Abstract
MED1/TRAP220, a subunit of the transcriptional Mediator/TRAP complex, is crucial for various biological events through its interaction with distinct activators, such as nuclear receptors and GATA family activators. In hematopoiesis, MED1 plays a pivotal role in optimal nuclear receptor-mediated myelomonopoiesis and GATA-1-induced erythropoiesis. In this study, we present evidence that MED1 in stromal cells is involved in supporting hematopoietic stem and/or progenitor cells (HSPCs) through osteopontin (OPN) expression. We found that the proliferation of bone marrow (BM) cells cocultured with MED1 knockout (Med1−/−) mouse embryonic fibroblasts (MEFs) was significantly suppressed compared to the control. Furthermore, the number of long-term culture-initiating cells (LTC-ICs) was attenuated for BM cells cocultured with Med1−/− MEFs. The vitamin D receptor (VDR)- and Runx2-mediated expression of OPN, as well as Mediator recruitment to the Opn promoter, was specifically attenuated in the Med1−/− MEFs. Addition of OPN to these MEFs restored the growth of cocultured BM cells and the number of LTC-ICs, both of which were attenuated by the addition of the anti-OPN antibody to Med1+/+ MEFs and to BM stromal cells. Consequently, MED1 in niche appears to play an important role in supporting HSPCs by upregulating VDR- and Runx2-mediated transcription on the Opn promoter.
The specialized microenvironmental niches in the bone marrow (BM), namely, the osteoblastic (or endosteal) and vascular niches, host and interface with hematopoietic stem cells (HSCs) and are the sites where their size and fate are strictly regulated (15, 29; reviewed in references 1, 16, 28, 30, and 34). HSCs and their niches produce diverse molecules, whose interactions control HSC self-renewal and differentiation. In the osteoblastic niche, almost 75% of the HSCs are in a quiescent (slowly cycling or G0) state. In a physiological condition, HSCs migrate from the osteoblastic niche toward the vascular niche, enter the cell cycle, and undergo symmetric cell division or asymmetric division, accompanied by differentiation and final maturation. In this manner, a defined set of mature differentiated progeny is continuously produced without HSC depletion.
The transcriptional Mediator complex, originally isolated as a thyroid hormone receptor-associated protein (TRAP) complex and subsequently identified as a mammalian counterpart of the yeast Mediator complex (i.e., a subcomplex of the RNA polymerase II holoenzyme), appears to serve as a bridge between diverse activators and the general transcriptional machinery (reviewed in references 4, 13, 18, and 21). This complex contains approximately 25 polypeptides, among which the MED1/TRAP220 subunit is responsible for specific binding of the complex to several activators, which include nuclear receptors (13), GATA family members (22, 27), C/EBPβ (20), and BRCA1 (33). Mediator conveys the specific signals of the activators to the recruited general transcriptional machinery to activate transcription by direct communication between MED1 and the activators (5).
Through the interaction with MED1, nuclear receptors are involved in various hematocytic differentiations. For example, the vitamin D receptor (VDR) and retinoic acid receptor (RAR) are members of the nuclear hormone receptor superfamily, whose interaction with MED1 is crucial for ligand-dependent monopoiesis and granulopoiesis, respectively, as well as for peroxisome proliferator-activated receptor γ (PPARγ)-mediated adipogenesis (7, 31). GATA-1, for which MED1 was recently shown to be a specific coactivator, mediates erythropoiesis through its interaction with MED1 (27). However, as Med1 null mice die early during embryogenesis (12, 22), it is difficult to determine the physiological role of MED1 in BM hematopoiesis in vivo.
Osteopontin (OPN), an acidic glycosylated phosphoprotein present in the bone extracellular matrix, is synthesized by osteoblasts, osteoclasts, and monocytes or macrophages. It has a well-described role in cell adhesion, inflammatory responses, angiogenesis, and tumor metastasis (reviewed in references 1, 10, 16, and 34). In the BM, it exists either as full-length OPN (flOPN) or as thrombin-cleaved truncated N-terminal OPN (trOPN), both of which appear to be restricted to the endosteal surface (9, 23). OPN is bound by CD44 and diverse integrins that include αvβ3, αvβ5, αvβ6, αvβ1, and α5β1, which recognize the RGD-binding sequence of OPN. Among these receptors, αvβ3 integrin and CD44 are responsible for bone cell attachment to the bone surface through the interaction with OPN. In contrast, trOPN, unlike flOPN, is a specific ligand for α9β1 and a much better ligand for α5β1 than flOPN (23, 35). The receptors, including CD44, α5β1, and α9β1, are also expressed on HSCs and mediate the adhesion of these cells to OPN, which possibly regulates HSC self-renewal, quiescence, and differentiation. Two independent studies utilizing Opn null mice have suggested a role for OPN in restricting the excessive expansion of the HSC pool that may result from niche activation (23, 26). However, in view of the existence of different forms of OPN, whose functions might differ, and the altered gene expression in Opn null mice, which might affect niche function, the precise role of OPN in the osteoblastic niche remains to be elucidated.
In this study, we investigated the previously unexamined role of MED1 in stromal cells by using mouse embryonic fibroblasts (MEFs) and BM stromal cells as a niche model. We show that MED1 in stromal cells is involved in supporting hematopoietic stem and/or progenitor cells (HSPCs) and that OPN, the downstream direct target of MED1, is responsible for this action.
MATERIALS AND METHODS
Mice.
Med1−/− mice were described previously (12). Mice backcrossed at least 10 times with C57BL6 were used for experiments. All animal experiments were performed according to the institutional guidelines of the Animal Research Center, Kobe University, Japan.
Cell culture.
Stable lines of Med1+/+ p53−/− and Med1−/− p53−/− MEFs were established from embryonic day 10.0 (E10.0) embryos derived from a single crossing of Med1+/− p53−/− male and Med1+/− p53+/− female mice in the C57BL6 background. Two lines of these MEFs were analyzed in all experiments. The MEFs were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C. The osteoblast-like MC3T3-E1 cells and OP-9 BM stromal cells, distributed by RIKEN BRC through the National Bio-Resource of the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), and MS-5 cells (14) were maintained in α-modified Eagle's medium (αMEM) supplemented with 10% FBS, 20% FBS, and 20% horse serum, respectively.
BM culture and colony-forming cell assay.
MEFs and MS-5 and OP-9 cells (2 × 105), treated with 100 μg/ml mitomycin C (MMC) to arrest the cell cycle, were plated on 0.1% gelatin-coated 12-well plates. On the next day, 1 × 106 BM cells, harvested from the femurs of congenic wild-type mice, were added to each well and cultured in Myelocult M5300 (Stem Cell Technologies, Canada) or Iscove's MDM (IMDM) supplemented with 20% BIT9500 (Stem Cell Technologies) and 0.222% low-density lipoprotein (Calbiochem) in the absence or presence of various amounts of recombinant mouse flOPN (R&D Systems), 0.2 μg/ml anti-mOPN (N terminus) rabbit polyclonal IgG (P-18; Santa Cruz Biotechnology), or normal rabbit IgG (Sigma) at 33°C.
