Abstract
The cytoplasmic replication of positive-sense RNA viruses is associated with a dramatic rearrangement of host cellular membranes. These virus-induced changes result in the induction of vesicular structures that envelop the virus replication complex (RC). In this study, we have extended our previous observations on the intracellular location of West Nile virus strain Kunjin virus (WNVKUN) to show that the virus-induced recruitment of host proteins and membrane appears to occur at a pre-Golgi step. To visualize the WNVKUN replication complex, we performed three-dimensional (3D) modeling on tomograms from WNVKUN replicon-transfected cells. These analyses have provided a 3D representation of the replication complex, revealing the open access of the replication complex with the cytoplasm and the fluidity of the complex to the rough endoplasmic reticulum. In addition, we provide data that indicate that a majority of the viral RNA species housed within the RC is in a double-stranded RNA (dsRNA) form.
West Nile virus (WNV) belongs to the Flaviviridae, which is a large family of enveloped, positive-strand RNA viral pathogens that are responsible for causing severe disease and mortality in humans and animals each year. The Australian WNV strain Kunjin virus (WNVKUN) is a relatively low-pathogenic virus that is closely related to the pathogenic WNV strain New York 99 (WNVNY99), the causative agent of the 1999 epidemic of encephalitis in New York City (11).
It has become increasingly known that the replication of most, if not all, positive-sense RNA viruses, whether they infect plants, insects, or humans, is associated with dramatic membrane alterations resulting in the formation of membranous microenvironments that facilitate efficient virus replication. In most cases the induced membrane structures house the actively replicating viral RNA and comprise 70- to 100-nm membrane “vesicles” (sometimes referred to as spherules). Although this distinct morphology is shared across virus families, the cellular origins of these membranes is diverse: the endoplasmic reticulum (ER), mitochondria, peroxisomes, and trans-Golgi membranes have been implicated in different viral systems (1, 8, 13, 23, 31, 38, 41, 45). This diversity implies that the processes involved in inducing the membrane vesicles/spherules are shared, rather than the composition of the membrane itself, although the exact purpose for utilizing membranes derived from different cellular compartments is still not completely resolved or understood.
The replication of the flavivirus WNVKUN is associated with the induction of morphologically distinct membrane structures that have defined roles during the WNVKUN replication cycle. Three well-defined structures can be seen as large convoluted membranes (CM), paracrystalline arrays (PC), or membrane sacs containing small vesicles, termed vesicle packets (VP) (18, 20, 48). Based on localization studies with viral proteins of specific functions, we observed that components of the virus protease complex (namely, nonstructural protein 3 [NS3] with cofactor NS2B) localize specifically to the CM/PC, whereas viral double-stranded RNA (dsRNA) and the viral RNA-dependent RNA polymerase (RdRp) NS5 localized primarily to VP (20-22, 47, 48). Additionally, we observed that the CM and PC originate from and are modified membranes of the intermediate compartment (IC) and rough endoplasmic reticulum (RER), whereas the VP appear to be derived from trans-Golgi network (TGN) membranes (19). Recently, we have found that the WNVKUN NS4A protein by itself has the intrinsic capacity to induce the CM and PC structures (35), a property also subsequently shown for Dengue virus (DENV) NS4A (29). Additionally, we have shown that upon WNV infection cellular cholesterol and cholesterol-synthesizing proteins are redistributed to the virus-induced membranes and that this redistribution severely disrupted the formation of cholesterol-rich microdomains (23). Furthermore, we have shown that the membranous structures induced during WNV replication provide partial protection of the WNV replication components from the interferon (IFN)-induced antiviral MxA protein, suggesting that the distinct compartmentalization of viral replication and components of the cellular antiviral response may be an evolutionary mechanism by which flaviviruses can protect themselves from host surveillance (6).
In this study we focused on three-dimensional (3D) modeling to give insight into the 3D structure of the VP and provide evidence of how these complexes are organized and formed within the RER membrane. These results add valuable information to our understanding of how the WNV replication complex (RC) functions.
MATERIALS AND METHODS
Viruses and cells.
Cells were infected with WNVKUN strain MRM61C at an approximate multiplicity of infection (MOI) of 3, as was described previously (46). Vero cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS) (Lonza, Basel, Switzerland) and penicillin-streptomycin (100 U/ml and 100 μg/ml, respectively; Gibco-BRL) at 37°C with 5% CO2. A WNVKUN trans-packaging BHK21 cell line (tetKUN-CprME [3]) was maintained in DMEM supplemented with 5% FCS; penicillin-streptomycin, G418 sulfate (500 μg/ml; Gibco), and puromycin (3 mg/ml, Sigma-Aldrich) for maintenance; and doxycycline (5 mg/ml; Sigma-Aldrich) to suppress the expression of WNVKUN structural proteins until required.
Antibodies.
