Abstract
CCR5 antagonists inhibit HIV entry by binding to a coreceptor and inducing changes in the extracellular loops (ECLs) of CCR5. In this study, we analyzed viruses from 11 treatment-experienced patients who experienced virologic failure on treatment regimens containing the CCR5 antagonist maraviroc (MVC). Viruses from one patient developed high-level resistance to MVC during the course of treatment. Although resistance to one CCR5 antagonist is often associated with broad cross-resistance to other agents, these viruses remained sensitive to most other CCR5 antagonists, including vicriviroc and aplaviroc. MVC resistance was dependent upon mutations within the V3 loop of the viral envelope (Env) protein and was modulated by additional mutations in the V4 loop. Deep sequencing of pretreatment plasma viral RNA indicated that resistance appears to have occurred by evolution of drug-bound CCR5 use, despite the presence of viral sequences predictive of CXCR4 use. Envs obtained from this patient before and during MVC treatment were able to infect cells expressing very low CCR5 levels, indicating highly efficient use of a coreceptor. In contrast to previous reports in which CCR5 antagonist-resistant viruses interact predominantly with the N terminus of CCR5, these MVC-resistant Envs were also dependent upon the drug-modified ECLs of CCR5 for entry. Our results suggest a model of CCR5 cross-resistance whereby viruses that predominantly utilize the N terminus are broadly cross-resistant to multiple CCR5 antagonists, whereas viruses that require both the N terminus and antagonist-specific ECL changes demonstrate a narrow cross-resistance profile.
Small-molecule CCR5 antagonists are a relatively new class of drugs that block HIV entry into target cells, with the first member of this class, maraviroc (MVC), having been approved for the treatment of HIV-infected patients. These drugs bind to a hydrophobic pocket formed by the transmembrane helices of CCR5, inducing conformational changes in the extracellular loops (ECLs) of the receptor (18, 31, 39, 40, 58, 62, 64). These conformational changes can vary with different drugs, as evidenced by differential chemokine binding and HIV resistance profiles, and block the ability of HIV to use drug-bound CCR5 as a coreceptor for entry (59, 64).
As with other antiretroviral agents, HIV can develop resistance to CCR5 antagonists. One pathway by which HIV can become resistant to CCR5 antagonists is via mutations in the viral envelope (Env) protein that enable it to recognize the drug-bound conformation of the coreceptor. Most of our information on this pathway has come from in vitro passaging of HIV-1 in the presence of increasing concentrations of inhibitor (2, 4, 5, 33, 41, 44, 61, 66). In most instances, the viral determinants of resistance are localized to the V3 loop of gp120 (5, 33, 41, 44, 46, 63, 66). This is as expected: the base of the V3 loop interacts with O-sulfated tyrosines in the N terminus of CCR5, while the tip of the V3 loop is thought to contact the ECLs of the receptor (14, 15, 17, 19, 26, 29, 37). Viral resistance to one CCR5 antagonist commonly results in cross-resistance to other drugs in this class, although this is not universally the case (33, 41, 60, 63, 66). Mechanistically, a number of CCR5 antagonist-resistant viruses have been shown to have increased dependence on the N-terminal domain of CCR5 (5, 34, 44, 45, 48), which is largely unaffected by drug binding and may allow viruses to tolerate drug-induced changes in ECL conformation.
In contrast to several well-characterized viruses that have evolved resistance to CCR5 antagonists in vitro, few examples of patient-derived CCR5 antagonist-resistant viruses have been reported. One mechanism of resistance that has been described in patients is the outgrowth of CXCR4-tropic HIV isolates that were present at low frequencies prior to the initiation of therapy (22, 23, 35, 36, 42, 65). Due to this finding, patients undergo tropism testing prior to treatment with CCR5 antagonists, with only those harboring exclusively R5-tropic viruses considered candidates for therapy. Patient-derived viruses capable of using drug-bound CCR5 have been reported in studies using vicriviroc and aplaviroc (45, 60, 63). The aplaviroc-resistant viruses were determined to utilize the drug-bound form of the receptor by interacting primarily with the N terminus of CCR5, similar to the viruses derived by serial in vitro passaging (48).
In the present study, we report the isolation of MVC-resistant Envs from a treatment-experienced patient who had a viral load rebound while on a regimen containing MVC. Viral Envs isolated from this patient at the time MVC therapy was initiated were fully sensitive to drug. However, resistance evolved over the course of 224 days, culminating in Envs that were completely resistant to inhibition but continued to use CCR5 for entry. The emergence of resistance was dependent upon changes within the V3 loop of the virus, while changes in the V4 loop modulated the magnitude of resistance. The MVC-resistant Envs studied here exhibited several unusual properties. First, while they were cross-resistant to TAK779, they remained sensitive to all other CCR5 antagonists tested, including vicriviroc and aplaviroc. Second, the Envs were particularly adept at utilizing low levels of CCR5 to mediate infection of cells. Third, and in contrast to several recent reports of CCR5 antagonist-resistant viruses, these Envs were dependent upon residues within both the N terminus and ECLs of CCR5 for efficient entry in the presence of drug. When considered in the context of other reports, our data suggest a model in which resistance to multiple CCR5 antagonists can arise if an Env protein becomes highly dependent upon the N-terminal domain of CCR5, the conformation of which appears to be unaffected by drug binding. A more narrow resistance profile results from changes in Env that enable it to use both the N-terminal domain of CCR5 as well as the drug-induced conformation of the CCR5 ECLs.
MATERIALS AND METHODS
Study population.
All subjects for this study were identified from the ongoing clinic-based cohort of HIV-infected persons followed at two academic clinics in San Francisco (the SCOPE cohort). This cohort was enriched for patients with highly resistant HIV who were initiating “salvage” regimens containing an integrase inhibitor and/or a CCR5 antagonist when these drugs became more widely available (24). From this cohort, we identified all individuals who initiated a regimen containing maraviroc who exhibited evidence of incomplete viral suppression (defined as failure to achieve an undetectable plasma HIV RNA level or persistently detectable plasma HIV RNA levels after having achieved undetectable levels). Subjects who received therapy in controlled clinical trials were eligible as long as their treatment assignment was subsequently determined. Plasma samples were collected and stored approximately every 4 months. All subjects provided written informed consent to have samples collected for these types of studies. This study was approved by the UCSF Institutional Review Board.
Cloning of patient env genes.
Cloning of env genes from patients' plasma was performed using multiple separate PCRs using a high-fidelity polymerase with 3′-to-5′ proofreading exonuclease activity, as previously described (30). Vectors were grown in Stbl-3 Escherichia coli at 30°C to minimize bacterially induced mutagenesis and recombination of env. Mutant and chimeric env clones were created by site-directed mutagenesis and overlap PCR, respectively, and were confirmed by sequencing.
Virus infection assays.
Patient env clones digested with KpnI and XbaI were subcloned into a pCI expression construct containing hepatitis B virus posttranscriptional regulatory element (PRE) to enable high-level, rev-independent Env expression. Pseudotyped viruses were produced from 293T/17 cells (30 μg pCI-PRE-env vector and 10 μg pNL-Luc-Env core [9, 12]), and 5 to 25 ng p24 equivalent were used to infect cell lines, amounts empirically determined to be in the linear range of the infection assay. Cells were spinoculated with virus at 450 × g for 2 h at 25°C. Three days postinfection, cells were lysed and luciferase activity was analyzed on a luminometer. For inhibition assays, cells were preincubated with CCR5 antagonists or enfuvirtide for 30 min at 37°C prior to spinoculation with virus.
454 pyrophosphate sequencing.
Viral RNA was extracted from 140 μl of human plasma from the day 1 time point from patient 6061 using the QIAamp viral RNA kit (Qiagen). cDNA synthesis was performed on isolated viral RNA using SuperScript III reverse transcriptase (Invitrogen) and the subtype B primer TTGCTACTTGTGATTGCTCCATGT. For V3 loop amplification, a nested PCR approach was used. First, the cDNA product was amplified in each of 8 to 16 different PCRs as mentioned previously. Multiple PCRs were performed simultaneously to ensure sequence diversity. One microliter of the first-round PCR product was then used for each of the 16 corresponding second-round PCRs. The second-round PCR was performed using subtype B-specific primers described by Rozera and colleagues that were modified to contain an 8-bp DNA barcode and the 454 adaptor sequences (25, 53). Consensus subtype B primers were used to reduce amplification bias.
Emulsion PCR was then performed, followed by pyrosequencing using a Roche/454 GS FLX sequencer at the University of Pennsylvania's DNA sequencing facility. Amplicons were run on 1/8 of a pico-titer plate yielding 55,050 reads. For sequencing analysis, only reads with 100% barcode and primer identity that were greater than 200 bp and without any ambiguous base calls were analyzed, yielding 47,202 sequences. The reverse complement of each sequence was then translated, and any read with a premature stop codon was discarded. Reads with intact V3 loop sequences, defined as cysteine-anchored loops of 35 ± 2 amino acids containing a GPGR motif in which three of the four amino acids were present, were identified, yielding 43,168 reads representing 3,466 unique V3 loop sequences.
