Abstract
Immunization of rhesus macaques with strains of simian immunodeficiency virus (SIV) that are limited to a single cycle of infection elicits T-cell responses to multiple viral gene products and antibodies capable of neutralizing lab-adapted SIV, but not neutralization-resistant primary isolates of SIV. In an effort to improve upon the antibody responses, we immunized rhesus macaques with three strains of single-cycle SIV (scSIV) that express envelope glycoproteins modified to lack structural features thought to interfere with the development of neutralizing antibodies. These envelope-modified strains of scSIV lacked either five potential N-linked glycosylation sites in gp120, three potential N-linked glycosylation sites in gp41, or 100 amino acids in the V1V2 region of gp120. Three doses consisting of a mixture of the three envelope-modified strains of scSIV were administered on weeks 0, 6, and 12, followed by two booster inoculations with vesicular stomatitis virus (VSV) G trans-complemented scSIV on weeks 18 and 24. Although this immunization regimen did not elicit antibodies capable of detectably neutralizing SIVmac239 or SIVmac251UCD, neutralizing antibody titers to the envelope-modified strains were selectively enhanced. Virus-specific antibodies and T cells were observed in the vaginal mucosa. After 20 weeks of repeated, low-dose vaginal challenge with SIVmac251UCD, six of eight immunized animals versus six of six naïve controls became infected. Although immunization did not significantly reduce the likelihood of acquiring immunodeficiency virus infection, statistically significant reductions in peak and set point viral loads were observed in the immunized animals relative to the naïve control animals.
Development of a safe and effective vaccine for human immunodeficiency virus type 1 (HIV-1) is an urgent public health priority, but remains a formidable scientific challenge. Passive transfer experiments in macaques demonstrate neutralizing antibodies can prevent infection by laboratory-engineered simian-human immunodeficiency virus (SHIV) strains (6, 33, 34, 53, 59). However, no current vaccine approach is capable of eliciting antibodies that neutralize primary isolates with neutralization-resistant envelope glycoproteins. Virus-specific T-cell responses can be elicited by prime-boost strategies utilizing recombinant DNA and/or viral vectors (3, 10, 11, 16, 36, 73, 77, 78), which confer containment of viral loads following challenge with SHIV89.6P (3, 13, 66, 68). Unfortunately, similar vaccine regimens are much less effective against SIVmac239 and SIVmac251 (12, 16, 31, 36, 73), which bear closer resemblance to most transmitted HIV-1 isolates in their inability to utilize CXCR4 as a coreceptor (18, 23, 24, 88) and inherent high degree of resistance to neutralization by antibodies or soluble CD4 (43, 55, 56). Live, attenuated SIV can provide apparent sterile protection against challenge with SIVmac239 and SIVmac251 or at least contain viral replication below the limit of detection (20, 22, 80). Due to the potential of the attenuated viruses themselves to cause disease in neonatal rhesus macaques (5, 7, 81) and to revert to a pathogenic phenotype through the accumulation of mutations over prolonged periods of replication in adult animals (2, 35, 76), attenuated HIV-1 is not under consideration for use in humans.
As an experimental vaccine approach designed to retain many of the features of live, attenuated SIV, without the risk of reversion to a pathogenic phenotype, we and others devised genetic approaches for producing strains of SIV that are limited to a single cycle of infection (27, 28, 30, 38, 39, 45). In a previous study, immunization of rhesus macaques with single-cycle SIV (scSIV) trans-complemented with vesicular stomatitis virus (VSV) G elicited potent virus-specific T-cell responses (39), which were comparable in magnitude to T-cell responses elicited by optimized prime-boost regimens based on recombinant DNA and viral vectors (3, 16, 36, 68, 73, 78). Antibodies were elicited that neutralized lab-adapted SIVmac251LA (39). However, despite the presentation of the native, trimeric SIV envelope glycoprotein (Env) on the surface of infected cells and virions, none of the scSIV-immunized macaques developed antibody responses that neutralized SIVmac239 (39). Therefore, we have now introduced Env modifications into scSIV that facilitate the development of neutralizing antibodies.
Most primate lentiviral envelope glycoproteins are inherently resistant to neutralizing antibodies due to structural and thermodynamic properties that have evolved to enable persistent replication in the face of vigorous antibody responses (17, 46, 47, 64, 71, 75, 79, 83, 85). Among these, extensive N-linked glycosylation renders much of the Env surface inaccessible to antibodies (17, 48, 60, 63, 75). Removal of N-linked glycans from gp120 or gp41 by mutagenesis facilitates the induction of antibodies to epitopes that are occluded by these carbohydrates in the wild-type virus (64, 85). Consequently, antibodies from animals infected with glycan-deficient strains neutralize these strains better than antibodies from animals infected with the fully glycosylated SIVmac239 parental strain (64, 85). Most importantly with regard to immunogen design, animals infected with the glycan-deficient strains developed higher neutralizing antibody titers against wild-type SIVmac239 (64, 85). Additionally, the removal of a single N-linked glycan in gp120 enhanced the induction of neutralizing antibodies against SHIV89.6P and SHIVSF162 in a prime-boost strategy by 20-fold (50). These observations suggest that potential neutralization determinants accessible in the wild-type Env are poorly immunogenic unless specific N-linked glycans in gp120 and gp41 are eliminated by mutagenesis.
The variable loop regions 1 and 2 (V1V2) of HIV-1 and SIV gp120 may also interfere with the development of neutralizing antibodies. Deletion of V1V2 from HIV-1 gp120 permitted neutralizing monoclonal antibodies to CD4-inducible epitopes to bind to gp120 in the absence of CD4, suggesting that V1V2 occludes potential neutralization determinants prior to the engagement of CD4 (82). A deletion in V2 of HIV-1 Env-exposed epitopes was conserved between clades (69), improved the ability of a secreted Env trimer to elicit neutralizing antibodies (9), and was present in a vaccine that conferred complete protection against SHIVSF162P4 (8). A deletion of 100 amino acids in V1V2 of SIVmac239 rendered the virus sensitive to monoclonal antibodies with various specificities (41). Furthermore, three of five macaques experimentally infected with SIVmac239 with V1V2 deleted resisted superinfection with wild-type SIVmac239 (51). Thus, occlusion of potential neutralization determinants by the V1V2 loop structure may contribute to the poor immunogenicity of the wild-type envelope glycoprotein.
Here we tested the hypothesis that antibody responses to scSIV could be improved by immunizing macaques with strains of scSIV engineered to eliminate structural features that interfere with the development of neutralizing antibodies. Antibodies to Env-modified strains were selectively enhanced, but these did not neutralize the wild-type SIV strains. We then tested the hypothesis that immunization might prevent infection in a repeated, low-dose vaginal challenge model of heterosexual HIV-1 transmission. Indeed, while all six naïve control animals became infected, two of eight immunized animals remained uninfected after 20 weeks of repeated vaginal challenge. Relative to the naïve control group, reductions in peak and set point viral loads were statistically significant in the immunized animals that became infected.
MATERIALS AND METHODS
Animals.
The animals included in this study were all female Indian-origin rhesus macaques (Macaca mulatta). They were housed in a biosafety level 3 containment facility at the New England Primate Research Center (NEPRC) and given care in accordance with standards of the Association for Assessment and Accreditation of Laboratory Animal Care and the Harvard Medical School Animal Care and Use Committee. These experiments and procedures were approved by the Harvard Medical Area Standing Committee on Animals and were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (4).
The animals in this study were typed for the major histocompatibility complex (MHC) class I alleles Mamu-A*01, -A*02, -A*08, -A*11, -B*01, -B*03, -B*04, -B*08, -B*17, and -B*29, as well as the MHC class II alleles DRB1*w201, DRB1*0401/06, and DPB1*06. The MHC typing results are summarized in Table 1. Typing was performed in David Watkins' laboratory at the Wisconsin National Primate Research Center (WNPRC), as previously described by Kaizu et al. (42).
TABLE 1.
Typing for MHC class I and II alleles of the rhesus macaques selected for this studya
| Group | Animal | MHC class I allele |
MHC class II allele |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A*01 | A*02 | A*08 | A*11 | B*01 | B*03 | B*04 | B*08 | B*17 | B*29 | DRB1*w201 | DRB1*0401/06 | DPB1*06 | ||
| Immunized | Mm 158-02 | − | + | − | − | + | − | − | − | − | − | − | + | + |
| Mm 234-99 | − | − | + | − | − | − | − | − | + | + | + | − | + | |
| Mm 241-01 | − | − | − | − | + | − | − | − | − | − | − | − | − | |
| Mm 259-03 | + | − | − | − | − | − | − | − | − | − | − | − | + | |
| Mm 261-00 | + | − | − | − | − | − | − | − | − | − | − | + | − | |
| Mm 284-99 | + | + | − | − | − | − | − | − | − | − | + | − | − | |
| Mm 305-99 | + | − | − | − | − | − | − | − | − | − | − | + | + | |
| Mm 316-98 | − | − | − | − | + | − | − | − | − | − | + | − | − | |
| Naïve | Mm 358-01 | + | − | − | − | − | − | − | − | − | − | − | − | − |
| Mm 418-00 | + | − | + | − | + | − | − | − | − | − | + | + | − | |
| Mm 128-01 | − | − | + | − | − | − | − | − | + | + | − | − | + | |
| Mm 215-98 | − | − | − | − | + | − | − | − | − | − | − | − | + | |
| Mm 214-04 | + | + | − | − | − | − | − | − | − | |||||
| Mm 230-04 | − | − | + | − | + | − | − | − | − | |||||
Allele-specific PCR was performed as described by Kaizu et al. (42).
Env-modified strains of scSIV.
Mutations were introduced into the 3′ half of the SIVmac239 genome (40, 63, 85). A fragment containing a stop codon followed by two single-nucleotide deletions in Nef (28) plus a stop codon at position E767 of Env was cloned into the NheI and SphI sites of the 3′ halves of SIV genomes containing the M5, g123, and ΔV1V2 Envs. Sequence tags gsa, cao, and ggr were previously introduced into scSIV constructs containing the mutated frameshift region and deletions in pol (23). SphI-SphI fragments of the 5′ halves containing the three sequence tags were cloned into the SphI sites of the 3′ halves containing the M5, g123, and ΔV1V2 Envs and stop codons in Env and Nef.
Preparation of scSIV.
Virus stocks of scSIV were produced by cotransfection of 293T cells with the Gag-Pol expression product pGPfusion and the proviral DNA for each strain of scSIV (27). 293T cells were seeded on day 0 at a density of 3 × 106 cells per 100-mm dish in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), l-glutamine, penicillin, and streptomycin. Cells were transfected on day 1 with 5 μg of each plasmid, using the GenJet transfection reagent according to the manufacturer's instructions (SignaGen Laboratories, Gaithersburg, MD). Medium was removed on day 2, and cells were washed twice with serum-free DMEM, which was replaced with DMEM supplemented with 10% rhesus serum (Equitech-Bio, Kerrville, TX). Cell culture supernatant was collected on day 3. Cellular debris was removed by centrifugation at 2,095 × g, and supernatants were concentrated by repeated, low-speed centrifugation in YM-50 ultrafiltration units (Millipore, Bedford, MA), according to the manufacturer's instructions. Aliquots of scSIV were cryopreserved at −80°C until use. The virus concentration was determined by SIV p27 antigen-capture enzyme-linked immunosorbent assay (ELISA) (Advance BioScience Laboratories, Kensington, MD).
Immunization of macaques with scSIV.
Eight animals were inoculated intravenously with identical doses containing 5 μg p27 of scSIVmac239M5, scSIVmac239g123, and scSIVmac239ΔV1V2 on weeks 0, 6, and 12. The animals were boosted with 15 μg p27 scSIVmac239 trans-complemented with VSV GNJ (New Jersey serotype) on week 18 and 15 μg p27 scSIVmac239 trans-complemented with VSV GI (Indiana serotype) on week 24. Animals were anesthetized by 15 mg/kg intramuscular injection of ketamine-HCl, and the concentrated scSIV stock was injected through a 22-gauge catheter aseptically placed into the saphenous vein.
