Abstract
2G12 is a broadly neutralizing anti-HIV-1 monoclonal human IgG1 antibody reactive with a high-mannose glycan cluster on the surface of glycoprotein gp120. A key feature of this very highly mutated antibody is domain exchange of the heavy-chain variable region (VH) with the VH of the adjacent Fab of the same immunoglobulin, which assembles a multivalent binding interface composed of two primary binding sites in close proximity. A non-germ line-encoded proline in the elbow between VH and CH1 and an extensive network of hydrophobic interactions in the VH/VH′ interface have been proposed to be crucial for domain exchange. To investigate the origins of domain exchange, a germ line version of 2G12 that behaves as a conventional antibody was engineered. Substitution of 5 to 7 residues for those of the wild type produced a significant fraction of domain-exchanged molecules, with no evidence of equilibrium between domain-exchanged and conventional forms. Two substitutions not previously implicated, AH14 and EH75, are the most crucial for domain exchange, together with IH19 at the VH/VH′ interface and PH113 in the elbow region. Structural modeling gave clues as to why these residues are essential for domain exchange. The demonstration that domain exchange can be initiated by a small number of substitutions in a germ line antibody suggests that the evolution of a domain-exchanged antibody response in vivo may be more readily achieved than considered to date.
Protein oligomers are able to exchange or swap an element of their secondary structure or an entire protein domain. The functional unit in domain-exchanged proteins thereby stays preserved, as only the linking hinge loop changes conformation significantly (4, 17, 27). Analogous to other domain-swapped proteins, antibodies can exchange an entire domain, in this case the heavy-chain variable region (VH), with an equivalent heavy-chain variable region of an adjacent Fab (VH′) within the same immunoglobulin (Ig) molecule (11). The advantages of domain-exchanged proteins, including antibodies, are higher local concentrations of active sites, a larger binding surface, and a potential secondary active site at the new subunit interface (27, 45). The one and only antibody shown to be domain exchanged to date is 2G12 (7, 11), but this arrangement is potentially possible for any Ig and could have been overlooked at least in some instances.
2G12 is one of only a few high-affinity monoclonal antibodies with broad neutralizing activity against different subtypes of HIV-1 (5, 30, 40, 43). The antibody binds a dense cluster of N-linked high-mannose glycans (Man8-9GlcNAc2) on the envelope surface glycoprotein gp120 (10, 35, 36, 41). The domain-exchanged arrangement forms a multivalent binding site composed of two primary binding sites in close proximity and a proposed secondary binding site formed by the novel VH/VH′ interface (11). 2G12 provides protection against infection in animal models (19, 31) and has been shown to induce neutralization escape following passive immunization in humans (39).
Consensus has grown that a successful HIV-1 vaccine will need to include a component that elicits broadly neutralizing antibodies (8, 18, 21, 26, 32, 42). All attempts to elicit 2G12-like antibodies with the desired specificity and neutralization activity have failed to date (22, 29, 44), conceivably due to difficulties in generating adequate mimicry of the glycan cluster and tolerance mechanisms or, very likely, the inability to induce domain exchange (1). Unraveling the mechanism of domain exchange and how this conformation might have evolved is highly desirable to direct future HIV-1 vaccine design to elicit 2G12-like antibodies.
By comparison with other domain-exchanged proteins (27), the following three mechanisms have been proposed to contribute to the unique structure of 2G12 compared to the structure of a conventional antibody: destabilization of the “closed” VH/VL interface, conformational change in the elbow between VH and CH1, and an energetically favorable “open” VH/VH′ interface (11). Key residues involved in promoting domain exchange were predicted based on examination of interacting residues at the two interfaces and by the effects of alanine substitutions on the binding of wild-type 2G12 to gp120. However, the importance of these key residues for domain exchange was not directly demonstrated experimentally (11).
Here, we explored the minimal requirements for domain exchange of 2G12, starting with a germ line version of the antibody that adopts a conventional antibody structure. Although wild-type 2G12 is heavily somatically mutated, only five to seven substitutions in the germ line version of the antibody were shown to produce a significant fraction of domain-exchanged molecules. The results suggest the evolution of domain-exchanged antibody responses may be more facile than considered to date.
MATERIALS AND METHODS
Germ line 2G12 determination.
The germ line 2G12 sequence was derived from the original 2G12 nucleotide sequence (provided by Hermann Katinger, Vienna, Austria) using the sequence alignment tool IMGT/V-QUEST (6). Amino acids derived from N nucleotides were inserted into the germ line sequence unchanged. The Kabat numbering scheme (23) is used throughout this paper.
Antibody expression and purification.
Variable regions were cloned and assembled from human genomic DNA (germ line 2G12 light chain) or synthesized de novo (germ line 2G12 heavy chain; GenScript, Piscataway, NJ). Amino acid substitutions were introduced by QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA). All constructs were verified by sequence analysis (Eton Bioscience, San Diego, CA). Antibody genes were cloned into full-length IgG1 expression vectors pDR12 (3, 9) or pγ1HC and pκLC (38) and transiently expressed with the FreeStyle 293 expression system (Invitrogen, Carlsbad, CA). Antibodies were purified using affinity chromatography (protein A Sepharose Fast Flow; GE Healthcare, United Kingdom), and purity and integrity were checked with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Assessment of domain exchange.