Antibody neutralization assays.
Five nanograms of virus was incubated with serial dilutions of monoclonal antibody (MAb) for 1 h at 37°C and then spinoculated onto NP2/CD4/CCR5 cells. MAbs 17b and IgG b12 were obtained from the IAVI Neutralizing Antibody Consortium repository. Cells were assayed for luciferase expression after 3 days.
Affinofile assay.
Infection of the HEK293-based CD4/CCR5 dual-inducible cell line (293-Affinofile) with luciferase-based pseudoviruses was performed as previously described (28). Briefly, 96-well plates were seeded with 1.0 × 104 inducible cells 2 days prior to CD4 and CCR5 induction. Cells were induced using 2-fold serial dilutions from 0.156 to 5 ng/ml of minocycline (resulting in 6 induction conditions for CD4) and from 0.0156 to 2 μM ponasterone A (resulting in 8 induction conditions for CCR5). Cells were incubated for 18 h at 37°C, after which induction media was removed and half the plates were preincubated with 10 μM maraviroc for 30 min and then were infected with pseudoviruses normalized for p24 content. Infection was quantified after 3 days as described above.
Mutant CCR5 assays.
293T cells stably transfected with human CD4 were transfected with the wild-type or mutant CCR5 genes using lipofectamine (Invitrogen). The CCR5 receptors transfected included the wild type, Y3A, Q4A, Y10A, D11A, Y14A, Y15A, N24A, Q27A, H88A, Q93A, N98A, S179A, H181A, F182A, P183A, Q186A, Y187A, F189A, W190A, E262A, N267A, N268A, C269A, and N273A. Expression of CD4 and CCR5 was quantified by flow cytometry 24 h posttransfection using anti-CD4 (BD) and anti-CCR5 antibodies (CTC5, R&D Systems, and 2D7, BD). Cells were infected with luciferase-based pseudoviruses 24 h posttransfection via spinoculation (450 × g, 2 h), and infection was quantified after 3 days as detailed above.
Monoclonal antibody binding to CCR5 in the presence of antagonists.
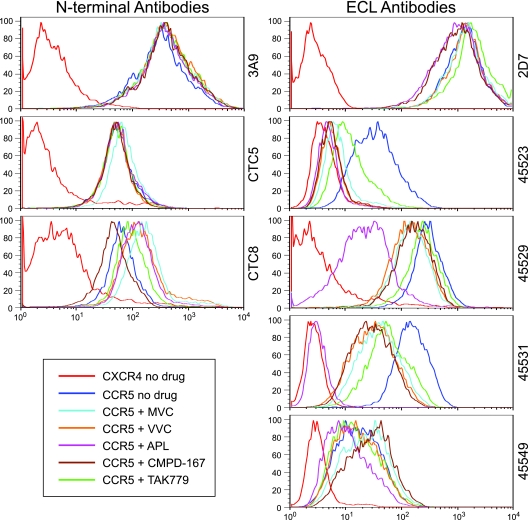
NP2/CD4 cells stably expressing CCR5 were preincubated for 30 min at 37°C with the CCR5 antagonists maraviroc, aplaviroc, vicriviroc, CMPD-167, or TAK779 or were left untreated. NP2/CD4 cells expressing CXCR4 were utilized as a negative control. Cells were placed into suspension by treatment with Versene and probed with the anti-CCR5 N-terminal monoclonal antibodies 3A9 (Becton Dickinson), CTC5, or CTC8 (R&D Systems) or with the anti-CCR5 ECL monoclonal antibodies 2D7 (Becton Dickinson), 45523, 45529, 45531, or 45549 (R&D Systems) for 30 min at 4°C. Cells were washed twice, resuspended, and run on a FACSCalibur flow cytometer (Becton Dickinson). At least 25,000 events per condition were collected and analyzed using FlowJo software (TreeStar).
RESULTS
In vivo resistance to maraviroc.
In the SCOPE cohort of treatment-experienced patients who received maraviroc (MVC) in combination with optimized background therapy, 11 exhibited evidence of virologic failure. To investigate the mechanisms of MVC failure in these patients, we obtained HIV env clones from plasma samples by isolating viral RNA, synthesizing cDNA, and performing amplification of full-length env sequences using a nested PCR strategy. Viral env V3 loop sequences were examined for evidence of CXCR4 use, which is a well-characterized mechanism of escape from CCR5 antagonists (22, 23, 35, 36, 42, 65). Six of 11 patients were found to have basic residues at positions 11, 24, and/or 25 of the V3 loop, which are highly predictive of CXCR4 use (8, 16, 20, 21, 27, 51). The remaining 5 patients had very few or no CXCR4-using viruses defined by sequencing. The env clones from these patients were subcloned into expression vectors, and pseudoviral particles were produced. The viruses pseudotyped with Envs from these 5 patients were strictly R5 tropic, as determined by their ability to infect NP2 cells expressing CD4 and CCR5 but not CD4 and CXCR4 (data not shown).
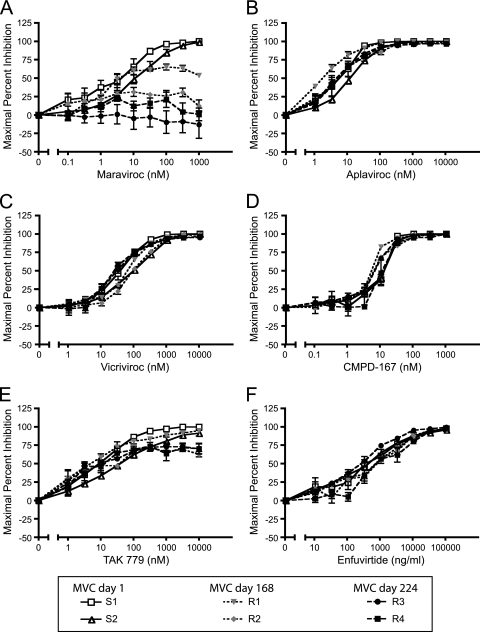
To determine the MVC susceptibility of the cloned Envs, pseudotype infection assays were performed on NP2/CD4/CCR5 cells in the presence or absence of increasing concentrations of MVC. Viruses from 4 of 5 patients with strictly R5-tropic HIV were found to be completely inhibited by MVC in this pseudoviral infection assay, with a maximal percent inhibition (MPI) of >95% (data not shown). However, in patient 6061, viral clones isolated after 224 days of therapy were not inhibited by MVC (MPI range, −13.2 to 0.5%). In contrast, env clones isolated from this patient at the beginning of treatment were efficiently inhibited by MVC (MPI range, 98.7 to 99.8%). At an intermediate time point 168 days into MVC therapy, the patient was found to harbor Envs with partial as well as complete resistance to MVC (MPI range, 0 to 53.4%). Results obtained with two viruses from each time point are shown in Fig. 1A; these were selected based on robust infection in the pseudoviral assay and on the absence of any unusual sequence motifs compared with Envs isolated from the same time points. The viral loads for this patient in the month preceding initiation of MVC ranged between 191,000 to 281,000 copies/ml, with the highest value recorded concurrently with starting treatment. Although there was an initial decrease to 8,960 copies/ml in the first 2 weeks of MVC therapy, the viral load rebounded by week 4 and returned to set-point values or slightly above, ranging from 193,000 to 319,000 copies/ml. These data indicate that in the context of this patient, the MVC-resistant viruses were able to replicate with equivalent efficiencies to the MVC-sensitive pretreatment viruses prior to the initiation of MVC therapy.
FIG. 1.
Viruses from patient 6061 evolved resistance to maraviroc but remained sensitive to most other CCR5 antagonists and enfuvirtide. Viruses pseudotyped with Envs cloned from patient 6061 at days 1, 168, and 224 on maraviroc therapy were used to infect NP2/CD4/CCR5 cells in the absence or presence of various concentrations of maraviroc (A), aplaviroc (B), vicriviroc (C), CMPD-167 (D), TAK779 (E), and enfuvirtide (F), and maximal percent inhibition was calculated. Viral Envs cloned after 168 and 224 days on maraviroc therapy are resistant to maraviroc and partially cross-resistant to TAK779 but remain sensitive to all other CCR5 antagonists and the fusion inhibitor enfuvirtide. Data represent results from six independent experiments ± standard errors of the mean.