WNVKUN-specific anti-NS1 (clone 4G4 [17]) monoclonal antibodies were generously provided by Roy Hall (University of Queensland, Brisbane, Australia). Rabbit anti-β-1,4-galactosyltransferase (GalT) polyclonal antibodies (44) were generously provided by Eric Berger (University of Zurich, Zurich, Switzerland). Mouse anti-dsRNA (clone J2) antibodies were purchased from English & Scientific Consulting Bt. (Hungary). Mouse anti-protein disulfide isomerase (PDI) (clone 1D3 [43]) monoclonal antibodies were generously provided by Steve Fuller (Oxford University, Oxford, United Kingdom). Rabbit anti-calnexin and anti-giantin antibodies were purchased from Merck-Calbiochem (Germany). Alexa Fluor 488-conjugated concanavalin A (ConA), Alexa Fluor 594-conjugated wheat germ agglutinin (WGA), and Alexa Fluor 488- and 594-conjugated anti-rabbit- and anti-mouse-specific IgG were purchased from Molecular Probes (Invitrogen, Leiden, Netherlands).
Immunofluorescence (IF) analysis.
Vero cell monolayers on coverslips were infected with WNVKUN and incubated at 37°C for 24 h. The cells were subsequently washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO), and permeabilized with 0.1% Triton X-100 as previously described (23). Primary and secondary antibodies were incubated within blocking buffer (PBS containing 1% bovine serum albumin [BSA]) and washed with PBS containing 0.1% BSA between incubation steps. After a final wash with PBS the coverslips were drained and mounted onto glass slides with a quick-dry mounting medium (United Biosciences, Brisbane, Australia) before visualization on a Leica TCS SP2 confocal microscope. Images were collected by using a Leica digital camera and Leica 3D software before processing for publication using Adobe Photoshop software.
Resin thin sections for electron microscopy.
Cells were fixed with 3% glutaraldehyde in 0.1 M cacodylate buffer for 2 h at room temperature. Cells were washed several times in 0.1 M cacodylate buffer followed by fixation with 1% OsO4 in 0.1 M cacodylate buffer for 1 h. After washing of the cells in 0.1 M cacodylate buffer, specimens were dehydrated in graded acetones for 10 to 20 min each. Subsequently, samples were infiltrated with Epon resin and polymerized in molds for 2 days at 60°C. Thin sections (50 to 60 nm) were cut on a Leica Ultracut ultramicrotome using a Diatome diamond knife and collected on Formvar-coated copper mesh grids. Before viewing in a Jeol 2010 transmission electron microscope (TEM), cells were poststained with 2% aqueous uranyl acetate (UA) and Reynold's lead citrate.
ET.
tetKUN-CprME cells were transfected with a WNVKUN replicon expressing the human MxA protein (KUNrepMxA) and processed for conventional resin embedding as described above (A. Hoenen et al., submitted for publication). These cells were utilized for this study, as the collection of images for the analysis was already being performed in the complementary experiment. Ribbons of thin (40- to 60-nm) or thick (300- to 400-nm) sections were cut on a microtome (Leica Microsystems) for a conventional two-dimensional (2D) survey at 80 to 100 keV to assess the quality of cell preservation and for 3D studies by electron tomography (ET) at 300 keV on Tecnai T12 and F30 microscopes, respectively (FEI Company). Thick sections were collected onto Formvar-coated copper (2- by 1-mm) slot grids (Electron Microscopy Sciences) and poststained with 2% aqueous UA and Reynold's lead citrate followed by carbon coating. To facilitate the image alignment that is required for the subsequent image reconstruction step, a suspension of 10-nm gold particles was layered on top of the thick sections as fiducial markers.
For tomography and 3D reconstruction, tilt series were obtained at 2° intervals over a range of 120° about two orthogonal axes. The tilt series were aligned by means of the fiducial markers, and dual-axis 3D density distributions (tomograms) were calculated by using IMOD software from the Boulder Laboratory for 3-D Electron Microscopy of Cells (http://bio3d.colorado.edu/imod/). Images of 3D reconstructions were produced by using the IMOD software package.
RNase treatments.
Vero cell monolayers were infected with WNVKUN at an MOI of 3 and incubated at 37°C for 24 h. The cells were subsequently permeabilized with 0.05% Triton X-100 on ice for 5 min, washed with PBS, and fixed with 4% paraformaldehyde-0.1% glutaraldehyde on ice for 30 min. Cells were then washed, equilibrated in either 0.1× SSC (1× SSC is 150 mM NaCl plus 15 mM sodium citrate [pH 7.0]) (low salt) or 2× SSC (high salt), and incubated with or without 50 μg/ml RNase A (Sigma-Aldrich) at 37°C for 1 h. The cells were subsequently washed and embedded in Epon resin as described above. Visible threads within the virus-induced vesicles were scored as a percentage within each frame, and results were collated from duplicate experiments.
RESULTS
The WNVKUN RNA replication complex appears to be derived at a pre-Golgi step.
Previously, we provided evidence that the membranes comprising the WNVKUN RC were derived from the trans-Golgi membrane and/or trans-Golgi network (TGN) (19). This was based largely on our immunolocalization studies that showed a distinct redistribution and colocalization of the TGN marker β-1,4-galactosyltransferase (GalT) with individual vesicles within the VP. Our previous observations implicating an additional intimate role for the RER (19) and those recently made by Welsch et al. (45) suggest an intriguing situation where the biogenesis of the VP could occur via two mechanisms: (i) the sequestration of the retrograde flow of the TGN proteins via the RER during normal steady-state recycling or (ii) the recruitment of proteins and membrane at an early stage of the secretory pathway, a direct pathway from the ER. To investigate these different hypotheses, we utilized two different fluorescently labeled lectins to identify the carbohydrate groups present on the cellular and viral proteins accumulating within the VP. We used an Alexa Fluor 488-conjugated concanavalin A (ConA) lectin to detect the presence of high-mannose glycans or an Alexa Flour 594-cojugated wheat germ agglutinin (WGA) lectin to detect complex glycans; the former can potentially label the whole ER and Golgi pathway, while the latter detects only glycol components starting with the medial Golgi membrane and the rest of the secretory pathway (2).
