Abstract
A screen of microRNA (miRNA) expression following differentiation in human foreskin keratinocytes (HFKs) identified changes in several miRNAs, including miRNA 203 (miR-203), which has previously been shown to play an important role in epithelial cell biology by regulating p63 levels. We investigated how expression of human papillomavirus type 16 (HPV16) oncoproteins E6 and E7 affected miR-203 expression during proliferation and differentiation of HFKs. We demonstrated that miR-203 expression is reduced in HFKs where p53 function is compromised, either by the viral oncoprotein E6 or by knockout of p53 using short hairpin RNAs (p53i). We show that the induction of miR-203 observed during calcium-induced differentiation of HFKs is significantly reduced in HFKs expressing E6 and in p53i HFKs. Induction of miR-203 in response to DNA damage is also reduced in the absence of p53. We report that proliferation of HFKs is dependent on the level of miR-203 expression and that overexpression of miR-203 can reduce overproliferation in E6/E7-expressing and p53i HFKs. In summary, these results indicate that expression of miR-203 is dependent on p53, which may explain how expression of HPV16 E6 can disrupt the balance between proliferation and differentiation, as well as the response to DNA damage, in keratinocytes.
MicroRNAs (miRNAs) are small, noncoding RNA molecules that can regulate protein expression at the posttranscriptional level by targeting mRNAs for degradation or translational repression (13). Over the past decade, a growing body of evidence has shown that they play a fundamental role in the development, function, and maintenance of tissues and cells in various organisms. Their importance is perhaps best demonstrated by studies on the RNase Dicer, which plays an integral role in the processing and generation of mature miRNA molecules, as mutations in Dicer cause severe developmental defects in Caenorhabditis elegans (22), while knockout of Dicer in mice is embryonically lethal (3). Moreover, it is now known that many miRNAs are implicated in several disease states, including heart disease (2, 23), viral infection (38), and many different cancers (7, 44), leading to increased interest in the biology and function of individual miRNAs in various cell processes.
In skin physiology, miRNAs have been shown to be involved both in normal processes, including epidermal development and hair follicle morphogenesis (48), and in skin-specific pathologies, such as psoriasis (37), systemic lupus erythematous (SLE) (8), and skin carcinogenesis (14). The importance of miRNAs in skin epithelial development is particularly emphasized when their expression is repressed by epidermal cell-specific deletion of Dicer in mouse models, which results in several defects, such as epidermal evagination and abnormal hair follicle development, although it is worth noting that epidermal differentiation is apparently unaffected (1, 48). To date, several individual miRNAs that are highly expressed in the skin epithelium have been identified, including microRNA 203 (miR-203), which has emerged as a potentially key player in epithelial cell biology.
miR-203 is of particular interest because one significant target of miR-203 is the transcription factor p63, a p53 family member which is known to be critical in the development of stratifying epithelia in human (35) and mouse (32, 47) and which is now considered to be a key player in controlling the proliferation and differentiation of keratinocytes (41). Recent studies have shown that miR-203 is capable of directly repressing the expression of ΔNp63α, the predominant isoform of p63 in keratinocytes. This is demonstrated in vivo where miR-203 expression is detected in the differentiating layers of normal mouse epidermis but not in the basal layer, inversely correlating with the pattern of ΔNp63α expression in these cells (49). In vitro studies on both human and mouse keratinocytes show that miR-203 is strongly induced during calcium-induced differentiation, concomitant with a downregulation of ΔNp63α expression (25). Furthermore, both these studies also showed that miR-203 exerts a clear effect on keratinocyte proliferation. Transgenic mice expressing miR-203 demonstrate a repression of the proliferative capacity, while inhibition of miR-203 using a specific antagonist molecule (antagomir) results in increased epidermal proliferation (49). Similarly, in vitro overexpression of miR-203 in human keratinocytes inhibited cell proliferation, whereas treatment with an miR-203 antagomir results in increased proliferation (25). Together, these studies suggested that miR-203, through the regulation of p63 levels, plays a key role in switching between the maintenance of proliferative capacity in keratinocytes and their commitment to differentiation.
However, both these reports also acknowledged that the mechanisms underlying miR-203 control of p63 levels were likely to be complex, may include other miRNAs, and may be subject to other factors. One such factor may be p53, since there is evidence to suggest that p63 effects on keratinocyte proliferation are p53 dependent. Several studies have demonstrated that the inhibition of epidermal proliferation associated with loss of p63 expression is rescued by p53 knockdown in human and mouse keratinocytes (20, 42), as well as in a zebrafish model (24), with ΔNp63α displaying dominant negative activity against p53 (24, 46). Since p53 and p63 demonstrate little association with each other in vitro (10, 15), their opposing functions seem more likely to depend upon direct competition for similar binding sites. Indeed, several genes have been shown to be targeted by both proteins, such as the cell cycle inhibitor p21, which is repressed by p63 and induced by p53 (11), a mechanism which may be central to maintenance of keratinocyte proliferation, although the specifics of this process remain unclear. With regard to miRNAs, investigation of the involvement of p53 has focused primarily on its regulation of the miR-34 family, which several studies have shown to be direct targets of p53 in different cellular systems (4, 6, 17, 40). However, miR-203 has also been identified among a subset of p53-regulated miRNAs in breast (5) and colon (39) cancer cell lines, although to date this connection has not been investigated further.
This becomes important in considering the effect of human papillomavirus type 16 (HPV16) infection of keratinocytes, since impaired p53 function due to E6 activity may have an impact upon miRNA expression and could contribute to the initiation of the tumorigenic process. Therefore, in this study, we investigated how miR-203 expression is affected by expression of HPV oncoproteins in keratinocytes during proliferation and differentiation.
MATERIALS AND METHODS
Cell culture, infections, and transfections.