Neutralizing antibodies.
Neutralizing antibody titers were determined by measuring the inhibition over serial, 2-fold plasma dilutions of the activity of a secreted alkaline phosphatase (SEAP) reporter gene under transcriptional control of the SIV long terminal repeat (LTR) promoter in a C8166-derived T-cell line (55). SEAP activity was measured using the Phospha-Light SEAP detection kit (Applied Biosystems, Foster City, CA). Cells incubated without virus or without plasma were used to determine the minimum and maximum SEAP activities, respectively. The nearest values above and below 50% SEAP activity were used to calculate what plasma dilution would intercept the 50% inhibition line.
Virus was incubated with plasma dilutions in 100-μl volumes for 1 h at 37°C before addition of a 100-μl volume containing the C8166-SEAP reporter cells. Due to differences in infectivities between Env mutants, different amounts of input virus and C8166-SEAP cells were used per well for each strain. Minimizing the amount of input virus and time to reading SEAP activity maximizes the apparent 50% neutralization titer (data not shown). These amounts and incubation times were as follows: 12.5 ng p27 SIVmac239M5 and 50,000 cells, read on day 5; 10 ng p27 SIVmac239g123 and 50,000 cells, read on day 4; 2.5 ng p27 SIVmac239ΔV1V2 and 40,000 cells, read on day 3; 2 ng p27 SIVmac251LA and 15,000 cells, read on day 3; 0.5 ng p27 SIVmac239 and 15,000 cells, read on day 3; and 0.5 ng p27 SIVmac251UCD and 15,000 cells, read on day 3. SIVmac239M5, SIVmac239g123, and SIVmac239 stocks were produced by transfection of 293T cells. An uncloned SIVmac239ΔV1V2 stock that was passed in herpesvirus saimiri-transformed macaque 221 cells (40) and expanded in CD8-depleted, phytohemagglutinin (PHA)-activated rhesus peripheral blood mononuclear cells (PBMCs) was selected for the neutralizing antibody assays due to its higher infectivity. SIVmac251LA was grown in MT4 cells. The SIVmac251UCD used for neutralization assays was an uncloned stock, expanded from the challenge stock for 5 days in CD8-depleted, PHA-activated rhesus PBMCs.
Collection and processing of mucosal specimens.
Cervicovaginal and rectal secretions were collected atraumatically with premoistened Weck-Cel sponges (Medtronics, Minneapolis, MN) and extracted by centrifugation as previously described (44). Vaginal biopsies containing both epithelium and underlying tissue were collected using sterile pinch biopsy forceps. Lymphocytes were isolated from biopsy specimens as previously described (29, 72). Biopsies were incubated in 1 mM EDTA for 30 min at 37°C and then twice incubated for an hour in fresh medium containing collagenase type IV while shaking vigorously. A cell suspension was obtained by mechanical dispersion of collagenase-treated samples with a blunt-ended 18-G needle, followed by filtration using a 70-μm-pore nylon cell strainer. Cells were layered over a 35%/60% discontinuous Percoll gradient and centrifuged for 20 min at 1,000 × g to enrich for lymphocytes.
Mucosal antibodies.
Concentrations of total IgG, total IgA, and antibodies to gp120 and viral lysates were measured by chromagenic ELISA as previously described (74), using microtiter plates coated with goat anti-monkey IgG (MP BioMedicals, Solon, OH), goat anti-monkey IgA (Rockland, Gilbertsville, PA), SIV mac251 rgp120 (ImmunoDiagnostics, Woburn, MA), or a 500-fold dilution of SIV mac251 viral lysate (Advanced Biotechnologies Inc, Columbia, MD). The viral lysate preparation contains undetectable levels of Env and is therefore predominantly a measure of antibodies to Gag. Preparations of rhesus macaque serum containing known quantities of each immunoglobulin or gp120-specific antibody were used as standards. Prior to analyses for IgA antibodies, specimens were depleted of IgG, using protein G-Sepharose as described previously (44). Plates were developed with biotinylated goat anti-monkey IgA (Alpha Diagnostics, San Antonio, TX) or anti-human IgG (Southern Biotech, Birmingham, AL) polyclonal antibodies. The concentration of SIV-specific IgG or IgA in secretions was normalized relative to the total IgG or IgA concentration by calculating the specific activity (SA) (ng gp120-specific antibody per μg total IgG or IgA). SA values were considered to be significant if greater than the mean plus 3 standard deviations of samples from naïve macaques.
IFN-γ ELISPOT assays.
Longitudinal T-cell responses to Gag, Tat, Rev, Vif, Vpr, Vpx, Env, and Nef were measured using pools of 15-mer peptides overlapping by 11 residues at 2.5 μg/ml. PBMCs were plated at 3 × 105 and 1 × 105 cells per well in duplicate wells at each density on Multiscreen 96-well plates (Millipore, Bedford, MA) and incubated overnight, and gamma interferon (IFN-γ) was detected using the Mabtech enzyme-linked immunospot (ELISPOT) kit for monkey/human IFN-γ (Mabtech, Mariemont, OH). Spots were enumerated by an automated ELISPOT reader (Zellnet Consulting, New York, NY). The number of spot-forming cells (SFCs) per million PBMCs was calculated by subtracting the number of background spots in wells that received cells but not peptide.
Full-proteome epitope mapping was expedited through the use of a deconvolution matrix. Each animal was mapped using deconvolution matrices consisting of one 96-well ELISPOT plate containing 92 matrix wells, three dimethyl sulfoxide (DMSO)-only negative control wells, and one concanavalin A (ConA)-positive control well. Peptides covering Gag, Pol, Tat, Rev, Vif, Vpr, Vpx, Env, and Nef were each present in two of the 92 matrix wells. The matrix was designed to minimize the number of potentially positive peptides that would require individual testing. Mapping of CD4+ T-cell epitopes was performed using PBMCs depleted of CD8+ cells by Dynal anti-CD8 magnetic beads (Invitrogen, Carlsbad, CA). CD8+ T-cell epitopes were mapped using PBMCs depleted of CD4+ cells. Depletions were conducted at a three-to-one bead-to-cell ratio for 45 min on a rotator at 4°C and confirmed by flow cytometry to have reduced the target population to a maximum of 0.1% of lymphocytes. Cells were seeded to the matrix plate at 1 × 105 cells per well. Surplus CD4- or CD8-depleted cells were rested at 37°C overnight. After processing and enumeration of spots, peptides present in two wells deemed positive, defined as greater than 50 SFCs per million PBMCs and 3 standard deviations above background, were selected for individual testing on the rested CD4- or CD8-depleted PBMCs.
MHC class I tetramer staining.
MHC class I tetramer staining of virus-specific CD8+ T cells was conducted for the Mamu-A*01- and -A*02-positive macaques. A 200-μl volume of whole blood was incubated for 30 min at 37°C with one of the allophycocyanin (APC)-conjugated tetramers Mamu-A*01 Gag181-189 (CM9), Mamu-A*01 Tat28-35 (SL8), Mamu-A*02 Nef159-167 (YY9), or Mamu-A*02 Gag71-79 (GY9). These tetramers were provided by David Watkins' laboratory (WNPRC, Madison, WI). Samples were then stained for 30 min at room temperature with fluorescein isothiocyanate (FITC)-conjugated anti-CD3 (clone SP34; BD Biosciences, San Jose, CA) and peridinin chlorophyll protein (PerCP)-conjugated anti-CD8 (clone SK1; BD Biosciences). Erythrocytes were then eliminated by treatment with fluorescence-activated cell sorter (FACS) lysing solution (BD Biosciences, San Jose, CA). Samples were washed and fixed in 2% formaldehyde in phosphate-buffered saline (PBS). A FACSCalibur flow cytometer was used for data collection (BD Biosciences, San Jose, CA). Data were analyzed using the FlowJo, version 8.7.1, software package (Tree Star, San Carlos, CA).
Repeated, low-dose vaginal challenge with SIVmac251UCD.
The SIVmac251UCD virus stock used for challenges was prepared at the California National Primate Research Center in June 2004. We have adopted the “UCD” designation to indicate that this virus stock has its own passage history, in PBMCs and in monkeys, although it was derived from a SIVmac251 stock originally provided by Ronald Desrosiers.
Fresh vials of virus were thawed at 37°C, transferred immediately to ice, and diluted 1 to 100 in RPMI tissue culture medium (Invitrogen, Carlsbad, CA) containing no additional additives. Each 1-ml dose contained 1 ng SIV p27 equivalents or 1,000 50% tissue culture infective doses (TCID50). Doses were stored on ice until inoculation. Animals were anesthetized by a 15-mg/kg intramuscular injection of ketamine-HCl and positioned with their hindquarters raised for vaginal inoculation and for the following 30 min. Four hours later, the above challenge procedure was repeated. Challenge days were spaced 1 week apart and were discontinued once viral RNA was detectable in plasma for two consecutive weeks.
Plasma viral RNA loads.
Plasma samples were collected in 0.5- to 1.5-ml volumes of sodium citrate anticoagulant and ultracentrifuged at 20,000 × g for 1 h to pellet virus. RNA was extracted, reverse transcribed into cDNA, and quantified by real-time PCR as previously described (19). The limit of detection for this assay is 30 copies of RNA per ml. The primer/probe sets for the quantitative, multiplex real-time reverse transcription (RT)-PCR assay for the unique sequence tags ggr, cao, and gsa have previously been described by DeGottardi et al. (23).
CD4+ T-cell subsets.
The maintenance or destruction of naïve, central memory, effector memory, CCR5+ memory, and total CD4+ T-cell populations was monitored by flow cytometry. Total CD4+ T-cell counts and the concentrations of each cell population per μl whole blood were calculated by enumerating lymphocytes per μl of whole blood by complete blood count (CBC) and multiplying by the percentage belonging to each CD4+ T-cell subpopulation as determined by flow cytometry. Whole blood was stained with fluorescein isothiocyanate (FITC)-conjugated anti-CD3 (clone SP34; BD Biosciences, San Jose, CA), PerCP-conjugated anti-CD4 (clone L200; BD Biosciences, San Jose, CA), APC-conjugated anti-CD95 (clone DX2; BD Biosciences, San Jose, CA), and phycoerythrin (PE)-conjugated anti-CD28 (clone CD28.2; BD Biosciences, San Jose, CA) or PE-conjugated anti-CCR5 (clone 3A9; BD Biosciences, San Jose, CA). Erythrocytes were eliminated with FACS lysing solution (BD Biosciences, San Jose, CA), and cells were washed and fixed in 2% formaldehyde in PBS. Flow cytometry data for CD4+ T cells were collected and analyzed as described for tetramer data.
Statistical methods.
All statistical evaluations were conducted using SPSS 15.0 (SPSS, Inc., Chicago, IL) and Strata MP 10.0 (Strata Corp., College Station, TX). The differences in the ratio of anti-gp120 IgG SA in CVS to plasma were compared between the uninfected immunized animals (n = 2) and those immunized and infected (n = 6) using a generalized estimation equation (GEE) model. Correlations between repeated measurements within the same subject were accounted for by use of unstructured covariance as well as first order autocorrelation. Both approaches lead to a similar result. In total, 31 measurements from eight animals were included.
A two-tailed Mann-Whitney U test on the highest measured log-transformed values between weeks 1 and 5 postinfection was used to determine the significance of differences in peak viral loads. A linear mixed model analysis was used to determine the significance of differences in CD4+ T-cell counts and log-transformed set point viral loads between the immunized and naïve groups (25). Set point viral loads were evaluated for the period between weeks 5 and 67 postinfection, centered on week 28.
RESULTS
Immunization of macaques with Env-modified strains of scSIV.
