Abstract
Chikungunya virus (CHIKV) is an emerging human pathogen transmitted by mosquitoes. Like that of other alphaviruses, CHIKV replication causes general host shutoff, leading to severe cytopathicity in mammalian cells, and inhibits the ability of infected cells to respond to interferon (IFN). Recent research, however, suggests that alphaviruses may have additional mechanisms to circumvent the host's antiviral IFN response. Here we show that CHIKV replication is resistant to inhibition by interferon once RNA replication has been established and that CHIKV actively suppresses the antiviral IFN response by preventing IFN-induced gene expression. Both CHIKV infection and CHIKV replicon RNA replication efficiently blocked STAT1 phosphorylation and/or nuclear translocation in mammalian cells induced by either type I or type II IFN. Expression of individual CHIKV nonstructural proteins (nsPs) showed that nsP2 was a potent inhibitor of IFN-induced JAK-STAT signaling. In addition, mutations in CHIKV-nsP2 (P718S) and Sindbis virus (SINV)-nsP2 (P726S) that render alphavirus replicons noncytopathic significantly reduced JAK-STAT inhibition. This host shutoff-independent inhibition of IFN signaling by CHIKV is likely to have an important role in viral pathogenesis.
Chikungunya virus (CHIKV) is a mosquito-borne arthrogenic member of the Alphavirus genus (family Togaviridae) causing current epidemics in the Indian Ocean region (25). The first reported CHIKV outbreak was in 1952 to 1953 in Tanzania. In the local Makonde language, chikungunya means “that which bends up” and refers to the body posture of infected individuals who suffer from associated arthralgia (29). CHIKV is transmitted mainly by Aedes mosquito species and is endemic in most of Central Africa and Southern Asia (18). From 2001 onwards, several major outbreaks have occurred affecting the islands of Mauritius, Madagascar, Mayotte, and Reunion Island. On Reunion Island, CHIKV affected up to one-third of the population, and CHIKV-associated deaths were recorded (25). Due to an acquired mutation in the viral glycoprotein E1 (39) and the concurrent expanding distribution of its novel mosquito vector Aedes albopictus, CHIKV is rapidly spreading to other parts of the world, including Europe (9). In 2006, mainland India suffered a major outbreak in which more than 1.4 million individuals were infected, after which more outbreaks occurred throughout the rest of Southern Asia (38). The first outbreak of CHIKV on the European continent occurred in Italy in 2007 (27). Currently, no licensed CHIKV vaccine and no effective antiviral treatment are available.
CHIKV is a plus-strand RNA virus with a genome of almost 12 kb and replicates in the cytoplasm of infected cells within virus-induced membranous vesicles (17). CHIKV produces two polyproteins, of which the first encodes nonstructural proteins (nsPs) 1, 2, 3, and 4. The nsP123 precursor and nsP4 function in a complex for viral negative-strand RNA synthesis, after which sequential processing of nsP123 into its individual proteins results in positive-strand RNA transcription and the production of subgenomic RNA (sgRNA). CHIKV nsPs serve functions needed for viral replication, e.g., methyltransferase and guanylyltransferase (nsP1), protease and helicase (nsP2), and RNA-dependent RNA polymerase (nsP4) (37). The second, structural polyprotein is translated from this sgRNA and contains capsid and envelope glycoproteins that constitute the virus particle (37). In mosquito cells, alphaviruses can replicate in a persistent manner, whereas alphavirus replication in mammalian cells usually results in severe cytopathicity, mainly caused by a dramatic shutoff of host gene expression, resulting in the suppression of innate immunity (16).
Cellular sensors, including the cytoplasmic RNA helicase MDA5, are able to detect alphavirus replication in infected mammalian cells (4). Downstream signal transduction ultimately leads to interferon regulatory factor 3 (IRF-3) activation and beta interferon (IFN-β) production. After secretion from the infected cell, IFN-β binds to the IFN-α/β receptor IFNAR in an autocrine or paracrine manner to amplify the signal or to prime uninfected cells to establish an antiviral state, respectively. Subsequently, the Janus kinases JAK1 and TYK2 are phosphorylated and, in turn, phosphorylate signal transducers and activators of transcription 1 and 2 (STAT1 and STAT2) (26). Heterodimers of STAT1/STAT2 are then translocated in an IRF-9-dependent manner from the cytoplasm into the nucleus, where they bind IFN-stimulated response elements (ISRE). STAT1 activation causes cells to produce and secrete IFN-α to further amplify the signal via the same signaling cascade. In addition, the expression of an array of antiviral proteins, including protein kinase R (PKR), 2′,5′-oligoadenylate synthetase (OAS), and Mx proteins, is then induced to ultimately clear the infection (26). In addition to the type I IFNs (IFN-α/β) expressed by most cells, type II IFN (IFN-γ) is also produced early in CHIKV infection, probably by NK cells (12), to promote the transition from innate to adaptive immunity. IFN-γ activates STAT1 via binding to the IFN-γ receptor, upon which the latter in the form of homodimers translocates to the nucleus, where it binds gamma-activating sequence (GAS) elements to transactivate antiviral gene expression (26).
Given the potency of IFNs in fighting viral infection, many viruses have evolved specific strategies to counteract or evade the antiviral IFN response (26). While alphaviruses are known to cause dramatic host protein synthesis shutoff (16), recent research has shown that this alone is not sufficient to ensure productive infection and that the IFN response is also antagonized in a more direct manner (35). Whether or not CHIKV counteracts the IFN response is unknown; however, it is clear that robust IFNAR-dependent type I IFN signaling is required in order to limit CHIKV replication in animals (5, 32). IFN-α was recently shown to inhibit CHIKV replication in mice if given before infection, but not when given 3 days after infection (12).
In this paper, we show that CHIKV replication is resistant to IFN treatment and inhibits IFN-induced JAK-STAT signaling and downstream gene transcription independently of host shutoff. We also show for the first time that alphavirus nsP2 alone is sufficient for JAK-STAT inhibition. A P726S substitution in a conserved region of Sindbis virus (SINV) nsP2 was previously reported to reduce SINV cytopathicity (8). Here we show that this substitution and the corresponding P718S substitution in CHIKV reversed the ability of CHIKV and SINV replicons to block the JAK-STAT pathway.
MATERIALS AND METHODS
Cells and virus.
African green monkey kidney (Vero) and baby hamster kidney (BHK-21J) cells were cultured in Dulbecco's modified Eagle medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen) at 37°C in an atmosphere with 5% CO2 in tissue culture flasks (Greiner). Chikungunya virus isolate 06113879 (Mauritius strain) was obtained from the Victorian Infectious Diseases Reference Laboratory (VIDRL) and was supplied via Queensland Health Forensic and Scientific Services (QHFSS). The isolate was titrated on Vero cells via plaque assay.
Construction of alphavirus replicons and expression plasmids.
