patterns. The crimp pattern was missing in Mkx−/− tail tendons (H), whereas the crimp bands were much more condensed in Mkx−/− limb tendons (J).
Although Mkx−/− mice did not have apparent impairment in their limb movement, their limb tendons, while all formed, also appeared smaller and less vibrant than those in their wild-type littermates (Fig. 3C to F). In contrast to the tail tendons, however, limb tendons in Mkx−/− mice did show a crimp pattern under direct illumination, but the bands were much more condensed than those in the wild-type littermates (Fig. 3I and J). These data indicate that Mkx function is required for normal morphogenesis of both axial and appendicular tendons.
Mkx−/− mutant mice show impairment in postnatal tendon collagen fibrillogenesis.
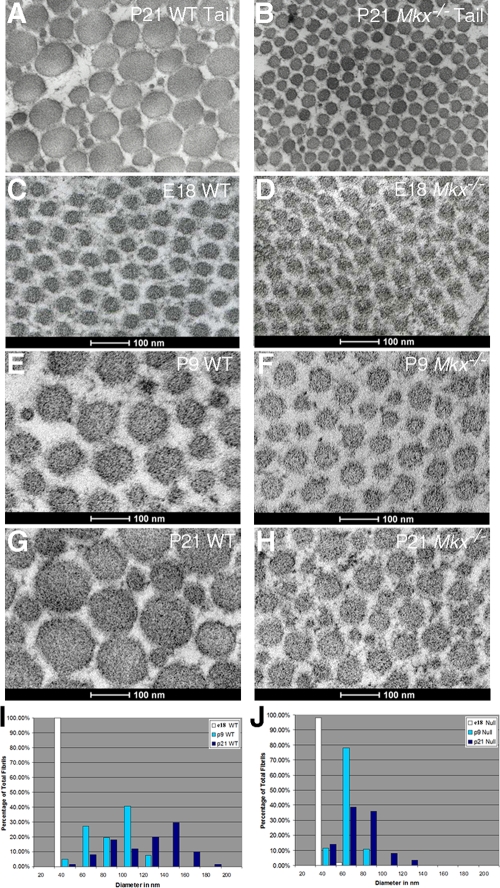
To investigate the effects of the Mkx mutation on tendon morphogenesis, cross sections of limb and tail tendons at matching positions from Mkx−/− mutant and wild-type littermates at various stages during tendon collagen fibrillogenesis, including embryonic stages E17 and E18 and postnatal stages P5, P9, and P21, were examined using transmission electron microscopy (TEM). At P21, the diameters of the collagen fibrils in Mkx−/− mutant tail tendons were dramatically smaller than those in the wild-type littermates (Fig. 4A and B). The reduction in fibril diameter was seen in all section levels through the tail. To avoid the effects of possible fibril size differences between tendons or in different positions along the tendon, all TEM analyses were subsequently performed on the forelimb flexor digitorium profundus (FDP) tendon at the site of the tendon vinculum, characterized by a distinctive tendon morphology that could be accurately identified at the TEM level (42). The collagen fibrils of the FDP tendon appeared uniform, with a diameter of ∼30 nm in both wild-type and Mkx−/− limbs at E18.5 (Fig. 4C, D, I, and J). Differences between wild-type and Mkx−/− littermates in collagen fibril diameter were apparent by P5 (data not shown) and became quite distinct by P9 (Fig. 4E and F). At P9, the fibrils in the wild-type FDP tendon had grown to up to 120 nm in diameter and showed significant heterogeneity in size (Fig. 4E and I), but the collagen fibrils in the FDP tendons of Mkx−/− mutants continued a slow and uniform rate of growth, so that close to 80% of the fibrils at this stage had a diameter of about 60 nm (Fig. 4F and J). These differences persisted at P21, when the wild-type FDP tendon had highly heterogeneous fibrils with diameters of up to 200 nm (Fig. 4A, G, and I) while the FDP tendons of Mkx−/− mutants had fibrils that extended only up to 100 nm in diameter (Fig. 4B, H, and J). Although the mutant tendon showed some heterogeneity in the sizes of collagen fibrils by P21, the spread was much more limited than that in the wild-type tendon, and the diameters of 80% of the fibrils were still between 60 and 80 nm. Mkx−/− mice thus showed a specific disruption of the unique features of postnatal collagen fibril growth.
FIG. 4.
Mkx−/− mutant mice had defects in postnatal growth of tendon collagen fibrils. (A and B) TEM of transverse sections of tail tendons from P21 wild-type and Mkx−/− littermates. (C to H) High-magnification TEM images of transverse sections through the FDP tendon at the level of the flexor vinculum from wild-type (C, E, and G) and Mkx−/− mutant (D, F, and H) mice at E18, P9, and P21, respectively. (I and J) Distribution patterns of the diameters of collagen fibrils in the FDP tendons of wild-type (I) and Mkx−/− mutant (J) mice. The diameters of 500 fibrils were measured at each developmental stage and binned at 20-nm increments.
Expression of Mkx during tendon development and maturation.