Primary human foreskin keratinocytes (HFKs) were isolated from neonatal foreskin, cultured under low-calcium conditions, and transduced with retrovirus produced in the ΦNYX-GP packaging cell line (ATCC) as previously described (18). Mutagenesis of E6 or E7 was performed as previously described to generate a stop codon at the 16th amino acid in the E7 gene of HPV16 (E6/E7s) (16), a stop codon at the 15th amino acid in the E6 gene of HPV16 (E6s/E7) (16), amino acid substitution F125L in the E6 gene (E6.F125L/E7) (9), amino acid substitution G134V in the E6 gene(E6.G134V/E7) (27), and amino acid deletion Δ123-7 in the E6 gene (E6.Δ123-7/E7) (21). Differentiation of HFK cell lines in organotypic raft cultures was carried out as previously described for transduced lines (30). The raft cultures were harvested, fixed in 4% paraformaldehyde (PFA), and then embedded in paraffin for subsequent sectioning and staining with hematoxylin and eosin (H&E). In order to label DNA-synthesizing cells, 20 μM bromodeoxyuridine (BrdU) was added to the raft culture 12 h prior to harvest. Cells in culture were pulsed for 20 min with 10 μm BrdU prior to fixation. For calcium-induced differentiation, confluent monolayers of HFKs were induced to differentiate by withdrawal of growth factors and addition of 1.5 mM CaCl2. Transfection of HFKs with anti-miR-203, pre-miR-203, and negative controls (all from Ambion, Warrington, United Kingdom) was performed using FuGene HD (Roche, Mannheim, Germany) following the manufacturer's protocols. Cells were transfected for 48 h with a final concentration of 50 nM. For small interfering RNA (siRNA) transfection, siRNAs were designed and synthesized (Dharmacon, CO) to target the p63 3′ untranslated region (UTR), adjacent to the region targeted by short hairpin RNA (shRNA), p53 (53i), and a green fluorescent protein (GFP)-Scr control (Scr). Transfection was performed at a final concentration of 200 nM using FuGene HD for up to 48 h. For DNA damage experiments, cycling HFKs were treated with doxorubicin (Dox) (2 μg/ml) for up to 24 h. H1299 cells were cultured in RPMI containing 10% fetal bovine serum (FBS). Transfection of H1299 cells with 3 μg wild-type and mutant p53 vectors (Clontech, France), with GFP vector as a control (Clontech), was performed using GeneJuice (Merck Biosciences, Nottingham, United Kingdom) following the manufacturer's protocols.
Immunofluorescence staining and analysis.
Indirect immunofluorescence staining of paraffin-embedded sections was performed following antigen retrieval with citrate buffer (Dako, Cambridgeshire, United Kingdom), using anti-BrdU (BD Biosciences, Oxford, United Kingdom), anti-p63(4A4) (Santa Cruz, CA), and Alexa-488-conjugated secondary reagents (Molecular Probes, TX). BrdU-pulsed cells on coverslips were fixed for 10 min with 4% paraformaldehyde, washed three times with phosphate-buffered saline (PBS), submitted to antigen retrieval, and stained as described above. For raft sections, number of BrdU-positive cells were counted for a minimum of 10 fields of view (1,000 μm). For coverslips, a minimum of 10 fields of view and >500 cells were counted for each slide. BrdU graphs represent means ± standard errors (SE) from three independent experiments, expressed relative to number of BrdU-incorporating cells in control experiments. In all cases immunofluorescent images were captured on a Leica AF6000 inverted fluorescence microscope with Leica AF imaging software.
Western blot analysis.
Protein lysates were electrophoresed and equal loading assessed by Ponceau Red staining following transfer to nitrocellulose membranes. Primary antibodies used for blotting were anti-p63(4A4) (Santa Cruz), anti-p53(DO-1) (Santa Cruz), anti-p21Cip1 (BD Biosciences), anti-K1 (Covance, Cambridge, United Kingdom), anti-K10 (Covance), and anti-β-actin (Sigma, Poole, United Kingdom) as a loading control. Secondary antibodies were goat anti-mouse- and anti-rabbit-horseradish peroxidase (HRP) (Santa Cruz). Luminescence was revealed by incubation with either Western Lightning Plus (Perkin-Elmer) or ECL Western blotting substrate (Pierce) and signal detected on an Alpha Innotech FluorChem SP imaging system.
Reverse transcription-PCR (RT-PCR) analysis.
RNA extraction was carried out with the High Pure RNA isolation kit (Roche) according to the manufacturer's instructions. One microgram of RNA was treated with RQ1 RNase-free DNase (Promega, Southampton, United Kingdom) prior to first-strand cDNA synthesis using random primers with the Transcriptor high-fidelity cDNA synthesis kit (Roche) according to manufacturer's instructions. For real-time PCR, amplification of PCR products was quantified using FastStart SYBR green Master (Roche) according to the manufacturer's instructions and fluorescence monitored on a DNA Engine Peltier thermal cycler (Bio-Rad) equipped with a Chromo4 real-time PCR detection system (Bio-Rad). Melting curve analysis was also performed. The following cycling conditions were used: initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 58°C for 15 s, and 60°C for 60 s using primer sets for p63α (forward, 5′-CAGCACCAGCACTTACTTCAGA-3′; reverse, 5′-CCACCTCGGTAAGAAACTGA-3′), p53 (forward, 5′-TCAACAAGATGTTTTGCCAACTG-3′; reverse, 5′-ATGTGCTGTGACTGCTTGTAGATG-3′), and RPLPO (ribosomal protein, large, PO) (forward, 5′-ATCAACGGGTACAAACGAGTC-3′; reverse, 5′-CAGATGGATCAGCCAAGAAGG-3′). Expression levels were assessed in triplicate and normalized to RPLPO levels, and graphs represent the combined results for three independent biological replicates. For semiquantitative PCR, amplification of PCR products was performed using the GoTaq Master Mix kit (Promega). Cycling conditions were as follows: initial denaturation at 95°C for 5 min, followed by 28 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 60 s using primer sets for E6 (forward, 5′-GAGAGGATCCATGTTTCAGGACCCACAGG-3′; reverse, 5′-CATGAATCCTTACAGCTGGGTTTCTCTAC-3′), E7 (forward, 5′-CGGGATCCATCATGCATGGAGATAC-3′; reverse, 5′-GTGGATCCGGTTTCTGAGAACAGATGG-3′), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (forward, 5′-ATGGTGAAGGTCGGTGTGAA-3′; reverse, 5′-ATGACAAGCTTCCCATTCTC-3′).
microRNA real-time PCR.