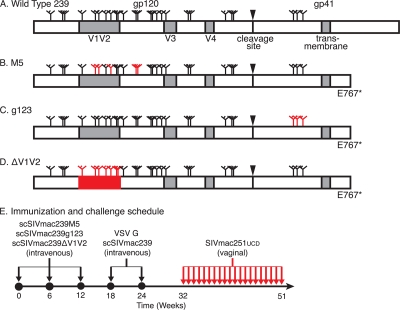
Rhesus macaques were immunized with three strains of scSIV, each of which had different mutations that deprive the SIV envelope glycoprotein of specific countermeasures against host antibody responses. The wild-type SIVmac239 Env has 23 potential N-linked glycosylation sites in gp120 and 3 potential N-linked glycosylation sites in gp41 (Fig. 1 A). The 5th, 6th, 8th, 12th, and 13th potential N-linked glycosylation sites in gp120 were eliminated by mutagenesis in the Env-modified scSIV strain scSIVmac239M5 (Fig. 1B) (64). All three potential N-linked glycosylation sites in gp41 were eliminated in strain scSIVmac239g123 (Fig. 1C) (85). The third Env-modified scSIV strain, scSIVmac239ΔV1V2, had 100 amino acids in the V1V2 loop structure deleted (Fig. 1D) (40). These three strains of Env-modified scSIV also had a glutamate-to-stop codon change to truncate the cytoplasmic tail of gp41, which maximizes Env incorporation onto virions, viral infectivity, and the Env-specific antibody response (Fig. 1B to D) (23, 86). In order to maximize CD8+ T-cell responses, each scSIV strain contained a premature stop codon in nef that eliminates 26 amino acids from the C terminus of the protein required for MHC class I downregulation (Fig. 1B to D) (70).
FIG. 1.
Animals were immunized with Env-modified single-cycle SIV and challenged by repeated, low-dose vaginal inoculation. A schematic representation of the wild-type SIVmac239 Env appears at the top (A). Positions of potential sites of N-linked glycosylation (N-X-S/T motifs) are indicated by tree-like symbols. Features removed from each Env-modified strain are indicated (red). The M5 Env has asparagine-to-glutamine substitutions that eliminate the 5th, 6th, 8th, 12th, and 13th potential N-linked glycosylation sites in gp120 (B). The g123 Env has all three potential N-linked glycosylation sites in gp41 similarly eliminated (C). Variable loops 1 and 2 of the ΔV1V2 envelope are deleted (D). All of the modified envelopes have a glutamate-to-stop codon change at position 767 (E767*). Eight macaques received an intravenous injection consisting of a mixture of 5 μg p27 equivalents of scSIVmac239M5, scSIVmac239g123, and scSIVmac239ΔV1V2 on weeks 0, 6, and 12 (E). All eight animals also received 15 μg p27 equivalents of VSV G trans-complemented scSIVmac239 intravenously on weeks 18 and 24. Beginning on week 32, the animals were challenged vaginally with 1,000 TCID50 (1 ng p27) of SIVmac251UCD twice per day on the same day each week for 20 weeks or until viral RNA was detected in plasma on two consecutive weeks.
Eight female rhesus macaques received three intravenous inoculations consisting of a mixture of 5 μg p27 equivalents of each of the three Env-modified scSIV strains on weeks 0, 6, and 12 to prime antibody responses to Env surfaces with limited accessibility in the native trimer (Fig. 1E). These immunogens would be expected to also prime antibody responses to novel surfaces unique to the mutant Envs. Truncation of the Env cytoplasmic tail can also alter the conformation of the ectodomain (21, 26, 84, 86). Therefore, to focus the antibody response on surfaces exposed in the native trimer, the animals were boosted on weeks 18 and 24 with 15 μg p27 equivalents of scSIVmac239, which expresses the wild-type SIVmac239 Env with a full-length cytoplasmic tail (Fig. 1E). These two booster doses were trans-complemented with VSV G to maximize infectivity. To prevent neutralization of the second boost by VSV G-specific antibodies (39, 67), scSIV was trans-complemented with the New Jersey serotype (VSV GNJ) on week 18, and the Indiana serotype (VSV GI) on week 24. This immunization regimen was designed to maximize the stimulation of both Env-specific antibody responses and the magnitude of virus-specific T-cell responses.
To determine whether this immunization regimen might reduce the likelihood of viral transmission by the vaginal route, the eight immunized animals and six naïve controls were repeatedly challenged for 20 weeks by low-dose vaginal inoculation of SIVmac251UCD, beginning on week 32 (Fig. 1E). This challenge regimen was expected to be sufficient to infect all the naïve control animals (52). Thus, significant differences in the number of doses required to establish infection or in the final number of infections for the immunized versus naïve groups might indicate that immunization reduced vaginal transmission.
Transient viremia following each dose of Env-modified and VSV G trans-complemented scSIV.
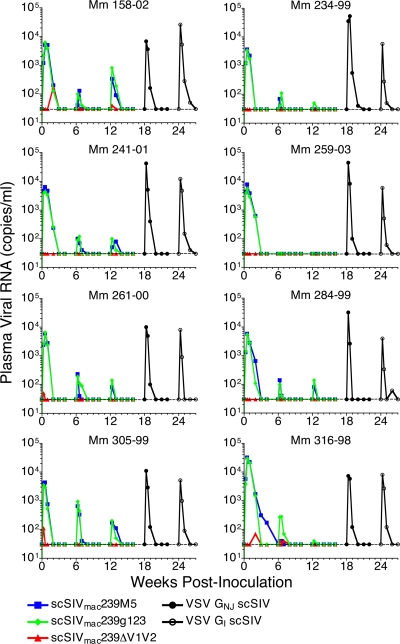
Transient viral loads in plasma representing the progeny virions released by cells infected with each Env-modified strain of scSIV were measured independently using a quantitative, multiplex real-time RT-PCR assay specific for unique sequence tags cloned into the pol gene of each strain (23). Since cell-free virus is cleared from plasma with a half-life on the order of just a few minutes (37, 87), these viral RNA load measurements reflect the life span of cells productively infected with scSIV in vivo and thus the duration of antigenic stimulation. The peaks of viremia for scSIVmac239M5 and scSIVmac239g123 were nearly identical, with geometric mean peak viral loads after the first inoculation of 6.8 × 103 and 6.1 × 103 RNA copies per ml (Fig. 2). However, peak viral loads for scSIVmac239ΔV1V2 were considerably lower than those of the two other Env-modified strains, resulting in detectable viral loads in only four of the eight immunized animals, the highest of which peaked at 170 copies of viral RNA per ml in Mm 158-02. Except for the first dose of scSIVmac239M5 in Mm 316-98, all single-cycle viral loads for each strain were cleared to below the limit of detection by 3 weeks after inoculation. Relative to the first dose, peak viral loads were lower after the second inoculation for both scSIVmac239M5 (P = 0.0002) and scSIVmac239g123 (P = 0.0002). Following the booster inoculations with VSV GNJ and VSV GI trans-complemented scSIV on weeks 18 and 24, peak geometric mean viral loads were 1.8 × 104 and 7.8 × 103 copies of viral RNA per ml, consistent with the infectivity enhancement afforded by VSV G (39). These viral load measurements confirm the productive infection of cells in vivo by the Env-modified and VSV G trans-complemented strains of scSIV.
FIG. 2.
Plasma viral RNA following each inoculation with single-cycle SIV. The level of viral RNA in plasma was measured independently for each of these strains using a quantitative, multiplex real-time RT-PCR assay for the unique sequence tags ggr, cao, and gsa cloned into each of the three scSIV strains (23). This assay has a threshold of detection of 30 copies of viral RNA per ml plasma (dotted line).
Neutralization of Env-modified and lab-adapted SIV, but not wild-type SIVmac239 or SIVmac251UCD.
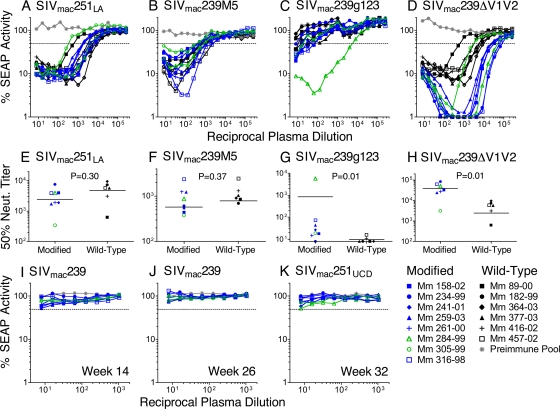
To determine whether immunization with the Env-modified strains of scSIV significantly enhanced antibody responses to surfaces occluded by the N-linked glycans and V1V2 loops, neutralization of the Env-modified viruses by plasma samples from this study was compared to neutralization by plasma samples from a previous study, in which animals were immunized with strains of scSIV expressing wild-type envelope glycoproteins from SIVmac239, SIVmac316, and SIVmac155T3 (Fig. 3 A to H) (39). The plasma samples utilized for this comparison were collected 2 weeks after the third intravenous inoculation with scSIV in both studies. Neutralization assays using plasma samples from both studies were performed in parallel.
FIG. 3.
Plasmas neutralized Env-modified and lab-adapted SIV but not wild-type SIV. Neutralizing antibody titers were compared between the animals immunized with Env-modified scSIV that subsequently became infected (blue) or remained uninfected (green) and historical control plasmas from animals immunized with scSIV expressing wild-type Envs (black). Pooled preimmune plasma served as a negative control (gray). The dotted line indicates 50% inhibition of SEAP activity. Plasma samples collected 2 weeks after the third dose of scSIV were tested for neutralization (neut.) of SIVmac251LA (A), SIVmac239M5 (B), SIVmac239g123 (C), SIVmac239ΔV1V2 (D), and SIVmac239 (I). Fifty percent neutralization titers are shown for SIVmac251LA (E), SIVmac239M5 (F), SIVmac239g123 (G), and SIVmac239ΔV1V2 (H). P values were determined by two-tailed Mann-Whitney U tests. Week 26 plasmas were tested for neutralization of SIVmac239 (J). Plasma samples drawn on week 32 were tested for neutralization of SIVmac251UCD (K).
No significant difference was detected for neutralization of SIVmac251LA by plasmas from the two groups (P = 0.30; two-tailed Mann-Whitney U test) (Fig. 3A and E). This suggests that any differences in neutralizing antibody titers against the Env-modified strains do not reflect nonspecific differences in overall antibody titers between the two studies. All of the animals immunized with Env-modified or wild-type strains of scSIV neutralized SIVmac239M5, consistent with previous observations that elimination of these carbohydrate attachment sites increases the susceptibility of this virus to neutralization (41, 64). However, neutralizing antibody titers to SIVmac239M5 were not significantly different for animals immunized with Env-modified versus wild-type strains of scSIV (P = 0.37; two-tailed Mann-Whitney U test) (Fig. 3B and F), indicating that scSIVmac239M5 did not specifically facilitate the induction of antibodies to epitopes occluded by the 5th, 6th, 8th, 12th, and 13th N-linked glycans in gp120. In contrast, SIVmac239g123 was detectably neutralized at low titers by only two of the animals immunized with wild-type scSIV but was neutralized by all of the animals immunized with the Env-modified strains of scSIV and at titers five times higher (P = 0.01; two-tailed Mann-Whitney U test) (Fig. 3C and G). The difference in 50% neutralization titers between the groups remains statistically significant even if the outlier, Mm 284-99, with a titer of 1:5,763 (nearly 200-fold higher than the average for the rest of the group), is excluded from the analysis (P = 0.02). These results suggest that the N-linked glycans in gp41 interfere with the induction of Env-specific antibody responses and that the elimination of these glycans by mutagenesis can enhance the induction of antibodies to surfaces accessible in this N-linked glycan-deficient strain.