For long-term culture, one-half of the medium was replaced with fresh medium each week. After a 6- to 8-week culture period, trypsinized cells containing HSPCs (adherent and nonadherent) were collected and cultured in complete methylcellulose medium (Methocult M3434; Stem Cell Technologies) for all types of colonies, methylcellulose medium without erythropoietin (EPO) (Methocult M3534) for myeloid colonies, or methylcellulose medium with EPO but without other cytokines (Methocult M3334) for erythroid colonies at 37°C for 10 days, and the colonies were counted.
Cell growth, DNA content, MTT assay, and DNA synthesis.
For cell growth, the cells (1 × 106) on 24-well plates were counted after trypsinization. For quantitation of genomic DNAs, high-molecular-weight DNAs were extracted and optically quantitated. For the MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay to measure mitochondrial function of live cells, the cells were purged with 0.5 mg/ml MTT for 3 h. Then 0.15 volume of acid isopropanol (0.04 M HCl in isopropanol) was added, and the optical density at 570 nm (OD570) was measured. For DNA synthesis, the incorporation of bromodeoxyuridine (BrdU) into the cells in 24-well plates, after purging for 6 h, was measured by using a cell proliferation enzyme-linked immunosorbent assay (ELISA) for BrdU (chemiluminescence) (Roche).
Apoptosis assay.
For quantitation of apoptosis, after the BM cells were cocultured with MMC-treated MEFs for a week, the incorporation of fluorescein isothiocyanate (FITC)-dUTP into floating cells by terminal deoxynucleotidyltransferase (TdT) was measured using the Mebstain Apoptosis Kit Direct (MBL International). Floating cells were also treated with annexin V-FITC and propidium iodide (PI), and the incorporation of annexin V was measured by using an annexin V-FITC apoptosis detection kit (Sigma).
Stable transfection.
For stable transfection, Med1−/− p53−/− MEFs (5 × 105) were transfected with 100 ng of human MED1 (hMED1) cDNA in the pIREShyg2 expression vector (Clontech) and Lipofectamine (Invitrogen) on a 6-cm dish and cultured in DMEM with 10% FBS and 180 μg/ml hygromycin B (Wako Chemicals) for 4 weeks. The resultant colonies were harvested and mixed.
Luciferase reporter assay and mammalian two-hybrid assay.
The cDNAs for activators (20 ng of hVDR, 100 ng of hRunx2) and wild-type or mutant hMED1 (100 ng), cloned into cytomegalovirus (CMV) promoter-driven mammalian expression vector pcDNA3.1 (Invitrogen) or pIRESneo (Clontech), and the Opn promoter (−793 to +79), amplified from genomic DNA by using KOD FX (Toyobo, Japan) and cloned into firefly luciferase reporter pGL4.10 (300 ng; Promega), were transfected together with the Renilla control luciferase vector (5 ng) into MEFs using Lipofectamine. After 48 h, the reporter activities were measured by using the dual-luciferase reporter assay system (Promega) and normalized to the control Renilla luciferase activity (25, 31). Mammalian two-hybrid assays were similarly performed by transfection with the Gal4-fused hMED1 expression vector, the VP16-fused hVDR or hRunx2 expression vector, and the reporter (pGL3; Promega) containing five Gal4 binding sites (100 ng each).
Quantitation of mRNA.
For the genome-wide gene expression analysis by microarray, the GeneChip Mouse Genome 430A 2.0 array (Affymetrix) was used for comparison of expression profiling between Med1+/+ p53−/− and Med1−/− p53−/− MEFs.
For semiquantitative PCR and quantitative PCR (qPCR), total RNAs (1 μg) were used to prepare cDNAs with the ReverTra Ace qPCR reverse transcription (RT) kit (Toyobo). The expression of various mouse genes was identified by either semiquantitative PCR or qPCR (7300 real-time PCR system; Applied Biosystems). Mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the reference marker. The sequences of the primers used and the condition of PCR for amplification are available upon request.
For Northern blot analysis, total RNAs (10 μg) were electrophoresed, transferred onto a nitrocellulose membrane, and probed with [32P]dCTP-labeled cDNA probes (12).
Western blot analysis and ELISA.
For the Western blot analysis, total cell lysates were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and probed with polyclonal antibodies (12). For quantitation of mouse OPN (mOPN), ELISA was performed by using the Quantikine mouse osteopontin immunoassay (R&D Systems).
ChIP assay.
For the chromatin immunoprecipitation (ChIP) assay, MEFs in 10-cm dishes, with or without transient transfection with FLAG-tagged hMED10/NUT2 in the pIREShyg2 expression vector, were fixed with 1% formalin for 10 min and used to prepare chromatin by enzymatic shearing according to the ChIP-IT Express manual (Active Motif). The samples were immunoprecipitated with anti-FLAG (M2; Sigma), -mVDR (C-20), and -mRunx2 (M-70) antibodies (Santa Cruz) and used to amplify the following Opn regions: promoter region, −211 to +79; 5′ remote region, −5781 to −5583.
Assay of ALP activity.
For alkaline phosphatase (ALP) activity, cell homogenates were incubated in 0.1 M 2-amino-2-methy-1-propanol, 1 mM MgCl2, and 8 mM p-nitrophenylphosphate disodium at 37°C for 30 min. Then the reaction was stopped with 0.1 M NaOH and the OD405 was measured. A standard curve was prepared with p-nitrophenol.
Statistical analysis.
The significance of differences between independent means was assessed by Student's t test. We considered a P value of <0.05 to be statistically significant.
Microarray data accession number.
The raw microarray data have been deposited in a minimum information about a microarray experiment (MIAME)-compliant database (GEO) under accession number GSE22471.
RESULTS
MEFs as an in vitro niche model.
Since the use of Med1−/− BM stromal cells was unfeasible (12), MEFs were employed as an in vitro model to analyze the role of MED1 in the niche. MEFs are mesenchymal cells with features reminiscent of osteoblastic precursors and are known to support HSPCs and thus mimic the osteoblastic niche. Stable MEF lines prepared from Med1+/+ p53−/− and Med1−/− p53−/− embryos derived from a single female were used for the subsequent experiments unless otherwise specified.
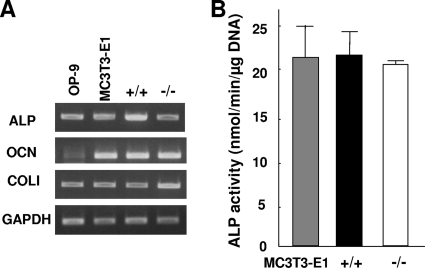
The Med1+/+ and Med1−/− MEFs (p53−/−), as well as MC3T3-E1 and OP-9 cells, expressed comparable levels of mRNAs for the osteoblastic markers, including ALP and type 1 collagen α1 (COLI) (Fig. 1A). The ALP activities of both the Med1+/+ and Med1−/− MEFs were also comparable to that of the MC3T3-E1 cells (Fig. 1B). The primary MEFs of each genotype prepared by Med1 heterozygous crossing (p53+/+) exhibited similar mRNA expression patterns (data not shown). Thus, the Med1+/+ and Med1−/− MEFs showed expression signatures attributable to the osteoblastic lineage.