Real-time quantitative PCR (RQ-PCR) of miRNAs was performed using the miRCURY LNA microRNA PCR system (Exiqon, Vedbaek, Denmark). Ten nanograms of template RNA was used in each first-strand cDNA synthesis reaction. PCR was performed over 40 amplification cycles and fluorescence monitored as described above. Analysis was performed using the Opticon real-time PCR detection system (Bio-Rad). For all RQ-PCR analysis, normalization was against U6snRNA and error bars represent means ± SE from three independent experiments.
Northern blotting for microRNAs.
Total RNA was extracted from cells using Trizol (Invitrogen, Paisley, United Kingdom) and quantified by spectophotometric analysis. Northern blotting was performed by resolving 10 μg total RNA on denaturing Tris-borate-EDTA (TBE)-urea 15% polyacrylamide gels (Invitrogen), transferring onto BrightStar-Plus positively charged nylon membranes (Ambion), and UV cross-linking. The membrane was hybridized overnight at 42°C with digoxigenin (DIG)-labeled LNA probe specific for miR-203 (0.1 nM) (Exiqon) or DIG-labeled antisense probe to U2snRNA (GGGTGCACCGTTCCTGGAGGTAC) (100 ng/ml). Following posthybridization washing, signal detection was performed using the DIG luminescence detection kit (Roche). Signal was detected on an Alpha Innotech FluorChem SP imaging system.
RESULTS AND DISCUSSION
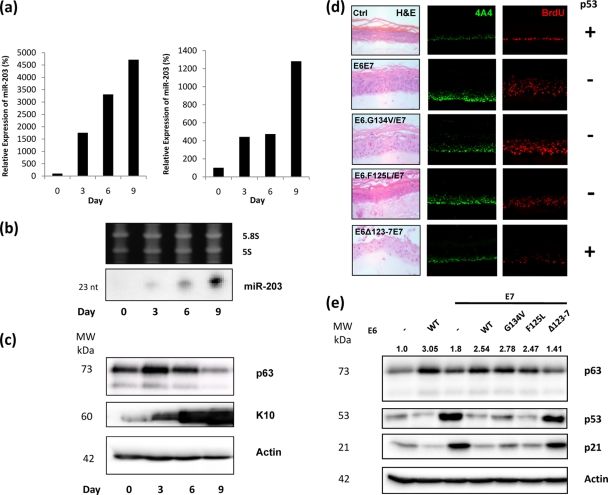
From an initial miRNA screen, carried out in collaboration with Eric Miska (University of Cambridge), we had noted that the expression levels of several miRNAs were changed following differentiation in HFKs (see Table S1 in the supplemental material). Among these was miR-203, which has previously been shown to be upregulated during differentiation of mouse epidermis and in cultured keratinocytes and which targets the 3′ UTRs of TAp63α and ΔNp63α, the latter being the predominant isoform of p63 in keratinocytes (25, 49). In this study, we showed by both real-time quantitative PCR (RQ-PCR) (Fig. 1 a) and Northern blotting (Fig. 1b) that miR-203 is also strongly upregulated in organotypic rafts, which are three-dimensional skin equivalents (30), derived from normal HFKs. This increase in miR-203 expression corresponds with induction of HFK differentiation in the rafts and seemingly contributes to the reduction in p63 levels over time (Fig. 1c). These results provide further confirmatory evidence that miR-203 plays a critical role in the differentiation of keratinocytes by regulating the expression of p63.
FIG. 1.
p63 and miR-203 expression in organotypic rafts. (a) RQ-PCR analysis of RNA samples harvested from organotypic rafts at different time points. Results from two sets of raft samples, independently generated from separate HFK batches, demonstrate that miR-203 expression increases significantly over time. (b) This observation is confirmed by Northern blotting analysis. (c) Western blot analysis of matched protein samples demonstrates increased K10 expression, indicating that differentiation of HFKs is occurring in the rafts. p63 levels initially increase slightly and are then reduced. (d) p63 expression is dependent on the ability of E6 to degrade p53. p63 expression in Ctrl organotypic rafts is confined to basal cells (row 1). In rafts expressing E6 and E7, increased p63 expression is observed in upper layers of the raft, together with evidence of increased cell proliferation (row 2). This increase in both p63 expression and proliferation is also apparent in rafts expressing E7 and mutant E6 which can still degrade p53 (rows 3 and 4). However, in rafts expressing E7 and mutant E6 which cannot degrade p53, p63 expression and proliferation are similar to those for the control (row 5). (e) Western blot analysis confirms that p63 levels are significantly elevated in HFKs expressing E6 (lane 2), both E6 and E7 (lane 4), and both E6 mutants capable of degrading p53 and E7 (lanes 5 and 6). A more moderate increase is observed in HFKs expressing E7 alone (lane 3). HFKs expressing both E6 mutants incapable of degrading p53 and E7 show no increase in p63 expression (lane 7). Numbers above the p63 panel represent expression signals of p63 bands after normalization to actin expression. With the exception of panel a (n = 2), all images and blots are representative of three independent experiments performed on separate batches of HFKs.
In light of these results, we were interested in investigating whether levels of miR-203 expression could help explain observations in our laboratory that showed upregulation of p63 in primary HFKs expressing E6/E7, an upregulation that is apparently dependent on the ability of E6 to degrade p53 (Fig. 1d and e). HFKs were retrovirally transduced to express the E6 and E7 oncoproteins. In addition, HFKs were also transfected to express E7 along with one of three mutant E6 proteins, two of which are capable of degrading p53 (F125L and G134V) (9, 27) and one which is not (Δ123-7) (21). Organotypic rafts generated from these cells demonstrated that p63 localization is confined to basal cells in the presence of p53 (Fig. 1d, row 1). In rafts where p53 function is impaired by the presence of either E6 (Fig. 1d, row 2) or the G134V or F125L mutants (Fig. 1d, rows 3 and 4), p63 expression is detected in the upper layers of the raft, along with increased cell proliferation, as shown by increased BrdU incorporation. However, in rafts expressing the Δ123-7 mutant, which cannot degrade p53, p63 expression is similar to that for the control and proliferation is reduced but not to the control levels due to activity of E7 (Fig. 1d, row 5). Western blot analysis correlates with these observations, showing that p63 levels are elevated in HFKs where p53 function is impaired but not in HFKs where p53 is not degraded (Fig. 1e).