Although MVC is the only CCR5 antagonist approved for treatment of HIV-infected patients, additional CCR5 antagonists have been developed, and several are currently in clinical trials. A major question for the use of CCR5 antagonists is whether viral resistance to one drug will result in cross-resistance to other compounds in this class. To determine the degree of cross-resistance to other entry inhibitors, pseudotype infection assays were performed in the presence of increasing concentrations of the CCR5 antagonists aplaviroc, vicriviroc, CMPD-167, and TAK779 as well as with the fusion inhibitor enfuvirtide (Fig. 1B to F). The clones with high MVC resistance were partially cross-resistant to TAK779 (MPI range, 62.7 to 70.2%) but remained sensitive to all other CCR5 antagonists tested as well as to enfuvirtide (MPI ≥ 95%). Thus, the MVC-resistant Envs derived from this patient have a more restricted resistance profile than many other CCR5 antagonist-resistant viruses described to date.
The V3 loop is required for MVC resistance, while the V4 loop modulates the magnitude of resistance.
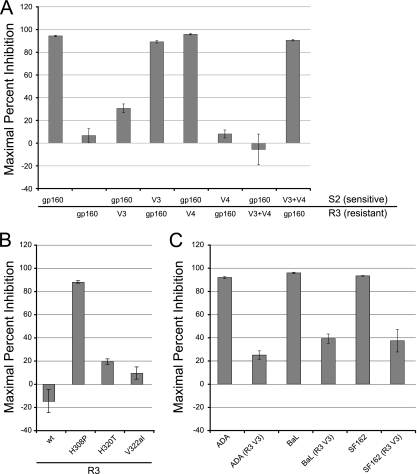
Amino acid mutations that might play major roles in conferring MVC resistance to Envs derived from patient 6061 should occur in all fully resistant clones and be absent in all sensitive clones. Six potential candidate mutations fulfilled these criteria: 3 within the V3 loop, 2 in the V4 loop, and 1 in the C5 domain. There were no mutations in the gp41 subunit that met the screening criteria. Using amino acid numbers corresponding to the HXB2 reference strain, the V3 loop mutations present in all resistant clones included P/T308H, T320H, and I322aV (I322aV occurs at an amino acid not present in HXB2 located between residues 322 and 323 and is designated 322a), the V4 mutations consisted of D407G and a loss of a glycosylation site at residue 386, and the C5 mutation was V489I. Because changes in the V3 loop can impart resistance to CCR5 antagonists, including the I322aV mutation observed in our clones (66), we exchanged the V3 loops between a resistant and a sensitive Env clone obtained from the pretreatment time point. Pseudoviral particles bearing these chimeric Envs were constructed and found to infect NP2/CD4/CCR5 cells efficiently. Introduction of the V3 loop from the sensitive Env into the resistant Env strongly reduced resistance to 10 μM MVC (MPI = 89.2 ± 1.1%) (Fig. 2A). The converse chimera, in which the V3 loop from the resistant clone was inserted into the sensitive clone, resulted in significant but incomplete resistance to MVC (MPI = 30.6 ± 3.9%). To test whether the two mutations present in the V4 loop influenced the magnitude of resistance, chimeras were created in which the V4 loop was exchanged between sensitive and resistant clones, either alone or in combination with V3. As expected, the resistant virus containing both the V3 and V4 loops from the sensitive clone was largely inhibited by MVC (MPI = 90.6 ± 0.6%). In addition, introducing both the V3 and V4 loops from the resistant clone into the sensitive clone completely reconstituted the resistance phenotype (MPI = −5.6 ± 13.6%). In contrast, chimeras containing the V4 loops alone exhibited resistance profiles similar to those of the parental strains. These data suggest that the V4 loops from this patient do not confer resistance to MVC but modulate the magnitude of resistance.
FIG. 2.
Viral resistance to maraviroc is dependent upon residues in the V3 loop of gp120 and is modulated by the V4 loop. (A) Chimeric viruses were created between the maraviroc-sensitive clone S2 and the maraviroc-resistant clone R3. The V3 loop is required for resistance to maraviroc, while the V4 loop can modulate the magnitude of resistance conferred by V3 but does not impart resistance by itself. (B) Point mutations present in the V3 loop of clone S2 were introduced into clone R3 to determine which residues were critical for maraviroc resistance. The H308P revertant greatly increased the sensitivity of clone R3 to maraviroc, while the H320T and V322aI mutations had more modest effects. (C) The V3 loop from resistant clone R3 was introduced into the ADA, BaL, and SF162 Envs. Transferring the V3 loop conferred a significant degree of maraviroc resistance to these heterologous Envs. Data are representative of results from four independent experiments ± standard errors of the mean.
To map the MVC resistance determinants with higher precision, we individually changed the three V3 loop mutations back to the baseline sequences in the resistant clone using site-directed mutagenesis. Pseudoviral particles bearing the three revertants, H308P, H320T, and V322aI all infected NP2/CD4/CCR5 cells efficiently. In the presence of 10 μM MVC, pseudoviruses bearing the resistant Env with the H308P mutation were strongly inhibited by MVC (MPI = 88.3%), whereas the H320T and V322aI revertants retained most of their resistance to MVC (MPI = 17.3% and 9.4%, respectively) (Fig. 2B). Together, these data indicate that the V3 loop played a major role in conferring resistance to MVC, with the P/T308H mutation playing the most important role, but additional mutations outside the V3 loop contributed to the extent of resistance.
Resistance determinants within the V3 loop can transfer resistance to heterologous viruses.
Previous studies of CCR5 antagonist-resistant viruses have identified resistance determinants that were dependent upon the parental Env context and did not transfer resistance when introduced into other viruses (2, 46, 48). To test whether the V3 loop alone from the MVC-resistant Envs would confer resistance to heterologous viral Env proteins, chimeric viruses were created in which the V3 loop was inserted into ADA, BaL, or SF162 Envs. Pseudotypes bearing the parental BaL and SF162 Envs were both inhibited by MVC, with MPIs of >95%, while those bearing the ADA Env were slightly less sensitive to MVC (MPI = 91.9%) (Fig. 2C). In marked contrast, chimeric viruses with the V3 loop from the MVC-resistant Env were highly resistant to MVC (MPI range, 25.0 to 39.2%). These data indicate that the V3 loop alone from the MVC-resistant virus can confer use of MVC-bound CCR5 to heterologous viruses.
Origins of MVC-resistant viruses.
To determine whether MVC resistance emerged due to the outgrowth of a preexisting resistant virus or whether it resulted from viral evolution induced by the selective pressure of MVC treatment, we employed deep sequencing of the V3 loop from patient plasma isolated prior to treatment with MVC. To assess the amino acid frequency of each of the V3 loop positions, we generated 43,168 high-quality sequences representing 3,466 unique V3 sequences. None of these reads contained all three of the mutations found in MVC-resistant clones (H308, H320, and V322a). In addition, of the three residues sufficient to confer partial MVC resistance, we detected only seven sequences with the major MVC resistance-conferring mutation, H308 (0.016%), no sequences with H320, and 5,966 sequences with V322a (13.8%) (Table 1). Notably, the seven sequences with H308 had the MVC-sensitive residues at all other V3 loop positions. While we cannot exclude the possibility that clones containing all resistance mutations existed either below a frequency of 1 in 40,000 or in sites other than plasma, it appears more likely that the mutations associated with MVC resistance emerged during the course of treatment. Consistent with this hypothesis, two of the Envs we analyzed at the intermediate time point (Fig. 1A) contained the H308 and V322a mutations but not the H320 mutation. In addition, sequences consistent with CXCR4 usage existed at a frequency of approximately 0.5% in the pretreatment viral reservoir, yet viral resistance to MVC emerged due to the evolution of drug-bound CCR5 usage rather than the outgrowth of CXCR4-using viruses.
TABLE 1.
Patient 6061 pretreatment viral V3 sequences
| Position in V3 | Amino acid no.a | Amino acid in patient: |
No. of sequences with: |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6061.d0.cl2 | 6061.d224.cl1 | Ala (A) | Arg (R) | Asn (N) | Asp (D) | Cys (C) | Glu (E) | Gln (Q) | Gly (G) | His (H) | Ile (I) | Leu (L) | Lys (K) | Met (M) | Phe (F) | Pro (P) | Ser (S) | Thr (T) | Try (W) | Tyr (Y) | Val (V) | Deletion (−) | ||
| 11 | 306 | S | S | 0 | 17 | 11 | 1 | 4 | 1 | 0 | 6,080 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 37,044 | 0 | 0 | 0 | 1 | 8 |
| 13 | 308 | P | H | 31 | 173 | 5,907 | 4 | 0 | 0 | 0 | 0 | 7 | 0 | 1 | 0 | 778 | 0 | 4,542 | 9 | 31,710 | 0 | 5 | 1 | 0 |
| 19 | 316 | A | S | 9,092 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 1 | 0 | 39 | 33,992 | 6 | 0 | 0 | 23 | 4 |
| 21 | 318 | Y | I | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 42,973 | 173 | 5 |
| 23 | 320 | T | H | 67 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 188 | 0 | 3 | 0 | 42,904 | 0 | 0 | 1 | 4 |
| 24 | 321 | G | G | 0 | 22 | 0 | 0 | 0 | 9 | 0 | 43,132 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 |
| 25 | 322 | D | D | 6 | 0 | 98 | 36,796 | 0 | 5,906 | 15 | 160 | 0 | 0 | 0 | 174 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 6 | 4 |
| 26 | 322a | I | V | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 36,989 | 0 | 0 | 0 | 27 | 0 | 0 | 5,966 | 177 |
Amino acid number is relative to the HXB2 reference strain.
