Abstract
Adenovirus isolate NADC-1, a strain of porcine adenovirus type 4, has a fiber containing an N-terminal virus attachment region, shaft and head domains, and a C-terminal galectin domain connected to the head by an RGD-containing sequence. The crystal structure of the head domain is similar to previously solved adenovirus fiber head domains, but specific residues for binding the coxsackievirus and adenovirus receptor (CAR), CD46, or sialic acid are not conserved. The structure of the galectin domain reveals an interaction interface between its two carbohydrate recognition domains, locating both sugar binding sites face to face. Sequence evidence suggests other tandem-repeat galectins have the same arrangement. We show that the galectin domain binds carbohydrates containing lactose and N-acetyl-lactosamine units, and we present structures of the galectin domain with lactose, N-acetyl-lactosamine, 3-aminopropyl-lacto-N-neotetraose, and 2-aminoethyl-tri(N-acetyl-lactosamine), confirming the domain as a bona fide galectin domain.
Adenoviridae are nonenveloped viruses with a linear double-stranded DNA genome that can infect all five major vertebrate classes. They may be used as vectors for gene or cancer therapy and as vaccination agents (1, 2, 45). Adenoviruses have an icosahedral (T=25) capsid consisting of the trimeric hexon, forming the facets of the particle, the pentameric penton base, which forms the vertices, and the trimeric fiber protein, which extends from the penton base at the vertex positions (38). The distal tip of each fiber is composed of a globular head domain, which serves as the major viral attachment site for a variety of cellular receptors, such as coxsackievirus and adenovirus receptor (CAR) (42), CD46 (15), and sialic acid (6). The structural characterization of different fiber proteins from human and nonhuman adenoviruses has been critical for understanding adenovirus tropism and developing new vectors with modified tropisms. Animal adenoviruses are of particular interest, as they may be less immunogenic to humans and may have novel receptor-binding properties. Recently, canine and fowl adenovirus fiber heads have been crystallized and their structures determined (44, 17, 18, 13).
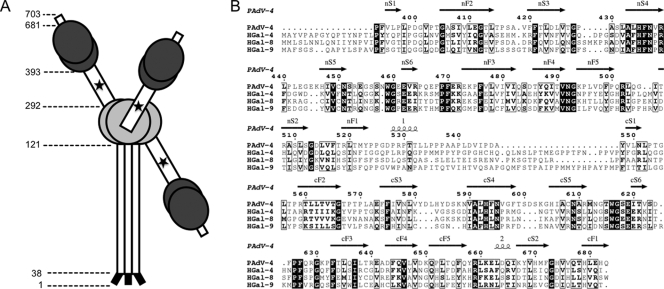
Porcine adenovirus is classified within the genus Mastadenovirus of the Adenoviridae family and has a genome of approximately 34 kb. It is generally regarded as a low-grade pathogen, at least in immunocompetent hosts. Porcine adenovirus types 1 to 3 are closely related, while types 4 and 5 are less similar, both to this group and to each other. All five are distinct from human, murine, canine, bovine, and fowl adenoviruses (21). The porcine adenovirus NADC-1 isolate is thought to be a strain of type 4 (25). Its 703-residue fiber protein is unique among adenovirus strains and contains, from N to C termini, a tail domain (residues 1 to 37; where the interaction sequence with the penton base is found), a short shaft domain with four clear triple beta-spiral repeats (residues 38 to 120 [47]), and what was originally predicted to be a large head domain (residues 121 to 703 [26]). The first 167 amino acids of the head domain (121 to 287) are homologous to other adenovirus head domains; therefore, we propose to rename this the head domain and to identify the adjacent C-terminal segments as follows (Fig. 1 A). The sequence containing residues 288 to 392 contains an RGD sequence and is rich in alanine and glutamate residues, suggesting that this is an integrin interaction site. Additionally, sequence analysis predicts a module composed of tandem carbohydrate recognition domains between residues 393 and 681. These putative carbohydrate recognition domains are closely related to those found in the galectin family of beta-galactoside-binding lectins (3, 10). This galectin domain is unique among adenovirus fibers. The porcine adenovirus galectin module is composed of tandem carbohydrate recognition domains linked by a 23-residue sequence rich in prolines.
FIG. 1.
(A) Schematic drawing of the domain organization of porcine adenovirus type 4 NADC-1 strain fiber. The predicted virus-binding tail is shown in black, the shaft domain in white, the head domain in light gray, the RGD-containing domain in white, and the tandem-repeat galectin domain in dark gray. The putative integrin-binding RGD sequence is indicated with an asterisk. Electron microscopy images of the fiber indicate flexibility between the head and C-terminal domains; therefore, the C-terminal domains are drawn in different orientations. (B) Sequence alignment of tandem-repeat galectins. Human galectin 4 (HGal-4; UNIPROT code P56470), human galectin 8 (H-Gal8; O00214), and human galectin 9 (HGal-9; O00182) are shown. Secondary structural elements of the porcine adenovirus 4 galectin domain are indicated.
Lectins of the galectin family specifically bind beta-galactoside sugars (10). Their carbohydrate recognition domains consist of two beta-sheets with a jelly roll topology. Strands are designated F1 to F5 and S1 to S6, in which the S sheet faces the carbohydrate. Galectins have been assigned to prototype, chimera-type, or tandem-repeat-type subfamilies (22). Prototype galectins consist of a single-carbohydrate recognition domain and usually form homodimers with the two carbohydrate recognition domains in a back-to-back orientation and sugar binding sites placed at opposite ends (31). Tandem-repeat-type galectins contain two nonidentical carbohydrate recognition domains separated by a linker peptide. The organization of the carbohydrate recognition domains in tandem-repeat galectins has not been determined; only structures of single carbohydrate recognition domains are known (37). However, by analogy to the prototype galectins, a back-to-back orientation, and thus bivalent binding, is generally accepted. Eight highly conserved residues, localized within a pocket formed by three adjacent beta strands (S4, S5, and S6), are involved in carbohydrate binding. Other, less-conserved residues, situated in beta strands S2 and S3, are implicated in the recognition of specific sugars.
Here, we report the structures of the porcine adenovirus type 4 NADC1 isolate fiber head domain and of the galectin domain, the first tandem-repeat-type galectin in which both carbohydrate recognition domains are simultaneously observed, revealing a face-to-face orientation of the carbohydrate-binding domains. Furthermore, we demonstrate that the galectin domain binds N-acetyl-lactosamine-containing carbohydrates.
MATERIALS AND METHODS
Adenoviruses.
Ad5Luc1 is a replication-defective E1-deleted adenovirus vector containing a firefly luciferase reporter gene driven by a cytomegalovirus promoter, and it encodes the native adenovirus type 5 fiber protein (27). The Ad5Luc1-PK vector is isogenic to Ad5Luc1, except that the adenovirus type 5 fiber head domain (residues 404 to 581) is replaced with the head, RGD, and galectin domains (residues 124 to 703) of the fiber protein from the NADC-1 isolate of porcine adenovirus type 4 (40). Adenovirus vectors were propagated in 293 cells and purified by equilibrium centrifugation in cesium chloride gradients using a standard protocol (16). Viral particle concentration was determined at 260 nm by the method of Maizel et al. (34) using a conversion factor of 1.1 × 1012 viral particles per absorbance unit.
Glycan array.
Glycan screening was performed using a high-throughput glycan array developed by cores D and H of the Consortium for Functional Glycomics (a National Institutes of Health, National Institute of General Medical Sciences initiative, Emory University School of Medicine, Atlanta, GA). The printed glycan array (version 3.2) contained 406 different natural and synthetic glycans printed on glass slides that contain six individual addresses per glycan or glycoconjugate, as described previously (5). A printed slide was incubated with a 70-μl volume of Ad5Luc1 or Ad5Luc1-PK virions at a concentration of 5.5 × 106 viral particles/μl in sample buffer comprised of 20 mM Tris-HCl, pH 7.4, 150 mM sodium chloride, 2 mM calcium chloride, 2 mM magnesium chloride, 0.05% (vol/vol) Tween 20, and 1% (wt/vol) bovine serum albumin. Following the binding of the virions to the slide, a fluorescein-conjugated anti-hexon primary monoclonal antibody was overlaid on the bound virions. The fluorescence intensity was detected using a ScanArray 5000 confocal scanner (Perkin-Elmer, Waltham, MA). The image was analyzed using the IMAGENE image analysis software (BioDiscovery, El Segundo, CA).