A CHIKV strain 37997 replicon (CHIKrep-EGFP) (see Fig. 4A) expressing EGFP was constructed by removing the structural genes from CHIKV infectious clone 5′-pCHIKic (40) and inserting enhanced green fluorescent protein (EGFP). Next, a firefly luciferase (Fluc) gene was generated by PCR (Phusion DNA polymerase; Finnzymes) from pGL3 (Promega) using primers AscI-Luc-F and BssHII-Luc-R and was cloned into CHIKrep-EGFP, in frame and upstream of the EGFP gene, to generate CHIKrep-FlucEGFP (see Fig. 1C). The red fluorescent marker gene mCherry (33) was amplified by PCR using primers AscI-mCherry-F and EcoRI-mCherry-R and was cloned into CHIKrep-EGFP in place of EGFP to generate CHIKrep-mCherry (see Fig. 5A). A puromycin acetyltransferase gene fused to the foot-and-mouth disease virus (FMDV) 2A autoprotease was generated by PCR from repPAC-βGal (21) using primers MluI-PAC2A-F and -R and was cloned into CHIKrep-EGFP in place of EGFP to generate CHIKrep-pac2AEGFP (see Fig. 6A). An MluI fragment from CHIKrep-pac2AEGFP was subcloned into pBluescript and was reinserted after nsP2 was mutated by QuikChange PCR using primers CHIK-nsP2-P718S-F and -R, generating CHIKrep-pac2AEGFP-nsP2m. A cytopathic, “wild-type” Sindbis virus replicon was generated from the noncytopathic replicon SINrepGFP (pHY213; pToto1101 derivative) by mutating the nsP2 serine at position 726 into a proline with primers SINnsP2-726P-V426 and SINnsP2-726P-V427 to generate SINrepGFP-wt (see Fig. 6A). Individual CHIKV nsPs were PCR amplified from CHIKrep-EGFP using the AttB1 and AttB2 primers listed (Table 1) and were cloned into expression plasmids downstream of a cytomegalovirus (CMV) immediate-early promoter (see Fig. 6A) via traditional cloning or Gateway technology using pDONR207 and pcDNA-DEST40 (Invitrogen). The mCherry gene was fused to the FMDV 2A autoprotease using PCR with primers EcoRI-mCherry-F and EcoRI-2A-mCherry-R and was cloned as an EcoRI fragment in frame and upstream of CHIKV nsPs for live visualization of transfected cells (see Fig. 5A). Autocleavage of the red fluorescent mCherry2A protein from the nsPs results in the expression of CHIKV nsP1 to nsP4 with nearly authentic N termini to retain biological activity. All constructs were verified by sequencing (Eurofins MWG Operon, Germany).
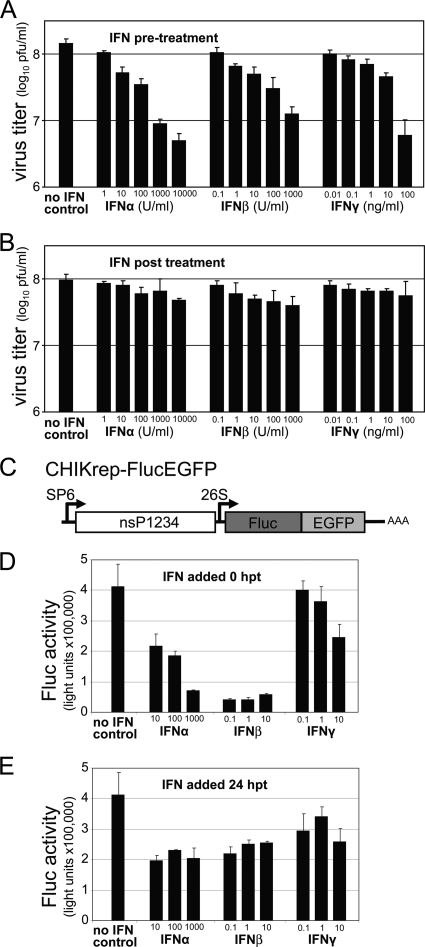
FIG. 1.
Resistance of CHIKV to type I/II IFN treatment. (A and B) Sensitivity of CHIKV infection to IFN treatment. IFNs were added as indicated to CHIKV-infected Vero cells 6 h prior to infection (A) or 4 h p.i. (B). Supernatants were collected 24 h p.i., and virus titers were determined by plaque assays. Error bars represent standard deviations of duplicates. (C) Schematic representation of the CHIKrep-FlucEGFP replicon expressing an Fluc-EGFP fusion protein. (D and E) Sensitivity of the replication of CHIKV replicon RNA to IFN treatment. Different concentrations of IFNs were added to CHIKV replicon-transfected Vero cells in 96-well plates directly posttransfection (0 h p.t.) (D) or 24 h p.t. (E), and Fluc activity was measured 48 h p.t. Concentrations of IFN-α are expressed in international units (IU) per ml, and IFN-β/γ concentrations are expressed in ng per ml. Error bars represent standard deviations.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence (5′-3′)a |
|---|---|
| AscI-Luc-F | TTGGGCGCGCCATGGAAGACGCCAAAAACATAA |
| BssHII-Luc-R | TTGTGCGCGCTCCACGGCGATCTTTCCGCC |
| MluI-PAC2A-F | TTGGACGCGTCATGACCGAGTACAAGCCCACG |
| MluI-PAC2A-R | TTGTACGCGTTCGGGCCCTGGGTTGGACTCG |
| AscI-mCherry-F | CGGGCGCGCCACCATGGTGAGCAAGGGCGAGGAG |
| EcoRI-mCherry-R | CGGAATTCCTTGTACAGCTCGTCCATG |
| CHIK-nsP2-P718S-F | GCTCAAGTCGGGTGGTTCATTACTG |
| CHIK-nsP2-P718S-R | CCACCCGACTTGAGCAGTCTCAGGG |
| SINnsP2-726P-V426 | CTGAATTGTTTAAACCCAGGAGGCACCCTC |
| SINnsP2-726P-V427 | GAGGGTGCCTCCTGGGTTTAAACAATTCAG |
| attB1-CHIK-nsP1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAATTCACCATGGATCCCGTGTACGTGG |
| attB1-CHIK-nsP2 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAATTCACCATGGGAATAATTGAAACTCCAAGAG |
| attB1-CHIK-nsP3 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAATTCACCATGGCACCGTCGTACCGGGTT |
| attB1-CHIK-nsP4 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAATTCACCATGTACATATTCTCATCTGACACC |
| attB2-CHIK-nsP1 | GGGGACCACTTTGTACAAGAAAGCTGGGTACTATGCCCCAGCTCTGTCTTC |
| attB2-CHIK-nsP2 | GGGGACCACTTTGTACAAGAAAGCTGGGTACTAGCACCCTGCTCGGGTGG |
| attB2-CHIK-nsP3 | GGGGACCACTTTGTACAAGAAAGCTGGGTACTACCCACCTGCCCTATCTAG |
| attB2-CHIK-nsP4 | GGGGACCACTTTGTACAAGAAAGCTGGGTACTATTTAGGACCACCGTACAG |
| EcoRI-mCherry-F | CGGAATTCACCATGGTGAGCAAGGGCGAGGAG |
| EcoRI-2A-mCherry-R | CGGAATTCGGGCCCTGGGTTGGACTCGACGTCGCCGGCCAACTTGAGCAGGTCAAAGTTAACCTTGTACAGCTCGTCCATG |
| HuOAS2-F | CGGTGTATGCCTGGGAACAGG |
| HuOAS2-R | GGGTCAACTGGATCCAAGATTAC |
| HuRPL13A-F | CATCGTGGCTAAACAGGTACTG |
| HuRPL13A-R | CGCACGACCTTGAGGGCAGC |
Restriction sites are italicized, start codons are underlined, and mutations are shown in boldface.
IFN sensitivity assay. (i) CHIKV.
For IFN pretreatment, Vero cells grown in 24-well plates were treated with various doses of IFN-α (I-4276; Sigma), IFN-β (I-4151; Sigma), and IFN-γ (I-1520; Sigma) for 6 h. The cells were washed and infected with CHIKV at a multiplicity of infection (MOI) of 1 PFU per cell. Three hours after viral absorption, the cells were washed; then they were incubated for an additional 21 h. For IFN posttreatment, Vero cells were infected with CHIKV at an MOI of 1 PFU/cell. Four hours after viral absorption, cells were treated with various doses of IFN as indicated (Fig. 1) and were left for an additional 21 h. The supernatants were collected, and viral titers were determined by plaque assays on Vero cells.
(ii) CHIKV replicon.