Pretreatment and MVC-resistant Envs use CCR5 efficiently.
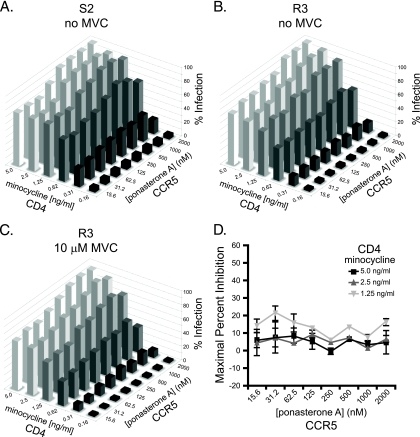
Most CCR5 antagonist-resistant viruses described to date use drug-bound receptors less efficiently than drug-free receptors (44, 50, 60, 66). However, the resistant viruses isolated from patient 6061 demonstrated an MPI of −13.2 to 0.5%, indicating that they could utilize drug-bound CCR5 with very high efficiency. To begin to probe the mechanisms responsible for this phenotype, we asked whether Envs derived from this patient before and during MVC treatment were unusually efficient at mediating infection of cells expressing low levels of CD4 or CCR5, a trait we previously observed in viruses obtained from a patient who exhibited partial resistance to CCR5 antagonists even prior to therapy (48). To do this, we tested the ability of virus pseudotypes bearing MVC-sensitive or MVC-resistant Envs from patient 6061 to infect the Affinofile cell line, a human HEK293 cell line in which CD4 and CCR5 expression can be independently regulated via the use of separate, inducible promoters (28). Cells in a 96-well plate were treated in duplicate with 8 different concentrations of ponasterone A, which induces CCR5 expression, and 6 different concentrations of minocycline, which induces CD4 expression, in all 48 possible combinations. Using quantitative fluorescence-activated cell sorting (FACS), we have found that CCR5 expression ranges from approximately 5,500 to 88,000 copies per cell, and CD4 expression ranges from 5,000 to 350,000 copies per cell. Cells were challenged with MVC-sensitive and -resistant pseudotypes, and infection was determined as normal. Results from this experimental system can be mathematically modeled as a single vector whose magnitude reflects the efficiency of virus entry and the angle of which represents the relative dependence on CD4 or CCR5 (Viral Entry Receptor Sensitivity Analysis [VERSA] computational platform, http://versa.biomath.ucla.edu). Viruses that are highly sensitive to changes in CD4 surface expression but are not impacted by various levels of CCR5 have a vector angle of 0°, while viruses that are independent of CD4 expression but sensitive to CCR5 levels have a vector angle of 90°. In our study, most primary R5 virus strains exhibited vector angles of between 20 and 60°.
Infection with pseudotyped viruses bearing an MVC-sensitive Env demonstrated that this viral Env was highly dependent on CD4 expression levels but used CCR5 very efficiently, even at very low concentrations (Fig. 3A). The sensitivity of this virus to CD4 but not CCR5 levels was reflected in the VERSA angle of 6.7 ± 1.0°. Infection with this sensitive Env was completely abrogated in the presence of 10 μM MVC, as expected (data not shown). Infection of Affinofile cells by pseudoviruses bearing an MVC-resistant Env displayed a similar profile: high dependence on CD4 expression levels but ability to use all CCR5 levels for efficient infection, reflected by the VERSA angle of 7.6 ± 0.7° (Fig. 3B). Viral requirements for CD4 and CCR5 levels were not strongly impacted by 10 μM MVC (Fig. 3C and D), as indicated by a VERSA angle of 7.2 ± 1.1°. These data indicate that the Envs from patient 6061 are unusually adept at infecting cells expressing very low levels of CCR5, provided that CD4 expression levels are adequate.
FIG. 3.
Cloned Envs from patient 6061 utilize CCR5 very efficiently, even in the presence of maraviroc. (A) Infection of pseudotyped viruses bearing Env clone S2 on the Affinofile cell line in the absence of drug. Clone S2 is sensitive to low levels of CD4 but can utilize low levels of CCR5 very efficiently. Infection is normalized to 100% at the highest CD4 and CCR5 levels. Pseudoviruses bearing Env clone S2 were completely inhibited by maraviroc (not shown). Infection of Affinofile cells with pseudotypes bearing maraviroc-resistant Env R3 in the absence (B) or presence (C) of 10 μM maraviroc. Clone R3 is also sensitive to low levels of CD4 but can infect cells bearing very low CCR5 levels both in the absence and presence of maraviroc. (D) Maximal percent inhibition of clone R3 by 10 μM maraviroc at different CCR5 densities was calculated from the data in panels B and C. Inhibition of clone R3 by maraviroc was not strongly impacted by cell CCR5 levels. Data are representative of results from four independent experiments ± standard errors of the mean.
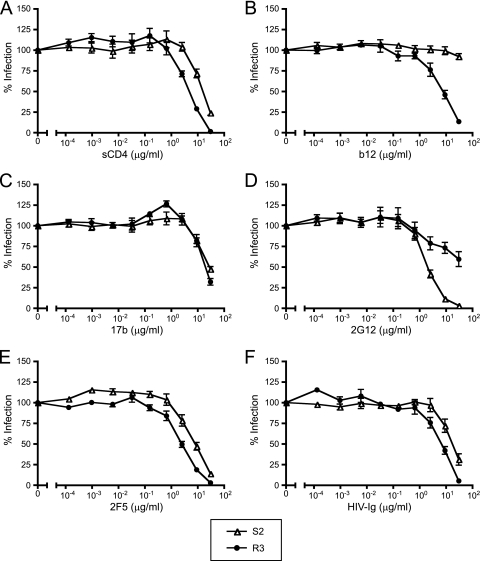
MVC resistance is associated with increased sensitivity to soluble CD4 and b12.
To provide insight into the exposure of the CD4 and coreceptor binding sites that would account for the unusual ability of these MVC-resistant viruses to infect cells expressing low CCR5 levels, as well as to determine whether MVC resistance was associated with increased sensitivity to neutralization, we performed infection assays in which pseudotypes bearing an MVC-resistant or an MVC-sensitive Env were preincubated with soluble CD4 (sCD4), monoclonal antibodies, or HIV-Ig and used to infect NP2/CD4/CCR5 cells (Fig. 4A to F). In these assays, the pseudotypes bearing the MVC-resistant Env were significantly more sensitive to inhibition by sCD4 and b12 (P < 0.02 for both), equally sensitive to 17b, and significantly less sensitive to 2G12 (P = 0.04). 2F5 and HIV-Ig both inhibited MVC-resistant Env to a greater degree than MVC-sensitive Env, but these did not reach statistical significance after corrections for multiple comparisons (P = 0.06 and P = 0.2, respectively). The decreased susceptibility of the MVC-resistant virus to inhibition by 2G12 is likely related to the loss of a glycosylation site at position 386, which has previously been identified as a peripheral glycan contributing to the 2G12 epitope (56). The increased sensitivity of MVC-resistant virus to sCD4 and b12 may reflect differential exposure of the CD4 binding site due to changes within the V3 or V4 loops, possibly including the loss of a shielding glycan at position 386 (55). Alternatively, the resistant virus may have slightly slower fusion kinetics than the sensitive virus, as evidenced by a small difference in susceptibility to inhibition by enfuvirtide (50% inhibitory concentration [IC50] of 252 versus 439 nM/ml). The latter hypothesis would also account for the trend toward increased neutralization by 2F5 and HIV Ig. The equivalent sensitivities of these Envs to 17b indicate that major differences in the exposure of the coreceptor binding site do not exist between the two viruses. Overall, the MVC-resistant virus appears slightly more neutralization sensitive, with the exception of the increased resistance to 2G12 due to the loss of a glycosylation site at residue 386.
FIG. 4.
Maraviroc-resistant clone R3 is more sensitive to sCD4 and b12 and more resistant to 2G12 than sensitive clone S2. Pseudotyped viruses bearing Env clones S2 and R3 were preincubated with various concentrations of sCD4 (A), b12 (B), 17b (C), 2G12 (D), 2F5 (E), and HIV Ig (F) for 1 h and spinoculated with NP2/CD4/CCR5 cells. The maraviroc-resistant clone R3 is significantly more sensitive to sCD4 and b12 and more resistant to 2G12 than the maraviroc-sensitive clone S2. Data are representative of results from three independent experiments ± standard errors of the mean.
MVC-resistant virus is sensitive to changes in both the N terminus and extracellular loops of CCR5 in the presence of drug.