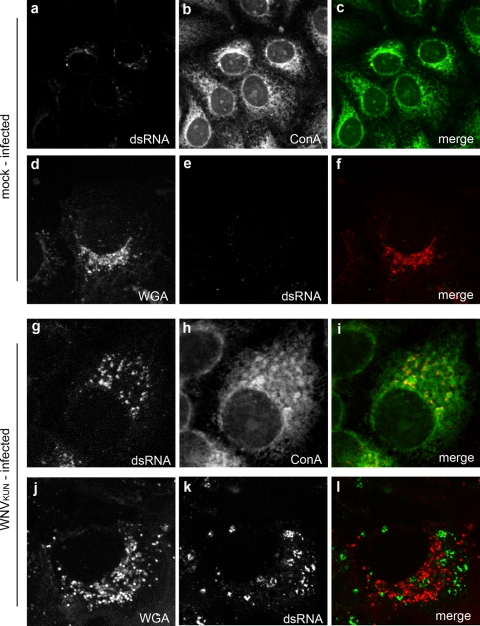
Our staining with both of the lectins ConA and WGA displayed a typical localization to the ER and Golgi membrane, respectively, in mock-infected Vero cells (Fig. 1A). ConA was observed to label a reticular network within the cytoplasm, whereas WGA stained as a “crescent-like” accumulation in the perinuclear region. The costaining of the lectins with the ER- and Golgi-resident proteins PDI and calnexin as well as giantin and GalT, respectively, in mock-infected cells revealed that the lectins labeled the proposed organelles (see Fig. S1 and S2 in the supplemental material). As glycoproteins within the ER consist mainly of high-mannose glycans and contain more trimmed and processed complex glycans in the Golgi apparatus, this labeling pattern is what is predicted. In WNVKUN-infected Vero cells, anti-dsRNA antibodies clearly detected individual foci scattered throughout the cytoplasm (Fig. 1B). In some cases these foci resembled “doughnut-like” structures but more often than not stained as complete foci. The vast majority of these foci were coincident with ConA; however, they were completely distinct for staining with WGA. The WGA staining was typically “Golgi-like,” but the dsRNA foci were interspersed within this staining pattern but not coincident. We believe that these observations provide evidence that the proteins residing within the WNVKUN VP, including potentially host cell glycoproteins, contain high-mannose glycans, implying that their trafficking to the VP has occurred at a pre-Golgi step. This suggests that the WNVKUN-mediated recruitment of host cell membranes and associated proteins to initiate the biogenesis of the VP is a very early event along the secretory pathway. Note that the secretory pathway remains intact within WNVKUN-infected cells, as we have observed that virion morphogenesis and maturation are dependent on the secretory pathway (25).
FIG. 1.
The WNVKUN RC contains only high-mannose conjugated glycans. IF analysis of Vero cells mock infected (a to f) or WNVKUN infected (g to l), immunolabeled with antibodies to dsRNA, and costained with Alexa Fluor 488-conjugated ConA or Alexa Fluor 594-conjugated WGA is shown. Colocalization is observed only in i as a yellow hue.
Glycoproteins residing within the VP contain high-mannose glycans.
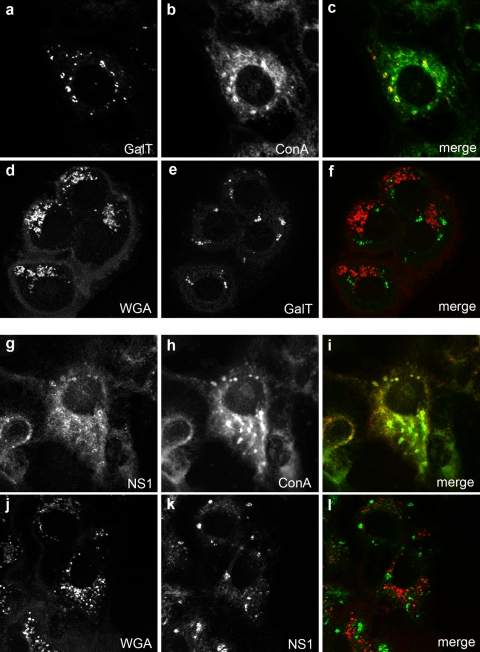
The host protein GalT is a trans-Golgi-resident glycoprotein that posttranslationally acquires complex glycans during transit through the Golgi apparatus (39). Additionally, the flavivirus NS1 glycoprotein contains both high-mannose and complex glycans during maturation and secretion from infected cells (27, 49). Both of these proteins have been implicated in playing roles during flavivirus RNA replication (15, 16, 19, 20, 48), and both are localized to the VP during WNVKUN and DENV infection (19, 20, 48). Thus, we aimed to confirm our above-described results and determine the glycosylation status of the GalT and NS1 proteins within the VP. As shown in Fig. 2, GalT confined within the WNVKUN RC could be colabeled only with ConA but not WGA (Fig. 2a to f). This was also observed for the WNVKUN NS1 protein, where again, costaining was observed only with ConA (Fig. 2g to l). In contrast, GalT was efficiently costained with WGA in mock-infected cells, as expected (see Fig. S2 in the supplemental material).