Surface plasmon resonance measurements.
Sensor chips were purchased from Xantec Bioanalytics (Muenster, Germany), and neutravidin was from Pierce Protein Research Products (Rockford, IL). Surface plasmon resonance binding assays were carried out using an SR7000DC optical biosensor spectrometer (Reichert Analytical Systems, Depew, NY). All experiments were performed at 25°C using 10 mM N-2-hydroxyethylpiperazine-N-2-ethanesulphonic acid-NaOH, pH 7.4, 150 mM sodium chloride, 3 mM ethylenediaminetetra-acetic acid, and 1 mM dithiothreitol as the running buffer. Data were processed with SCRUBBER 2.0b software (BioLogic Software, Campbell, Australia). Lactose was obtained from Sigma-Aldrich Quimica (Madrid, Spain); N-acetyl-lactosamine and lacto-N-neotetraose were from Dextra Laboratories (Reading, United Kingdom); and 3-aminopropyl-lacto-N-neotetraose, 2-azidoethyl-di(N-acetyl-lactosamine), and 2-aminoethyl-tri(N-acetyl-lactosamine) were from the Glycan Array Synthesis Core D of the Consortium for Functional Glycomics (The Scripps Research Institute, Department of Molecular Biology, La Jolla, CA). The porcine adenovirus 4 galectin domain was covalently immobilized on a flow cell of a polycarboxylated hydrogel-coated gold surface (HC1000 sensor chip) at a flow rate of 0.01 ml/min. Surface activation was performed with 0.05 M N-hydroxysuccinimide and 0.2 M N-ethyl-N′-(3-dimethyl-aminopropyl)-carbodiimide hydrochloride (10 min), and a 1.3-mg/ml solution of porcine adenovirus 4 galectin domain in 10 mM sodium acetate, pH 5, was injected (10 min); unreacted N-hydroxysuccinimide esters were deactivated with 1 M ethanolamine, pH 8.5 (10 min). The control flow cell was coated with neutravidin using the same coupling protocol. Two sensor chips were prepared with galectin/neutravidin immobilization levels of 12 × 103 μRiU/14 × 103 μRiU (sensor chip 1) and 10.5 × 103 μRiU/6.4 × 103 μRiU (sensor chip 2); 1 μRiU corresponds to 0.73 ng/mm2 coverage in terms of mass (RiU stands for refractive index unit). For binding assays, solutions of each oligosaccharide were prepared in running buffer [1.4 to 45 mM for lactose, 0.12 to 2.6 mM for N-acetyl-lactosamine, 0.030 to 1.3 mM for lacto-N-neotetraose, 0.039 to 1.5 mM for 3-aminopropyl-lacto-N-neotetraose, 0.040 to 2.0 mM for 2-azidoethyl-di(N-acetyl-lactosamine), and 0.010 to 1.9 mM for 2-aminoethyl-tri(N-acetyl-lactosamine)]. The samples were injected over the galectin-coated and control surfaces at 0.05 ml/min for 120 s in duplicate. As analytes fully dissociated after injection, regeneration steps were not necessary. Experimental data were corrected for instrumental and bulk artifacts by double referencing to the control sensor chip surface and buffer injections. In all cases, sensorgrams showed on and off binding profiles that were too fast for kinetic analysis, so the equilibrium analysis of the sensorgrams was performed to obtain the dissociation constants of the complexes.
Crystallization.
The porcine adenovirus type 4 isolate NADC-1 head domain and ligand-free galectin domain were independently expressed, purified, and crystallized as described previously (19). Sitting drop vapor diffusion cocrystallization methods also were employed to yield crystals of the complexes. Lactose was obtained from Sigma-Aldrich Quimica (Madrid, Spain), N-acetyl-lactosamine was from Dextra Laboratories (Reading, United Kingdom), and 3-aminopropyl-lacto-N-neotetraose and 2-aminoethyl-tri(N-acetyl-lactosamine) were from the Glycan Array Synthesis Core D of the Consortium for Functional Glycomics (The Scripps Research Institute, Department of Molecular Biology, La Jolla, CA). Crystals of the lactose complex were obtained using 28% (wt/vol) polyethylene glycol 3350, 100 mM sodium nitrate, 5 mM dithiothreitol as the reservoir solution, and 70 mM lactose in the protein solution. Crystals of the N-acetyl-lactosamine complex were obtained using 35% (wt/vol) polyethylene glycol 3350, 500 mM sodium nitrate, 5 mM dithiothreitol as the reservoir solution, and 40 mM N-acetyl-lactosamine in the protein solution. Crystals of the lacto-N-neotetraose complex were obtained using 27% (wt/vol) polyethylene glycol 3350, 200 mM lithium nitrate, 5 mM dithiothreitol as the reservoir solution, and 5 mM lacto-N-neotetraose in the protein solution. Crystals of the 2-aminoethyl-tri(N-acetyl-lactosamine) complex were obtained using 28% (wt/vol) polyethylene glycol 3350, 300 mM lithium nitrate, 5 mM dithiothreitol as the reservoir solution, and 5 mM 2-aminoethyl-tri(N-acetyl-lactosamine) in the protein solution. Crystallization experiments were performed using the sitting drop vapor diffusion method at 18°C. All crystals appeared after 1 to 2 weeks and were cryoprotected by raising the polyethylene glycol 3350 concentration to 35% (wt/vol) if necessary.
Structure solution and analysis.
Data collection and processing for the head and ligand-free galectin domains were performed as described previously (19); data for the galectin domain cocrystallized with carbohydrates was collected and processed similarly. Crystallographic models were examined and corrected manually using the program COOT (14). Restrained refinement was performed using the program REFMAC (36). The program MOLPROBITY (8) was used to evaluate the stereochemistry. Figures were prepared with Pymol (DeLano Scientific, Palo Alto, CA). Multiple-sequence alignments were performed with CLUSTALW (9). Interfaces were analyzed using PISA (28). The structure solution of the head domain was achieved by molecular replacement using the adenovirus 5 head trimer (Protein Data Bank [PDB] code 1KNB) as a search model with the program MOLREP (46); a clear solution consisting of two trimers in the asymmetric unit was obtained. Despite the moderate resolution (3.2 Å), the high solvent content and the 6-fold noncrystallographic symmetry contribute to generate electron density maps of high quality. Refinement was performed with noncrystallographic symmetry restraints throughout. Toward the end, TLS (translation/libration/screw) refinement was included (48), defining each protein chain as a TLS group. The use of a model refined against high-resolution data in molecular replacement, the aforementioned high solvent content and 6-fold noncrystallographic symmetry, and the use of TLS refinement led to unusually low R factors for a 3.2-Å resolution structure (Table 1).
TABLE 1.