Recent studies have demonstrated that viral Envs resistant to most or all CCR5 antagonists often have an increased reliance upon the N-terminal domain of CCR5 for entry (34, 43, 44, 48). To investigate how the MVC-resistant virus is able to recognize the MVC-CCR5 complex but not other drug-CCR5 complexes, we performed pseudoviral infection assays of 293T cells stably expressing CD4 and transiently expressing either wild-type CCR5 or one of a panel of CCR5 mutants. Mutants within the N terminus (Y3A, Q4A, Y10A, D11A, Y14A, Y15A, N24A, and Q27A), ECL1 (H88A, Q93A, and N98A), ECL2 (S179A, H181A, F182A, P183A, Q186A, Y187A, F189A, and W190A), and ECL3 (E262A, N267A, N268A, C269A, and N273A) were analyzed for their ability to support infection in the absence or presence of 10 μM MVC. All CCR5 mutants were able to support robust infection by MVC-sensitive and -resistant clones in the absence of MVC, with relative light unit (RLU) values >1,000-fold higher than background levels in this assay. The presence of 10 μM MVC completely inhibited infection with the MVC-sensitive clones (MPI of >97% for the wild-type CCR5 and all mutants), indicating that none of these amino acid substitutions prevented drug from binding to the receptor.
On wild-type CCR5, the MVC-resistant clone exhibited an MPI of −5.9 ± 15.7% (Table 2). Consistent with previous studies, residues within the N terminus of CCR5 were critical for the ability of the MVC-resistant virus to utilize the drug-bound receptor. The tyrosine mutants Y10A, Y14A, and Y15A all strongly inhibited the ability of the resistant Env to use the drug-bound receptor. The D11A mutation also strongly reduced infection, while the remaining N-terminal mutations reduced infection in the presence of MVC to variable extents. In addition, this Env also demonstrated sensitivity to several residues within the ECLs of CCR5, albeit to a lesser degree than the N-terminal mutants. The single amino acid changes H88A, H181A, F182A, P183A, W190A, and C269A all significantly increased MVC sensitivity, with MPIs of 48.7 ± 10.5%, 65.2 ± 8.5%, 67.2 ± 9.0%, 68.0 ± 9.4%, and 73.8 ± 3.4%, respectively. Taken together, these results indicate that amino acid residues within the N-terminal domain of CCR5 play an essential role in supporting infection by the MVC-resistant viruses studied here, while interactions with individual amino acids in the ECLs of CCR5, particularly ECL2, are also important but not absolutely required for infection in the presence of drug.
TABLE 2.
Sensitivity of clone R3 to inhibition by MVC on mutant CCR5 receptorsa
| CCR5 receptor | MPI (%) | % resistance of wild-type CCR5 |
|---|---|---|
| Wild-type CCR5 | −5.9 ± 15.7 | 100 |
| N-terminal mutants | ||
| Y3A | 47.3 ± 11.0 | 48.9 ± 4.9 |
| Q4A | 6.7 ± 1.5 | 92.8 ± 15.8 |
| Y10A | 94.3 ± 1.9 | 5.8 ± 2.2 |
| D11A | 92.1 ± 2.7 | 7.6 ± 2.1 |
| Y14A | 92.8 ± 3.3 | 6.3 ± 2.3 |
| Y15A | 96.6 ± 1.5 | 3.1 ± 1.1 |
| N24A | 33.1 ± 7.6 | 65.1 ± 8.4 |
| Q27A | 8.3 ± 8.9 | 89.6 ± 11.9 |
| ECL1 mutants | ||
| H88A | 48.7 ± 10.5 | 47.6 ± 4.4 |
| Q93A | 23.3 ± 2.8 | 75.9 ± 11.9 |
| N98A | 38.7 ± 2.7 | 61.9 ± 13.6 |
| ECL2 mutants | ||
| S179A | 33.1 ± 11.4 | 62.9 ± 2.8 |
| H181A | 65.2 ± 8.5 | 31.7 ± 3.8 |
| F182A | 67.2 ± 9.0 | 29.6 ± 5.0 |
| P183A | 50.1 ± 2.1 | 50.2 ± 10.6 |
| Q186A | 28.0 ± 14.7 | 67.1 ± 6.9 |
| Y187A | 5.5 ± 15.3 | 88.8 ± 3.1 |
| F189A | 23.4 ± 25.0 | 70.0 ± 17.7 |
| W190A | 68.0 ± 9.4 | 28.9 ± 5.1 |
| ECL3 mutants | ||
| E262A | 14.1 ± 20.0 | 79.3 ± 10.6 |
| N267A | 28.7 ± 21.2 | 65.4 ± 12.1 |
| N268A | −0.7 ± 15.7 | 95.0 ± 4.7 |
| C269A | 73.8 ± 3.4 | 25.8 ± 4.4 |
| N273A | −1.3 ± 17.5 | 96.7 ± 11.7 |
All values represent the means ± standard errors of the means.
CCR5 antagonists induce drug-specific conformational changes in the extracellular loops but not the N terminus of CCR5.
To gain further insight into how different CCR5 antagonists disrupt the interactions between gp120 and CCR5 and how resistant viruses may recognize drug-bound receptors, we utilized a panel of N-terminal- and ECL-directed MAbs to probe drug-bound CCR5. NP2/CD4 cells stably expressing CCR5 were preincubated with the CCR5 antagonists maraviroc, aplaviroc, vicriviroc, CMPD-167, and TAK779 and then were stained with the N-terminal MAb clones 3A9, CTC5, and CTC8 or with the ECL MAb clones 2D7, 45523, 45529, 45531, and 45549. NP2/CD4/CCR5 cells stained in the absence of drug were utilized as a positive control, while NP2/CD4/CXCR4 cells served as a negative control.
The N-terminal antibodies 3A9 and CTC5 bound equivalently to unbound CCR5 and to all of the drug-bound CCR5s (Fig. 5). The binding of the N-terminal antibody CTC8 to CCR5 was slightly reduced by preincubation with CMPD-167 and slightly enhanced by maraviroc, aplaviroc, vicriviroc, or TAK779. In contrast, the ECL MAbs 45523, 45529, and 45531 demonstrated diminished binding to CCR5 in the presence of all of the antagonists tested. The ECL antibodies 2D7 and 45549 showed variable results, with some of the drugs inhibiting and some enhancing antibody binding. A common feature of ECL loop antibodies was their differential sensitivity to CCR5 antagonists. For example, binding of 45531 to CCR5 was moderately diminished by maraviroc but was completely blocked by aplaviroc. These data suggest several important features of CCR5 antagonists. First, the conformation of the N terminus of CCR5 does not appear to be dramatically altered by drug, as judged by the ability of N terminus-specific MAbs to bind with equivalent efficiencies to drug-free or drug-bound receptors. Second, the conformation of the ECLs of CCR5 is altered by the presence of drug, as measured by diminished or completely abrogated binding of several ECL-specific MAbs. Third, different CCR5 antagonists cause different changes to the ECLs of CCR5, suggesting that the conformational changes induced are drug specific.
FIG. 5.
CCR5 antagonists disrupt the extracellular loops of CCR5 but have minor effects upon the N terminus. NP2/CD4/CCR5 cells were incubated with medium alone or medium containing saturating concentrations of maraviroc (MVC), aplaviroc (APL), vicriviroc (VVC), CMPD-167, or TAK779. NP2/CD4/CXCR4 cells were utilized as controls. Cells were probed with the N terminus-specific anti-CCR5 monoclonal antibodies (MAbs) 3A9, CTC5, and CTC8 or the extracellular loop (ECL)-specific anti-CCR5 MAbs 2D7, 45523, 45529, 45531, or 45549. CCR5 antagonists had no effect on the binding of N terminus-specific MAbs 3A9 and CTC5 and minor effects on CTC8 binding. In contrast, CCR5 antagonists caused significantly reduced binding of the ECL-specific MAbs 45523, 45529, and 45531 and minor disruption of 2D7 and 45549 binding. Different CCR5 antagonists reduced binding by ECL-specific MAbs to various degrees, suggesting that these drugs induce inhibitor-specific alterations to the ECLs. Data are representative of results from three independent experiments.
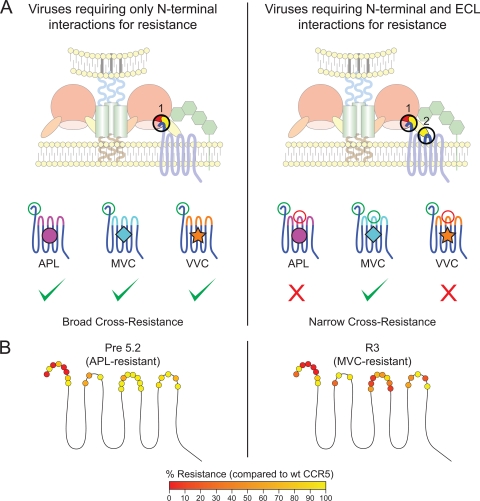
Based on these findings and the results from the studies examining infection of cells expressing mutant CCR5 receptors, we propose a model of cross-resistance for this class of drug (Fig. 6A). Viruses that are heavily dependent upon the N terminus but not the ECLs of CCR5 could be able to recognize multiple drug-bound conformations of CCR5 and demonstrate broad cross-resistance. In contrast, viruses that are sensitive to alterations in ECLs of CCR5—the region of the receptor most greatly affected by small-molecule antagonists—may be restricted by their ability to recognize drug-specific ECL changes and may display a much more narrow cross-resistance profile.