Antibodies at a concentration of 4 mg/ml were digested with 160 μg/ml papain (Sigma, St. Louis, MO) for 4 h at 37°C, and Fabs were purified using protein A affinity chromatography. Domain exchange was assessed by size exclusion chromatography (SEC) of 100 μg purified Fab in phosphate-buffered saline (PBS) on a Superdex 200 10/300 column (GE Healthcare) at a flow rate of 0.5 ml/min (ÄKTA FPLC; GE Healthcare). The molecular weights of the Fab monomer and dimer were checked using a gel filtration standard (Bio-Rad, Hercules, CA). To determine the percentage of domain exchange, the area under the peaks was integrated using Unicorn 5.11 software (GE Healthcare). Duplicate batches of selected antibodies were independently expressed and digested and showed similar percentages of domain exchange.
Enzyme-linked immunosorbent assays.
Binding to gp120 and M1G1 was assessed by coating high-binding microtiter plates (Corning Life Sciences, Lowell, MA) with 5 μg/ml gp120 JR-FL (Progenics, Tarrytown, NY) or 2.5 μg/ml M1G1, respectively, overnight at 4°C. For all enzyme-linked immunosorbent assays (ELISAs), plates were blocked with 3% bovine serum albumin (BSA) and washed after each step five times with PBS-0.05% Tween. Serial dilutions of antibodies in PBS were incubated for 2 h at room temperature. Binding was detected with an anti-human Fab-alkaline phosphatase (AP) conjugate (1:1,000 or 1.3 μg/ml; Jackson ImmunoResearch, West Grove, PA) and phosphatase substrate (Sigma). Absorption was read at 405 nm. Mean and standard deviation values of duplicate measurements are shown.
Molecular modeling.
Substituted side chains were modeled in their most favorable rotamers while avoiding steric clashes where possible with surrounding side chains, using COOT (14). Molecular graphics images were generated using the PyMOL molecular graphics system (2002; DeLano Scientific, Palo Alto, CA).
RESULTS
Construction of a germ line version of 2G12.
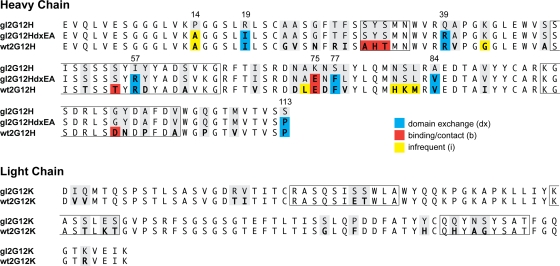
The original nucleotide sequence of 2G12 (provided by Hermann Katinger) was analyzed by the sequence alignment tool IMGT/V-QUEST (6). The best matches for putative germ line sequences resulted in IGHV3-21, IGHD6-25, and IGHJ3 for the heavy chain and IGKV1-5 and IGKJ1 for the light chain (Fig. 1). In this alignment, the D segment is heavily truncated, and 18 N nucleotides have been added between the V and D segments, leading to an HCDR3 length of 14. N nucleotide-encoded amino acids were incorporated unchanged into our germ line version. The IGHV3-21 gene is used in 2.7% of all rearranged sequences in the IMGT database (median usage of germ line IGHV genes is 1.8%), IGHJ3 in 15.9% (median usage of germ line IGHJ genes is 13.8%), IGKV1-5 in 7.5% (median usage of germ line IGKV genes is 1.2%), and IGKJ1 in 30.2% (median usage of germ line IGKJ genes is 24.3%). We would therefore consider the usage of the germ line 2G12 genes to be unremarkable.
FIG. 1.
Wild-type 2G12 is heavily somatically mutated. The germ line 2G12 amino acid sequence was derived from the original nucleotide sequence using IMGT/V-QUEST (6). The wild-type 2G12 heavy chain (wt2G12H) has 38 somatic mutations, and the wild-type 2G12 light chain has 16 (wt2G12K). Residues differing from those in germ line 2G12 (gl2G12) are in boldface. Residues proposed to promote domain exchange (dx) are shaded in blue, binding/contact residues (b) are shaded in red, and infrequent residues (i) are shaded yellow (see the text). CDRs according to the Kabat definition are boxed.
The putative germ line sequence used in our study differs from a previously published alignment, where a nonconventional D segment (HSIGMMDL 67829) without the addition of N nucleotides was used (25). In another study of germ line 2G12, no D segment was assigned, and HCDR3 was left in the mutated form (46). Compared to the latter study, our germ line version has two additional residues in HCDR3 (DH100b and NH100c) and one at the D/J junction (PH100e) that were mutated back to those in the germ line. 2G12 shows extreme somatic mutation. In the heavy chain, 38/123 amino acids are mutated, and in the light chain, 16/109 amino acids are mutated (corresponding to 71/369 and 43/321 at the nucleotide level, respectively). This makes 2G12 one of the most mutated antibodies described to date (20, 37, 46).
Germ line 2G12 is not domain exchanged and does not detectably bind gp120.