Crystallographic data, refinement, and validation statisticsa
| Statistic | Head domain | Galectin domain |
||||
|---|---|---|---|---|---|---|
| Ligand free | Lactose | N-acetyl-lactosamine | 3-Aminopropyl-lacto-N-neotetraose | 2-Aminoethyl-tri(N-acetyl-lactosamine) | ||
| Data | ||||||
| Space group | P212121 | C2 | C2 | C2 | C2 | C2 |
| Cell parameters a, b, c (Å) | 125.7, 145.4, 147.6 | 167.6, 77.3, 94.1 | 114.6, 38.8, 70.2 | 108.5, 43.7, 63.9 | 95.4, 38.8, 84.4 | 176.1, 38.3, 86.5 |
| Cell parameters α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 101.5, 90.0 | 90.0, 93.5, 90.0 | 90.0, 105.8, 90.0 | 90.0, 102.2, 90.0 | 90.0, 92.1, 90.0 |
| No. of unique observations | 45,035 (4,367) | 89,027 (12,129) | 21,094 (3,019) | 22,025 (3,229) | 23,751 (3,155) | 20,441 (2,926) |
| Resolution range (Å) | 40.0-3.2 (3.32-3.2) | 40.0-1.9 (2.0-1.9) | 40.0-2.0 (2.1-2.0) | 35.0-1.9 (2.0-1.9) | 40.0-1.9 (2.0-1.9) | 40.0-2.5 (2.6-2.5) |
| Multiplicity | 3.8 (3.8) | 4.1 (3.8) | 3.5 (3.5) | 2.7 (2.7) | 3.4 (2.7) | 3.6 (3.6) |
| Completeness (%) | 98.9 (97.5) | 96.1 (90.1) | 99.6 (99.3) | 98.0 (98.5) | 98.5 (91.7) | 99.9 (99.8) |
| Mean I/σ(I) | 17.5 (2.9) | 10.0 (3.7) | 10.4 (3.3) | 5.9 (2.7) | 12.6 (3.2) | 8.8 (3.1) |
| Rsymmb (%) | 7.2 (30.7) | 7.5 (35.2) | 6.9 (32.2) | 9.8 (31.3) | 5.8 (30.3) | 9.5 (43.2) |
| Refinement | ||||||
| Resolution range | 29.2-3.2 (3.37-3.20) | 35-1.9 (2.00-1.90) | 40-2.0 (2.11-2.00) | 35-1.91 (2.01-1.91) | 40-1.9 (2.00-1.90) | 40-2.5 (2.63-2.50) |
| No. of protein molecules/ asymmetric unit | 6 | 4 | 1 | 1 | 1 | 2 |
| R factorc | 0.173 (0.264) | 0.206 (0.298) | 0.183 (0.244) | 0.182 (0.270) | 0.187 (0.275) | 0.199 (0.324) |
| Rfree | 0.201 (0.299) | 0.259 (0.327) | 0.228 (0.296) | 0.238 (0.337) | 0.223 (0.310) | 0.268 (0.355) |
| Final model | ||||||
| RMSD bond lengths (Å) | 0.009 | 0.010 | 0.009 | 0.012 | 0.011 | 0.009 |
| RMSD bond angles (°) | 1.3 | 1.3 | 1.3 | 1.3 | 1.4 | 1.3 |
| Ramachandran plotd (%) | 93.9/6.0/0.1 | 97.0/2.4/0.6 | 96.6/3.4/0.0 | 97.7/2.0/0.3 | 98.0/2.0/0.0 | 97.5/2.5/0.0 |
| PDB accession code | 2WST | 2WSU | 2WSV | 2WT0 | 2WT1 | 2WT2 |
One crystal was used per structure; values in parentheses are for the highest-resolution shell.
Rsymm = ∑h∑i|Ihi−<Ih>|/∑h∑i|Ihi|, where Ihi is the intensity of the ith measurement of the same reflection, and <Ih> is the mean observed intensity for that reflection.
R factor = Σ||Fobs(hkl)| − |Fcalc(hkl)||/Σ|Fobs(hkl)|. Fobs(hkl) and Fcalc(hkl) are observed and calculated structure factors at indices hkl, respectively.
The percentages are of residues in the favored, allowed, and outlier regions, respectively.
The structure of the ligand-free galectin domain was solved by molecular replacement using the galectin-3 carbohydrate recognition domain (PDB code 1A3K) as a search model with the program PHASER (35). A clear solution composed of eight carbohydrate recognition domains, forming four molecules in the asymmetric unit, was obtained. Refinement without noncrystallographic restraints was carried out. Four TLS groups were defined for each molecule, representing the vestige of the purification tag (GGQQGRI; numbered 386 to 391), the N-terminal carbohydrate recognition domain (amino acids 392 to 525), the linker that connects them (residues 526 to 543), and the C-terminal carbohydrate recognition domain (residues 544 to 685). In chains B and C the C-terminal tail (residues 686 to 691) was defined as a fifth group. There are seven outliers in the Ramachandran plot: Ala539 in all four chains and Pro540 in chain A, which are in the linker region, and Pro429 of chains A and C, which are in a surface loop. The structures of the galectin module cocrystallized in the presence of sugars were solved by molecular replacement using chain A of the ligand-free structure as a search model. Refinement was carried out as described for the ligand-free form. In the model with lactose, lactose bound to the N-terminal carbohydrate recognition domain has clear density. Lactose bound to the C-terminal carbohydrate recognition domain has density for the galactose ring, and some disperse clouds of positive density for the glucose moiety; it was refined with occupancy at 0.5. In the model of the galectin domain in complex with N-acetyl-lactosamine, Pro540, which is placed within the rather disordered region 539 to 542, is the unique outlier in the Ramachandran plot.
RESULTS
To obtain structural information on the porcine adenovirus type 4 isolate NADC-1 fiber protein, we constructed expression vectors for amino acids 40 to 703, encoding shaft, head, RGD-containing, and galectin domains; amino acids 116 to 703, encoding head, RGD-containing, and galectin domains; amino acids 40 to 392, encoding shaft, head, and RGD-containing domains; amino acids 40 to 291, encoding shaft-plus-head domains; amino acids 116 to 291, encoding the head domain; amino acids 292 to 392, encoding the RGD-containing domain; and amino acids 392 to 703, encoding the galectin domain. All contained N-terminal six-histidine purification tags. The first two constructs expressed only small amounts of protein, while for the others milligram amounts of protein could be obtained for extensive crystallization trials. From these, crystals were obtained for the shaft-plus-head domain construct (residues 40 to 291), the head domain alone, and the galectin domain (19). However, only the structures of the head and galectin domains could be determined by molecular replacement and refined (Table 1). The shaft domain was disordered in the crystals of the shaft-plus-head domain. Transmission electron microscopy images of negatively stained specimens of the almost-full-length fiber containing residues 40 to 703 (data not shown) suggest that the galectin domains do not adopt a fixed orientation relative to the head and shaft domains, rather the linker consisting of residues 292 to 392 appears to allow flexibility.
Structure of the head domain.
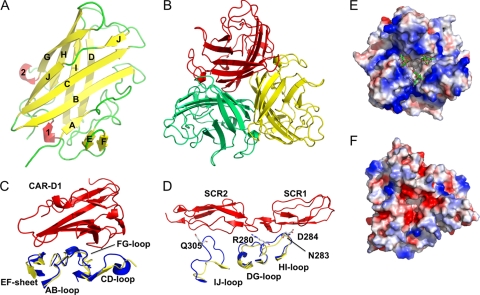
The head domain of each monomer forms an antiparallel beta sandwich of eight beta strands (Fig. 2A), which associate to form a beta propeller trimer (Fig. 2B). When the structure is compared to that of the fiber head of human adenovirus type 5 (50), the most prominent secondary structural variations occur in the region of the CD and IJ loops, which are located at the top of the trimer and are notably shorter in porcine adenovirus type 4; the HI loop at the side of the trimer also is shorter in porcine adenovirus type 4. Many human adenoviruses bind CAR as a primary attachment receptor (42). Structural analyses of domain D1 of CAR in complex with human adenovirus type 12 fiber head (4) have revealed the interaction residues, which are conserved in the fibers of types that use CAR as a receptor. It also has been suggested that the fiber heads that bind CAR have a specific conformation of the AB loop (43). The AB loop of porcine adenovirus type 4 (yellow) structurally resembles AB loops found in human adenovirus type 12 fiber and other CAR binding fibers (Fig. 2C). However, none of the CAR binding residues are conserved in porcine adenovirus type 4 fiber, apart from Pro-222. Additionally, we could not detect any interaction between the CAR D1 domain and porcine adenovirus 4 head domain in size-exclusion chromatography or surface plasmon resonance experiments; the CAR binding adenovirus type 5 fiber head also did not block the infection of adenovirus containing NADC-1 fiber (data not shown). CD46 is used as a receptor by human adenoviruses of species B as well as some of species D (15, 49). Interaction regions involve the DG, HI, and IJ loops of the adenovirus head domain (41). Structural superposition shows that the conformation of loops DG and HI are different, and the IJ loop is shorter in porcine adenovirus type 4 (Fig. 2D); furthermore, binding residues Asn-283, Asp-284, Arg-280, and Gln-305 are not conserved in porcine adenovirus 4 fiber head. Finally, cell surface glycoconjugates containing alpha-(2,3)- or alpha-(2,6)-linked sialic acid residues have been reported to serve as receptors for human adenovirus types 37 and 19p (6), which is consistent with a positively charged patch on their surfaces (Fig. 2E). The relative absence of positive charge on the porcine adenovirus type 4 head domain (Fig. 2F) suggests that porcine adenovirus type 4 head domain does not bind sialic acid. Taken together, these findings suggest that the fiber head domain does not bind common adenovirus receptors.