Immunization with Env-modified strains also elicited higher neutralizing antibody titers to the virus with V1V2 deleted than did scSIV expressing wild-type Envs. While plasma from animals immunized with either the Env-modified or wild-type strains of scSIV both neutralized SIVmac239ΔV1V2, titers for the animals immunized with Env-modified strains were seven times higher (P = 0.01; two-tailed Mann-Whitney U test) (Fig. 3D and H). Indeed, the animals immunized with Env-modified scSIV were all able to neutralize SIVmac239ΔV1V2 infectivity to <2% and had 50% neutralization titers that ranged from 3.1 × 103 to 8.4 × 104. Thus, in spite of the comparably poor take of infection by scSIVmac239ΔV1V2 (Fig. 2), immunization with Env-modified scSIV enhanced the induction of antibodies to determinants revealed by deletion of the V1V2 loops.
Although immunization with Env-modified strains of scSIV enhanced the induction of antibodies that neutralize SIVmac239g123 and SIVmac239ΔV1V2, plasma samples collected 2 weeks after the third dose of Env-modified scSIV (week 14) and 2 weeks after the second VSV G scSIV boost (week 26) failed to detectably neutralize SIVmac239 (Fig. 3I and J). Likewise, plasma collected on the first day of challenge (week 32) did not detectably neutralize the SIVmac251UCD challenge strain (Fig. 3K).
SIV-specific antibodies in mucosal secretions.
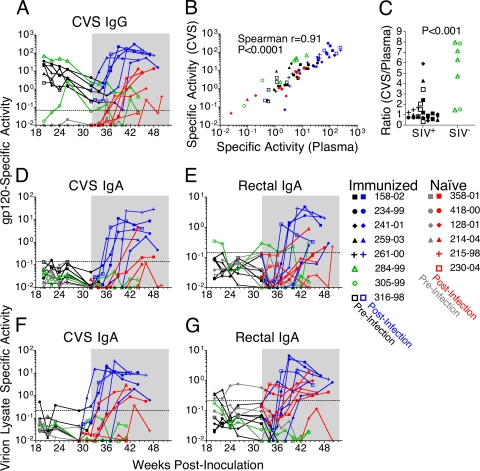
Mucosal secretions were tested for SIV-specific antibodies before and after challenge (Fig. 4). Vaginal and rectal secretions were collected using premoistened Weck-Cel sponges to avoid abrasion of the mucosal epithelium (44). This approach permits the nearly exclusive collection of secretions, without drawing fluid from the underlying tissue (44). To control for significant variability in the concentrations of total IgG and IgA in mucosal secretions over time and over the menstrual cycle, the specific activity (SA) of antibody titers to gp120 or viral lysates was calculated by dividing the μg amount of SIV-specific antibody by the mg amount of total IgG or IgA in the sample (Fig. 4).
FIG. 4.
Virus-specific antibody responses were detected in mucosal secretions. Anti-gp120 IgG specific activity (SA) was monitored longitudinally in CVS of immunized animals prior to infection (black), from the week each became infected (blue), and for the two that did not become infected (green) (A). Mucosal antibody responses in the naïve animals were measured prior to infection (gray) and postinfection (red). The 20-week challenge phase is shaded. The dashed line indicates the limit of detection, defined as the mean plus 3 standard deviations of negative samples. Anti-gp120 IgG SAs in CVS and plasma are correlated (Spearman's r = 0.91; P < 0.0001) (B). The ratios of anti-gp120 SA in CVS versus plasma were significantly higher at the five prechallenge time points in the two animals that remained SIV negative (C) (ratio, 3.6; 95% CI, 3.1 to 4.0; P < 0.001). IgG samples containing readings below the limit of detection, defined as 3 standard deviations above the average SA for naïve controls, were excluded from correlative and ratio analyses (B and C). IgA SA was monitored against gp120 in CVS (D) and rectal secretions (E) and against viral lysate in CVS (F) and rectal secretions (G).
Gp120-specific IgG was detectable in the cervicovaginal secretions (CVS) of all the immunized animals prior to challenge (Fig. 4A). Comparison of the specific activity of IgG in CVS versus plasma revealed a positive correlation (Fig. 4B) (Spearman's r = 0.91; P < 0.0001). This correlation demonstrates that the proportion of total IgG specific for gp120 in the vaginal mucosa closely mirrors the proportion found in plasma. Indeed, the median ratio of the specific activities in CVS versus plasma was 1:1 (Fig. 4C). However, the two immunized animals that remained SIV negative after the 20-week challenge period had significantly higher prechallenge ratios of specific activity in CVS versus plasma than the six animals that subsequently became infected (Fig. 4C): ratio, 3.6 (95% confidence interval [CI], 3.1 to 4.0) (P < 0.001).
gp120-specific IgA was measured in CVS, rectal secretions, and plasma (Fig. 4D and E). Prior to challenge, none of the animals had detectable gp120-specific IgA titers in plasma (data not shown) or CVS (Fig. 4D). Likewise, with the exception of Mm 305-99, none of the animals had detectable Env-specific IgA titers in rectal secretions (Fig. 4E). High-titer gp120-specific IgA responses were observed postinfection in both CVS and rectal secretions for all six of the scSIV-immunized animals that became infected (Fig. 4D and E).
IgA in CVS, rectal secretions, and plasma was tested for reactivity against a SIVmac251 viral lysate preparation (Fig. 4F and G). No IgA specific for viral lysates was detectable in plasma prior to challenge (data not shown), but low levels were detectable in CVS from Mm 234-99 (Fig. 4F) and in rectal secretions from Mm 259-03 and Mm 261-00 (Fig. 4G). High levels of viral lysate-specific IgA were detectable in CVS and rectal secretions after infection of all of the immunized animals (Fig. 4F and G). Thus, the virus-specific IgA detected in some animals prior to challenge and the high-titer IgA responses that developed in immunized animals that became infected may have contributed to the control of viral replication.
Breadth of virus-specific T-cell responses elicited by immunization with scSIV.
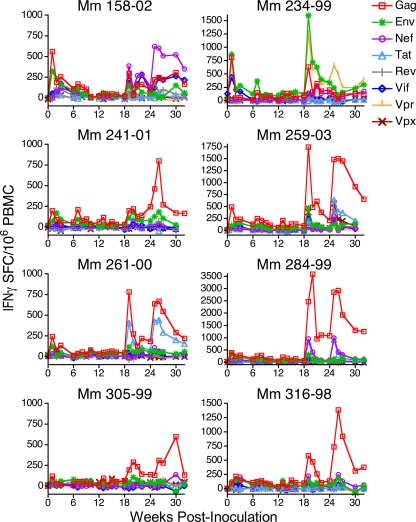
T-cell responses to the eight viral antigens expressed by scSIV were measured longitudinally during the immunization phase of this experiment by IFN-γ ELISPOT assays (Fig. 5). Each animal responded to multiple viral antigens. These responses were boosted by inoculation with VSV G scSIV on weeks 18 and 24.
FIG. 5.
T-cell responses were detectable against all proteins expressed by scSIV. IFN-γ ELISPOT responses against peptide pools of overlapping 15-mers representing the Gag, Tat, Rev, Vif, Vpr, Vpx, Env, and Nef proteins were measured over the immunization period (weeks 0 to 32). The limit of detection was 50 spot-forming cells (SFCs) per 106 PBMCs.
The breadth of the CD4+ and CD8+ T-cell responses elicited by scSIV was assessed by whole-proteome, deconvolution epitope mapping. CD4+ T-cell epitopes were defined on week 25 (Table 2), and CD8+ T-cell epitopes were defined on week 26 (Table 3). CD4+ T-cell epitopes were analyzed with PBMCs depleted of CD8+ cells by anti-CD8 antibody-coated magnetic beads, and CD8+ T-cell epitopes were analyzed with PBMCs depleted of CD4+ cells by anti-CD4 antibody-coated magnetic beads. In cases where consecutive 15-mer peptides both scored positive, the overlapping 11-amino-acid sequence is shown due to the high probability that a single epitope exists within this region (Tables 2 and 3). Due to routinely low PBMC yields from Mm 241-01, a comprehensive analysis of T-cell epitopes was not possible for this animal. The seven immunized animals that were mapped recognized an average of 5.4 CD4+ and 4.3 CD8+ T-cell epitopes. The same methodology was used to determine that animals chronically infected with live, attenuated SIVmac239Δnef recognized an average of 5.2 CD4+ T-cell epitopes (n = 5 animals) and 5.2 CD8+ T-cell epitopes (n = 17 animals) (data not shown). Thus, the breadths of CD4+ and CD8+ T-cell responses elicited by scSIV were comparable to the breadth of T-cell responses elicited by persistent infection with live, attenuated SIV.
TABLE 2.
CD4+ T-cell epitopesa
| Macaque and protein | Sequence | No. of SFCs/106 PBMCs |
|---|---|---|
| Mm 158-02 | ||
| Rev | RLRLIHLLHQT | 209 |
| Gag | CGKMDHVMAKC | 161 |
| Env | IVKHPRYTGTN | 161 |
| Gag | PGQKARLMAEA | 67 |
| Gag | TLNAWVKLIEEKKFG | 67 |
| Nef | RRHRILDIYLEKEEG | 67 |
| Rev | RQRKRRWRRRWQQLL | 51 |
| Total | 7 | 783 |
| Mm 234-99 | ||
| Vif | LQEGSHLEVQGYWHL | 372 |
| Env | IVKHPRYTGTN | 182 |
| Gag | PGQKARLMAEA | 170 |
| Gag | TNILDVKQGPK | 155 |
| Rev | ELRKRLRLIHLLHQT | 152 |
| Gag | DRQAGFLGLGPWGKK | 139 |
| Env | KTVLPVTIMSG | 102 |
| Env | YVPCHIRQIINTWHK | 102 |
| Gag | CGKMDHVMAKCPDRQ | 93 |
| Env | QAWCWFGGKWKDAIK | 92 |
| Vif | SKNFWTDVTPNYADI | 62 |
| Vpx | TIGEAFEWLNRTVEE | 62 |
| Rev | IYSFPDPPTDTPLDL | 62 |
| Env | YCKMNWFLNWVEDRN | 52 |
| Env | YVPCHIRQIINTWHK | 52 |
| Total | 15 | 1,849 |
| Mm 259-03 | ||
| Rev | RLRLIHLLHQT | 313 |
| Gag | CGKMDHVMAKC | 182 |
| Nef | ILDIYLEKEEGIIPD | 146 |
| Rev | RRWQQLLALAD | 99 |
| Gag | QNANPDCKLVLKGLG | 75 |
| Total | 5 | 815 |
| Mm 261-00 | ||
| Gag | CTPYDINQMLNCVGD | 85 |
| Tat | CISEADASTPESANL | 50 |
| Nef | AYRKQNMDDIDEEDD | 50 |
| Total | 3 | 185 |
| Mm 284-99 | ||
| Gag | CTPYDINQMLNCVGD | 139 |
| Gag | MYNPTNILDVKQGPK | 100 |
| Tat | CISEADASTPESANL | 87 |
| Vif | MEEEKRWIAVPTWRI | 74 |
| Total | 4 | 400 |
| Mm 305-99 | ||
| Env | IVKHPRYTGTN | 59 |
| Gag | GCWKCGKMDHVMAKC | 54 |
| Total | 2 | 113 |
| Mm 316-98 | ||
| Nef | YKLAIDMSHFI | 135 |
| Env | YVPCHIRQIINTWHK | 81 |
| Total | 2 | 216 |
CD4+ T-cell epitopes were determined on week 25 by whole-proteome, deconvolution epitope mapping of CD8-depleted PBMCs. The total numbers of CD4+ T-cell epitopes and spot-forming cells (SFCs)/106 PBMCs for each animal are indicated. The overlapping 11-residue sequence is shown for two consecutive peptides with the higher of the two SFC/106-PBMC numbers.
TABLE 3.