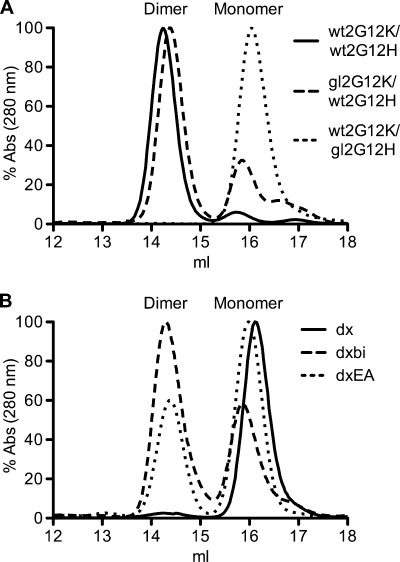
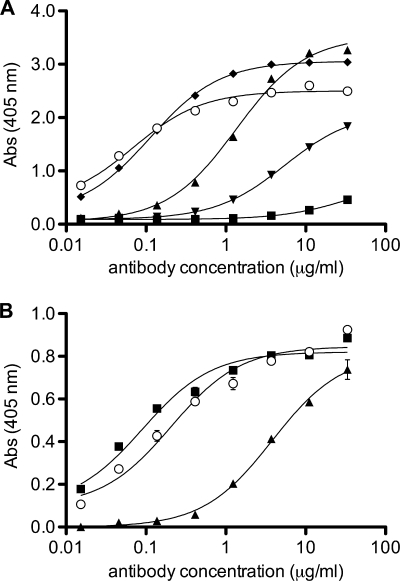
Antibodies were expressed as IgGs in FS293 cells and purified using affinity chromatography. All antibodies were expressed well and folded correctly into ∼150-kDa IgGs, as assessed by SDS-PAGE. Domain exchange was assessed by size exclusion chromatography (SEC) of Fabs prepared by papain digest, and chromatograms showed distinct monomer and dimer peaks (Fig. 2). Wild-type Fab 2G12 (wt2G12K/wt2G12H) is domain exchanged and runs mostly as a dimer (>95%), while 2G12 with both VH and VL reverted to those of the germ line (gl2G12K/gl2G12H) is not domain exchanged and runs as a monomer (Fig. 2A). Additionally, gl2G12K/gl2G12H showed no detectable binding to gp120 in extensive binding studies, including virus capture assays, glycan arrays, and binding to yeast (data not shown) (46). If only the light chain was reverted to that of the germ line, the antibody retained binding to gp120 and was still largely domain exchanged (80%) (Fig. 2A). However, if the heavy chain was reverted, then the antibody did not bind to gp120 and did not exchange domains. We, therefore, conclude that the contribution of the light chain to domain exchange is small, and we retained the wild-type 2G12 light chain for the following studies.
FIG. 2.
A germ line 2G12 variant with 8 wild-type substitutions (gl2G12dxEA) is notably domain exchanged. Domain exchange was assessed by size exclusion chromatography of Fabs prepared by papain digest, and chromatograms show distinct monomer and dimer peaks. (A) Wild-type 2G12 (solid line) and the gl2G12K/wt2G12H chimeric antibody (dashed line) run predominantly as domain-exchanged dimers; wt2G12K/gl2G12H (dotted line) and germ line 2G12 (not shown) are not domain exchanged. (B) Gl2G12dx (solid line) is not domain exchanged, while both gl2G12dxbi (dashed line) and gl2G12dxEA (dotted line) are domain exchanged by 55% and 37%, respectively. Abs, absorbance.
Significant domain exchange can be induced by substituting only five to seven residues in germ line 2G12.
We attempted to promote domain exchange beginning with germ line 2G12 by introducing increasing numbers of substitutions corresponding to the wild-type sequence by site-directed mutagenesis. First, we focused on the six residues that were proposed to be crucial for domain exchange based on the 2G12 crystal structure (IH19, RH39, RH57, FH77, VH84, and PH113; gl2G12dx) (Fig. 1 and 2B) (11). Second, as we did not know if domain exchange and binding are obligatorily linked, we added a further seven additional wild-type residues that were found by alanine scanning to be essential for binding (AH31, HH32, TH33, TH55, EH75, DH100B, and NH100C; gl2G12dxb) (11). Third, we added a further six residues that are infrequently observed at otherwise conserved sites (HH82a, KH82b, and MH82c), are positioned in conserved turns in close proximity to the elbow region or the VH/VL interface (AH14 and GH43), or could further stabilize the VH/VH′ interface (EH75; gl2G12dxbi, 19 substitutions). All these mutated heavy chains were expressed with the wild-type 2G12 light chain, as we found that the contribution of the light chain to domain exchange is small, and the light chain does not contribute directly to the newly formed “open” VH/VH′ interface and the hinge region. SEC analysis of purified Fabs revealed that gl2G12dx and gl2G12dxb are not domain exchanged, while gl2G12dxbi is more than 50% domain exchanged (Fig. 2B).
Starting with these results, extensive site-directed mutagenesis was performed. By expressing more than 60 variants with different numbers and combinations of the above-mentioned residues, we were able to narrow down the number of residues that are essential for domain exchange (see Fig. 7; see also Table S1 in supplemental material elsewhere [http://www.scripps.edu/ims/burton/supplemental/Huber_JVI_2010.pdf]). We found that variant gl2G12dxEA, which contains all six residues previously proposed to be important for domain exchange as well as AH14 and EH75 is still considerably domain exchanged (37%) (Fig. 2B).
IH19, AH14, PH113, and EH75 are the most important residues for domain exchange.