FIG. 2.
Structure of the porcine adenovirus 4 (NADC-1 strain) fiber head. (A) The monomer is depicted with beta strands colored yellow, helices red, and loops green. Beta strands are labeled with letters, and alpha helices are numbered. (B) A view from the top along the 3-fold axis of the trimer. Monomers are colored red, green, and yellow. (C) Comparison of the porcine adenovirus type 4 fiber head structure (yellow) to that of the human adenovirus type 12 fiber head (blue) bound to the CAR D1 domain (red). (D) Comparison of the porcine adenovirus type 4 fiber head structure (yellow) to that of the human adenovirus type 11 fiber head (blue) bound to the CD46 short consensus repeats 1 and 2 (red). The DG, HI, and IJ loops of the adenovirus fiber head are shown and labeled; residues of the human adenovirus type 11 fiber head involved in the interaction also are labeled. (E and F) Electrostatic potential surfaces (blue, positive; red, negative) of human adenovirus type 37 fiber head in complex with sialyl-lactose (E; shown as sticks) and the porcine adenovirus type 4 fiber head (F).
Overall structure of the galectin domain.
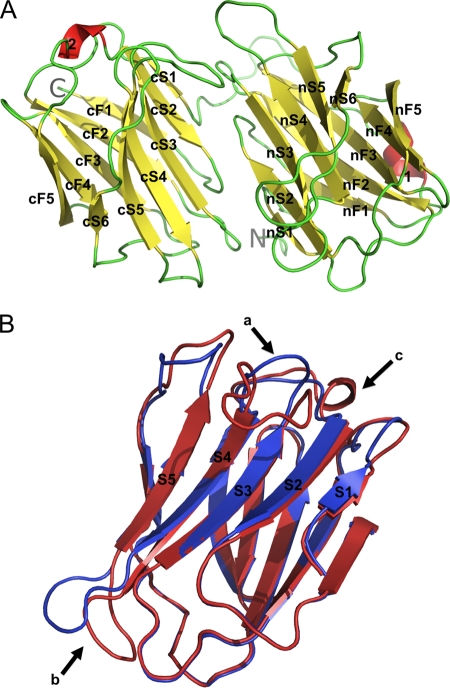
We determined the crystal structure of the porcine adenovirus type 4 galectin domain (Fig. 3). It shows two carbohydrate recognition domains connected by a proline-rich linker and interacting through a new interaction interface. Both carbohydrate recognition domains are arranged such that the sugar-binding pockets are positioned face to face, allowing them to interact simultaneously with a sugar. Both carbohydrate recognition domains are folded in a beta sandwich with a jelly roll topology typical of galectins. Strands are designated F1 to F5 and S1 to S6, and the prefix n or c is added to distinguish between the N- and C-terminal carbohydrate recognition domain. In addition, a 310-helix (helix 2) can be observed in the loop between strands cF5 and cS2 (Fig. 3A). The peptide linking the two carbohydrate recognition domains runs parallel to the nF sheet. It is 23 residues long and rich in prolines, and it has a small 310-helix (helix 1). In the ligand-free form, it was possible to trace the short C-terminal tail in the chains B and C containing a short beta strand interacting with strand nS6 of a neighboring molecule. In all crystal forms a short vestige of the N-terminal purification tag is present, which is not discussed and not included in subsequent analyses. The N- and C-terminal carbohydrate recognition domains superimpose well, with a root mean square difference (RMSD) of 1.3 Å (128 superimposed residues). The superposition (Fig. 3B) shows that the main differences between the domains occur in loop S3-S4, which is longer in the C-terminal carbohydrate recognition domain, in loop S4-S5, which adopts a different conformation, and the presence of the 310-helix in the loop cF5-cS2, which is absent from the N-terminal domain.
FIG. 3.
Structure of the porcine adenovirus type 4 (NADC-1 isolate) fiber galectin domain. (A) The monomer is depicted with beta strands colored yellow, helices in red, and loops in green. N and C termini are labeled. (B) Superposition of the N-terminal (blue) onto the C-terminal (red) carbohydrate recognition domain. The S3-S4 (a), S4-S5 (b), and cF5-cS2 (c) loops are labeled.
Identification of carbohydrate ligands for the galectin domain.
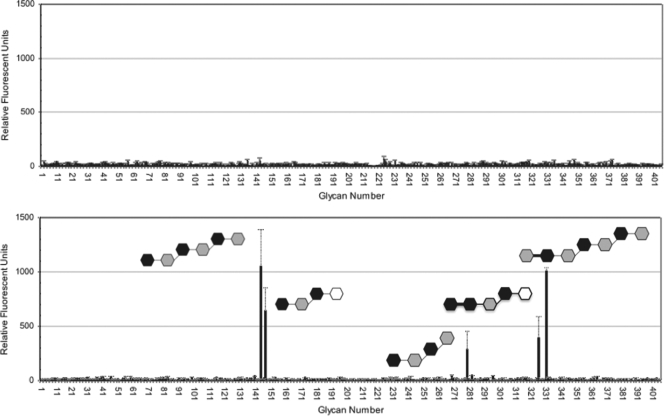
To determine the potential carbohydrate binding specificity of the porcine adenovirus type 4 NADC-1 isolate fiber, we used a glycan microarray approach, wherein 406 potential carbohydrate ligands could be evaluated simultaneously. Figure 4 shows the carbohydrate binding profiles of purified adenovirus-based vectors Ad5Luc1, containing the native human adenovirus type 5 fiber, and Ad5Luc1-PK, which contains a modified fiber wherein the adenovirus type 5 head domain (residues 404 to 581) is replaced by the fiber head, RDG, and galectin domains (residues 124 to 703) of the porcine adenovirus type 4 NADC-1 isolate.
FIG. 4.
Glycan binding specificity of adenovirus vectors. Screening of a glycan array with an adenovirus type 5-based vector (upper) and the same vector modified with the porcine adenovirus type 4, head, RGD, and galectin domains (lower). The plot shows the average relative fluorescence units (RFU; y axis) for the six addresses of each glycan versus the glycan number (x axis) as bars. Standard errors of the means in the fluorescence for the six addresses are indicated for each glycan. Carbohydrates giving a clear signal above the background level are drawn schematically as follows: black hexagons represent galactose, white hexagons represent glucose, and gray hexagons represent N-acetyl-glucosamine (the anomeric carbon is placed at the right side, and the numbering is clockwise). Thick and thin bars represent alpha and beta bonds, respectively.
The control Ad5Luc1 vector showed no specific binding to the glycan microarray. In contrast, the Ad5Luc1-PK vector specifically recognized five structurally related carbohydrates: Galß1-4GlcNAcß1-3Galß1-4GlcNAcß1-3Galß1-4GlcNAc [tri(N-acetyl-lactosamine)], GlcNAcα1-4Galß1-4GlcNAcß1-3Galß1-4GlcNAcß1-3Galß1-4GlcNAc, Galß1-4GlcNAcß1-3Galß1-4Glc (lacto-N-neotetraose), Galα1-4Galß1-4GlcNAcß1-3Galß1-4Glc, and Galß1-4GlcNAcß1-3Galß1-3GlcNAc. Each glycan contains at least one unit of N-acetyl-lactosamine (Galß1-4GlcNAc). Since the Ad5Luc1-PK vector is isogenic to the control Ad5Luc1 vector save for the NADC-1 head and galectin domains, it is likely that the galectin domain mediates these carbohydrate interactions via the galactoside-binding residues in the N-terminal carbohydrate recognition domain. We also performed glycan array screening with purified head and galectin domains, but neither of these proteins yielded positive results. The lack of binding of galectin domain may be due to low-affinity binding to monomeric protein, while binding to the virus results in a higher effective affinity due to avidity effects (32). However, the glycan array results defined specific carbohydrates for our subsequent cocrystallization studies, where low-affinity binding can be overcome by adding the carbohydrate ligands in sufficient concentrations.
Structure of the carbohydrate recognition site and interaction with ligands.