FIG. 2.
Both host and viral glycoproteins residing within the WNVKUN RC appear to be sequestered at a pre-Golgi step. Costaining with Alexa Fluor 488-conjugated ConA (b and h) or Alexa Fluor 594-conjugated WGA (d and j) and either the host glycoprotein GalT (a and e) or the viral glycoprotein NS1 (g and k) reveal that both marker proteins are dually labeled only with Alexa Fluor 488-conjugated ConA and not Alexa Fluor 594-conjugated WGA. These results suggest that the RC-resident glycoproteins have not transited through the Golgi apparatus, where further processing and the addition of complex glycans would have occurred.
These results support a hypothesis where the sequestration of host proteins and membranes within the VP and its biogenesis occur at a pre-Golgi step, likely via interactions that occur within the ER membrane platform. This has profound implications for the processes that regulate protein and membrane trafficking at this site within WNVKUN-infected cells, in particular the mechanism by which host glycoproteins bound for the Golgi apparatus may be sequestered early in the secretory pathway and then transit en bloc to their sites of residency within the Golgi stack. As we have recently shown that cholesterol is a major factor during WNVKUN replication (23), a potential role for lipid-based sorting may be exploited by WNVKUN whereupon the cholesterol-rich membrane platform is sequestered and, in doing so, additionally “captures” host proteins that have been collected within these platforms for transport.
3D electron tomography reveals open access between the WNVKUN RC and the cytoplasm.
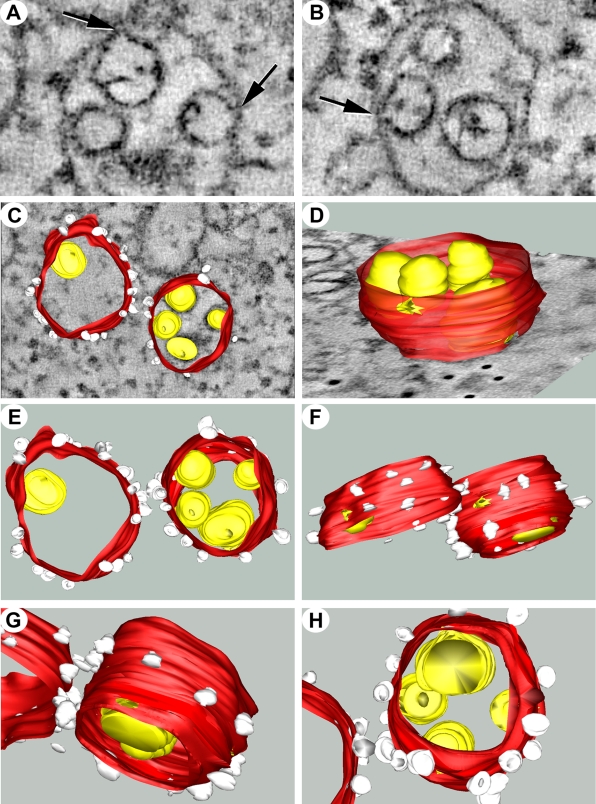
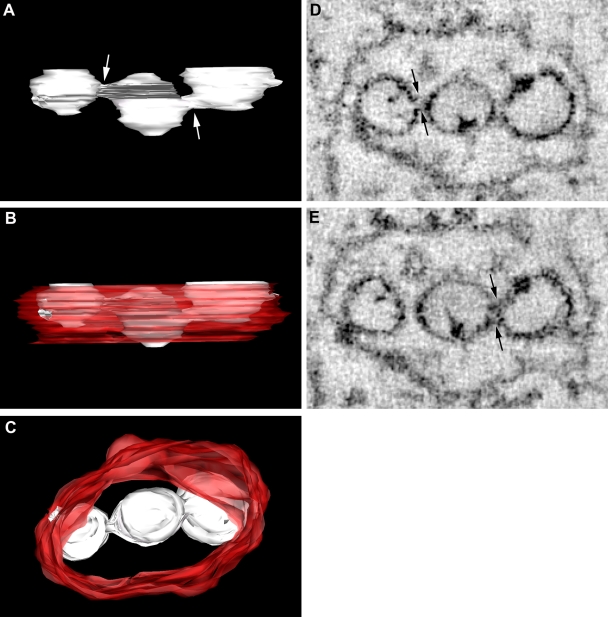
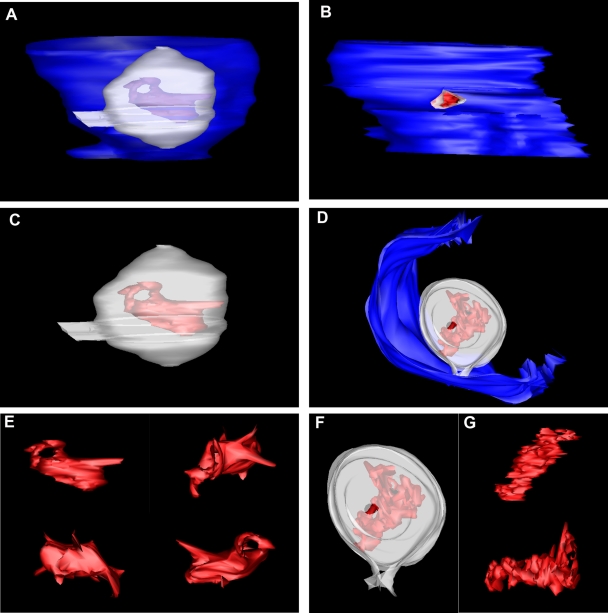
Our previous studies investigating the ultrastructural localization of the WNVKUN RC have relied upon the immunolabeling of thawed thin sections from cryofixed infected cells. Although these studies have revealed many insights into our current understanding of the composition and functionality of the RC, thin sectioning still provides only multiple 2D snapshots of the events occurring intracellularly. The technique of electron tomography has become an increasingly powerful tool to reveal 3D information on cellular structures, including structures involved in the intracellular replication of positive-sense RNA viruses (7, 8, 45). To extend our studies further, we also utilized the technique of electron tomography to visualize the WNVKUN RC. However, we modeled the RC from a WNV packaging cell line transfected with WNV replicon RNA (see Materials and Methods). We have previously shown that the formation of the WNVKUN RC in replicon-transfected cells is morphologically analogous to that during virus infection (24). In short, a 300-nm-thick section was collected from conventional Epon-embedded cells and visualized in a 300-kV TEM. The section was tilted over 120°, and images were collected at every 2° interval. With the help of computer software the collected images were aligned and tomograms were calculated, followed by the construction of 3D models of the intracellular structures. As shown in Fig. 3, the most striking feature was the intimate association of the RER that comprises the bounding membrane of the VP. This supports our