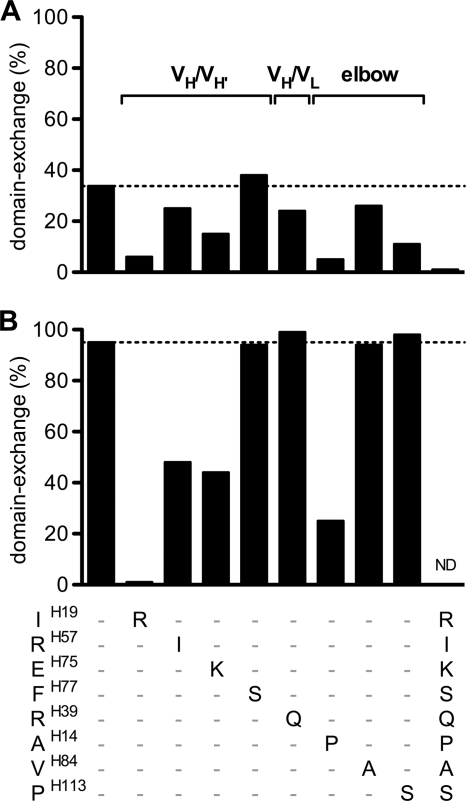
To determine individual residue contributions to domain exchange, the eight wild-type residues identified above were reverted back independently to the germ line residues in the background of the gl2G12dxEA or wt2G12 heavy-chain sequence. In the dxEA background, AH14P, IH19R, PH113S, and EH75K reduced domain exchange the most, while the reversion FH77S had no effect on domain exchange at all (Fig. 3 A). Taking a wt2G12 background and mutating the same eight residues back to germ line showed that the IH19R, AH14P, RH57I, and EH75K substitutions lead to a significant reduction of domain exchange (Fig. 3B). The single point mutation IH19R completely reverted the antibody back to its conventional form, which was also shown by the determination of its crystal structure (13). Overall, the effects on domain exchange of substitutions in the context of dxEA or wild-type antibodies were well correlated. One exception was the PH113S substitution, which had no effect on domain exchange in the context of wild-type 2G12 but reduced domain exchange in the dxEA variant.
FIG. 3.
Residues IH19, AH14, PH113, and EH75 are most important for domain exchange in 2G12. (A) The eight wild-type residues identified to be most important for domain exchange were reverted back individually to the germ line residues, starting with germ line 2G12dxEA (first bar; 37% domain exchange). Substitutions IH19R, AH14P, PH113S, and EH75K most reduced domain exchange. The last bar shows complete reversion to germ line 2G12. (B) Same reversion to germ line residues as described in the legend to panel A, but starting with wild-type 2G12 (first bar; 95% domain exchange). In the wild-type setting, the IH19R, AH14P, RH57I, and EH75K substitutions lead to a significant reduction of domain exchange, and IH19R completely disrupted domain exchange. ND, not determined.
Structural modeling of PH14A and KH75E substitutions.
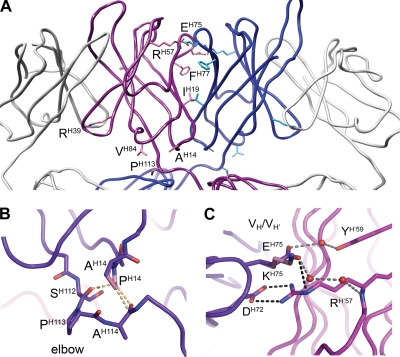
The possible contributions to domain exchange of six residues have previously been discussed (11, 16). Briefly, PH113 in the elbow region is proposed to facilitate domain exchange and stabilize the structure by hydrophobic interaction with VH84; the three residues IH19, RH57, and FH77 are proposed to contribute to the favorable VH/VH′ interface through hydrophilic interactions (RH57) and an extensive hydrophobic patch (IH19 and FH77); and RH39 is proposed to weaken the conventional VH/VL interface (Fig. 4 A). For the two residues newly assigned as important for domain exchange, AH14 and EH75, molecular modeling analyses were performed using the high-resolution structure of wild-type 2G12 (Protein Data Bank accession no. 1OP3) (11) as the template in COOT (14). A proline at position H14 (as found in germ line 2G12) was found to introduce steric clashes with residues that help form the elbow region in the domain-exchanged structure (AH114 and SH112) (Fig. 4B), suggesting that this residue would not be tolerated at this position in domain-exchanged 2G12. A lysine substitution at position H75 (as found in germ line 2G12) was found to remove a charged interaction (between EH75 and RH′57) and water-mediated hydrogen bonds (from EH75 to RH′57 and YH′59) at the VH/VH′ interface (Fig. 4C). A glutamate at position H75 (as found in wild-type 2G12) is thus likely to significantly stabilize the VH/VH′ interface.
FIG. 4.
Structural factors that promote domain exchange in 2G12 are located in the VH/VH′ interface and the elbow region. (A) The variable domains of Fab 2G12, with the domain-exchanged heavy chains shown in blue and purple and the light chains shown in gray (produced with coordinates from Protein Data Bank accession no. 1OP3) (11). Residues important for domain exchange are shown in stick format and are located in the VH/VH′ interface (IH19, RH57, and EH75), the elbow region (AH14, VH84, and PH113), and the VH/VL interface (RH39). (B) Modeling of proline at position H14 (as found in germ line 2G12) introduces steric clashes (steric clashes are represented as dashed orange lines). (C) Modeling of a lysine substitution at position H75 (as found in germ line 2G12) leads to the loss of a charged interaction (between EH75 and RH′57; charged interactions are shown as dashed black lines) as well as water-mediated hydrogen bonds (between EH75, RH′57, and YH′59; hydrogen bonds are shown as dashed gray lines, and waters are shown as spheres) at the VH/VH′ interface. DH72 forms two electrostatic interactions with mutated residue RH′57 in domain-exchanged wild-type 2G12.
Anti-2G12 idiotypic antibody M1G1 can be used to assess domain exchange.
In order to assess domain exchange by other means than SEC, we tested the anti-2G12 idiotypic antibody M1G1 (34) for binding to different versions of 2G12. It was speculated that M1G1 binds to the elbow region of the light chain and that the epitope contains a pronounced conformational component (34). ELISA data showed that M1G1 bound well to the domain-exchanged wt2G12 but not to a conventional antibody, as represented by wt2G12K/gl2G12H, and with intermediate affinity to different 2G12 variants depending on the percentage of domain exchange (Fig. 5 A). Purified Fab monomers and dimers were also bound differentially (Fig. 5B). Anti-2G12 idiotypic antibody M1G1 was thus found to be domain exchange sensitive and could be used to predict the domain exchange of 2G12 variants.