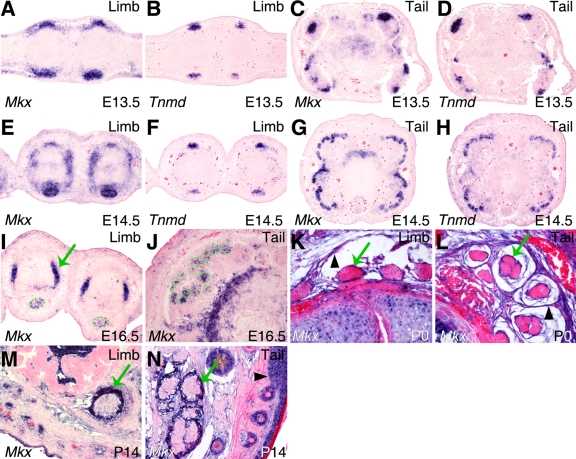
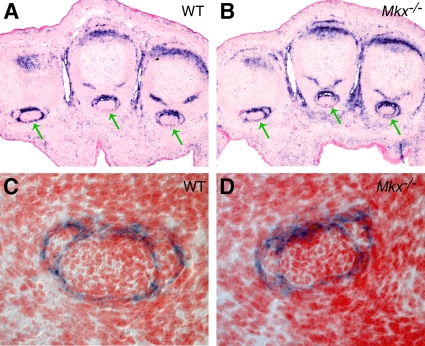
The tendon defects in Mkx−/− mutant mice prompted us to carry out a detailed analysis of Mkx expression during tendon development. Since previous studies have shown expression patterns of Mkx mRNA in the developing somites and in tendon precursor cells (2, 26), we focused on examining Mkx mRNA expression in later stages of tendon differentiation in order to understand the tendon defects in the Mkx−/− mice. In the developing limb, tendon progenitor cells align between the differentiating muscles and cartilage condensations at E12.5, and tendon condensation and differentiation ensue by E13.5 (32-34). The onset of tendon differentiation is accompanied by expression of the tendon-specific transmembrane protein tenomodulin (Tnmd) (10, 36). Mkx mRNA expression was detected in tendon progenitors at the onset of tendon differentiation at E12.5 (2). In situ hybridization on transverse sections of limbs and tails showed robust Mkx mRNA expression in the differentiating tendons of the limbs and tail at E13.5 and E14.5, stages at which the tendon progenitors undergo condensation and differentiation (Fig. 5A to H). The domain of Mkx expression at these stages was broader than that of Tnmd expression (Fig. 5A to H). Mkx mRNA expression decreased in differentiated tendon cells in both limbs and tails by E16.5 (Fig. 5I and J). Interestingly, robust expression of Mkx mRNA was detected in the same limb sections in the collateral ligaments (Fig. 5I), which differentiate at E16.5, highlighting a restriction of high levels of Mkx expression to early stages in tendon and ligament differentiation. At postnatal stages P0 and P14, the expression of Mkx mRNA in the tendon cells was much weaker than that at embryonic stages (Fig. 5K to N). However, strong expression of Mkx mRNA was detected in the tendon sheath cells in both limbs and tail at postnatal stages P0 and P14 (Fig. 5K to N).
FIG. 5.
Expression patterns of Mkx mRNA in developing mouse tendons. mRNA signals were detected by section in situ hybridization and are shown in blue/purple. All samples were briefly counterstained with eosin (red). (A to H) Expression patterns of Mkx and Tnmd mRNAs were compared on adjacent transverse sections of E13.5 and E14.5 limb or tail tissues. Strong Mkx mRNA expression was detected in the tendon progenitors at E13.5 and E14.5. (I and J) Mkx mRNA expression was significantly decreased in differentiated tendon cells in limb (I) and tail (J) tissues at E16.5. The dashed green circles mark the tendon structures. In contrast, robust expression of Mkx mRNA was detected in the collateral ligaments (green arrow in panel I) in the limb. (K to N) Expression of Mkx mRNA was very weak in tendon cells at postnatal stages P0 and P14. However, strong expression of Mkx mRNA was detected in tendon sheaths (green arrows) in both the limb and tail tissues at postnatal stages P0 and P14. In addition, Mkx expression was detected in the fibrous connective tissue surrounding the tendons at P0 (arrowheads in panels K and L) and in the epidermal cells in the Malpighian layer by P14 (arrowhead in panel N).
Effects of Mkx loss of function on tendon cell differentiation and gene expression.