In vitro-transcribed, capped CHIKrep-FlucEGFP replicon RNA (400 ng/well) was transfected into Vero cells in 96-well plates by using Lipofectamine 2000 (Invitrogen) and Opti-MEM medium (Invitrogen) according to the manufacturer's recommendations. The transfection mixture was removed after 4 h of incubation and was replaced with DMEM plus 10% FBS. Directly after transfection (0 h posttransfection [p.t.]) or 24 h p.t., type I IFNs (IFN-α and IFN-β; Calbiochem, Nottingham, United Kingdom) and type II IFN (IFN-γ; AbD Serotec, Düsseldorf, Germany) were added to the wells in increasing concentrations. Two days after transfection, cells were lysed in 100 μl passive lysis buffer (Promega Benelux, Leiden, Netherlands), and luciferase expression was measured on a Fluostar Optima microplate reader (BMG Labtech, Germany) using d-luciferin (Synchem OHG, Germany) as a substrate basically as described previously (14).
IFN reporter assay.
Vero cells grown in 24-well plates were cotransfected with 40 ng pRL-TK (Promega) plasmid DNA expressing Renilla luciferase (Rluc) and with 200 ng of either the IFN-α/β-responsive (ISRE) firefly luciferase (Fluc) reporter plasmid p(9-27)4th (−39)Lucter or the IFN-γ-responsive (GAS) luciferase reporter plasmid p(IRF-1*GAS)6tk (−39)Lucter (15) by using the Genejammer (Stratagene) transfection reagent. Briefly, at 24 h p.t., cells were infected with CHIKV at an MOI of 5 PFU/cell. At 4, 8, and 12 h postinfection (p.i.), cells were treated with 1,000 IU of IFN-α (Intron A Redipen) per ml or 100 ng of IFN-γ (BD Pharmingen) per ml for 6 h and were then assayed for Fluc and Rluc activities using the Dual luciferase reporter assay system (Promega) as described previously (22).
Real-time RT-PCR.
Vero cells grown in 24-well plates were infected with CHIKV at an MOI of 5 PFU/cell. Healthy or infected cells were subsequently incubated at 4, 8, or 12 h p.i. with 1,000 IU of IFN-α (Intron A Redipen) per ml or 100 ng of IFN-γ (BD Pharmingen) per ml for 10 h. Total RNA was purified using Trizol reagent (Invitrogen), and real-time reverse transcription-PCR (real-time RT-PCR) was carried out on a Rotor-Gene 3000 PCR machine (Corbett Research) using Superscript III (Invitrogen) and SYBR green (Invitrogen) basically as described previously (12). Primers for amplification of OAS2 transcripts were HuOAS2-F and -R, and primers for the housekeeping gene RPL13A (24) were HuRPL13A-F and -R. Each sample was analyzed in duplicate and normalized to RPL13A mRNA levels. OAS2 mRNA transcription levels were expressed relative to levels in mock-infected, IFN-treated samples.
Immunofluorescence and Western blotting. (i) CHIKV virus.
Vero cells grown on glass coverslips in 24-well plates were infected with CHIKV at an MOI of 1 PFU/cell. Twenty-four hours after infection, cells were treated with 1,000 IU/ml of IFN-α (I-4276; Sigma) or 50 ng/ml of IFN-γ (I-1520; Sigma) for 30 min at 37°C. Cells were fixed in 4% formaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature, permeabilized with ice-cold acetone-methanol (1:1) for 30 min at −20°C, and stained sequentially with cross-reacting monoclonal antibodies specific for CHIKV envelope protein and with polyclonal antibodies against STAT1 (SC-345; SantaCruz Biotechnology, Santa Cruz, CA) or STAT2 (SC-476; Santa Cruz) at concentrations of 1 μg/ml essentially as described by the manufacturer. Secondary antibodies (GaM-AF546 and GaR-AF488) were obtained from Invitrogen, and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Microscopy was performed using a Zeiss LSM 510 Meta confocal microscope. For Western blot analysis, Vero cells in 6-well plates were infected with CHIKV at an MOI of 1 PFU/cell. Twenty-four hours p.i., cells were either treated with IFN-α (I-4276; Sigma) or IFN-γ (I-1520; Sigma) for 30 min or left untreated as indicated (see Fig. 3). Western blotting was performed on Vero cell lysates as described previously (22) using antibodies against phosphorylated STAT1 (pSTAT1) (Pharmingen, San Diego, CA), STAT1 (SC-345; Santa Cruz), and tubulin (T2200; Sigma), and analysis was performed with an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE).
(ii) Replicons and single nsPs.
Vero cells grown in 96-well plates were transfected with capped, in vitro-transcribed CHIKrep-EGFP, CHIKrep-mCherry, CHIKrep-pac2AEGFP, or CHIKrep-pac2AEGFP-nsP2m replicon RNA, one of the four pCMV-nsP constructs, or the SINrepGFP construct (400 ng of RNA or DNA per well) using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, cells were treated for 30 min with 100 IU IFN-α (Calbiochem), 2.5 ng IFN-β (Calbiochem), or 1 ng IFN-γ (AbD Serotec) per well (100 μl). For the host shutoff experiment, cells were transfected with the CHIKrep-EGFP replicon in normal medium or medium containing 0.5 μg/ml cycloheximide. Twelve hours p.t., cells received a similar IFN-β treatment. Cells were fixed with 4% paraformaldehyde in PBS and were permeabilized with 0.1% sodium dodecyl sulfate (SDS) in PBS to retain EGFP and/or mCherry fluorescence. Nuclei were stained with Hoechst 3342. STAT1 nuclear translocation was visualized either with an anti-pSTAT1 primary antibody (phospho-Tyr701; SAB Signalway Antibody, Pearland, TX) and the secondary antibody GaR-rhodamine (Nordic Immunology, Tilburg, Netherlands) or GaR-AF488 (Molecular Probes, Leiden, Netherlands) or with an anti-STAT1 primary antibody (SC-417; Santa Cruz) and the secondary antibody GaM-AF546 (Molecular Probes), using an Olympus IX71 inverted microscope with an X-Cite 120 series lamp.
RESULTS
CHIKV replication confers resistance to type I/II IFN treatment.
Since an intact IFN response is a requirement for limiting CHIKV infection in animals (5), we first investigated to what degree CHIKV replication could be inhibited in cells by (pre)treatment with type I and type II IFNs. Vero cells have an intact IFN signaling pathway and respond to IFN treatment; however, they cannot produce IFN (7) and thus lack the autocrine IFN amplification loop. These characteristics allow accurate measurement of the effects of different, exogenous IFNs on viral RNA amplification and virus production. When cells were primed for 6 h with IFN prior to virus infection, CHIKV production was decreased in an IFN concentration-dependent manner (Fig. 1A). IFN-α was most effective, followed by IFN-β and IFN-γ. Although pretreatment with 10,000 U/ml of IFN-α could reduce virus production approximately 25-fold (from 8.1 × 108 to 6.7 × 106 PFU/ml), viral titers were not reduced further than 6.7 × 107 PFU/ml, indicating that CHIKV was rather insensitive to IFN pretreatment under the experimental conditions used and still replicated to relatively high titers. When IFN was applied 4 h p.i., viral titers were not significantly decreased (maximum reduction from 1 × 108 to 7.7 × 107 PFU/ml) (Fig. 1B), indicating that virus production was not greatly affected by high concentrations of IFN when IFN was added after the establishment of infection.
Next, the effect of IFN treatment on CHIKV RNA replication, independently of virus production and/or secondary infection, was tested. A CHIKV replicon was constructed in which the structural genes were replaced by a firefly luciferase (Fluc)-enhanced green fluorescent protein (EGFP) fusion gene (CHIKrep-FlucEGFP [Fig. 1C]). In this way, transfected cells could be visualized by fluorescence microscopy and replication measured by luminometry. In vitro-transcribed, capped CHIKrep-FlucEGFP replicon RNA was transfected into Vero cells. Directly after transfection or 24 h posttransfection (p.t.), type I/II IFNs were added to the wells in increasing concentrations, and luciferase expression was measured 2 days after transfection. In results similar to those obtained with CHIKV infection, when IFN was added directly after RNA transfection (pretreatment), CHIKV replication was negatively affected in a concentration-dependent manner (Fig. 1D). In the concentrations used, IFN-β was most effective (∼10% of transgene expression retained), followed by IFN-α and IFN-γ. This is similar to what was reported for SINV, another Old World alphavirus (41). When IFN was added 24 h p.t., however, Fluc expression could not be reduced further than approximately 50%, even with the highest IFN concentrations (Fig. 1E). Collectively, these results suggest that CHIKV is insensitive to IFN once viral RNA replication has been established.