FIG. 1.
MEFs express molecules attributable to the osteoblastic lineage. (A) Both Med1+/+ and Med1−/− MEFs (p53−/−), as well as MC3T3-E1 and OP-9 cells, express mRNAs of ALP, osteocalcin (OCN), and COLI. (B) The ALP activities of Med1+/+ and Med1−/− MEF lysates are comparable to those of MC3T3-E1 cells. The values are means ± standard deviations (SD) of a representative experiment performed in duplicate.
MEF MED1 mediates the mitogenic signal(s) to BM cells.
If MED1 in MEFs is required for hematopoietic support in culture, the maintenance or proliferation of BM cells on Med1−/− MEFs might be affected. To test this possibility, we first performed short-term BM coculture on Med1+/+ and Med1−/− MEFs.
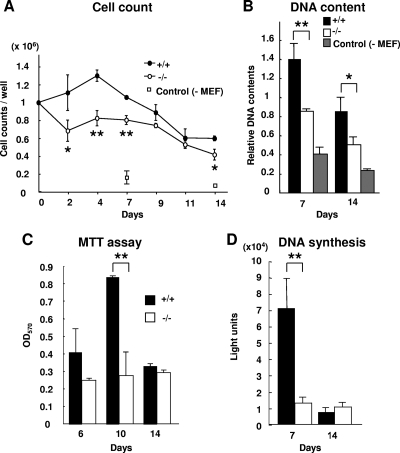
When normal mouse BM cells were cocultured on MMC-treated Med1+/+ or Med1−/− MEFs for 2 weeks, the numbers of cells on the Med1−/− MEFs were lower than the numbers of cells on the Med1+/+ MEFs but were higher than the number of cells cultured without MEFs (Fig. 2A). The DNA content of the total BM cells on the Med1−/− MEFs was less than that of cells on the Med1+/+ MEFs but more than that of the cells grown without MEFs, confirming the influence of MEFs and MED1 on the number of BM cells (Fig. 2B). The MTT assay disclosed attenuation of live cells on the Med1−/− MEFs (Fig. 2C). Thus, it appeared that the MEFs conveyed a signal(s) to the BM cells that either enhanced cell growth or inhibited cell death and that the putative signal(s) mediated by the Med1+/+ MEFs was stronger than that by the Med1−/− MEFs.
FIG. 2.
MED1 in MEFs mediates mitogenic stress to cocultured BM cells. (A and B) The numbers (A) and DNA content (B) of the total BM cells cultured on the Med1−/− MEFs are less than those of cells cultured on Med1+/+ MEFs but more than those of cells cultured without MEFs. (C and D) The number of live BM cells, measured by the MTT assay (C), and the level of BM cell DNA synthesis, measured by BrdU incorporation (D), were less when cells were cocultured on Med1−/− MEFs than when they were cultured on Med1+/+ MEFs. The values are means ± SD of a representative experiment performed in triplicate (*, P < 0.05; **, P < 0.01).
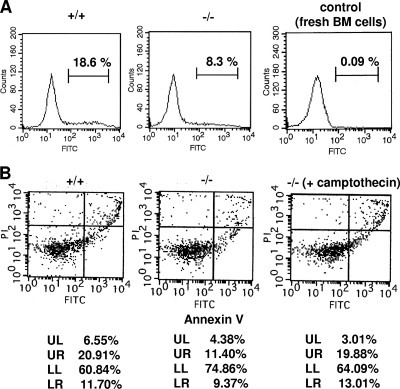
To assess the reason for the difference in the numbers of BM cells on the Med1+/+ and Med1−/− MEFs, DNA synthesis and cell death were measured. The incorporation of BrdU into the BM cells was lower on the Med1−/− MEFs than on the Med1+/+ MEFs after 1 week and prominently decreased on both types of MEFs after 2 weeks, possibly due to terminal differentiation and/or senescence of (most of) the dividing cells (Fig. 2D). TdT-mediated dUTP incorporation demonstrated that the amount of apoptosis of BM cells on the Med1−/− MEFs was less than that of BM cells on the Med1+/+ MEFs (8.3% versus 18.6%) (Fig. 3A). Annexin V and PI double staining revealed that the amounts of both apoptotic cells (9.37% versus 11.70%) (Fig. 3B, lower right quadrant) and necrotic cells (11.40% versus 20.91%) (Fig. 3B, upper right quadrant) were less on the Med1−/− MEFs than on the Med1+/+ MEFs. These results indicated that the BM cells on the Med1+/+ MEFs, in contrast to those on the Med1−/− MEFs, actively proliferated and subsequently died as a result of the mitogenic signal(s) that the MEFs transmitted to the BM cells.
FIG. 3.
BM cells on Med1−/− MEFs are less susceptible to cell death. (A) Incorporation of FITC-dUTP into BM cells by TdT. Normal BM cells were used as the negative control. (B) Annexin V-FITC and PI double staining. Cells treated with 50 nM camptothecin for 4 h were used as the positive control. BM cells cocultured for a week on Med1+/+ MEFs are more susceptible to apoptosis (A and B; LR) and necrosis (B; UR).
MEF MED1 mediates support of long-term culture-initiating cells.
We next conducted a long-term BM culture. After BM coculture on Med1+/+ or Med1−/− MEFs for 6 weeks, long-term culture-initiating cells (LTC-ICs), which quantified HSPCs and had the ability to form discrete colonies, were counted by colony-forming cell (CFC) assays using methylcellulose differentiation medium.
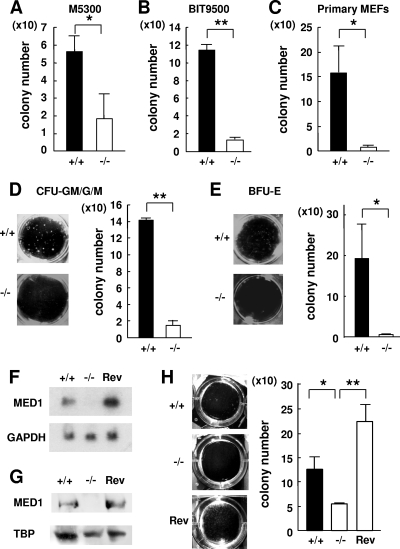
There were fewer CFCs on the Med1−/− MEFs than on the Med1+/+ MEFs (p53−/−) after BM coculture in both the Myelocult M5300 (Fig. 4A)- and BIT9500-based (Fig. 4B) long-term culture media. To exclude the possibility that the p53−/−-based MEFs might be affected by the additional phenotype(s) caused by p53 deficiency, we repeated the same experiments thrice by using primary MEFs prepared from a Med1+/− (p53+/+) crossing and confirmed the results (Fig. 4C). Therefore, the decreased number of LTC-ICs on the Med1−/− MEFs was attributed to the intrinsic MED1 deficiency. Moreover, the numbers of both the myeloid and the erythroid CFU on the Med1−/− MEFs were profoundly attenuated (Fig. 4D and E). However, the numbers of colonies on the Med1−/− MEFs into which MED1 was reintroduced (Rev-Med1−/− MEFs) (Fig. 4F and G) were restored to the control level (Fig. 4H). These results strongly indicated that MED1 in MEFs has a crucial role in activating the transcription of an mRNA-encoding molecule(s) having the potency of HSPC support.