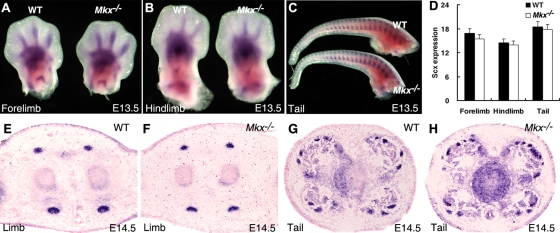
Although the tendon collagen fibrils were comparable in size in the Mkx−/− and wild-type littermates at E18.5, the robust expression of Mkx mRNA at the onset of tendon differentiation suggested that Mkx might be involved in tendon cell differentiation. The bHLH transcription factor Scx is expressed in tendon cells throughout tendon development, and Scx−/− mutant mice showed severe disruption of tendon differentiation (32). We therefore examined the expression of Scx mRNA in Mkx−/− mutant embryos at E13.5 and E14.5, when the condensation and differentiation of tendon progenitors take place. No obvious differences in either the levels or spatiotemporal patterns of expression of Scx in the axial and limb tendon cells were observed between wild-type and Mkx−/− mutant embryos (Fig. 6). These results indicate that Mkx does not regulate Scx gene expression and that tendon cell differentiation initiated normally in Mkx−/− embryos.
FIG. 6.
Scx mRNA expression in developing tendons is not dependent on Mkx function. (A to C) Whole-mount in situ hybridization assays detected similar patterns of Scx mRNA expression during tendon formation in the developing limb and tail regions in wild-type and Mkx−/− mutant embryos at E13.5. mRNA signals are shown in blue/purple. (D) No significant changes in the levels of Scx mRNA expression in the E13.5 wild-type and Mkx−/− mutant embryonic limb and tail tissues were detected using real-time RT-PCR analyses. The error bars represent standard deviations. (E to H) Expression of Scx mRNA in the limb and tail tissues of E14.5 wild-type and Mkx−/− mutant littermates detected by section in situ hybridization. The patterns and levels of Scx mRNA expression were similar in the wild-type and Mkx−/− mutant embryos.
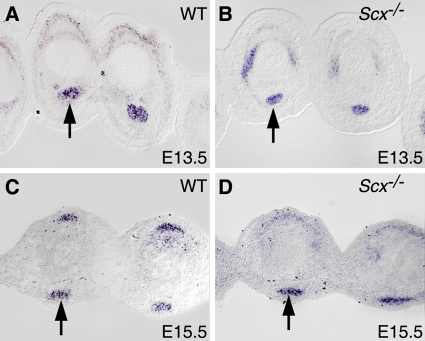
We also tested if Mkx expression was regulated by Scx. We found that Mkx expression persisted in tendon cells in Scx−/− embryos (Fig. 7), indicating that Mkx expression is not dependent on Scx.
FIG. 7.
Mkx mRNA is expressed in tendon cells in Scx−/− mutant embryos. In situ hybridization analysis of Mkx mRNA expression (shown in purple) was carried out on cross sections through the digits of the developing forelimbs of E13.5 (A and B) and E15.5 (C and D) wild-type (A and C) and Scx−/− mutant (B and D) embryos. The arrows point to corresponding tendons in wild-type and mutant embryos.
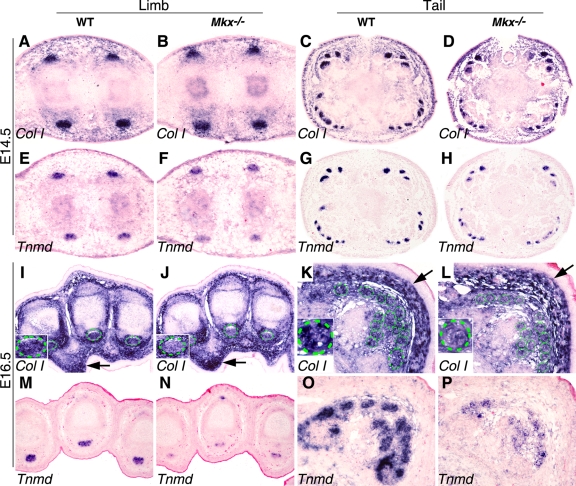
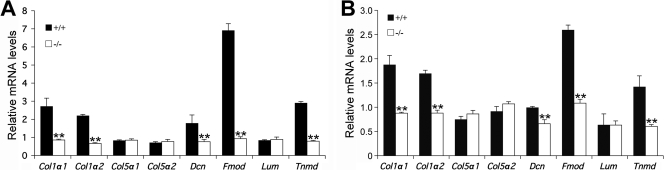
We next examined the expression of molecular markers of tenocyte differentiation. Collagen I is the major component of the tendon extracellular matrix and is abundantly expressed by the tendon cells by E14.5. Although the expression of Col1a1 mRNA was not changed at E14.5 (Fig. 8A to D), it was significantly reduced in tendon cells in both the limbs and tails of Mkx−/− mutant embryos by E16.5 compared with their wild-type littermates (Fig. 8I to L). In contrast, expression levels of Col1a1 mRNA in other connective tissues were comparable in E16.5 Mkx−/− mutant and control embryos (Fig. 8I to L), indicating that the effects of Mkx on Col1a1 expression are tendon specific. Tnmd, a type II transmembrane glycoprotein, is predominantly expressed in tendons and is involved in collagen fibril maturation (10, 19). The expression of Tnmd mRNA was slightly reduced by E14.5 (Fig. 8E to H) and dramatically downregulated by E16.5 in the tendon cells of Mkx−/− mutant embryos compared with their wild-type littermates (Fig. 8M to P). To quantitate these differences in gene expression and to examine potential differences in the expression of other genes involved in tendon collagen fibrillogenesis, we isolated tendon tissues from neonatal mice by microdissection and carried out quantitative real-time RT-PCR assays. As shown in Fig. 9, the expression levels of Col1a1, Col1a2, fibromodulin (Fmod), and Tnmd mRNAs were each reduced by more than 50% in both limb and tail tendons in the Mkx−/− mutant mice in comparison with their wild-type littermates. Expression of decorin (Dcn) mRNAs was also significantly reduced, by about 35%, in the limb and tail tendons in Mkx−/− mice compared with wild-type littermates. In contrast, the expression levels of Col5a1, Col5a2, and lumican (Lum) were not significantly different in the mutant and control tendon samples. These data indicate that Mkx plays specific roles in the regulation of tendon cell gene expression.