CHIKV infection inhibits type I/II IFN signaling.
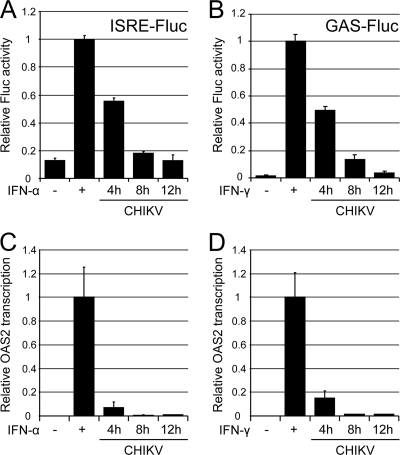
Since CHIKV replication is partially sensitive to the priming of cells with type I IFNs (and to a lesser extent with type II IFN) but is largely resistant to IFN treatment after viral RNA replication is well under way, it is likely that CHIKV blocks downstream IFN signaling and expression of IFN-stimulated genes (ISGs) with antiviral activity. To test this hypothesis, the effect of CHIKV RNA replication on downstream IFN-induced gene transcription was investigated. Vero cells were transfected with type I IFN-responsive (ISRE) or type II IFN-responsive (GAS) Fluc reporter plasmids and were subsequently infected with CHIKV. Fluc expression was induced by stimulation with type I/II IFNs at 4, 8, and 12 hpi and was normalized to Renilla luciferase (Rluc) activity expressed from a constitutive promoter on a cotransfected pRL-TK plasmid (Fig. 2 A and B). Rluc activity decreased approximately 1.5-fold, 2.5-fold, and 4-fold at 4, 8, and 12 hpi, respectively, compared to that in mock-infected cells (not shown), indicating that CHIKV infection resulted in some host shutoff within this time frame. However, the inhibition by CHIKV of IFN-stimulated gene transcription was more pronounced. Relative Fluc expression from the responsive element ISRE or GAS (normalized to Rluc expression) in response to treatment with IFN-α (Fig. 2A) or IFN-γ (Fig. 2B), respectively, was substantially inhibited in Vero cells infected with CHIKV. This inhibition was apparent at 4 hpi (2-fold) and 8 hpi (5 to 8-fold) and was essentially 100% at 12 hpi (Fig. 2A and B). In the absence of CHIKV infection, a >7-fold or 58-fold induction of normalized Fluc expression in response to treatment with IFN-α (Fig. 2A) or IFN-γ (Fig. 2B), respectively, was observed. These results clearly indicated that CHIKV infection efficiently blocks IFN signaling beyond the inhibition mediated by host shutoff.
FIG. 2.
Inhibition of type I/II IFN signaling and ISG induction by CHIKV infection. (A and B) Vero cells were transfected with a pRL-TK plasmid expressing Rluc and either a type I IFN-responsive (ISRE) or a type II IFN-responsive (GAS) Fluc reporter plasmid. At 24 h p.t., cells were infected with CHIKV at an MOI of 5 PFU/ml. At 4, 8, and 12 h p.i., cells were treated with IFN-α at 1,000 IU/ml (A) or with IFN-γ at 100 ng/ml (B) for 6 h; then they were assayed for Fluc and Rluc activities. Activities in mock-infected (uninfected) cells with/without IFN induction were also measured. Fluc values were divided by Rluc readings to compensate for virus-induced downregulation of transcription/translation and were expressed relative to values for mock-infected, IFN-treated samples. Average values from triplicate samples are shown. Error bars represent standard deviations. (C and D) Vero cells, either healthy or infected with CHIKV for 4, 8, or 12 h, were incubated with 1,000 IU of IFN-α (C) or 100 ng of IFN-γ (D) per ml for 10 h. Real-time RT-PCR values for the IFN-stimulated gene OAS2 were normalized to those for the housekeeping gene RPL13A. OAS2 mRNA transcription levels were expressed relative to those of mock-infected, IFN-treated samples. Average values from duplicate samples are shown. Error bars represent standard deviations.
To illustrate that CHIKV infection also inhibited the induction of ISG expression, an RT-PCR assay was used to monitor the expression of 2′-5′-oligoadenylate synthetase 2 (OAS2) transcripts. As expected (31), large increases in OAS mRNA levels were seen in Vero cells after treatment with IFN-α or IFN-γ (Fig. 2C and D, first two bars). However, in cells infected with CHIKV and treated with type I and II IFNs at various time points p.i., OAS mRNA levels were substantially reduced relative to levels of the housekeeping gene RPL13A (Fig. 2C and D). These results demonstrated that CHIKV infection efficiently blocks ISG expression beyond that mediated by host shutoff.
CHIKV infection and CHIKV replicon RNA replication block type I/II IFN-induced STAT1 nuclear translocation.
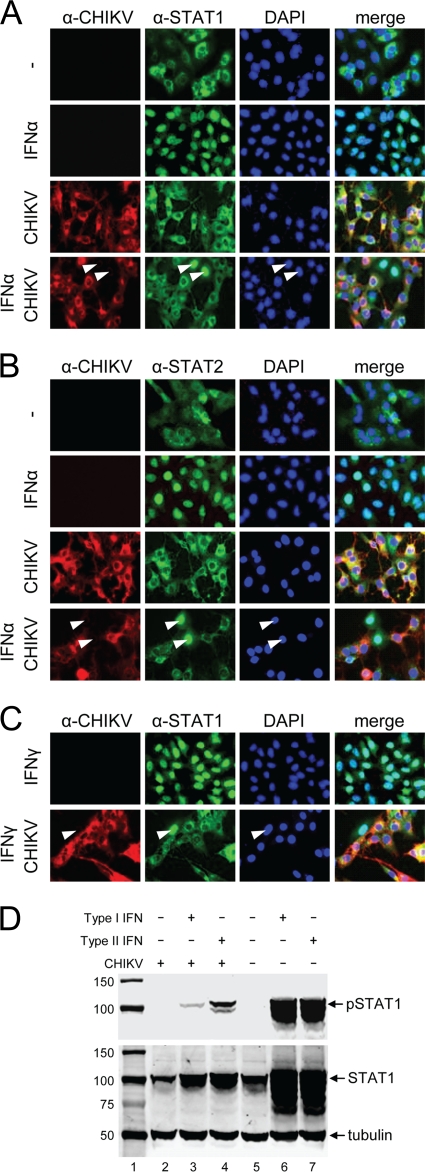
In order to investigate whether CHIKV could block IFN signaling by specifically interfering with the JAK-STAT pathway, Vero cells were infected with CHIKV at an MOI of 1 PFU/cell and were subsequently induced with type I IFN. Induction with type I IFNs should result in STAT1/STAT2 phosphorylation/heterodimerization and subsequent nuclear translocation. As expected, STAT1 in normal Vero cells was localized in the cytoplasm but translocated to the nucleus upon induction with type I IFN (Fig. 3A). In contrast, when cells were infected with CHIKV 12 h prior to IFN induction, STAT1 nuclear translocation was completely blocked (Fig. 3A). The same result was obtained for STAT2 (Fig. 3B). Similarly, type II IFN stimulation should lead to STAT1 phosphorylation/homodimerization and nuclear translocation in normal Vero cells, and this was indeed observed in uninfected cells (Fig. 3C). Again, CHIKV infection effectively blocked STAT1 nuclear translocation (Fig. 3C). Taken together, these results indicate that CHIKV infection blocks both type I and type II IFN-induced JAK-STAT signaling.