FIG. 4.
MED1 in MEFs mediates support of LTC-ICs. (A and B) After a 6-week coculture, the numbers of CFCs on Med1+/+ MEFs (p53−/−) in M5300 medium (A) and BIT9500-based medium (B) are higher than the numbers of CFCs on Med1−/− MEFs (p53−/−). (C) After a 6-week coculture, the number of CFCs on primary Med1+/+ MEFs (p53+/+) is higher than numbers of CFCs on primary Med1−/− MEFs (p53+/+). (D and E) The numbers of myeloid (D) and erythroid (E) colonies on Med1+/+ MEFs (p53−/−) in M5300 medium are higher than the numbers of colonies on Med1−/− MEFs (p53−/−). (F and G) Northern (F) and Western (G) blot analyses of the Med1+/+, Med1−/−, and Rev-Med1−/− MEFs. GAPDH (F) and TATA-binding protein (TBP) (G) were used as a control. (H) The CFCs on the Rev-Med1−/− MEFs recovered to the level on the Med1+/+ MEFs. The values are means ± SD of a representative experiment performed in triplicate (*, P < 0.05; **, P < 0.01).
Attenuated expression of Opn mRNA in Med1−/− MEFs.
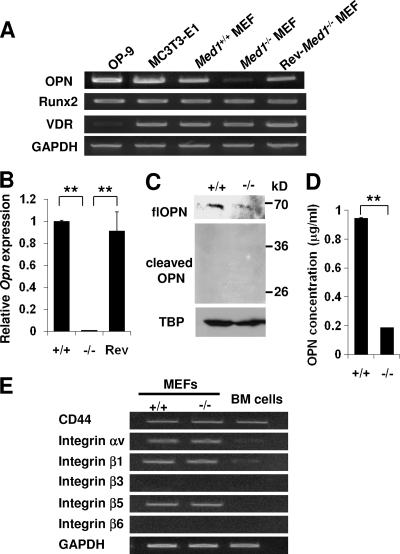
A microarray analysis of mRNA comparing the Med1+/+ and Med1−/− MEFs disclosed approximately 15 genes whose expression levels were profoundly attenuated (data deposited in GEO, accession number GSE22471). Among the molecules encoded by these genes, only OPN was a known molecule that has activity in niche (2, 16). The level of Opn mRNA was high in Med1+/+ MEFs, similar to that in the MC3T3-E1 and OP-9 cells; it was significantly downregulated in the Med1−/− MEFs and restored to the control level in the Rev-Med1−/− MEFs (Fig. 5 A and B). The transcripts of Runx2 and VDR, which activate transcription on the Opn promoter (25), were comparable. Western blot analysis of the Med1+/+ MEFs using a polyclonal antibody that recognized the N terminus of mOPN disclosed abundant expression of flOPN (∼70 kDa), which was much less in the Med1−/− MEFs. In contrast, trOPN was barely visible in both types of MEFs (Fig. 5C). The quantity of OPN secreted to the culture media by the Med1−/− MEFs was also much less (Fig. 5D). These data indicated that MED1 might act as a positive cofactor on the Opn promoter and that OPN might play a role in niche function in MEFs.
FIG. 5.
Expression of flOPN is attenuated in Med1−/− MEFs. (A and B) Semiquantitative (A) and quantitative (B) PCR. Opn mRNA is suppressed in the Med1−/− MEFs but recovers in the Rev-Med1−/− MEFs. (C) Western blot analysis. Expression of flOPN is reduced in the Med1−/− MEFs; trOPN is invisible. (D) ELISA of OPN. OPN concentrations in culture media during LTC-IC assays were measured. OPN produced by the Med1−/− MEFs is reduced. (E) Expression of various receptors for OPN in MEFs and BM cells was analyzed by semiquantitative PCR.
Next, the expression levels of various OPN receptors in both MEFs and BM cells were analyzed. Semiquantitative PCR analyses disclosed that MEFs express both αvβ1 and αvβ5 integrins and CD44, while BM cells express mainly CD44 (Fig. 5E).
MEF OPN-mediated mitogenicity of BM cells.
We next asked whether OPN is responsible for the phenotypes of the Med1−/− MEFs. To answer this question, we first analyzed the effect of OPN on the mitogenicity of cocultured BM cells.
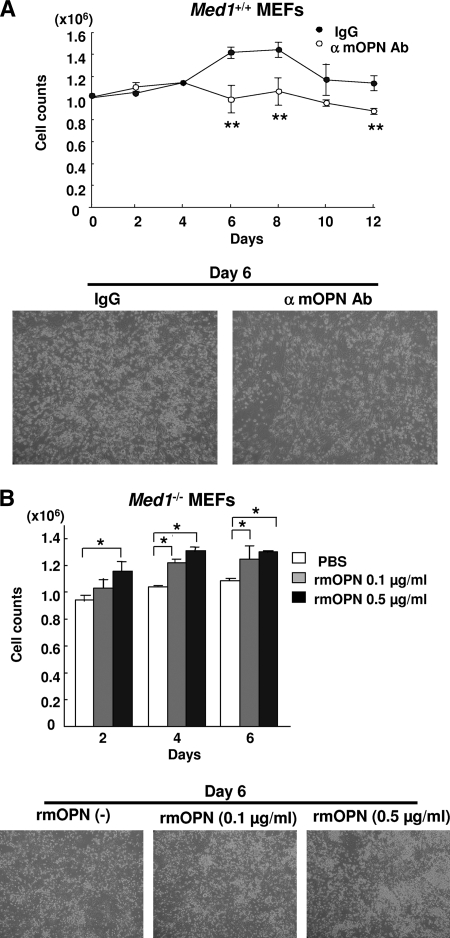
When BM cells on Med1+/+ MEFs were cocultured in the presence of either anti-OPN IgG or control IgG, the number of BM cells cultured with anti-OPN IgG was significantly lower than the control (Fig. 6A). Prompted by this observation, which could be interpreted to have resulted from a specific blocking effect of the antibody, we employed a BM culture on Med1−/− MEFs in the presence or absence of recombinant murine OPN (rmOPN). As expected, the number of BM cells increased in an rmOPN dose-dependent manner (Fig. 6B). Taken together, these data strongly suggested that OPN produced by MEFs had mitogenic activity on the cocultured BM cells.
FIG. 6.
OPN mediates mitogenicity of BM cells on MEFs. (A) The number of BM cells cocultured on Med1+/+ MEFs in the presence of the anti-mOPN antibody (Ab) is attenuated. α, anti. (B) The number of BM cells cocultured on Med1−/− MEFs increases in an rmOPN dose-dependent manner. PBS, phosphate-buffered saline. The values are the means ± SD of a representative experiment performed in triplicate (*, P < 0.05; **, P < 0.01).
OPN in MEFs mediated LTC-IC support.