We cocrystallized the galectin domain with four different carbohydrates: lactose, N-acetyl-lactosamine, 3-aminopropyl-lacto-N-neotetraose, and 2-aminoethyl-tri(N-acetyl-lactosamine), and solved the structures by molecular replacement. Clear unbiased density was observed for all ligands bound to the carbohydrate recognition site of the N-terminal domain (Fig. 5A to D), and average temperature factors of the ligands were comparable to those of the protein. No electron density was visible for the respective 3-aminopropyl and 2-aminoethyl substituents of lacto-N-neotetraose and tri(N-acetyl-lactosamine), thus these were not modeled. No significant differences were observed between the ligand-free protein structure and ligand-bound ones; they superimpose with an overall RMSD of between 0.3 and 0.9 Å. Local differences are observed at amino acid Leu541 of the linker region, and we believe this is a consequence of contacts in the crystal. Small differences also are observed in the ligand binding pocket, where the side chains of residues Tyr585 and Asp586 adopt different positions upon interacting with the sugars. The structural analysis of other galectins cocrystallized with lactose or derivates have identified the residues involved in carbohydrate binding. In the N-terminal carbohydrate recognition domain, residues His435, Asn437, Arg439, Val449, Asn451, Trp459, Glu462, and Arg464 make up the canonical carbohydrate binding site (see the alignment in Fig. 1B). In the C-terminal domain only five of these residues are conserved, the valine is replaced by Ala607, and the arginines are replaced by Gly597 and Thr621, respectively. Concordantly, lactose binding to the C-terminal carbohydrate recognition domain was observed only with high concentrations of lactose (70 mM); density for this molecule was less clear, and it was modeled with an occupancy of 0.5.
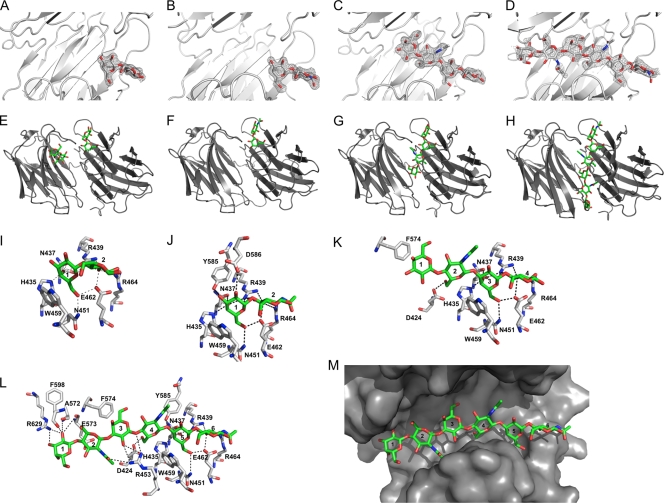
FIG. 5.
Crystal structures of porcine adenovirus 4 galectin domain bound to lactose (A, E, I), N-acetyl-lactosamine (B, F, J), 3-aminopropyl-lacto-N-neotetraose (C, G, K), and 2-aminoethyl-tri(N-acetyl-lactosamine) (D, H, L, M). Panels A to D show unbiased electron density after molecular replacement contoured at 2σ (only the lactose molecule bound to the N-terminal carbohydrate domain is shown), panels E to H show overviews of the structures [only chain A of the structure with 2-aminoethyl-tri(N-acetyl-lactosamine) model is shown], and panels I to L show stick representations of the carbohydrates (carbon, green) and their interaction residues (carbon, white). Hydrogen bonds are shown as black dashed lines. Residues involved in the interaction and carbohydrate residues are labeled. (M) Surface representation of the galectin binding site for 2-aminoethyl-tri(N-acetyl-lactosamine).
The structures of the complexes with lactose and N-acetyl-lactosamine show that both sugars bind the canonical binding site in the N-terminal carbohydrate recognition domain (Fig. 5E and F). We propose that this site functions as the primary carbohydrate recognition site. In both sugars, a stacking interaction occurs between the side chain of Trp459 and the sugar ring of the galactose moiety (Fig. 5I and J). The side chains of His435, Asn437, and Arg439 form hydrogen bonds with galactose O4, the Arg439 side chain with galactose O5, and the side chains of Asn451 and Glu462 with galactose O6. The glucose moiety of lactose and the N-acetyl-glucosamine moiety of N-acetyl-lactosamine occupy the same position and make similar contacts. The O3 atom forms hydrogen bonds with the side chains of Arg439, Glu462, and Arg464 and the O4 atom with the Arg439 side chain. In the complex with lactose, a hydrogen bond is formed between the glucose O2 atom. The side chain of Arg464 and in the complex with N-acetyl-lactosamine, the galactose O3 and O4 atoms form hydrogen bonds with the side chain of Tyr585, and the O2 atom interacts with the side chain of Asp586. Both residues are located in the long cS3-cS4 loop of the C-terminal carbohydrate binding domain. N-acetyl-lactosamine was found only in the C-terminal carbohydrate recognition domain; some density for lactose also could be detected in the canonical binding site of the N-terminal carbohydrate recognition domain.
The structure of the complex with 3-aminopropyl-lacto-N-neotetraose (Fig. 5G and K) shows the lactose moiety occupying the primary recognition site with the same interactions as those described for lactose. However, the N-acetyl-lactosamine moiety occupies a new binding pocket in which the O6 atom of the N-acetyl-glucose forms a hydrogen bond with the side chain of Asp424, and the side chain of Phe574 stacks onto the sugar ring of the galactose residue. In the structure of the complex with 2-aminoethyl-tri(N-acetyl-lactosamine), the N-acetyl-lactosamine at the reducing end (i.e., residues 5 and 6 in the hexasaccharide) is located at the primary recognition site (Fig. 5H and L) and, with the exception of the interaction with the side chain of Asp586, we observed the same interactions as those seen with N-acetyl-lactosamine alone (Fig. 5J). The central N-acetyl-lactosamine unit (residues 3 and 4) binds in the same place as the N-acetyl-lactosamine unit of 3-aminopropyl-lacto-N-neotetraose, and, besides the interactions with Phe574 and Asp424 described above, the N-acetyl-glucose O4 and galactose O3 atoms form hydrogen bonds with the side chain of Arg453. Finally, the N-acetyl-lactosamine unit at the nonreducing end (residues 1 and 2) is located in a binding site made up of residues of the C-terminal carbohydrate recognition domain, where the galactose O2 and O3 atom hydrogen bonds interact with the side chain of Arg629, and the carbonyl oxygen of Ala572, Glu573, and Phe598 interacts with the galactose O2 atom.
The tri(N-acetyl-lactosamine) ligand fills the groove between both carbohydrate recognition domains (Fig. 5M). The lower affinity of the C-terminal carbohydrate recognition domain as well as steric hindrance from the long loop cS3-cS4 prevent this interaction from occurring with the sugar in the opposite orientation. Our structures suggest that the porcine adenovirus type 4 galectin domain would not make additional interactions with longer N-acetyl-lactosamine polymers, therefore an enhanced affinity for longer poly-N-acetyl-lactosamine polymers may be explained by avidity contributions. However, branched sugars may facilitate additional interactions with the protein, possibly involving the C-terminal carbohydrate recognition domain (one might even speculate that the second lactose-binding site is mimicking such an interaction).
Surface plasmon resonance experiments.