FIG. 5.
Anti-2G12 idiotypic antibody M1G1 can be used to assess domain exchange. (A) Binding of IgG variants of 2G12 to anti-2G12 idiotypic antibody M1G1 is dependent on the conformation of 2G12. The following heavy chains were all expressed with the wild-type 2G12 light chain: wt2G12H (open dots), gl2G12H (squares), gl2G12Hdx (upright triangles), gl2G12Hdxb (inverted triangles), and gl2G12Hdxbi (diamonds). (B) Fab wt2G12dxEA (open dots; 37% domain exchanged) and corresponding purified dimeric (squares) and monomeric (triangles) Fabs.
No evidence for equilibrium between domain-exchanged and conventional forms of IgG.
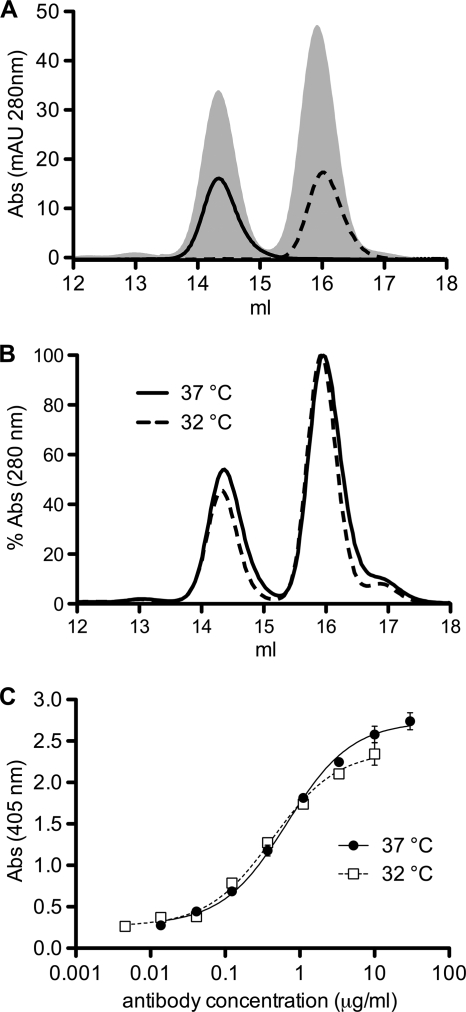
To test if there is an equilibrium between the domain-exchanged and the nondomain-exchanged forms of 2G12 in solution, purified gl2G12dxEA Fab monomers and dimers were incubated separately for approximately 2 weeks at 4°C and 3 h at 37°C or for 10 min at up to 60°C. No evidence of a change in the proportions of domain-exchanged or conventional antibodies in any preparation was detected, as assessed by SEC or binding to the domain exchange-sensitive anti-2G12 antibody M1G1 (Fig. 6 A and data not shown). To test if the percentage of domain exchange is already determined during folding and assembly of the IgG inside the producer cells (15, 24), we expressed gl2G12dxEA at both 32 and 37°C to vary the rate of protein synthesis. Although protein expression was much less at the lower temperature, there was no difference in the percentages of domain exchange of the purified Fabs, as measured by SEC (Fig. 6B). As the whole purification and papain digestion process could impact the amount of domain-exchanged protein, we also tested the binding of the crude supernatant to M1G1 (Fig. 6C). There was no difference in binding, showing that the domain-exchanged antibody fractions remain stable throughout the entire purification process.
FIG. 6.
No evidence for an equilibrium between domain-exchanged and conventional forms of germ line 2G12dxEA. (A) Purified Fabs were analyzed and separated in monomer and dimer configurations with SEC (shaded area). After storage at 4°C for approximately 2 weeks and incubation for 3 h at 37°C, the distribution of dimers (solid line) and monomers (dashed line) remained unchanged. (B) Expression of germ line 2G12HdxEA at 37°C (solid line) or 32°C (dashed line) does not change the percentage of domain exchange of purified Fabs. (C) Expression of germ line 2G12HdxEA at 37°C (solid dots) or 32°C (open squares) does not change the M1G1 binding of crude supernatant.
Domain exchange could have evolved with only a few somatic mutations.
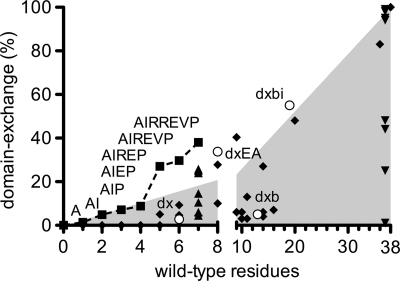
To explore the question of how domain exchange might have evolved in vivo, we made a series of additive substitutions in germ line 2G12 in the order of decreasing importance to domain exchange, as estimated from Fig. 3. Domain-exchanged molecules are detected, even with very few substitutions, and their fractions gradually increase with every additional substitution (Fig. 7). By adding the fifth substitution (to give AH14IH19RH39EH75PH113), the percentage of domain-exchanged gl2G12 rises substantially to 27% (Fig. 7). Although this proposed path of the evolution of the domain exchange is purely experimental and not necessarily how it arose in vivo, it shows that a domain-exchanged antibody could be achieved with a reasonably small number of mutations.
FIG. 7.