FIG. 6.
Model of cross-resistance of Envs to CCR5 antagonists. (A) CCR5 antagonist-resistant viruses that only require the N terminus for binding in the presence of drug recognize a region of the coreceptor that is relatively unaffected by different drugs. As a consequence, these viruses can utilize wild-type CCR5 or CCR5 bound to a variety of different CCR5 antagonists, resulting in broad cross-resistance. In contrast, other resistant viruses require both the N terminus and the extracellular loops (ECLs) of CCR5. Unlike the N terminus, the ECLs of CCR5 are disrupted in an inhibitor-specific manner. Thus, the maraviroc (MVC)-resistant virus can recognize MVC-bound CCR5 but not aplaviroc (APL)- or vicriviroc (VVC)-bound conformations, resulting in narrow cross-resistance. (B) Effect of N-terminal ECL1, ECL2, and ECL3 mutations on drug resistance of the APL-resistant Env Pre5.2 (data from reference 48) and the MVC-resistant Env R3 (data from Table 2). Mutations are shaded according to the degree to which they eliminate drug resistance, with those abolishing resistance shaded red and those having no effect on drug resistance shaded yellow. Data are representative of results from three independent experiments ± standard errors of the mean.
DISCUSSION
HIV can evolve resistance to CCR5 antagonists by utilizing CXCR4 for entry or by adapting to recognize drug-bound CCR5. Treatment of patients with the CCR5 antagonists maraviroc, vicriviroc, and aplaviroc often results in virologic failure due to the emergence of viruses that utilize CXCR4 as a coreceptor for entry (23, 35, 65). In most cases, this appears to be due to the selective outgrowth of minor CXCR4-utilizing variants that were preexisting in the patients' viral reservoirs prior to treatment and which gain a selective advantage during CCR5 antagonist therapy. In other cases, viruses replicating in the presence of CCR5 antagonists either in vitro or in vivo have evolved resistance by acquiring the ability to utilize the drug-bound conformation of CCR5 for entry while still retaining the ability to efficiently utilize the drug-free conformation of CCR5 (2, 4, 33, 41, 44, 60, 61, 63, 66). In most instances described to date, such viruses exhibit cross-resistance to multiple CCR5 antagonists (33, 41, 60). Less commonly, a more narrow drug resistance profile is observed (66). For example, the MVC-resistant viruses studied here remain sensitive to most other CCR5 antagonists. Together, these observations indicate that resistance to CCR5 antagonists entails a variable degree of conformational plasticity with regard to CCR5 recognition. In all cases, drug-resistant viruses are able to efficiently utilize the drug-free conformation of CCR5, and in addition at least one and more often multiple drug-bound conformations are recognized with variable efficiency. In this study, we sought to explore the mechanisms that account for these different phenotypes.
There is ample precedent for variable Env-coreceptor recognition. Mutations introduced into CCR5 can have differential effects on infection by different HIV-1 strains (3, 6, 17, 29, 32, 38, 49, 54). In many ways, R5X4 viruses represent an extreme example of this property, as they are able to efficiently utilize both CCR5 and CXCR4 despite significant differences in the ectodomain sequences of these coreceptors. A large number of studies suggest that Env-coreceptor interactions involve at least two regions in Env and two coreceptor domains. One interaction occurs between residues within the bridging sheet and at the base of the V3 loop of gp120, which bind to the N-terminal domain of CCR5, with O-sulfated tyrosines playing a critical role (15, 19, 26, 29, 52). A second interaction is thought to occur between the crown of the V3 loop and the extracellular loops, particularly ECL2, of CCR5 (14, 17, 29, 37). The small-molecule antagonists of CCR5 do not act by reducing surface expression of CCR5 but rather by binding to a hydrophobic pocket within the transmembrane helices and inducing conformational changes that prevent HIV from utilizing drug-bound receptors for entry (18, 39, 40, 58, 62). That CCR5 antagonists alter coreceptor conformation in different ways is documented by differential effects of various drugs on chemokine and antibody binding to CCR5 (59, 64). We explored the impact of this drug class on CCR5 conformation in a more systematic way and found that while the various drugs had little to no impact on the recognition of CCR5 by three different MAbs to the N-terminal domain of the receptor, there was striking variability in how antibodies directed to the ECLs recognized drug-bound forms of CCR5. The implication of these findings, when coupled with previous work, is that CCR5 antagonists alter ECL conformation while not necessarily altering the conformation of the N-terminal domain and that different CCR5 antagonists induce different conformational changes in the ECLs. Although the MVC-resistant Envs from patient 6061 were also partially cross-resistant to TAK779, the MAbs directed against the N terminus or ECLs did not reveal obvious structural differences between MVC- or TAK779-bound CCR5 compared with other antagonist-CCR5 complexes which still inhibit infection. The mechanism of cross-resistance between MVC and TAK779 is under investigation and may be the result of structural similarities between the drug-bound receptors that were not probed by the MAbs utilized or the ability of the Envs to independently recognize two structurally different drug-receptor complexes.
We can propose a model to account for how the viruses studied here can utilize drug-free and MVC-bound CCR5 for infection but not CCR5 bound to other antiviral drugs, while other viruses can utilize CCR5 in the presence of any CCR5 antagonist. The viruses studied to date that exhibit resistance to multiple CCR5 antagonists appear to be exquisitely sensitive to mutations in the N-terminal domain of CCR5 in the presence of drug but relatively insensitive to mutations within the ECL domains (Fig. 6B). As the conformation of the N-terminal domain of CCR5 appears to not be greatly altered by binding of different CCR5 antagonists to the receptor, a plausible model posits that these viruses have adapted to recognize a portion of the receptor that is relatively unaffected by drug binding and thus can utilize drug-bound forms of CCR5 that differ in ECL conformation as well as drug-free receptors. In contrast, the narrowly cross-resistant Envs reported here require residues within both the N-terminal domain and the ECLs of CCR5 in the presence of drug. Unlike the N terminus, the conformations of the ECLs are disrupted by drug binding in a manner that appears to be compound specific. We speculate that the mutations in the V3 loop found in this virus enable it to interact with the MVC-bound conformation of the ECLs but not with conformations induced by other CCR5 antagonists. Unusually, transfer of the V3 mutations responsible for drug resistance into drug-sensitive Env backgrounds conferred the ability to use MVC-bound CCR5 with three diverse subtype B viruses. Thus, these viruses appear to be uniquely adapted to recognize the MVC-bound conformation of CCR5 yet are still able to utilize the drug-free receptor via interactions with the N-terminal domain of the coreceptor and perhaps the ECLs as well.
It is not clear why virologic failure following CCR5 antagonist therapy results sometimes from outgrowth of CXCR4-using viruses, sometimes from viruses that are resistant to multiple CCR5 antagonists, and sometimes from viruses that exhibit a narrow drug resistance profile. There are clearly selective pressures against the emergence of CXCR4-using viruses, since these typically fail to arise until years after infection and often fail to evolve to detectable levels (1, 7, 10, 11, 13, 47, 57). In the patient studied here, deep sequencing revealed the presence of V3 sequences that are consistent with the use of CXCR4 for entry at a frequency of approximately 0.5% in the pretreatment sample. These findings indicate that the presence of X4-tropic HIV variants at baseline does not necessarily predict that patients will fail CCR5 antagonist therapy by the outgrowth of these X4 viruses and raises the possibility that these agents may have some efficacy in certain patients harboring dual/mixed viral populations. In contrast, no sequences containing all three of the mutations associated with resistance in this patient were present at baseline, suggesting that de novo viral evolution to use MVC-bound CCR5 was favored over outgrowth of CXCR4-using viruses. Using the newly developed Affinofile assay by Johnston and colleagues (28), which allows detailed characterization of viral requirements for CD4 and CCR5 levels, we found that the MVC-resistant viruses described here as well as aplaviroc-resistant Envs derived from a second patient were unusual in their ability to efficiently utilize low levels of CCR5 for virus infection (48). Although speculative, it is possible that the ability to utilize very low levels of CCR5 for entry reflects highly efficient use of CCR5 and that viruses that utilize CCR5 very efficiently may be able to tolerate mutations within the V3 loop without abolishing infectivity. Some of these mutations, in turn, may enable the virus to utilize drug-bound CCR5 more efficiently and give rise to drug-resistant variants. Further studies with resistant viruses and their requirements for CCR5 levels will be necessary to test this hypothesis.