FIG. 3.
(A to C) CHIKV infection blocks STAT1/STAT2 nuclear translocation without depleting endogenous STAT1 levels. Vero cells were infected by CHIKV and were treated with IFN-α (A and B) or IFN-γ (C) for 30 min. Cells were fixed and stained with monoclonal antibodies specific for CHIKV envelope protein and STAT1 (A and C) or STAT2 (B). (C) Block in nuclear translocation of STAT1 in CHIKV infection in response to treatment with IFN-γ. Arrowheads indicate cells negatively infected with CHIKV but with nuclear STAT1/2. (D) CHIKV infection blocks STAT1 phosphorylation in Vero cells in response to IFN treatment. pSTAT1, STAT1, and tubulin were detected by Western blotting in CHIKV-infected or mock-infected Vero cells that were either left untreated or induced with type I or type II IFNs. Lane 1, protein size marker (in kilodaltons).
It is well known that alphavirus replication leads to host protein synthesis shutoff (16). However, based on the immunofluorescence detection of similar levels of endogenous STAT1 and STAT2 in infected and uninfected cells (Fig. 3A to C), it is unlikely that CHIKV infection depletes/degrades STAT1/2 proteins. To confirm that the absence of nuclear phospho-STAT1 in cells infected with CHIKV was not the result of depletion of STAT1 protein, Western blotting was performed to detect endogenous STAT1. It is apparent that cells infected with CHIKV (Fig. 3D, lane 2) have levels of endogenous STAT1 similar to those in uninfected cells (Fig. 3D, lane 5), suggesting that CHIKV does not degrade endogenous STAT1 but may act via the inhibition of STAT1 phosphorylation and/or nuclear translocation. As expected, STAT1 was highly upregulated by IFN induction in uninfected cells, likely through signaling via the JAK-STAT pathway (22). In contrast, this was not the case in CHIKV-infected cells, suggesting that CHIKV also blocks the IFN-induced upregulation of STAT1. Importantly, Western blot analysis performed with antibodies against phospho-STAT1 showed that CHIKV infection causes a major reduction in the amount of phospho-STAT1 in induced cells (Fig. 3D, lanes 3 and 4) compared to that in IFN-induced, uninfected cells (Fig. 3D, lanes 6 and 7). These data support the observations from the immunofluorescence experiments and indicate that CHIKV infection inhibits STAT phosphorylation.
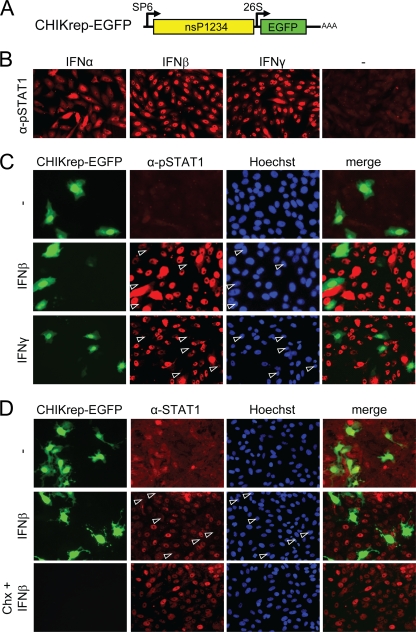
Some so-called New World alphaviruses need expression of their capsid gene to modulate the IFN response (1). CHIKV is an Old World alphavirus and therefore is not expected to need capsid expression for the suppression of IFN signaling. To determine whether RNA replication and expression of CHIKV nsPs are sufficient to block the JAK-STAT pathway, a CHIKV replicon in which the structural genes were deleted and replaced by EGFP was constructed (CHIKrep-EGFP [Fig. 4A]). In vitro-transcribed CHIKrep-EGFP RNA was transfected into Vero cells, and the cells were then stimulated with type I and type II IFNs 24 h p.t. As expected, in untransfected cells, phospho-STAT1 was found in the nuclei of Vero cells after 30 min of induction with IFN-α, and this process occurred even more efficiently with IFN-β or IFN-γ (Fig. 4B). In contrast, however, cells transfected with CHIKrep-EGFP (green; expressing EGFP) and induced with IFN-β or IFN-γ lacked nuclear STAT1 (Fig. 4C, arrowheads), indicating that CHIKV replication blocks type I and type II IFN-induced STAT1 phosphorylation and/or nuclear translocation.
FIG. 4.
A CHIKV replicon efficiently inhibits type I/II IFN-induced JAK-STAT signaling independently of host shutoff. (A) Schematic representation of CHIKrepEGFP, expressing EGFP. (B) pSTAT1 nuclear translocation in Vero cells upon induction with type I and type II IFNs. (C) A CHIKV replicon blocks pSTAT1 nuclear translocation upon type I/II IFN induction. Vero cells were immunostained with an anti-pSTAT1 antibody 24 h p.t. (D) CHIKV RNA replication, but not translational shutoff, blocks STAT1 nuclear translocation. Vero cells were transfected with CHIKrep-EGFP replicon RNA in the absence or presence of cycloheximide (Chx). Cells were induced for 30 min with IFN-β at 12 h p.t. and were stained with an anti-STAT1 antibody. Open arrowheads indicate CHIKV replicon-positive cells lacking nuclear STAT1.
There is a possibility that the lack of nuclear STAT1 translocation in replicon cells could still be due to host shutoff resulting from CHIKV replicon RNA replication, although Fig. 3D showed that endogenous STAT1 levels were not decreased by CHIKV infection. Nevertheless, to rule out this possibility, cells were treated with cycloheximide to inhibit translation. This method of pharmacologically induced host cell protein synthesis shutoff was recently used in experiments with Venezuelan equine encephalitis virus (VEEV) to show that JAK-STAT signaling was blocked by VEEV and not by host shutoff (35). As expected, STAT1 fluorescence in control cells not treated with cycloheximide was cytoplasmic, with no apparent difference in localization or fluorescence intensity between untransfected cells and green CHIKV replicon-transfected cells (Fig. 4D, top row). After IFN-β treatment, STAT1 was translocated into the nucleus in all cells except those expressing the CHIKV replicon (Fig. 4D, open arrowheads). In cells treated with cycloheximide, CHIKV replicon-encoded EGFP was absent due to effective inhibition of protein synthesis (Fig. 4D, bottom). However, STAT1 nuclear translocation upon IFN-β induction was still clearly apparent, despite effective inhibition of translation by cycloheximide (Fig. 4D, bottom). Taken together, these experiments clearly show that CHIKV infection and the replication of CHIKV replicon RNA efficiently inhibit IFN-stimulated JAK-STAT signaling independently of host shutoff.
CHIKV nsP2 inhibits IFN-induced STAT1 nuclear translocation.
Since the CHIKV replicon could efficiently inhibit JAK-STAT signaling, the next question was whether any of the CHIKV nsPs could be found to be responsible for this activity. Previous reports suggested that alphavirus nsP2 may be an important modulator of the IFN response (3, 11); however, direct inhibition of the JAK-STAT pathway by an individual alphaviral nsP2 has not been reported.