FIG. 8.
Mkx is required for maintenance of expression of Col1a and Tnmd mRNAs in developing tendons. mRNA signals were detected by section in situ hybridization and are shown in blue/purple. All samples were briefly counterstained with eosin (red). (A to H) Matching transverse sections of limb (A, B, E, and F) and tail (C, D, G, and H) regions of E14.5 wild-type (A, C, E, and G) and Mkx−/− mutant (B, D, F, and H) embryos showing Col1a1 and Tnmd mRNA expression. While Col1a1 expression levels were similar in the developing tendon tissues in wild-type and Mkx−/− mutant littermates at this stage, Tnmd expression was reduced in the mutant tendons in comparison with the wild-type littermates. (I to L) At E16.5, Col1a1 mRNA expression was reduced in the mutant tendon cells in both the limb and tail in comparison with wild-type littermates. In contrast, Col1a1 mRNA expression in other connective tissues, such as in the dermis (arrows), was not significantly altered in the mutant. The green circles mark tendons. The inset in the lower left corner of each panel shows a higher-magnification image of a representative tendon section of that sample. (M to P) At E16.5, expression of Tnmd mRNA was dramatically reduced in both the limb and tail tendon cells in Mkx−/− mutant embryos in comparison with their wild-type littermates.
FIG. 9.
Quantitative real-time RT-PCR analysis of changes of gene expression in the tendon tissues in Mkx−/− mutant mice. (A) Relative expression levels of Col1a1, Col1a2, Col5a1, Col5a2, decorin (Dcn), fibromodulin (Fmod), lumican (Lum), and Tnmd mRNAs in the hind-limb tendons from P0 Mkx−/− mice and wild-type littermates. (B) Relative expression levels of Col1a1, Col1a2, Col5a1, Col5a2, Dcn, Fmod, Lum, and Tnmd mRNAs in the tail tendons from P0 Mkx−/− mice and wild-type littermates. mRNA levels of wild-type (+/+) and mutant (−/−) mice were normalized to that of Hprt in the same experiments. The error bars represent standard deviations. **, statistically significant difference (P < 0.05) between the mRNA levels in the mutant and wild-type samples.
It has been reported that Tnmd-deficient mice had reduced tenocyte proliferation (19). We thus investigated whether tendon growth was affected in Mkx−/− mutant embryos. To facilitate the characterization of tendon developmental defects in mutant mouse strains, we recently reported a detailed pattern and organization of tendons in the developing mouse forelimb (42). To measure possible differences in tendon sizes in Mkx−/− mutant and control littermates, we carried out in situ hybridization of serial forelimb sections with a cRNA probe to Tppp3, whose expression specifically marks the tendon-ensheathing tissues and therefore provides an outline of the tendon circumference (37) (Fig. 10A and B). Tendon measurements were performed on sections at distinct anatomic positions that could be matched with great precision in wild-type and mutant limbs based on the patterns of tendon distribution (42). For example, the FDP tendon sections were measured at matched digit levels, whereas the extensor digitorium communis (EDC) tendon sections were measured at matched metacarpal levels (Table 1). Three FDP tendons and three EDC tendons in each limb sample were measured to yield two distinct tendons with a larger sample size. The sizes of both the FDP and EDC tendons were reduced in mutant embryos to approximately 70% of that of the wild-type controls (Table 1). In measurements of other tendons, however, we found that the change in tendon size was variable. For example, the sizes of the flexor carpi radialis tendon were similar in the control and mutant samples, while the extensor carpi radialis brevis and extensor digiti quinti tendons showed a more limited reduction in size in the mutant samples (Table 1). Similarly, we found that tendon cell numbers were significantly reduced in the FDP and EDC tendons, but the reduction in cell numbers in the other tendons was not as pronounced (Table 2). Moreover, the reduction in the sizes of the FDP and EDC tendons in the Mkx−/− mutant embryos was more pronounced than the reduction in cell numbers, which correlated with the higher density of tendon cells in the cross sections of these tendons in the mutant embryos (Fig. 10C and D and data not shown).
FIG. 10.