CD8+ T-cell epitopesa
| Macaque and protein | Sequence | Epitope | No. of SFCs/106 PBMCs |
|---|---|---|---|
| Mm 158-02 | |||
| Nef | QDYTSGPGIRY | A*02 Nef YY9 | 1,919 |
| Vif | WTDVTPNYADI | A*02 Vif VI8 | 529 |
| Gag | PTGSENLKSLY | A*02 Gag GY9 | 386 |
| Pol | WMGYELWPTKWKLQK | Undefined | 90 |
| Total | 4 | 2,924 | |
| Mm 234-99 | |||
| Env | VFGNWFDLASW | Undefined | 1,478 |
| Vpr | FRGGCIHSRIG | Undefined | 934 |
| Nef | SGPGIRYPKTFGWLW | A*02 Nef YY9 | 314 |
| Vpr | DEWVVEVLEELKEEA | Undefined | 73 |
| Env | KERDGGEGGGNSSWP | Undefined | 73 |
| Vif | TPNYADILLHSTYFP | A*02 Vif YY10 | 61 |
| Vif | WAWWTCSRVIFPLQE | Undefined | 61 |
| Total | 7 | 2,994 | |
| Mm 259-03 | |||
| Gag | CTPYDINQMLN | A*01 Gag CM9 | 1,939 |
| Tat | ADASTPESANL | A*01 Tat SL8 | 590 |
| Vpx | SGEETIGEAFE | Undefined | 305 |
| Env | CLPDNGDYSEVALNV | Undefined | 118 |
| Env | GTTQCLPDNGDYSEV | Undefined | 80 |
| Total | 5 | 3,032 | |
| Mm 261-00 | |||
| Gag | CTPYDINQMLN | A*01 Gag CM9 | 1,305 |
| Tat | ADASTPESANL | A*01 Tat SL8 | 1,259 |
| Env | CAPPGYALLRCNDTN | A*01 Env CL9 | 119 |
| Total | 3 | 2,683 | |
| Mm 284-99 | |||
| Gag | CTPYDINQMLN | A*01 Gag CM9 | 2,227 |
| Nef | QDYTSGPGIRY | A*02 Nef YY9 | 235 |
| Tat | ADASTPESANL | A*01 Tat SL8 | 178 |
| Total | 3 | 2,640 | |
| Mm 305-99 | |||
| Gag | CTPYDINQMLN | A*01 Gag CM9 | 295 |
| Rev | TDTPLDLAIQQLQNL | Undefined | 145 |
| Gag | QRHLVVETGTTETMP | Undefined | 123 |
| Env | VPWPNASLTPKWNNE | A*01 Env TL9 | 91 |
| Total | 4 | 654 | |
| Mm 316-98 | |||
| Gag | LDRFGLAESLL | Undefined | 1,599 |
| Gag | DRQAGFLGLGPWGKK | Undefined | 216 |
| Gag | AKCPDRQAGFLGLGP | Undefined | 146 |
| Nef | RSRPSGDLRQRLLRA | Undefined | 76 |
| Total | 4 | 2,037 |
CD8+ T-cell epitopes were determined on week 26 by whole-proteome, deconvolution epitope mapping of CD4-depleted PBMCs. The total number of CD8+ T-cell epitopes and spot-forming cells (SFCs) per 106 PBMCs for each animal are indicated. The overlapping 11-residue sequence is shown for two consecutive peptides with the higher of the two SFC/106-PBMC numbers.
Virus-specific CD8+ T cells in peripheral blood and vaginal mucosa.
Immunization with scSIV elicited virus-specific CD8+ T cells that were detectable in peripheral blood and the vaginal mucosa by MHC class I tetramer staining (Fig. 6). Tetramer-positive CD8+ T cells were detected in peripheral blood following the first inoculation in each of the Mamu-A*01- and -A*02-positive animals, except Mm 305-99. These responses were boosted by immunization with VSV G scSIV on weeks 18 and 24. Virus-specific CD8+ T-cell responses identified by tetramer staining mirror the longitudinal IFN-γ ELISPOT results (Fig. 5 and 6).
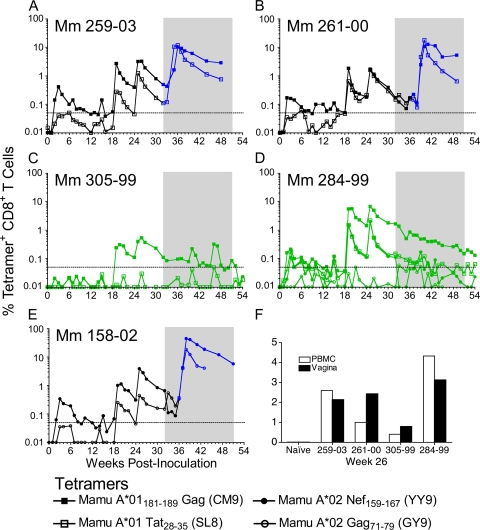
FIG. 6.
Virus-specific CD8+ T cells were detected in peripheral blood and the vaginal mucosa. CD8+ T cells binding the MHC class I tetramers Mamu-A*01 Gag181-189 (CM9), Mamu-A*01 Tat28-35 (SL8), Mamu-A*02 Nef159-167 (YY9), and Mamu A*02 Gag71-79 (GY9) were detected in peripheral blood (A to E). The limit of detection indicated by the dotted line is 0.05% of CD8+ T cells. The 20-week challenge period is shaded. Tetramer-positive CD8+ T-cell frequencies are shown prior to infection (black) and from the week each animal became infected (blue) (A, B, and E). Frequencies of tetramer-positive CD8+ T cells are shown for the two immunized animals that remained uninfected (green) (C and D). Similar frequencies of CD8+ T cells were tetramer positive among lymphocytes isolated from vaginal biopsies versus peripheral blood (F).
The magnitude of MHC class I tetramer-positive CD8+ T-cell responses elicited by immunization with scSIV varied from animal to animal. Mm 284-99 had the highest prechallenge peak responses, with 6.7% of CD8+ T cells recognizing Mamu-A*01 Gag181-189 (CM9) and 2% each of CD8+ T cells recognizing Mamu-A*01 Tat28-35 (SL8) and Mamu-A*02 Nef159-167 (YY9) (Fig. 6D). Mm 305-99 had the lowest-magnitude response to Mamu-A*01 Gag181-189 (CM9), which became detectable only after boosting with VSV G scSIV and reached a maximal frequency of 0.5% of CD8+ T cells (Fig. 6C). The CD8+ T-cell response to Mamu-A*01 Tat28-35 (SL8) never exceeded the limit of detection in this animal. This is consistent with the absence of T-cell responses to peptides containing this epitope for both the longitudinal analysis of IFN-γ ELISPOT responses to the Tat peptide pool (Fig. 5) and to the individual 15-mer peptides containing this epitope (Table 3). Thus, even among Mamu-A*01-positive macaques, there was considerable variation in the magnitude of CD8+ T-cell responses elicited by immunization with scSIV.
SIV-specific CD8+ T-cell frequencies were also measured in lymphocytes isolated from biopsies of the vaginal mucosa on week 26, 2 weeks after boosting with VSV G scSIV. Tetramer-positive cell frequencies among lymphocytes isolated from these biopsies were similar to the frequencies observed in peripheral blood samples processed in parallel (Fig. 6F). Frequencies of Mamu-A*01 Gag181-189 (CM9)-specific CD8+ T cells in the biopsies ranged from 0.8% in Mm 305-99 to 3.1% in Mm 284-99. These biopsy results confirm that intravenous immunization with scSIV elicited SIV-specific CD8+ T cells that trafficked to the vaginal mucosa and were therefore present at the site of challenge.
Extraordinarily high anamnestic CD8+ T-cell responses ensued following infection of immunized animals. Cumulative Mamu-A*01 Gag181-189 (CM9) plus Mamu-A*01 Tat28-35 (SL8) responses peaked at 22% and 28% of CD8+ T cells in peripheral blood from Mm 259-03 and Mm 261-00, respectively (Fig. 6A and B). Likewise, an astonishing 44% of CD8+ T cells recognized the Mamu-A*02 Nef159-167 (YY9) epitope in Mm 158-02 (Fig. 6E). In contrast, the percentage of Mamu-A*01 Gag181-189 (CM9)-specific CD8+ T cells declined steadily in Mm 284-99 from 1.7% to 0.13% of CD8+ T cells during the 20-week challenge period (Fig. 6D). In the case of Mm 305-99, Mamu-A*01 Gag181-189 (CM9)-specific CD8+ T cells increased in frequency on weeks 40 to 41 and 46 to 47, perhaps reflecting the simulation of recall responses by the challenge virus (Fig. 6C). Robust anamnestic T-cell responses were observed in the six immunized animals that became infected, but not in the two that remained uninfected.
Outcome of repeated, low-dose vaginal challenge.
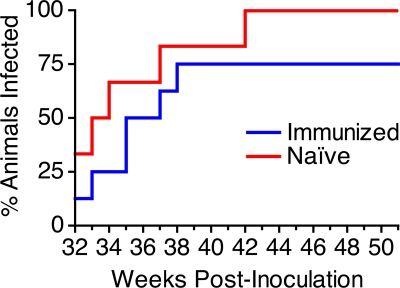
Repeated, low-dose vaginal inoculation with SIVmac251UCD was conducted to model heterosexual transmission of HIV-1. Beginning on week 32, each of the eight immunized animals and six naïve control animals was challenged vaginally with 1,000 TCID50 (1 ng p27) of SIVmac251UCD. Inoculations were administered in the morning and again in the afternoon of the same day each week, for 20 consecutive weeks. Challenge doses were discontinued when viral RNA was detected in plasma for two consecutive weeks for any given animal. All naïve control animals were expected to become infected (52), and a significant difference in the number of doses required to establish infection or in the final number of animals becoming infected in the immunized group versus the naïve controls would indicate that immunization prevented transmission. All six naïve control animals and six of the eight immunized animals became infected (Fig. 7). Four of the six naïve animals were infected by a single clone, as determined by single-genome amplification (SGA) and analysis of sequences encoding Env at the first viral RNA-positive time point (Eric Hunter, Emory University, personal communication). A Weibull regression analysis was performed to determine whether the immunized animals required significantly more doses than the naïve controls to establish infection. The number of doses required to establish infection in each group computed a hazard (risk) ratio of 0.38 (95% CI, 0.12 to 1.21), where a ratio of 1 would mean the groups are at equal risk of becoming infected. Although there was a trend toward greater resistance to infection for the scSIV-immunized animals, this analysis did not reveal a statistically significant difference in the risk of infection relative to the control animals (P = 0.10).
FIG. 7.
Animals were challenged by repeated, low-dose vaginal inoculation. The eight immunized animals (blue) and six naïve controls (red) were challenged vaginally with 1 ng p27 (1,000 TCID50) of SIVmac251UCD for 20 weeks, beginning on week 32. Virus was administered twice on the days of challenge—once in the morning and again in the afternoon—on the same day each week. All six naïve control animals became infected by the 11th week of challenge, but two of the eight immunized animals remained uninfected after the 20th week.
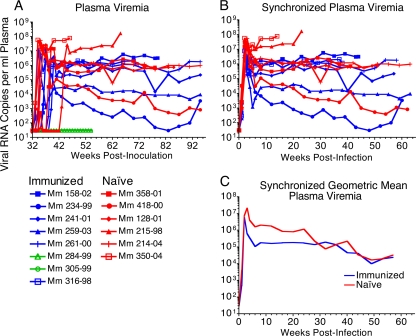
The repeated, low-dose challenge regimen resulted in animals becoming infected on different weeks. Viral load measurements were therefore synchronized to the last week in which viral RNA was undetectable in each animal to compare differences in peak and set point viral loads (Fig. 8B). All viral load analyses excluded the two immunized animals that remained uninfected. Peak viral loads were reduced 0.72 log for the immunized group relative to the control group (Fig. 8C). The blunting of peak viral loads in the immunized group was statistically significant (P = 0.0038; two-tailed Mann-Whitney U test). Geometric mean viral loads were 1.1, 1.2, 1.1, and 1.0 log lower on weeks 4, 6, 8, and 12 postinfection in the immunized group relative to the naïve controls. A linear mixed model analysis of chronic-phase log viral loads also revealed a statistically significant difference in set point viral loads for the period between 5 and 67 weeks postinfection, centered on week 28 (P = 0.004) (Fig. 8C). Despite the convergence of viral loads after week 23 due to the loss of two rapidly progressing naïve control animals on weeks 16 and 23, viral loads for the immunized group were 0.96 log lower for this period (95% CI, 0.31 to 1.6). Thus, immunization of macaques according to a prime-boost regimen with Env-modified and VSV G trans-complemented scSIV resulted in statistically significant containment of viral replication.