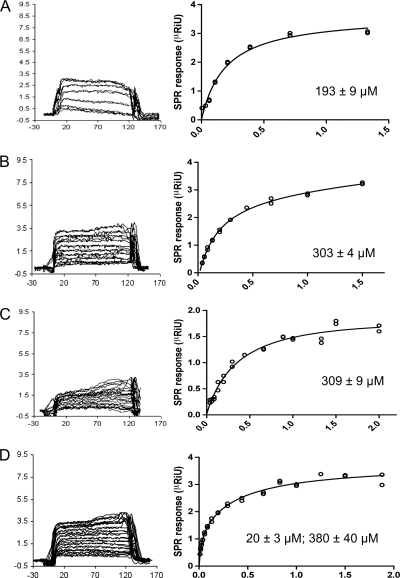
The affinity of porcine adenovirus 4 galectin domain for lactose, N-acetyl-lactosamine, 3-aminopropyl-lacto-N-neotetraose, 2-azidoethyl-di(N-acetyl-lactosamine), and 2-aminoethyl-tri(N-acetyl-lactosamine) was assessed by surface plasmon resonance experiments (Fig. 6). The galectin domain was bound to the sensor chip, and the oligosaccharides were injected at various concentrations to monitor their binding to the galectin domain. Sensorgrams were evaluated via steady-state analysis to yield the corresponding binding isotherms from which the dissociation constants were calculated. Lactose and N-acetyl-lactosamine bound very weakly to the galectin domain, and only very rough estimates of the dissociation constants could be made; even so, the affinity of N-acetyl-lactosamine seems to be greater than that of lactose (estimated dissociation constants of around 10 and 100 mM, respectively). The crystal structures of the complex with N-acetyl-lactosamine show additional interactions with Tyr585 and Asp586, which may explain this enhanced affinity. Due to the weak binding of both disaccharides, the saturation region of the binding isotherms could not be experimentally reached, hindering the estimation of binding stoichiometries. The lactose and N-acetyl-lactosamine used are mixtures of alpha and beta anomers. Although the beta anomer should be favored and is the one we observe in our crystal structures, the affinities we measured may be underestimated. Two of the tetrasaccharides and the hexasaccharide used below were derivatized compounds locked in their beta anomeric form. The tetrasaccharides lacto-N-neotetraose, 3-aminopropyl-lacto-N-neotetraose, and 2-azidoethyl-di(N-acetyl-lactosamine) bound the galectin with similar affinities, displaying dissociation constants of 200 to 300 μM, with a calculated 1:1 stoichiometry. The dissociation constants obtained for lacto-N-neotetraose and 3-aminopropyl-lacto-N-neotetraose were comparable, which suggests that additional small glycan groups do not influence the binding much.
FIG. 6.
Binding affinities of the porcine adenovirus type 4 galectin domain for carbohydrates obtained by surface plasmon resonance experiments. Sensorgrams and binding curves are shown for lacto-N-neotetraose (A), 3-aminopropyl-lacto-N-neotetraose (B), 2-azidoethyl-di(N-acetyl-lactosamine) (C), and 2-aminoethyl-tri(N-acetyl-lactosamine) (D). On the y axis, the SPR response in μRIU is plotted (normalized by division by the molecular weight of the carbohydrate and multiplied by 100); on the horizontal axis, the time in seconds for the sensorgrams and the carbohydrate concentration in mM for the binding curves are plotted. Calculated dissociation constants also are shown.
The surface plasmon resonance data for the binding of the hexasaccharide 2-aminoethyl-tri(N-acetyl-lactosamine) to galectin provided a binding isotherm that was best fit using an alternative binding site model. We propose that the high-affinity binding mode, with an estimated dissociation constant of 20 μM and a 1:1 stoichiometry, corresponds to the crystal structure of the complex with 2-aminoethyl-tri(N-acetyl-lactosamine), where the six sugar residues are involved in interactions with the galectin domain. The second binding mode, with an estimated dissociation constant of 380 μM (and also a 1:1 stoichiometry), may correspond to a binding mode in which the primary binding site recognizes the central N-acetyl-lactosamine residue, and only the first four sugar residues from the nonreducing end are involved in interactions with the protein. A similar binding mode has been reported for the N-terminal domain of human tandem-repeat-type galectin 9 (37). Although the measured affinities toward the monomeric galectin are moderate, one has to remember that adenovirus fiber is trimeric and thus contains three independent galectin domains. Furthermore, each viral particle contains 12 trimeric fibers. This multimeric arrangement would enable the establishment of multivalent interactions between the viral particles and target surface-bound oligosaccharides on cell surfaces, resulting in higher functional affinities (32). Furthermore, oligosaccharides of sufficient length may bind more than one galectin domain (33). Another thing that should be kept in mind is that more-complex, branched sugars not present in the glycan array (and not used in our binding or cocrystallization studies) may make even more contacts with galectin and bind with higher affinity than the carbohydrates mentioned.
Comparison to other tandem-repeat galectins.
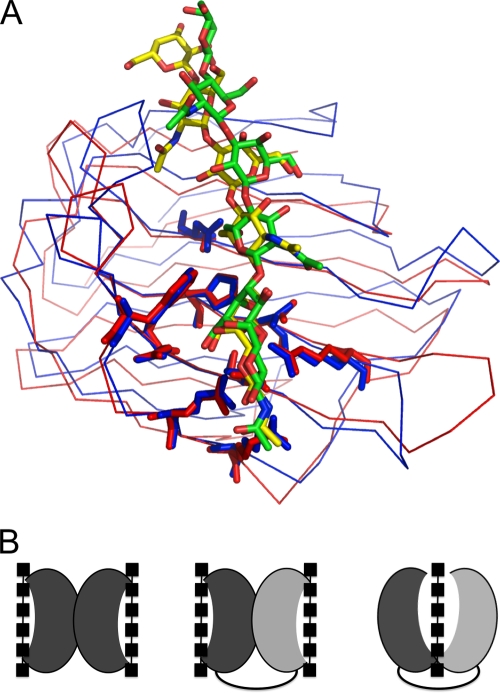
Multiple-sequence alignments of tandem-repeat galectins (Fig. 1B) show that most of the residues involved in the interaction with tri(N-acetyl-lactosamine) are conserved in human galectin 9. A structural superposition (Fig. 7A) of the N-terminal carbohydrate recognition domain of human galectin 9 in complex with tri(N-acetyl-lactosamine) (37) on the C-terminal carbohydrate recognition domain of porcine adenovirus 4 galectin in complex with the same sugar reveals that both N-terminal domains have a mode of interaction that is similar to that of the sugar. Although a back-to-back orientation is currently favored for tandem-repeat galectins, it is possible that the C-terminal domain of human galectin 9 adopts a position similar to that observed in porcine adenovirus type 4 galectin. This would explain the observation that although di- and tri(N-acetyl-lactosamine) make the same interactions with the N-terminal domain of galectin 9 (37), affinity chromatography indicates that tri(N-acetyl-lactosamine) has a higher affinity for galectin 9 containing both domains (23).
FIG. 7.
Comparison to other tandem-repeat galectins. (A) Structural superposition of the N-terminal carbohydrate recognition domain of human galectin-9 (blue) on the N-terminal carbohydrate recognition domain of porcine adenovirus 4 galectin (red), both in complex with tri(N-acetyl-lactosamine). The oxygen and nitrogen atoms of the carbohydrates are colored in red and blue, respectively, and the carbon atoms are green (for the carbohydrate bound to human galectin-9) or yellow (for the carbohydrate bound to porcine adenovirus 4 galectin). (B) Schematic drawing of homodimeric prototype galectin domains (left) of the current model for tandem-repeat galectin domains (middle) and our new model for tandem-repeat-type galectins (right) based on the structures determined.
These models represent the first complete structures of a tandem-repeat galectin with both carbohydrate recognition domains observed in the same structure, although the crystallization of the nematode tandem-repeat galectin LEC-1 structure was reported (24) and a structure containing both carbohydrate recognition domains was reported in a conference abstract (39). It has been suggested previously that tandem-repeat galectins do not crystallize readily due to disorder introduced by the linker peptides (11). Perhaps the relatively short linker peptide of porcine adenovirus type 4 galectin was a factor in allowing us to circumvent this problem. The structures reported here allow an analysis of the interactions between the two domains. The interface between the N- and C-terminal carbohydrate recognition domains buries about 12% of the total solvent-accessible area of both domains (830 to 900 Å2 for each). The interface is of a mixed nature, involving salt bridges, hydrogen bonds, and multiple van der Waals interactions. The interaction interface can be mapped into two distinct regions. One of these regions is comprised of residues located in the nS4-nS5 loop and at the end of the nS4 strand and interacts with residues in the cS3-cS4 loop. A second interaction region is formed by residues located just before strand nS1, the nF2-nS3 loop, the end of strand nS2, and in the nS2-nF1 loop, which interact with the structurally equivalent amino acids of the C-terminal domain. Aromatic residues from each domain (Phe394 and Tyr548, respectively) contribute to a hydrophobic core between the carbohydrate recognition domains, which may be important for interface stability. These aromatic residues are universally conserved as Phe or Tyr in tandem-repeat galectins (Fig. 1B), but they are absent from prototype galectins, an observation that may be taken as an argument for a face-to-face orientation in other tandem-repeat galectins. Structures of other tandem-repeat galectins will have to be analyzed to determine how general our observed face-to-face orientation is. It may even be possible that the relatively long linkers between carbohydrate recognition domains of eukaryotic tandem-repeat galectins allow the protein the flexibility to adopt both back-to-back and face-to-face orientations.