observations of a role for the RER in the biogenesis of the WNVKUN VP (see above) and recent observations made during DENV replication (45). We have additionally shown the localization of PDI within the VP, as described previously (19), and the colocalization of both PDI and calnexin with dsRNA by IF analysis (see Fig. S3 in the supplemental material). The other striking feature was the visualization of individual necks on the vesicles within the VP that are open to the cytoplasm (arrows in Fig. 3A and B). This was not always obvious by standard thin-section analysis of resin-embedded material, but we are now able to visualize this via sequential planes within the 2-nm tomogram slices. The presence of an open neck is a common observation for other positive-strand RNA viruses, including alphaviruses (i.e., Semliki Forest virus [1]) and alphanodaviruses (i.e., Flock House virus [8]); however, it has been observed only once within flavivirus-infected cells (45). The presence of a neck more than likely allows the transfer of cytoplasmic constituents (i.e., nucleotides) into the vesicles and the release of newly synthesized viral genomic RNA to the cytoplasm. Interestingly, the individual vesicles themselves can be continuous with each other, as “internally confined” vesicles within the VP display direct connections with adjoining vesicles that have access to the cytoplasm (Fig. 4 and see Movie S3 in the supplemental material). This suggests that there is a great range of dynamic movement and, perhaps, the recycling of constituents within the VP.
FIG. 3.
The WNVKUN RC is intimately associated with the RER membranes and is open to the cytoplasm via a membranous neck. (A) At 48 h posttransfection, KUNrepMxA-transfected tetKUN-CprME cells were fixed for resin embedding and analyzed by electron tomography. A VP utilized for the subsequent 3D surface model is shown. (B) Visualization of the vertical stacks revealed the presence of “neck-like” structures (indicated by arrows) that tether the individual vesicles to the membrane. (C) 3D surface model of the WNVKUN RC reveals the intimate association of the individual vesicles housing the viral RNA (indicated in yellow) with the RER membrane (indicated in red) that is decorated with associated ribosomes (indicated in white and additionally highlighted with asterisks in A and B). (D to H) Rotational views of the WNVKUN RC highlighting the pores connecting the vesicles to the cytoplasm and the spatial arrangement of the vesicles within the VP. The 3D model was constructed by using median filtering and automatic threshold segmentation using IMOD software.
FIG. 4.
Vesicles within the WNVKUN RC are connected to each other directly via pores. (A to C) 3D surface modeling revealed that vesicles within the internal space of the VP, not directly in connection with the bounding membrane, are fused to neighboring vesicles via pores (highlighted by arrows in A). The individual vesicles are indicated in white, and the ER membrane is depicted in red. C is a 90° rotation (or top view) of the VP in B. (D and E) Snapshots collected from the tomogram shown to highlight the “necks” or “pores” that link individual vesicles within the RC. Connections are indicated by arrows.
A majority of the viral RNA within the RC is dsRNA.