FIG. 8.
Postchallenge plasma viral RNA loads. Viral loads were measured at weekly intervals beginning on week 32 using a real-time RT-PCR assay with a limit of detection of 30 copies of RNA per ml of plasma for the immunized animals that became infected (blue) or remained uninfected (green) and for the naïve controls (red) (A) (19). Viral load measurements were synchronized to the last week in which viral RNA was undetectable in each animal that became infected (B). Comparison of geometric mean viral loads for the infected animals revealed acute peak viremia was reduced by 0.72 log (P = 0.0038; two-tailed Mann-Whitney U test) and was lower by 1.1, 1.2, 1.1, and 1.0 log on weeks 4, 6, 8, and 12 in the immunized group (C). Set point viral loads were significantly reduced for the period between weeks 5 and 67 postinfection, centered on week 28, as determined by a mixed linear model analysis (P = 0.004; 0.96 log lower; 95% CI, 0.31 to 1.6). Due to progression to AIDS, naïve control animals Mm 230-04, Mm 215-98, Mm 358-01, and Mm 128-01 were euthanized weeks 16, 23, 43, and 51 postinfection, and immunized animals Mm 316-98 and Mm 158-02 were euthanized 38 and 44 weeks postinfection.
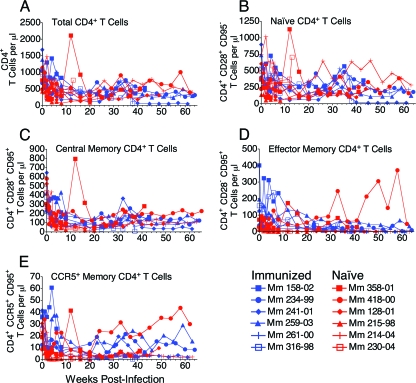
Despite statistically significant differences in viral loads, immunization did not result in significant preservation of CD4+ T cells. CD4+ T-cell counts in peripheral blood were monitored beginning on the first day of challenge to determine whether the targets of SIV infection were better protected in the scSIV-immunized animals (Fig. 9A). Frequencies of naïve, central memory, and effector memory subsets were defined by staining for CD28 and CD95 (Fig. 9B to D). CD4+ T cells were also stained for CCR5 to identify this specific target population (Fig. 9E). Although memory CD4+ T cells in the immunized animals appeared to persist at higher levels during acute infection, differences in CD4+ T-cell counts between the groups were not significant (Fig. 9C, D, and E). Therefore, although immunization reduced viral loads, this statistically significant reduction did not appear to translate into significant differences in CD4+ T-cell counts in peripheral blood.
FIG. 9.
CD4+ T-cell populations postinfection. CD4+ T-cell counts were synchronized to the last week in which viral RNA was undetectable for each animal. The numbers of total CD3+ CD4+ (A), CD3+ CD4+ CD28+ CD95− naïve (B), CD3+ CD4+ CD28+ CD95+ central memory (C), CD3+ CD4+ CD28− CD95+ effector memory (D), and CD3+ CD4+ CCR5+ CD95+ memory (E) T cells per μl of whole blood were monitored.
DISCUSSION
Stimulation of effective T-cell and antibody responses is likely to be essential for achieving protection against HIV-1. As an experimental vaccine approach designed to elicit broad T-cell responses plus antibodies to the native conformation of the viral envelope glycoprotein as it exists on virions, we developed a technique for producing strains of SIV that are limited to a single cycle of infection (27, 28, 39). Immunization of rhesus macaques with scSIV stimulated robust virus-specific T-cell responses and significantly reduced viral replication following an intravenous challenge with SIVmac239 (39). While scSIV also elicited low-titer Env-specific antibodies capable of neutralizing lab-adapted SIVmac251LA, these antibody responses were unable to detectably neutralize SIVmac239 (39). In an effort to enhance Env-specific antibody responses and the extent of protection afforded by scSIV, we immunized macaques with Env-modified strains of scSIV lacking specific structural features thought to interfere with the induction of neutralizing antibodies.
Immunization with Env-modified scSIV facilitated the development of antibodies to epitopes accessible in Envs lacking the three N-linked glycans in gp41 or 100 amino acids in the V1V2 region of gp120. However, none of the animals detectably neutralized SIVmac239 or SIVmac251UCD. Therefore, the antibodies responsible for the enhanced neutralization of the g123 and ΔV1V2 Envs may have recognized surfaces that are not accessible on wild-type virions. Alternatively, a subset of these antibodies may have interacted with the wild-type Envs but bound with an affinity too low to detect neutralization. Likewise, antibodies capable of binding wild-type Envs may have been present but were produced at concentrations too low to block infectivity. However, the induction of antibodies that detectably neutralize these strains, in the absence of persistent infection by a replicating virus, would have been unprecedented. Although no animals detectably neutralized the challenge strain, we cannot exclude the possibility that antibodies may have contributed to protection through mechanisms other than neutralization, such as antibody-dependent cell-mediated cytotoxicity (ADCC), antibody-dependent cell-mediated viral inhibition (ADCVI) (32), complement fixation, and clearance of opsonized virus.
Antibodies present in mucosal secretions were measured due to their potential importance in preventing sexual transmission of HIV-1. Immunization elicited gp120-specific IgG that was detectable in the CVS of all of the animals and appeared to be in equilibrium with the plasma in most cases. Therefore, virus-specific IgG would be expected to be present in CVS for any immunization strategy that elicits virus-specific IgG in plasma. In contrast, virus-specific IgA was not efficiently elicited. Prior to challenge, none of the animals had detectable gp120-specific IgA in CVS, and low-titer gp120-specific IgA was detectable in the rectal secretions of only one immunized animal, Mm 305-99. Despite having low to undetectable levels of virus-specific IgA prior to challenge, all of the immunized animals that became infected developed high-titer IgA responses capable of binding to gp120 and to whole-virus lysate. These high IgA titers are consistent with postchallenge anamnestic responses primed by immunization. Alternatively, the capacity of the immunized animals to mount high-titer virus-specific IgA responses after becoming infected may be an effect of immunological containment of viral replication.
T-cell responses measured by IFN-γ ELISPOT and MHC class I tetramer staining were similar in magnitude to responses elicited by prime-boost vaccine regimens employing recombinant DNA and/or viral vectors (3, 16, 36, 68, 73, 78). Virus-specific CD8+ T-cell frequencies among lymphocytes isolated from the vaginal mucosa were similar to frequencies in peripheral blood. Thus, immunization with scSIV elicited T-cell responses that trafficked to the vaginal mucosa and were present at the site of challenge. Deconvolution epitope mapping identified means of 5.4 CD4+ and 4.3 CD8+ T-cell epitopes per animal, which was similar to the breadth of responses elicited by persistent infection with live, attenuated SIV. Therefore, immunization with scSIV elicited high-frequency virus-specific T cells that recognized multiple viral antigens and trafficked to the vaginal mucosa.
Protection was assessed by repeated, low-dose vaginal inoculation with SIVmac251UCD to model heterosexual transmission of HIV-1. Although there appeared to be a trend toward a greater number of challenge doses required to establish infection for the immunized animals, this trend was not statistically significant (P = 0.10; hazard ratio of 0.38, 95% CI, 0.12 to 1.21). However, peak and set point viral loads were significantly lower in the six immunized animals that became infected relative to the naïve controls. Peak viral loads were reduced by 0.72 log (P = 0.0038), and set point viral loads were reduced by approximately 1 log for the period between 5 and 67 weeks postinfection (P = 0.004; 0.96 log lower; 95% CI, 0.31 to 1.6). Geometric mean viral loads converged after 23 weeks postinfection, due to the loss and subsequent exclusion of the two control animals with the highest viral loads.
The SIVmac251UCD challenge strain appears to be particularly difficult to protect against by immunization, perhaps even more so than SIVmac239. Differences in peak and set point viral loads in the immunized animals relative to the naïve controls were not as great as those previously observed following intravenous challenge with SIVmac239 (39). Moreover, despite the partial containment of viral replication in the immunized animals, we did not observe better preservation of CD4+ T cells. It is possible that genetic differences between SIVmac239 and the SIVmac251UCD challenge stock (52) may have been responsible for this difference in protection. The virus stock of SIVmac251UCD utilized for repeated, low-dose vaginal challenge in this study was derived from the original SIVmac251 isolate by passage of acute-phase virus through animals and selection of virus cultures with the fastest growth kinetics on rhesus macaque PBMCs (52). This process may have favored the selection of more fit or more pathogenic variants. Furthermore, two amino acids in Env that consistently revert in animals infected with SIVmac239 (15) differ from the SIVmac251UCD challenge stock (data not shown). These, plus additional differences including suboptimal nucleotides at positions elsewhere in the SIVmac239 genome (1), might explain why it appears to be more difficult to protect against SIVmac251UCD than SIVmac239.
Two immunized animals, Mm 284-99 and Mm 305-99, remained uninfected after 20 weeks of repeated vaginal challenge with SIVmac251UCD. Inherent genetic differences in the capacity to support SIV replication could explain why these two animals remained uninfected. However, there is no evidence that these animals had reduced inherent capacity to support SIV infection, since peak viral loads following the first dose of scSIV were similar in magnitude among all of the immunized animals. Furthermore, these animals were not resistant to a subsequent intravenous challenge with SIVmac251UCD. Nevertheless, it is possible that genetic factors underlie differences in the T-cell and antibody responses stimulated in these animals by immunization with scSIV. Both animals possessed the protective MHC class I allele Mamu-A*01 (57, 58), and Mm 284-99 had the highest virus-specific T-cell responses in the vaginal mucosa at week 26 and in the peripheral blood at the beginning of the challenge period. Antibody responses in these two animals also appeared to differ from those in the six immunized animals that became infected. Mm 284-99 neutralized SIVmac239g123 at a titer nearly 200-fold higher than the average for the rest of the group and 80-fold higher than the next highest animal. Moreover, neutralization of SIVmac251UCD approached a 50% reduction in infectivity at a 1:8 dilution of plasma from Mm 284-99 collected at the beginning of the challenge period. In addition, Mm 284-99 also had the highest anti-gp120 IgG-specific activity in cervicovaginal secretions at four of five time points prior to challenge. Both Mm 284-99 and Mm 305-99 also had higher prechallenge ratios of anti-gp120 IgG-specific activity in cervicovaginal secretions versus plasma (ratio, 4.0; 95% CI, 2.6 to 6.0; P < 0.01), perhaps reflecting local overproduction of gp120-specific IgG by B cells residing in the cervicovaginal mucosa (14, 61, 62). Also, the only animal that had detectable gp120-specific IgA prior to challenge was Mm 305-99. Hence, there were a number of qualitative and quantitative differences in the T-cell and antibody responses that may have contributed to the absence of SIV infection in Mm 284-99 and Mm 305-99.