DISCUSSION
The natural receptor of porcine adenovirus type 4 has not been identified but does not appear to be CAR, CD46, or sialic acid (40 and this paper). The analysis of the sequence C terminal to the head domain revealed an RGD-containing sequence and a galectin-like sequence consisting of two putative carbohydrate recognition domains. In many adenoviruses, the RGD-containing sequence is located in the penton base protein, the capsid protein into which the fiber is anchored. There, it has been shown to be important for interaction with integrins (30). It is plausible that porcine adenovirus type 4 has the ability to interact with integrins via the RGD sequence of its fiber (the sequence of the penton protein is not known). The results presented in this paper demonstrate the ability of the porcine adenovirus 4 fiber C-terminal galectin domain to bind sugars containing N-acetyl-lactosamine units, thus confirming it as a bona fide galectin. Three linearly connected N-acetyl-lactosamine units may bind at the same time. The location of the galectin domain at the C-terminal end of the NADC-1 fiber suggests that this domain projects away from the virus, making it probable that sugars containing repeating N-acetyl-lactosamine units are natural receptors for this virus. However, the relative importance of N-acetyl-lactosamine-containing carbohydrates and integrins as receptors for porcine adenovirus type 4 remains to be determined in detail, although preliminary experiments (not shown) indicate that the isolated galectin domain partially blocks gene transfer by the vector Ad5Luc1-PK, containing the porcine adenovirus type 4 fiber C-terminal domains.
Structural studies have revealed the structural basis of the dimeric organization in some prototype galectins, in which carbohydrate-binding sites are projected away from each other, and it is unlikely that the two sites cooperatively bind one carbohydrate molecule (3) (Fig. 7B, left). The dimeric prototype galectin structure has been used to build models of tandem-repeat galectins (51) (Fig. 7B, middle), although experimental evidence suggests cooperative interactions between the two domains and the necessity of the linker peptide for proper functioning (29). Furthermore, glycan array experiments carried out with other tandem-repeat galectins point to the necessity of both domains for the recognition of specific sugars, and studies with human galectin 8 have shown that although the specificity of both domains is additive in solution, they bind synergistically to carbohydrates (7). Here, we report the first structure of a tandem-repeat galectin in which both carbohydrate recognition domains are present and organized in a way not resembling the prototype galectin dimer. The domains are arranged such that they can interact cooperatively with a single glycan (Fig. 7B, right); the cocrystal structure with 2-aminoethyl-tri(N-acetyl-lactosamine) reported here confirms they do. This structure may represent a new paradigm in the organization of tandem-repeat galectins and explains the importance of the linker peptide as an organizer of both domains. The face-to-face orientations of the carbohydrate recognition domains allow residues of both domains to interact with the same oligosaccharide and, in principle, may allow for the recognition of more complex sugars than that of one domain alone. Future structures of other tandem-repeat galectins will reveal how general the face-to-face orientation we observe is.
Porcine adenovirus type 4 has been associated with encephalitis (12). The results reported here will be of use in future efforts focused on elucidating the early steps of the porcine adenovirus 4 replication cycle and may lead to therapeutic applications for swine diseases. Incorporating the galectin domain in human gene therapy vectors may allow the targeting of these vectors to specific cells. The cocrystal structures presented here identify the interactions of the galectin domain with the N-acetyl-lactosamine repeats, opening up the possibility of generating site-directed mutants to change the specificity of the galectin domain and the targeting of adenovirus-based vectors to specific disease-related carbohydrates (20).
Acknowledgments
We thank José M. Otero, Sergio Galán-Bartual, and Bruno Dacunha-Marinho for help with data collection and Patricia Ferraces Casais for technical assistance. We gratefully acknowledge David F. Smith and Jamie Heimburg-Molinaro of the Protein-Glycan Interaction Core Facility (Core H) of the Consortium for Functional Glycomics, funded by NIGMS GM62116, Emory University School of Medicine (Atlanta, GA) for performing the glycan array analysis, and the staff of the ESRF beam lines ID14-2 and BM30A and EMBL-DESY beam line X11 for help with data collection.
This research was sponsored by research grants BFU2005-02974, BFU2005-24982-E, and BFU2008-01588 and by a predoctoral FPU fellowship to P.G.-C. from the Spanish Ministry of Education and Science. This work also was supported by funds from the European Commission under contract NMP4-CT-2006-033256 (BeNatural-coordinated project), by NIH grant 1R01HL092941 to D.T.C., and by the Xunta de Galicia via an Isidro Parga Pondal fellowship to E.M.M.
Footnotes
Published ahead of print on 4 August 2010.
REFERENCES
- 1.Arnberg, N. 2009. Adenovirus receptors: implications for tropism, treatment and targeting. Rev. Med. Virol. 19:165-178. [DOI] [PubMed] [Google Scholar]
- 2.Bachtarzi, H., M. Stevenson, and K. Fisher. 2008. Cancer gene therapy with targeted adenoviruses. Expert Opin. Drug Deliv. 5:1231-1240. [DOI] [PubMed] [Google Scholar]
- 3.Barondes, S. H., D. N. Cooper, M. A. Gitt, and H. Leffler. 1994. Galectins. Structure and function of a large family of animal lectins. J. Biol. Chem. 269:20807-20810. [PubMed] [Google Scholar]
- 4.Bewley, M. C., K. Springer, Y. B. Zhang, P. Freimuth, and J. M. Flanagan. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579-1583. [DOI] [PubMed] [Google Scholar]
- 5.Blixt, O., S. Head, T. Mondala, C. Scanlan, M. E. Huflejt, R. Alvarez, M. C. Bryan, F. Fazio, D. Calarese, J. Stevens, N. Razi, D. J. Stevens, J. J. Skehel, I. van Die, D. R. Burton, I. A. Wilson, R. Cummings, N. Bovin, C. H. Wong, and J. C. Paulson. 2004. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U. S. A. 101:17033-17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burmeister, W. P., D. Guilligay, S. Cusack, G. Wadell, and N. Arnberg. 2004. Crystal structure of species D adenovirus fiber knobs and their sialic acid binding sites. J. Virol. 78:7727-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlsson, S., C. T. Oberg, M. C. Carlsson, A. Sundin, U. J. Nilsson, D. Smith, R. D. Cummings, J. Almkvist, A. Karlsson, and H. Leffler. 2007. Affinity of galectin-8 and its carbohydrate recognition domains for ligands in solution and at the cell surface. Glycobiology 17:663-676. [DOI] [PubMed] [Google Scholar]
- 8.Chen, V. B., W. B. Arendall, J. J. Headd, D. A. Keedy, R. M. Immormino, G. J. Kapral, L. W. Murray, J. S. Richardson, and D. C. Richardson. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66:12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, D. N. 2002. Galectinomics: finding themes in complexity. Biochim. Biophys. Acta 1572:209-231. [DOI] [PubMed] [Google Scholar]
- 11.Cummings, R. D., and F. Liu. 2009. S-type lectins (galectins), p. 475-479. In A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, G. W. Hart, and J. Marth (ed.), Essentials of glycobiology, 2nd ed. Cold Spring Harbor Laboratory Press, Woodbury, NY. [PubMed]
- 12.Edington, N., L. Kasza, and G. J. Christofinis. 1972. Meningo-encephalitis in gnotobiotic pigs inoculated intransally and orally with porcine adenovirus 4. Res. Vet. Sci. 13:289-291. [PubMed] [Google Scholar]