Previous reports suggested that the observed threads within the virus-induced vesicles are the replicating viral RNA. This has been based largely on the postembedding labeling of rubella virus-infected cells (12) and RNase treatment of tombusvirus-infected (37) and comovirus-infected (5) plant cells. Additionally, our previous studies indicated that the WNVKUN VP are the sole induced membranes decorated with antibodies to dsRNA, strongly implying that a majority of the dsRNA is housed within (19, 21, 22, 48). Thus, we sought to additionally model the threads present in the WNVKUN VP in our tomograms, based on our proposal that these structures are the viral RNA. As shown in Fig. 5, these structures were highly diverse in their structures and were quite dynamic in their appearances. Although we are aware that the structure of the RNA molecule has more than likely been affected by the fixation protocol, we believe that this representation still provides arguably the first structural visualization of replicating viral RNA in infected cells. Note that in most cases, the viral RNA spanned the breadth of the vesicles and was juxtaposed to the necks open to the cytoplasm.
FIG. 5.
Structural modeling of the viral RNA within the vesicles reveals a complex structure closely aligned to the vesicle pore. Two individual vesicles were 3D surface modeled (A, C, and E and B, D, F, and G, respectively), and the visible RNA was also surface rendered. The analyses showed that the viral RNA is present in a dynamic state and may reveal the presence of individual tertiary stem-loop structures present on the RNA backbone. Our analyses have also revealed that the viral RNA is juxtaposed to the pore-like opening tethering the individual vesicles.
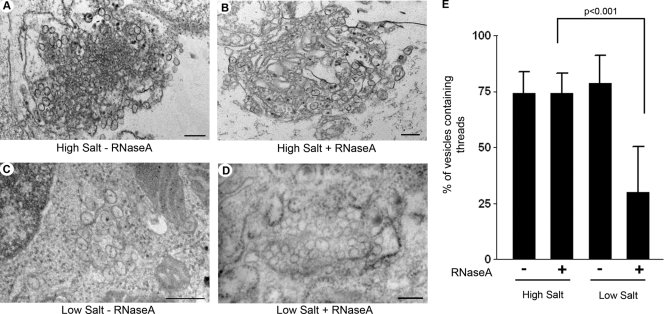
To identify the nature of the viral RNA within the VP, we treated WNVKUN-infected cells with RNase A in the presence of high- or low-salt concentrations, which will digest single-stranded RNA (ssRNA) or all RNA, respectively. We have previously utilized this treatment during the in vivo labeling of WNVKUN with bromouridine (47). High- or low-salt-treated cells were processed for resin embedding, and a quantitative analysis was performed to determine the fraction of vesicles that contain threads within the VP under the different treatment conditions (Fig. 6). If the threads were solely dsRNA, then we would expect that these threads would be digested under low-salt but not under high-salt conditions. As shown in Fig. 6, this was indeed the case. The analyses revealed that under high-salt conditions without RNase A, 74.08% (±9.86%; 452 of 618 vesicles) of vesicles contained visible threads, and under high-salt conditions plus RNase A, 74.15% (±9.02%; 661 of 890 vesicles) of vesicles contained visible threads, strongly suggesting that only a very small proportion of vesicles contained ssRNA. Under low-salt conditions without RNase A treatment, 78.65% (±12.58%; 488 of 590 vesicles) of vesicles contained visible threads, but under low-salt conditions plus RNase A, only 29.99% (±20.57%; 396 of 1,221 vesicles) of vesicles were observed to have a visible thread. Although we are aware that some RNA structures may not be accessible to RNase under these conditions, we believe that these analyses suggest that the vast majority of RNA within the VP is dsRNA and that newly transcribed genomic viral ssRNA is efficiently transported out of the VP, presumably via the open neck, for translation and/or packaging.
FIG. 6.
The majority of viral RNA within the VP is dsRNA. (A and B) Representative images of WNVKUN-infected Vero cells without (A) or with (B) RNase A treatment under high-salt conditions. As can be observed, the majority of vesicles within both samples contain visible threads. (C and D) Representative images of WNVKUN-infected Vero cells without (C) or with (D) RNase A treatment under low-salt conditions. As can be observed, the majority of vesicles within C contain visible threads, which is in stark contrast to the vesicles in D, where very few threads are observed. In all cases, the magnification bars represent 200 nm. (E) Quantitative analysis of the percentage of vesicles containing visible threads under all conditions. The results were calculated from duplicate experiments, and the statistical analysis was performed by using an unpaired t test with a 95% confidence interval.
DISCUSSION
Using 3D tomography we have observed that the formation of the vesicles containing the WNVKUN RC most likely occurs via the sequestration of host cell membranes from an early stage within the secretory pathway and via the invagination of the ER membrane giving rise to open necks within the individual vesicles within the “packet.” These necks most likely provide a means whereby cytoplasmic constituents (such as nucleotides) gain access to the internally replicating viral RNA and conversely provide an avenue for the replicated RNA to enter the cytoplasm for translation or packaging into progeny virus particles.
Our previous studies (19), studies from others (29, 30, 45), and the results presented here have revealed that the flavivirus RC is built upon a membrane scaffold rich in the ER-resident protein chaperones. In addition, we provide evidence to suggest that the biogenesis and recruitment of host proteins and/or membranes occur during a pre-Golgi step. Combined with our recent observations indicating a crucial role for cholesterol in facilitating efficient WNVKUN replication (23), we suggest that a highly dynamic lipid-based sorting mechanism is in place to enable the biogenesis of the viral RC. This feature could explain the presence of GalT (and other identified trans-Golgi proteins) within the VP, as it may well be coincidental rather than functional. We are postulating that GalT and probably other TGN-resident proteins are “sorted” within the ER via a lipid-based mechanism before transit to the Golgi apparatus, possibly within cholesterol-rich domains within the ER. It is therefore probable that during the sequestering of intracellular cholesterol by WNVKUN, these host proteins are additionally redistributed merely due to their locale. Further studies will be required to determine whether host proteins (like GalT) are functionally required by WNVKUN or not.