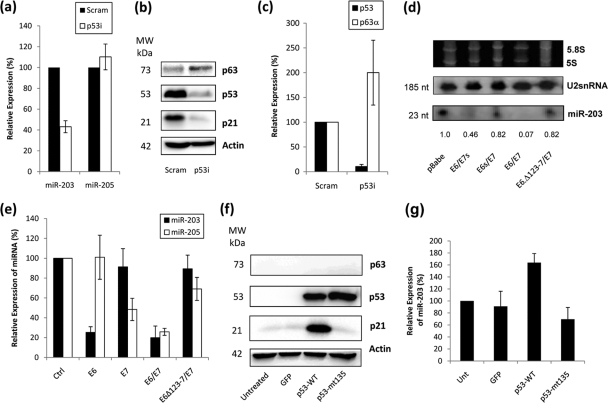
In light of these observations, we proceeded to determine if the modulation of p63 by E6/E7 was due to an association between p53 and miR-203 by investigating the levels of miR-203 in HFKs with p53 knockdown. For comparison, we also investigated the levels of miR-205, which was identified in our initial miRNA screen as being highly expressed in keratinocytes and was also upregulated during differentiation (see Table S1 in the supplemental material). RQ-PCR analysis revealed a reduction in miR-203 expression in the presence of p53 knockdown (Fig. 2 a), whereas levels of miR-205 were not significantly altered, demonstrating that p53 does not affect all miRNAs in a similar manner. In p53i HFKs, the downregulation of miR-203 corresponded with an increase in p63α expression at both the protein (Fig. 2b) and mRNA (Fig. 2c) levels. Since p63 was known to be a direct target of miR-203 (25, 49), we wanted to investigate whether the levels of p63 observed in HFKs were related to the effect of p53 upon expression of miR-203 in these cells. Northern blotting demonstrated decreased levels of miR-203 when p53 function was impaired by the presence of E6, regardless of the presence of E7 (Fig. 2d). In contrast, HFKs expressing E7 alone or with mutant E6 which cannot degrade p53 showed only a slight decrease in miR-203 levels compared to controls (Fig. 2d). This was confirmed by RQ-PCR analysis, which showed a clear reduction in miR-203 expression when HFKs expressed E6 (Fig. 2e). It should also be noted that E7 has been shown to increase p53 levels, but it is not transcriptionally active (12), thereby offering an explanation for why miR-203 levels are not increased by p53 levels in E7-expressing HFKs (Fig. 2e). These results show that the ability of E6 to degrade p53 determines the levels of miR-203. Interestingly, miR-205 seems to be dependent on E7 expression, suggesting that it may be affected by levels of pRb within the cell (Fig. 2e), and this is being investigated further. We also showed that the transcriptional activity of p53 also has an impact upon miR-203 levels in another cell type by introducing wild-type p53 into p53-null H1299 cells (Fig. 2f), which resulted in an increase of miR-203 levels, whereas mutant p53 did not (Fig. 2g). Expression of E6 and E7 alone in H1299 cells had no apparent effect on miR-203 expression, indicating that the effect of E6 on miR-203 must depend on p53 to some degree (see Fig. S1 in the supplemental material).
FIG. 2.
miR-203 expression is p53 dependent. (a) RQ-PCR analysis demonstrates a reduction in miR-203 expression in HFKs when p53 was knocked down (p53i) compared to scramble controls (Scram). In comparison, the expression of miR-205, another highly expressed miRNA in the skin, did not change. (b) Western blot analysis confirms the knockdown of p53 in these cells and also demonstrates an increase in p63 expression. (c) RQ-PCR analysis confirms that knockdown of p53 at the mRNA level results in an increase in expression of p63α mRNA in these cells. (d) Northern blotting demonstrates that miR-203 expression is reduced in HFKs expressing E6 alone (lane 2) and both E6 and E7 (lane 4). In HFKs expressing E7 alone (lane 3) and E7 and mutant E6 (Δ123-7) (lane 5), which cannot degrade p53, miR-203 expression is only slightly decreased. Numbers below the panel represent the expression signal of miR-203 after normalization to the U2snRNA signal. (e) RQ-PCR analysis confirms that miR-203 expression is decreased in HFKs expressing E6 and both E6 and E7 compared to the control, but is not significantly reduced in HFKs expressing E7 alone and both E7 and mutant E6 (Δ123-7). In contrast, miR-205 levels are reduced in the presence of E7, regardless of the presence or function of E6. (f) Western blotting shows equal levels of wild-type and mutant p53 when expressed in a p53-null H1299 cell line, but upregulation of p21 is observed only with the former. Control experiments demonstrate that no expression of p53 is detected in untreated cells or cells transfected with a GFP-expressing vector. (g) Expression of wild-type p53 in H1299 cells results in an increase of miR-203 levels, as measured by RQ-PCR. In contrast, expression of dominant negative p53 mut135 does not induce miR-203, with expression levels similar to those for controls. All blots are representative of three independent experiments performed on separate batches of cells.