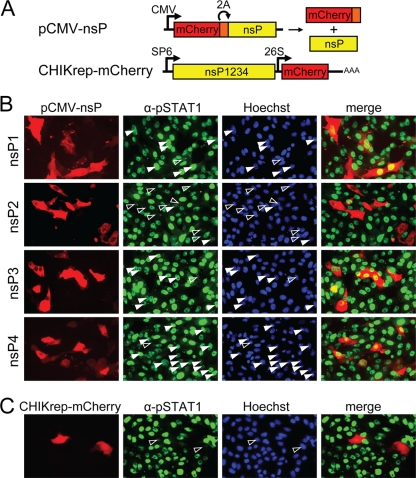
In order to identify the CHIKV-encoded protein responsible for blocking STAT1 nuclear translocation, Vero cells were transfected with plasmids expressing individual nonstructural proteins fused to self-cleaving mCherry2A; as a control, cells were transfected with a CHIKV replicon expressing mCherry (CHIKrep-mCherry) (Fig. 5A). Two days p.t., cells were incubated with IFN-β, and nuclear localization of phospho-STAT1 was visualized using anti-pSTAT1 antibodies (note that pSTAT1 appears green in these images). IFN-β induction of transfected Vero cells showed that STAT1 efficiently translocated to the nucleus in cells expressing nsP1, nsP3, or nsP4 (Fig. 5B, solid arrowheads). Only very few cells were found to lack nuclear phospho-STAT1 (Fig. 5B, open arrowheads), suggesting that nsP1, -3, and -4 were not capable of efficiently blocking STAT1 nuclear translocation. In sharp contrast, however, STAT1 nuclear translocation was absent in the vast majority of cells expressing nsP2 (Fig. 5B, nsP2, open arrowheads) and the positive control CHIKrep-mCherry (Fig. 5C, open arrowheads). In the few nsP2-expressing cells that did display nuclear pSTAT1, the fluorescence intensity was much lower than that in untransfected cells (Fig. 5B, nsP2, solid arrowheads). As expected, the CHIKrep-mCherry-transfected cells also showed no nuclear translocation after IFN-β treatment (Fig. 5C, open arrowheads).
FIG. 5.
Inhibition of IFN-β-induced STAT1 nuclear translocation by individual CHIKV nsPs. (A) Schematic representation of the pCMV-nsP1, -2, -3, and -4 expression plasmids and the CHIKrep-mCherry replicon, expressing mCherry. CMV, cytomegalovirus immediate-early promoter; 2A, foot-and-mouth disease virus 2A autoprotease. The bacteriophage SP6 and CHIKV 26S promoters are indicated. (B) pSTAT1 nuclear translocation upon IFN-β induction in Vero cells transfected with pCMV-nsP1, -2, -3, or -4. Cells were immunostained with an anti-pSTAT1 antibody. (C) pSTAT1 nuclear translocation upon IFN-β induction in CHIKrep-mCherry-transfected Vero cells. Open arrowheads indicate cells positive for nsP1, -2, -3, or -4- or for the CHIKV replicon that lack nuclear pSTAT1; solid arrowheads indicate nsP1- to nsP4-positive cells with nuclear pSTAT1.
These results clearly indicate that individually expressed CHIKV nsP2 is capable of inhibiting JAK-STAT signaling.
Mutation of a conserved proline in the C terminus of nsP2 abolishes the inhibitory effect of CHIKV and SINV replicons on JAK-STAT signaling.
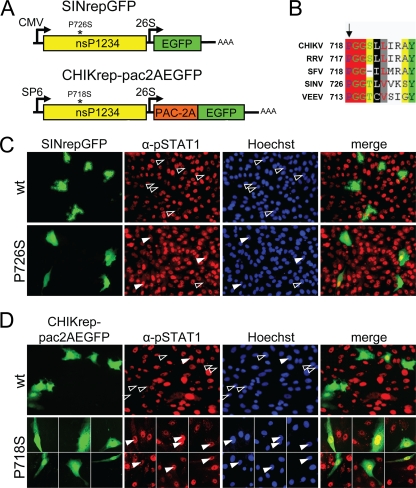
Mutations in alphavirus nsP2 can have significant effects on the IFN response (3, 11). For example, a mutation of a conserved proline (Fig. 6B) at position 726 in SINV was previously shown to result in noncytopathic RNA replication (8) and reduced viral titers associated with higher IFN production (11). We hypothesized that this mutation could render the replicon unable to block JAK-STAT signaling. This possibility was investigated by transfecting Vero cells with cytopathic “wild-type” SINrepGFP-wt (with proline restored at position 726) and the noncytopathic SINV replicon SINrepGFP (containing the P726S mutation in nsP2) (Fig. 6A). Transfected cells were induced 24 h p.t. with IFN-β for 30 min and were stained with phospho-STAT1 antibodies as before. According to the hypothesis, the cytopathic “wild-type” SIN replicon was able to effectively block STAT1 nuclear translocation, whereas the noncytopathic SIN replicon with the nsP2 P726S mutation (8) was not (Fig. 6C).
FIG. 6.
Mutation of a conserved proline in nsP2 abolishes the inhibitory effect of CHIKV and SINV replicons on JAK-STAT signaling. (A) Schematic representation of the CHIKrep-pac2AEGFP and SINrepLuc replicons. nsP2 mutations P718S and P726S are indicated with asterisks; pac, puromycin acetyltransferase. (B) Partial amino acid alignment of alphavirus nsP2s. RRV, Ross River virus; VEEV, Venezuelan equine encephalitis virus. The conserved proline and amino acid numbers within nsP2 proteins are indicated. (C) pSTAT1 nuclear translocation upon IFN-β induction in SINrepGFP (wild type and mutant nsP2-P726S)-transfected Vero cells. Cells were immunostained with an anti-pSTAT1 antibody. Open arrowheads indicate replicon-positive cells lacking nuclear pSTAT1; solid arrowheads indicate replicon-positive cells with nuclear pSTAT1. (D) Nuclear translocation of phospho-STAT1 upon IFN-β induction in CHIKrep-pac2AEGFP (wild type and mutant nsP2-P718S)-transfected Vero cells. Cells were immunostained with an anti-pSTAT1 antibody.
We then investigated for CHIKV whether an analogous mutation of the conserved proline in CHIKV nsP2 at position 718 (Fig. 6B) could also be linked to a reduced ability to block JAK-STAT signaling. A puromycin-selectable CHIKV replicon designated CHIKrep-pac2AEGFP (Fig. 6A) and the same construct with a nsP2 P718S mutation (CHIKrep-pac2AEGFP-nsP2m) were constructed and tested for their abilities to block the JAK-STAT pathway in transient transfection experiments. The replication efficiency in Vero cells of CHIKrep-pac2AEGFPnsP2m (5 to 10 EGFP-expressing cells per well in a 96-well plate) was severely reduced in comparison to that of CHIKrep-pac2AEGFP (∼10% EGFP-expressing cells). In contrast, the replication efficiency in BHK-21J cells of CHIKrep-pac2AEGFP-nsP2m compared to CHIKrep-pac2AEGFP was only slightly reduced (∼10% versus ∼20% EGFP-expressing cells), but with notable differences in the induction of cytopathic effect (CPE). BHK-21J cells transfected with CHIKrep-pac2AEGFP-nsP2m retained normal cell morphology, in contrast to cells transfected with CHIKrep-pac2AEGFP, which lost adherence and showed cell rounding 48 h p.t. (data not shown).
In order to investigate the effect of the CHIKV nsP2 P718S mutation on JAK-STAT signaling, Vero cells transfected with CHIKrep-pac2AEGFP or CHIKrep-pac2AEGFP-nsP2m were induced with IFN-β at 24 h p.t. and were stained with an anti-STAT1 antibody as before. In results similar to those obtained with SINV, the CHIKV replicon expressing nsP2-P718S was indeed unable of blocking IFN-β-induced STAT1 nuclear translocation, in contrast to its parental “wild-type” CHIKV replicon (Fig. 6D). This observation suggests that SINV and CHIKV most likely employ similar mechanisms of blocking the JAK-STAT pathway and that the conserved proline in nsP2 at positions 726 and 718, respectively, is essential for this activity.