Our tomographic analyses revealed that the WNV RC is open to the cellular cytosol via necks that tether the individual vesicles within the VP to the ER membrane. Such structures were also recently observed for the VP induced during DENV replication using a similar approach (45). Our study and a previous study by Welsch et al. (45) have visualized the continuity of the rough ER membrane with the VP, and we provide a clear indication of the topology of the replicating RNA within the VP. This new model and this proposal are also in stark contrast to the model proposed previously by Uchil and Satchidanandam (40), who suggested closed vesicles encasing the viral RNA. Our observations of WNV and the DENV study provide compelling evidence indicating the open access of the replicating RNA with cellular cytosolic components (Fig. 3 to 5) (45).
The induction of vesicles tethered to a cellular membrane is in fact a shared trait of arguably all positive-sense RNA viruses infecting animals, plants, and insects (1, 4, 9, 10, 12, 13, 20, 26, 28, 32, 36, 38). This commonality implies a conserved mechanism for the biogenesis of such structures and a shared purpose for inducing these membrane invaginations. Some of our recent studies have implied a role for these induced vesicles in protecting the viral RNA and associated components from immune surveillance (6). It would be of interest to determine whether this is also true of the other virus families. Although morphologically similar, the cellular membrane origins of the virus-induced vesicles can differ markedly. The alpha- and togaviruses utilize membranes derived from modified endosomes/lysosomes (1, 12), members of the Nodaviridae utilize membranes of the mitochondria (28), and the plant virus tombusvirus utilizes peroxisomes (33), whereas the similar plant virus Brome mosaic virus, like the flaviviruses, nidoviruses, and coronaviruses, utilizes membranes derived from the ER (7, 19, 34, 42). Based on these observations it is apparent that the specific lipid compositions of the members of the above-described organelles play an important role in replicating the viral RNA, conclusions that have been emphasized by studies of Brome mosaic virus replication (14).
Thus, in summary, we believe that our results have provided additional information on the biogenesis of the WNVKUN RC and have indicated the process by which WNVKUN recruits both cellular membrane and associated proteins to the RC. Our tomographic analysis also supports the recent observations made with DENV that reveal an open neck that tethers the RC to the ER membrane. Our analyses also reveal a greater degree of fluidity that exists within the RC, as we have observed that individual vesicles within the VP are also intimately connected via pores to allow the transfer of viral RNA and/or associated components during replication.
Supplementary Material
Acknowledgments
We thank Alexander Khromykh (University of Queensland) for generously providing the WNVKUN replicons and tetKUN-CprME packaging cell line. We also thank Roy Hall, Steve Fuller, and Eric Berger for generously providing antibodies. We also thank Gareth Griffiths for critical review of the manuscript.
Footnotes
Published ahead of print on 4 August 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffiths, G., R. Brands, B. Burke, D. Louvard, and G. Warren. 1982. Viral membrane proteins acquire galactose in trans Golgi cisternae during intracellular transport. J. Cell Biol. 95:781-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey, T. J., W. J. Liu, X. J. Wang, R. Linedale, M. Jacobs, A. Davidson, T. T. Le, I. Anraku, A. Suhrbier, P. Y. Shi, and A. A. Khromykh. 2004. Tetracycline-inducible packaging cell line for production of flavivirus replicon particles. J. Virol. 78:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatta, T., S. Bullivant, and R. E. Matthews. 1973. Fine structure of vesicles induced in chloroplasts of Chinese cabbage leaves by infection with turnip yellow mosaic virus. J. Gen. Virol. 20:37-50. [DOI] [PubMed] [Google Scholar]
- 5.Hatta, T., and R. I. Francki. 1978. Enzyme cytochemical identification of single-stranded and double-stranded RNAs in virus-infected plant and insect cells. Virology 88:105-117. [DOI] [PubMed] [Google Scholar]
- 6.Hoenen, A., W. Liu, G. Kochs, A. A. Khromykh, and J. M. Mackenzie. 2007. West Nile virus-induced cytoplasmic membrane structures provide partial protection against the interferon-induced antiviral MxA protein. J. Gen. Virol. 88:3013-3017. [DOI] [PubMed] [Google Scholar]
- 7.Knoops, K., M. Kikkert, S. H. E. van den Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kujala, P., T. Ahola, N. Ehsani, P. Auvinen, H. Vihinen, and L. Kaariainen. 1999. Intracellular distribution of rubella virus nonstructural protein P150. J. Virol. 73:7805-7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 12.Lee, J. Y., J. A. Marshall, and D. S. Bowden. 1994. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology 200:307-312. [DOI] [PubMed] [Google Scholar]
- 13.Lee, J. Y., J. A. Marshall, and D. S. Bowden. 1992. Replication complexes associated with the morphogenesis of rubella virus. Arch. Virol. 122:95-106. [DOI] [PubMed] [Google Scholar]