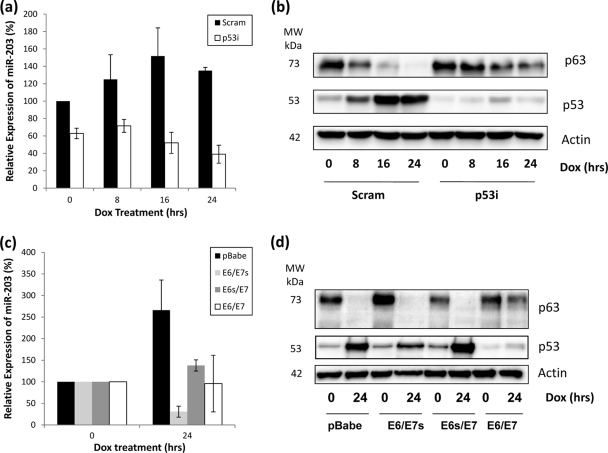
Given this apparent link between p53 and miR-203, we wanted to investigate how induction of p53 activity following DNA damage would affect miR-203 and p63 expression. In normal HFKs, miR-203 expression was induced in the 24 h following doxorubicin treatment, correlating with an increase in p53 levels and a decrease in p63 levels during the same period (Fig. 3 a and b). However, in p53i HFKs, miR-203 induction was not observed. Since we have proposed that miR-203 expression is p53 dependent, this result correlates with that hypothesis, as does the observation that p63 is not degraded to the same extent as in control HFKs (Fig. 3b). When we carried out a similar experiment with HFKs expressing E6 alone and in tandem with E7 (Fig. 3c), we noted the same lack of miR-203 induction compared to that in control cells. However, we also noted that miR-203 induction was decreased in HFKs expressing E7 alone (Fig. 3c), indicating that E6 and E7 can both apparently have an impact upon miR-203 levels following DNA damage. Interestingly, only when both are expressed do we observe that residual protein levels of p63 remain after 24 h compared to the control (Fig. 3d), suggesting that p63 levels following DNA damage are not wholly determined by miR-203 expression.
FIG. 3.
Induction of miR-203 following DNA damage is lost in p53i HFKs and HFKS expressing E6 and/or E7. (a) RQ-PCR shows that miR-203 expression increases following DNA damage induced by doxorubicin. However, this induction is not observed in p53i HFKs. (b) Western blotting demonstrates that p53 levels increase and p63 levels decrease in control cells after DNA damage. However, in the absence of p53 induction in p53i HFKs, this decrease in p63 levels is not as great, with detection still apparent at 24 h. (c) RQ-PCR shows that induction of miR-203 expression is not observed in HFKs expressing E6 alone (E6/E7s), E7 alone (E6s/E7), or both together (E6/E7) following DNA damage induced by doxorubicin. (d) Western blotting demonstrates that p53 levels increase and p63 levels decrease in control cells after DNA damage. A similar decrease is noted in E6/E7s and E6s/E7 HFKs, but higher residual p63 levels are apparent in E6/E7 HFKs after 24 h. All blots are representative of three independent experiments performed on different batches of primary HFKs.
The potential link between p53 and miR-203 expression may be important in relation to cancer and cancer therapy, since the cellular response to DNA damage is disrupted when p53 function is compromised, which in turn limits the effectiveness of radiotherapy and chemotherapy in the treatment of cancer. A link between miR-203 and the DNA damage response in cancer cells has previously been suggested by Lena et al. (25), who observed that miR-203 expression is increased in head and neck squamous cell carcinoma (HNSCC) cells in response to UVC radiation, corresponding with a decrease in ΔNp63α levels. However, the function of p63 in the cellular response to damage is not well understood, although it now seems clear that complex interactions occur between all the members of the p53 family in determining cell fate following exogenous insult (43). For example, p63 is overexpressed in many HNSCCs, where it is thought to antagonize the proapoptotic activity of p73 (34). In keratinocytes, the complexity is furthered by the observation that p63 isoforms apparently behave differently under cellular stress, with ΔNp63α reported to decrease following UV radiation (29, 45), while TAp63α (33) and TAp63γ (19) increase. In this study, we have now shown that the normal induction of miR-203 does not occur in the absence of p53 in HFKs. This implies that the cellular response to damage may be compromised in cells which have impaired p53 activity due to E6 expression or due to mutation, since subsequent overexpression of p63 may function as a prosurvival factor in cancerous cells. To ascertain whether p53 was capable of directly regulating miR-203, we analyzed the promoter and the upstream regions of miR-203 for potential p53 binding sites using the rVISTA 2.0 transcription factor binding prediction tool (28). However, only two candidate sites were identified, and chromatin immunoprecipitation showed no evidence of p53 binding at either site (data not shown).
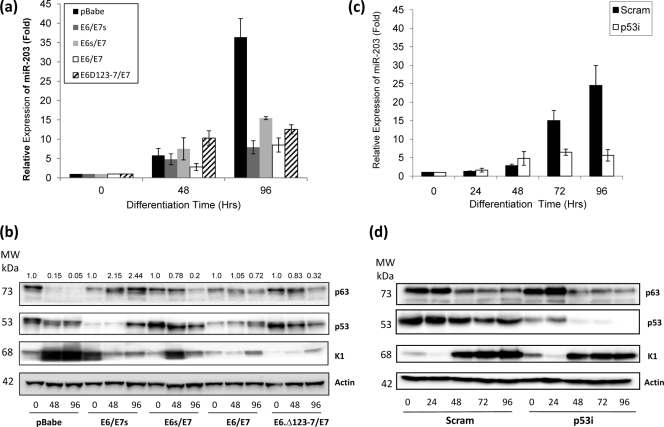
Yi et al. (49) and Lena et al. (25) both contend that miR-203, through its targeting of p63, acts as a molecular switch between keratinocyte proliferation and differentiation. Since this balance is disrupted in E6/E7-infected keratinocytes, this may be partly due to the effect on miR-203 function caused by decreased activity of p53. Therefore, we investigated whether miR-203 expression differed during differentiation of E6- and/or E7-expressing and p53i HFKs compared to controls. We observed a strong upregulation of miR-203 expression in control HFKs following calcium-induced differentiation as measured by RQ-PCR (Fig. 4 a). This induction is significantly reduced in HFKs expressing E6 alone, E7 alone, and E6/E7 together (Fig. 4a). In these cells, we observe that p63 expression remains elevated compared to that in controls during differentiation, which would be expected since miR-203 controls p63 levels (Fig. 4b). When we repeated the experiment with p53i HFKs, we observed a similar lack of induction of miR-203 expression compared to that in controls (Fig. 4c). However, in p53i cells, p63 levels are not significantly different from those in controls (Fig. 4d). Together, these results indicate that p53 function is important for the induction of miR-203 in HFKs during differentiation. However, since K1 induction is not significantly affected in p53i HFKs or in E6-expressing HFKs (Fig. 4d), it also suggests that factors other than miR-203 must also contribute to the failure of the terminal differentiation program in E6/E7-expressing HFKs. This agrees with observations of Lena et al. (25), who concluded that miR-203 alone is not sufficient to induce differentiation, and it is also worth remembering that other proteins are known to promote p63 degradation during differentiation as well, such as the ubiquitin ligases Itch (36) and WWP1 (26). How all these factors interact in the control of p63 levels is still poorly understood and may even involve other, as-yet-unidentified, proteins or miRNAs. Nevertheless, it seems reasonable to conclude that the decrease in miR-203 induction observed when p53 function is compromised has an impact upon p63 expression and therefore offers an explanation for how HPV infection disrupts the delicate balance between proliferation and differentiation in these cells.