The partial containment of SIV replication afforded by T-cell-based vaccines has thus far failed to provide adequate protection against immunodeficiency virus infection (12, 16, 36, 49, 54, 58, 73). Neutralizing antibodies can prevent the acquisition of viral infection (6, 33, 34, 53, 59), but eliciting such antibodies is a daunting challenge due to structural and thermodynamic properties of the envelope glycoprotein that render it resistant to antibodies (17, 46, 47, 64, 71, 75, 79, 83, 85). However, a strategy that elicits effective antibody responses to the viral envelope glycoprotein may be a necessary component of any protective vaccine against HIV-1. In this study, we tested the hypothesis that removal of specific structural features thought to interfere with the induction of neutralizing antibodies might facilitate their development. Despite significantly enhancing antibody responses to epitopes revealed by the removal of the V1V2 loops or the three N-linked glycans in gp41, neutralizing activity against SIVmac239 and SIVmac251UCD remained undetectable. Nevertheless, two immunized animals remained uninfected after a 20-week period of repeated, low-dose vaginal challenge in which all of the naïve control animals became infected. The six immunized animals that ultimately became infected had significantly lower peak and set point viral loads relative to the naïve control animals. The two that remained uninfected appeared to differ in several prechallenge measures of T-cell and antibody responses. These results are particularly intriguing in light of recent evidence that Env-specific antibodies contributed to the modest decrease in the incidence of HIV-1 infection observed among participants in an HIV-1 vaccine phase III clinical trial of a prime-boost regimen based on a recombinant canarypox vector and soluble gp120 protein (65).
Acknowledgments
We thank Nancy Wilson, Gretta Borchardt, and David Watkins at the University of Wisconsin—Madison for providing MHC class I tetramers and for MHC typing the animals. In addition, we thank Jackie Gillis and Michelle Connole in the Division of Immunology, NEPRC, for flow cytometry services.
This work was supported by grants AI063993, AI071306, and RR000168 from the National Institutes of Health and by federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. D.T.E. is an Elizabeth Glaser Scientist supported by the Elizabeth Glaser Pediatric AIDS Foundation.
Footnotes
Published ahead of print on 11 August 2010.
REFERENCES
- 1.Alexander, L., L. Denekamp, S. Czajak, and R. C. Desrosiers. 2001. Suboptimal nucleotides in the infectious, pathogenic simian immunodeficiency virus clone SIVmac239. J. Virol. 75:4019-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, L., P. O. Illyinskii, S. M. Lang, R. E. Means, J. Lifson, K. Mansfield, and R. C. Desrosiers. 2003. Determinants of increased replicative capacity of serially passaged simian immunodeficiency virus with nef deleted in rhesus monkeys. J. Virol. 77:6823-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. 1996. Guide for the care and use of laboratory animals, p. 86-123. The Institute of Laboratory Animal Resources, National Research Council, Washington, DC.
- 5.Baba, T. W., Y. S. Jeong, D. Pennick, R. Bronson, M. F. Greene, and R. M. Ruprecht. 1995. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science 267:1820-1825. [DOI] [PubMed] [Google Scholar]
- 6.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 7.Baba, T. W., V. Liska, A. H. Khimani, N. B. Ray, P. J. Dailey, D. Penninck, R. Bronson, M. F. Greene, H. M. McClure, L. N. Martin, and R. M. Ruprecht. 1999. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat. Med. 5:194-203. [DOI] [PubMed] [Google Scholar]
- 8.Barnett, S. W., B. Burke, Y. Sun, E. Kan, H. Legg, Y. Lian, K. Bost, F. Zhou, A. Goodsell, J. Zur Megede, J. Polo, J. Donnelly, J. Ulmer, G. R. Otten, C. J. Miller, M. Vajdy, and I. K. Srivastava. Antibody-mediated protection against mucosal simian-human immunodeficiency virus challenge of macaques immunized with alphavirus replicon particles and boosted with trimeric envelope glycoprotein in MF59 adjuvant. J. Virol. 84:5975-5985. [DOI] [PMC free article] [PubMed]
- 9.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barouch, D. H., A. Craiu, M. J. Kuroda, J. E. Schmitz, X. X. Zheng, S. Santra, J. D. Frost, G. R. Krivulka, M. A. Lifton, C. L. Crabbs, G. Heidecker, H. C. Perry, M. E. Davies, H. Xie, C. E. Nickerson, T. D. Steenbeke, C. I. Lord, D. C. Montefiori, T. B. Strom, J. W. Shiver, M. G. Lewis, and N. L. Letvin. 2000. Augmentation of immune responses to HIV-1 and simian immunodeficiency virus DNA vaccines by IL-2/Ig plasmid administration in rhesus monkeys. Proc. Natl. Acad. Sci. U. S. A. 97:4192-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barouch, D. H., A. Craiu, S. Santra, M. A. Egan, J. E. Schmitz, M. J. Kuroda, T. M. Fu, J. H. Nam, L. S. Wyatt, M. A. Lifton, G. R. Krivulka, C. E. Nickerson, C. I. Lord, B. Moss, M. G. Lewis, V. M. Hirsch, J. W. Shiver, and N. L. Letvin. 2001. Elicitation of high-frequency cytotoxic T-lymphocyte responses against both dominant and subdominant simian-human immunodeficiency virus epitopes by DNA vaccination of rhesus monkeys. J. Virol. 75:2462-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 13.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 14.Belec, L., T. Dupre, T. Prazuck, C. Tevi-Benissan, J. M. Kanga, O. Pathey, X. S. Lu, and J. Pillot. 1995. Cervicovaginal overproduction of specific IgG to human immunodeficiency virus (HIV) contrasts with normal or impaired IgA local response in HIV infection. J. Infect. Dis. 172:691-697. [DOI] [PubMed] [Google Scholar]
- 15.Burns, D. P., and R. C. Desrosiers. 1991. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J. Virol. 65:1843-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casimiro, D. R., F. Wang, W. A. Schleif, X. Liang, Z. Q. Zhang, T. W. Tobery, M. E. Davies, A. B. McDermott, D. H. O'Connor, A. Fridman, A. Bagchi, L. G. Tussey, A. J. Bett, A. C. Finnefrock, T. M. Fu, A. Tang, K. A. Wilson, M. Chen, H. C. Perry, G. J. Heidecker, D. C. Freed, A. Carella, K. S. Punt, K. J. Sykes, L. Huang, V. I. Ausensi, M. Bachinsky, U. Sadasivan-Nair, D. I. Watkins, E. A. Emini, and J. W. Shiver. 2005. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with DNA and recombinant adenoviral vaccine vectors expressing Gag. J. Virol. 79:15547-15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, Z., A. Gettie, D. D. Ho, and P. A. Marx. 1998. Primary SIVsm isolates use the CCR5 coreceptor from sooty mangabeys naturally infected in west Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2, and SIVmac. Virology 246:113-124. [DOI] [PubMed] [Google Scholar]
- 19.Cline, A. N., J. W. Bess, M. Piatak, Jr., and J. D. Lifson. 2005. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J. Med. Primatol. 34:303-312. [DOI] [PubMed] [Google Scholar]
- 20.Connor, R. I., D. C. Montefiori, J. M. Binley, J. P. Moore, S. Bonhoeffer, A. Gettie, E. A. Fenamore, K. E. Sheridan, D. D. Ho, P. J. Dailey, and P. A. Marx. 1998. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J. Virol. 72:7501-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crooks, E. T., P. Jiang, M. Franti, S. Wong, M. B. Zwick, J. A. Hoxie, J. E. Robinson, P. L. Moore, and J. M. Binley. 2008. Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization. Virology 377:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniel, M. D., F. Kirchhoff, S. C. Czajak, P. K. Sehgal, and R. C. Desrosiers. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938-1941. [DOI] [PubMed] [Google Scholar]
- 23.DeGottardi, M. Q., S. K. Lew, M. Piatak, Jr., B. Jia, Y. Feng, S. J. Lee, J. M. Brenchley, D. C. Douek, T. Kodama, J. D. Lifson, and D. T. Evans. 2008. Comparison of plasma viremia and antibody responses in macaques inoculated with envelope variants of single-cycle simian immunodeficiency virus differing in infectivity and cellular tropism. J. Virol. 82:321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del Prete, G. Q., B. Haggarty, G. J. Leslie, A. P. Jordan, J. Romano, N. Wang, J. Wang, M. C. Holmes, D. C. Montefiori, and J. A. Hoxie. 2009. Derivation and characterization of a simian immunodeficiency virus SIVmac239 variant with tropism for CXCR4. J. Virol. 83:9911-9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diggle, P., P. Heagerty, K. Liang, and S. Zeger. 2002. Analysis of longitudinal data. Oxford University Press, New York, NY.
- 26.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hoxie, R. W. Doms, and F. Baribaud. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans, D. T., J. E. Bricker, and R. C. Desrosiers. 2004. A novel approach for producing lentiviruses that are limited to a single cycle of infection. J. Virol. 78:11715-11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans, D. T., J. E. Bricker, H. B. Sanford, S. Lang, A. Carville, B. A. Richardson, M. Piatak, Jr., J. D. Lifson, K. G. Mansfield, and R. C. Desrosiers. 2005. Immunization of macaques with single-cycle simian immunodeficiency virus (SIV) stimulates diverse virus-specific immune responses and reduces viral loads after challenge with SIVmac239. J. Virol. 79:7707-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans, D. T., L. M. Chen, J. Gillis, K. C. Lin, B. Harty, G. P. Mazzara, R. O. Donis, K. G. Mansfield, J. D. Lifson, R. C. Desrosiers, J. E. Galan, and R. P. Johnson. 2003. Mucosal priming of simian immunodeficiency virus-specific cytotoxic T-lymphocyte responses in rhesus macaques by the Salmonella type III secretion antigen delivery system. J. Virol. 77:2400-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falkensammer, B., B. Rubner, A. Hiltgartner, D. Wilflingseder, C. S. Hennig, S. Kuate, K. Uberla, S. Norley, A. Strasak, P. Racz, and H. Stoiber. 2009. Role of complement and antibodies in controlling infection with pathogenic simian immunodeficiency virus (SIV) in macaques vaccinated with replication-deficient viral vectors. Retrovirology 6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feinberg, M. B., and J. P. Moore. 2002. AIDS vaccine models: challenging challenge viruses. Nat. Med. 8:207-210. [DOI] [PubMed] [Google Scholar]
- 32.Forthal, D. N., G. Landucci, K. S. Cole, M. Marthas, J. C. Becerra, and K. Van Rompay. 2006. Rhesus macaque polyclonal and monoclonal antibodies inhibit simian immunodeficiency virus in the presence of human or autologous rhesus effector cells. J. Virol. 80:9217-9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hessell, A. J., L. Hangartner, M. Hunter, C. E. Havenith, F. J. Beurskens, J. M. Bakker, C. M. Lanigan, G. Landucci, D. N. Forthal, P. W. Parren, P. A. Marx, and D. R. Burton. 2007. Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449:101-104. [DOI] [PubMed] [Google Scholar]
- 34.Hessell, A. J., E. G. Rakasz, P. Poignard, L. Hangartner, G. Landucci, D. N. Forthal, W. C. Koff, D. I. Watkins, and D. R. Burton. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann-Lehmann, R., J. Vlasak, A. L. Williams, A. L. Chenine, H. M. McClure, D. C. Anderson, S. O'Neil, and R. M. Ruprecht. 2003. Live attenuated, nef-deleted SIV is pathogenic in most adult macaques after prolonged observation. AIDS 17:157-166. [DOI] [PubMed] [Google Scholar]
- 36.Horton, H., T. U. Vogel, D. K. Carter, K. Vielhuber, D. H. Fuller, T. Shipley, J. T. Fuller, K. J. Kunstman, G. Sutter, D. C. Montefiori, V. Erfle, R. C. Desrosiers, N. Wilson, L. J. Picker, S. M. Wolinsky, C. Wang, D. B. Allison, and D. I. Watkins. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 76:7187-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Igarashi, T., C. Brown, A. Azadegan, N. Haigwood, D. Dimitrov, M. A. Martin, and R. Shibata. 1999. Human immunodeficiency virus type 1 neutralizing antibodies accelerate clearance of cell-free virions from blood plasma. Nat. Med. 5:211-216. [DOI] [PubMed] [Google Scholar]