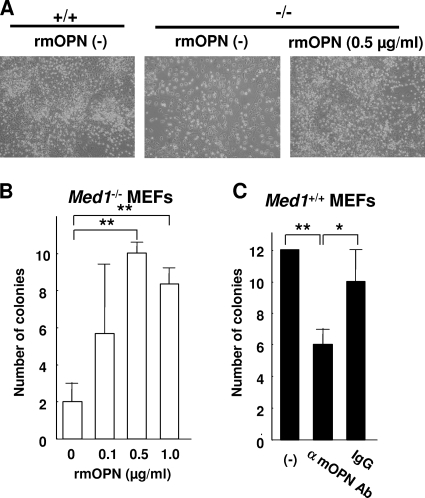
To verify the possibility that OPN produced by MEFs plays a role in HSPC support in BM culture, we first cultured BM cells on Med1−/− MEFs in Myelocult M5300 medium in the absence or presence of rmOPN for 8 weeks and counted the resulting LTC-ICs by colony formation assay. The cells on the Med1−/− MEFs were scarce when rmOPN was not present but increased to the control level when it was present (Fig. 7A). Trypan blue staining showed that these BM cells were viable. More importantly, the number of LTC-ICs increased in an rmOPN dose-dependent manner (Fig. 7B). A similar BM culture conducted on Med1+/+ MEFs in the absence or presence of either rabbit anti-mOPN IgG or whole IgG disclosed the attenuation of LTC-ICs specifically in the presence of anti-mOPN IgG, possibly through its blocking effect on OPN secreted by MEFs (Fig. 7C). Together, these results confirmed the role of OPN in HSPC support in a MEF-based long-term BM culture.
FIG. 7.
OPN mediates the support of LTC-ICs on MEFs. (A) The number of BM cells on Med1−/− MEFs in the presence of rmOPN recovers to the control level. (B) The number of CFCs on Med1−/− MEFs increases in the presence of rmOPN in a dose-dependent manner. (C) The number of CFCs on Med1+/+ MEFs is reduced in the presence of the anti-mOPN antibody. The values are the means ± SD of a representative experiment performed in triplicate (*, P < 0.05; **, P < 0.01).
BM stromal cell OPN-mediated growth of BM cells and LTC-IC support.
To exclude the possibility that the phenotypes described so far are specific to MEFs, we performed similar experiments by using MS-5 and OP-9 mouse BM stromal cells, established independently from the mouse BM stroma, because these cells are considered to represent a BM niche and have the capability for long-term HSPC support in vitro (6, 14).
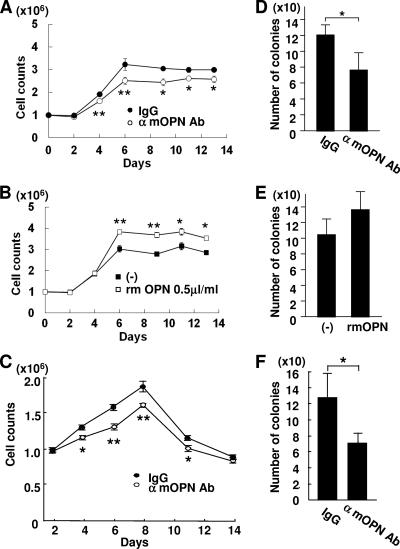
As expected, when BM cells were cocultured with MMC-treated MS-5 or OP-9 BM stromal cells for 2 weeks, the addition of the anti-OPN antibody to these cells attenuated the growth of the cocultured BM cells and the addition of rmOPN to the MS-5 cells enhanced growth (Fig. 8A, B, and C). Similarly, when BM cells were likewise cocultured for 6 weeks, the number of LTC-ICs was attenuated by the addition of the anti-OPN antibody to these stromal cells (Fig. 8D and F). These data further supported the MEF-based conclusion that OPN has a role in BM cell growth and HSPC support in vitro.
FIG. 8.
OPN depletion attenuates the mitogenicity of BM cells and LTC-ICs cocultured on BM stromal cells. (A to C) BM cells on MS-5 (A and B) and OP-9 (C) BM stromal cells during a 2-week period were counted in the presence of the anti-mOPN antibody (A and C) or rmOPN (B). (D to F) CFCs after a 6-week coculture on MS-5 (D and E) and OP-9 (F) BM stromal cells were counted in the presence of the anti-mOPN antibody (D and F) or rmOPN (E). The numbers of BM cells and CFCs on stromal cells are reduced in the presence of the anti-mOPN antibody, and the number of BM cells increases in the presence of rmOPN. The values are the means ± SD of a representative experiment performed in triplicate (*, P < 0.05; **, P < 0.01).
MED1-mediated transcription initiation on the Opn promoter.
To assess the role of MED1 on the Opn promoter, luciferase reporter assays were employed using the Opn promoter (−793 to +79), which includes consensus sequences for two well-described activators: a VDR-responsive element (VERE; −757 to −743) and a binding site for Runx2 (−136 to −130) (25). As both VDR and Runx2 are known to play an important role in Opn transcription within osteoblasts and bone biology, we hypothesized that these activators also have a role in niche function and analyzed the coactivation function of MED1 on the Opn promoter in the context of these activators.
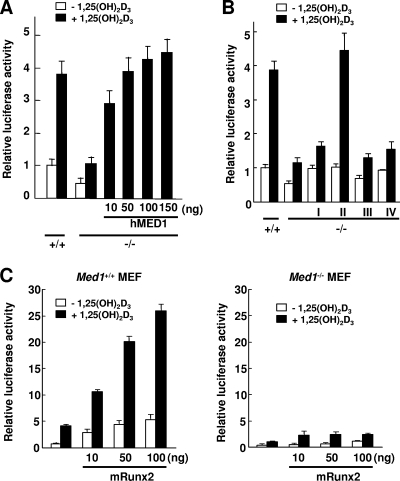
The reporter assays showed that the basal level of transcription in the Med1−/− MEFs in the presence of VDR but in the absence of its ligand was one-half of the level in the Med1+/+ MEFs (Fig. 9A). After the addition of 10−7 M 1,25-dihydroxyvitamin D3 [1,25(OH)2D3], approximately 4-fold activation of reporter activity was observed in the Med1+/+ MEFs whereas only a little (2-fold, one-fourth of the control level) activation was observed in the Med1−/− MEFs. Thus, both ligand-dependent and -independent activation exists in VDR- and MED1-mediated transcription on the Opn promoter. The introduction of exogenous hMED1 into the Med1−/− MEFs rescued ligand-dependent activation in a hMED1 dose-dependent manner (Fig. 9A). Various hMED1 mutants were cotransfected to dissect the MED1 domain(s) responsible for ligand-dependent and -independent activation (Fig. 9B). MED1 has two closely located LXXLL nuclear receptor recognition motifs (NR boxes) (13). Mutant I [MED1(1-602)], lacking NR boxes, was capable of restoring basal transcription to the level of that in the wild-type control but lacked recovery of ligand-dependent activation. In contrast, mutant II, having NR boxes [MED1(1-703)] showed full recovery of both basal activity and ligand-induced activation. However, mutant III [MED1(592-1587)], incapable of complex formation (8), was unable to recover any activity. Finally, mutant IV, the full-length MED1 with two inactivated NR boxes (LXXLL to LXXAA), did recover basal transcription but not ligand dependency. These data suggested that the basal level of Opn transcription depends on MED1(1-602) and that ligand-dependent activation is critically dependent on the NR boxes.