Domain exchange could have evolved with only a few somatic mutations. Extensive site-directed mutagenesis was performed, and each substitution was analyzed for domain exchange. The percentage of domain exchange was graphed versus the number of germ line residues of the 2G12 heavy chain that were changed into wild-type ones (i.e., 0 equals germ line, and 38 equals wild-type including all somatic hypermutations). About five residues are necessary to get a fraction of domain exchange significantly higher than a linear trend (shaded area). The variants gl2G12dx, gl2G12dxb, gl2G12dxbi, and gl2G12dxEA are depicted as open circles. The panels of individual germ line reversion in the gl2G12dxEA (Fig. 3A) and wt2G12 (Fig. 3B) backgrounds are shown with upright and inverted triangles, respectively. A potential pathway for evolution into a domain-exchanged antibody is shown (squares). Diamonds show other ineffective and unrelated variants that were tested (see Table S1 in supplemental material elsewhere [http://www.scripps.edu/ims/burton/supplemental/Huber_JVI_2010.pdf]).
DISCUSSION
Although the unique, domain-exchanged antibody 2G12 is heavily somatically mutated, we found that only five to seven crucial amino acid mutations in the VH-VH′ interface (IH19, RH57, and EH75), the VH-VL interface (RH39), and the elbow region (AH14, VH84, and PH113) are necessary to effect considerable domain exchange when starting with a germ line version of 2G12. The most important residues are IH19 and AH14, located at the VH/VH′ interface and in close proximity to the elbow region, respectively, yet the contributions to overall domain exchange from individual residues are small and lead only to incremental increases in the percentage of domain exchange (Fig. 7).
Key residues proposed earlier to promote domain exchange are not sufficient by themselves (11, 16), as we have shown that two additional residues (AH14 and EH75) (Fig. 3A) contribute significantly. Proline is the most frequent residue (95% [n = 768] of all paired human antibody sequences in the 2010 Abysis database [www.abysis.org]) at position H14 (Fig. 3B) and participates in a highly conserved β turn. Structural modeling, however, suggests that substitution by alanine (as in wild-type 2G12) might be necessary for domain exchange to avoid steric clashes. Moreover, H19 is usually a large basic residue (arginine or lysine; together, 74%), with isoleucine (as in wild-type 2G12) being very infrequent (1%) at this site. Apart from 2G12, only five other antibodies in the Abysis database do not have a proline at position H14 or a large basic residue at H19. The variation at position H75 is much greater, with 4% of heavy chains having a glutamate at this site. On the other hand, one of the residues proposed earlier to be essential for domain exchange, FH77, was found here not to be critical (Fig. 3A). This is also supported by the fact that wild-type 2G12 FH77A still binds to gp120 (data not shown), contrary to data from crude Fabs published previously (11). The amount of domain-exchanged antibody could differ if antibodies are expressed as Fabs or IgGs. Moreover, some of the essential residues might have been difficult to uncover in earlier studies (11, 16), as domain exchange was assessed only from structural interactions and changes in the binding affinity of variants in a wild-type background and not a germ line 2G12 background.
Introduction of domain exchange into a conventional germ line antibody and disruption of domain exchange in wild-type 2G12 are two different processes. In the germ line background, the loss of a single important residue can have a profound effect on domain exchange (Fig. 3A), whereas substitution of a single residue in wild-type 2G12 could be compensated by the presence of several other domain exchange-supporting residues (Fig. 3B), and the importance of individual residues in the two different backgrounds might, therefore, not be obligatorily linked. We found though that IH19 and AH14 are the most important residues in both backgrounds and that the contribution to domain exchange is overall well correlated, with the exception of the elbow proline PH113 (Fig. 3A and B). Furthermore, residues that are not somatically mutated and substitutions to residues differing from the wild type have not been studied so far. Also, while it may be that a domain-exchanged conformation can be achieved for a number of antibodies, i.e., with a variety of combinations of heavy- and light-chain germ line genes, different residues might be responsible for the key interactions that drive domain exchange in different antibodies.
Once folded, both conventional and domain-exchanged forms of 2G12 seem to be equally stable. While, in other domain-exchanged molecules, an equilibrium is often observed between monomeric and dimeric forms, e.g., RNase A (27, 28, 33), domain exchange in 2G12 appears to have a high kinetic barrier, as we found no evidence for equilibrium between domain-exchanged and conventional forms nor any effect on the ratio of domain exchange by expressing proteins at different temperatures (Fig. 6). At what time point domain exchange occurs, whether during or after folding and assembly of whole Ig (15, 24), needs to be further investigated. All of the antibodies used in this study were expressed as IgGs, as expression of Fabs might bias the percentage of domain exchange.
Assessment of the binding of germ line 2G12 variants to gp120 was not carried out here, as the requirements for binding exceed those for domain exchange and include mutations in the complementarity-determining regions (CDRs). 2G12 domain exchange may have appeared independently of binding, although most likely selection occurred first for binding to mannose epitopes and the molecule subsequently became domain exchanged. This notion is supported by other experiments showing that nondomain-exchanged 2G12 in its conventional form still binds mannose epitopes with essentially unchanged binding interactions (13). Binding to antigen could have evolved initially in the context of a conventional antibody, and domain exchange then could have occurred later without disturbing the conformation of the antigen binding sites.
Our results show that the evolutionary pathway to a domain-exchanged antibody could be surprisingly short, as substitution of only five to seven residues resulted in at least a partially domain-exchanged antibody. Once enough mutations have accumulated to permit significant domain exchange, in vivo, a B cell expressing a domain-exchanged receptor with favorable avidity to highly clustered antigens on a pathogen due to the unique arrangement of primary binding sites would be selected and activated. This initial activation would result in an avidity-driven competition for survival and clonal expansion and, thereby, increase the percentage of domain exchange, comparable to selection for high-affinity clones. Although no binding of germ line 2G12 to gp120 was detectable in our hands, this antibody could still be selected in vivo, as affinities needed for B-cell selection can be very low (in the micromolar range or even undetectable) (2, 12).