FIG. 4.
Differentiation-specific upregulation of miR-203 expression is lost in E6/E7 and p53i HFKs. (a and c) RQ-PCR shows that the upregulation of miR-203 expression in control HFKs following calcium-induced differentiation is significantly reduced in HFKs expressing functional E6 and/or functional E7 (a) and p53i HFKs (c). RQ-PCR results are expressed relative to control values at T = zero. (b and d) Protein expression analysis from matched protein samples demonstrates that K1 induction is reduced in HFKs expressing E6 and/or E7, indicating lack of proper differentiation (b), but is not significantly altered in p53i HFKs (d). p63 levels are higher in HFKs expressing E6 and/or E7 than in controls, but reduction is not significantly affected in p53i HFKs compared to controls. Numbers above the p63 panel in panel b represent expression signals of p63 bands after normalization to actin expression. All blots are representative of three independent experiments performed on separate batches of HFKs.
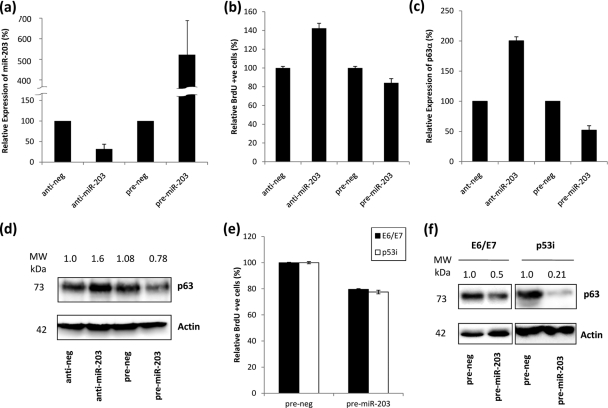
To specifically examine and verify the effect of miR-203 expression on proliferation in cycling HFKs, miR-203 levels were knocked down and overexpressed by transfection with antago-miR-203 and pre-miR-203 molecules, respectively (Fig. 5 a). Knockdown resulted in increased HFK proliferation, with ∼40% more cells staining for BrdU incorporation than control cells, while overexpression resulted in a decrease in proliferation, with ∼19% fewer cells staining for BrdU (Fig. 5b). This correlated with changes in p63 expression at both the mRNA (Fig. 5c) and protein (Fig. 5d) levels, which were elevated when miR-203 was knocked down and reduced when miR-203 was overexpressed. These observations also relate to the increase in miR-203 expression that occurs in normal HFKs during differentiation, emphasizing that the reduction of p63 levels caused by elevation of miR-203 expression results in an inhibition of the proliferative capacity of the cells, thereby encouraging the cells the commit to the differentiation pathway.
FIG. 5.
Effects of miR-203 expression on HFK proliferation. (a) RQ-PCR demonstrating knockdown and overexpression of miR-203 in HFKs compared to the respective controls. (b) Proliferation, as measured by BrdU incorporation, is increased in HFKS in which miR-203 is inhibited and decreased in HFKs which overexpress miR-203. (c) RQ-PCR analysis showing that expression of p63α mRNA is increased in HFKs in which miR-203 is inhibited and decreased in HFKs which overexpress miR-203, relative to the respective controls, indicating that miR-203 levels control expression of the predominant isoform of p63 in HFKs. (d) Western blot confirming that protein levels of p63 are increased in HFKs in which miR-203 is inhibited and decreased in HFKs which overexpress miR-203, relative to the respective controls. Numbers above the panel represent expression signals of p63 bands after normalization to actin expression and are representative of three independent experiments performed on different batches of primary HFKs. (e) Proliferation, as measured by BrdU incorporation, is reduced in E6/E7 and p53i HFKs which overexpress miR-203. (f) Western blot demonstrating that levels of p63 are significantly reduced in E6/E7 and p53i HFKs which overexpress miR-203. Numbers above panels represent expression signal of p63 bands after normalization to actin expression and are representative of three independent experiments performed on different batches of primary HFKs.
If the influence of E6 upon p53 results in decreased miR-203 levels, then it would be reasonable to suggest that overexpressing miR-203 in cells where p53 function is compromised should render E6 activity redundant and restore to some extent the balance which influences HFK proliferation. Therefore, we investigated whether exogenous expression of miR-203 in highly proliferative HFKs would result in decreased proliferative potential, again due to a reduction in p63 levels. We found that the number of BrdU-incorporating cells was decreased by ∼21% among E6/E7 cells and ∼23% among p53i cells, relative to controls, when miR-203 was overexpressed (Fig. 5e). This correlated with a significant decrease in p63 levels compared to controls in matched protein samples (Fig. 5f). These findings further highlight the importance of p63 in maintaining proliferative potential in HFKs.
While this paper was being prepared, a paper by Melar-New and Laimins (31) demonstrating that expression of E7 is primarily responsible for the downregulation of miR-203 levels during differentiation of keratinocytes was published. Our results confirm that E7 can have an impact upon miR-203 levels (Fig. 4a and b), but in addition we show that E6 expression also downregulates miR-203 and demonstrate that this effect is more significant than that of E7 in both proliferating and differentiating HFKs. We suggest that the impact of E6 on p53 activity affects miR-203 expression, while Melar-New and Laimins propose that the effect of E7 on miR-203 is via the mitogen-activated protein kinase (MAPK)/protein kinase C (PKC) pathway. Given the results of both studies, it seems apparent that the expression of both oncoproteins, whether alone or in tandem, has a significant effect on miR-203 levels and that the subsequent impact upon p63 levels will disrupt the balance between proliferation and differentiation in keratinocytes.