Expression of Tppp3 mRNA marks the tendon sheath and provides an outline of the tendon circumference for measurements of tendon size and tendon cell numbers. (A and B) In situ hybridization with a Tppp3 probe on transverse sections of limbs from wild-type (A) and Mkx−/− mutant (B) embryos. Tppp3 mRNA signals were detected as a blue color. The arrows point to the flexor profundus tendon tissues. (C and D) DAPI staining (shown in red) following in situ hybridization detection of Tppp3 revealed more densely packed tendon cells in the flexor profundus tendon in the Mkx−/− mutant (D) than in the wild-type littermate (C).
TABLE 1.
Comparison of tendon sizes in Mkx−/− and wild-type embryos
| Tendon namea | No. of samples/genotype | Locationb | Sizec |
Mkx−/−/wild-type ratio (%) | P value | |
|---|---|---|---|---|---|---|
| Wild type | Mkx−/− | |||||
| FDP | 15 | 6 | 5.28 ± 0.69 | 3.84 ± 0.60 | 72.4 | <0.05 |
| EDC | 15 | 11 | 1.89 ± 0.19 | 1.32 ± 0.21 | 69.9 | <0.05 |
| Brevis | 5 | 14 | 3.73 ± 0.35 | 3.13 ± 0.29 | 83.9 | <0.05 |
| Quinti | 5 | 14 | 2.18 ± 0.21 | 2.00 ± 0.46 | 91.7 | >0.05 |
| Radialis | 5 | 14 | 4.02 ± 0.16 | 4.14 ± 0.69 | 102.9 | >0.05 |
The tendon names are according to Watson et al. (42): Brevis, extensor carpi radialis brevis; Quinti, extensor digiti quinti; Radialis, flexor carpi radialis.
The section location of each tendon in the wild-type and Mkx−/− mutant samples was matched to the published numeric location (42).
Cross section area in μm2.
TABLE 2.
Comparison of tendon cell number in cross sections of Mkx−/− and wildtype embryos
| Tendon namea | No. of samples/genotype | Locationb | No. of cells (SD) |
Mkx−/− /wild-type ratio (%) | P value | |
|---|---|---|---|---|---|---|
| Wild type | Mkx−/− | |||||
| FDP | 15 | 6 | 123.13 ± 7.7 | 99.2 ± 14.2 | 80.0 | <0.05 |
| EDC | 15 | 11 | 34.94 ± 4.2 | 25.6 ± 3.7 | 73.6 | <0.05 |
| Brevis | 5 | 14 | 58 ± 12.5 | 47.8 ± 5.5 | 82.0 | >0.05 |
| Quinti | 5 | 14 | 38.4 ± 6.5 | 34.6 ± 9.2 | 90.1 | >0.05 |
| Radialis | 5 | 14 | 68.2 ± 6.9 | 66.6 ± 6.9 | 97.7 | >0.05 |
Taken together, these data indicate that the reduction in tendon size in postnatal Mkx−/− mutant mice (Fig. 3) resulted from a combination of defects in prenatal tendon development and postnatal tendon fibril growth.
Abnormality of tendon sheaths in Mkx−/− mutant mice.
Since Mkx mRNA was strongly expressed in tendon sheath cells after birth, we carried out detailed histological analyses of tendon sheaths in Mkx−/− mutant mice. Tendon sheaths appeared normal at embryonic stages and P0 in Mkx−/− mutant mice (data not shown). However, in comparison with wild-type littermates, the tendon sheath was much thicker and contained more cell layers in Mkx−/− mutant mice by P14 (Fig. 11). Thus, in addition to regulating tendon growth and tendon cell differentiation, Mkx plays an important role in the maintenance of the tendon sheath.
FIG. 11.
Histology of the tendon sheath in 2-week-old wild-type and Mkx−/− mutant mice. In comparison with wild-type littermates (A and C), the Mkx−/− mutant (B and D) mice had more cell layers in the tendon sheath in the limb (A and B) and tail (C and D) tissues. The arrows point to the tendon sheath.
DISCUSSION
Despite the essential roles of tendons in the patterning and function of the musculoskeletal system, a molecular framework for the regulation of tendon differentiation has only recently begun to emerge (32, 41). This report presents the first characterization of a critical role for the atypical homeodomain transcription factor Mkx in tendon morphogenesis. We found that Mkx mRNA was strongly expressed in tendon cells during the early stages of tendon development and was downregulated following tenocyte differentiation. The roles of Mkx in tendon morphogenesis were manifested in two distinct stages. During embryogenesis, while the pattern of tendon formation was not disrupted in Mkx−/− mutants, expression of collagen I, fibromodulin, and tenomodulin was significantly reduced in the mutant tendon cells; in postnatal stages, the collagen fibrils in Mkx−/− mutant tendons were significantly smaller and more uniform in diameter than those of wild-type littermates. Collagen fibril size appeared normal during embryogenesis, and smaller fibril diameters emerged due to failure to initiate the fast postnatal phase of collagen fibril growth. In postnatal stages, Mkx expression was also detected in the sheaths of limb and tail tendons, and in Mkx−/− mutants, the tendon sheaths appeared thicker, suggesting a separate role for Mkx in the differentiation of the tendon sheath, as well.