FIG. 9.
MED1 mediates VDR- and Runx2-mediated transcription on the Opn promoter. Luciferase reporter assays were performed in the presence of VDR. (A) Ligand-dependent activation is attenuated in the Med1−/− MEFs but recovers in a MED1 dose-dependent manner. (B) Reintroduction of mutants I [MED1(1-602)]) and IV (MED1 with NR box mutations) into Med1−/− MEFs recovers the basal level of transcription but not ligand-dependent activation, whereas reintroduction of mutant II [MED1(1-703)] recovers both basal transcription and ligand-induced activation. Mutant III [MED(592-1587)] has no effect. (C) Addition of Runx2 activates both ligand-independent and -dependent activation in the Med1+/+ MEFs but not in the Med1−/− MEFs. The values (means ± SD of a representative experiment performed in triplicate) are plotted as a fold increases against the value for Med1+/+ MEFs without a ligand.
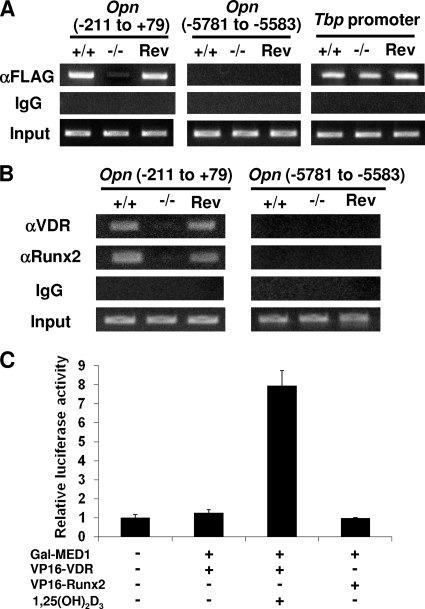
VDR and Runx2 cooperate in Opn transcription (25). We next analyzed the role of Runx2 on the Opn promoter in the context of VDR by luciferase reporter assays. Both the basal level of transcriptional activation and 1,25(OH)2D3-dependent transcriptional activation were prominently promoted in a Runx2 dose-dependent manner in the Med1+/+ MEFs. However, in the Med1−/− MEFs, Runx2-driven activation was significantly attenuated in the presence of VDR (Fig. 9C). A ChIP assay of MEFs proved that Mediator recruitment was attenuated in the Med1−/− MEFs but was recovered in the Rev-Med1−/− MEFs (Fig. 10A), confirming MED1 (or Mediator) as a bona fide coactivator on this promoter. These data suggested that VDR and Runx2 cooperate to regulate Opn transcriptional activation and that MED1 is crucial for this activity.
FIG. 10.
Recruitment of MED1, VDR, and Runx2 on the Opn promoter. (A and B) ChIP assays. (A) FLAG-MED10 was transiently expressed, and sheared chromatin was immunoprecipitated with anti-FLAG (M2) IgG or mouse IgG as the control. On the Opn promoter proximal to the transcription initiation point, Mediator recruitment is attenuated in the Med1−/− MEFs but recovered in the Rev-Med1−/− MEFs. Mediator is not recruited to the 5′ remote region. Mediator recruitment on the Tbp promoter is comparable. (B) VDR and Runx2 recruitment is attenuated in the Med1−/− MEFs but recovers in the Rev-Med1−/− MEFs. (C) Mammalian two-hybrid assays. Luciferase activities of the reporter with 5 Gal4-binding sites were measured. Gal4-fused MED1 interacts with VP16-fused VDR in a ligand-dependent manner, but not with VP16-fused Runx2.
The binding of VDR and Runx2 to the Opn promoter and to the MED1 complex in these MEFs was then examined. ChIP assays confirmed the recruitment of both these activators onto the Opn promoter (Fig. 10B). Mammalian two-hybrid assays using Med1+/+ MEFs disclosed the direct interaction of MED1 with VDR in a ligand-dependent manner, but not with Runx2, in these cells (Fig. 10C).
MED1 domain that is responsible for BM cell growth.
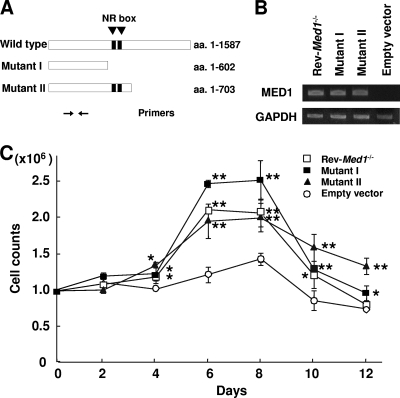
We then aimed to determine whether the attenuated cell growth stress on the Med1−/− MEFs (above) can be rescued by reintroducing MED1 as well, and if so, which MED1 domain is responsible for this function. To address this issue, we stably transfected Med1−/− MEFs expressing either full-length hMED1 (Rev-Med1−/−) or its mutant I or II (Fig. 11A and B).
FIG. 11.
MED1(1-602) in MEFs is sufficient for the mitogenicity of BM cells. (A) Primer positions for PCR. (B) Semiquantitative PCR shows stable expression of the MED1 mutants in the Med1−/− MEFs. (C) Mutants I and II as well as the Rev-Med1−/− MEFs recover the mitogenicity of BM cells. The values are the means ± standard errors (SE) of a representative experiment performed in triplicate (*, P < 0.05; **, P < 0.01).
The Rev-Med1−/− MEFs mediated more growth of BM cells compared to the Med1−/− MEFs to which the empty expression vector was transfected (Fig. 11C). More intriguingly, recovery of BM cell growth was observed when the cells were cultured on the Med1−/− MEFs to which mutant I or II was transfected (Fig. 11C). These data indicated that N-terminal MED1 in MEFs is sufficient for BM cell growth in this setting.
DISCUSSION
We determined the role of MED1 in a hematopoietic niche model. Our findings suggest that MED1 in stromal cells may have activity to support HSPCs, that Opn is the target gene of MED1, and that OPN may be responsible for the activity of MED1. This is the first demonstration that the general transcriptional coactivator complex Mediator has a role in niche cells for maintenance of stem cells.
OPN as a candidate for mitogenic stress of BM cells and support of HSPCs.
Although HSPCs in the endosteal niche are maintained in the G0 or slowly cycling state by various cell adhesion molecules and chemokines in a physiological setting (28), they must enter the cell cycle under conditions where the reservoir of HSPCs becomes smaller. The osteoblastic niche might have a mechanism for producing (symmetric) proliferative stress on HSPCs in such a situation. The attenuated BM cell growth and LTC-ICs on the Med1−/− MEFs suggest that MED1 in MEFs is involved in the proliferation of HSPCs by producing some molecule(s) that promotes cell cycling of HSPCs either directly or indirectly through another molecule(s) in a paracrine or autocrine manner. OPN might be a candidate molecule for this action. OPN is significantly downregulated in the Med1−/− MEFs, whereas other niche molecules, including angiopoietin-1, Jagged-1, N-cadherin, Wnt, and BMP-4, remain unchanged (data not shown). This contrasts with the Opn knockouts, where angiopoietin-1 and Jagged-1 are overexpressed and the number of HSPCs increases. The fact that flOPN in the niche both potentiates BM cell growth and supports HSPCs in vitro (this study) indicates that OPN can induce mitogenic stress for differentiating both BM cells and more immature (stem/precursor) cells either directly or indirectly. The candidate direct target on BM cells may be CD44, while CD44 and some integrins expressed on MEFs might function in an autocrine or paracrine manner.