- 13.El Bakkouri, M., E. Seiradake, S. Cusack, R. W. Ruigrok, and G. Schoehn. 2008. Structure of the C-terminal head domain of the fowl adenovirus type 1 short fibre. Virology 378:169-176. [DOI] [PubMed] [Google Scholar]
- 14.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 15.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 16.Graham, F. L., and L. Prevec. 1995. Methods for construction of adenovirus vectors. Mol. Biotechnol. 3:207-220. [DOI] [PubMed] [Google Scholar]
- 17.Guardado Calvo, P., A. L. Llamas-Saiz, P. Langlois, and M. J. van Raaij. 2006. Crystallization of the C-terminal head domain of the avian adenovirus CELO long fibre. Acta Crystallogr. F 62:449-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guardado-Calvo, P., A. L. Llamas-Saiz, G. C. Fox, P. Langlois, and M. J. van Raaij. 2007. Structure of the C-terminal head domain of the fowl adenovirus type 1 long fiber. J. Gen. Virol. 88:2407-2416. [DOI] [PubMed] [Google Scholar]
- 19.Guardado-Calvo, P., A. L. Llamas-Saiz, G. C. Fox, J. N. Glasgow, and M. J. van Raaij. 2009. Crystallization of the head and galectin-like domains of porcine adenovirus isolate NADC-1 fibre. Acta Crystallogr. F 65:1149-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hakomori, S. 1984. Tumor-associated carbohydrate antigens. Annu. Rev. Immunol. 2:103-126. [DOI] [PubMed] [Google Scholar]
- 21.Hammond, J. M., and M. A. Johnson. 2005. Porcine adenovirus as a delivery system for swine vaccines and immunotherapeutics. Vet. J. 169:17-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirabayashi, J., and K. Kasai. 1993. The family of metazoan metal-independent beta-galactoside-binding lectins: structure, function and molecular evolution. Glycobiology 3:297-304. [DOI] [PubMed] [Google Scholar]
- 23.Hirabayashi, J., T. Hashidate, Y. Arata, N. Nishi, T. Nakamura, M. Hirashima, T. Urashima, T. Oka, M. Futai, W. E. Muller, F. Yagi, and K. Kasai. 2002. Oligosaccharide specificity of galectins: a search by frontal affinity chromatography. Biochim. Biophys. Acta 1572:232-254. [DOI] [PubMed] [Google Scholar]
- 24.Itagaki, T., S. Nishizaki, K. Sekihashi, H. Kobayashi, S. Kidokoro, Y. Kezuka, Y. Arata, J. Hirabayashi, K. Kasai, and T. Nonaka. 2008. Crystallization and preliminary X-ray crystallographic analysis of galectin LEC-1 from Caenorhabditis elegans. Protein Pept. Lett. 15:419-422. [DOI] [PubMed] [Google Scholar]
- 25.Kleiboeker, S. B., B. S. Sealm, and W. L. Mengeling. 1993. Genomic cloning and restriction site mapping of a porcine adenovirus isolate: demonstration of genomic stability in porcine adenovirus. Arch. Virol. 133:357-368. [DOI] [PubMed] [Google Scholar]
- 26.Kleiboeker, S. B. 1995. Sequence analysis of the fiber genomic region of a porcine adenovirus predicts a novel fiber protein. Virus Res. 39:299-309. [DOI] [PubMed] [Google Scholar]
- 27.Krasnykh, V., N. Belousova, N. Korokhov, G. Mikheeva, and D. T. Curiel. 2001. Genetic targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J. Virol. 75:4176-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krissinel, E., and K. Henrick. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774-797. [DOI] [PubMed] [Google Scholar]
- 29.Levy, Y., S. Auslender, M. Eisenstein, R. R. Vidavski, D. Ronen, A. D. Bershadsky, and Y. Zick. 2006. It depends on the hinge: a structure-functional analysis of galectin-8, a tandem-repeat type lectin. Glycobiology 16:463-476. [DOI] [PubMed] [Google Scholar]
- 30.Lindert, S., M. Silvertry, T. M. Mullen, G. Nemerow, and P. L. Stewart. 2009. Cryo-electron microscopy structure of an adenovirus-integrin complex indicates conformational changes in both penton base and integrin. J. Virol. 83:11491-11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lobsanov, Y. D., M. A. Gitt, H. Leffler, S. H. Barondes, and J. M. Rini. 1993. X-ray crystal structure of the human dimeric S-Lac lectin L-14-II, in complex with lactose at 2.9 Å resolution. J. Biol. Chem. 268:27034-27038. [DOI] [PubMed] [Google Scholar]
- 32.Lortat-Jacob, H., E. Chouin, S. Cusack, and M. J. van Raaij. 2001. Kinetic analysis of adenovirus fiber binding to its receptor reveals an avidity mechanism for trimeric receptor-ligand interactions. J. Biol. Chem. 276:9009-9015. [DOI] [PubMed] [Google Scholar]
- 33.Lundquist, J. J., and E. J. Toone. 2002. The cluster glycoside effect. Chem. Rev. 102:555-578. [DOI] [PubMed] [Google Scholar]
- 34.Maizel, J. V., Jr., D. O. White, and M. D. Scharff. 1968. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36:115-125. [DOI] [PubMed] [Google Scholar]
- 35.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 37.Nagae, M., N. Nishi, T. Murata, T. Usui, T. Nakamura, S. Wakatsuki, and R. Kato. 2009. Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain. Glycobiology 19:112-117. [DOI] [PubMed] [Google Scholar]
- 38.Nemerow, G. R., L. Pache, V. Reddy, and P. L. Stewart. 2009. Insights into adenovirus host cell interactions from structural studies. Virology 384:380-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nonaka, T., K. Sekihashi, S. Nishizaki, Y. Arata, J. Hirabayashi, K. Kasai, and Y. Mitsui. 1999. Crystal structure of the tandem-repeat galectin of the nematode Caenorhabditis elegans. Glycoconj. J. 16:S119. [Google Scholar]
- 40.Paul, C. P., M. Everts, J. N. Glasgow, P. Dent, P. B. Fisher, I. V. Ulasov, M. S. Lesniak, M. A. Stoff-Khalili, J. C. Roth, M. A. Preuss, C. M. Dirven, M. L. Lamfers, G. P. Siegal, Z. B. Zhu, and D. T. Curiel. 2008. Characterization of infectivity of knob-modified adenoviral vectors in glioma. Cancer Biol. Ther. 7:786-793. [DOI] [PubMed] [Google Scholar]
- 41.Persson, B. D., D. M. Reiter, M. Marttila, Y. F. Mei, J. M. Casasnovas, N. Arnberg, and T. Stehle. 2007. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat. Struct. Mol. Biol. 14:164-166. [DOI] [PubMed] [Google Scholar]
- 42.Roelvink, P. W., A. Lizonova, J. G. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seiradake, E., and S. Cusack. 2005. Crystal structure of enteric adenovirus serotype 41 short fiber head. J. Virol. 79:14088-14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seiradake, E., H. Lortat-Jacob, O. Billet, E. J. Kremer, and S. Cusack. 2006. Structural and mutational analysis of human Ad37 and canine adenovirus 2 fiber heads in complex with the D1 domain of coxsackie and adenovirus receptor. J. Biol. Chem. 281:33704-33716. [DOI] [PubMed] [Google Scholar]
- 45.Thacker, E. E., L. Timares, and Q. L. Matthews. 2009. Strategies to overcome host immunity to adenovirus vectors in vaccine development. Expert Rev. Vaccines 8:761-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vagin, A., and A. Teplyakov. 2010. Molecular replacement with MOLREP. Acta Crystallogr. D 66:22-25. [DOI] [PubMed] [Google Scholar]
- 47.van Raaij, M. J., A. Mitraki, G. Lavigne, and S. Cusack. 1999. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401:935-938. [DOI] [PubMed] [Google Scholar]
- 48.Winn, M. D., M. N. Isupov, and G. N. Murshudov. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D 57:122-133. [DOI] [PubMed] [Google Scholar]
- 49.Wu, E., S. A. Trauger, L. Pache, T. M. Mullen, D. J. von Seggern, G. Siuzdak, and G. R. Nemerow. 2004. Membrane cofactor protein is a receptor for adenoviruses associated with epidemic keratoconjunctivitis. J. Virol. 78:3897-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia, D., L. J. Henry, R. D. Gerard, and J. Deisenhofer. 1994. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure 2:1259-1270. [DOI] [PubMed] [Google Scholar]
- 51.Zick, Y., M. Eisenstein, R. A. Goren, Y. R. Hadari, Y. Levy, and D. Ronen. 2004. Role of galectin-8 as a modulator of cell adhesion and cell growth. Glycoconj. J. 19:517-526. [DOI] [PubMed] [Google Scholar]