Transcriptional regulation of tendon differentiation by Mkx.
Tendon progenitors, identified as Scx-expressing cells, are first detected at E9.5 and by E12.5 align between the differentiating muscles and skeletal condensations (32-34). The loosely organized progenitors condense and organize into morphologically distinct tendons by E13.5, and collagen fibrillogenesis initiates at E14.5 (14, 43). The onset of Mkx expression is close to that of Scx in the autopod and in the somites, but Mkx expression in more proximal parts of the limbs coincides with tendon cells only at the onset of tendon differentiation at E12.5 (2). While Scx expression persists in differentiated tenocytes, Mkx expression was downregulated following tenocyte differentiation (Fig. 5). Robust Mkx expression is thus associated with the early stages of tendon differentiation. The timing of Mkx expression suggested a connection with Scx, whose function is required for tenocyte differentiation at E13 (32). We found, however, that expression of these two transcription factors in tendon cells is not interdependent, Scx expression is maintained in Mkx−/− embryos, and Mkx expression is not affected in tendons that do differentiate in Scx−/− embryos.
The morphological consequences of the loss of Mkx function in the Mkx−/− mice are not coincident with the peak of Mkx mRNA expression but rather follow in later stages. Mkx function is apparently not required for the ability of tendon progenitors to differentiate into tenocytes, but rather, for subsequent tendon growth during embryogenesis and for the regulation of postnatal growth and maturation of collagen fibrils. While the low levels of Mkx at these stages may still be directly responsible for the phenotypic consequences we identified in Mkx−/− mutants, it is also possible that Mkx function is required at the time of its peak expression during tenocyte differentiation and that in Mkx−/− mutants, early disruption of tenocyte differentiation is manifested at later stages. The decreases in expression of Col1a1 and Tnmd from E14.5 to E16.5 in the tendons of Mkx−/− mutant embryos compared with their wild-type littermates suggest that there is indeed an early effect of Mkx on tenocyte differentiation. An involvement of Mkx in tenocyte differentiation is intriguing in light of a recent report of Mkx capacity to block myogenic conversion of C3H10T1/2 cells (3). The essential developmental stage for Mkx function will need to be addressed in future studies using the Mkxflox conditional allele.
Mkx is essential for the postnatal growth of collagen fibrils in tendons.
Studies of collagen fibril growth have thus far focused on extracellular matrix proteins that regulate fibril growth. In particular, several small leucine-rich repeat proteoglycans (SLRPs), including decorin, fibromodulin, and lumican, have been shown to bind to the surfaces of collagen I fibrils and to regulate fibril growth (1). Mice lacking either decorin or lumican had irregular and thicker-than-normal collagen fibrils in their tendons (15, 16). Mice lacking fibromodulin also had irregular collagen fibrils in tendons, but with a thinner average diameter than in their wild-type littermates (38). The highly irregular collagen fibril sizes in these mutant mice indicated that these SLRPs regulate collagen fibril assembly and growth, in part by preventing aberrant fusion of collagen fibrils (13). Although the expression of both decorin and fibromodulin was significantly reduced in the tendons of Mkx−/− mice compared with the wild-type controls, the collagen fibril defects in the Mkx−/− mice were distinct from those in the mouse strains lacking any of the SLRPs. Thus, the alterations in expression of these extracellular matrix proteins in Mkx−/− mice likely reflect critical roles of Mkx in the regulation of tenocyte maturation rather than modulating their specific roles in fibril assembly. The downstream targets of Mkx responsible for the postnatal growth defect of the tendon collagen fibrils in Mkx−/− mice remain to be identified.
The orchestration of the production of building blocks required for fibril assembly and the regulation of the rate of fibril growth are functions of the tenocytes and therefore are likely controlled by the transcriptional network that governs tendon cell differentiation. Identifying the transcriptional regulators of this process may therefore be crucial both for deciphering the mechanism of the regulation of fibril growth and for the development of approaches to tendon repair. Scx has been implicated in the regulation of Collagen I and Collagen XIV expression (32), but the severe tendon differentiation defects in Scx−/− mutant mice precluded a comprehensive analysis of Scx functions in collagen fibrillogenesis (32). Mkx is thus the first transcription factor associated directly with a positive role in collagen fibril growth and therefore offers a unique opportunity to identify the cellular and molecular mechanisms that drive this process.
Acknowledgments
This work was supported by Public Health Service grants DE013681 and DE015207 from the National Institute of Dental and Craniofacial Research to R.J. and by Public Health Service grant AR055640 from the National Institute of Arthritis, Musculoskeletal and Skin Diseases to R.S.
Footnotes
Published ahead of print on 9 August 2010.