- 14.Lee, W. M., and P. Ahlquist. 2003. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J. Virol. 77:12819-12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindenbach, B. D., and C. M. Rice. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 73:4611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindenbach, B. D., and C. M. Rice. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macdonald, J., J. Tonry, R. A. Hall, B. Williams, G. Palacios, M. S. Ashok, O. Jabado, D. Clark, R. B. Tesh, T. Briese, and W. I. Lipkin. 2005. NS1 protein secretion during the acute phase of West Nile virus infection. J. Virol. 79:13924-13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackenzie, J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackenzie, J. M., M. K. Jones, and P. R. Young. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232-240. [DOI] [PubMed] [Google Scholar]
- 21.Mackenzie, J. M., M. T. Kenney, and E. G. Westaway. 2007. West Nile virus strain Kunjin NS5 polymerase is a phosphoprotein localized at the cytoplasmic site of viral RNA synthesis. J. Gen. Virol. 88:1163-1168. [DOI] [PubMed] [Google Scholar]
- 22.Mackenzie, J. M., A. A. Khromykh, M. K. Jones, and E. G. Westaway. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203-215. [DOI] [PubMed] [Google Scholar]
- 23.Mackenzie, J. M., A. A. Khromykh, and R. G. Parton. 2007. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2:229-239. [DOI] [PubMed] [Google Scholar]
- 24.Mackenzie, J. M., A. A. Khromykh, and E. G. Westaway. 2001. Stable expression of noncytopathic Kunjin replicons simulates both ultrastructural and biochemical characteristics observed during replication of Kunjin virus. Virology 279:161-172. [DOI] [PubMed] [Google Scholar]
- 25.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magliano, D., J. A. Marshall, D. S. Bowden, N. Vardaxis, J. Meanger, and J. Y. Lee. 1998. Rubella virus replication complexes are virus-modified lysosomes. Virology 240:57-63. [DOI] [PubMed] [Google Scholar]
- 27.Mason, P. W. 1989. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology 169:354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock House virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, S., S. Kastner, J. Krijnse-Locker, S. Buhler, and R. Bartenschlager. 2007. Non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873-8882. [DOI] [PubMed] [Google Scholar]
- 30.Miller, S., S. Sparacio, and R. Bartenschlager. 2006. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 281:8854-8863. [DOI] [PubMed] [Google Scholar]
- 31.Nagy, P. D., A. Dzianott, P. Ahlquist, and J. J. Bujarski. 1995. Mutations in the helicase-like domain of protein 1a alter the sites of RNA-RNA recombination in Brome mosaic virus. J. Virol. 69:2547-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng, M. L. 1987. Ultrastructural studies of Kunjin virus-infected Aedes albopictus cells. J. Gen. Virol. 68:577-582. [DOI] [PubMed] [Google Scholar]
- 33.Panavas, T., C. M. Hawkins, Z. Panaviene, and P. D. Nagy. 2005. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 338:81-95. [DOI] [PubMed] [Google Scholar]
- 34.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roosendaal, J., E. G. Westaway, A. Khromykh, and J. M. Mackenzie. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 80:4623-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo, M., A. Di Franco, and G. P. Martelli. 1987. Cytopathology in the identification and classification of tombusviruses. Intervirology 28:134-143. [DOI] [PubMed] [Google Scholar]
- 37.Russo, M., A. Di Franco, and G. P. Martelli. 1983. The fine structure of Cymbidium ringspot virus infections in host tissues. III. Role of peroxisomes in the genesis of multivesicular bodies. J. Ultrastruct. Res. 82:52-63. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 39.Strous, G. J., and E. G. Berger. 1982. Biosynthesis, intracellular transport, and release of the Golgi enzyme galactosyltransferase (lactose synthetase A protein) in HeLa cells. J. Biol. Chem. 257:7623-7628. [PubMed] [Google Scholar]
- 40.Uchil, P. D., and V. Satchidanandam. 2003. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 278:24388-24398. [DOI] [PubMed] [Google Scholar]
- 41.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Meer, Y., H. van Tol, J. K. Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaux, D., J. Tooze, and S. Fuller. 1990. Identification by anti-idiotype antibodies of an intracellular membrane protein that recognizes a mammalian endoplasmic reticulum retention signal. Nature 345:495-502. [DOI] [PubMed] [Google Scholar]
- 44.Watzele, G., R. Bachofner, and E. G. Berger. 1991. Immunocytochemical localization of the Golgi apparatus using protein-specific antibodies to galactosyltransferase. Eur. J. Cell Biol. 56:451-458. [PubMed] [Google Scholar]
- 45.Welsch, S., S. Miller, I. Romero-Brey, A. Merz, C. K. E. Bleck, P. Walther, S. D. Fuller, C. Antony, J. Krijnse-Locker, and R. Bartenschlager. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. 5:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westaway, E. G., A. A. Khromykh, M. T. Kenney, J. M. Mackenzie, and M. K. Jones. 1997. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 234:31-41. [DOI] [PubMed] [Google Scholar]
- 47.Westaway, E. G., A. A. Khromykh, and J. M. Mackenzie. 1999. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology 258:108-117. [DOI] [PubMed] [Google Scholar]
- 48.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler, G., S. E. Maxwell, C. Ruemmler, and V. Stollar. 1989. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology 171:302-305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.