- 38.Jia, B., M. Q. DeGottardi, and D. T. Evans. 2006. Single-cycle SIV: a novel AIDS vaccine approach. Future Virol. 1:747-758. [Google Scholar]
- 39.Jia, B., S. K. Ng, M. Q. DeGottardi, M. Piatak, E. Yuste, A. Carville, K. G. Mansfield, W. Li, B. A. Richardson, J. D. Lifson, and D. T. Evans. 2009. Immunization with single-cycle SIV significantly reduces viral loads after an intravenous challenge with SIV(mac)239. PLoS Pathog. 5:e1000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson, W. E., J. Morgan, J. Reitter, B. A. Puffer, S. Czajak, R. W. Doms, and R. C. Desrosiers. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson, W. E., H. Sanford, L. Schwall, D. R. Burton, P. W. Parren, J. E. Robinson, and R. C. Desrosiers. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 77:9993-10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaizu, M., G. J. Borchardt, C. E. Glidden, D. L. Fisk, J. T. Loffredo, D. I. Watkins, and W. M. Rehrauer. 2007. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics 59:693-703. [DOI] [PubMed] [Google Scholar]
- 43.Keele, B. F., E. E. Giorgi, J. F. Salazar-Gonzalez, J. M. Decker, K. T. Pham, M. G. Salazar, C. Sun, T. Grayson, S. Wang, H. Li, X. Wei, C. Jiang, J. L. Kirchherr, F. Gao, J. A. Anderson, L. H. Ping, R. Swanstrom, G. D. Tomaras, W. A. Blattner, P. A. Goepfert, J. M. Kilby, M. S. Saag, E. L. Delwart, M. P. Busch, M. S. Cohen, D. C. Montefiori, B. F. Haynes, B. Gaschen, G. S. Athreya, H. Y. Lee, N. Wood, C. Seoighe, A. S. Perelson, T. Bhattacharya, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozlowski, P. A., R. M. Lynch, R. R. Patterson, S. Cu-Uvin, T. P. Flanigan, and M. R. Neutra. 2000. Modified wick method using Weck-Cel sponges for collection of human rectal secretions and analysis of mucosal HIV antibody. J. Acquir. Immune Defic. Syndr. 24:297-309. [DOI] [PubMed] [Google Scholar]
- 45.Kuate, S., C. Stahl-Hennig, P. ten Haaft, J. Heeney, and K. Uberla. 2003. Single-cycle immunodeficiency viruses provide strategies for uncoupling in vivo expression levels from viral replicative capacity and for mimicking live-attenuated SIV vaccines. Virology 313:653-662. [DOI] [PubMed] [Google Scholar]
- 46.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 47.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 49.Letvin, N. L., J. R. Mascola, Y. Sun, D. A. Gorgone, A. P. Buzby, L. Xu, Z. Y. Yang, B. Chakrabarti, S. S. Rao, J. E. Schmitz, D. C. Montefiori, B. R. Barker, F. L. Bookstein, and G. J. Nabel. 2006. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science 312:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, Y., B. Cleveland, I. Klots, B. Travis, B. A. Richardson, D. Anderson, D. Montefiori, P. Polacino, and S. L. Hu. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82:638-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mansfield, K., S. M. Lang, M. C. Gauduin, H. B. Sanford, J. D. Lifson, R. P. Johnson, and R. C. Desrosiers. 2008. Vaccine protection by live, attenuated simian immunodeficiency virus in the absence of high-titer antibody responses and high-frequency cellular immune responses measurable in the periphery. J. Virol. 82:4135-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marthas, M. L., D. Lu, M. C. Penedo, A. G. Hendrickx, and C. J. Miller. 2001. Titration of an SIVmac251 stock by vaginal inoculation of Indian and Chinese origin rhesus macaques: transmission efficiency, viral loads, and antibody responses. AIDS Res. Hum. Retroviruses 17:1455-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 54.McDermott, A. B., D. H. O'Connor, S. Fuenger, S. Piaskowski, S. Martin, J. Loffredo, M. Reynolds, J. Reed, J. Furlott, T. Jacoby, C. Riek, E. Dodds, K. Krebs, M. E. Davies, W. A. Schleif, D. R. Casimiro, J. W. Shiver, and D. I. Watkins. 2005. Cytotoxic T-lymphocyte escape does not always explain the transient control of simian immunodeficiency virus SIVmac239 viremia in adenovirus-boosted and DNA-primed Mamu-A*01-positive rhesus macaques. J. Virol. 79:15556-15566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Means, R. E., T. Greenough, and R. C. Desrosiers. 1997. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J. Virol. 71:7895-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Means, R. E., T. Matthews, J. A. Hoxie, M. H. Malim, T. Kodama, and R. C. Desrosiers. 2001. Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J. Virol. 75:3903-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mothe, B. R., J. Weinfurter, C. Wang, W. Rehrauer, N. Wilson, T. M. Allen, D. B. Allison, and D. I. Watkins. 2003. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 77:2736-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pal, R., D. Venzon, N. L. Letvin, S. Santra, D. C. Montefiori, N. R. Miller, E. Tryniszewska, M. G. Lewis, T. C. VanCott, V. Hirsch, R. Woodward, A. Gibson, M. Grace, E. Dobratz, P. D. Markham, Z. Hel, J. Nacsa, M. Klein, J. Tartaglia, and G. Franchini. 2002. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J. Virol. 76:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parren, P. W., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pikora, C., C. Wittish, and R. C. Desrosiers. 2005. Identification of two N-linked glycosylation sites within the core of the simian immunodeficiency virus glycoprotein whose removal enhances sensitivity to soluble CD4. J. Virol. 79:12575-12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raux, M., L. Finkielsztejn, D. Salmon-Ceron, H. Bouchez, J. L. Excler, E. Dulioust, J. M. Grouin, D. Sicard, and C. Blondeau. 1999. Comparison of the distribution of IgG and IgA antibodies in serum and various mucosal fluids of HIV type 1-infected subjects. AIDS Res. Hum. Retroviruses 15:1365-1376. [DOI] [PubMed] [Google Scholar]
- 62.Raux, M., L. Finkielsztejn, D. Salmon-Ceron, H. Bouchez, J. L. Excler, E. Dulioust, J. M. Grouin, D. Sicard, and C. Blondeau. 2000. IgG subclass distribution in serum and various mucosal fluids of HIV type 1-infected subjects. AIDS Res. Hum. Retroviruses 16:583-594. [DOI] [PubMed] [Google Scholar]
- 63.Reitter, J. N., and R. C. Desrosiers. 1998. Identification of replication-competent strains of simian immunodeficiency virus lacking multiple attachment sites for N-linked carbohydrates in variable regions 1 and 2 of the surface envelope protein. J. Virol. 72:5399-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 65.Rerks-Ngarm, S., P. Pitisuttithum, S. Nitayaphan, J. Kaewkungwal, J. Chiu, R. Paris, N. Premsri, C. Namwat, M. de Souza, E. Adams, M. Benenson, S. Gurunathan, J. Tartaglia, J. G. McNeil, D. P. Francis, D. Stablein, D. L. Birx, S. Chunsuttiwat, C. Khamboonruang, P. Thongcharoen, M. L. Robb, N. L. Michael, P. Kunasol, and J. H. Kim. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209-2220. [DOI] [PubMed] [Google Scholar]
- 66.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 67.Rose, N. F., A. Roberts, L. Buonocore, and J. K. Rose. 2000. Glycoprotein exchange vectors based on vesicular stomatitis virus allow effective boosting and generation of neutralizing antibodies to a primary isolate of human immunodeficiency virus type 1. J. Virol. 74:10903-10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 69.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Swigut, T., A. J. Iafrate, J. Muench, F. Kirchhoff, and J. Skowronski. 2000. Simian and human immunodeficiency virus Nef proteins use different surfaces to downregulate class I major histocompatibility complex antigen expression. J. Virol. 74:5691-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 72.Veazey, R. S., I. C. Tham, K. G. Mansfield, M. DeMaria, A. E. Forand, D. E. Shvetz, L. V. Chalifoux, P. K. Sehgal, and A. A. Lackner. 2000. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4+ T cells are rapidly eliminated in early SIV infection in vivo. J. Virol. 74:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vogel, T. U., M. R. Reynolds, D. H. Fuller, K. Vielhuber, T. Shipley, J. T. Fuller, K. J. Kunstman, G. Sutter, M. L. Marthas, V. Erfle, S. M. Wolinsky, C. Wang, D. B. Allison, E. W. Rud, N. Wilson, D. Montefiori, J. D. Altman, and D. I. Watkins. 2003. Multispecific vaccine-induced mucosal cytotoxic T lymphocytes reduce acute-phase viral replication but fail in long-term control of simian immunodeficiency virus SIVmac239. J. Virol. 77:13348-13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, S. W., P. A. Kozlowski, G. Schmelz, K. Manson, M. S. Wyand, R. Glickman, D. Montefiori, J. D. Lifson, R. P. Johnson, M. R. Neutra, and A. Aldovini. 2000. Effective induction of simian immunodeficiency virus-specific systemic and mucosal immune responses in primates by vaccination with proviral DNA producing intact but noninfectious virions. J. Virol. 74:10514-10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 76.Whatmore, A. M., N. Cook, G. A. Hall, S. Sharpe, E. W. Rud, and M. P. Cranage. 1995. Repair and evolution of nef in vivo modulates simian immunodeficiency virus virulence. J. Virol. 69:5117-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilson, N. A., B. F. Keele, J. S. Reed, S. M. Piaskowski, C. E. Mac Nair, A. J. Bett, X. Liang, F. Wang, E. Thoryk, G. J. Heidecker, M. R. Citron, L. Huang, J. Lin, S. Vitelli, C. D. Ahn, M. Kaizu, N. J. Maness, M. R. Reynolds, T. C. Friedrich, J. T. Loffredo, E. G. Rakasz, S. Erickson, D. B. Allison, M. Piatak, Jr., J. D. Lifson, J. W. Shiver, D. R. Casimiro, G. M. Shaw, B. H. Hahn, and D. I. Watkins. 2009. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J. Virol. 83:6508-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilson, N. A., J. Reed, G. S. Napoe, S. Piaskowski, A. Szymanski, J. Furlott, E. J. Gonzalez, L. J. Yant, N. J. Maness, G. E. May, T. Soma, M. R. Reynolds, E. Rakasz, R. Rudersdorf, A. B. McDermott, D. H. O'Connor, T. C. Friedrich, D. B. Allison, A. Patki, L. J. Picker, D. R. Burton, J. Lin, L. Huang, D. Patel, G. Heindecker, J. Fan, M. Citron, M. Horton, F. Wang, X. Liang, J. W. Shiver, D. R. Casimiro, and D. I. Watkins. 2006. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 80:5875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 80.Wyand, M. S., K. H. Manson, M. Garcia-Moll, D. Montefiori, and R. C. Desrosiers. 1996. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J. Virol. 70:3724-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wyand, M. S., K. H. Manson, A. A. Lackner, and R. C. Desrosiers. 1997. Resistance of neonatal monkeys to live attenuated vaccine strains of simian immunodeficiency virus. Nat. Med. 3:32-36. [DOI] [PubMed] [Google Scholar]
- 82.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 84.Wyss, S., A. S. Dimitrov, F. Baribaud, T. G. Edwards, R. Blumenthal, and J. A. Hoxie. 2005. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J. Virol. 79:12231-12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yuste, E., J. Bixby, J. Lifson, S. Sato, W. Johnson, and R. Desrosiers. 2008. Glycosylation of gp41 of simian immunodeficiency virus shields epitopes that can be targets for neutralizing antibodies. J. Virol. 82:12472-12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yuste, E., J. D. Reeves, R. W. Doms, and R. C. Desrosiers. 2004. Modulation of Env content in virions of simian immunodeficiency virus: correlation with cell surface expression and virion infectivity. J. Virol. 78:6775-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang, L., P. J. Dailey, T. He, A. Gettie, S. Bonhoeffer, A. S. Perelson, and D. D. Ho. 1999. Rapid clearance of simian immunodeficiency virus particles from plasma of rhesus macaques. J. Virol. 73:855-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang, Y., B. Lou, R. B. Lal, A. Gettie, P. A. Marx, and J. P. Moore. 2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J. Virol. 74:6893-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]