REFERENCES
- 1.Ameye, L., and M. F. Young. 2002. Mice deficient in small leucine-rich proteoglycans: novel in vivo models for osteoporosis, osteoarthritis, Ehlers-Danlos syndrome, muscular dystrophy, and corneal diseases. Glycobiology 12:107R-116R. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, D. M., J. Arredondo, K. Hahn, G. Valente, J. F. Martin, J. Wilson-Rawls, and A. Rawls. 2006. Mohawk is a novel homeobox gene expressed in the developing mouse embryo. Dev. Dyn. 235:792-801. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, D. M., B. J. Beres, J. Wilson-Rawls, and A. Rawls. 2009. The homeobox gene Mohawk represses transcription by recruiting the sin3A/HDAC co-repressor complex. Dev. Dyn. 238:572-580. [DOI] [PubMed] [Google Scholar]
- 4.Banos, C. C., A. H. Thomas, and C. K. Kuo. 2008. Collagen fibrillogenesis in tendon development: current models and regulation of fibril assembly. Birth Defects Res. C Embryo Today 84:228-244. [DOI] [PubMed] [Google Scholar]
- 5.Benjamin, M., E. Kaiser, and S. Milz. 2008. Structure-function relationships in tendons: a review. J. Anat. 212:211-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamin, M., and J. R. Ralphs. 2000. The cell and developmental biology of tendons and ligaments. Int. Rev. Cytol. 196:85-130. [DOI] [PubMed] [Google Scholar]
- 7.Benjamin, M., and J. R. Ralphs. 1997. Tendons and ligaments—an overview. Histol. Histopathol. 12:1135-1144. [PubMed] [Google Scholar]
- 8.Bilioni, A., G. Craig, C. Hill, and H. McNeill. 2005. Iroquois transcription factors recognize a unique motif to mediate transcriptional repression in vivo. Proc. Natl. Acad. Sci. U. S. A. 102:14671-14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birk, D. E., J. F. Southern, E. I. Zycband, J. T. Fallon, and R. L. Trelstad. 1989. Collagen fibril bundles: a branching assembly unit in tendon morphogenesis. Development 107:437-443. [DOI] [PubMed] [Google Scholar]
- 10.Brandau, O., A. Meindl, R. Fassler, and A. Aszodi. 2001. A novel gene, tendin, is strongly expressed in tendons and ligaments and shows high homology with chondromodulin-I. Dev. Dyn. 221:72-80. [DOI] [PubMed] [Google Scholar]
- 11.Brent, A. E., R. Schweitzer, and C. J. Tabin. 2003. A somitic compartment of tendon progenitors. Cell 113:235-248. [DOI] [PubMed] [Google Scholar]
- 12.Bürglin, T. R. 1997. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25:4173-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canty, E. G., and K. E. Kadler. 2002. Collagen fibril biosynthesis in tendon: a review and recent insights. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 133:979-985. [DOI] [PubMed] [Google Scholar]
- 14.Canty, E. G., Y. Lu, R. S. Meadows, M. K. Shaw, D. F. Holmes, and K. E. Kadler. 2004. Coalignment of plasma membrane channels and protrusions (fibripositors) specifies the parallelism of tendon. J. Cell Biol. 165:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravarti, S., T. Magnuson, J. H. Lass, K. J. Jepsen, C. LaMantia, and H. Carroll. 1998. Lumican regulates collagen fibril assembly: skin fragility and corneal opacity in the absence of lumican. J. Cell Biol. 141:1277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danielson, K. G., H. Baribault, D. F. Holmes, H. Graham, K. E. Kadler, and R. V. Iozzo. 1997. Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J. Cell Biol. 136:729-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Campos Vidal, B. 2003. Image analysis of tendon helical superstructure using interference and polarized light microscopy. Micron 34:423-432. [DOI] [PubMed] [Google Scholar]
- 18.Diamant, J., A. Keller, E. Baer, M. Litt, and R. G. Arridge. 1972. Collagen; ultrastructure and its relation to mechanical properties as a function of ageing. Proc. R. Soc. Lond. B Biol. Sci. 180:293-315. [DOI] [PubMed] [Google Scholar]
- 19.Docheva, D., E. B. Hunziker, R. Fassler, and O. Brandau. 2005. Tenomodulin is necessary for tenocyte proliferation and tendon maturation. Mol. Cell. Biol. 25:699-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farley, F. W., P. Soriano, L. S. Steffen, and S. M. Dymecki. 2000. Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28:106-110. [PubMed] [Google Scholar]
- 21.Gong, S. G., N. J. White, and A. Y. Sakasegawa. 2000. The Twirler mouse, a model for the study of cleft lip and palate. Arch. Oral Biol. 45:87-94. [DOI] [PubMed] [Google Scholar]
- 22.Kannus, P. 2000. Structure of the tendon connective tissue. Scand. J. Med. Sci. Sports 10:312-320. [DOI] [PubMed] [Google Scholar]