In summary, we have provided further evidence that miR-203 is a critical factor in the control of p63 levels in keratinocytes. Moreover, we have also shown that the expression of miR-203 is dependent on E6 activity upon p53, thereby providing new evidence as to how HPV infection can lead to deregulation of proliferation and differentiation in keratinocytes during the development of cervical cancer. This also offers an association which may explain the relationship between p53 and p63 during epidermal homeostasis and potentially in the cellular response to damage as well.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Medical Research Council, United Kingdom (MRC G0700754).
We declare no conflict of interest.
Footnotes
Published ahead of print on 11 August 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Andl, T., E. P. Murchison, F. Liu, Y. Zhang, M. Yunta-Gonzalez, J. W. Tobias, C. D. Andl, J. T. Seykora, G. J. Hannon, and S. E. Millar. 2006. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 16:1041-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauersachs, J., and T. Thum. 2007. MicroRNAs in the broken heart. Eur. J. Clin. Invest. 37:829-833. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, E., S. Y. Kim, M. A. Carmell, E. P. Murchison, H. Alcorn, M. Z. Li, A. A. Mills, S. J. Elledge, K. V. Anderson, and G. J. Hannon. 2003. Dicer is essential for mouse development. Nat. Genet. 35:215-217. [DOI] [PubMed] [Google Scholar]
- 4.Bommer, G. T., I. Gerin, Y. Feng, A. J. Kaczorowski, R. Kuick, R. E. Love, Y. Zhai, T. J. Giordano, Z. S. Qin, B. B. Moore, O. A. MacDougald, K. R. Cho, and E. R. Fearon. 2007. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 17:1298-1307. [DOI] [PubMed] [Google Scholar]
- 5.Brosh, R., R. Shalgi, A. Liran, G. Landan, K. Korotayev, G. H. Nguyen, E. Enerly, H. Johnsen, Y. Buganim, H. Solomon, I. Goldstein, S. Madar, N. Goldfinger, A. L. Børresen-Dale, D. Ginsberg, C. C. Harris, Y. Pilpel, M. Oren, and V. Rotter. 2008. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol. Syst. Biol. 4:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, T. C., E. A. Wentzel, O. A. Kent, K. Ramachandran, M. Mullendore, K. H. Lee, G. Feldmann, M. Yamakuchi, M. Ferlito, C. J. Lowenstein, D. E. Arking, M. A. Beer, A. Maitra, and J. T. Mendell. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowland, J. B., C. Hother, and K. Grønbaek. 2007. MicroRNAs and cancer. APMIS 115:1090-1106. [DOI] [PubMed] [Google Scholar]
- 8.Dai, Y., Y. S. Huang, M. Tang, T. Y. Lv, C. X. Hu, Y. H. Tan, Z. M. Xu, and Y. B. Yin. 2007. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus 16:939-946. [DOI] [PubMed] [Google Scholar]
- 9.Dalal, S., Q. Gao, E. J. Androphy, and V. Band. 1996. Mutational analysis of human papillomavirus type 16 E6 demonstrates that p53 degradation is necessary for immortalization of mammary epithelial cells. J. Virol. 70:683-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davison, T. S., C. Vagner, M. Kaghad, A. Ayed, D. Caput, and C. H. Arrowsmith. 1999. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J. Biol. Chem. 274:18709-18714. [DOI] [PubMed] [Google Scholar]
- 11.De Laurenzi, V., A. Rossi, A. Terrinoni, D. Barcaroli, M. Levrero, A. Costanzo, R. A. Knight, P. Guerrieri, and G. Melino. 2000. p63 and p73 transactivate differentiation gene promoters in human keratinocytes. Biochem. Biophys. Res. Commun. 273:342-346. [DOI] [PubMed] [Google Scholar]
- 12.Eichten, A., M. Westfall, J. A. Pietenpol, and K. Münger. 2002. Stabilization and functional impairment of the tumor suppressor p53 by the human papillomavirus type 16 E7 oncoprotein. Virology 295:74-85. [DOI] [PubMed] [Google Scholar]
- 13.Erson, A. E., and E. M. Petty. 2008. MicroRNAs in development and disease. Clin. Genet. 74:296-306. [DOI] [PubMed] [Google Scholar]
- 14.Feramisco, J. D., R. L. Cayce, and H. Tsao. 2008. Recent updates on genetics: teaching old dogmas new tricks. Pediatr. Dermatol. 25:99-108. [DOI] [PubMed] [Google Scholar]
- 15.Gaiddon, C., M. Lokshin, J. Ahn, T. Zhang, and C. Prives. 2001. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21:1874-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guess, J. C., and D. J. McCance. 2005. Decreased migration of Langerhans precursor-like cells in response to human keratinocytes expressing human papillomavirus type 16 E6/E7 is related to reduced macrophage inflammatory protein-3alpha production. J. Virol. 79:14852-14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, L., X. He, L. P. Lim, E. de Stanchina, Z. Xuan, Y. Liang, W. Xue, L. Zender, J. Magnus, D. Ridzon, A. L. Jackson, P. S. Linsley, C. Chen, S. W. Lowe, M. A. Cleary, and G. J. Hannon. 2007. A microRNA component of the p53 tumour suppressor network. Nature 447:1130-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Incassati, A., D. Patel, and D. J. McCance. 2006. Induction of tetraploidy through loss of p53 and upregulation of Plk1 by human papillomavirus type-16 E6. Oncogene 25:2444-2451. [DOI] [PubMed] [Google Scholar]
- 19.Katoh, I., K. I. Aisaki, S. I. Kurata, S. Ikawa, and Y. Ikawa. 2000. p51A (TAp63gamma), a p53 homolog, accumulates in response to DNA damage for cell regulation. Oncogene 19:3126-3130. [DOI] [PubMed] [Google Scholar]
- 20.Keyes, W. M., Y. Wu, H. Vogel, X. Guo, S. W. Lowe, and A. A. Mills. 2005. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 19:1986-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 380:79-82. [DOI] [PubMed] [Google Scholar]