DISCUSSION
The IFN response is the first line of defense against invading pathogens, and therefore it is no surprise that many viruses actively suppress this antiviral mechanism to promote virus replication and spread (reviewed by Randall and Goodbourn [26]). In this research, we have shown that once established, CHIKV replication is largely resistant to treatment with type I and II IFNs. While IFN-α has been proposed as an antiviral drug to control CHIKV replication (36), our results suggest that IFN may have limited use in antiviral therapy. Recent experiments with mice support this view, showing that IFN-α treatment before, but not after, CHIKV infection inhibits disease and viremia (12). Next, we demonstrated that CHIKV infection and CHIKV replicon RNA replication both efficiently blocked IFN-induced JAK-STAT signaling. This activity was mapped to the nsP2 gene by the expression of nsP2 alone and in the context of an attenuated CHIKV replicon harboring an nsP2 mutation from a conserved proline to a serine at position 718.
nsP2 had earlier been recognized as an important player in modulating the IFN response associated with host shutoff (10). Recently, it has become clear that host shutoff and suppression of the IFN response by alphaviruses can be regarded as separate activities (35). In Old World alphaviruses, nsP2 has been found to be the most important viral protein in modulating the IFN response, with an additional role for the capsid protein in the New World alphaviruses (1, 13). Through the generation of adaptive mutants, nsP2 has been identified as the main viral factor to establish persistent replication in mammalian cells. Noncytopathic variants of SINV and Semliki Forest virus with different mutations in nsP2 display severe defects in counteracting the IFN response (3, 11) and result in high IFN production. This leads to the hypothesis that nsP2 has an essential role in the modulation of the IFN response, likely via interference with downstream JAK-STAT signaling. We show here for the first time that alphavirus nsP2 alone is able to block the JAK-STAT pathway.
Whether or not the other nsPs or their intermediate precursors could possibly contribute to the activity displayed by nsP2 was not further investigated. However, given the potency of the individual protein nsP2 in blocking STAT1 nuclear translocation, any contributory activity by other viral proteins may not be required to establish a productive infection. Selection of Vero or BHK-21J cell lines harboring persistently replicating, attenuated CHIKV replicon RNA was unfortunately not accomplished. It might be possible that for CHIKV replicons, additional mutations in nsP2 or other locations are required to support persistent replication in mammalian cells, as was previously reported for noncytopathic SINV (8).
Previous research has suggested important roles for nsP2 and a host-encoded cellular endoribonuclease, RNase L, in initiating the transition from minus- to plus-strand RNA synthesis (30, 34). Since RNase L is activated by OAS, which itself is an interferon-stimulated gene (ISG), this seems at odds with the inhibitory role of nsP2 on the JAK/STAT pathway. However, the switch from the minus-strand replication complex (RC−) to RC+ occurs at a later stage during infection, and only after cleavage of the nsP2/3 precursor. In CHIKV-infected cells, we have observed inhibition of OAS induction by IFN treatment at later time points (>8 hpi). This correlates with the current view that nsP2 is released in its free form after early replication has been established and creates an environment where host transcription/translation is reduced and the IFN response is actively suppressed.
We have shown by several different experimental approaches that CHIKV replication blocks the JAK-STAT pathway, yet the exact mechanism at the molecular level remains to be elucidated in follow-up experiments. We have ruled out the possibility that the observed blockage of JAK-STAT signaling was due to host shutoff, since signaling in these settings was unaffected in cells treated with cycloheximide. We have also ruled out the possibility that CHIKV reduces endogenous STAT1 levels, similar to what was reported for VEEV- and SINV-infected cells (41).
During dengue virus infection, STAT1 nuclear translocation is inhibited by dengue virus nonstructural protein NS5 as an indirect result of the prevention of STAT2 phosphorylation and STAT1-STAT2 heterodimer formation (2, 23). Consequently, dengue virus is not capable of inhibiting IFN-γ-induced STAT1 phosphorylation/homodimer formation. In contrast to dengue virus, however, incubation with IFN-γ of cells infected with CHIKV or transfected with a CHIKV replicon demonstrates that STAT1 activation is blocked (Fig. 3C and 4C), suggesting that the inhibitory mechanism is different in the case of CHIKV.
The increased STAT1 levels upon IFN induction in normal but not in CHIKV-infected cells (Fig. 3D) may be the result of signal transduction via the JAK-STAT pathway, as was suggested earlier (22). In this scenario, STAT1 upregulation in CHIKV-infected cells is prevented by active inhibition of JAK-STAT signaling, which is supported by the observed decreased luciferase production from the IFN-responsive plasmids in infected cells (Fig. 2).
We showed that a SINV replicon containing nsP2 with a serine at position 726 was not able to efficiently block phospho-STAT1 nuclear translocation, in contrast to the “wild-type” SINV replicon containing nsP2 with a restored proline at position 726. Others have previously claimed that wild-type SINV infection does not impair the ability to respond to IFN-α, as judged by similar levels of STAT1 phosphorylation in infected and uninfected cells (20). The reason for this apparent discrepancy in results is not clear, but an explanation may be the timing of the experiment or the genetic background of the SINV constructs. In our studies, we induced Vero cells with IFN 24 h after transfection with a pToto1101-derived replicon (28), whereas Lin et al. (20) used a dsTE12Q recombinant Sindbis virus vector (19) and induced Vero cells with IFN 6 h p.i. It would be interesting to map the putative differences between these SINV vectors, within nsP2 or elsewhere in the genome, and to identify the domain(s) or amino acid(s) responsible.
Taken together, the inability of alphaviruses with mutated nsP2 proteins to efficiently block STAT1 nuclear translocation may now provide an explanation for the reported overall increased IFN production by such mutants. In this light, it is noteworthy that in preliminary studies, Ross River virus (RRV), another arthrogenic alphavirus and a close relative of CHIKV, does not appear to antagonize STAT1 activation (6), although this finding awaits confirmation. In future research, it may be interesting to investigate whether this apparent difference between CHIKV and RRV could be due to differences of their respective nsP2 proteins. Mapping the functional domains within CHIKV nsP2 and deciphering the exact mechanism by which nsP2 blocks the JAK-STAT pathway, possibly by preventing STAT1 phosphorylation and/or prohibiting the nuclear import of phosphorylated STAT1, will be the focus of future studies in our laboratories. Our results may also provide insights into the development of live-attenuated vaccines to control CHIKV and other alphavirus infections.
Acknowledgments
We thank the VIDRL (Australia) for supplying the chikungunya virus isolate. We acknowledge Klaske Schippers and Dirk Martens (Department of Process Engineering, Wageningen University) for use of the fluorescence microscope. We thank Julia Eekels (AMC, Amsterdam, Netherlands) for sending us the pcDNA-DEST40 plasmid, Brigitte Biesinger (University of Erlangen-Nürnberg) for sharing native STAT1 antibody, Peter de Haan (Amarna Therapeutics B.V., Leiden, Netherlands) for the SINrepGFP construct, and Peter Bredenbeek (LUMC, Leiden, Netherlands) for BHK-21J cells. We thank Konstantin Tsetsarkin and Stephen Higgs for their roles in the construction of CHIKrep-EGFP. Joël van Mierlo, Teije Kleikamp, Jason Leung, and Jan Vermond were involved in the construction of SINrepGFP-wt, CHIKrep-FlucEGFP, CHIKrep-pac2AEGFP, and CHIKrep-pac2AEGFP-nsP2m, respectively.
Footnotes
Published ahead of print on 4 August 2010.
Dedicated to Rob W. Goldbach (1949-2009).