In summary, among individuals who are exhibiting failure of an MVC-based regimen, the vast majority appeared to fail with either a CXCR4-utilizing virus or a CCR5-utilizing virus that lacked any in vitro evidence of CCR5 inhibitor resistance. CCR5 resistance can occur, however, and at least in the case described here, resistance to MVC requires the emergence of multiple mutations with the V3 and perhaps V4 loops. The relative fitness of this resistant virus remains to be defined but is likely to be high given its remarkable ability to replicate in vivo (high viral load) and its ability to replicate in vitro in the presence of very low levels of CCR5. It remains to be determined whether patients with viruses capable of utilizing low CCR5 levels are more apt to fail MVC therapy due to mutations in Env that enable the use of drug-bound CCR5. If so, cross-resistance to other CCR5 inhibitors may not be inevitable due to differences in how Env can interact with distinct CCR5 domains.
Footnotes
Published ahead of print on 11 August 2010.
REFERENCES
- 1.Abebe, A., D. Demissie, J. Goudsmit, M. Brouwer, C. L. Kuiken, G. Pollakis, H. Schuitemaker, A. L. Fontanet, and T. F. Rinke de Wit. 1999. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and coreceptor usage among Ethiopian patients with AIDS. AIDS 13:1305-1311. [DOI] [PubMed] [Google Scholar]
- 2.Anastassopoulou, C. G., T. J. Ketas, P. J. Klasse, and J. P. Moore. 2009. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc. Natl. Acad. Sci. U. S. A. 106:5318-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atchison, R. E., J. Gosling, F. S. Monteclaro, C. Franci, L. Digilio, I. F. Charo, and M. A. Goldsmith. 1996. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science 274:1924-1926. [DOI] [PubMed] [Google Scholar]
- 4.Baba, M., H. Miyake, X. Wang, M. Okamoto, and K. Takashima. 2007. Isolation and characterization of human immunodeficiency virus type 1 resistant to the small-molecule CCR5 antagonist TAK-652. Antimicrob. Agents Chemother. 51:707-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berro, R., R. W. Sanders, M. Lu, P. J. Klasse, and J. P. Moore. 2009. Two HIV-1 variants resistant to small molecule CCR5 inhibitors differ in how they use CCR5 for entry. PLoS Pathog. 5:e1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieniasz, P. D., R. A. Fridell, I. Aramori, S. S. Ferguson, M. G. Caron, and B. R. Cullen. 1997. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 16:2599-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorndal, A., A. Sonnerborg, C. Tscherning, J. Albert, and E. M. Fenyo. 1999. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res. Hum. Retroviruses 15:647-653. [DOI] [PubMed] [Google Scholar]
- 8.Cardozo, T., T. Kimura, S. Philpott, B. Weiser, H. Burger, and S. Zolla-Pazner. 2007. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res. Hum. Retroviruses 23:415-426. [DOI] [PubMed] [Google Scholar]
- 9.Chen, B. K., K. Saksela, R. Andino, and D. Baltimore. 1994. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J. Virol. 68:654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cilliers, T., J. Nhlapo, M. Coetzer, D. Orlovic, T. Ketas, W. C. Olson, J. P. Moore, A. Trkola, and L. Morris. 2003. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J. Virol. 77:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connell, B. J., K. Michler, A. Capovilla, W. D. Venter, W. S. Stevens, and M. A. Papathanasopoulos. 2008. Emergence of X4 usage among HIV-1 subtype C: evidence for an evolving epidemic in South Africa. Aids 22:896-899. [DOI] [PubMed] [Google Scholar]
- 12.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 13.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1 infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cormier, E. G., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cormier, E. G., D. N. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doranz, B. J., Z. H. Lu, J. Rucker, T. Y. Zhang, M. Sharron, Y. H. Cen, Z. X. Wang, H. H. Guo, J. G. Du, M. A. Accavitti, R. W. Doms, and S. C. Peiper. 1997. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J. Virol. 71:6305-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dragic, T., A. Trkola, D. A. Thompson, E. G. Cormier, F. A. Kajumo, E. Maxwell, S. W. Lin, W. Ying, S. O. Smith, T. P. Sakmar, and J. P. Moore. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. U. S. A. 97:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farzan, M., H. Choe, L. Vaca, K. Martin, Y. Sun, E. Desjardins, N. Ruffing, L. Wu, R. Wyatt, N. Gerard, C. Gerard, and J. Sodroski. 1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol. 72:1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouchier, R. A., M. Brouwer, S. M. Broersen, and H. Schuitemaker. 1995. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J. Clin. Microbiol. 33:906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulick, R. M., J. Lalezari, J. Goodrich, N. Clumeck, E. DeJesus, A. Horban, J. Nadler, B. Clotet, A. Karlsson, M. Wohlfeiler, J. B. Montana, M. McHale, J. Sullivan, C. Ridgway, S. Felstead, M. W. Dunne, E. van der Ryst, and H. Mayer. 2008. Maraviroc for previously treated patients with R5 HIV-1 infection. N. Engl. J. Med. 359:1429-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulick, R. M., Z. Su, C. Flexner, M. D. Hughes, P. R. Skolnik, T. J. Wilkin, R. Gross, A. Krambrink, E. Coakley, W. L. Greaves, A. Zolopa, R. Reichman, C. Godfrey, M. Hirsch, and D. R. Kuritzkes. 2007. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-infected, treatment-experienced patients: AIDS clinical trials group 5211. J. Infect. Dis. 196:304-312. [DOI] [PubMed] [Google Scholar]
- 24.Hatano, H., H. Lampiris, S. Fransen, S. Gupta, W. Huang, R. Hoh, J. N. Martin, J. Lalezari, D. Bangsberg, C. Petropoulos, and S. G. Deeks. 2010. Evolution of integrase resistance during failure of integrase inhibitor-based antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 54:389-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann, C., N. Minkah, J. Leipzig, G. Wang, M. Q. Arens, P. Tebas, and F. D. Bushman. 2007. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 35:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, C. C., S. N. Lam, P. Acharya, M. Tang, S. H. Xiang, S. S. Hussan, R. L. Stanfield, J. Robinson, J. Sodroski, I. A. Wilson, R. Wyatt, C. A. Bewley, and P. D. Kwong. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, M. A., F. S. Li, A. B. van 't Wout, D. C. Nickle, D. Shriner, H. X. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston, S. H., M. A. Lobritz, S. Nguyen, K. Lassen, S. Delair, F. Posta, Y. J. Bryson, E. J. Arts, T. Chou, and B. Lee. 2009. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J. Virol. 83:11016-11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajumo, F., D. A. Thompson, Y. Guo, and T. Dragic. 2000. Entry of R5X4 and X4 human immunodeficiency virus type 1 strains is mediated by negatively charged and tyrosine residues in the amino-terminal domain and the second extracellular loop of CXCR4. Virology 271:240-247. [DOI] [PubMed] [Google Scholar]
- 30.Keele, B. F., E. E. Giorgi, J. F. Salazar-Gonzalez, J. M. Decker, K. T. Pham, M. G. Salazar, C. Sun, T. Grayson, S. Wang, H. Li, X. Wei, C. Jiang, J. L. Kirchherr, F. Gao, J. A. Anderson, L. H. Ping, R. Swanstrom, G. D. Tomaras, W. A. Blattner, P. A. Goepfert, J. M. Kilby, M. S. Saag, E. L. Delwart, M. P. Busch, M. S. Cohen, D. C. Montefiori, B. F. Haynes, B. Gaschen, G. S. Athreya, H. Y. Lee, N. Wood, C. Seoighe, A. S. Perelson, T. Bhattacharya, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondru, R., J. Zhang, C. Ji, T. Mirzadegan, D. Rotstein, S. Sankuratri, and M. Dioszegi. 2008. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol. Pharmacol. 73:789-800. [DOI] [PubMed] [Google Scholar]
- 32.Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat. 1997. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J. Virol. 71:8642-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhmann, S. E., P. Pugach, K. J. Kunstman, J. Taylor, R. L. Stanfield, A. Snyder, J. M. Strizki, J. Riley, B. M. Baroudy, I. A. Wilson, B. T. Korber, S. M. Wolinsky, and J. P. Moore. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 78:2790-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laakso, M. M., F. H. Lee, B. Haggarty, C. Agrawal, K. M. Nolan, M. Biscone, J. Romano, A. P. Jordan, G. J. Leslie, E. G. Meissner, L. Su, J. A. Hoxie, and R. W. Doms. 2007. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 3:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lalezari, J., M. Thompson, P. Kumar, P. Piliero, R. Davey, K. Patterson, A. Shachoy-Clark, K. Adkison, J. Demarest, Y. Lou, M. Berrey, and S. Piscitelli. 2005. Antiviral activity and safety of 873140, a novel CCR5 antagonist, during short-term monotherapy in HIV-infected adults. AIDS 19:1443-1448. [DOI] [PubMed] [Google Scholar]