Another explanation is that MEFs secrete into the microenvironment either a single molecule that has dual roles or multiple molecules that have two distinct roles, namely, roles in (i) cell growth stress on differentiating BM cells and (ii) quiescence or maintenance of HSPCs. OPN might influence both these roles either directly or indirectly through action on MEFs in an autocrine manner and activation of another molecule(s) with these roles. Several chemokines and growth factors abundantly expressed in the presence of MED1, including CXCL5, CXCL15, and fibroblast growth factor 7 (FGF7) (GEO accession number GSE22471), might be responsible for the mitogenicity. The action of these chemokines on BM cells in the presence of OPN must be carefully examined in the future.
The expression of both flOPN and trOPN is reportedly restricted to the endosteal region and plays a critical role in regulating the physical location and maintenance of HSCs (9, 23, 26). It is reported that trOPN, but not flOPN, has a restricting effect on the number of LTC-ICs (9, 23). These forms of OPN use distinct sets of receptors and probably have different biological outputs (9). For instance, production of more quiescence molecules, such as angiopoietin-I, in Opn null mice might reflect the lack of trOPN. In contrast, OPN is known to underscore mitogenicity in other settings, particularly in tumor biology. OPN produced by stromal cells serves as a niche molecule, has mitogenic activity, and promotes preneoplastic cell growth and tumorigenesis (3, 19, 24).
Another important question is the role of OPN produced by osteoclasts and macrophages. Osteoclasts produce abundant OPN and might partially contribute to the niche function, possibly explaining the fact that genetic ablation of osteoblasts does not deplete HSCs immediately (32).
MED1 is a specific coactivator on the Opn promoter.
The findings of downregulated Opn mRNA in the Med1−/− MEFs and the results of the luciferase and ChIP assays strongly suggest the direct involvement of MED1 on the Opn promoter. In terms of the mechanism, our data clearly show that (i) MED1 is a bona fide important ligand-dependent coactivator for VDR, (ii) Runx2 acts synergistically as an activator with VDR in a MED1-dependent manner, and (iii) MED1(1-602), missing the VDR interaction site, is a coactivator for a putative activator(s) that maintains basal ligand-independent transcription. A candidate molecule(s) for the putative activator(s) might be CCAR1, which was recently reported to bypass nuclear receptor signaling (17), and/or an unknown activator(s); the presence of such an activator might explain the role of MED1(1-602) in MEFs in growth stress on BM cells (this study). Further molecular and functional dissection may resolve this issue in the future. The next issue is whether MED1 has the same role in the osteoblastic niche within a living animal. An osteoblastic-lineage-specific Med1 conditional knockout model will help answer this question in the future.
Role of MED1 in hematopoiesis.
MED1 plays an important role in multiple lineages of hematopoietic cell differentiation, namely, RAR-mediated myelopoiesis (31), VDR-mediated monopoiesis (31), GATA-1-mediated erythropoiesis (27), and megakaryopoiesis (22). MED1 interacts with GATA-3 as well (22); therefore, it is reasonable to predict that MED1 also acts as a coactivator for GATA-3 and plays a role in GATA-3-meditated T-cell differentiation. In addition, a recent study has defined MED1 as a specific coactivator for C/EBPβ, which is required for “emergency” granulopoiesis (11). Thus, MED1 apparently has a pivotal role in virtually all types of hematopoietic cell differentiation. In this regard our current study suggests a conceptually novel role for MED1 in hematopoiesis, namely, in the mitogenicity of hematopoietic cells and support of HSPCs through the action of MED1 in stromal cells as a coactivator on the Opn promoter. All these lines of evidence suggest a novel model for transcriptional control of hematopoiesis: MED1 as a key coactivator for the regulation of hematopoiesis both in hematopoietic cells and the microenvironment.
Acknowledgments
We thank T. Kasukawa and J. Nishio for performing the DNA microarray analysis, H. Kaji for MC3T3-E1 cells, K. Ito for MS-5 cells, J. Zhang for a VP16-based expression vector, I. Kashiwakura, A. Iwama, and S. Shiozawa for valuable advice and support, and H. Kato, M. Makishima, S. Asano, and members in our laboratory for helpful discussion.
This study was supported by grants from the MEXT, the Global Center for Excellence Program Global Center of Excellence for Education and Research on Signal Transduction Medicine in the Coming Generation from MEXT, the Takeda Science Foundation, the Sagawa Foundation for Promotion of Cancer Research, the Suzuken Memorial Foundation, and the ONO Medical Research Foundation (to M.I.) and by a grant (DK071900) from the NIH (to R.G.R.).
Footnotes
Published ahead of print on 16 August 2010.