REFERENCES
- 1.Aguilar, P. V., S. C. Weaver, and C. F. Basler. 2007. Capsid protein of Eastern equine encephalitis virus inhibits host cell gene expression. J. Virol. 81:3866-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashour, J., M. Laurent-Rolle, P. Y. Shi, and A. Garcia-Sastre. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breakwell, L., P. Dosenovic, G. B. Karlsson Hedestam, M. D'Amato, P. Liljestrom, J. Fazakerley, and G. M. McInerney. 2007. Semliki Forest virus nonstructural protein 2 is involved in suppression of the type I interferon response. J. Virol. 81:8677-8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke, C. W., C. L. Gardner, J. J. Steffan, K. D. Ryman, and W. B. Klimstra. 2009. Characteristics of alpha/beta interferon induction after infection of murine fibroblasts with wild-type and mutant alphaviruses. Virology 395:121-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couderc, T., F. Chretien, C. Schilte, O. Disson, M. Brigitte, F. Guivel-Benhassine, Y. Touret, G. Barau, N. Cayet, I. Schuffenecker, P. Despres, F. Arenzana-Seisdedos, A. Michault, M. L. Albert, and M. Lecuit. 2008. A mouse model for Chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog. 4:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruz, C. C., M. S. Suthar, S. A. Montgomery, R. Shabman, J. Simmons, R. E. Johnston, T. E. Morrison, and M. T. Heise. 2010. Modulation of type I IFN induction by a virulence determinant within the alphavirus nsP1 protein. Virology 399:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desmyter, J., J. L. Melnick, and W. E. Rawls. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 2:955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dryga, S. A., O. A. Dryga, and S. Schlesinger. 1997. Identification of mutations in a Sindbis virus variant able to establish persistent infection in BHK cells: the importance of a mutation in the nsP2 gene. Virology 228:74-83. [DOI] [PubMed] [Google Scholar]
- 9.Enserink, M. 2007. Epidemiology. Tropical disease follows mosquitoes to Europe. Science 317:1485. [DOI] [PubMed] [Google Scholar]
- 10.Frolov, I. 2004. Persistent infection and suppression of host response by alphaviruses. Arch. Virol. Suppl. 2004:139-147. [DOI] [PubMed] [Google Scholar]
- 11.Frolova, E. I., R. Z. Fayzulin, S. H. Cook, D. E. Griffin, C. M. Rice, and I. Frolov. 2002. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner, J., I. Anraku, T. T. Le, T. Larcher, L. Major, P. Roques, W. A. Schroder, S. Higgs, and A. Suhrbier. 2010. Chikungunya virus arthritis in adult wild-type mice. J. Virol. 84:8021-8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garmashova, N., R. Gorchakov, E. Volkova, S. Paessler, E. Frolova, and I. Frolov. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J. Virol. 81:2472-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinemann, S., B. Biesinger, B. Fleckenstein, and J. C. Albrecht. 2006. NF-κB signaling is induced by the oncoprotein Tio through direct interaction with TRAF6. J. Biol. Chem. 281:8565-8572. [DOI] [PubMed] [Google Scholar]
- 15.King, P., and S. Goodbourn. 1994. The beta-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 269:30609-30615. [PubMed] [Google Scholar]
- 16.Kuhn, R. J. 2007. Togaviridae: the viruses and their replication, p. 1001-1022. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 17.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam, S. K., K. B. Chua, P. S. Hooi, M. A. Rahimah, S. Kumari, M. Tharmaratnam, S. K. Chuah, D. W. Smith, and I. A. Sampson. 2001. Chikungunya infection—an emerging disease in Malaysia. Southeast Asian J. Trop. Med. Public Health 32:447-451. [PubMed] [Google Scholar]
- 19.Levine, B., J. E. Goldman, H. H. Jiang, D. E. Griffin, and J. M. Hardwick. 1996. Bc1-2 protects mice against fatal alphavirus encephalitis. Proc. Natl. Acad. Sci. U. S. A. 93:4810-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, R. J., B. L. Chang, H. P. Yu, C. L. Liao, and Y. L. Lin. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 80:5908-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, W. J., P. L. Sedlak, N. Kondratieva, and A. A. Khromykh. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J. Virol. 76:10766-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, W. J., X. J. Wang, V. V. Mokhonov, P. Y. Shi, R. Randall, and A. A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzon, M., M. Jones, A. Davidson, B. Chain, and M. Jacobs. 2009. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J. Infect. Dis. 200:1261-1270. [DOI] [PubMed] [Google Scholar]
- 24.Mogal, A., and S. A. Abdulkadir. 2006. Effects of Histone Deacetylase Inhibitor (HDACi); Trichostatin-A (TSA) on the expression of housekeeping genes. Mol. Cell. Probes 20:81-86. [DOI] [PubMed] [Google Scholar]
- 25.Powers, A. M., and C. H. Logue. 2007. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J. Gen. Virol. 88:2363-2377. [DOI] [PubMed] [Google Scholar]
- 26.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1-47. [DOI] [PubMed] [Google Scholar]
- 27.Rezza, G., L. Nicoletti, R. Angelini, R. Romi, A. C. Finarelli, M. Panning, P. Cordioli, C. Fortuna, S. Boros, F. Magurano, G. Silvi, P. Angelini, M. Dottori, M. G. Ciufolini, G. C. Majori, and A. Cassone. 2007. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370:1840-1846. [DOI] [PubMed] [Google Scholar]
- 28.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson, M. C. 1955. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952-53. I. Clinical features. Trans. R. Soc. Trop. Med. Hyg. 49:28-32. [DOI] [PubMed] [Google Scholar]
- 30.Sawicki, D. L., R. H. Silverman, B. R. Williams, and S. G. Sawicki. 2003. Alphavirus minus-strand synthesis and persistence in mouse embryo fibroblasts derived from mice lacking RNase L and protein kinase R. J. Virol. 77:1801-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scagnolari, C., S. Trombetti, A. Alberelli, S. Cicetti, D. Bellarosa, R. Longo, A. Spano, E. Riva, M. Clementi, and G. Antonelli. 2007. The synergistic interaction of interferon types I and II leads to marked reduction in severe acute respiratory syndrome-associated coronavirus replication and increase in the expression of mRNAs for interferon-induced proteins. Intervirology 50:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schilte, C., T. Couderc, F. Chretien, M. Sourisseau, N. Gangneux, F. Guivel-Benhassine, A. Kraxner, J. Tschopp, S. Higgs, A. Michault, F. Arenzana-Seisdedos, M. Colonna, L. Peduto, O. Schwartz, M. Lecuit, and M. L. Albert. 2010. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J. Exp. Med. 207:429-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 34.Silverman, R. H. 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 81:12720-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simmons, J. D., L. J. White, T. E. Morrison, S. A. Montgomery, A. C. Whitmore, R. E. Johnston, and M. T. Heise. 2009. Venezuelan equine encephalitis virus disrupts STAT1 signaling by distinct mechanisms independent of host shutoff. J. Virol. 83:10571-10581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock, I. 2009. Chikungunya fever—expanded distribution of a re-emerging tropical infectious disease. Med. Monatsschr. Pharm. 32:17-26. [PubMed] [Google Scholar]
- 37.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Townson, H., and M. B. Nathan. 2008. Resurgence of chikungunya. Trans. R. Soc. Trop. Med. Hyg. 102:308-309. [DOI] [PubMed] [Google Scholar]
- 39.Tsetsarkin, K. A., D. L. Vanlandingham, C. E. McGee, and S. Higgs. 2007. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 3:e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanlandingham, D. L., K. Tsetsarkin, C. Hong, K. Klingler, K. L. McElroy, M. J. Lehane, and S. Higgs. 2005. Development and characterization of a double subgenomic chikungunya virus infectious clone to express heterologous genes in Aedes aegypti mosquitoes. Insect Biochem. Mol. Biol. 35:1162-1170. [DOI] [PubMed] [Google Scholar]
- 41.Yin, J., C. L. Gardner, C. W. Burke, K. D. Ryman, and W. B. Klimstra. 2009. Similarities and differences in antagonism of neuron alpha/beta interferon responses by Venezuelan equine encephalitis and Sindbis alphaviruses. J. Virol. 83:10036-10047. [DOI] [PMC free article] [PubMed] [Google Scholar]