Here, we have determined which residues are crucial for domain exchange of 2G12 and have successfully domain exchanged a germ line version of 2G12. The demonstration that domain exchange can be initiated by a small number of substitutions in a germ line antibody may have important consequences for the evolution of a domain-exchanged antibody response in vivo. Although 2G12 is the only antibody known so far to be domain exchanged, it would be very surprising if there were not more domain-exchanged antibodies in nature, given the few mutations necessary to induce this configuration and the apparent stability of the arrangement. Further studies will pinpoint the requirements for inducing or increasing domain exchange in other antibodies and may provide clues as to how this exchange may be promoted through immunization. A potential strategy would include a prime immunization with a regularly arrayed immunogen to get specific conventional antibodies, followed by boosts with the same immunogen in small discrete clusters to favor domain exchange.
Acknowledgments
We thank Hermann Katinger for the original 2G12 nucleotide sequence and antibody M1G1 and Rena Astronomo for helpful discussions.
This work was supported by the International AIDS Vaccine Initiative (IAVI) through the Neutralizing Antibody Consortium; the National Institutes of Allergy and Infectious Diseases, NIH (grants AI33292 and U01 AI078224 to D.R.B. and grants GM46192 and AI84817 to I.A.W.); the Ragon Institute of MGH, MIT, and Harvard (support given to D.R.B., M.H., and K.J.D.); the Swiss National Science Foundation (grant PBEZB-119694 to M.H.); and the Skaggs Institute for Chemical Biology.
Footnotes
Published ahead of print on 11 August 2010.
REFERENCES
- 1.Astronomo, R. D., and D. R. Burton. 2010. Carbohydrate vaccines: developing sweet solutions to sticky situations? Nat. Rev. Drug Discov. 9:308-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batista, F. D., and M. S. Neuberger. 1998. Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity 8:751-759. [DOI] [PubMed] [Google Scholar]
- 3.Bebbington, C. R., G. Renner, S. Thomson, D. King, D. Abrams, and G. T. Yarranton. 1992. High-level expression of a recombinant antibody from myeloma cells using a glutamine synthetase gene as an amplifiable selectable marker. Biotechnology (N. Y.) 10:169-175. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, M. J., M. R. Sawaya, and D. Eisenberg. 2006. Deposition diseases and 3D domain swapping. Structure 14:811-824. [DOI] [PubMed] [Google Scholar]
- 5.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brochet, X., M.-P. Lefranc, and V. Giudicelli. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36:W503-W508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses 10:359-369. [DOI] [PubMed] [Google Scholar]
- 8.Burton, D. R., R. Desrosiers, R. Doms, W. Koff, P. Kwong, J. Moore, G. Nabel, J. Sodroski, I. Wilson, and R. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233-236. [DOI] [PubMed] [Google Scholar]
- 9.Burton, D. R., J. Pyati, R. Koduri, S. Sharp, G. Thornton, P. Parren, L. Sawyer, R. Hendry, N. Dunlop, and P. Nara. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 10.Calarese, D., H. Lee, C. Huang, M. Best, R. D. Astronomo, R. Stanfield, H. Katinger, D. R. Burton, C. Wong, and I. Wilson. 2005. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. U. S. A. 102:13372-13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 12.Dal Porto, J. M., A. M. Haberman, M. J. Shlomchik, and G. Kelsoe. 1998. Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J. Immunol. 161:5373-5381. [PubMed] [Google Scholar]
- 13.Doores, K. J., Z. Fulton, M. Huber, I. A. Wilson, and D. R. Burton. 2010. Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged. J. Virol. 84:10690-10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 15.Feige, M. J., S. Groscurth, M. Marcinowski, Y. Shimizu, H. Kessler, L. M. Hendershot, and J. Buchner. 2009. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol. Cell 34:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gach, J. S., P. G. Furtmüller, H. Quendler, P. Messner, R. Wagner, H. Katinger, and R. Kunert. 2010. Proline is not uniquely capable of providing the pivot point for domain swapping in 2G12, a broadly neutralizing antibody against HIV-1. J. Biol. Chem. 285:1122-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gronenborn, A. M. 2009. Protein acrobatics in pairs—dimerization via domain swapping. Curr. Opin. Struct. Biol. 19:39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haynes, B., and D. Montefiori. 2006. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev. Vaccines 5:347-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hessell, A. J., E. G. Rakasz, P. Poignard, L. Hangartner, G. Landucci, D. N. Forthal, W. C. Koff, D. I. Watkins, and D. R. Burton. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, C.-C., M. Venturi, S. Majeed, M. J. Moore, S. Phogat, M.-Y. Zhang, D. S. Dimitrov, W. A. Hendrickson, J. Robinson, J. Sodroski, R. Wyatt, H. Choe, M. Farzan, and P. D. Kwong. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl. Acad. Sci. U. S. A. 101:2706-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston, M. I., and A. S. Fauci. 2007. An HIV vaccine—evolving concepts. N. Engl. J. Med. 356:2073-2081. [DOI] [PubMed] [Google Scholar]
- 22.Joyce, J. G., I. J. Krauss, H. C. Song, D. W. Opalka, K. M. Grimm, D. D. Nahas, M. T. Esser, R. Hrin, M. Feng, V. Y. Dudkin, M. Chastain, J. W. Shiver, and S. J. Danishefsky. 2008. An oligosaccharide-based HIV-1 2G12 mimotope vaccine induces carbohydrate-specific antibodies that fail to neutralize HIV-1 virions. Proc. Natl. Acad. Sci. U. S. A. 105:15684-15689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabat, E. A., T. T. Wu, H. Bilofsky, M. Reid-Miller, and H. M. Perry. 1983. Sequences of proteins of immunological interest. National Institutes of Health, Bethesda, MD.