REFERENCES
- 1.Adams, G. B., and D. T. Scadden. 2006. The hematopoietic stem cell in its place. Nat. Immunol. 7:333-337. [DOI] [PubMed] [Google Scholar]
- 2.Askmyr, M., N. A. Sims, T. J. Martin, and L. E. Purton. 2009. What is the true nature of the osteoblastic hematopoietic stem cell niche? Trends Endocrinol. Metab. 20:303-309. [DOI] [PubMed] [Google Scholar]
- 3.Chang, P. L., M. Cao, and P. Hicks. 2003. Osteopontin induction is required for tumor promoter-induced transformation of preneoplastic mouse cells. Carcinogenesis 24:1749-1758. [DOI] [PubMed] [Google Scholar]
- 4.Conaway, R. C., S. Sato, C. Tomomori-Sato, T. Yao, and J. W. Conaway. 2005. The mammalian mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30:250-255. [DOI] [PubMed] [Google Scholar]
- 5.Fan, X., D. M. Chou, and K. Struhl. 2006. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 13:117-120. [DOI] [PubMed] [Google Scholar]
- 6.Feugier, P., N. Li, D. Y. Jo, J. H. Shieh, K. L. MacKenzie, J. F. Lesesve, V. Latger-Cannard, D. Bensoussan, R. G. Crystal, S. Rafii, J. F. Stoltz, and M. A. Moore. 2005. Osteopetrotic mouse stroma with thrombopoietin, c-kit ligand, and flk-2 ligand supports long-term mobilized CD34+ hematopoiesis in vitro. Stem Cells Dev. 14:505-516. [DOI] [PubMed] [Google Scholar]
- 7.Ge, K., M. Guermah, C. X. Yuan, M. Ito, A. E. Wallberg, B. M. Spiegelman, and R. G. Roeder. 2002. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature 417:563-567. [DOI] [PubMed] [Google Scholar]
- 8.Ge, K., Y. W. Cho, H. Guo, T. B. Hong, M. Guermah, M. Ito, H. Yu, M. Kalkum, and R. G. Roeder. 2008. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor γ-stimulated adipogenesis and target gene expression. Mol. Cell. Biol. 28:1081-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grassinger, J., D. N. Haylock, M. J. Storan, G. O. Haines, B. Williams, G. A. Whitty, A. R. Vinson, C. L. Be, S. Li, E. S. Sørensen, P. P. L. Tam, D. T. Denhardt, D. Sheppard, P. F. Choong, and S. K. Nilsson. 2009. Thrombin-cleaved osteopontin regulates hemopoietic stem and progenitor cell functions through interactions with α9β1 and α4β1 integrins. Blood 114:49-59. [DOI] [PubMed] [Google Scholar]
- 10.Haylock, D. N., and S. K. Nilsson. 2006. Osteopontin: a bridge between bone and blood. Br. J. Haematol. 134:467-474. [DOI] [PubMed] [Google Scholar]
- 11.Hirai, H., P. Zhang, T. Dayaram, C. J. Hetherington, S. Mizuno, J. Imanishi, K. Akashi, and D. G. Tenen. 2006. C/EBPβ is required for ‘emergency’ granulopoiesis. Nat. Immunol. 7:732-739. [DOI] [PubMed] [Google Scholar]
- 12.Ito, M., C. X. Yuan, H. J. Okano, R. B. Darnell, and R. G. Roeder. 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5:683-693. [DOI] [PubMed] [Google Scholar]
- 13.Ito, M., and R. G. Roeder. 2001. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol. Metab. 12:127-134. [DOI] [PubMed] [Google Scholar]
- 14.Itoh, K., H. Tezuka, H. Sakoda, M. Konno, K. Nagata, T. Uchiyama, H. Uchino, and K. J. Mori. 1989. Reproducible establishment of hemopoietic supportive stromal cell lines from murine bone marrow. Exp. Hematol. 17:145-153. [PubMed] [Google Scholar]
- 15.Kiel, M. J., O. H. Yilmaz, T. Iwashita, O. H. Yilmaz, C. Terhorst, and S. J. Morrison. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121:1109-1121. [DOI] [PubMed] [Google Scholar]
- 16.Kiel, M. J., and S. J. Morrison. 2008. Uncertainty in the niches that maintain haematopoietic stem cells. Nat. Rev. Immunol. 8:290-301. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. H., C. K. Yang, K. Heo, R. G. Roeder, W. An, and M. R. Stallcup. 2008. CCAR1, a key regulator of Mediator complex recruitment to nuclear receptor transcription complexes. Mol. Cell 31:510-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornberg, R. D. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30:235-239. [DOI] [PubMed] [Google Scholar]
- 19.Krause, D. S., K. Lazarides, U. H. von Andrian, and R. A. Van Etten. 2006. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat. Med. 12:1175-1180. [DOI] [PubMed] [Google Scholar]
- 20.Li. H., P. Gade, S. C. Nallar, A. Raha, S. K. Roy, S. Karra, J. K. Reddy, S. P. Reddy, and D. V. Kalvakolanu. 2008. The Med1 subunit of transcriptional mediator plays a central role in regulating CCAAT/enhancer-binding protein-β-driven transcription in response to interferon-γ. J. Biol. Chem. 283:13077-13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik, S., and R. G. Roeder. 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30:256-263. [DOI] [PubMed] [Google Scholar]
- 22.Misra, P., E. D. Owuor, W. Li, S. Yu, C. Qi, K. Meyer, Y. J. Zhu, M. S. Rao, A. N. Kong, and J. K. Reddy. 2002. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J. Biol. Chem. 277:48745-48754. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson, S. K., H. M. Johnston, G. A. Whitty, B. Williams, R. J. Webb, D. T. Denhardt, I. Bertoncello, L. J. Bendall, P. J. Simmons, and D. N. Haylock. 2005. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood 106:1232-1238. [DOI] [PubMed] [Google Scholar]
- 24.Pazolli, E., X. Luo, S. Brehm, K. Carbery, J. J. Chung, J. L. Prior, J. Doherty, S. Demehri, L. Salavaggione, D. Piwnica-Worms, and S. A. Stewart. 2009. Senescent stromal-derived osteopontin promotes preneoplastic cell growth. Cancer Res. 69:1230-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen, Q., and S. Christakos. 2005. The vitamin D receptor, Runx2, and the Notch signaling pathway cooperate in the transcriptional regulation of osteopontin. J. Biol. Chem. 280:40589-40598. [DOI] [PubMed] [Google Scholar]
- 26.Stier, S., Y. Ko, R. Forkert, C. Lutz, T. Neuhaus, E. Grünewald, T. Cheng, D. Dombkowski, L. M. Calvi, S. R. Rittling, and D. T. Scadden. 2005. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J. Exp. Med. 201:1781-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stumpf, M., C. Waskow, M. Krötschel, D. van Essen, P. Rodriguez, X. Zhang, B. Guyot, R. G. Roeder, and T. Borggrefe. 2006. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc. Natl. Acad. Sci. U. S. A. 103:18504-18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suda, T., F. Arai, and A. Hirao. 2005. Hematopoietic stem cells and their niche. Trends Immunol. 26:426-433. [DOI] [PubMed] [Google Scholar]
- 29.Sugiyama, T., H. Kohara, M. Noda, and T. Nagasawa. 2006. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25:977-988. [DOI] [PubMed] [Google Scholar]
- 30.Taichman, R. S. 2005. Blood and bone: two tissues whose fates are intertwined to create the hematopoietic stem-cell niche. Blood 105:2631-2639. [DOI] [PubMed] [Google Scholar]
- 31.Urahama, N., M. Ito, A. Sada, K. Yakushijin, K. Yamamoto, A. Okamura, K. Minagawa, A. Hato, K. Chihara, R. G. Roeder, and T. Matsui. 2005. The role of transcriptional coactivator TRAP220 in myelomonocytic differentiation. Genes Cells 10:1127-1137. [DOI] [PubMed] [Google Scholar]
- 32.Visnjic, D., Z. Kalajzic, D. W. Rowe, V. Katavic, J. Lorenzo, and H. L. Aguila. 2004. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood 103:3258-3264. [DOI] [PubMed] [Google Scholar]
- 33.Wada, O., H. Oishi, I. Takada, J. Yanagisawa, T. Yano, and S. Kato. 2004. BRCA1 function mediates a TRAP/DRIP complex through direct interaction with TRAP220. Oncogene 23:6000-6005. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, A., and A. Trumpp. 2006. Bone-marrow hematopoietic-stem-cell niches. Nat. Rev. Immunol. 6:93-106. [DOI] [PubMed] [Google Scholar]
- 35.Yokosaki, Y., K. Tanaka, F. Higashikawa, K. Yamashita, and A. Eboshida. 2005. Distinct structural requirements for binding of the integrins αvβ6, αvβ3, αvβ5, α5β1 and α9β1 to osteopontin. Matrix Biol. 24:418-427. [DOI] [PubMed] [Google Scholar]