- 36.Landovitz, R. J., J. B. Angel, C. Hoffmann, H. Horst, M. Opravil, J. Long, W. Greaves, and G. Fatkenheuer. 2008. Phase II study of vicriviroc versus efavirenz (both with zidovudine/lamivudine) in treatment-naive subjects with HIV-1 infection. J. Infect. Dis. 198:1113-1122. [DOI] [PubMed] [Google Scholar]
- 37.Lee, B., M. Sharron, C. Blanpain, B. J. Doranz, J. Vakili, P. Setoh, E. Berg, G. Liu, H. R. Guy, S. R. Durell, M. Parmentier, C. N. Chang, K. Price, M. Tsang, and R. W. Doms. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617-9626. [DOI] [PubMed] [Google Scholar]
- 38.Lu, Z., J. F. Berson, Y. Chen, J. D. Turner, T. Zhang, M. Sharron, M. H. Jenks, Z. Wang, J. Kim, J. Rucker, J. A. Hoxie, S. C. Peiper, and R. W. Doms. 1997. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc. Natl. Acad. Sci. U. S. A. 94:6426-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maeda, K., D. Das, H. Ogata-Aoki, H. Nakata, T. Miyakawa, Y. Tojo, R. Norman, Y. Takaoka, J. Ding, G. F. Arnold, E. Arnold, and H. Mitsuya. 2006. Structural and molecular interactions of CCR5 inhibitors with CCR5. J. Biol. Chem. 281:12688-12698. [DOI] [PubMed] [Google Scholar]
- 40.Maeda, K., D. Das, P. D. Yin, K. Tsuchiya, H. Ogata-Aoki, H. Nakata, R. B. Norman, L. A. Hackney, Y. Takaoka, and H. Mitsuya. 2008. Involvement of the second extracellular loop and transmembrane residues of CCR5 in inhibitor binding and HIV-1 fusion: insights into the mechanism of allosteric inhibition. J. Mol. Biol. 381:956-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marozsan, A. J., S. E. Kuhmann, T. Morgan, C. Herrera, E. Rivera-Troche, S. Xu, B. M. Baroudy, J. Strizki, and J. P. Moore. 2005. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D). Virology 338:182-199. [DOI] [PubMed] [Google Scholar]
- 42.Moore, J. P., and D. R. Kuritzkes. 2009. A piece de resistance: how HIV-1 escapes small molecule CCR5 inhibitors. Curr. Opin. HIV AIDS 4:118-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nolan, K. M., G. Q. Del Prete, A. P. Jordan, B. Haggarty, J. Romano, G. J. Leslie, and J. A. Hoxie. 2009. Characterization of a human immunodeficiency virus type 1 V3 deletion mutation that confers resistance to CCR5 inhibitors and the ability to use aplaviroc-bound receptor. J. Virol. 83:3798-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogert, R. A., L. Ba, Y. Hou, C. Buontempo, P. Qiu, J. Duca, N. Murgolo, P. Buontempo, R. Ralston, and J. A. Howe. 2009. Structure-function analysis of human immunodeficiency virus type 1 gp120 amino acid mutations associated with resistance to the CCR5 coreceptor antagonist vicriviroc. J. Virol. 83:12151-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogert, R. A., Y. Hou, L. Ba, L. Wojcik, P. Qiu, N. Murgolo, J. Duca, L. M. Dunkle, R. Ralston, and J. A. Howe. 2010. Clinical resistance to vicriviroc through adaptive V3 loop mutations in HIV-1 subtype D gp120 that alter interactions with the N-terminus and ECL2 of CCR5. Virology 400:145-155. [DOI] [PubMed] [Google Scholar]
- 46.Ogert, R. A., L. Wojcik, C. Buontempo, L. Ba, P. Buontempo, R. Ralston, J. Strizki, and J. A. Howe. 2008. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120. Virology 373:387-399. [DOI] [PubMed] [Google Scholar]
- 47.Peeters, M., R. Vincent, J. L. Perret, M. Lasky, D. Patrel, F. Liegeois, V. Courgnaud, R. Seng, T. Matton, S. Molinier, and E. Delaporte. 1999. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:115-121. [DOI] [PubMed] [Google Scholar]
- 48.Pfaff, J., C. B. Wilen, J. E. Harrison, J. Demarest, B. Lee, R. W. Doms, and J. C. Tilton. 2010. HIV-1 resistance to CCR5 antagonists associated with highly efficient use of CCR5 and altered tropism on primary CD4+ T cells. J. Virol. 84:6505-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picard, L., G. Simmons, C. A. Power, A. Meyer, R. A. Weiss, and P. R. Clapham. 1997. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J. Virol. 71:5003-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pugach, P., N. Ray, P. J. Klasse, T. J. Ketas, E. Michael, R. W. Doms, B. Lee, and J. P. Moore. 2009. Inefficient entry of vicriviroc-resistant HIV-1 via the inhibitor-CCR5 complex at low cell surface CCR5 densities. Virology 387:296-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Resch, W., N. Hoffman, and R. Swanstrom. 2001. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288:51-62. [DOI] [PubMed] [Google Scholar]
- 52.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 53.Rozera, G., I. Abbate, A. Bruselles, C. Vlassi, G. D'Offizi, P. Narciso, G. Chillemi, M. Prosperi, G. Ippolito, and M. R. Capobianchi. 2009. Massively parallel pyrosequencing highlights minority variants in the HIV-1 env quasispecies deriving from lymphomonocyte subpopulations. Retrovirology 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rucker, J., M. Samson, B. J. Doranz, F. Libert, J. F. Berson, Y. Yi, R. J. Smyth, R. G. Collman, C. C. Broder, G. Vassart, R. W. Doms, and M. Parmentier. 1996. Regions in beta-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell 87:437-446. [DOI] [PubMed] [Google Scholar]
- 55.Sanders, R. W., E. van Anken, A. A. Nabatov, I. M. Liscaljet, I. Bontjer, D. Eggink, M. Melchers, E. Busser, M. M. Dankers, F. Groot, I. Braakman, B. Berkhout, and W. A. Paxton. 2008. The carbohydrate at asparagine 386 on HIV-1 gp120 is not essential for protein folding and function but is involved in immune evasion. Retrovirology 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seibert, C., W. Ying, S. Gavrilov, F. Tsamis, S. E. Kuhmann, A. Palani, J. R. Tagat, J. W. Clader, S. W. McCombie, B. M. Baroudy, S. O. Smith, T. Dragic, J. P. Moore, and T. P. Sakmar. 2006. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology 349:41-54. [DOI] [PubMed] [Google Scholar]
- 59.Strizki, J. M., S. Xu, N. E. Wagner, L. Wojcik, J. Liu, Y. Hou, M. Endres, A. Palani, S. Shapiro, J. W. Clader, W. J. Greenlee, J. R. Tagat, S. McCombie, K. Cox, A. B. Fawzi, C. C. Chou, C. Pugliese-Sivo, L. Davies, M. E. Moreno, D. D. Ho, A. Trkola, C. A. Stoddart, J. P. Moore, G. R. Reyes, and B. M. Baroudy. 2001. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 98:12718-12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tilton, J. C., H. Amrine-Madsen, J. L. Miamidian, K. M. Kitrinos, J. Pfaff, J. F. Demarest, N. Ray, J. L. Jeffrey, C. C. Labranche, and R. W. Doms. 2010. HIV type 1 from a patient with baseline resistance to CCR5 antagonists uses drug-bound receptor for entry. AIDS Res. Hum. Retroviruses 26:13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. U. S. A. 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsamis, F., S. Gavrilov, F. Kajumo, C. Seibert, S. Kuhmann, T. Ketas, A. Trkola, A. Palani, J. W. Clader, J. R. Tagat, S. McCombie, B. Baroudy, J. P. Moore, T. P. Sakmar, and T. Dragic. 2003. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 77:5201-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsibris, A. M., M. Sagar, R. M. Gulick, Z. Su, M. Hughes, W. Greaves, M. Subramanian, C. Flexner, F. Giguel, K. E. Leopold, E. Coakley, and D. R. Kuritzkes. 2008. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J. Virol. 82:8210-8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watson, C., S. Jenkinson, W. Kazmierski, and T. Kenakin. 2005. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol. Pharmacol. 67:1268-1282. [DOI] [PubMed] [Google Scholar]
- 65.Westby, M., M. Lewis, J. Whitcomb, M. Youle, A. L. Pozniak, I. T. James, T. M. Jenkins, M. Perros, and E. van der Ryst. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80:4909-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Westby, M., C. Smith-Burchnell, J. Mori, M. Lewis, M. Mosley, M. Stockdale, P. Dorr, G. Ciaramella, and M. Perros. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 81:2359-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]