- 24.Kaloff, C. R., and I. G. Haas. 1995. Coordination of immunoglobulin chain folding and immunoglobulin chain assembly is essential for the formation of functional IgG. Immunity 2:629-637. [DOI] [PubMed] [Google Scholar]
- 25.Kunert, R., F. Ruker, and H. Katinger. 1998. Molecular characterization of five neutralizing anti-HIV type 1 antibodies: identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res. 14:1115-1128. [DOI] [PubMed] [Google Scholar]
- 26.Letvin, N. L. 2006. Progress and obstacles in the development of an AIDS vaccine. Nat. Rev. Immunol. 6:930-939. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Y., and D. Eisenberg. 2002. 3D domain swapping: as domains continue to swap. Protein Sci. 11:1285-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López-Alonso, J. P., G. Gotte, and D. V. Laurents. 2009. Kinetic analysis provides insight into the mechanism of ribonuclease A oligomer formation. Arch. Biochem. Biophys. 489:41-47. [DOI] [PubMed] [Google Scholar]
- 29.Luallen, R. J., C. Agrawal-Gamse, H. Fu, D. F. Smith, R. W. Doms, and Y. Geng. 2010. Antibodies against Manα1,2-Manα1,2-Man oligosaccharide structures recognize envelope glycoproteins from HIV-1 and SIV strains. Glycobiology 20:280-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mascola, J. R., and D. C. Montefiori. 2010. The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 28:413-444. [DOI] [PubMed] [Google Scholar]
- 31.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 32.McMichael, A. J. 2006. HIV vaccines. Annu. Rev. Immunol. 24:227-255. [DOI] [PubMed] [Google Scholar]
- 33.Rousseau, F., J. W. Schymkowitz, H. R. Wilkinson, and L. S. Itzhaki. 2001. Three-dimensional domain swapping in p13suc1 occurs in the unfolded state and is controlled by conserved proline residues. Proc. Natl. Acad. Sci. U. S. A. 98:5596-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roux, K. H., P. Zhu, M. Seavy, H. Katinger, R. Kunert, and V. Seamon. 2004. Electron microscopic and immunochemical analysis of the broadly neutralizing HIV-1-specific, anti-carbohydrate antibody, 2G12. Mol. Immunol. 41:1001-1011. [DOI] [PubMed] [Google Scholar]
- 35.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1->2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheid, J. F., H. Mouquet, N. Feldhahn, M. S. Seaman, K. Velinzon, J. Pietzsch, R. G. Ott, R. M. Anthony, H. Zebroski, A. Hurley, A. Phogat, B. K. Chakrabarti, Y. Li, M. Connors, F. Pereyra, B. D. Walker, H. Wardemann, D. Ho, R. T. Wyatt, J. R. Mascola, J. V. Ravetch, and M. C. Nussenzweig. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636-640. [DOI] [PubMed] [Google Scholar]
- 38.Tiller, T., E. Meffre, S. Yurasov, M. Tsuiji, M. C. Nussenzweig, and H. Wardemann. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329:112-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trkola, A., H. Kuster, P. Rusert, B. Joos, M. Fischer, C. Leemann, A. Manrique, M. Huber, M. Rehr, A. Oxenius, R. Weber, G. Stiegler, B. Vcelar, H. Katinger, L. Aceto, and H. F. Gunthard. 2005. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat. Med. 11:615-622. [DOI] [PubMed] [Google Scholar]
- 40.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas III, D. R. Burton, D. D. Ho, et al. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Virgin, H. W., and B. D. Walker. 2010. Immunology and the elusive AIDS vaccine. Nature 464:224-231. [DOI] [PubMed] [Google Scholar]
- 43.Walker, L. M., S. K. Phogat, P.-Y. Chan-Hui, D. Wagner, P. Phung, J. L. Goss, T. Wrin, M. D. Simek, S. Fling, J. L. Mitcham, J. K. Lehrman, F. H. Priddy, O. A. Olsen, S. M. Frey, P. W. Hammond, P. G. P. Investigators, S. Kaminsky, T. Zamb, M. Moyle, W. C. Koff, P. Poignard, and D. R. Burton. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, L.-X. 2006. Toward oligosaccharide- and glycopeptide-based HIV vaccines. Curr. Opin. Drug Discov. Devel. 9:194-206. [PubMed] [Google Scholar]
- 45.West, A. P., Jr., R. P. Galimidi, C. P. Foglesong, P. N. Gnanapragasam, K. E. Huey-Tubman, J. S. Klein, M. D. Suzuki, N. E. Tiangco, J. Vielmetter, and P. J. Bjorkman. 2009. Design and expression of a dimeric form of human immunodeficiency virus type 1 antibody 2G12 with increased neutralization potency. J. Virol. 83:98-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao, X., W. Chen, Y. Feng, Z. Zhu, P. Prabakaran, Y. Wang, M.-Y. Zhang, N. S. Longo, and D. S. Dimitrov. 2009. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 390:404-409. [DOI] [PMC free article] [PubMed] [Google Scholar]