- 22.Knight, S. W., and B. L. Bass. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293:2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latronico, M. V., D. Catalucci, and G. Condorelli. 2007. Emerging role of microRNAs in cardiovascular biology. Circ. Res. 101:1225-1236. [DOI] [PubMed] [Google Scholar]
- 24.Lee, H., and D. Kimelman. 2002. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev. Cell 2:607-616. [DOI] [PubMed] [Google Scholar]
- 25.Lena, A. M., R. Shalom-Feuerstein, P. Rivetti di Val Cervo, D. Aberdam, R. A. Knight, G. Melino, and E. Candi. 2008. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 15:1187-1195. [DOI] [PubMed] [Google Scholar]
- 26.Li, Y., Z. Zhou, and C. Chen. 2008. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 5:1941-1951. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Y., J. J. Chen, Q. Gao, S. Dalal, Y. Hong, C. P. Mansur, V. Band, and E. J. Androphy. 1999. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J. Virol. 73:7297-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loots, G., and I. Ovcharenko. 2004. rVista 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32:W217-W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchbank, A., L. J. Su, P. Walsh, J. DeGregori, K. Penheiter, T. B. Grayson, R. P. Dellavalle, and L. A. Lee. 2003. The CUSP DeltaNp63alpha isoform of human p63 is downregulated by solar-simulated ultraviolet radiation. J. Dermatol. Sci. 32:71-74. [DOI] [PubMed] [Google Scholar]
- 30.McCance, D. J., R. Kopan, E. Fuchs, and L. A. Laimins. 1988. Human papillomavirus type 16 alters human epithelial cell differentiation in vitro. Proc. Natl. Acad. Sci. U. S. A. 85:7169-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melar-New, M., and L. A. Laimins. 2010. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J. Virol. 84:5212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mills, A. A., B. Zheng, X. J. Wang, H. Vogel, D. R. Roop, and A. Bradley. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708-713. [DOI] [PubMed] [Google Scholar]
- 33.Okada, Y., M. Osada, S. Kurata, S. Sato, K. Aisaki, Y. Kageyama, K. Kihara, Y. Ikawa, and I. Katoh. 2002. p53 gene family p51(p63)-encoded, secondary transactivator p51B(TAp63alpha) occurs without forming an immunoprecipitable complex with MDM2, but responds to genotoxic stress by accumulation. Exp. Cell Res. 276:194-200. [DOI] [PubMed] [Google Scholar]
- 34.Ratovitski, E., B. Trink, and D. Sidransky. 2006. p63 and p73: teammates or adversaries? Cancer Cell 9:1-2. [DOI] [PubMed] [Google Scholar]
- 35.Rinne, T., H. G. Brunner, and H. van Bokhoven. 2007. p63-associated disorders. Cell Cycle 6:262-268. [DOI] [PubMed] [Google Scholar]
- 36.Rossi, M., R. I. Aqeilan, M. Neale, E. Candi, P. Salomoni, R. A. Knight, C. M. Croce, and G. Melino. 2006. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc. Natl. Acad. Sci. U. S. A. 103:12753-12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonkoly, E., T. Wei, P. C. Janson, A. Sääf, L. Lundeberg, M. Tengvall-Linder, G. Norstedt, H. Alenius, B. Homey, A. Scheynius, M. Ståhle, and A. Pivarcsi. 2007. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One 2:e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan, C. S., and D. Ganem. 2005. MicroRNAs and viral infection. Mol. Cell 20:3-7. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki, H. I., K. Yamagata, K. Sugimoto, T. Iwamoto, S. Kato, and K. Miyazono. 2009. Modulation of microRNA processing by p53. Nature 460:529-534. [DOI] [PubMed] [Google Scholar]
- 40.Tarasov, V., P. Jung, B. Verdoodt, D. Lodygin, A. Epanchintsev, A. Menssen, G. Meister, and H. Hermeking. 2007. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6:1586-1593. [DOI] [PubMed] [Google Scholar]
- 41.Truong, A. B., and P. A. Khavari. 2007. Control of keratinocyte proliferation and differentiation by p63. Cell Cycle 6:295-299. [DOI] [PubMed] [Google Scholar]
- 42.Truong, A. B., M. Kretz, T. W. Ridky, R. Kimmel, and P. A. Khavari. 2006. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 20:3185-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vilgelm, A., W. El-Rifai, and A. Zaika. 2008. Therapeutic prospects for p73 and p63: rising from the shadow of p53. Drug Resist. Updat. 11:152-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visone, R., and C. M. Croce. 2009. MiRNAs and cancer. Am. J. Pathol. 174:1131-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westfall, M. D., A. S. Joyner, C. E. Barbieri, M. Livingstone, and J. A. Pietenpol. 2005. Ultraviolet radiation induces phosphorylation and ubiquitin-mediated degradation of DeltaNp63alpha. Cell Cycle 4:710-716. [DOI] [PubMed] [Google Scholar]
- 46.Yang, A., M. Kaghad, Y. Wang, E. Gillett, M. D. Fleming, V. Dötsch, N. C. Andrews, D. Caput, and F. McKeon. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2:305-316. [DOI] [PubMed] [Google Scholar]
- 47.Yang, A., R. Schweitzer, D. Sun, M. Kaghad, N. Walker, R. T. Bronson, C. Tabin, A. Sharpe, D. Caput, C. Crum, and F. McKeon. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398:714-718. [DOI] [PubMed] [Google Scholar]
- 48.Yi, R., D. O'Carroll, H. A. Pasolli, Z. Zhang, F. S. Dietrich, A. Tarakhovsky, and E. Fuchs. 2006. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 38:356-362. [DOI] [PubMed] [Google Scholar]
- 49.Yi, R., M. N. Poy, M. Stoffel, and E. Fuchs. 2008. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 452:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.