- 23.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U. S. A. 93:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan, Y., P. D. Kingsley, E. S. Cho, and R. Jiang. 2001. Osr2, a new mouse gene related to Drosophila odd-skipped, exhibits dynamic expression patterns during craniofacial, limb, and kidney development. Mech. Dev. 107:175-179. [DOI] [PubMed] [Google Scholar]
- 25.Léjard, V., G. Brideau, F. Blais, R. Salingcarnboriboon, G. Wagner, M. H. Roehrl, M. Noda, D. Duprez, P. Houillier, and J. Rossert. 2007. Scleraxis and NFATc regulate the expression of the pro-alpha1(I) collagen gene in tendon fibroblasts. J. Biol. Chem. 282:17665-17675. [DOI] [PubMed] [Google Scholar]
- 26.Liu, H., W. Liu, K. M. Maltby, Y. Lan, and R. Jiang. 2006. Identification and developmental expression analysis of a novel homeobox gene closely linked to the mouse Twirler mutation. Gene Expr. Patterns 6:632-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyon, M. F. 1958. Twirler: a mutant affecting the inner ear of the house mouse. J. Embryol. Exp. Morphol. 6:105-116. [PubMed] [Google Scholar]
- 28.Markiewicz, M., Y. Asano, S. Znoyko, Y. Gong, D. K. Watson, and M. Trojanowska. 2007. Distinct effects of gonadectomy in male and female mice on collagen fibrillogenesis in the skin. J. Dermatol. Sci. 47:217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, J. F., A. Bradley, and E. N. Olson. 1995. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 9:1237-1249. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto, K., S. Nishihara, M. Kamimura, T. Shiraishi, T. Otoguro, M. Uehara, Y. Maeda, K. Ogura, A. Lumsden, and T. Ogura. 2004. The prepattern transcription factor Irx2, a target of the FGF8/MAP kinase cascade, is involved in cerebellum formation. Nat. Neurosci. 7:605-612. [DOI] [PubMed] [Google Scholar]
- 31.McLeod, M. J. 1980. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology 22:299-301. [DOI] [PubMed] [Google Scholar]
- 32.Murchison, N. D., B. A. Price, D. A. Conner, D. R. Keene, E. N. Olson, C. J. Tabin, and R. Schweitzer. 2007. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development 134:2697-2708. [DOI] [PubMed] [Google Scholar]
- 33.Pryce, B. A., S. S. Watson, N. D. Murchison, J. A. Staverosky, N. Dunker, and R. Schweitzer. 2009. Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development 136:1351-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schweitzer, R., J. H. Chyung, L. C. Murtaugh, A. E. Brent, V. Rosen, E. N. Olson, A. Lassar, and C. J. Tabin. 2001. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128:3855-3866. [DOI] [PubMed] [Google Scholar]
- 35.Shinohara, H. 1999. The musculature of the mouse tail is characterized by metameric arrangements of bicipital muscles. Okajimas Folia Anat. Jpn. 76:157-169. [DOI] [PubMed] [Google Scholar]
- 36.Shukunami, C., A. Takimoto, M. Oro, and Y. Hiraki. 2006. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev. Biol. 298:234-247. [DOI] [PubMed] [Google Scholar]
- 37.Staverosky, J. A., B. A. Pryce, S. S. Watson, and R. Schweitzer. 2009. Tubulin polymerization-promoting protein family member 3, Tppp3, is a specific marker of the differentiating tendon sheath and synovial joints. Dev. Dyn. 238:685-692. [DOI] [PubMed] [Google Scholar]
- 38.Svensson, L., A. Aszodi, F. P. Reinholt, R. Fassler, D. Heinegard, and A. Oldberg. 1999. Fibromodulin-null mice have abnormal collagen fibrils, tissue organization, and altered lumican deposition in tendon. J. Biol. Chem. 274:9636-9647. [DOI] [PubMed] [Google Scholar]
- 39.Swiatek, P. J., and T. Gridley. 1993. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 7:2071-2084. [DOI] [PubMed] [Google Scholar]
- 40.Takeuchi, J. K., and B. G. Bruneau. 2007. Irxl1, a divergent Iroquois homeobox family transcription factor gene. Gene Expr. Patterns 7:51-56. [DOI] [PubMed] [Google Scholar]
- 41.Tozer, S., and D. Duprez. 2005. Tendon and ligament: development, repair and disease. Birth Defects Res. C Embryo Today 75:226-236. [DOI] [PubMed] [Google Scholar]
- 42.Watson, S. S., T. J. Riordan, B. A. Pryce, and R. Schweitzer. 2009. Tendons and muscles of the mouse forelimb during embryonic development. Dev. Dyn. 238:693-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, G., B. B. Young, Y. Ezura, M. Favata, L. J. Soslowsky, S. Chakravarti, and D. E. Birk. 2005. Development of tendon structure and function: regulation of collagen fibrillogenesis. J. Musculoskelet. Neuronal Interact. 5:5-21. [PubMed] [Google Scholar]
- 44.Zhang, Y., X. Zhao, Y. Hu, T. St Amand, M. Zhang, R. Ramamurthy, M. Qiu, and Y. Chen. 1999. Msx1 is required for the induction of Patched by Sonic hedgehog in the mammalian tooth germ. Dev. Dyn. 215:45-53. [DOI] [PubMed] [Google Scholar]